Graphical abstract
Keywords: COVID-19 control, SARS-CoV-2 virus detection, Electrochemical biosensors, Electrochemical detection, Au-based nanomaterials, Carbon-based nanomaterials
Abstract
The SARS-CoV-2 virus is still causing a dramatic loss of human lives worldwide, constituting an unprecedented challenge for the society, public health and economy, to overcome. The up-to-date diagnostic tests, PCR, antibody ELISA and Rapid Antigen, require special equipment, hours of analysis and special staff. For this reason, many research groups have focused recently on the design and development of electrochemical biosensors for the SARS-CoV-2 detection, indicating that they can play a significant role in controlling COVID disease. In this review we thoroughly discuss the transducer electrode nanomaterials investigated in order to improve the sensitivity, specificity and response time of the as-developed SARS-CoV-2 electrochemical biosensors. Particularly, we mainly focus on the results appeard on Au-based and carbon or graphene-based electrodes, which are the main material groups recently investigated worldwidely. Additionally, the adopted electrochemical detection techniques are also discussed, highlighting their pros and cos. The nanomaterial-based electrochemical biosensors could enable a fast, accurate and without special cost, virus detection. However, further research is required in terms of new nanomaterials and synthesis strategies in order the SARS-CoV-2 electrochemical biosensors to be commercialized.
1. Introduction
Currently, SARS-CoV-2 can be diagnosed by two different ways: i) antigen tests (point-of-care, POC) and ii) molecular tests (nucleic acid, RNA or PCR-polymerase chain reaction) [1], [2], [3]. Antigen tests can detect parts of SARS-CoV-2 proteins, known as antigens, via a nasopharyngeal or nasal swab sampling method [4]. The main advantages of POC-test include the high specificity, quick response (less than an hour) and portability, with no need of fixed laboratory facilities. On the other hand, in a molecular diagnostic test, a reverse transcriptase polymerase chain reaction (RT-PCR) is evolved, also known as nucleic acid amplification method [3], which requires expensive laboratory equipment. According to the chain reaction, virus RNA is converted to its DNA and then is amplified, using polymerase enzymes, by producing millions of DNA copies until they become detectable [3]. This process is characterized of high sensitivity and specificity, while the sample required can be taken from the saliva, or the respiratory system or the nasal cavity (nasopharyngeal or nasal swab sampling method). However, this method is time-consuming, and it requires several hours for the analysis οf the results [5].
Antibody or serology tests are searching for antibodies in human blood to determine if there were a past infection with the virus that causes COVID-19. Antibodies are proteins created by human body's immune system, soon after the infection or vaccination. These tests cannot be classified as diagnostic tools. However, they can be used to struggle the COVID-19 disease. It is suggested the antibody tests to be complementary used to the PCR ones, as their specificity for the SARS-CoV-2 has been reported higher than 99.5% and their sensitivity higher than 91%. Especially for patients exhibiting prolonged symptoms or for those who are asymptomatic, the antibody test can offer an extra appeasement of being cleared from the virus. However, the challenge that must be overcome is the unknown lasting period of immunity, as well as the degree of a body’s immunity [6]
Meanwhile, despite the great efforts of the scientific community towards the development of diagnostic tools and the achievement of high specificity and sensitivity of the molecular tests, the concern about the control and detection of the SARS-CoV-2 remains until the complete vaccination of the population will be achieved. This concern begins from the complexity the diagnostic tools present, as well as the time and the specialised personnel they demand, however the main roots are localized in the SARS-CoV-2 behaviour (e.g. continuous mutations - changes to new variants - like the S. African and the Indian ones etc.) and its rapid spread.
