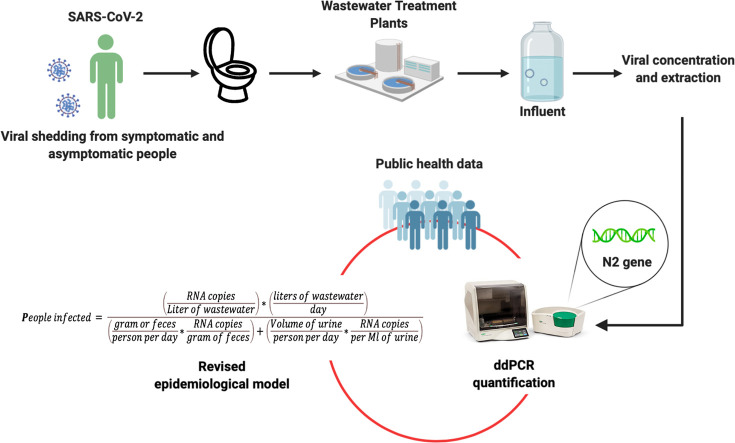
Abstract
Monitoring of COVID-19 infections within communities via wastewater-based epidemiology could provide a cost-effective alternative to clinical testing. This approach, however, still requires improvement for its efficient application. In this paper, we present the use of wastewater-based epidemiology in monitoring COVID-19 infection dynamics in the KwaZulu-Natal province of South Africa, focusing on four wastewater treatment plants for 14 weeks. The SARS-CoV-2 viral load in influent wastewater was determined using droplet digital PCR, and the number of people infected was estimated using published models as well as using a modified model to improve efficiency. On average, viral loads ranged between 0 and 2.73 × 105 copies/100 ml, 0–1.52 × 105 copies/100 ml, 3 × 104–7.32 × 105 copies/100 ml and 1.55 × 104–4.12 × 105 copies/100 ml in the four wastewater treatment plants studied. The peak in viral load corresponded to the reported COVID-19 infections within the districts where these catchments are located. In addition, we also observed that easing of lockdown restrictions by authorities corresponded with an increase in viral load in the untreated wastewater. Estimation of infection numbers based on the viral load showed that a higher number of people could potentially be infected, compared to the number of cases reported based on clinical testing. The findings reported in this paper contribute to the field of wastewater-based epidemiology for COVID-19 surveillance, whilst highlighting some of the challenges associated with this approach, especially in developing countries.
Keywords: SARS-CoV-2, COVID-19, Wastewater-based epidemiology, Droplet digital PCR, Infection prediction models
Graphical abstract
1. Introduction
COVID-19 remains a major threat to public health and the economy globally, more than a year after the first cases were reported in China. A key challenge associated with this disease is the difficulty in identifying infected persons based on symptoms as well as the spread in the community (Hart and Halden, 2020). Curtailing its spread and impact is still a major problem with several countries experiencing a second wave of infections and are back on lockdown, despite an earlier reduction in infections (Walker and Bisserbe, 2020). Rapid identification and isolation of infected individuals are key; however, the prolonged incubation period (2–14 days) of the virus and its transmission in the absence of clinical signs and symptoms have contributed to its community spread (Daughton, 2020; Le et al., 2020).
According to Daughton (2020), the conventional approach for managing epidemics has always been the large-scale application of diagnostic testing at the individual case level. However, clinical diagnostic testing has proven to be inadequate for providing rapid surveillance of larger populations and keeping pace with the COVID-19 infections. As noted by Hart and Halden (2020), this approach of mass testing is time-consuming, expensive, and involves exposure risks for those administering the tests. According to Al-Tawfiq (2020), the contribution of asymptomatic individuals to SARS-CoV-2 transmission is still not well understood. Hence, identifying the disease hotspot based on clinical surveillance may be impossible, resulting in the ineffective deployment of public health interventions. Therefore, most countries are struggling to keep pace with the containment of the pandemic, whilst many potential hotspots remain unidentified.
Wastewater-based epidemiology (WBE) also known as environmental surveillance has been proposed by many authors as an alternative approach for a cost-effective and near real-time identification of COVID-19 hotspots and community spread (Tetteh et al., 2020; Venugopal et al., 2020). WBE is a relatively new tool that can be used in profiling human exposure to chemicals or biological agents at the population level (Daglioglu et al., 2020). This approach has gained prominence during the current pandemic. According to Kitajima et al. (2020), both viable SARS-CoV-2 and viral nucleic acid is shed via saliva, sputum, urine, and feces, which eventually end up in the wastewater (Kitajima et al., 2020). Hence, WBE could serve as a data source, for tracking COVID-19 hotspots and dynamics of the disease circulating in the community within the vicinity of the wastewater treatment plant (WWTP) catchment (Lodder and de Roda Husman, 2020). Based on the WBE principle, as described by Choi et al. (2018), the viral RNA copy numbers quantified in the untreated wastewater samples can be normalized to the per capita mass loads estimated from a combination of flow rate and total population within the WWTP catchment boundaries. Although WBE cannot be a replacement for clinical testing, it can serve as an early warning system indicating the presence of infected individuals in a particular community (i.e. district, town, or cities) or within a specific WWTP catchment boundary (Hart and Halden, 2020).
Several studies have demonstrated the applicability of WBE models for the identification of COVID-19 infection hotspots and to estimate the number of infected individuals (Ahmed et al., 2020; Gonzalez et al., 2020; Hart and Halden, 2020). With the application of appropriate models, WBE can be used to back-calculate exposure to or usage of chemicals or prevalence of infection (Hill et al., 2020). The accuracy of this back-calculation depends on several factors including populations, excretion rates, the stability of the indicators, sampling, and sample preparation (Hill et al., 2020; Mao et al., 2020; Sims and Kasprzyk-Hordern, 2020). According to Hart and Halden (2020), different parameters such as temperature, average in-sewer travel time, and per-capita water are critical variables needed for determining infection hotspots when using the WBE model. The addition of physicochemical parameters (such as shear rates, pH, and dissolved oxygen) and biological reactions, i.e. sewer biofilms could enhance the reliability of the model (Hill et al., 2020).
