Abstract
In times when herpesvirus genomic data were scarce, the cospeciation between these viruses and their hosts was considered to be common knowledge. However, as more herpesviral sequences were made available, tree reconciliation analyses started to reveal topological incongruences between host and viral phylogenies, indicating that other cophylogenetic events, such as intrahost speciation and host switching, likely played important roles along more than 200 million years of evolutionary history of these viruses. Tree reconciliations performed with undated phylogenies can identify topological differences, but offer insufficient information to reveal temporal incongruences between the divergence timing of host and viral species. In this study, we performed cophylogenetic analyses using time-resolved trees of herpesviruses and their hosts, based on careful molecular clock modelling. This approach enabled us to infer cophylogenetic events over time and also integrate information on host biogeography to better understand host–virus evolutionary history. Given the increasing amount of sequence data now available, mismatches between host and viral phylogenies have become more evident, and to account for such phylogenetic differences, host switches, intrahost speciations and losses were frequently found in all tree reconciliations. For all subfamilies in Herpesviridae, under all scenarios we explored, intrahost speciation and host switching were more frequent than cospeciation, which was shown to be a rare event, restricted to contexts where topological and temporal patterns of viral and host evolution were in strict agreement.
Keywords: tree reconciliation, herpesvirus, phylogenetics, host switch, host–virus evolution
1. Introduction
Herpesviridae is a diverse family of large double-stranded DNA viruses, subdivided in three subfamilies—Alpha-, Beta-, and Gammaherpesvirinae—that infect different groups of vertebrates, including birds, mammals, and reptiles (Davison et al. 2009). The temporal scale of the evolution of herpesviruses (HVs) is still a matter of debate, with some estimates dating their most recent common ancestors (MRCA) back to 180–220 million years ago (Ma), in the Triassic/Jurassic period (McGeoch et al. 1995), while others place their origins as far back in time as 374–420 Ma, in the Devonian period (McGeoch and Gatherer 2005). Up until recently, HVs were considered to evolve alongside their hosts mainly by cospeciation (McGeoch et al. 1995; Davison 2002; Jackson 2005; McGeoch, Rixon, and Davison 2006). Cospeciation, not to be confused with ‘coevolution’ (Jackson 2005; De Vienne et al. 2013) is a process of concomitant speciation of host and viral species, which leads the parasite phylogeny to mirror that of its host, to a certain degree (De Vienne et al. 2013) (Fig. 1). Figure 1C shows some examples of cospeciation, which are mainly observed when the topology and divergence times of hosts and viruses strictly agree. As more sequence data and flexible molecular clock models became available, we can now detect more mismatches between host and viral trees than in decades ago, and the discovery of new herpesviruses now challenges the predominance of cospeciation as the main type of event driving their evolution alongside their hosts. These topological and temporal disagreements evoke alternative hypotheses to explain HV evolution, such as host switches (transfers) and intrahost speciations (Davison 2002; Escalera-Zamudio et al. 2016).
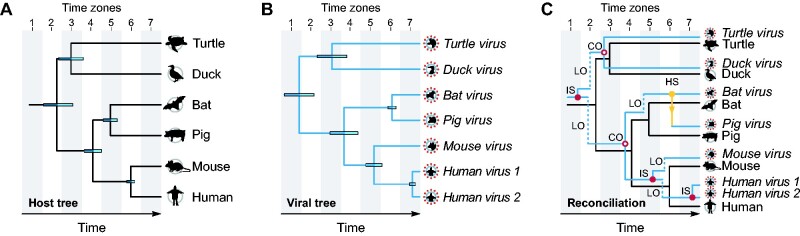
Figure 1.
Tree reconciliation. (A) Hypothetical host tree showing divergence times (node height intervals, as horizontal bars). While some divergence time intervals span a single time zone, as seen for the mammalian MRCA in zone 4, others can span multiple time zones, as observed for the MRCA of all host species in the host tree, which divergence intervals span the time zones 2 and 3. (B) Hypothetical viral tree, also with divergence time intervals shown as horizontal bars. While this tree resembles the host tree, some mismatches can be observed, especially in terms of divergence times, which in many instances do not coincide. (C) By reconciling both trees, topological and temporal properties of the trees are compared, and cophylogenetic events can be inferred, such as cospeciations (CO), intrahost speciations (IS), host switches (HS), and losses (LO).
Host switching takes place when viruses succeed in infecting a new host still unexplored by their ancestors (De Vienne et al. 2013). An example of host switching is depicted in Fig. 1C, where a hypothetical virus infecting bats, closely related to a virus infecting pigs, diverged much later (zone 6) than the point of divergence of their hosts (zone 5). This exemplifies a context where a host switch likely took place. Recent studies have been suggesting that host switches involving herpesviruses are probably more frequent than previously thought (Escalera-Zamudio et al. 2016; Geoghegan, Duchêne and Holmes 2017), but detecting host switches can be difficult, as extinctions of viruses transmitted to new hosts may occur frequently (Johnson et al. 2003; Geoghegan, Duchêne, and Holmes 2017). Intrahost speciation (also known as duplication) is another evolutionary process playing an important role in host-virus evolution (Johnson et al. 2003; De Vienne et al. 2013). By means of intrahost speciations, multiple species of viruses can explore a single host species. Examples of intrahost speciation are shown in Fig. 1C, where viral lineages underwent speciation while infecting the same host, some earlier in the evolution (zones 1 and 5), and others in more recent times, leading to two species of hypothetical ‘Human viruses’. It is known that related viral lineages are more likely to persist infecting the same host if they occupy distinct biological niches (tissues) within the hosts (Davison 2002). Another event playing an important role in host-virus evolution is the loss of viral lineages, events that in a cophylogenetic context may represent: 1, symbiotic extinction (Lovisolo, Hull and Rösler 2003); 2, sorting events (Johnson et al. 2003); or 3) rare/undiscovered species, a result of undersampling (Page and Charleston 1998).
The aforementioned events can be inferred using cophylogenetic analyses, such as tree reconciliations, which help us understand the relationship between hosts and parasites over time (Page and Charleston 1998). These analyses can identify differences and similarities between the topologies of host and parasite trees, where congruences may indicate points of cospeciation, while incongruences may imply host switches or intrahost speciations followed by losses (Fig. 1) (Johnson et al. 2003; De Vienne et al. 2013). Topological congruence is not always caused by cospeciation events: similar topologies of host and parasite phylogenies can happen by chance, as a result of repeated host switches (De Vienne et al. 2013). To better understand the intricate evolutionary processes of HVs, cophylogenetic analyses can be applied to elucidate the pathways taken by viruses as their hosts and the environment evolve.
By reconciling time-resolved phylogenies, we here present detailed scenarios unravelling the evolution of herpesviruses with their hosts, showing the main cophylogenetic events that played important roles in the evolution of herpesviruses, and how members of Herpesviridae achieved their current broad host range, infecting reptiles, birds and mammals.
2. Materials and methods
2.1 Host/virus species and sequence datasets
To include the maximum number of herpesviruses in this study, while also allowing consistent phylogenetic analyses, we downloaded from NCBI all DNA sequences encoding UL27 (gB) and UL30 (DNApol) for which host information was available, which is essential for reconciliation analysis. Following these criteria, we found sequences related to 121 herpesviruses (family: Herpesviridae, subfamilies Alpha-, Beta-, and Gammaherpesvirinae), which were associated with sixty-seven host species (mammals, birds and reptiles; see Supplementary Table S2). The inclusion of members of Herpesvirales from other families was not possible, as UL30 is the only gene found in all members of this viral order, data that would not be sufficient for inferring robust time-resolved phylogenetic analyses. We translated UL27 and UL30 to amino acid sequences (gB and DNApol, respectively), and combined the sequences in two ways: 1, including all gB protein sequences from all taxa in Herpesviridae, in a single dataset, proceeding in the same way for DNApol and 2, separating gB and DNApol protein sequences per herpesvirus subfamily, making gene datasets specific for members of Alpha-, Beta-, and Gammaherpesvirinae. Each gene dataset was individually aligned using MAFFT (Katoh and Standley 2013), columns with more than 50 per cent of missing data were excluded, and the multiple sequence alignments (MSAs) were then concatenated to create four alignments containing: all ‘Herpesviridae’ taxa, ‘Only Alphaherpesvirinae’, ‘Only Betaherpesvirinae’, and ‘Only Gammaherpesvirinae’ taxa. These MSAs were analysed in twelve independent Bayesian phylogenetic analyses described in the next section.
2.2 Phylogenetic analyses
We first performed Bayesian model selection on a set of three molecular clock models namely, the strict clock model, the relaxed (uncorrelated lognormal) clock model, and the random local clock model, for each of the four gene sets mentioned above. In doing so, we used a Yule speciation model (Udny Yule 1925) as the tree-generative process, the LG general amino acid replacement matrix (Le and Gascuel 2008) in combination with among-site rate heterogeneity (Yang 1994). We assumed monophyly for the normally distributed fossil calibration priors (see below) and otherwise employed the default parameter priors in BEAST (Suchard et al. 2018) including those for the different molecular clock models.
