Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) uses subgenomic RNA (sgRNA) to produce viral proteins for replication and immune evasion. We apply long-read RNA and cDNA sequencing to in vitro human and primate infection models to study transcriptional dynamics. Transcription-regulating sequence (TRS)-dependent sgRNA upregulates earlier in infection than TRS-independent sgRNA. An abundant class of TRS-independent sgRNA consisting of a portion of open reading frame 1ab (ORF1ab) containing nsp1 joins to ORF10, and the 3′ untranslated region (UTR) upregulates at 48 h post-infection in human cell lines. We identify double-junction sgRNA containing both TRS-dependent and -independent junctions. We find multiple sites at which the SARS-CoV-2 genome is consistently more modified than sgRNA and that sgRNA modifications are stable across transcript clusters, host cells, and time since infection. Our work highlights the dynamic nature of the SARS-CoV-2 transcriptome during its replication cycle.
Keywords: SARS-CoV-2, COVID-19, coronavirus, Nanopore sequencing, direct RNA sequencing, direct cDNA sequencing, discontinuous transcription, RNA modification, differential expression, poly(A) tail
Graphical abstract
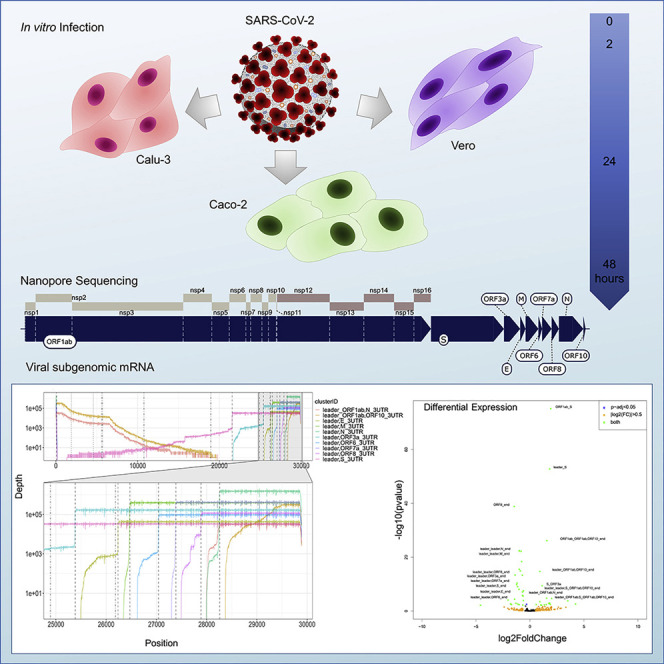
SARS-CoV-2 is the pathogen that is responsible for the global COVID-19 pandemic. Chang et al. demonstrate that the transcriptome of SARS-CoV-2 is dynamic and complex, with expression and relative proportions of viral mRNA changing to reflect the stage of infection in vitro. In contrast, the epi-transcriptome is stable throughout infection.
Introduction
SARS-CoV-2, a positive-strand RNA beta-coronavirus, is the causative agent of coronavirus disease 2019 (COVID-19) (Zhou et al., 2020). As with all identified coronaviruses, the replicative and infectious cycle of SARS-CoV-2 is characterized by a process termed discontinuous minus-strand extension, which occurs during replication of viral RNA by the viral replication and transcription complex (RTC) within the host cell. The RTC halts synthesis of negative sense RNA when it encounters a 6 to 8 nucleotide (nt) transcription-regulating sequence (TRS) in the body of the genome (TRS-B) and reinitiates synthesis via a template switching event with a homologous TRS present in the 5′ leader sequence (TRS-L) (V’Kovski et al., 2021). This results in a set of nested negative-strand templates (shown in Figure S1), which are utilized for expression of subgenomic mRNA (sgRNA). Each sgRNA includes the 3′ polyadenylated (poly(A)) untranslated region (UTR), a truncated set of 3′ open reading frames (ORFs), and a common 5′ leader sequence. The production of subgenomic transcripts alleviates pressure on the primary viral genome for protein synthesis and enables the translation of proteins at greater speed and concentration. Major SARS-CoV-2 TRS-dependent mRNAs have been previously described (Davidson et al., 2020; Kim et al., 2020; Taiaroa et al., 2020). However, the changes in the viral transcriptome and epi-transcriptome across the course of cellular infection have not yet been explored.
Long-read sequencing platforms can generate reads spanning the length of these sgRNA and are thus better suited to transcriptomic characterization of its highly nested transcriptome. One such platform is the MinION (Oxford Nanopore Technologies [ONT]), which can sequence either native RNA or cDNA directly without requirement for PCR amplification, therefore reducing PCR-induced biases in estimation of expression levels (Garalde et al., 2018; Batista et al., 2020). Furthermore, RNA modifications induce changes in ONT signal, which enable exploration of the epi-transcriptome using direct RNA (dRNA) sequencing (Garalde et al., 2018; Kim et al., 2020).
In this manuscript, we carried out a comprehensive assessment of SARS-CoV-2 transcription. We generated more than 8 million long-read viral dRNA sequences and direct cDNA reads across multiple time points (2, 24, and 48 h post-infection [hpi]) with infected African green monkey (Vero) and human (Calu-3, Caco-2) cell lines. Our dataset was supplemented with publicly available virion (Taiaroa et al., 2020) and HCoV-229E (Viehweger et al., 2019) datasets. We have developed a site to explore the dynamic SARS-CoV-2 transcriptome in an interactive web app: http://coinlab.mdhs.unimelb.edu.au/. Our dataset provides an expansive overview of the SARS-CoV-2 transcriptome and its changes throughout the course of infection. Our work will enable the development of new diagnostic tests for monitoring the progression of SARS-CoV-2 infectious cycle both in vitro and in vivo. This will assist in better understanding the mechanism of action of therapeutic agents and in monitoring the efficiency of the immune response to SARS-CoV-2 in vaccination studies.
Results
Infection dynamics are represented by changes in proportion of sgRNA
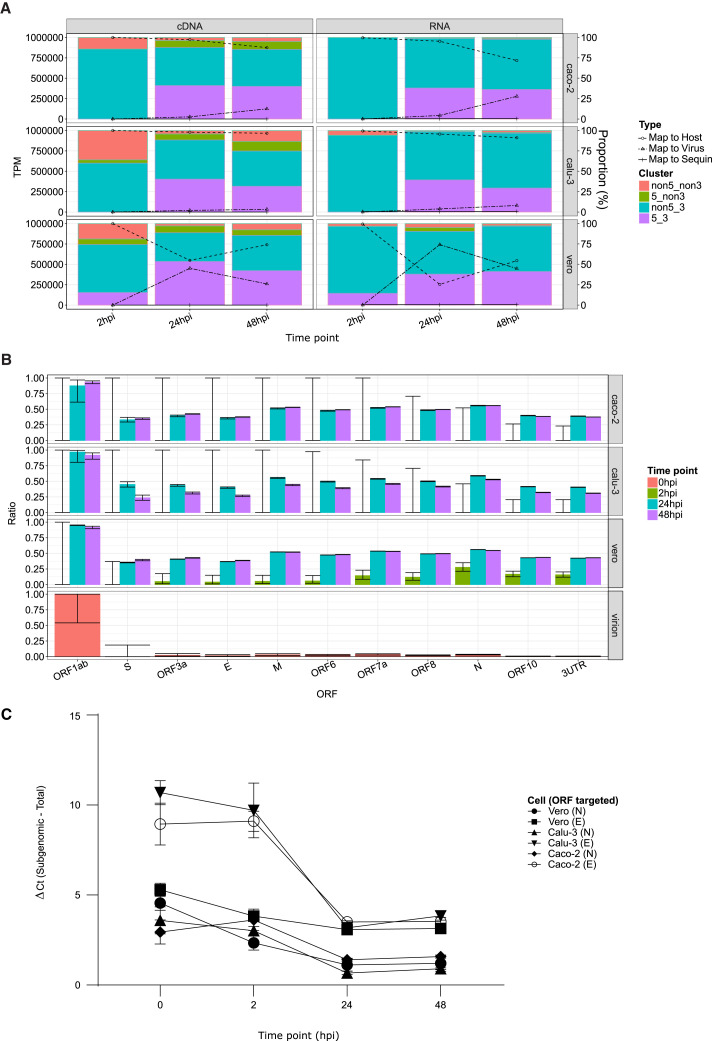
Viral RNA load was substantially higher in African green monkey Vero cells in comparison with human Caco-2 and Calu-3 cell lines, reaching a maximum of 74% of all sequenced RNA at 24 hpi. In comparison, a maximum of 4% of all sequenced RNA mapped to SARS-CoV-2 in infected human cell lines at 48 hpi (Figure 1 A). Even as early as 2 hpi, substantially more viral reads were detectable in Vero compared with Caco-2 and Calu-3 cells (Figure S2), suggesting a faster course of infection in Vero cells.
Figure 1.
Infection dynamics are represented by changes in proportion of sgRNA
Time points sequenced: 2, 24, and 48 hpi; cells infected: Caco-2, Calu-3, and Vero.
(A) Bar chart (left axis) indicate classification of viral mapping reads based on whether they include 5′ (e.g., leader), as well as 3′ (i.e., UTR and poly(A) tail), in terms of transcripts per million (TPMs) mapped viral reads. Line graph (right axis) indicates proportion of host, viral, or sequin mapping reads.
(B) Proportion of reads covering each ORF, which are sgRNA by virtue of containing 5′ leader sequence for direct RNA sequencing datasets. Error bar indicates 95% binomial confidence interval (CI) of proportion estimate using the logistic parameterization.