Today, the results of the as developed vaccines are visible, but most of the population has still not been vaccinated, so people must be repeatedly screened for COVID-19 disease. Even those proven not infected, taking negative results from molecular tests, it is possible the same day to be infected by the virus and so the test should be repeated, and many more tests are recommended. Moreover, according to the U.S. food and drug administration [7], the amplification process of the RT-PCR test should not exceed the forty cycles (one cycle contain the heating and cooling process to make copies of a specific region of DNA); even after so many cycles the available amount of genetic material is very low to be detectable. Thus, the difficulty to achieve the as called herd immunity the spread of the virus’ infection, the continuous mutations of the virus, the complexity of the diagnostic tools, create a vicious cycle that would be interrupted only when appropriate diagnostic tools and efficient therapy protocols will be developed.
The evolution of the electrochemical biosensors over the years and their involvement into medical diagnostic field, for diabetes [8], [9], [10], [11], [12], [13], Alzheimer [14], [15], [16] and other diseases, show that they could be a successful virus diagnostic tool of high sensitivity, specificity, low cost, quick response, requiring no special personnel and offering the advantage of the portability.
Recently, Ji et al. [17] reviewed the already developed detection techniques of SARS-CoV-2, confirming our abovementioned notification that the lack of the appropriate detection technology for patients with low load of virus and the necessity of early detection, pointing out the role the biosensors could play in struggling the virus. Furthermore, according the Mahshid et al. [18] survey, the electrochemical biosensors offer the alternative of using different analytes, thus detecting the virus at any stage of its evolution without worrying about its load. Among them, the electrochemical biosensors detecting viral nucleic acids exhibited the highest sensitivity. The limited results quality of commercially available kits (e.g. self-tests) and the need for early SARS-CoV-2 diagnosis through a simple process, rightly point out that electrochemical biosensors could offer a safer solution.
The advances of the electrochemical biosensors (EBs) and the importance of viruses electrochemical sensing were recently reviewed by Ruiz de Eguilaz et al. [19]. They reported that the research interest, concerning the development of these biosensors, focuses mainly on the sample preparation strategies, the point-of-care platforms and the target analyte, while the low sensitivity (compared to molecular tests) is the main challenge that remains to be overcome.
Furthermore, as reported by Khan et al. [20], the sensitivity and the lifetime of an electrochemical biosensor depend: i) on the method of virus immobilization on the electrode and ii) on the electrode’s material. Asif et al. [21] also recognized, among other biosensors, the important role of the electrochemical ones for the detection of acute respiratory syndromes, like SARS-CoV-2, highlighting the role nanoscience should play for the improvement of virus antigens selectivity and sensitivity. Srivastava et al. [22] discussed the significant role and the contribution of various developed nanomaterials for respiratory infections sensing, emphasizing on the different biosensing tools. They recognised that Au, lanthanide-doped polysterene nanoparticles, graphene and iron oxide nanoparticles-based biosensors that detect RNA or DNA and protein (antigen or antibody), are considered the main nanomaterials under investigation. In a more extensive review by Bukkitgar et al. [23], about electrochemical biosensors for viruses (including SARS-CoV-2) detection, suggested graphene nanomaterials as the most promising ones for such application. They specifically focused on the preparation method and the surface functionalization strategies that could help to eliminate the contamination, caused by other bioreceptors bound on them.
Moreover, as Kudr et al. [24] and Kotru et al. [25] highlighted that the electrochemical biosensors are of the most ambitious contestants in the fight against COVID-19 disease. According to Kotru et al. the use of the optimum biomarker can further enhance virus electrochemical sensing process, providing also a picture of the disease evolution. Considering the above works devoted to the kind of electrode, the analysis method, the kind of biomarker and the basic principle of analysis, one might conclude that they mainly offer a good qualitative comparison.
With the current review we aim to thoroughly summarize and discuss the materials recently investigated as electrode transducers for the development of SARS-CoV-2 electrochemical biosensors, emphasizing on their quantitative characteristics, such as sensitivity, lower limit of detection, specificity and response time. More precisely, we seek to examine the operating parameters of the electrode materials investigated for SARS-CoV-2 detection. In addition, this review deals with the electrochemical techniques being used for the SARS-CoV-2 electrochemical biosensors evaluation, which as we noticed, constitute a controversial subject in the scientific community.