South Africa serves as an intersection between developed and developing countries in respect of the wastewater treatment infrastructure. South Africa is always at the forefront concerning the wastewater treatment infrastructure; however, recent reports highlight the challenges with the operation and management of some plants due to the failing infrastructure. Furthermore, there is an additional discharge of fecal sludge from non-sewered sanitation systems into the centralized wastewater treatment facilities which add additional pressure to the existing treatment plants. It, therefore, gives a unique scenario for the development of the WBE approach for infection surveillance for South African WWTPs. In this study, we present the first report of the use of WBE in determining COVID-19 infection dynamics in South Africa. Additionally, we propose a modified predictive model that may give a better estimation of the number of infected people within the catchment of WWTPs. We believe that this contributes significantly to the growing field of WBE, especially concerning COVID-19 surveillance and potential application for other outbreaks in the future.
2. Methodology
2.1. Sampling methodology
Four municipal wastewater treatment plants (WWTPs) primarily treating domestic wastewater in the province of KwaZulu- Natal (KZN) of South Africa were selected for this study. In addition to treating primarily domestic wastewater, the Darvil and Central WWTPs also receive industrial wastewater. Within the Umgungundlovu district, the Darvill and Howick WWTPs, treating an average of 70 ml/d and 6 ml/d of wastewater per day respectively, were chosen. Within the eThekwini district, the Isipingo and Central WWTPs were selected, treating an average of 14 ml/d and 80 ml/d of wastewater respectively. The eThekwini district is largely made up of Durban, a commercial and industrial hub of the KZN province. It is a coastal city along the Indian Ocean and is a major tourist destination in South Africa. Pietermaritzburg, the capital of the KZN province, is also a major industrial hub, mainly producing aluminium, timber, and dairy products, and is the main city in the Umgungundlovu district (Fig. 1 ). Grab samples (2 l) of raw sewage were collected at the head of works (post-primary screening) for each of the WWTPs weekly between the peak hours (7:00–11:00 am). Sampling was carried out for approximately four months, from July to October 2020. Full personal protective equipment (PPE) (Face shield, FFP2 face mask, waterproof coveralls, and safety boots) was worn during each sampling event.
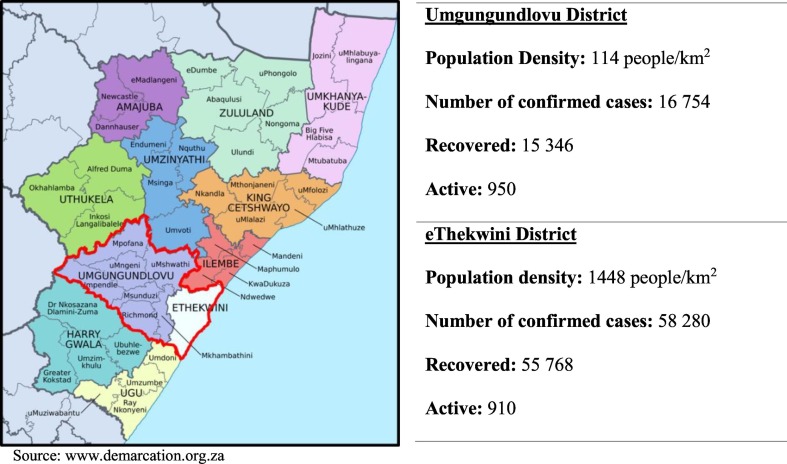
Fig. 1.
Covid-19 statistics for Umgungundlovu and eThekwini districts as of 16 November 2020.
2.2. Viral recovery and RNA extraction
Samples were heat-treated at 60 °C for 90 min immediately upon arrival in the laboratory (within two hours of sampling) and were left to cool to room temperature before further analysis was done. Thereafter, samples were mixed thoroughly, and 250 ml aliquots were removed for processing and the rest of the samples were stored at −80 °C. The 250 ml sample from each WWTP was then equally divided into 50 ml centrifuge tubes and clarified by centrifugation at 3500 xg for 10 min. The supernatants from each tube were then pooled and used for viral concentration while the pellets were stored at −80 °C for future analysis. The method of ultrafiltration was used to concentrate the virus particles as previously described by Medema et al. (2020). Briefly, 60 ml of supernatant was filtered through a Centricon® Plus-70 centrifugal ultrafilter with a cut-off of 10 kDa at 3500 x g for 30 min. The volume of the resulting concentrate for each sample varied due to the composition of the sample matrix. In instances where the concentrate was less than 140 μl, (the minimum amount required for RNA extraction) the concentrate was topped up to 140 μl using PBS. The volumes of the concentrate recovered as well as the volume of PBS added (if required) were recorded to account for the dilution effect.
The RNA was extracted from 140 μl of viral concentrate using the QiAmp Viral RNA MiniKit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. RNA was eluted in 80 μl of sterile nuclease-free water and then quantified using the Implen Nanophotometer®. The extracted RNA was then stored at −80 °C for further analysis.
2.3. Droplet digital PCR
Detection of SARS-CoV-2 in the wastewater samples was performed as described by Amoah et al. (2020: preprint) with some modifications. The current method involved the use of the One-Step RT-ddPCR Advanced Kit for Probes from Biorad (USA) together with primers and probes targeting the N2 region of the viral (SARS-CoV-2) genome. The N2 gene was chosen as it is the most widely used target gene in SARS-CoV-2 detection assays and it had the lowest limit of detection during preliminary tests in our laboratory. Other researchers have identified N2 as a good target for amplification compared to either N1 or N3 (Randazzo et al., 2020; Shirato et al., 2020). The 22 μl reaction mixture contained: 5 μl supermix, 2 μl reverse transcriptase, 1 μl dithiothreitol, 1.98 μl each of the forward and reverse primers (10 μM), 0.55 μl of 10 μM probe, 2.49 μl nuclease-free water, and 7 μl template RNA (1 ng). The 96-well plates containing the reaction mixtures were then sealed, vortexed, and centrifuged. Droplets were generated using the QXDx Automated Droplet Generator (Biorad, USA). The primer and probe sequences, together with the thermal cycling conditions can be found in Table 1 . The results after thermal cycling were read in the QX200 Droplet Reader, using the QuantaSoft 1.7 software (Biorad, USA) while further analysis was carried out using the QuantaSoft Analysis Pro 1.0 software (Biorad, USA).
Table 1.