(C) sgRNA activity of SARS-CoV-2 measured by comparing mean differences in Ct values between subgenomic and total N and E genes across technical replicate wells of infected cells (n = 2–3) from all cell lines and across four time points (0, 2, 24, and 48 hpi), shown with ± SD error bars. The mean difference between subgenomic and total transcripts decreases over time and reaches a minimum at 24 hpi, indicating that sgRNA reaches its peak transcriptional activity at 24 hpi across all cell lines.
See also Figures S1–S3 and S5 and Table S1.
The proportion of sgRNA (i.e., reads containing both 5′ leader and 3′ UTR) among all viral mapping reads peaked at around 40% in all three cell lines at 24 hpi (Figure 1A). In the non-replicating virion sample (Taiaroa et al., 2020), as well as the 2 hpi samples, most reads were sequenced from the viral genome, because they had complete 3′ UTR but no 5′ leader (labeled as non5_3; Figure S2). This indicated that transcriptional activity had yet to accelerate at this early time point. Vero cells showed a greater proportion of sgRNA at 2 hpi compared with the Caco-2 and Calu-3 (Fisher’s exact test p = 0.02), suggesting that transcriptional activity is able to commence earlier during infection of Vero cells.
To further investigate the relationship between production of sgRNA and progression of infection, we calculated, for each ORF, the proportion of reads spanning the ORF, which also contained the leader sequence (Figure 1B). We observed that the RNA derived from the virion sample had the least sgRNA, followed by 2 hpi, whereas the 24 hpi samples had the highest proportion of sgRNA in Vero and Calu-3 cell lines, with maximum discrimination between the virion RNA and 24 hpi obtained for the N ORF.
We then designed primers to measure both subgenomic and total N ORF expression and used quantitative reverse transcription PCR (qRT-PCR) with primers targeting these regions. For comparison, we applied the same approach for both subgenomic and total E ORF (Wölfel et al., 2020; Corman et al., 2020). In all three cell lines, the difference between subgenomic and total N and E ORFs was smallest at 24 hpi (1.1 and 3.1 cycle threshold [Ct] difference, respectively, in Vero) with a slight increase at the final 48 hpi time point (Figure 1C). This suggests that SARS-CoV-2 reaches its peak rate of transcriptional activity at the 24 hpi time point. By calculating expected Ct differences between subgenomic and total E and N ORFs from sequence data, we further confirmed that qRT-PCR results captured the same dynamics (Figure S3).
Overall, these results reveal the changing proportions of sgRNA during the SARS-CoV-2 virus infectious cycle. Our analysis using qRT-PCR to compare total and sgRNA demonstrates the potential to track viral transcriptional activity using PCR. Our data indicate that the slower rate of infection in human compared with monkey Vero cell lines may arise because of both differences in viral entry and differences in rate of early viral genome replication.
Coronaviruses produce classes of TRS-independent sgRNA, which are abundantly expressed
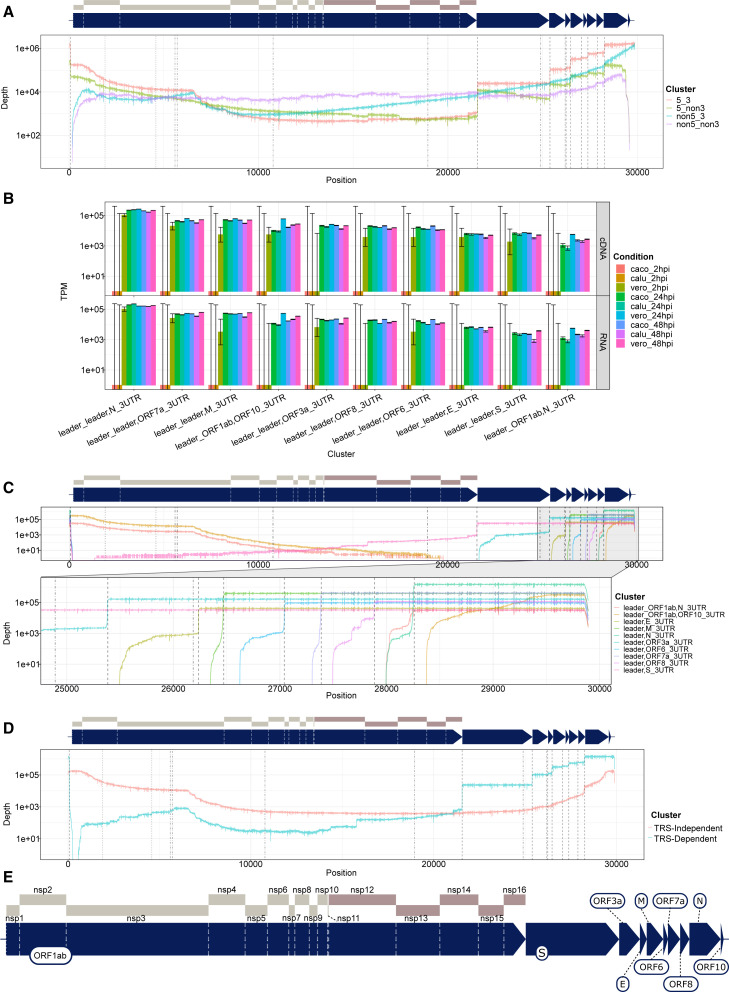
Although all coronaviruses use a repetitive 6 nt TRS throughout the genome to generate a nested set of TRS-dependent sgRNA, the breadth of data generated in this study reveals a more detailed transcriptome that is also constituted by transcripts generated through other, unknown genome mechanisms. The depth profile of sgRNA showed sharp changes in read depth, corresponding to negative-strand disjunction mediated by TRS immediately upstream of the ORF (Figure 2 A). To better quantify different classes of sgRNA, we developed a new tool, npTranscript, which assigns reads to transcript clusters (see STAR Methods). Using npTranscript, we could calculate the abundance of the sgRNA at various stages of infection. At the peak of infection in Vero cells (24 hpi), the most abundant sgRNA in terms of transcripts per million (TPMs) mapped viral reads were ORFs N (266,000), 7a/7b (63,000), M (62,000), ORF1ab,ORF10 (60,000), ORF3a (26,000), ORF8 (16,000), ORF6 (13,000), S (7,500), E (6,100), and ORF1ab,N (5,700) (Figure 2B). Of these transcripts, formations of ORFs N,7a/7n, M, 3a, 8, 6, and S were all mediated by a TRS-dependent homology (Figure 2C). The remaining two transcripts, ORF1ab,ORF10 and ORF1ab,N, were abundant in all 24 and 48 hpi datasets and did not have breakpoints at TRS motifs (Figure 2C). Further inspection of TRS-independent sgRNA indicated that the majority included the first polypeptide in ORF1ab (Figure 2D). Taking into account polypeptide boundaries, these transcripts contained leader nsp1 and a variable 3′ trailer incorporating a segment of the genome upstream of ORF10 and continuing until the terminus. The exclusion of the ORF1ab stop codon will allow translation to continue into a portion of the 3′ ORF downstream of the junction site before a stop codon is reached, which has the potential to produce truncated proteins of unknown function.
Figure 2.
SARS-CoV-2 produces classes of TRS-independent sgRNA, which are abundantly expressed
The transcript nomenclature W_X,Y_Z indicates that the transcript consists of the continuous segment from W to X joined with the segment from Y to Z.
(A) Total depth of coverage summed over all cDNA sequencing runs by categorization of reads based on mapping to 5′ and 3′ end of virus (within 10 base pairs [bp]) plotted on a log y scale. Dashed lines indicate location of TRS motifs, with alternative motifs detected using FIMO shown in dotted and dot-dash lines.
(B) Transcript abundance of major classes of sgRNA in TPMs mapped viral reads, plotted on a log scale for dRNA experiments (bottom row) and direct cDNA (top line). 95% CIs estimated from binomial model using the logistic parameterization.
(C) Transcript coverage of major classes of sgRNA in terms of total read depth across all cDNA samples, shown on log scale. Dashed lines indicate positions of TRS motif.
(D) Coverage of TRS-dependent sgRNA (blue) versus TRS-independent RNA (orange), summed over all cDNA sequencing experiments. Black dashed lines indicate position of TRS motifs. y axis is on log scale.
(E) Enlarged schematic of genome annotation for SARS-CoV-2. Regions are to scale.
See also Figure S1.