To assess which clock model yields the best fit to each data set, we estimated the log marginal likelihood for each of these models using generalized stepping-stone sampling (Baele, Lemey, and Suchard 2016) as implemented in BEAST v1.10.4 (Suchard et al. 2018) and making use of the BEAGLE v3 high-performance computational library (Ayres et al. 2019). Each log marginal likelihood estimate was obtained by first running an initial chain of twenty million iterations, followed by collecting samples from 101 power posteriors—spread according to a Beta(1.0, 0.3) distribution—that connect the posterior to a collection of working priors for the models under consideration (Fan et al. 2011). Each power posterior was run for 500,000 iterations and sampled every 1,000th iteration. The resulting log marginal likelihood estimates can then be used to compute the log Bayes factor between each competing set of models, and we employ the log Bayes factor cut-offs proposed by (Kass and Raftery 1995) to assess its significance.
We subsequently performed Bayesian phylogenetic inference in BEAST to obtain the viral phylogeny using the uncorrelated relaxed clock model with an underlying lognormal distribution, which was estimated to offer the highest relative model fit to the data of the clock models considered (see Supplementary Table S1). These analyses were run until all relevant parameters acquired an effective sample size higher than 200, as assessed using Tracer v1.7 (Rambaut et al. 2018). After removing 10 per cent of the samples as burn-in, the maximum clade credibility trees were summarized using TreeAnnotator v1.10.4, and visualized using FigTree v1.4.4.
During Bayesian model selection and subsequent phylogenetic inference, the tree was calibrated by pegging one node per subfamily: the MRCA of viruses from the genera Simplexvirus, Cytomegalovirus, and Lymphocryptovirus. These viruses infect Old World and New World Monkeys, and assuming they diverged alongside their ancestral Simiiformes hosts (approximately 42.9 Ma, parameterized as a normal prior distribution with mean 43.5 Ma and SD of 1.25 million years) (Steiper and Young 2006), we inferred the time scale of the viral phylogeny, following similar approaches employed in previous studies (McGeoch et al. 1995; Wertheim et al. 2014; Murthy et al. 2019). The inferred tree fully agreed with the current taxonomic classification provided by International Committee on Taxonomy of Viruses (ICTV) (King et al. 2018). To obtain the host tree, we downloaded a validated tree topology from timetree.org (Kumar et al. 2017), alongside information about confidence intervals, and median divergence times of host ancestors, which we combined in a nexus file using a Python script (see Data Availability).
2.3 Tree reconciliation
To find low cost, historic associations between host and viral ancestors, we performed dated tree reconciliations individually for each HV subfamily using Jane 4 (Conow et al. 2010). Taking advantage of the divergence time credibility interval associated with each internal node of the trees, we converted the phylogenies into a Jane timed tree in nexus format using a Python script available on GitHub (see Data Availability). At this step, we performed discretization of the continuous time scales of the phylogenies as bins of five million years (‘time zones’), shared by viral and host trees. This enabled the internal nodes of host and viral trees to be assigned to specific time zones, ensuring that only nodes belonging to the same time zone could be associated for inferring potential host switches and cospeciation events, in this way avoiding chronological inconsistencies. The algorithm implemented in Jane allows internal nodes to be assigned to more than one time zone and requires that all zones are populated with at least one host node. To meet this requirement, we added an outgroup clade to the original host tree, containing artificial taxa, ensuring in this way that its internal nodes could span time zones not originally covered by the original host nodes. We added a similar outgroup to the viral tree, allowing the pairing of artificial host-virus pairs. Since it is not possible to ascertain what are the most appropriate relative costs of events such as cospeciations, intrahost speciations, host switches and losses, we reconciled trees under multiple combinations of relative costs, in which those events were weighted with cost values varying from 0 to 3. For each HV subfamily, we explored a total of 256 cophylogenetic cost regimes, generating several parsimonious reconstructions, which differed in terms of overall cost and number of inferred events (see Supplementary Tables S2–S5). In this solution space, we calculated the median number of inferred events for each event type, and we selected an optimal cost regime based on its ability to: 1, reconstruct the median number of events for each event type, and 2, produce a solution with a median total cost. Following these criteria, the selected cost regime had the following relative costs of events: cospeciations = 0; intrahost speciation = 0; host switches = 2; and losses = 2 (see Supplementary Tables S2–S5).
3. Results
3.1 The evolutionary time scale of Herpesviridae
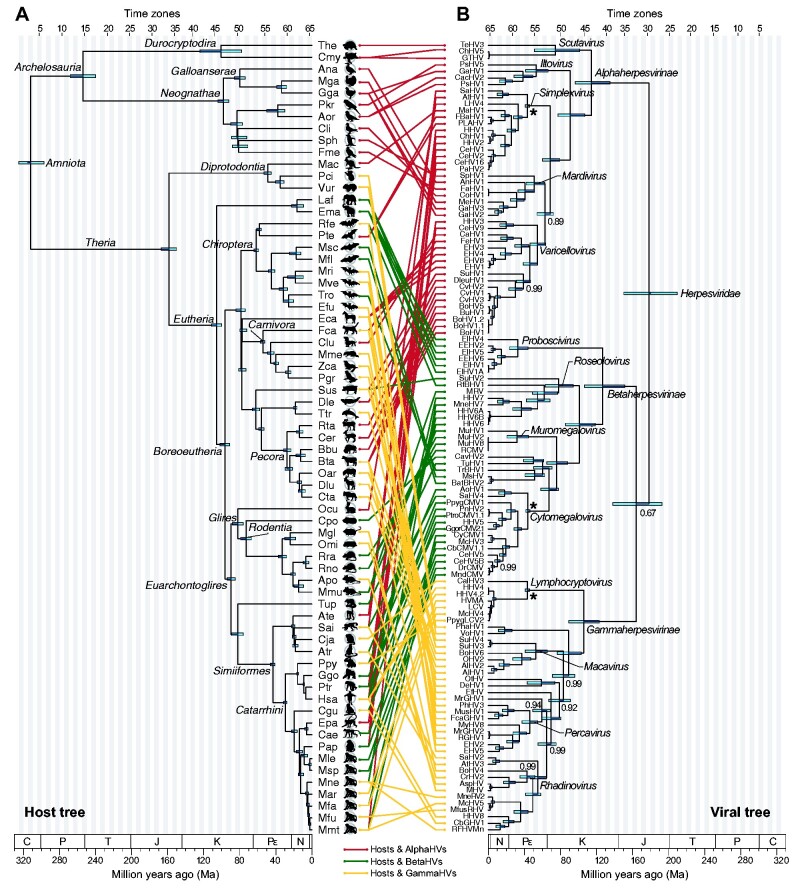
To understand how herpesvirus evolved alongside their hosts, both phylogenies need to be placed in a common time scale. While host species have plenty of fossil evidence to calibrate the internal nodes of their phylogeny, the same is not true for herpesviruses. Currently, two competing hypotheses trace the origins of Herpesviridae back to distinct time periods: 1, McGeoch et al. (1995) suggest that the MRCA of viruses in this family existed between 180 and 220 million years ago (between the Jurassic and Triassic periods); but 2, a decade later, a study by McGeoch and Gatherer (2005) doubled that estimate, suggesting a much earlier origin of Herpesviridae, dating back to a period between 374 and 420 Ma (Devonian). Instead of choosing one of these two estimates to infer the time to the most recent common ancestor (TMRCA) of Herpesviridae and its subfamilies, we calibrated the viral tree pegging one node per subfamily, assuming that ancestors of Simplexvirus, Cytomegalovirus, and Lymphocryptovirus infecting Old World and New World Monkeys diverged along the same time frame as their ancestral Simiiformes hosts, around 42.9 Ma (CI = 41–46 Ma) (Kumar et al. 2017), as done in other studies (McGeoch et al. 1995; Wertheim et al. 2014; Murthy et al. 2019). Using this approach, we performed twelve independent Bayesian phylogenetic analyses, using distinct MSAs and molecular clock models (see Section 2), which produced comparable results (Supplementary Fig. S1). In analyses including all available sequences, the estimates of the Herpesviridae TMRCA had median values varying between 177.3 and 209 Ma (Supplementary Fig. S1). By using alignments including all HV sequences, and those including only sequences specific to each subfamily, the median values of the origins of Alpha-, Beta-, and Gammaherpesvirinae were as follows: Alpha = 113.8–140.6 Ma; Beta = 102.1–144.4 Ma, and Gammaherpesvirinae = 112.2–145.5 Ma (Supplementary Fig. S1). Across all the alignments and molecular clock models we tested, we found that an uncorrelated relaxed clock model consistently yielded the best fit to the data (see Supplementary Table S1). The analyses that used subfamily-specific alignments failed to reconstruct the expected topology within specific HV genera, and the distribution of TMRCA estimates for each subfamily was much broader than that obtained with all Herpesviridae taxa in a single MSA (Supplementary Fig. S1). For these reasons, among all analyses we ran, we opted to use the results obtained using the full Herpesviridae alignment, under a relaxed clock model (Supplementary Fig. S1A and Table S1). This analysis yielded a time-resolved herpesvirus phylogeny that placed the TMRCA of Herpesviridae in the Jurassic period, around 177.3 Ma (credibility interval: 150.1–209.5 Ma), in a time range that mostly matches the hypothesis 1, proposed by McGeoch et al (1995). By comparing virus and host trees using a tanglegram (Fig. 2), we observed that some herpesviruses-infecting closely related hosts are grouped together in the viral tree. However, we also see several topological disagreements involving closely related viruses associated with distantly related hosts in the tanglegram.