Primer and probe sequences used in this study.
| Gene target | Sequence | Cycling conditions | Manufacturer | Reference |
|---|---|---|---|---|
| N2-F | TTACAAACATTGGCCGCAAA | Reverse transcription at 50 °C for 1 h, enzyme activation at 95 °C for 10 min, 40 cycles of denaturation at 94 °C for 30 s and annealing at 55 °C for 60 s. Deactivation of enzymes at 98 °C for 10 min and stabilization of the droplets at 4 °C for 30 min with a ramp rate of 2 °C/s | Inqaba Biotechnology (South Africa) | Giri et al. (2020) |
| N2-R | GCGCGACATTCCGAAGAA | |||
| N2-P | ACAATTTGC(ZEN)CCCCAGCGCTTCAG 5′ Modification: FAM 3′ Modification: Iowa Black® FQ | Integrated DNA Technologies | Barra et al. (2020) |
2.4. Assessment of viral recovery
To assess the efficiency of the concentration and extraction methods employed in this study, a 200 μl suspension (corresponding to 3360 copies of N2 gene) of inactivated SARS-CoV-2 strain USA/WA1/2020 (Microbiologics, USA) was seeded into 400 ml of raw wastewater and mixed thoroughly. Thereafter, the samples were separated into 8 × 50 ml centrifuge tubes (each containing approximately 420 copies of N2) and processed according to the methods described above and thereafter assessed for the presence of SARS-CoV-2. Additionally, unseeded wastewater was analysed in the same manner to determine the background concentrations of SARS-CoV-2. The ddPCR assay to determine the concentration of the N2 gene was conducted on the same day as RNA extraction to avoid any losses in RNA integrity that may result from storing and/or freeze thawing of the RNA. The entire process was also tested to determine the effect of possible inhibitors present in the wastewater matrix. This was done by spiking 120 ml of sterile Milli-Q water with 60 μl of inactivated SARS-CoV-2 suspension and processing it under the same conditions. The recovery efficiency obtained using Milli-Q water was then compared with that of wastewater.
The recovery efficiency of SARS-CoV-2 for wastewater and Milli-Q water was calculated using the following equation:
Where C SW is the concentration of SARS-CoV-2 detected in spiked wastewater or Milli-Q waterC UW is the concentration of SARS-CoV-2 detected in un-spiked wastewater or Milli-Q water.C SC is the concentration of SARS-CoV-2 that was spiked into the wastewater or Milli-Q water.
Taking into consideration that 60 ml of a spiked sample contains 504 copies of N2, the recovery percentage for wastewater was calculated at 62.86 (±12.84) % as the concentration of N2 quantified via ddPCR was 144.32 (±14.93) copies/60 ml and 461.12 (±79.64) copies/60 ml in un-spiked and spiked samples respectively. For Milli-Q water, the recovery percentage was 78.62 (±1.79) % as there was 396.23 (±9.96) copies/60 ml of N2 gene detected in the spiked sample while no target genes were present in the un-spiked (raw) sample. The differences in the recovery efficiency between the wastewater and Milli-Q possibly indicate the presence of inhibitors in the wastewater sample.
2.5. Quality control
Quality checks for the method used in this study were performed with the addition of positive, negative, and no-template controls, which were added to each plate/run during the ddPCR process. The positive controls contained synthetic RNA targeting 5 regions (E, N, ORF1ab, RdRP, and S genes) of the SARS-CoV-2 viral genome. Human genomic DNA and RNA contained in a synthetic matrix were used as negative control. These positive and negative controls were supplied by Exact Diagnostics (USA). No template controls were sterile nuclease-free water.
2.6. Prediction of the number of people infected
Estimation of the number of people infected within the communities connected to the wastewater treatment plants sampled was done using the prediction model published by Ahmed et al. (2020):
We used the same input data as published by Ahmed et al. (2020), except for the RNA copies per liter of wastewater, which was taken from the data generated from our study using the ddPCR protocol described above. Furthermore, we used different input data for the daily stool mass per person. This was because Ahmed et al. (2020) used daily stool mass per person representing high-income countries. The daily stool mass produced per person specific for South Africa was therefore used, and this was taken from Burkitt et al. (1972). These were modeled as a normal distribution with a mean of 2.07 log10 and a standard deviation of 1.08 log10. The amount of SARS-CoV-2 shed per gram of feces was modeled as a log-uniform distribution, with minimum and maximum values of 2.56 and 7.67respectively (Wölfel et al., 2020). To reduce variability, and improve on the outputs of the models, Monte Carlo Simulations using 10,000 iterations were performed. All models were built with @Risk (Palisade Corp, USA) addon to Excel (Microsoft Corp, USA).
2.7. Modification of prediction models
To improve the accuracy of the prediction model in estimating the number of people infected, we added the viral load shed per ml of urine in infected persons, as well as the recovery percentage of viral particles in the wastewater. The viral load shed per ml of urine was taken to be 2.50 Log10 (Peng et al., 2020). The volume of urine produced per person per day was modeled as a log-uniform distribution with a minimum of 2.78 and a maximum of 3.76 (Lemann et al., 1996).
In this study we measured the recovery efficiency to be 62.86 (±12.84) % of inactivated SARS-CoV-2 spiked into untreated wastewater. We, therefore, modeled the recovery efficiency as a uniform distribution with a minimum recovery of 50.02% and a maximum of 75.7%. The revised model used to estimate the number of people infected is represented by the equation;
2.8. Statistical analysis
Data entry was done in Excel and Origin (OriginLab Corp, USA) was used in plotting all the graphs. Comparison of SARS-CoV-2 viral load over the four months was done using the Kruskal-Wallis tests (Bethea et al., 1995). The difference in the number of infected people estimated with the published model and the modified model developed as part of this study was determined using the Mann-Whitney tests (Bethea et al., 1995). All statistical comparisons were done with Origin (OriginLab Corp, USA).