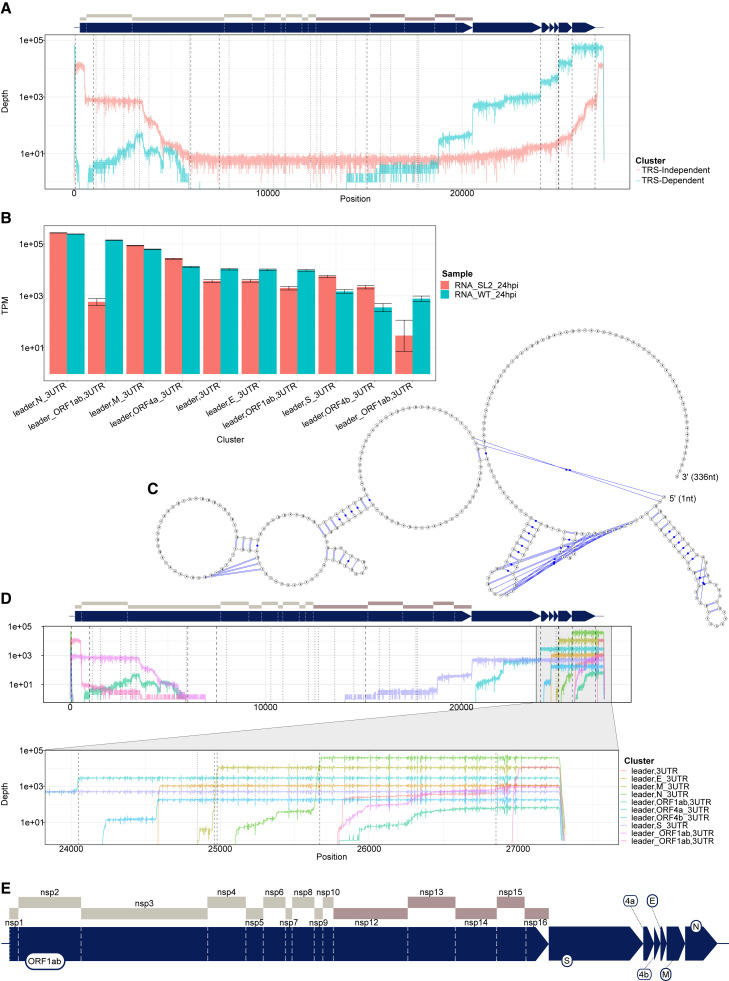
To investigate whether this unusual transcript is unique to the SARS-CoV-2 (which is part of the beta-coronavirus family), we re-analyzed ONT dRNA sequence data from the alpha-coronavirus HCoV-229E (Viehweger et al., 2019). We found a similar pattern of TRS-dependent and TRS-independent sgRNA (Figures 3A and 3D), in which the first polypeptide nsp1 was joined to a portion of the 3′ UTR. Using npTranscript to quantify abundance of these transcripts, we found that TRS-independent transcripts were substantially more abundant in wild-type HCoV-229E compared with a mutant form of 229E in which the conserved 5′ stem loop 2 (SL2) in HCoV-229E is replaced with that from SARS-CoV and B-CoV 3′ UTR (Figure 3B). This finding is suggestive of a role for the SL2 of the leader sequence in the creation of these transcripts in 229E, perhaps via long-range RNA-RNA interaction, and may be relevant to the similar extended leader mRNAs found in SARS-CoV-2. Inspection of the RNA secondary structure of ORF10 + 3′ UTR indicates that ORF10 forms a bulged stem loop (BSL) structure, upstream of the hypervariable BSL region of 3′ UTR (Figure 3C). The BSL is a conserved feature of beta-coronavirus genomes and thought to be essential for viral replication (Madhugiri et al., 2016). Taken together, this evidence supports the role of ORF10 as part of the 3′ UTR of SARS-CoV-2.
Figure 3.
Alphacoronavirus HCoV-229E produces classes of TRS-independent sgRNA that are abundantly expressed
The transcript nomenclature W_X,Y_Z indicates that the transcript consists of the segment from W to X joined with the segment from Y to Z.
(A) Total depth of TRS-independent versus TRS-dependent sgRNA in HCoV-229E. Dashed vertical lines indicate positions of TRS motifs, with alternative motifs detected using FIMO shown in dotted and dot-dash lines.
(B) Normalized transcript counts (TPM viral-mapped reads) of major sgRNA from HCoV-229E for wild-type (WT) or with stem loop 2 replaced (SL2). Error bars indicate 95% binomial CI of TPM estimate using the logistic parameterization.
(C) Predicted secondary structure of ORF10 + 3′ UTR from SARS-CoV-2 showing bulged stem loop in ORF10. Prediction calculated with IPknot software.
(D) Transcript coverage of major classes of sgRNA in terms of total read depth across all cDNA samples, shown on log scale. Dotted lines indicate positions of TRS.
(E) Enlarged schematic of genome annotation for HCoV-229E. Regions are to scale.
See also Figure S1.
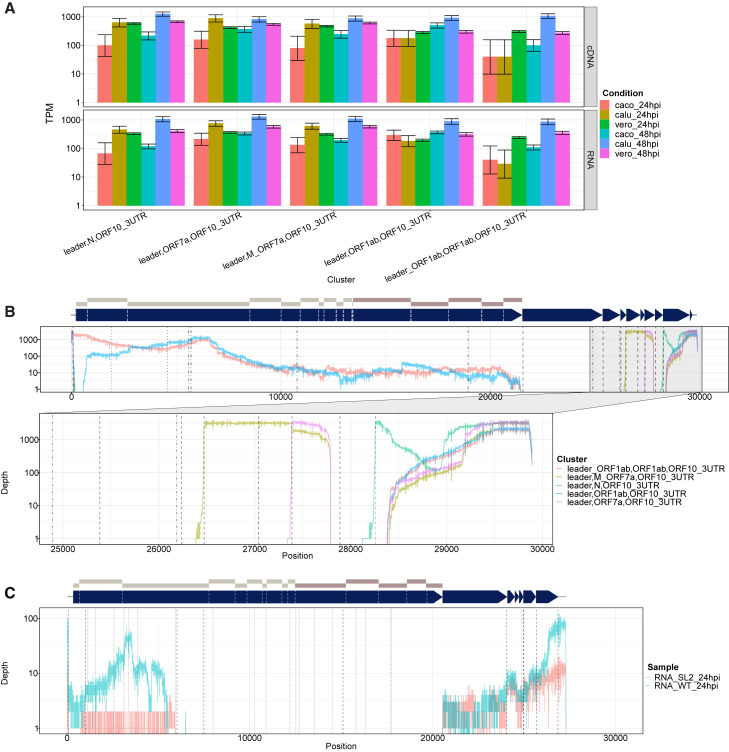
SARS-CoV-2 produces double-junction sgRNA
We also identified a persistent “double-junction” pattern in SARS-CoV-2 transcripts. This category featured sgRNA that showed two patterns of disjunction present at low concentrations across both dRNA and cDNA datasets (Figure 4 A). ORF10 was the most frequently added terminal 3′ ORF in double-junction sgRNA (Figure 4B). Most first disjunction events were TRS dependent, although 10% used the TRS-independent ORF1ab breakpoint as described in the previous section (Figure S4). In contrast, most second disjunctions were non-TRS dependent, and the 3′ breakpoint mirrored the ORF1ab,ORF10 breakpoint, suggestive of shared joining mechanism controlling this second junction that differs from TRS-mediated discontinuous minus-strand extension (Figure S4A). Double-junction sgRNAs were greatest in the Calu-3 48 hpi dataset, in which we observed leader,N,ORF10 and leader,ORF7a,ORF10 as most abundant, with 1,241 and 811 TPMs, respectively. We also observed triple-disjunction clusters at very low levels of expression, such as ORF1ab,ORF1ab,ORF1ab,ORF10, which had an estimated 60 TPMs in the Calu-3 48 hpi dataset. The majority of final junctions of these triple-junction reads includes the ORF10 breakpoint (Figure S4B).
Figure 4.
SARS-CoV-2 produces double-junction sgRNA
The transcript nomenclature U_V,W_X,Y_Z indicates that the transcript consists of the segments U to V, W to X, and Y to Z.
(A) Normalized counts (in TPM mapped viral reads) of double-junction reads in SARS-CoV-2 cDNA datasets. Error bars indicate 95% binomial CI of TPM estimate using the logistic parameterization.
(B) Depth of coverage of double-junction sgRNA in SARS-CoV-2 (summed over all cDNA sequencing experiments), shown on log scale. Dashed lines indicate positions of TRS motifs.
(C) Coverage of double-junction reads in 229E for WT and samples with modified SL2.
See also Figures S1 and S4.
In comparison, HCoV-229E appeared to have a smaller proportion of double-junction reads. Nevertheless, we observe a leader,ORF1ab,3UTR double-junction cluster (Figure 3D). This cluster was observed only in the WT HCoV-229E strain, and hence highly dependent on SL2 in the 3′ UTR (Figure 4C).
Viral transcript polyadenylation patterns consistent with templating from negative strand
We detected reads with no region mapping to the 3′ end of the viral genome in both cDNA and dRNA datasets (Figure 1A). Upon inspection with Nanopolish 'polya,' we found that no poly(A) tail is detected in most of these reads (Figure S5A), and that more than half of these reads also lacked detectable sequencing adaptor. This observation was consistent regardless of whether the transcripts mapped to the viral 5′ terminus and contrasted with transcript categories that mapped to the 3′ end and possessed clearly segmented poly(A) tails. The pattern observed was also consistent with polyadenylation being produced by templating from the negative strand, rather than from host polyadenylation factors. The quantity of non-3′ reads varied from 2% to 4% (median = 3.3%) of viral reads in all the dRNA datasets we analyzed except for Vero 24 hpi in which 10% of reads lacked the expected 3′ viral segment (Table S1). We found a non-random distribution of terminal breaks for non-3′ reads; however, the sequence composition of their end segments does not support the idea that it is driven by runs of internal poly(A) (Figures S5B and S5C). Given the requirement for poly(A) tails for ONT sequencing, we considered that these reads may arise from incorrect segmentation of a single read into multiple reads, only one of which possessed a poly(A) tail.