Figure 2.
Host–virus tanglegram. The host phylogeny and divergence intervals were obtained from timetree.org (Kumar et al. 2017), and the viral tree was reconstructed using a Bayesian phylogenetic approach implemented on BEAST v1.10.4 (Suchard et al. 2018), using amino acid alignments (see Section 2). Node height 95 per cent highest posterior density (HPD) intervals are shown as blue horizontal bars, and labels are provided for key taxonomic groups. Nodes with posterior probabilities below 1 have their support values highlighted. The asterisks (*) in the viral tree highlight the nodes used for calibration, as described in Section 2. Hosts (A) are connected to their respective viruses (B) with lines coloured to represent the three herpesviral subfamilies: Alpha- (red); Beta- (green), and Gammaherpesvirinae (yellow). Both trees are divided in time zones of five million years, as shown by the scale at the top, and many node HPD intervals span more than one time zone. The geologic time scale is set according to (Gradstein et al. 2012), where D = Devonian period, C = Carboniferous, P = Permian, T = Triassic, J = Jurassic, K = Cretaceous, Pε = Paleogene, N = Neogene. Host acronyms are defined as follows: Ana = Anas sp., Aor = Amazona oratrix, Apo = Apodemus sp., Ate = Ateles sp., Atr = Aotus trivirgatus, Bbu = Bubalus bubalis, Bta = Bos taurus, Cae = Chlorocebus aethiops, Cer = Cervus sp., Cgu = Colobus guereza, Cja = Callithrix jacchus, Cli = Columba livia, Clu = Canis lupus familiaris, Cmy = Chelonia mydas, Cpo = Cavia porcellus, Cta = Connochaetes taurinus, Dle = Delphinapterus leucas, Dlu = Damaliscus lunatus, Eca = Equus caballus, Efu = Eptesicus fuscus, Ema = Elephas maximus, Epa = Erythrocebus patas, Fca = Felis catus, Fme = Falco mexicanus, Gga = Gallus gallus, Ggo = Gorilla gorilla, Hsa = Homo sapiens, Laf = Loxodonta africana, Mac = Macropodidae, Mar = Macaca arctoides, Mfa = Macaca fascicularis, Mfl = Miniopterus fuliginosus, Mfu = Macaca fuscata, Mga = Meleagris gallopavo, Mgl = Myodes glareolus, Mle = Mandrillus leucophaeus, Mme = Meles meles, Mmt = Macaca mulatta, Mmu = Mus musculus, Mne = Macaca nemestrina, Mri = Myotis ricketti, Msc = Miniopterus schreibersii, Msp = Mandrillus sphinx, Mve = Myotis velifer, Oar = Ovis aries, Ocu = Oryctolagus cuniculus, Omi = Oligoryzomys microtis, Pap = Papio sp., Pci = Phascolarctos cinereus, Pgr = Pagophilus groenlandicus, Pkr = Psittacula krameri, Pte = Pteropus sp., Ppy = Pongo pygmaeus, Ptr = Pan troglodytes, Rfe = Rhinolophus ferrumequinum, Rno = Rattus norvegicus, Rra = Rattus rattus, Rta = Rangifer tarandus, Sai = Saimiri sp., Sph = Spheniscus sp., Sus = Sus sp., The = Testudo hermanni, Tro = Tylonycteris robustula, Ttr = Tursiops truncatus, Tup = Tupaiidae, Vur = Vombatus ursinus, Zca = Zalophus californianus. For more details about host and viral taxonomy, accession numbers, and other metadata, see Table Supplementary S2.
3.2 Dated tree reconciliations and cost regimes
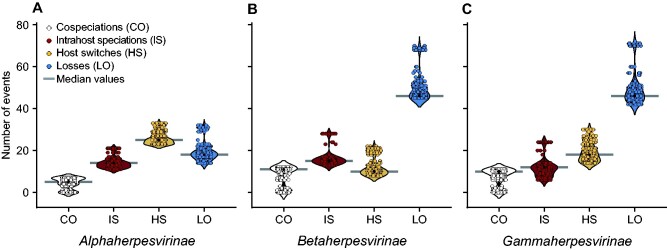
To investigate which events may explain the topological disagreements revealed in the tanglegram in Fig. 2, we performed dated tree reconciliations of 121 herpesviral species and their 67 hosts. Viruses belonging to the main HV subfamilies and their hosts were reconciled in separate runs, using several event costs. We tested 256 cost regimes, which favoured or penalized events differently (see Supplementary Tables S3–S5). Since the association of internal nodes in viral and host trees was constrained by their time zones, the total number of possible solutions was more restricted, and certain events, especially host switches and intrahost speciations, were inferred under all selected cost regimes, being inescapable to explain the evolution of herpesviruses (Supplementary Figs S3–S5). As shown in Figs 4–6, the numbers of cospeciations, intrahost speciations, host switches, and losses inferred by most solutions varied around a common range. Under all tested cost regimes, cospeciations were among the least common events along the evolution of all HV subfamilies (Supplementary Tables S3–S5). Losses were particularly common in beta- and gammaherpesviruses, but among alphaherpesviruses, host switching was the most common event.
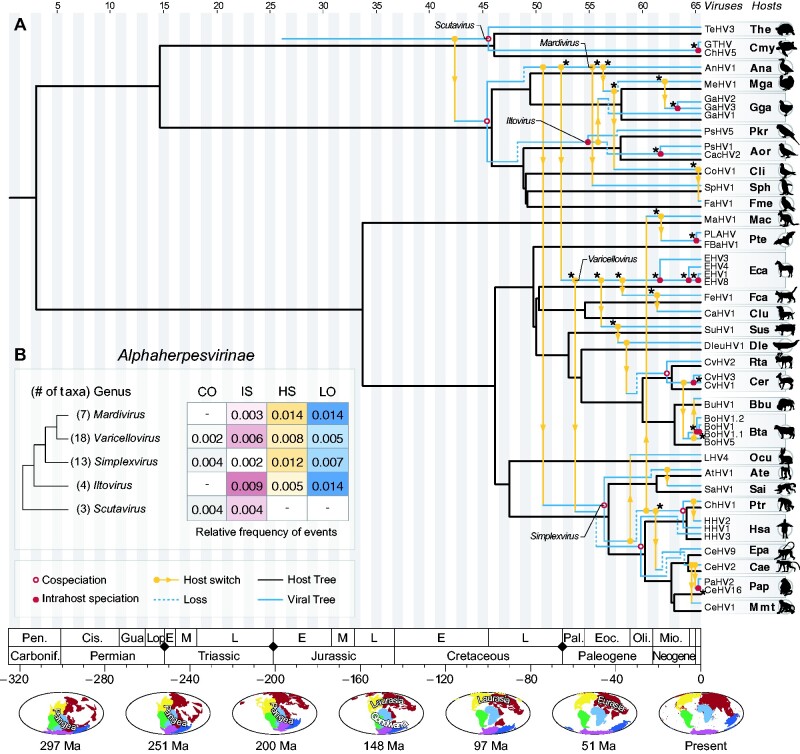
Figure 4.
Tree reconciliation of alphaherpesviruses and their hosts. (A) In this representation, the host tree is shown in black, and the viral tree is shown in blue and yellow, twisted while keeping its original topology, with its nodes assigned to time zones, as shown in Fig. 2. Cophylogenetic events found in more than 90 per cent of the cost regimes employed in this study are marked with an asterisk (*) (see Supplementary Fig. S3). (B) Relative frequency of cophylogenetic events in distinct genera of Alphaherpesvirinae, considering the optimal cost regime (see Supplementary Table S3). The frequencies represent the number of events normalized by the total number of taxa in each genus, and by their respective TMRCA. Along the time scale at the bottom, black diamonds denote major events of mass extinction, as described in (Raup 1993). The maps show changes of landmasses (continental drift) over time, and were retrieved from Paleobiology Database (PBDB) (Peters and McClennen 2016). Cis. = Cisuralian; E = Early; Eoc. = Eocene; Gua. = Guadalupian; L = Late; Lop. = Lopingian; M = Middle; Mio. = Miocene; Oli. = Oligocene; P = Pliocene; Pal. = Paleocene; Pen. = Pennsylvanian.