3. Results and discussion
3.1. COVID-19 in South Africa and regulatory policies introduced by authorities
On the 5th of March 2020, the first confirmed case of COVID-19 in South Africa was reported in the province of KwaZulu-Natal. Ten days later, a national state of disaster was declared, restricting interprovincial and international travel. By the 23rd of March 2020, a 21-day national lockdown was announced by the authorities to contain the spread of the disease in the country and mitigate the negative impact of COVID-19. Based on a risk-adjusted strategy, easing off the lockdown levels began on 1 May 2020, moving from high-alert level 5 to level 4. On 1st June 2020, the country moved to level 3 which permitted inter-provincial travel for work and essential services, places of worship were opened, exercise was allowed, students could attend schools and universities and public transport services (bus, rail, and taxi) were resumed. When the country moved to level 2 lockdown on 17 August 2020, restaurants were opened and the bans on inter-provincial travel were lifted. The country moved to lockdown level 1 from 21st September 2020, which further allowed for limited restrictions on social gatherings and the opening of international borders. As of 16 November 2020, South Africa has recorded 751,000 coronavirus cases with 123,617 of them in the province of Kwa-Zulu Natal.
3.2. Detection of SARS-CoV-2 viral RNA in wastewater
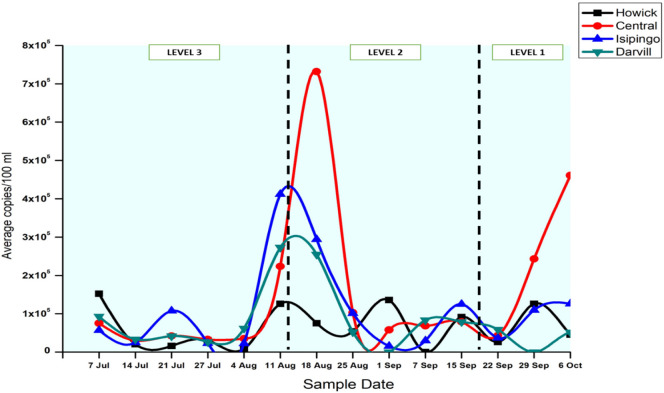
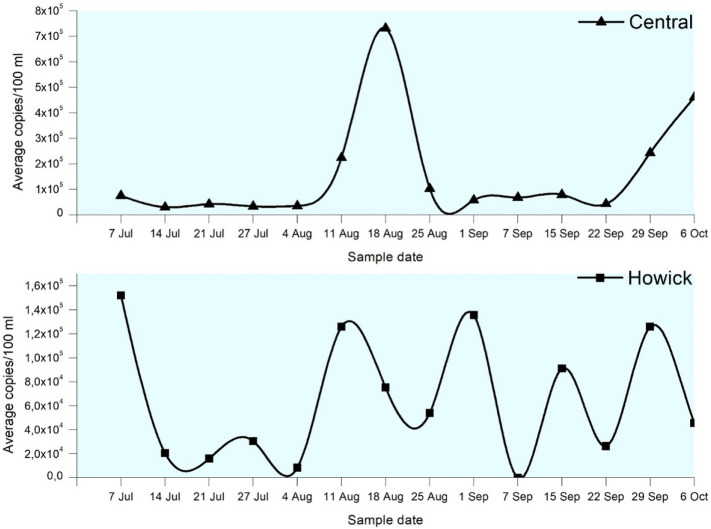
Globally, numerous studies have reported the presence of SARS-CoV-2 in wastewater and the implications it has on establishing a global WBE approach for COVID-19 (Ahmed et al., 2020; La Rosa et al., 2020; Lodder and de Roda Husman, 2020; Medema et al., 2020; Wu et al., 2020). While the majority of these proof-of-concept studies have been successful in demonstrating the applicability of wastewater-based testing for disease surveillance in communities, there remains an urgent need for larger-scale studies, a uniformed SARS-CoV-2 testing approach (for wastewater), proper collation of relevant clinical data, and consideration of the dynamic nature of each WWTP before national and global surveillance programs can be effectively implemented. In the present study, SARS-CoV-2 was detected in every sample tested for the Central and Isipingo WWTPs (14/14), while it was detected in 86% (12/14) and 93% (13/14) of the samples from Darvill and Howick WWTPs respectively. On average, viral loads ranged between 0 and 2.73 × 105 copies/100 ml, 0–1.52 × 105 copies/100 ml, 3 × 104–7.32 × 105 copies/100 ml and 1.55 × 104–4.12 × 105 copies/100 ml at the Darvill, Howick, Central and Isipingo WWTPs respectively. Comparatively, there was no statistically significant difference in the viral loads detected in the four WWTPs (P-value ≥0.05). The non-detection of the viral particles or RNA in some of the wastewater samples could be due to concentrations below the limit of detection of ddPCR, which was found to be 0.2 copies/μl (see Fig. S1 in Appendix I). The observed changes in the viral loads in the wastewater throughout the study, as described above, were statistically significant at a 95% confidence interval (p value ≤0.05). Viral loads recorded for the majority of this study are significantly higher than those reported by Ahmed et al. (2020), Serchan et al. (2020), Wu et al. (2020), and Randazzo et al. (2020) among many others. However, the discrepancies in results could be attributed to differences in disease prevalence and the efficiency of the various processes involved in the detection and quantification of SARS-CoV-2 from wastewater. It must also be noted that the higher viral loads observed in this study may also be partly attributed to the use of the ddPCR platform, which has been reported to have lower limits of detection (0.2 copies/μl) and is more sensitive and accurate than the RT-qPCR methods that are currently being used in majority of WBE studies (Lu et al., 2020; Dong et al., 2020; Zhou et al., 2020).