Viral sgRNA expression patterns change during the course of cellular infection
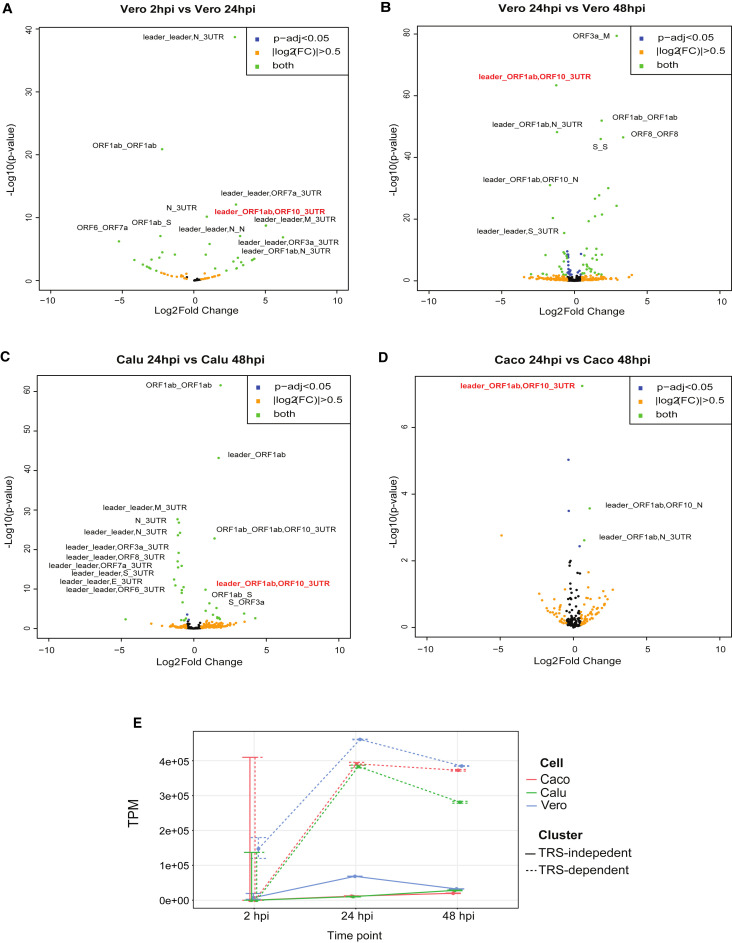
In order to interrogate the differential expression of SARS-CoV-2 transcriptional clusters during the time course, we analyzed ONT direct cDNA data, which were sequenced in triplicate for each time point (2, 24, and 48 hpi) and each cell line (Vero, Calu-3, Caco-2). We utilized npTranscript to generate a reference transcriptome of sgRNA produced by SARS-CoV-2 and to assign reads to transcript clusters (see STAR Methods), followed by DESeq2 for differential expression analyses. We normalized each library by the number of viral mapping reads, rather than the total number of viral and host mapping reads in order to establish changes in relative abundance, rather than simply track increase in overall viral RNA during the infection (which can be seen in Figure 1). From this analysis, we could identify differentially expressed transcripts between time points, 24 versus 48 hpi (late) in all three cell lines and 2 versus 24 hpi (early to late) in the Vero cell line only (because of extremely low abundance of viral mapping reads in human cell lines at 2 hpi).
Interestingly, in addition to differential expression of transcripts, which have both 5′ leader and 3′ UTR, we also found differential expression of transcripts that lacked the leader (non5_3) or the 3′ UTR (5_non3), or both (non5_non3) (Figures 5 A–5D). For the main analysis, we proceeded to analyze the 5_3 subset of the differential expression results (Table 1 ). From our data, we estimate that the general trajectory of differential expression of SARS-CoV-2 subgenomic transcripts during an infection presents an upregulation of TRS-dependent and TRS-independent transcripts between early and late infection, and then downregulation of TRS-dependent and TRS-independent transcripts, followed by an upregulation of fragmented non5_non3 transcripts at the final stage. Of note, the transcriptional activity of TRS-independent transcripts appeared to occur faster in Vero cells compared with the human cell lines, as seen by the delayed upregulation of TRS-independent transcript in human cell lines in relation to Vero cells (Figure 5E).
Figure 5.
Viral sgRNA expression patterns change during the course of cellular infection with delayed responses in TRS-independent transcripts
Volcano plots of differentially expressed SARS-CoV-2 transcripts from direct cDNA datasets (n = 3, where n is the number of technical replicates).
(A) Vero cells between 2 versus 24 hpi.
(B–D) Vero (B), Calu-3 (C), and Caco-2 (D) cell lines between 24 versus 48 hpi analyzed using DESeq2. Thresholds of p-adj < 0.05 and |log2FC| > 0.5 were applied to the data. Orange dots indicate transcripts that have |log2FC| > 0.5, blue dots indicate transcripts that have p-adj < 0.05, and green dots indicate transcripts that satisfy both criteria. Positive and negative log2FC indicate upregulation and downregulation at the latter time point, respectively. The transcript nomenclature W_X,Y_Z indicates that the transcript consists of the segment from W to X joined with the segment from Y_Z.
(E) Changes in TRS-dependent (dotted) and TRS-independent (continuous) TPM mapped viral reads across multiple time points (2, 24, and 48 hpi) in Caco-2 (orange), Calu-3 (green), and Vero (blue) cell lines. Error bars indicate 95% binomial CI of TPM estimate using the logistic parameterization.
See also Figures S1 andS7 and Table S2.
Table 1.
Viral sgRNAs are differentially expressed between 2 versus 24 and 24 versus 48 hpi in Vero cells and 24 versus 48 hpi in Calu-3 and Caco-2 cells
| Transcript | Log2FC | p-adj |
|---|---|---|
| Vero: 2 versus 24 hpi | ||
| leader_leader,N_3UTR | 2.87 | 9E−37 |
| leader_leader,ORF7a_3UTR | 2.95 | 1E−10 |
| leader_ORF1ab,ORF10_3UTR | 5.04 | 1E−7 |
| leader_leader,M_3UTR | 5.03 | 1E−7 |
| leader_leader,ORF3a_3UTR | 6.23 | 7E−6 |
| leader_leader,ORF8_3UTR | 3.08 | 0.001 |
| leader_ORF1ab,N_3UTR | 4.07 | 0.001 |
| leader_leader,ORF6_3UTR | 3.31 | 0.003 |
| leader_leader,S_3UTR | 3.45 | 0.006 |
| leader_leader,E_3UTR | 2.28 | 0.035 |
| Vero: 24 versus 48 hpi | ||
| leader_ORF1ab,ORF10_3UTR | −1.25 | 7E−62 |
| leader_ORF1ab,N_3UTR | −1.20 | 5E−47 |
| leader_leader,S_3UTR | −0.71 | 6E−15 |
| leader_3UTR | −1.99 | 5E−8 |
| leader_ORF1ab,3UTR_3UTR | −1.03 | 2E−4 |
| leader_ORF1ab,end | −0.92 | 0.001 |
| leader_ORF1ab,ORF3a_3UTR | −0.85 | 0.007 |
| leader_ORF1ab,ORF8_ORF8,ORF10_3UTR | −2.18 | 0.025 |
| leader_leader,ORF3a_ORF7a,end | −2.17 | 0.026 |
| leader_ORF1ab,ORF3a_ORF3a,ORF10_3UTR | −2.99 | 0.035 |
| leader_ORF1ab,S_3UTR | −1.91 | 0.042 |
| Caco-2: 24 versus 48 hpi | ||
| leader_ORF1ab,ORF10_3UTR | 0.60 | 3E−6 |
| leader_ORF1ab,N_3UTR | 0.74 | 0.029 |
| Calu-3: 24 versus 48 hpi | ||
| leader_leader,M_3UTR | −1.12 | 1E−26 |
| leader_leader,N_3UTR | −1.09 | 7E−23 |
| leader_leader,ORF3a_3UTR | −1.05 | 2E−18 |
| leader_leader,ORF8_3UTR | −1.11 | 2E−16 |
| leader_leader,ORF7a_3UTR | −0.84 | 2E−15 |
| leader_leader,S_3UTR | −1.36 | 6E−12 |
| leader_leader,E_3UTR | −1.27 | 2E−10 |
| leader_ORF1ab,ORF10_3UTR | 0.81 | 2E−9 |
| leader_leader,ORF6_3UTR | −0.83 | 1E−8 |
| leader_ORF1ab,S_ORF1ab,ORF10_3UTR | 3.47 | 0.001 |
| leader_leader,S_ORF1ab,ORF10_3UTR | 1.80 | 0.025 |
The differential expression results have been filtered by p-adj < 0.05 and |log2FC| > 0.5, and the transcript nomenclature W_X,Y_Z indicates that the transcript consists of the segment from W to X joined with the segment from Y_Z.
Among these results, one TRS-independent transcript, leader_ORF1ab,ORF10_3UTR, has been shown to be consistently differentially expressed across all cell types. This transcript was significantly upregulated (adjusted p value [p-adj] < 0.05) between 2 and 24 hpi in Vero cells and downregulated between 24 versus 48 hpi (Figures 5A–5D). In comparison, this transcript was upregulated between 24 versus 48 hpi in Caco-2 and Calu-3 cells, mirroring the viral counts over time (Figure 1A) as the level of these transcripts peaked at 24 hpi in Vero cells and at 48 hpi in the human cell lines. Collectively, these results suggest that the peak of TRS-independent transcriptional activity occurs earlier in Vero cells compared with human cell lines, and the presence of this TRS-independent transcript is of importance because it appears in all three cell types.
Additionally, we found that differentially expressed 5_3 transcripts (p-adj < 0.05) that were either genome mapped or transcriptome mapped revealed a positive linear correlation in log2FC between the two mapping methods (Table S2), with less transcripts being differentially expressed in transcriptome-mapped reads.