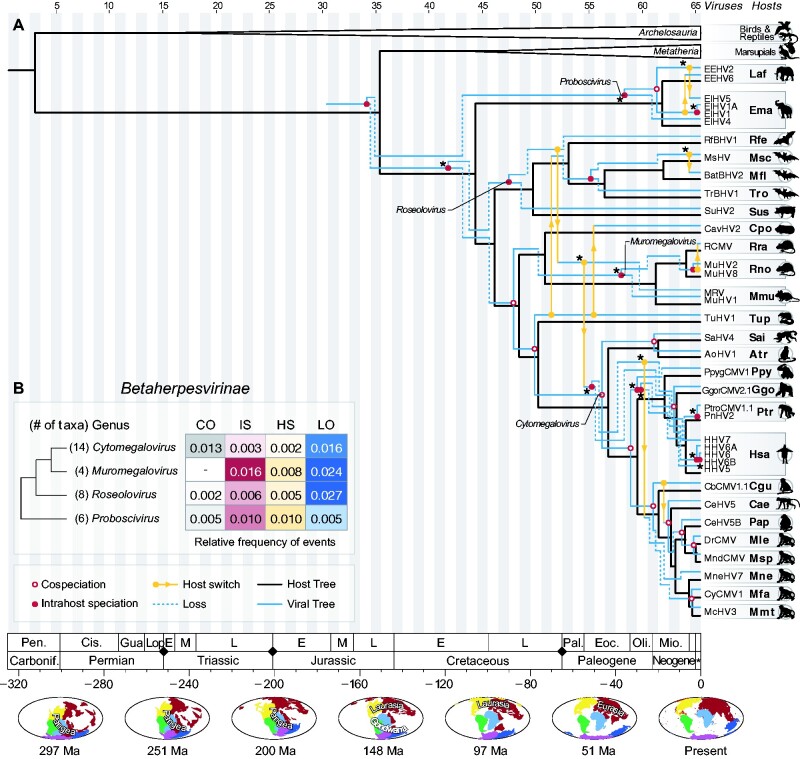
Figure 5.
Tree reconciliation of betaherpesviruses and their hosts. (A) As shown in Fig. 4, the host tree is shown in black, and the viral tree in blue and yellow. Cophylogenetic events found in more than 90 per cent of the cost regimes employed in this study are marked with an asterisk (*) (see Supplementary Fig. S4). (B) Relative frequency of cophylogenetic events in distinct genera of Betaherpesvirinae, considering the optimal cost regime (see Supplementary Table S4). The frequencies represent the number of events normalized by the total number of taxa in each genus, and by their respective TMRCA. Along the time scale, black diamonds denote major events of mass extinction, as described in (Raup 1993). The maps at the bottom show changes of landmasses (continental drift) over time and were retrieved from Paleobiology Database (PBDB) (Peters and McClennen 2016).
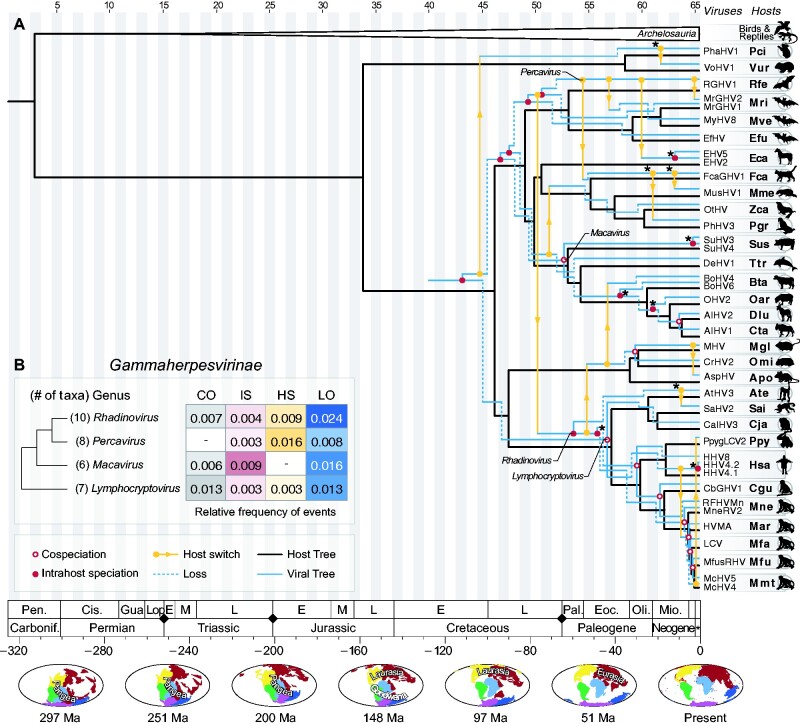
Figure 6.
Tree reconciliation of gammaherpesviruses and their hosts. (A) As shown in Fig. 4, the host tree is shown in black, and the viral tree in blue and yellow. Cophylogenetic events found in more than 90 per cent of the cost regimes employed in this study are marked with an asterisk (*) (see Supplementary Fig. S5). (B) Relative frequency of cophylogenetic events in distinct genera of Betaherpesvirinae, considering the optimal cost regime (see Supplementary Table S5). The frequencies represent the number of events normalized by the total number of taxa in each genus, and by their respective TMRCA. Along the time scale, black diamonds denote major events of mass extinction, as described in (Raup 1993). The maps at the bottom show changes of landmasses (continental drift) over time, and were retrieved from Paleobiology Database (PBDB) (Peters and McClennen 2016).
Following the criteria listed below, a single cost regime was selected, ensuring it was able to: 1, reconstruct the median number of events for each event type (as shown by the grey bars on Fig. 3) and 2, produce a solution with a median total cost (see Section 2). Following this rationale, reconciliations between HVs from different subfamilies were performed and compared (Table 1), allowing us to examine the predominance of each cophylogenetic event across different herpesvirus subfamilies.
Figure 3.
Cophylogenetic events inferred in reconciliations between viral and host trees. A total of 256 distinct cost regimes were tested, resulting in distinct solutions highlighting the agreements and disagreements between viral and host evolutionary histories. The panels above show the number of inferred events under distinct cost regimes, in host-virus tree reconciliations involving (A) alphaherpesviruses, (B) betaherpesviruses, and (C) gammaherpesviruses. The distinct inferred scenarios involved similar numbers of cophylogenetic events, with many losses frequently reported in beta- and gamma-HVs, while cospeciations were the least frequent type of event. Based on the median number of events for each event type (grey bars), an optimal cost regime was selected.
Table 1.
Overall statistics of cophylogenetic events inferred under the optimal cost regime used for host–virus tree reconciliations.
| Subfamily | Number of inferred events |
Overall cost | |||
|---|---|---|---|---|---|
| CO | IS | HS | LO | ||
| α | 6 | 12 | 25 | 17 | 84 |
| β | 12 | 14 | 10 | 43 | 106 |
| γ | 10 | 12 | 17 | 46 | 126 |
The number of inferred events per HV subfamily (α, β and γ) match the median values in Fig. 3. CO, cospeciation; IS, intrahost speciation; HS, host switch; LO, loss.
3.3 Cospeciations in herpesvirus evolution
To distinguish the concepts of ‘cospeciation’ and ‘coevolution’, we here refer to cospeciation as an event of co-divergence, that is the parallel cladogenesis of ancestral forms of hosts and parasites into distinct species along a common period of time, regardless of any causal relationship between the speciation of host and parasite (Jackson 2005; De Vienne et al. 2013). Coevolution between hosts and parasites, on the other hand, is a phenomenon that implies causality and takes place when genetic variations in the parasite species impose selection pressures for the fixation of genetic changes in the host species, and vice versa (Daugherty and Malik 2012). Coevolution is particularly observed in scenarios of molecular ‘arms race’ (Daugherty and Malik 2012), and we consider the investigation of such a phenomenon out of the scope of the present study. By reconciling time-resolved phylogenies, we found that cospeciations were the least common events along the evolution of Herpesviridae (Figs 3–6, Supplementary Figs S2–S5, Tables S3–S5).
Based on the available data, among alpha-HVs, we inferred up to seven cospeciations among the 256 cost regimes we tested (see Supplementary Table S3 and Fig. S3). Using an optimal cost regime, we inferred six of those events (Table 1). The oldest ones date back to the Cretaceous period and involved ancestors of Scutavirus infecting turtle ancestors and ancestors of avian HVs (Iltovirus and Mardivirus) infecting Neognathae birds (see Fig. 4). Among members of Varicellovirus, a cospeciation likely took place between ancestors of HVs infecting deers (CvHV1, CvHV2, and CvHV3) in the Miocene, and another one between ancestors of HVs infecting Catarrhini (Old World Monkeys and Apes) in the Oligocene. Finally, among members of Simplexvirus, we observed two other cospeciation events. One corresponds to the event used as a calibration point, which took place in the Eocene, and involved HVs infecting primate ancestors (Simiiformes), and another one likely took place in the Miocene, involving viruses infecting ancestors of Humans and Chimpanzees (Hominini) (Fig. 4).