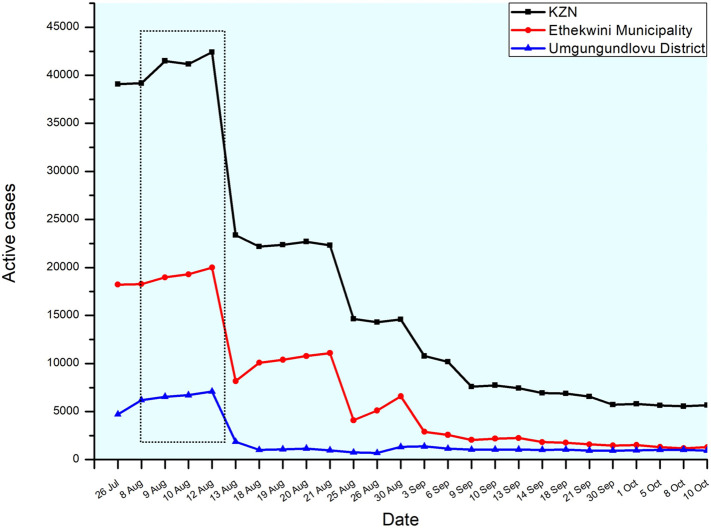
Fig. 3 shows the number of active clinical cases reported in KZN, eThekwini district, and Umgungundlovu district (KwaZulu-Natal Department of Health Covid-19 Situational Report: 7 July – 6 October 2020). Both districts followed a similar trend in terms of the reported number of active cases. During the period of 9th–12th August 2020, the highest number of active cases were recorded at both the provincial and district level. Interestingly, this increase in clinical cases also corroborated with an increase in the SARS-CoV-2 loads in wastewater. As seen from Fig. 2 , the samples collected on 11th August 2020 indicated a significant increase in the viral load. This trend continued to rise and reaching its peak on 18th August 2020 (Fig. 3 ). These results further confirmed that surveillance of wastewater could be a suitable method to track disease circulation within the associated catchments/communities of the WWTPs under investigation.
Fig. 3.
Number of active cases in KZN, eThekwini and Umgungundlovu districts from 26 July to 10 October 2020. *dotted line highlights the peak in active cases during the sample period.
Fig. 2.
SARS-CoV-2 loads detected in wastewater influent of 4 WWTPs over 3 months.
SARS-CoV-2 RNA load in untreated wastewater from three (3) of the four (4) WWTPs (Darvill, Isipingo, and Central) presented a typical bell-shaped curve, peaking in correlation with clinical data. However, results from the Howick WWTP presented an atypical scenario (Fig. 4 ). No observable trends were detected between viral counts and clinical data. The highest viral load (1.52 × 105 copies/100 ml) was recorded at the start of the study (7th July 2020). This could be attributed to the smaller size of the plant and the community served. The Howick WWTP is the smallest plant among the four studied, treating about 6 ml/d. Therefore, it can easily be impacted by slight changes in wastewater flows and shedding of the SARS-CoV-2. Small changes in the number of people infected could therefore potentially significantly affect the RNA loads detected. Additionally, small variations in water use patterns and industrial components could potentially impact loads of SARS-CoV-2 RNA detected due to dilution or inhibition.
Fig. 4.
SARS-CoV-2 viral loads in wastewater from Central and Howick WWTPs, showing variation in the trend of viral concentrations.
3.3. Effect of lockdown regulations
The current study began one month after the implementation of the level 3 lockdown. When stay-at-home regulations were eased on 17 August 2020 (level 3 to level 2) and 21 September 2020 (level 2 to level 1) people were now allowed to move freely across the country for recreational purposes and many returned to the city for work-related purposes. As social and economic activity resumed within the country, it was expected that this would reflect in the wastewater as many studies have already demonstrated how analysis of population pooled wastewater can represent the activity and lifestyle of a given community (Sims and Kasprzyk-Hordern, 2020). The results presented in Fig. 2 demonstrates this effect. Using the Central WWTP as an example, viral loads increased from 2.25 × 105 copies/100 ml (11 August 2020) to 7.32 × 105 copies/100 ml after the move from level 3 to level 2. Wastewater samples collected from the Central, Isipingo, and Howick WWTPs on 29 September 2020 (after the start of lockdown level 1) was indicative of this trend as well. Where viral loads of 2.43 × 105 copies/100 ml, 1.09 × 105 copies/100 ml and 1.26 × 105 copies/100 ml for Central, Isipingo, and Howick WWTPs respectively were recorded. The findings of our study are following that of Wurtzer et al. (2020) who evaluated the effect of lockdown regulations on SARS-CoV-2 dynamics in wastewater in Paris. Wurtzer et al. (2020) observed that the WBE approach was not necessarily an early warning system and concluded that quantitative monitoring of wastewater is a time-related indicator of the health status of the community. The findings of our study agree with this observation by Wurtzer et al. (2020).
3.4. Estimation of infected people based on SARS-CoV-2 load in wastewater
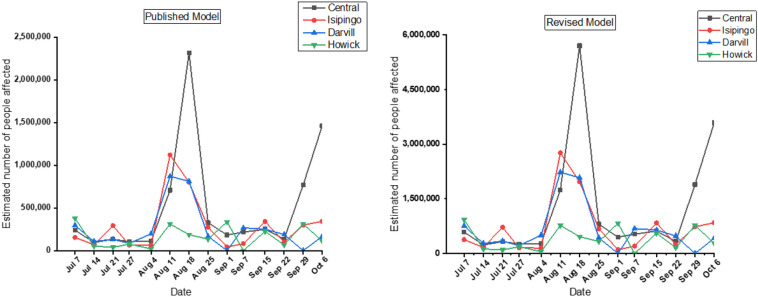
In addition to its utility as a surveillance tool, the potential of WBE as a predictive tool has been reported. For instance, it was used to assess the circulation of polio in populations (Ndiaye et al., 2014; Hovi et al., 2012), to retrospectively predict gastroenteritis caused by the Hepatitis A virus and norovirus (Hellmér et al., 2014). During the current COVID-19 pandemic, WBE has been used to estimate the number of infected people within the catchment of a WWTP based on viral load (Ahmed et al., 2020: Weidhaas et al., 2020; Vallejo et al., 2020). These reports use different mathematical approaches/models, for instance, Vallejo et al. (2020) developed a simple statistical regression model for the estimation of infection numbers based on viral load. They reported a 90% reliability in the use of this model to estimate COVID-19 cases in a metropolitan area of A Coruna in Spain. Application of the model proposed by Ahmed et al. (2020), resulted in varying estimates of infected people. For instance, the model predicted that between 95, 000 to 2.3 million people within the catchment of the Central WWTP were infected. This is high compared to the estimates from the catchment of the Howick WWTP, where estimated infection numbers ranged from 21, 000 to 377,000. Comparatively, our estimated numbers are also higher than those reported in the literature. This could be due to several reasons, such as higher infection number within our catchments as opposed to the clinical data, a difference in the efficiency of the methods used, or a higher viral load estimation based on the ddPCR platform. For instance, Falzone et al. (2020) reported that SYBR-Green RT-qPCR was unable to detect SARS-CoV-2 in samples with low viral load, whilst the ddPCR showed higher sensitivity and specificity. Therefore, the use of ddPCR in the determination of the SARS-CoV-2 viral load in our study could have accounted for the high viral load reported, which subsequently impacted the estimated number of people infected. In developing countries or regions where accurate data on the number of active cases of COVID-19 is absent, either due to poor record keeping or inadequate testing, the use of WBE could be an excellent tool. However, its application is limited, due to the uncertainties surrounding these predictive modeling applications. For instance, sensitivity analysis by Ahmed et al. (2020) indicated that the estimated number of people infected is greatly impacted by the input data such as SARS-CoV-2 RNA copies shed in the stool of infected individuals, accurate quantification of RNA copies in wastewater, and lastly the quantity of feces