1.1. Operational principles of biosensors
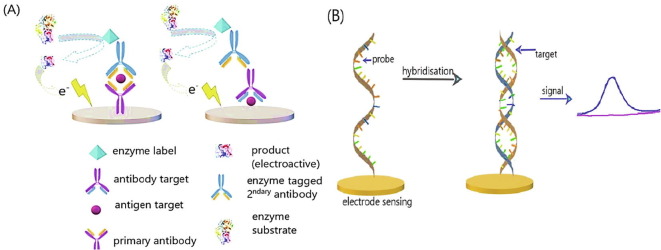
Generally, a typical sensor is a device used for measuring a physical quantity (input), which is converted into a signal (output, electrical or optical) that is then interpreted by the specialists. Accordingly, a biosensor consists of: i) a bioreceptor (a biomolecule, like enzyme or antibody or other) and its role is to capture the target analyte (e.g. a virus) and ii) a transducer, the role of which is to transform the signal produced from the biomolecule in electricity (current or potential) or other kind of signal depending on the type of transducer (Fig. 1Α). The recorded signal is usually amplified by a signal processing circuit and the interpreted results are displayed in a computer or a smartphone [26].
Fig. 1.
(Α) Operational principle of a biosensor; (B) Label-free and label-based detection process of biosensors; (C) Label-free aptamer detection process; (D) Antibody-aptamer sandwich type (a&b), aptamers pair sandwich type (c&d). Reproduced with permission.
Biological analytes, due to their intrinsic physical properties, are hard to be directly detected, so an element such as a radioactive or a fluorescent dye, nanoparticles or enzymes (known as label), can be either immobilized onto the bioreceptor surface (antibody) or onto the target molecule (Fig. 1 B). The main drawback of the labelling procedure is that it is time-consuming, as well as laborious and cost intensive. Furthermore, sometimes the labelling of biomolecules may block active binding sites, leading to the modification of binding properties attributed to the unknown degree of affinity between the target analyte and the bioreceptor [27], thus reducing biosensors sensitivity, selectivity and reproducibility.
On the contrary, label-free biosensors detect the analytes, utilizing intrinsic physical properties (electrical impedance, dielectric permittivity, or refractive index, etc.) of the analytes in their natural form (Fig. 1B). For example, when the electrical change is studied, the conduction or resistance or capacity is measured; in the case of mechanical change the mass and frequency changes are measured; in the case of optical elements the adsorption and emission of light are measured.
Label-free biosensing methods are rapid and of low cost, using small reaction volumes. Moreover, they can be integrated into lab-on-chip platforms, detecting in real time the concentration of the target analyte and reducing the adsorption of non-specific molecules [27]. Especially the new strategy in label free detection, using aptamers/oligonucleotides [28], offers the opportunity of analysing various target analytes, even those with small molecules, which otherwise - with the common label-free sensing method - would require more time and special attention. The aptamers, which are in vitro synthesized, usually form non-covalent bonds with the target analyte, increasing biosensor’s specificity and sensitivity towards a precise target. Generally, aptamers (usually a single-stranded DNA/RNA oligonucleotide sequence) are immobilised onto transducer solid surface (electrode transducer) and usually bind to the virus protein [29] (Fig. 1C). When the aptamer binds to the target-analyte acts as a barrier that increases the sensing surface resistance, preventing the electrons transfer to the electrode. The amount of the target sample is proportional to the charge resistance [30].
The main advantages of the aptamers are as follows: they can be chemically modified for increasing their stability and affinity towards the target analyte and can be synthesized in large amounts, when their nucleic sequence becomes known [31]. However, even though aptamers are considered as the next generation bioreceptors, the up-to-date commercialized ones do not use a single aptamer, since it presents instability as far as concerns the reproducibility and the signal generation. Thus, as an alternative, either the pair antibody-aptamer sandwich type [(Fig. 1D (a & b)] or the aptamers pair sandwich-type (aptamer1-target-aptamer2), were developed [(Fig. 1D (c & d)] [32].