Among Betaherpesviruses, at least fourteen cospeciations were inferred using all cost regimes we tested. Using the optimal cost regime, however, twelve cospeciations were reconstructed (Fig. 5, Table 1), most of them among members of Cytomegalovirus infecting primate ancestors, including the one used as a calibration node. The oldest cospeciations observed among beta-HVs date back to the Late Cretaceous, involving viruses infecting ancestors of Euarchontoglires.
Among Gammaherpesviruses, at least twelve cospeciations were found among the cost regimes explored, and ten of these events were found using the optimal cost regime (Fig. 6, Table 1). The earliest event involved Macavirus ancestors, in the Paleocene. Following this event, HVs infecting ancestors of Alcelaphines (gnus and tsessebes) likely co-diverged with their hosts during the Neogene (∼6 Ma). As observed for members of other subfamilies, in Gammaherpesvirinae, cospeciations were more common among viruses infecting primates, such as those of the genera Lymphocryptovirus and Rhadinovirus, and took place especially during the Neogene period (Fig. 6).
3.4 Intrahost speciations: duplications of viral lineages
Intrahost speciations (also known as duplications) occur when a parasite diverges and both lineages remain infecting the same host species (Jackson 2005; De Vienne et al. 2013). Intrahost speciations likely took place during early and late periods of the evolution of HVs (Figs 4–6). During the evolution of alpha-HVs, at least twelve events of intrahost speciation occurred, eleven of which were inferred in more than 90 per cent of the 256 cost regimes explored in this study (see Supplementary Fig. S3). The oldest event of this type likely took place in the Paleogene period, involving ancestors of Iltovirus, but most events occurred during the Neogene and the Quaternary periods, when duplications gave rise to multiple HV species sharing common hosts, as observed for Gallid (GaHV2, 3), Equid (EHV1, 3, 4, 8) and Bovine alphaherpesviruses (BHV1 and similar isolates) (Fig. 4).
The evolutionary histories of beta-HVs and gamma-HVs were also characterized by intrahost speciations (Fig. 3B, C), events that were detected throughout their evolution, since early times (Jurassic and Cretaceous periods). Among members of Betaherpesvirinae, at least fourteen intrahost speciations were identified, nine of which were inferred in all cost regimes (Fig. 5), most of them dating back to early periods of the evolution of beta-HVs, before the Neogene period. The relative frequencies of these events were particularly higher among members of Muromegalovirus and Proboscivirus (Fig. 5). In Gammaherpesvirinae, a similar pattern was found (Fig. 6): a total of twelve intrahost speciations were detected, most of them assigned to early periods. Half (six) of these events were inferred under all cost regimes tested in our analyses, and represent important events in the evolution of these viruses (see Supplementary Fig. S4). In this subfamily, the genus Macavirus had the highest relative frequency of intrahost speciations (Fig. 6).
3.5 Losses: extinctions, sorting events, and undiscovered herpesviruses
Among the four types of cophylogenetic events, losses were among the most frequent (Fig. 3). Losses are usually preceded by intrahost speciations and highlight host clades that lack viruses from certain lineages. At this point, it is essential to emphasize that, in the context of host–parasite tree reconciliations, losses can be interpreted in at least three distinct ways: 1, as lineage sorting events (‘missing the boat’), when a parasite fails to disperse to one of the new host species after their speciation (Johnson et al. 2003); 2, as undiscovered or rare parasites (undersampling) (Page and Charleston 1998); or 3, as genuine events of parasite extinction (Lovisolo, Hull, and Rösler 2003). In the latter scenario, if extinctions explain the absence of viruses infecting certain host clades, it is important to consider that points of losses in reconciliations do not reflect the exact period when extinctions occurred, but rather highlight a point after which such events could have happened at any subsequent time.
Throughout the tree reconciliations (Figs 4–6), viral losses are depicted as dashed lines, which point towards the opposite direction of host clades missing certain viral lineages. Along the evolutionary history of alpha-HVs, the oldest losses date back to the Late Cretaceous period, involving avian HVs, but losses are also observed in earlier periods, such as along the evolution of primates during the Neogene (Fig. 4). Losses were especially detected among beta-HVs (Fig. 5B). The large range of hosts that herpesviruses in this subfamily are associated with can only be explained with the assignment of multiple losses along their evolution. The highest frequency of losses was found in the genus Roseolovirus, a group of HVs that infect a diverse group of mammals, such as bats, rodents and mostly primates (Fig. 5). The gamma-HVs also show a high frequency of losses, which were detected throughout their evolution, since the Cretaceous period. The evolution of HVs of the genus Rhadinovirus, which also infect a diverse group of mammals, can only be explained by means of multiple losses (Fig. 6). Such losses may indicate viral lineages that went extinct in certain host groups, or viruses that may still exist as rare/undiscovered species in nature.
3.6 Host switches
Host switches (transfers) take place when a parasite species gets transferred and succeeds at establishing an infection in a new host not yet explored by their immediate ancestors (De Vienne et al. 2013). In all cost regimes investigated in this study, host switches were evoked to explain the existence of closely related viral lineages infecting distantly related hosts. In tree reconciliations, host switches are special events, since they have directionality: a take-off, and a landing branch in the host phylogeny. Just like in events of loss, the exact timing of the host transfers cannot be determined by phylogenetics itself, and the arrows highlighting the occurrence of such events (see Figs 4–6) indicate how early in time that host switch could have happened. While the direction of some host switches is clear, many were impossible to determine.
By assessing 256 cost regimes, we found that host switches were reported in all cophylogenetic scenarios, providing strong evidence of the crucial role of these events in the evolution of herpesviruses. Based on the optimal cost regime adopted in our analyses (Table 1), we found at least 25 host switches in alpha-HVs, and nine of them were found under all cost regimes we applied (Supplementary Fig. S3). The oldest host switch of alpha-HVs likely took place in the Cretaceous period, involving HVs infecting turtles and bird ancestors. Although the directionality of this event cannot be precisely determined using reconciliation alone, that early host switch along the Cretaceous is inescapable, and essential to reconcile virus and host evolutionary histories. After this first transfer, several other similar events took place along the evolution of alpha-HVs. During the Paleogene and Neogene periods, viruses belonging to the genera Iltovirus and Mardivirus switched between avian hosts on more than one occasion (Fig. 4). Our results also suggest that herpesviruses from the genera Varicellovirus and Simplexvirus, currently known to infect a wide range of mammalian hosts (Davison et al. 2009; Davison 2010), likely had an avian origin, and jumped into mammals at some point between the Late Cretaceous and the Paleogene period (Fig. 4). This interpretation is aligned with hypotheses previously raised in other studies (McGeoch and Cook 1994; McGeoch, Rixon, and Davison 2006). After their transfers to mammalian hosts, the chronological mismatches between host and viral cladogenesis, and the large number of distantly related hosts, infected by closely related viruses, prompt an evolutionary scenario that can only be explained by multiple host switches. Some undeniable host switches revealed in our analysis, for example, involve ancestors of Simplexvirus that now infect primates (humans and non-humans), such as bats (infected by PLAHV and FBaHV1) and marsupials (infected by MaHV1). Another important host switch likely took place after 2.2 Ma, when viruses infecting Chimpanzees (genus Pan) were transferred to humans, giving rise to HHV2, as reported in a previous study (Wertheim et al. 2014).
Among beta-HVs, host switches were less prominent, but still crucial during their evolution (Fig. 5). We inferred a total of ten events following the optimal cost regime, four of which are found in most cost regimes (Supplementary Fig. S3): a transfer involving members of Proboscivirus infecting elephants; a transfer involving HV infecting bats, such as MsHV and BatBHV2; a transfer that established Roseolovirus in primate hosts during the Paleogene; and a transfer that likely allowed HVs of great apes to infect macaques (Fig. 5A).
Finally, for gamma-HVs, we found a total of seventeen host switches, four of which were found in more than 90 per cent of the cost regimes we tested (Supplementary Fig. S4): a switch from marsupial hosts that likely took place in the Neogene, between Koala and Wombat ancestors; two transfers of HVs between ancestors of felines and other members of Carnivora (mustelids and phocids), and; a transfer involving HVs infecting New World Monkeys (Fig. 6A). Overall, transfers were particularly common among members of the genus Percavirus, which infect a broad range of mammals (Davison 2010) (Fig. 6B).
4. Discussion
To understand how herpesviruses succeeded at infecting a diverse group of animal hosts, cophylogenetic analyses of time-resolved trees can provide valuable insights about the dynamics of host-virus evolution. Exploiting the temporal data embedded in time-resolved phylogenies, we used dated tree reconciliations to uncover patterns of herpesvirus-host evolution by inferring events of cospeciation, host switches, intrahost speciations, and losses along millions of years of evolutionary history.