shed per person per day. Therefore, to improve on the models and address the limitations posed by these factors; we incorporated the efficiency of recovery of the SARS-CoV-2 in wastewater (described above under Methodology), the shedding of SARS-CoV-2 in the urine of infected individuals, and the amount or volume of urine produced per person per day. The revised model resulted in a higher number of estimated infections compared to the model published by Ahmed et al. (2020) (Fig. 5 ). The estimated number of people infected within the catchments of these WWTPs followed a similar trend as the SARS-CoV-2 viral loads measured, corresponding to peaks in reported clinical active cases as well. However, our estimated numbers were significantly higher than the total number of active cases for the two districts within which these WWTPs are located. This could be attributed to two factors, firstly under clinical testing, which may result in a lower number of active cases compared to the actual number of infected persons within the district. For instance, South Africa has a testing rate of 54, 224 tests per million population (Chitungo et al., 2020), which is lower than the testing rate in the UK, USA, and many other developed countries. However, comparatively, South Africa has a better testing rate than any other African country and most countries in Asia (Chitungo et al., 2020). It is estimated that for every 10 diagnosed infections, there are 7 undiagnosed infections (Ahmed et al., 2020). Therefore, with lower testing the number of infections reported by health authorities could be much lower; therefore, our estimates could be a true reflection of the actual number of people infected within the catchments. Secondly, the higher estimated number of infected individuals could be attributed to overestimation by the models employed. For instance, sensitivity analysis by Ahmed et al. (2020) indicated that these models are affected by the SARS-CoV-2 RNA shedding in stool and the copies of RNA detected in wastewater. Therefore, without local data on the number of SARS-CoV-2 shed by infected persons (both symptomatic and asymptomatic), the estimates could be either over or underestimated.
Fig. 5.
Estimation of the number of infected people within the catchments of the WWTPs using a published model and a revised model.
3.5. Limitations with predictive models for estimation of COVID-19 infections
Despite the usefulness of predictive models in the estimation of the number of people infected with COVID-19 based on the concentration of SARS-CoV-2 in wastewater, several limitations or challenges have been identified. These challenges are elaborated below;
-
1.
Viral shedding information: The concentration of SARS-CoV-2 in wastewater is largely dependent on the shedding of the virus in the stool of infected individuals. Therefore, for efficient estimation of infection numbers through the use of predictive models, accurate information on the shedding pattern within the catchment of the wastewater treatment plant is very crucial. For instance, information on the viral load per gram of feces shed by infected individuals is key. This is due to the variation in concentration of SARS-CoV-2 in stool reported in the literature. For instance, Han et al. (2020) reported viral loads of 1.7 × 106–4.1 × 107 gc/ml in contrast to the viral load reported by Lescure et al. (2020) (6.3× 106–1.26× 108 gc/g). Furthermore, studies by Xiao et al. (2020) and Wu et al. (2020), showed that fecal samples were positive for 11.2–35 days even after patients tested negative while Zhang et al. (2020) revealed that certain patients can shed the virus in stool for up to 22 days. It is also known that viral loads in stools are dependent on when in the infection cycle the sample is taken, (Wölfel et al., 2020), with some reports of shedding in asymptomatic individuals as well (Campioli et al., 2020; Xu et al., 2020; Long et al., 2020; Li et al., 2020). It is worth noting there is no information on the viral load shed in stool by infected individuals in South Africa. This creates an information gap and could potentially account for the disparity in the estimated infection number using the predictive models and the reported actives cases.
-
2.
Weight of stool produced per person per day: In addition to information on the concentration of SARS-CoV-2 shed by each infected person, an accurate estimation of the number of infected people will be affected by the weight of stool produced per person per day. This information varies from region to region and could be impacted by several factors (Rose et al., 2015). For instance, Ahmed et al. (2020) used stool production per person per day from high-income countries as reported by Rose et al. (2015). This data cannot be used for a middle-income country like South Africa. Therefore, the local supporting information is critical. In this study, we had to rely on old data from 1972 on the weight/quantity of stool produced per person per day in South Africa. These values could have changed over the years, therefore update information might have improved the accuracy of the predictive model.
3.6. Recommendations for the implementation of the WBE approach
Considering all of the above, correlating epidemiological data with viral loads in wastewater is currently difficult as not all COVID-19 carriers are included in epidemiological data, making the implementation of the WBE surveillance strategy complicated for many parts of the world (Polo et al., 2020). Based on this we will like to make the following recommendations:
-
1).
There is an urgent need for a univocal testing framework, which takes into consideration the different analytical sensitivities of each step in the testing process (especially with PCR assays and platforms) (Michael-Kordatou et al., 2020). This framework should include a sampling approach that will ensure an accurate representation of viral load within the catchment. This could involve a more frequent sampling regime or the use of autosamplers to make composite samples.
-
2).
Changes in environmental conditions and the unique signature of each WWTP are probably the most important contributing factors to the variability of the WBE approach. In the environment, rainfall events and temperature play an important role and affect the dilution and stability of the virus in water (La Rosa et al., 2020). While in WWTPs, it is imperative to know hydraulic retention times, peak flow rates, as well as the size and configuration of sewer networks in each community. In addition to macropollutant loads, which are used to calculate the population served by the WWTP, it is also important to consider the contribution of stormwater incursions, greywater input, septic tank discharge at the plant as well as the presence of industrial waste which are common challenges in South African WWTPs.