RNA modifications vary between genomic and sgRNA, but not throughout the course of infection
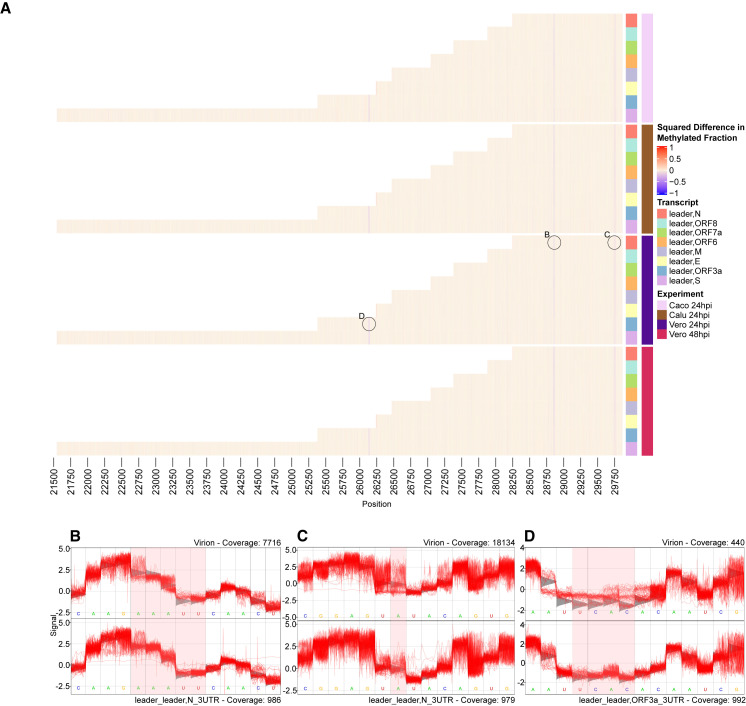
We used Tombo to determine de novo modification predictions on the various mRNA transcripts of the viral genome. Using virion dRNA as baseline (Taiaroa et al., 2020), we identified changes to modification of the genome throughout the course of infection, between individual transcripts, and across the three cell lines: Vero, Calu-3, and Caco-2. The vast majority (98.2%) of reads from the virion dataset included the 3′ UTR, but not the 5′ leader, and thus we inferred that it was almost entirely composed of reads from the viral genome rather than transcribed mRNA. The depth of coverage of this dataset was very low at the 5′ leader, and thus we are unable to report results of RNA modifications in the leader region (Figure S6).
The rapid infectibility of Vero cells allows a clear analysis of modifications at 24 and 48 hpi. The 2 hpi time point failed to produce adequate subgenomic expression for the analysis, and only 311 viral reads were detected in total.
In our analysis, predicted viral modification sites on specific sgRNA clusters did not change markedly throughout the infection time course (Figure 6 ). However, we saw differences on sgRNA as compared with the RNA genome. In particular, all analyzed sgRNA clusters displayed an absence of modification relative to virion genome in three regions as measured by the mean difference in methylated fraction (μ DMF; see STAR Methods): 26,130–26,135 in ORF3a (μ DMF = 0.62), 28,858–28,862 in ORFN (μ DMF = 0.6), and 29750A in 3′ UTR (μ DMF = 0.48) (Figure 6). We also observed that these modifications in the virion genome generated an artificially high rate of base-calling error at these positions.
Figure 6.
RNA modifications vary between genomic and sgRNA, but not throughout the course of infection
(A) Heatmap indicates (% age methylated reads cell line − % age methylated reads virion)2. sgRNAs are color coded on the y axis, and genome position is mapped on the x axis. 5′ leader sequence and final 30 bases of 3′ end are excluded because of insufficient coverage. Raw heatmap values are included in Data S1.
(B–D) Squiggle plots of selected significant locations from Vero 24 hpi as highlighted on heatmap. Bases of interest are highlighted in red windows. Gray triangles behind squiggle indicate expected signal distribution under the standard model (i.e., no modification). For (B), unmodified region of the N mRNA (bottom) is compared with squiggle from genomic RNA from the virion (top) signal information at genome position 28852–28862. For (C), unmodified base at N mRNA (bottom) position 29750A compared with predicted modification on virion. For (D), unmodified region 26310–26135 of ORF3a mRNA (bottom) compared with predicted modification on virion (top).
These same findings were repeated in data from Calu-3 and Caco-2 cells at 24 hpi, indicating that the different cell lines had little impact on modification changes (Figure 6). These results demonstrate that the viral genome carries RNA modifications that are not detectable on expressed mRNA.
The modifications reported here all consist of changes at >0.2 DMF2 (equivalent to DMF 0.44). A results summary including bases at >0.1 DMF2 (equivalent to 0.31) are included in Data S1.
Discussion
The use of long-read native RNA and direct cDNA sequencing allowed the identification of TRS-dependent and -independent transcripts in SARS-CoV-2. TRS-independent transcripts (sometimes referred to as non-canonical sgRNA) are formed without utilizing homologous TRS sequences and have been observed to occur in other SARS-CoV-2 transcriptome studies (Nomburg et al., 2020; Gribble et al., 2021; Kim et al., 2020; Taiaroa et al., 2020). Analysis of the time-course data presented in this manuscript shows a delayed increase of TRS-independent transcripts relative to TRS-dependent transcripts in two SARS-CoV-2-susceptible human cell lines. The most strongly upregulated of these included the leader_ORF1ab,ORF10_3UTR transcript (Table 1).
The function of SARS-CoV-2 ORF10 remains unclear. Some studies have reported evidence of ORF10 translation (Finkel et al., 2021), while others have not found conclusive evidence of its existence in proteome databases (Taiaroa et al., 2020). Pancer et al. (2020) identified single-nucleotide polymorphisms (SNPs) that cause premature stop codons in ORF10 but do not impact viability in vitro or in vivo. The active transcription of the leader ORF1ab,ORF10_3UTR transcript in our data (Figures 5A–5D) suggests a role for ORF10 distinct from its protein coding potential. This transcript contains the full-length nsp1 peptide, which is responsible for inhibiting host translation (Schubert et al., 2020), as well as the stabilizing stem loop structure from ORF10. Thus, the role of ORF10 in this context may be to stabilize the RNA molecule and enhance production of nsp1. The RNA family database RFAM (Kalvari et al., 2018) includes ORF10 in the Sarbecovirus-3UTR-annotated (RF03125) region of the SARS-CoV-2 genome.
We also observed ORF10 participating in the formation of the second junction in double-junction transcripts. These transcripts typically have a TRS-dependent first junction and a TRS-independent second junction to ORF10. The position of the second junction in the region upstream of ORF10 is variable, further supporting the notion that the ORF10 junction occurs in a homology-independent manner.
One explanation for the mechanism of TRS-independent sgRNA formation may be long-range RNA interactions. Long-range RNA interactions have previously been demonstrated as important for TRS-mediated leader-body joining in other coronaviruses (Mateos-Gomez et al., 2013) and may also be essential for non-TRS-mediated binding as seen in SARS-CoV-2. Ziv et al. (2020) explored cis-acting RNA-RNA interactions in SARS-CoV-2 and found several long-distance interactions within ORF1ab, including one that binds position 8357 nt with the 3′ UTR of the genome. This interaction may be responsible for promoting generation of the ORF1ab-ORF10 transcripts we describe.
Differential expression analyses were produced by mapping to the viral genome, as well as to the transcriptome (Table S2). Mapping to the genome allows novel transcripts to be found (Tombácz et al., 2016), whereas mapping to the transcriptome ensures the identity of the transcripts by clearly defining the junctions/breakpoints (Zhao, 2014). In this context of investigating the transcriptome of a novel coronavirus, there is more merit in mapping to the genome than the transcriptome because the transcriptome has not yet been extensively investigated and is most likely to be incomplete. The issues of using an incomplete reference transcriptome have been outlined previously (Pyrkosz et al., 2013). In our data, this is exemplified in transcripts that contain ORF1ab, because the breakpoint of ORF1ab is variable and cannot simply be defined by one breakpoint coordinate (Figure S7). This may explain why some transcripts are found differentially expressed in genome-mapped, but not transcriptome-mapped, analyses.
The data generated in this study are uniquely suited to studying the dynamics of viral epi-transcriptomics. Earlier studies have gained insights on methylation of 5′ capping for the escape of host immunity (Chen et al., 2011) and the impact of host epigenetics on disease outcome (Pinto et al., 2020). Kim et al. (2020) published the first bioinformatics analysis of base modifications on the SARS-CoV-2 viral genome in which they report 41 potential 5mC viral modification sites by contrasting signal-space information of the dRNA-sequenced viral genome against unmodified in-vitro-transcribed (IVT) sequence data. We used the virion-derived RNA as a control, which enabled us to focus on differences between modifications on the viral genome and transcriptome.
We find that the genomic RNA harbors more RNA modifications than the transcribed sgRNA. In particular, we report three regions that are more modified in genomic RNA than sgRNA. The strongest of these (position 28858–28862) was reported by Kim et al. (2020), who also reported that position 28859 is more modified among longer sgRNA. We extended this finding by showing that the modified state is representative of the genome RNA. We also report a remarkably stable pattern of modifications that showed very little change across cell lines and time points in transcribed sgRNA. This is the first evidence reported for the stability of SARS-CoV-2 epi-transcriptome throughout infection.
A deeper understanding of the SARS-CoV-2 transcriptome and how it changes during infection may lead to new avenues for therapeutic strategies. One example is development of strategies to disrupt the complex patterns of negative-strand disjunction to form sgRNA. Our work also highlights the importance of TRS-independent transcripts in the infectious cycle of SARS-CoV-2, which may also be an avenue for therapeutic development. Moreover, such knowledge also spurs the next generation of diagnostics for monitoring infection progression. The RNA genome modifications described here may also be a target for therapy, although further research is required to understand the role of the modifications described here.