From the four SARS-CoV-2 structural properties (spike, envelope, matrix and nucleocapsid), the spike protein (S1 and S2) is usually chosen as the domain receptor, since it constitutes a trans-membrane protein of the virus-cell with high immunogenicity, having the ability to induce immune responses and facilitating SARS detection [33]. Moreover, the spike protein has affinity with the angiotensin-converting enzyme 2 (hACE2), which is used as a receptor infecting human cells [34], [35]. Currently the aptamers, having affinity with the SARS-CoV-2 spike protein, were identified through the known Systematic Evolution of Ligands by Exponential enrichment system or “SELEX” system [36]. According to this process, a variety of polynucleotides (aptamer pool) are exposed to the target molecule and the aptamers with higher affinity are in vitro selected and then eluted, while the rest is disregarded (Fig. 2 A).
Fig. 2.
(A) SELEX procedure for aptamers production; (B) Direct (i) and indirect (ii) detection (enzyme-linked immunosorbent assay ELISA) [37]. Reproduced with permission.
This process is repeated many times until a small number of aptamers with high affinity and specificity against the desired target will remain.
Today, one of the SARS-CoV-2 diagnostic tools is based on an enzyme-linked immunosorbent assay (ELISA) biosensor. The operational key principle is based onto the target-analyte interaction with the enzyme-antibody complex, followed by an enzymatic reaction, which is responsible for the signal production. In Fig. 2(Bi and Bii) are depicted the direct and indirect detection processes, respectively. At the direct detection, the target-analyte is adsorbed directly on the microplates’ surface (Fig. 2Bi), with the risk that undesired proteins (albumin) will also be adsorbed. On the other hand, at the indirect detection process, the indirect adsorption (sandwich structure: antibody-analyte-antibody, seen in Fig. 2Bii), exhibits higher specificity as the adsorption is more selective. In both cases the analyte amount is specified via the colour change from the enzyme reaction [37].
1.2. Electrochemical biosensors
An electrochemical transducer is characterized of the lower cost, simpler construction, higher specificity, relatively lower sensitivity and portability. Their operational principle is based on the recording of the electrical signal, produced when the bioreceptor selectively reacts with the target-analyte (labelled or not labelled), which is proportional to the analyte’s concentration [38].
According to the identification process of the target-analyte, the electrochemical biosensors are classified as: i) biocatalytic and ii) affinity sensors. A biocatalytic electrochemical biosensor includes usually enzymes, the role of which is to identify the target-analyte producing electroactive species. On the other hand, the operation of an affinity electrochemical biosensor is based on the selective interaction between a bioreceptor (antibody, enzyme, nucleic acid or other receptor) and the consequent production of an electrical signal (Fig. 1A).
For the virus diagnosis, the affinity electrochemical biosensors are chosen. Usually the binding process includes antibody-antigen interaction (Fig. 3 A) or DNA hybridization (Fig. 3B). Immunosensors operation is based on the interaction between the antibody, immobilized on the sensing surface of the electrode, and the antigen of a specific analyte. When the binding process takes place, an electrical current is produced accordingly to the antigen concentration of target analyte, which then it is amplified and displayed [39]. The challenges of the immunosensors, however, led also to the development of bio recognition procedure, using DNA or RNA fragments [40]. It should be noted that among immunosensors, the electrochemical ones have been more perfectionized mainly because of their simplicity and, their ability to be portable, and for their in situ or automated detection.
Fig. 3.
(A) Antibody-antigen electrochemical immunosensors. (B) DNA-based electrochemical immunosensors. Reproduced with permission.
At the hybridization procedure, the single-stranded DNA segments, placed onto the surface of the sensing electrode (DNA probes), bind selectively with the complementary part of the target-analyte’s DNA (pair-base procedure), producing an electrical current/signal. Afterwards, the confirmation of a successful hybridization process occurs: i) using electroactive complexes (redox indicators), such as ferrocenyl naphthalene diimide that present different affinity towards a single stranded DNA, than to a double strand (successful hybridization) [41] or ii) using enzymatic labels, such as horseradish peroxidase; then from the occurring redox reactions the produced current is measured.