4. Conclusion
Our findings show that WBE can be used to give an indication of infection levels in connected communities. This is due to the correlation observed between high viral loads in the untreated wastewater and peak in active clinical cases within the province. We also observed that the transition between the levels of lockdown (from higher to lower levels of restriction) resulted in an increase in viral loads in the untreated wastewater. Additionally, we also showed that mathematical models estimated a higher number of infected people compared to data from clinical testing. However, this is not conclusive due to the scarcity of active clinical cases specific to the catchments of the WWTPs studied. Despite the challenges faced (highlighted above), we can conclude that WBE can be used to detect possible surges in COVID-19 infections in communities serviced by WWTPs. Additionally, with an improved predictive model, WBE will be useful in forecasting the potential number of people that could be infected, an approach that is important for risk reduction interventions.
The following is the supplementary data related to this article.
Limit of detection of the ddPCR protocol used for the detection of SARS-CoV-2 in wastewater.
CRediT authorship contribution statement
All authors were involved in the conceptualization of the study and L. Pillay, I.D.Amoah, O. Awolusi, N. Deepnarian, and K. Pillay performed data collection. The first draft of the manuscript was prepared by L. Pillay, I.D.Amoah and O. Awolusi under the supervision of S. Kumari and F. Bux. Final review and approval of the manuscript were done by all the authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
We are grateful for the financial support received from Umgeni Water and the Durban University of Technology. The support of the eThekwini Municipality and operators of the wastewater treatment plant is acknowledged. We also acknowledge the support of Prof. Saloshni Naidoo from the University of KwaZulu-Natal.
Editor: Damia Barcelo
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawfiq J.A. Asymptomatic coronavirus infection: MERS-CoV and SARS-CoV-2 (COVID-19) Travel Med. Infect. Dis. 2020;35:1–2. doi: 10.1016/j.tmaid.2020.101608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoah I.D., Pillay L., Deepnarain N., Awolusi O.O., Ramlal P., Kumari S., Bux F. 2020. Detection of SARS-CoV-2 on contact surfaces within shared sanitation facilities and assessment of the potential risks for COVID-19 infections. Researchsquare (preprint) [DOI] [Google Scholar]
- Barra G.B., Santa Rita T.H., Mesquita P.G., Jácomo R.H., Nery L.F.A. Analytical sensitivity and specificity of two RT-qPCR protocols for SARS-CoV-2 detection performed in an automated workflow. Genes. 2020;11 doi: 10.3390/genes11101183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethea R.M., Duran B.S., Boullion T.L. 3rd ed. Marcel Dekker; New York: 1995. Statistical Methods for Engineers and Scientists. [Google Scholar]
- Burkitt D.P., Walker A.R.P., Painter N.S. Effect of dietary fibre on stools and transit-times, and its role in the causation of disease. The Lancet. 1972;300(7792):1408–1411. doi: 10.1016/s0140-6736(72)92974-1. [DOI] [PubMed] [Google Scholar]
- Campioli C.C., Cevallos E.C., Assi M., Patel R., Binnicker M.J., O’Horo J.C. Clinical predictors and timing of cessation of viral RNA shedding in patients with COVID-19. J. Med. Virol. 2020;130 doi: 10.1016/j.jcv.2020.104577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitungo I., Dzobo M., Hlongwa M., Dzinamarira T. COVID-19: unpacking the low number of cases in Africa. Public Heal. Pract. 2020;1 doi: 10.1016/j.puhip.2020.100038. (100038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P.M., Tscharke B.J., Donner E., O’Brien J.W., Grant S.C., Kaserzon S.L., Mackie R., O’Malley E., Crosbie N.D., Thomas K.V., Mueller J.F. Wastewater-based epidemiology biomarkers: past, present and future. TrAC - Trends Anal. Chem. 2018;105:453–469. doi: 10.1016/j.trac.2018.06.004. [DOI] [Google Scholar]
- Daglioglu N., Atasoy A., Asadi A., Guzel E.Y., Dengiz H. Estimating alcohol consumption by using wastewater-based epidemiology in Adana Province, Turkey. Environ. Sci. Pollut. Res. 2020;27:31884–31891. doi: 10.1007/s11356-020-09056-w. [DOI] [PubMed] [Google Scholar]
- Daughton C.G. Wastewater surveillance for population-wide Covid-19: the present and future. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Wang X., Wang S., Du M., Niu C., Yang J., Li L., Zhang G., Fu B., Gao Y., Wang J. Interlaboratory assessment of droplet digital PCR for quantification of BRAF V600E mutation using a novel DNA reference material. Talanta. 2020;207 doi: 10.1016/j.talanta.2019.120293. [DOI] [PubMed] [Google Scholar]
- Falzone L., Musso N., Gattuso G., Bongiorno D., Palermo C.I., Scalia G., Libra M., Stefani S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020;46:957–964. doi: 10.3892/ijmm.2020.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri B., Pandey S., Shrestha R., Pokharel K., Ligler F.S., Neupane B.B. Review of analytical performance of COVID-19 detection methods. Anal. Bioanal. Chem. 2020 doi: 10.1007/s00216-020-02889-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.S., Seong M.W., Heo E.Y., Park J.H., Kim N., Shin S., Cho S.I., Park S.S., Choi E.H. Sequential analysis of viral load in a neonate and her mother infected with SARS-CoV-2. Clin. Infect. Dis. 2020;71:2236–2239. doi: 10.1093/cid/ciaa447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2 / COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis a virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80:6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Zamyadi A., Deere D., Vanrolleghem P.A., Crosbie N.D. SARS-CoV-2 known and unknowns, implications for the water sector and wastewater-based epidemiology to support national responses worldwide: early review of global experiences with the COVID-19 pandemic. Water Qual. Res. J. 2020:1–11. doi: 10.2166/wqrj.2020.100. [DOI] [Google Scholar]
- Hovi T., Shulman L.M., Van Der Avoort H., Deshpande J., Roivainen M., De Gourville E.M. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol. Infect. 2012;140:1–13. doi: 10.1017/S095026881000316X. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Bonadonna C.V.L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736:1625–1628. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, T.Q.M., Takemura, T., Moi, M.L., Nabeshima, T., Nguyen, L.K.H., Hoang, V.M.P., Ung, T.H.T., Le, T.T., Nguyen, V.S., Pham, H.Q.A., Duong, T.N., Nguyen, H.T., Ngu, D.N., Nguyen, C.K., Morita, K., Hasebe, F., Dang, D.A. Severe acute respiratory syndrome coronavirus 2 shedding by travelers, Vietnam, 2020. Emerg. Infect. Dis. Ther. 26, 1624–1626. doi: 10.3201/eid2607.200591. [DOI] [PMC free article] [PubMed]