4.1 The origins of Herpesviridae
In the 1990s, several studies focused on dating the origins of the family Herpesviridae using distinct sets of gene sequences and molecular clock models. Those studies employed neighbor-joining methods, and used host divergence times to peg internal nodes of the viral tree, assuming cospeciations between alpha-HVs and ancestors of Perissodactyla, Artiodactyla, New World and Old World Monkeys. Following this approach, they dated the TMRCA of Herpesviridae back to the Jurassic/Triassic periods, between 180 and 220 million years ago (McGeoch and Cook 1994; McGeoch et al. 1995; McGeoch, Dolan, and Ralph 2000). Years later, using more sequences from HVs infecting mammals, birds and reptiles, and also assuming cospeciations between HVs and their respective hosts, new studies placed the origin of Herpesviridae in the range of 374 to 420 Ma, in the Devonian period (McGeoch, Dolan, and Ralph 2000; McGeoch and Gatherer 2005), doubling the root age estimated in aforementioned studies. Differing from our previous approach in which we used estimates from these latest studies to peg the TMRCA of Herpesviridae back to the Devonian (Brito and Pinney 2020), we calibrated our viral phylogeny in this study assuming that New World and Old World Monkeys, and their respective HVs, diverged along a common period of time, matching the divergence time of Simiiformes ancestors (∼42.9 Ma) (Steiper and Young 2006). Doing so, we applied the same rationale used in those studies from the 1990s and 2000s, but adopted Bayesian phylogenetic inference. Using Bayesian model selection approaches and distinct molecular clock models implemented in BEAST v.1.10.4 (Suchard et al. 2018), we concluded that the relaxed molecular clock model fits our data better than a strict or random local clock model. Therefore, by applying a relaxed molecular model to the amino acid alignments of gB (UL27) and DNApol (UL30) including 121 herpesviruses from all subfamilies, we estimated the origin of Herpesviridae to range between 150.1 and 209.5 Ma. This timing is conditional on the calibration points that were used (McGeoch and Cook 1994; McGeoch et al. 1995; McGeoch, Dolan, and Ralph 2000) on the present study. However, unless further evidence is available to debunk its plausibility, the timescale proposed in this study remains as a reasonable hypothesis for the origins of Herpesviridae, which are in line with previous estimates (McGeoch and Cook 1994; McGeoch et al. 1995; McGeoch, Dolan, and Ralph 2000). As more HV sequences are made available, especially those from HV-infecting reptiles, the TMRCA of the root will likely be pushed further back. Nodes that are more proximal to the root may likely be pushed back as well, but we expect that the highest posterior density of their TMRCAs will likely fluctuate around similar time periods.
4.2 Consistency in tree reconciliations
Dated tree reconciliations are very nuanced analyses, subject to multiple levels of uncertainties, as they rely on two phylogenetic properties that frequently elicit debates: the tree topologies and their divergence times. For performing reconciliations, yet another element is of paramount importance: the host-parasite associations, i.e. who infects whom. If any of these elements disagrees with the current understanding about the ecology and evolution of the organisms under study, the analysis will likely render questionable results. To prevent such issues, we ensured the accuracy of the host tree topologies by using a host tree with divergence times validated by zoologists and paleontologists, available on timetree.org (Kumar et al. 2017). To achieve a similar level of accuracy in the viral side, we performed thorough phylogenetic analyses using BEAST v.1.10.4 (Suchard et al. 2018), testing distinct alignments and clock models, as detailed in the Methods section, while ensuring that viral clades match the taxonomic classification proposed by the ICTV (King et al. 2018) and those identified in independent studies. Finally, host–virus pairings were automatically extracted from NCBI (Brister et al. 2015), compared with data available on Virus-Host Database (Mihara et al. 2016), and manually curated.
Another important aspect that may also affect the accuracy of tree reconciliations is the choice of relative costs associated with each cophylogenetic event: cospeciation, intrahost speciation, host switching, and loss. To assess the level of uncertainty that this choice imposes, we explored 256 distinct cost regimes, where we tested all possible combinations of relative costs per event (from 0 to 3). This allowed us to choose a cost regime that would yield the median number of events per event type, with a median total cost (Table 1, Supplementary Tables S3–S5). In the absence of more objective evidence to choose a cost regime, using an approach aiming at median values constitutes a balanced approach, and future studies should account for this limitation while performing tree reconciliations. To assess how variable the reconciliation reconstructions were, we show (Supplementary Figs S3–S5) all possible events found in the best solutions obtained under all cost regimes. This analysis revealed that many events were inferred in all reconstructions, irrespectively of the relative cost of the events. This shows that, due to the constraints imposed by the temporal scale, many events are likely inescapable, essential to explain the evolution of herpesviruses alongside their hosts.
4.3 Interpreting topological and chronological incongruences in virus–host phylogenies
When the first sequences of herpesviruses were made available, the scarcity of sampled taxa did not allow accurate interpretations about the evolution of herpesviruses and their hosts. Originally, herpesviruses were thought to evolve mainly by cospeciation, with viral divergence contingent upon host speciation (McGeoch et al. 1995; Davison 2002; Jackson 2005; McGeoch, Rixon, and Davison 2006). For cospeciations to be accurately inferred, parasite and host trees must not only show topological congruence, but also similar divergence times (De Vienne et al. 2013). Tree reconciliations performed with undated trees and few taxa may overestimate the occurrence of cospeciations via chronologically inconsistent node pairings. In the present study we performed reconciliation analyses using time-resolved trees, which enabled us to observe that, due to temporal incompatibilities, cospeciations were in fact rare events in most herpesvirus genera, with other events playing more predominant roles. Our results show that members of Cytomegalovirus and Lymphocryptovirus are exceptions to this trend, with cospeciations playing a central role during their evolution (Figs 5 and 6).
As more herpesviruses were characterized and sequenced, topological disagreements between host and viral tree topology became evident. Since cospeciations alone cannot explain the evolution of HVs, intrahost speciations and host switches had already been proposed to account for such phylogenetic incongruences (McGeoch et al. 1995; McGeoch and Gatherer 2005; McGeoch, Rixon, and Davison 2006; Ehlers et al. 2008; Escalera-Zamudio et al. 2016). We revealed that herpesvirus and host phylogenies show chronological and topological disagreements, caused by closely related viruses associated with a diverse range of hosts, pattern that could only be explained by host switches or intrahost speciations, many of which were reported in all scenarios we investigated (Supplementary Figs S2–S5).
4.4 Losses in tree reconciliations: gaps in the natural history of viruses
The occurrence of losses along the viral evolution timeline provides another explanation for the phylogenetic disagreements detected during tree reconciliations. Viral losses do not necessarily mean extinctions, as they may also indicate undersampling or undiscovered viruses (De Vienne et al. 2013). Losses have shown higher frequency in specific genera, such as Muromegalovirus, Roseolovirus and Rhadinovirus, some of which are largely undersampled (Fig. 5B), an observation that makes the hypothesis of undiscovered viruses more plausible. If extinctions are invoked to explain these losses, some events linked to the evolution of the hosts can be assigned as potential causes of the elimination of viral clades, one of them being host extinction. Although each mass extinction could have wiped out up to 96 per cent of the ancient species, most extinctions in the last 500 million years were the result of minor events taking place in-between major events of mass extinctions (Raup 1993). Since the average duration of species is estimated to be four million years, with genera lasting for around 28 Myr (Raup 1993), symbiont extinction may also explain some of the losses inferred in the present study. Apart from host populations being wiped out, leading to viral elimination, cataclysmic events can also cause sharp decreases in host populations (Hesse and Buckling 2016), leading to bottleneck effects that may promote fixation of alleles linked to hosts’ resistance to pathogens, consequently affecting viral adaptation to those hosts (Hesse and Buckling 2016).
The high frequency of losses inferred in our tree reconciliation highlight how little we know about the diversity and natural history of viruses. In this study we made an effort to include as many taxa as possible, basing our analysis only on UL27 (gB) and UL30 (DNApol). Unfortunately, phylogenetic signal from single gene alignments (like DNApol) was not enough to resolve the evolutionary history of herpesviruses, and prevented the inclusion of amphibian, fish and invertebrate HVs, which would reveal even more interesting patterns of evolution. However, at least with the inclusion of more taxa based on UL27 and UL30 sequences, future studies will be able to resolve the large amount of losses highlighted in the present study and provide a more accurate view of herpesvirus-host evolution. As genome sequencing becomes more accessible, more herpesvirus species will be discovered, not only adding new pieces in the puzzle but also revealing new gaps in our knowledge about the ecology and evolution of herpesviruses.