Limitations of study
We used in vitro infection of mammalian cell lines, and as such our conclusions may not fully reflect in vivo SARS-CoV-2 infection. We included three time points in our study (2, 24, and 48 h), which may potentially miss key transcriptional changes in between these time points or after the final time point. We used a SARS-CoV-2 strain obtained from early (January 2020) in the pandemic, which is close to the original Wuhan strain and may not fully reflect infection dynamics of currently circulating strains. Our experimental design used replicates resulting from three separate infections of the same cell line performed at the same time and should therefore be regarded as technical rather than biological replicates. These replicates were sequenced separately for direct cDNA sequencing but were sequenced after pooling for the dRNA because of difficulties in multiplexing dRNA sequencing using ONT sequencing kits. The direct cDNA libraries were multiplexed and sequenced in batches of three infected and three control from the same time point on the same flow cell; thus, there is a possibility of batch effects in comparison between time points.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| SARS-CoV-2/Australia/VIC01/2020 | Laboratory of Kanta Subbarao | NCBI: MT007544.1 |
| Chemicals, peptides, and recombinant proteins | ||
| GlutaMAX | GIBCO | Cat#35050061 |
| TPCK-treated trypsin | Worthington | Cat#LS003740 |
| Critical commercial assays | ||
| MycoAlert Mycoplasma Detection Kit | Lonza | Cat#LT07-118 |
| RNeasy Mini Kit | QIAGEN | Cat#74106 |
| Homogenizer | Invitrogen | Cat#12183026 |
| Turbo DNA-free Kit | Invitrogen | Cat#AM1907 |
| RNAClean XP magnetic beads | Beckman Coulter | Cat#A63987 |
| Direct cDNA sequencing kit | Oxford Nanopore Technologies | Cat#SQK-DCS109 |
| Direct RNA sequencing kit | Oxford Nanopore Technologies | Cat#SQK-RNA002 |
| Native barcoding kit | Oxford Nanopore Technologies | Cat#EXP-NBD104&114 |
| PowerUp SYBR Green Master Mix (2X) | Applied Biosystems | Cat#A25742 |
| Deposited data | ||
| Raw FAST5 & FASTQ ONT data | This paper | NCBI repository BioProject Accession: PRJNA675370 |
| Virion ONT data | Taiaroa et al. (2020) | BioProject Accession: PRJNA608224 |
| 229E-HCoV ONT data | Viehweger et al. (2019) | European Nucleotide Archive (ENA) Accession: PRJEB33797 |
| R Code for methylation analysis | This paper | Github: https://github.com/dn-ra/SARS-CoV-2_Mods |
| Pipeline for breakpoint, differential expression analysis for viral reads (npTranscript) | This paper | Github: https://github.com/lachlancoin/npTranscripthttps://github.com/dn-ra/SARS-CoV-2_Mods |
| R code for DESeq2 | Github: https://gist.github.com/stephenturner/f60c1934405c | |
| Data S1 | This paper | Mendeley: https://dx.doi.org/10.17632/bpckrn3vtn.1 |
| Interactive web app | This paper | http://coinlab.mdhs.unimelb.edu.au/ |
| Experimental models: Cell lines | ||
| Vero | Laboratory of Kanta Subbarao | ATCC Cat# CCL-81; RRID: CVCL_0059 |
| Caco-2 | Laboratory of Kanta Subbarao | ATCC Cat# HTB-37; RRID: CVCL_0025 |
| Calu-3 | ATCC | ATCC Cat# HTB-55; RRID: CVCL_0609 |
| Oligonucleotides | ||
| Subgenomic N forward primer:CTTCC CAGGTAACAAACCAACC |
This paper | N/A |
| Subgenomic N reverse primer:CCATT CTGGTTACTGCCAGTTG |
This paper | N/A |
| Total N forward primer:TGCAA TCGTGCTACAACTTCCT |
This paper | N/A |
| Total N reverse primer:TGCCT GGAGTTGAATTTCTTGA |
This paper | N/A |
| Subgenomic E forward primer:CGAT CTCTTGTAGATCTGTTCTC |
Wölfel et al., 2020 | N/A |
| Subgenomic E/Total E reverse primer:ATATTGCAGCAGTACGCACACA | Corman et al., 2020 | N/A |
| Total E forward primer:TCATT CGTTTCGGAAGAGACAG |
This paper | N/A |
| Software and algorithms | ||
| Guppy v3.5.2 | Oxford Nanopore Technologies | https://community.nanoporetech.com/sso/login?next_url=%2Fdownloads |
| Minimap2 v2.11 & v2.17 | Li, 2018 | https://github.com/lh3/minimap2 |
| MEME-suite v5.1.1 | Bailey et al., 2009 | https://meme-suite.org/meme/doc/download.html |
| IPknot v1.3.1 | Sato et al., 2011 | http://rtips.dna.bio.keio.ac.jp/ipknot/ |
| Tombo v1.5 | Stoiber et al., 2016 | https://nanoporetech.github.io/tombo/ |
| SquiggleKit | Ferguson and Smith, 2019 | https://github.com/Psy-Fer/SquiggleKit |
| ComplexHeatmap | Gu et al., 2016 | https://www.bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html |
| Nanopolish v0.13.2 | Simpson et al., 2017 | https://nanopolish.readthedocs.io/en/latest/index.html |
| QuantStudio Real-Time PCR Software v1.3 | Applied Biosystems | https://www.thermofisher.com/us/en/home/global/forms/life-science/quantstudio-6-7-flex-software.html |
| GraphPad Prism v8 | GraphPad Software | https://www.graphpad.com/ |
| Samtools v1.9 | Li et al., 2009 | http://www.htslib.org/ |
| Salmon v0.13.1 | Patro et al., 2017 | https://github.com/COMBINE-lab/salmon |
| DESeq2 v1.28.1 | Love et al., 2014 | http://bioconductor.org/packages/release/bioc/html/DESeq2.html |
Resource availability
Lead contact
Further information and requests for resources, reagents, and code should be directed to and will be fulfilled by the lead contact, Lachlan Coin (lachlan.coin@unimelb.edu.au).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The datasets supporting the results presented here are available in the NCBI repository BioProject: PRJNA675370 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA675370). All code related to npTranscript is available on Github: https://github.com/lachlancoin/npTranscriptand and R code for methylation analysis is available on Github: https://github.com/dn-ra/SARS-CoV-2_Mods. Data S1 is available as a standalone zip file, and is available from Mendeley Data: https://dx.doi.org/10.17632/bpckrn3vtn.1. Transcript counts, coverage, and base-calling error rates can be explored and exported via an interactive web app: http://coinlab.mdhs.unimelb.edu.au/.
Experimental model and subject details
Cell lines were sourced from the American Type Culture Collection (ATCC) and included Vero (African green monkey kidney epithelial cells, ATCC Cat#CCL-81; RRID: CVCL_0059), Caco-2 (human intestinal epithelial cells, ATCC Cat#HTB-37; RRID: CVCL_0025) and Calu-3 (human lung epithelial cells, ATCC Cat#HTB-55; RRID: CVCL_0609) and maintained at 37°C, 5% (v/v) CO2. Vero cells were cultured in Minimum Essential Media (MEM) (Media Preparation Unit, Peter Doherty Institute) supplemented with 10% Fetal Bovine Serum (FBS) (Sigma-Aldrich), 1X penicillin/streptomycin, 1X GlutaMAX (GIBCO), and 15 mM HEPES (GIBCO). Caco-2 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Media Preparation Unit, Peter Doherty Institute) supplemented with 1X non-essential amino acids (Sigma-Aldrich), 20 mM HEPES, 2 mM L-glutamine, 1X GlutaMAX 2 μg/mL Fungizone solution, 26.6 μg/mL gentamicin, 100 IU/mL penicillin, 100 μg/mL streptomycin, and 20% FBS. Calu-3 cells were cultured in Advanced DMEM (GIBCO) supplemented with 10% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin, 1X GlutaMAX. All cell lines were seeded in 4 × 6-well tissue-culture plates and maintained at 37°C, 5% (v/v) CO2 for infection. The cell lines were tested for presence of mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza) and were not authenticated.
Method details
Infection
SARS-CoV-2 (Australia/VIC01/2020) virus was passaged in Vero cells at 37°C, 5% (v/v) CO2 and stored at −80°C. One 6-well plate per cell line was used for each time point (0, 2, 24, 48 hpi) with triplicate wells (n = 3) for mock controls and infected cells. All three cell lines were infected with SARS-CoV-2 (Australia/VIC01/2020), at a multiplicity of infection of 0.1 with infection inoculum composed of serum-free culture media and TPCK-treated trypsin (Worthington). The plates were incubated at 37°C for 30 minutes. The 0-hour time point plates were removed from incubation for harvesting, and 2 mL of serum-free media + TPCK trypsin mixture was added to the plates for the remaining time points (2, 24, 48 hpi). The 2, 24 and 48 hpi plates were placed in the incubator in 37°C, 5% (v/v) CO2 until harvesting time.
RNA extraction, treatment, and purification
The RNeasy Mini Kit (QIAGEN) was used to extract the RNA using the ‘Purification of Total RNA from Animal Cells Using Spin Technology’ protocol with minor modifications. The modifications include the following; 600 μL of RLT buffer was added to the cells, and the lysates were homogenized using the Homogenizer columns (Invitrogen) as per the manufacturer’s guidelines. RNA extracted using the RNeasy Mini Kit was treated with the DNase from the Turbo DNA-free Kit (Invitrogen) according to the manufacturer’s ‘rigorous DNase treatment’ protocol. The RNA in the supernatant was cleaned using RNAClean XP magnetic beads (Beckman Coulter) using the protocol ‘Agencourt RNAClean XP protocol 001298v001’. The magnetic beads were added to the RNA at 1.8X concentration and the final RNA was eluted in nuclease-free water.
ONT library preparation and sequencing
Direct cDNA sequencing libraries were prepared using an input of 3 μg of total RNA (equivalent to approximately 150 ng of poly(A) + RNA) for Vero cell infections, and 1-2 μg of total RNA (equivalent to approximately 50 – 100 ng of poly(A) + RNA) for Caco-2 and Calu-3 cell infections per triplicate (0, 2, 24, 48 hpi). RNA was converted to cDNA via the Direct cDNA sequencing kit (SQK-DCS109) and multiplexed using the native barcoding kit (EXP-NBD104 & 114). DRNA sequencing libraries were generated using an input of 6 μg of total RNA (2 μg per triplicate equivalent to ∼300 ng poly(A) + RNA) for Vero cell infections and 3 μg of total RNA (1 μg per triplicate equivalent to ∼150 ng poly(A) + RNA) for Caco-2 and Calu-3 cell infections via the SQK-RNA002 kit. Due to the absence of multiplexing, control or infected triplicates were pooled for dRNA sequencing per flow cell (2, 24, 48 hpi). Synthetic RNA controls (Hardwick et al., 2016) were spiked into samples at ∼10% of expected poly(A) + RNA content with Mix A used for infected samples and Mix B for uninfected controls. All libraries were sequenced with MinION R9.4.1 flow cells. Sequencing generated approximately 6 – 11 million reads for direct cDNA and roughly 1-3 million reads for dRNA sequencing. Raw data (FAST5 files) were basecalled using Guppy v3.5.2.