- Lemann J., Jr., Pleuss J.A., Worcester E.M., Hornick L., Schrab D., Hoffmann R.G. Urinary oxalate excretion increases with body size and decreases with increasing dietary calcium intake among healthy adults. Kidney Int. 1996;49:200–208. doi: 10.1038/ki.1996.27. [DOI] [PubMed] [Google Scholar]
- Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q., Enouf V. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Su, Y.Y., Zhi, S.S., Huang, J., Zhuang, C.L., Bai, W.Z., Wan, Y., Meng, X.R., Zhang, L., Zhou, Y.B. and Luo, Y.Y., 2020. Virus shedding dynamics in asymptomatic and mildly symptomatic patients infected with SARS-CoV-2. Clin. Microbiol. Infect. 26, 1556.e1–1556.e6. [DOI] [PMC free article] [PubMed]
- Lodder, W., de Roda Husman, A.M., 2020. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 1253, 30087. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed]
- Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Hu J.L., Xu W., Zhang Y., Lv F.J., Su K. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26,1200-1204 doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Lu, R., Wang, J., Li, M., Wang, Y., Dong, J. and Cai, W., 2020. SARS-CoV-2 detection using digital PCR for COVID-19 diagnosis, treatment monitoring and criteria for discharge. medRxiv. doi: 10.1101/2020.03.24.20042689. [DOI]
- Mao K., Zhang K., Du W., Ali W., Feng X., Zhang H. The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Curr. Opin. Environ. Sci. Heal. 2020;17:1–7. doi: 10.1016/j.coesh.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett. [DOI] [PubMed] [Google Scholar]
- Michael-Kordatou I., Karaolia P., Fatta-Kassinos D. Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020;10436 doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndiaye A.K., Diop P.A.M., Diop O.M. Environmental surveillance of poliovirus and non-polio enterovirus in urban sewage in Dakar, Senegal (2007-2013) Pan Afr. med. J. 2014;19 doi: 10.11604/pamj.2014.19.243.3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Liu J., Xu W., Luo Q., Chen D., Lei Z., Huang Z., Li X., Deng K., Lin B., Gao Z. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J. Med. Virol. 2020;92:1676–1680. doi: 10.1002/jmv.25936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., Romalde J.L. Making waves wastewater-based epidemiology for COVID-19 approaches and challenges for surveillance and prediction. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:1–9. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose C., Parker A., Jefferson B., Cartmell E. The characterization of feces and urine: a review of the literature to inform advanced treatment technology. Crit. Rev. Environ. Sci. Technol. 2015;45(17):1827–1879. doi: 10.1080/10643389.2014.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serchan S.P., Shahin S., Ward L.M., Tandukar Aw T.G., Schmitz B., Ahmed W., Kitajima M.K. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y., Kageyama T., Matsuyama S., Takeda M. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. J. Infect. Dis. 2020 doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh E.K., Amankwa Mark Opoku, Armah E.K., Rathilal S. Fate of COVID-19 occurrences in wastewater systems: emerging detection and treatment technologies—a review. Water. 2020;12:2680. doi: 10.3390/w12102680. [DOI] [Google Scholar]
- Vallejo J.A., Rumbo-Feal S., Conde-Pérez K., López-Oriona Á., Tarrío-Saavedra J., Reif R., Ladra S., Rodiño-Janeiro B.K., Nasser M., Cid Á., Veiga M.C., Acevedo A., Lamora C., Bou G., Cao R., Poza M. Highly predictive regression model of active cases of COVID-19 in a population by screening wastewater viral load. medRxiv. 2020 (2020.07.02.20144865) [Google Scholar]
- Venugopal A., Ganesan H., Sudalaimuthu Raja S.S., Govindasamy V., Arunachalam M., Narayanasamy A., Sivaprakash P., Rahman P.K.S.M., Gopalakrishnan A.V., Siama Z., Vellingiri B. Novel wastewater surveillance strategy for early detection of coronavirus disease 2019 hotspots. Curr. Opin. Environ. Sci. Heal. 2020;17:8–13. doi: 10.1016/j.coesh.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M., Bisserbe N. Europe imposes new Covid-19 restrictions as second wave accelerates [WWW document] Wall Str. J. 2020 https://www.wsj.com/articles/european-countries-announce-new-covid-19-restrictions-record-infections-11603638712 URL. (accessed 11.2.20) [Google Scholar]
- Weidhaas J., Aanderud Z., Roper D., VanDerslice J., Gaddis E., Ostermiller J., Hoffman K., Jamal R., Heck P., Zhang Y., Torgersen K. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Researchsquare. 2020 doi: 10.1016/j.scitotenv.2021.145790. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;58:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Moniz K., Erickson T., Chai P., Thompson J., Alm E. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv. 2020 doi: 10.1101/2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J., Moulin L. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases. medRxiv. 2020 doi: 10.1101/2020.04.12.20062679. 2020.04.12.20062679. [DOI] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;1-3 doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C.L., Raval M., Schnall J.A., Kwong J.C., Holmes N.E. Duration of respiratory and gastrointestinal viral shedding in children with SARS-CoV-2: a systematic review and synthesis of data. Pediatr. Infect. Dis. J. 2020;39:e249–e256. doi: 10.1097/INF.0000000000002814. [DOI] [PubMed] [Google Scholar]
- Zhang N., Gong Y., Meng F., Shi Y., Wang J., Mao P., Chuai X., Bi Y., Yang P., Wang F. Comparative study on virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. Sci. China Life Sci. 2020;63 doi: 10.1007/s11427-020-1783-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.B., Kong W.H., Wang S., Long Y.B., Dong L.H., He Z.Y., Liu M.Q. Potential transmission risk of SARS-CoV-2 through medical wastewater in COVID-19 outbreak cities. Researchsquare. 2020 (preprint) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Limit of detection of the ddPCR protocol used for the detection of SARS-CoV-2 in wastewater.