4.5 Genetic factors determining host switches
The main finding of our study was the important role of host switches in the evolution of herpesviruses, events previously proposed to play some role (McGeoch et al. 1995; McGeoch and Gatherer 2005; McGeoch, Rixon and Davison 2006), but considered to be less common than cospeciations and intrahost speciations (McGeoch et al. 1995; McGeoch, Dolan and Ralph 2000; McGeoch and Gatherer 2005). The importance of this type of event, however, could not be revealed up until recent years, with more extensive sampling, and more robust phylogenetic tools. Taking advantage of these resources, our study revealed the important contribution of host switches at defining the current host range of herpesviruses. As mutation rate and effective population size affect the likelihood of adaptation of a pathogen to a new host (Longdon et al. 2014), the extinction of viruses transmitted to new host species may occur frequently (Geoghegan, Duchêne and Holmes 2017). As a result, most host switches cannot be easily detected, and the number of transfers inferred in our cophylogenetic analyses is probably underestimated. Although viruses are more likely to switch between closely related hosts, which may have similar ecological and genetic characteristics, host switches can also occur over large phylogenetic distances (De Vienne et al. 2013; Geoghegan, Duchêne, and Holmes 2017). Hosts from distantly related clades can independently acquire or lose immunogenetic elements, such as protein motifs, domains or whole genes, which can eventually increase their levels of susceptibility to pathogens, making some host switches more feasible (Longdon et al. 2014). Along their evolutionary history, herpesviruses gained, duplicated and lost genomic regions encoding specific protein domains (Brito and Pinney 2020). Given the defensive and offensive strategies of evolution adopted by pathogens and their hosts in molecular arms races (Daugherty and Malik 2012), some herpesviruses likely succeeded at host switching by evading and/or neutralizing host immune factors after acquiring and/or losing elements from their domain repertoires (Brito and Pinney 2017, 2020). Comparing our results here with our previous study (Brito and Pinney 2020), it is possible to identify many instances of pronounced changes in protein domain repertoires along branches representing host transfers in Figs 4–6. Some notable examples are the major genomic reshaping that were observed in FBaHV1 and MaHV1 (Alphaherpesvirinae), likely after being transferred from primates: FBaHV1 gained and duplicated many envelope and modulatory protein domains, while MaHV1 lost domains related to envelope proteins (Brito and Pinney 2020). Similar patterns were also reported for GaHV2 and GaHV3, viruses that are likely the result of a transfer from other gallid, and now infect chickens. While GaHV2 gained and duplicated many accessory protein domains, and is highly pathogenic to their hosts, GaHV3 followed a distinct path, and is known to be non-pathogenic, being even used in vaccine formulations (López-Osorio et al. 2017; Brito and Pinney 2020). These changes in genomic composition following not only transfers, but also cospeciations and intrahost speciations are worth further investigations, in order to determine how herpesviruses have been adapting to their hosts along their evolution.
4.6 Ecological factors determining host switches
Looking at the interactions between viruses and hosts from a historical perspective, not only genetic factors, but also ecological factors changed over time, and may have affected the likelihood of certain host switches. Since direct or indirect contact is required for herpesviral transmission, the geological movement of landmasses split and/or merged host populations in the past, which may have allowed or prevented certain host switches (Lovisolo, Hull, and Rösler 2003). To acquire a better understanding of ancestral host transfers, it is also important to consider the biogeography (spatial distribution) of ancestral hosts, and the geological history of the Earth. As shown at the bottom of Figs 4–6, alongside the evolution of hosts and their associated viruses, the planet underwent drastic changes. Taking these ecological factors into consideration, most host switches inferred in this study are consistent with the historical biogeography of the hosts. For example, the origins of mammalian alpha-HVs (Simplexvirus and Varicellovirus) by means of host switches from avian ancestors are a plausible hypothesis, since ancestors of those animal species likely coexisted in time and space, and it is now clear that that event of host switch is a required step in the evolution of herpesviruses, given the currently available sequence data, and understanding about their host range. Among alpha-HVs infecting birds, those of the genus Mardivirus experienced relatively more host switches than other viruses of the same subfamily (Fig. 4). The ability of powered flight, and the widespread distribution of avian species since the early stages of their evolution (Claramunt and Cracraft 2015), may have favoured host transfers from and between these animals, due to their likely shared habitats, and as a consequence, more close contacts. Powered flight may also explain the close relation between HVs infecting bats and other mammalian hosts. Among alpha-HVs, for example, the viruses FBaHV1 and PLAHV, and MaHV1 are related to primate HVs, but infect distantly related hosts, such as bats of the genus Pteropus, and macropodids, respectively. Since ancestors of Pteropus sp. (Megabats) inhabited Europe and Asia alongside primate ancestors in the Paleogene and Neogene (Springer et al. 2011, 2012), transfers of HVs between primates and bats were likely to occur in that period. The subsequent host switch between megabats and ancestors of the Kangaroo and Wallaby can be explained by their current and ancestral distributions in Australia, and by the dispersal capabilities of bats using powered flight (Springer et al. 2011). Similar patterns of host switch involving bats are also observed among beta-HV and gamma-HV (Figs 5 and 6), subfamilies where herpesviruses infecting bats cluster closely together with viruses infecting a broad range of mammalian hosts, including primates (Fig. 2).
The prominent role of host switches among a diverse range of hosts was also observed in non-flying animals. Members of Varicellovirus, for example, infect a broad range of mammalian hosts, such as carnivores and ungulates, and the co-existence of their ancestors in Eurasia during the Paleogene (Springer et al. 2011) shows that host switches between these host groups were plausible from chronologic and geographic stand points (Fig. 4). Host switches involving primate HVs were observed in all herpesvirus subfamilies, most of them taking place during the Miocene, Pliocene and Quaternary. Our analyses reproduced the results by Wertheim et al. (2014), which suggested that HHV2, closely related to HHV1 and ChHV1, could have originated from a host switch of HVs from Chimpanzees around 1.6 Ma. The same study pointed out another possible host switch involving ancestors of CeHV2 that we independently discovered. Our results revealed that simplexviruses infecting Old World Monkeys, such as CeHV1, CeHV2, and CeHV16 (Fig. 2), are likely the result of host switches that took place around the Pliocene (Fig. 4). The highly social behaviour of primate species, and the shared ecological niches they occupy may favour not only the transmission of viruses within members of the same species but also cross-species transmission (Griffin and Nunn 2012; Karesh et al. 2012). Cophylogenetic analysis itself is not sufficient to uncover the directionality of host switches, and cannot tell us the impact of specific animal groups at spreading herpesviruses. Further studies related to the ecology of animal hosts, and their associations with zoonoses caused by herpesviruses are necessary (Tischer and Osterrieder 2010; Karesh et al. 2012).
In conclusion, we used time-resolved phylogenies of herpesviruses and their hosts to perform dated tree reconciliations. By means of this approach we were not only able to detect topological disagreements between viral and host tree topologies but also, more importantly, we revealed important chronological mismatches of divergence times between animal species and their herpesviruses. Our dated reconciliations highlighted the important roles of host switches and intrahost speciations in the evolution of herpesviruses. Losses were also common, but their meaning along the natural history of herpesviruses cannot be determined by tree reconciliation alone. However, they likely indicate the existence of undiscovered viruses, or even episodes of viral extinction. As more viral sequences are incorporated in tree reconciliations, the real nature of such losses will be revealed. Finally, cospeciations between herpesviruses and their hosts are uncommon, and mainly observed between specific host–virus pairs, such as herpesviruses-infecting primates.
Supplementary Material
Acknowledgements
AFB is funded by Ciência sem Fronteiras, a scholarship programme managed by the Brazilian federal government (CAPES, Ministry of Education, Grant number: 11911-13-1). GB acknowledges support from the Interne Fondsen KU Leuven/Internal Funds KU Leuven under grant agreement C14/18/094, and the Research Foundation—Flanders (‘Fonds voor Wetenschappelijk Onderzoek—Vlaanderen’, G0E1420N). KDN acknowledges support from the Research Foundation—Flanders (‘Fonds voor Wetenschappelijk Onderzoek—Vlaanderen’, 1S33020N). The computational resources and services used in this work were provided by the VSC (Flemish Supercomputer Center), funded by the Research Foundation—Flanders (FWO) and the Flemish Government—department EWI. NDG is supported by a start-up package provided by the Yale School of Public Health. JWP is supported by a University Research Fellowship from the Royal Society. The authors thank the Imperial College London Open Access Fund for the financial support.
Data availability
All data used in this study and codes generated for the analyses are deposited in the following repository on GitHub: https://github.com/andersonbrito/openData/tree/master/brito_2020_reconciliation.
Supplementary data
Supplementary data are available at Virus Evolution online.
Conflict of interest: None declared.
Contributor Information
Anderson F Brito, Department of Life Sciences, Imperial College London, South Kensington Campus. London SW7 2AZ, UK; Department of Epidemiology of Microbial Diseases, Yale School of Public Health, Yale University, New Haven, CT 06510, USA.
Guy Baele, Department of Microbiology, Immunology and Transplantation, Laboratory of Clinical and Epidemiological Virology, Rega Institute, KU Leuven, Leuven 3000, Belgium.
Kanika D Nahata, Department of Microbiology, Immunology and Transplantation, Laboratory of Clinical and Epidemiological Virology, Rega Institute, KU Leuven, Leuven 3000, Belgium.