Quantitative reverse transcription PCR
As a measure of infectivity, the differences between total and subgenomic transcripts which encode for the Nucleocapsid (N) and Envelope (E) ORFs were investigated using qRT-PCR (n = 2-3, where n is the number of infected replicate wells involved per time point per cell line). Barcoded cDNA from direct cDNA sequencing libraries were diluted and ∼0.17 ng was amplified in triplicate using four sets of primers (1 μM input each primer) (Sigma-Aldrich) via the PowerUp SYBR Green Master Mix (2X) (Applied Biosystems). Primer details are listed in the Key resources table. The amplification was carried out within the Quantstudio 7 Flex Real-Time PCR Systems (Applied Biosystems) with the standard cycling mode (50°C, 2 mins; 95°C, 2 mins; 50 cycles of 95°C, 15 s and 60°C x 1 min). qPCR was repeated to a total of two times for earlier time points – 0 and 2 hpi for cDNA from Caco-2 and Calu-3 cells, and 2 hpi for cDNA from Vero cells.
Quantification and statistical analysis
Transcript Discovery
FASTQ sequences were mapped to the reference SARS-CoV-2 genome from the first Australian case of COVID-19 (Australia/VIC01/2020, NCBI: MT007544.1) using Minimap2 v2.11 with the splice option ‘-x splice’ engaged and ignoring TRS-dependent splice signal ‘-un’. Mapped sequences in the resulting BAM file were passed through a transcript discovery pipeline which annotates reads with information on the location of splice breakpoints relative to the viral genome. CIGAR strings are used to determine splice regions by continuous sequence of the N (not mapped) operator. Any splice traversing longer than 1000 bp of the viral genome is treated as a valid break and the genomic sites of the break are recorded in a vector such as [read_start, break1_5′, break1_3′, read_end]. We then convert this to an annotation-based string array. The read_start, read_end and 5′ breakpoint ends are converted to the first annotation which starts 5′ upstream of its position, or within 10 bp downstream of its position to allow for sequencing error. The 3′ breakpoint ends are converted to text based on the next 3′ downstream annotation, or within 10 bp 5′ upstream. This captures the fact that the disjunction sites occur immediately upstream of the target ORF. We note that this is different to the way a eukaryotic or prokaryotic gene annotation program would work. Finally, we convert the string array into a string via concatenation, with 5′ break to 3′ break concatenated using a comma to indicate the break. The end result of this procedure is an assignment of string ID, such as leader_leader,N_3UTR indicating the read starts in the leader sequence, has its first break point starting in leader and going to upstream of N, and finally ending within the 3′UTR. The code for this analysis is available at [https://github.com/lachlancoin/npTranscript].
TRS Finding
Transcription Regulating Sequences (TRS) are required for leader-body joining during discontinuous minus-strand extension. TRS sites were located in the viral genome via a motif search using FIMO v5.1.1 from MEME-suite v5.1.1 (Bailey et al., 2009). The 6 bp segments of viral-mapping reads aligned to the TRS-dependent 5′ ACGAAC 3′ TRS were extracted from the BAM file and transformed into a Position Weight Matrix (PWM) to model variability in the sequence. The hexamer 5′ CTAAAC 3′ was used for locating TRSs in the 229E genome. The resulting PWM was converted into meme format using jaspar2meme from MEME-suite v5.1.1 (Bailey et al., 2009) and then used for scanning the full viral genome using FIMO from the same software suite. The code for this analysis is also available at [https://github.com/lachlancoin/npTranscript].
Methylation analysis
Signal-space FAST5 files were assessed to identify signal changes corresponding to RNA modifications using Tombo v1.5 (Stoiber et al., 2016). Having already been allocated a transcript cluster in npTranscript, read IDs from each of the 8 major subgenomes were down-sampled to 1000 reads. FAST5 reads were retrieved using the ‘fast5_fetcher_multi’ function in SquiggleKit (Ferguson and Smith, 2019) and resquiggled to the respective reference transcript. Transcript clusters with fewer than 1000 reads were abandoned for fear of generating an inaccurate assessment of methylation.
Resquiggled FAST5 reads were input into the ‘detect_modifications’ function using the ‘de_novo’ option which searches for any deviation from the TRS-dependent FAST5 signal. Outputs were converted to dampened_fraction wiggle files and exported for visualization and analysis in R. Quantification of modification changes was measured in Difference in Methylated Fraction (DMF), which is given by:
where t denotes the transcript of interest, b is the base position, f is the calculated methylated fraction from Tombo, and v is the SARS-CoV-2 virion used for comparison.
ComplexHeatmap (Gu et al., 2016) was used to produce heatmap plots of methylation data, and DMF was squared in plotting for ease of visual interpretation of results. Tombo ‘plot’ was used to generate squiggle plots at sites of interest. All R code for this analysis is available at [https://github.com/dn-ra/SARS-CoV-2_Mods].
Poly(A) analysis
FASTQ passed and failed reads from dRNA sequencing were merged and indexed via Nanopolish v0.13.2 ‘index’ (Simpson et al., 2017) using the default parameters ‘-d $FAST5 -s sequencing_summary.txt $FASTQ’. The poly(A) tails of each read were estimated using the ‘polya’ function with the parameters ‘–reads $FASTQ –bam $BAM–genome $REFERENCE GENOME > combined.tsv’. A merged reference genome containing the SARS-CoV-2 Australia virus (Australia/VIC01/2020, NCBI: MT007544.1), host genome from Ensembl (release 100) and RNA sequin decoy chromosome genome (Hardwick et al., 2016) was used.
Quantitative reverse transcription PCR
The mean differences in Ct (subgenomic - total) of N and E SARS-CoV-2 ORFs from cDNA libraries derived from replicate groups of infected cells (n = 2-3) per time point per cell line were calculated. The results were plotted with mean ± SD error bars using GraphPad Prism v8. As the Ct values of subgenomic E mRNA were undetectable (> 40 Ct) in 0 and 2 hpi time points in the human cell lines across duplicate runs, the Ct value was regarded as 40 for the purposes of measuring infectivity.
Counts and composition of mapped reads
Samtools v1.9 ‘view’ (Li et al., 2009) was used to generate the name and length of all the chromosomes in the host reference genome using the commands ‘–H $BAM | grep SQ | cut –f2-3 | sed ‘s/SN://g’ | sed ‘s/LN:?1\t/g’’. The number of host, virus, and sequin reads mapping to the combined genome was counted using parameters ‘-F4 -F2048 -F256 -L $LIST_OF_CHROMS_IN_HOST.txt $BAM | wc –l’, ‘-F4 -F2048 -F256 $BAM MT007544.1 | wc –l’ and ‘-F4 -F2048 –F256 $BAM chrIS | wc –l’, respectively.
Differential expression analysis
Passed and failed FASTQ files from direct cDNA sequencing were merged for each sequencing run and used for downstream differential expression analysis. Mapping was carried out with Minimap2 v2.17 (Li, 2018) with the parameters ‘-ax splice –secondary = no’ to a merged reference genome containing the SARS-CoV-2 Australia virus (Australia/VIC01/2020, NCBI: MT007544.1), host genome from Ensembl (release 100) and RNA sequin decoy chromosome genome (Hardwick et al., 2016). Using the npTranscript pipeline, the viral reads were extracted from the BAM files to separate out reads which had primary mapping to the viral genome. The extracted viral reads were re-mapped to the viral genome using Minimap2 with the parameters ‘-ax splice –un’ as these parameters account for TRS-independent splice sites within the viral genome. During this process, Featurecounts-like count files were generated for differential expression analysis as Featurecounts (Liao et al., 2014) was unable to be used to generate suitable counts tables for the virus, perhaps due to the viral annotations being generated in-house using the npTranscript pipeline which are based on the ORF start position downstream of the 3′ break point instead of the breakpoint being considered as the start of the ORF. The raw counts from npTranscript were analyzed using DESeq2 v1.28.1 (Love et al., 2014) as per described below, where thresholds of |log2FoldChange (FC)| > 0.5 and p-adj < 0.05 were applied. Transcript clusters with both 5′ leader and 3′ UTR sequences were retained in the results (Table 1). Furthermore, due to the low counts at 2 hpi, transcripts with direction of normalized counts conflicting with the direction of log2FC between 2 hpi and 24 hpi were regarded as false positives and flagged as being spurious.
In order to assess correlation of differential expression between genome-mapped and transcriptome-mapped transcripts, extracted viral transcripts from npTranscript which map to both the 5′ and 3′ ends of the full-length viral transcripts were isolated using a custom script. The new FASTQ reads were re-mapped to the viral genome using Minimap2 with the parameters ‘-ax splice –un’ and the transcriptome with the default ‘-ax ont-map’. The same Featurecounts-like files were generated for genome-mapped reads as above, which were used for DESeq2 analysis. For viral reads re-mapped with the viral 5_3 transcriptome generated by npTranscript, primary-mapped reads were isolated using Samtools ‘view -b -h -F 2308 $BAM > primary.bam’. Salmon v0.13.1 (Patro et al., 2017) was used for isoform quantification of alignments with the parameters ‘–noErrorModel –noLengthCorrection’ to obtain viral transcript counts which were input for differential transcript expression analysis in DESeq2. A threshold of p-adj < 0.05 was applied for this analysis.