Nathan D Grubaugh, Department of Epidemiology of Microbial Diseases, Yale School of Public Health, Yale University, New Haven, CT 06510, USA.
John W Pinney, Department of Life Sciences, Imperial College London, South Kensington Campus. London SW7 2AZ, UK.
References
- Ayres D. L. et al. (2019). ‘BEAGLE 3: improved performance, scaling, and usability for a high-performance computing library for statistical phylogenetics’, Systematic biology, 68: 1052–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G., Lemey P., Suchard M. A. (2016) ‘Genealogical Working Distributions for Bayesian Model Testing with Phylogenetic Uncertainty’, Systematic Biology, 65: 250–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brister J. R. et al. (2015) ‘NCBI Viral Genomes Resource’, Nucleic Acids Research, 43: D571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito A. F., Pinney J. W. (2017) ‘Protein–Protein Interactions in Virus–Host Systems’, Frontiers in Microbiology, 8: 1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito A. F., Pinney J. W. (2020) ‘The Evolution of Protein Domain Repertoires: Shedding Light on the Origins of the Herpesviridae Family’, Virus Evolution, 6: veaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claramunt S., Cracraft J. (2015) ‘A New Time Tree Reveals Earth History’s Imprint on the Evolution of Modern Birds’, Science Advances, 1: e1501005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conow C. et al. (2010) ‘Jane: A New Tool for the Cophylogeny Reconstruction Problem’, Algorithms for Molecular Biology : Amb, 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty M. D., Malik H. S. (2012) ‘Rules of Engagement: Molecular Insights from Host-Virus Arms Races’, Annual Review of Genetics, 46: 677–700. [DOI] [PubMed] [Google Scholar]
- Davison A. J. (2002) ‘Evolution of the Herpesviruses’, Veterinary Microbiology, 86: 69–88. [DOI] [PubMed] [Google Scholar]
- Davison A. J. (2010) ‘Herpesvirus Systematics’, Veterinary Microbiology, 143: 52–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J. et al. (2009) ‘The Order Herpesvirales’, Archives of Virology, 154: 171–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vienne D. M. et al. (2013) ‘Cospeciation vs Host-Shift Speciation: Methods for Testing, Evidence from Natural Associations and Relation to Coevolution’, The New Phytologist, 198: 347–85. [DOI] [PubMed] [Google Scholar]
- Ehlers B. et al. (2008) ‘Novel Mammalian Herpesviruses and Lineages within the Gammaherpesvirinae: Cospeciation and Interspecies Transfer’, Journal of Virology, 82: 3509–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalera-Zamudio M. et al. (2016) ‘Bats, Primates, and the Evolutionary Origins and Diversification of Mammalian Gammaherpesviruses’, MBio, 7:. DOI: 10.1128/mBio.01425-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y. et al. (2011) ‘Choosing among Partition Models in Bayesian Phylogenetics’, Molecular Biology and Evolution, 28: 523–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J. L., Duchêne S., Holmes E. C. (2017) ‘Comparative Analysis Estimates the Relative Frequencies of co-Divergence and Cross-Species Transmission within Viral Families’, PLoS Pathogens, 13: e1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradstein F. M. et al. (2012) The Geologic Time Scale 2012. Oxford, UK: Elsevier. [Google Scholar]
- Griffin R. H., Nunn C. L. (2012) ‘Community Structure and the Spread of Infectious Disease in Primate Social Networks’, Evolutionary Ecology, 26: 779–800. [Google Scholar]
- Hesse E., Buckling A. (2016) ‘Host Population Bottlenecks Drive Parasite Extinction during Antagonistic Coevolution’, Evolution; International Journal of Organic Evolution, 70: 235–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. (2005) ‘The Effect of Paralogous Lineages on the Application of Reconciliation Analysis by Cophylogeny Mapping’, Systematic Biology, 54: 127–45. [DOI] [PubMed] [Google Scholar]
- Johnson K. P. et al. (2003) ‘When Do Parasites Fail to Speciate in Response to Host Speciation? ’, Systematic Biology, 52: 37–47. [DOI] [PubMed] [Google Scholar]
- Karesh W. B. et al. (2012) ‘Ecology of Zoonoses: Natural and Unnatural Histories’, Lancet (London, England), 380: 1936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass R. E., Raftery A. E. (1995) ‘Bayes Factors’, Journal of the American Statistical Association, 90: 773–95. [Google Scholar]
- Katoh K., Standley D. M. (2013) ‘MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. M. Q. et al. (2018) ‘Changes to Taxonomy and the International Code of Virus Classification and Nomenclature Ratified by the International Committee on Taxonomy of Viruses (2018)’, Archives of Virology, 163: 2601–31. [DOI] [PubMed] [Google Scholar]
- Kumar S. et al. (2017) ‘TimeTree: A Resource for Timelines, Timetrees, and Divergence Times’, Molecular Biology and Evolution, 34: 1812–9. [DOI] [PubMed] [Google Scholar]
- Le S. Q., Gascuel O. (2008) ‘An Improved General Amino Acid Replacement Matrix’, Molecular Biology and Evolution, 25: 1307–20. [DOI] [PubMed] [Google Scholar]
- Longdon B. et al. (2014) ‘The Evolution and Genetics of Virus Host Shifts’, PLoS Pathogens, 10: e1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Osorio S. et al. (2017) ‘Molecular Characterization of Marek’s Disease Virus in a Poultry Layer Farm from Colombia’, Poultry Science, 96: 1598–608. [DOI] [PubMed] [Google Scholar]
- Lovisolo O., Hull R., Rösler O. (2003) ‘Coevolution of Viruses with Hosts and Vectors and Possible Paleontology’, Advances in Virus Research, 62: 325–79. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Cook S. (1994) ‘Molecular Phylogeny of the Alphaherpesvirinae Subfamily and a Proposed Evolutionary Timescale’, Journal of Molecular Biology, 238: 9–22. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J. et al. (1995) ‘Molecular Phylogeny and Evolutionary Timescale for the Family of Mammalian Herpesviruses’, Journal of Molecular Biology, 247: 443–58. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dolan A., Ralph A. C. (2000) ‘Toward a Comprehensive Phylogeny for Mammalian and Avian Herpesviruses’, Journal of Virology, 74: 10401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Gatherer D. (2005) ‘Integrating Reptilian Herpesviruses into the Family Herpesviridae’, Journal of Virology, 79: 725–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Rixon F. J., Davison A. J. (2006) ‘Topics in Herpesvirus Genomics and Evolution’, Virus Research, 117: 90–104. [DOI] [PubMed] [Google Scholar]
- Mihara T. et al. (2016) ‘Linking Virus Genomes with Host Taxonomy’, Viruses, 8: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy S. et al. (2019) ‘Cytomegalovirus Distribution and Evolution in Hominines’, Virus Evolution, 5: vez015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R. D., Charleston M. A. (1998) ‘Trees within Trees: Phylogeny and Historical Associations’, Trends in Ecology & Evolution, 13: 356–9. [DOI] [PubMed] [Google Scholar]
- Peters S. E., McClennen M. (2016) ‘The Paleobiology Database Application Programming Interface’, Paleobiology, 42: 1–7. [Google Scholar]
- Rambaut A. et al. (2018) ‘Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7’, Systematic Biology, 67: 901–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raup D. M. (1993) ‘Extinction from a Paleontological Perspective’, European Review (Chichester, England), 1: 207–16. [DOI] [PubMed] [Google Scholar]
- Springer M. S. et al. (2012) ‘Macroevolutionary Dynamics and Historical Biogeography of Primate Diversification Inferred from a Species Supermatrix’, PLoS One, 7: e49521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S. et al. (2011) ‘The Historical Biogeography of Mammalia’, Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 366: 2478–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiper M. E., Young N. M. (2006) ‘Primate Molecular Divergence Dates’, Molecular Phylogenetics and Evolution, 41: 384–94. [DOI] [PubMed] [Google Scholar]
- Suchard M. A. et al. (2018) ‘Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10’, Virus Evolution, 4: vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer B. K., Osterrieder N. (2010) ‘Herpesviruses—a Zoonotic Threat?’, Veterinary Microbiology, 140: 266–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udny Yule G. (1925) ‘A Mathematical Theory of Evolution, Based on the Conclusions of Dr. JC Willis’, Frs. Rsptb, 213: 21–87. [Google Scholar]
- Wertheim J. O. et al. (2014) ‘Evolutionary Origins of Human Herpes Simplex Viruses 1 and 2’, Molecular Biology and Evolution, 31: 2356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (1994) ‘Maximum Likelihood Phylogenetic Estimation from DNA Sequences with Variable Rates over Sites: Approximate Methods’, Journal of Molecular Evolution, 39: 306–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study and codes generated for the analyses are deposited in the following repository on GitHub: https://github.com/andersonbrito/openData/tree/master/brito_2020_reconciliation.