All DESeq2 analyses were performed as per the following methods. The counts from npTranscript and Salmon were input for gene and transcript level analysis respectively. Count matrices were filtered to remove very lowly expressed features (≤5 in total for each gene/transcript). Counts were normalized for sequencing depth within DESeq2 prior to statistical analysis. Log2FCs and adjusted p-values were calculated for each annotated gene or transcript and used to determine statistical significance. A regularized log transformation was subsequently performed on the normalized counts for visualization. The PCA and volcano plots were made using the following code: [https://gist.github.com/stephenturner/f60c1934405c127f09a6].
RNA Secondary Structure Prediction
RNA secondary structure for the ORF10 + 3′UTR region of SARS-CoV-2 was predicted using IPknot webserver v1.4.1 (Sato et al., 2011). Default settings of Level 2 prediction, McCaskill scoring model, and nil refinement were used.
Additional resources
The data from this study can be visualized using an interactive website: http://coinlab.mdhs.unimelb.edu.au/.
Acknowledgments
We would like to acknowledge Josh Lee and Uli Feltzmann for assistance with the Shiny app. We would like to thank Georgia Deliyannis for assistance with culturing cell lines, and Dr. Eva Maria Novoa Pardo for her advice regarding the analysis of SARS-CoV-2 modifications. L.J.M.C. was supported by a Career Development Fellowship from NHMRC (GNT1130084). This research was supported by a University of Melbourne “Driving research momentum” award (to L.J.M.C.) and NHMRC EU project grant (GNT1195743 to L.J.M.C.). K.S. is supported by an NHMRC Investigator grant. The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health. J.J.-Y.C was supported by the Miller Foundation and the Australian Government Research Training Programme (RTP) scholarship. Computational analysis of data was made possible with access to two server systems: Spartan at University of Melbourne (Lafayette et al., 2016) and Nectar from the Australia Research Data Commons.
Author contributions
J.J.-Y.C.: methodology, software, validation, formal analysis, investigation, data curation, writing – original draft, writing – review & editing, and visualization; D.R.: methodology, software, formal analysis, data curation, writing – original draft, writing – review & editing, and visualization; M.E.P.: methodology, investigation, validation, supervision, project administration, writing – original draft, and writing – review & editing; J.G.: software, formal analysis, data curation, and visualization; G.T.: writing – review & editing; C.Z.: software, formal analysis, data curation, visualization, and writing – review & editing; F.L.M.: investigation and methodology; R.D.P.-I.: investigation; L.C.: writing – review & editing; D.F.J.P.: writing – review & editing; D.A.W.: writing – review & editing; T.P.S.: writing – review & editing; S.L.L.: conceptualization, methodology, and resources; M.B.C.: conceptualization, resources, writing – review & editing, and supervision; K.S.: conceptualization, methodology, resources, writing – review & editing, and supervision; L.J.M.C.: conceptualization, methodology, software, validation, formal analysis, investigation, resources, data curation, writing – original draft, writing – review & editing, visualization, supervision, project administration, and funding acquisition.
Declaration of interests
L.J.M.C., M.E.P., J.G., R.D.P.-I., and M.B.C. have received support from ONT to present their findings at scientific conferences. ONT played no role in study design, execution, analysis, or publication. L.J.M.C. has received research funding from ONT unrelated to this project.
Published: May 11, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109108.
Supplemental information
Sheet 1 contains the raw data for values in the heatmap. Sheet 2 contains a subset of sheet 1 with all values for bases with any one value > 0.1 or < −0.1. Red highlighted base numbers indicate positions that are near a transcript breakpoint and therefore in question. Units for both sheets is squared DMF.
References
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista F.M., Stapleton T., Lowther J.A., Fonseca V.G., Shaw R., Pond C., Walker D.I., van Aerle R., Martinez-Urtaza J. Whole Genome Sequencing of Hepatitis A Virus Using a PCR-Free Single-Molecule Nanopore Sequencing Approach. Front. Microbiol. 2020;11:874. doi: 10.3389/fmicb.2020.00874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., Matthews D.A. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson J.M., Smith M.A. SquiggleKit: a toolkit for manipulating nanopore signal data. Bioinformatics. 2019;35:5372–5373. doi: 10.1093/bioinformatics/btz586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- Garalde D.R., Snell E.A., Jachimowicz D., Sipos B., Lloyd J.H., Bruce M., Pantic N., Admassu T., James P., Warland A. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods. 2018;15:201–206. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
- Gribble J., Stevens L.J., Agostini M.L., Anderson-Daniels J., Chappell J.D., Lu X., Pruijssers A.J., Routh A.L., Denison M.R. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 2021;17:e1009226. doi: 10.1371/journal.ppat.1009226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Hardwick S.A., Chen W.Y., Wong T., Deveson I.W., Blackburn J., Andersen S.B., Nielsen L.K., Mattick J.S., Mercer T.R. Spliced synthetic genes as internal controls in RNA sequencing experiments. Nat. Methods. 2016;13:792–798. doi: 10.1038/nmeth.3958. [DOI] [PubMed] [Google Scholar]
- Kalvari I., Argasinska J., Quinones-Olvera N., Nawrocki E.P., Rivas E., Eddy S.R., Bateman A., Finn R.D., Petrov A.I. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018;46(D1):D335–D342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafayette L., Sauter G., Vu L., Meade B. OpenStack Summit; 2016. Spartan Performance and Flexibility: An hpc-Cloud Chimera; p. 27. [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhugiri R., Fricke M., Marz M., Ziebuhr J. Elsevier; 2016. Coronavirus cis-Acting RNA Elements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gomez P.A., Morales L., Zuñiga S., Enjuanes L., Sola I. Long-distance RNA-RNA interactions in the coronavirus genome form high-order structures promoting discontinuous RNA synthesis during transcription. J. Virol. 2013;87:177–186. doi: 10.1128/JVI.01782-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomburg J., Meyerson M., DeCaprio J.A. Pervasive generation of non-canonical subgenomic RNAs by SARS-CoV-2. Genome Med. 2020;12:108. doi: 10.1186/s13073-020-00802-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer K., Milewska A., Owczarek K., Dabrowska A., Branicki W., Sanak M., Pyrc K. Cold Spring Harbor Laboratory; 2020. The SARS-CoV-2 ORF10 Is Not Essential In Vitro or In Vivo in Humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto B.G.G., Oliveira A.E.R., Singh Y., Jimenez L., Gonçalves A.N.A., Ogava R.L.T., Creighton R., Schatzmann Peron J.P., Nakaya H.I. ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19. J. Infect. Dis. 2020;222:556–563. doi: 10.1093/infdis/jiaa332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrkosz A.B., Cheng H., Brown C.T. RNA-Seq Mapping Errors When Using Incomplete Reference Transcriptomes of Vertebrates. arXiv. 2013 1303.2411v1. [Google Scholar]
- Sato K., Kato Y., Hamada M., Akutsu T., Asai K. IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. Bioinformatics. 2011;27:i85–i93. doi: 10.1093/bioinformatics/btr215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.-A., Leibundgut M., Thiel V., Mühlemann O., Ban N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- Simpson J.T., Workman R.E., Zuzarte P.C., David M., Dursi L.J., Timp W. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods. 2017;14:407–410. doi: 10.1038/nmeth.4184. [DOI] [PubMed] [Google Scholar]
- Stoiber M., Quick J., Egan R., Eun Lee J., Celniker S., Neely R.K., Loman N., Pennacchio L.A., Brown J. Cold Spring Harbor Laboratory; 2016. De Novo Identification of DNA Modifications Enabled by Genome-Guided Nanopore Signal Processing. [Google Scholar]
- Taiaroa G., Rawlinson D., Featherstone L., Pitt M., Caly L., Druce J., Purcell D., Harty L., Tran T., Roberts J. Direct RNA sequencing and early evolution of SARS-CoV-2. bioRxiv. 2020 2020.03.05.976167. [Google Scholar]
- Tombácz D., Csabai Z., Oláh P., Balázs Z., Likó I., Zsigmond L., Sharon D., Snyder M., Boldogkői Z. Full-Length Isoform Sequencing Reveals Novel Transcripts and Substantial Transcriptional Overlaps in a Herpesvirus. PLoS ONE. 2016;11:e0162868. doi: 10.1371/journal.pone.0162868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viehweger A., Krautwurst S., Lamkiewicz K., Madhugiri R., Ziebuhr J., Hölzer M., Marz M. Direct RNA nanopore sequencing of full-length coronavirus genomes provides novel insights into structural variants and enables modification analysis. Genome Res. 2019;29:1545–1554. doi: 10.1101/gr.247064.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Zhao S. Assessment of the impact of using a reference transcriptome in mapping short RNA-seq reads. PLoS One. 2014;9:e101374. doi: 10.1371/journal.pone.0101374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv O., Price J., Shalamova L., Kamenova T., Goodfellow I., Weber F., Miska E.A. The Short- and Long-Range RNA-RNA Interactome of SARS-CoV-2. Mol. Cell. 2020;80:1067–1077.e5. doi: 10.1016/j.molcel.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sheet 1 contains the raw data for values in the heatmap. Sheet 2 contains a subset of sheet 1 with all values for bases with any one value > 0.1 or < −0.1. Red highlighted base numbers indicate positions that are near a transcript breakpoint and therefore in question. Units for both sheets is squared DMF.
Data Availability Statement
The datasets supporting the results presented here are available in the NCBI repository BioProject: PRJNA675370 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA675370). All code related to npTranscript is available on Github: https://github.com/lachlancoin/npTranscriptand and R code for methylation analysis is available on Github: https://github.com/dn-ra/SARS-CoV-2_Mods. Data S1 is available as a standalone zip file, and is available from Mendeley Data: https://dx.doi.org/10.17632/bpckrn3vtn.1. Transcript counts, coverage, and base-calling error rates can be explored and exported via an interactive web app: http://coinlab.mdhs.unimelb.edu.au/.