Lately, nanoparticles are investigated as electrochemical labels, because compared to enzymes own higher stability and conductivity, offering faster electron transfer from the immobilized target to the electrode. At the DNA hybridization procedure, in absence of labels, the guanine base is oxidized on the electrode’s surface, while the produced current can be amplified by the aid of redox mediators [e.g. Ru(bpy)3 +2] [42]. At this method, the success of the hybridization procedure is checked by measuring the changes of transducer’s capacitance or conductivity [43].
The shape as well as the size of the binding site define the degree of affinity, sensitivity and selectivity and it is critical for each kind of biosensor. The main challenge of immunosensors is the antibody (Ab) production for a specific virus antigen (Ag) to a host organism, which is a time consuming and costly process. The immobilization of the antibody on the solid surface of the electrode is another challenge to overcome. In case the antibodies are immobilised with wrong orientation they may lose their binding ability being unable to provide a highly sensitive biosensor. Moreover, in the case of DNA biosensors the orientation and the amount of the DNA probe are very important, confirming the significant role the electrode material plays. For this reason, the DNA molecules should be immobilized in a specific orientation to maintain stability and activity, and the other characteristics as well [44]. Moreover, in order to avoid the binding of undesired species, the surface of the electrode should be covered with blocking agents.
When the aptamer production is adopted, a series of DNA or RNA oligonucleotides (aptamers) are produced, which detect and bind selectively to a three-dimensional surface, such as the one of proteins (Fig. 1C). The sequences of the aptamers are produced via a polymerase chain reaction (PCR).
Therefore, for achieving the optimal characteristics for biosensors operation, the choice of electrode material along with its treatment procedure is of great importance. Notably, the immobilization procedure has been improved by far due to the development of nanomaterials [45] and their use in electrochemical immunosensors. For this reason, the choice and the treatment of the electrode’s material of the up-to-date developed SARS-Co-V2 electrochemical biosensors are thoroughly discussed in the present review.
1.3. Electrochemical detection techniques
Today, most of the biosensors are fabricated using electrochemical transducers, as they are simple to be constructed, easy to use, require small reaction volumes and the measurement is not affected by foreign interferences, such as red-blood cells, etc. The principle of operation of an electrochemical transducer is based on the measurement either of current, or charge accumulation or alteration of the electrode conduction, caused by a “reaction”. Therefore, based on the measured property, the electrochemical detection techniques can be classified into: i) amperometric or voltametric (current is measured), ii) impedance biosensors (conduction or resistance is measured) and iii) potentiometric ones (potential is measured) [46], [47].
1.3.1. Amperometric or voltametric-based detection
By using the amperometric detection method, a constant potential is applied at the working electrode (vs the reference electrode) and an electrochemical oxidation or reduction takes place, delivering a current. The as produced current (measured vs a counter electrode) is proportional to the concentration of target analyte into the sample [48]. When the measurement is conducted applying the rotating disk electrode technique [49], the mass transport rate of the analyte onto the working electrode surface can be enhanced.
This method is mainly adopted for both the biocatalytic and affinity biosensors as they own the advantage of very low detection limit and very high selectivity towards target analyte. Moreover, the applying of an exact potential, which causes the electrochemical oxidation or reduction reaction, is a time-consuming detection procedure [50].
On the other hand, according to the voltametric detection method, the working electrode is imposed to work under a potential range (vs a reference electrode) into which an electrochemical oxidation or reduction reaction occurs, and the produced current is measured (vs a counter electrode). The current response is proportional to the analyte concentration when applying the techniques of differential pulse voltammetry, cyclic voltammetry, square-wave voltammetry and linear sweep voltammetry [50]. Nowadays, aptamer based biosensors [51], which are considered as the next generation biosensors, tend to adopt the voltammetric sensing techniques, exploiting the robustness and the very low detection limits they present even with small amounts of analyte volume.
1.3.2. Electrochemical impedance spectroscopy (EIS)-based detection (or impedimetric detection)
According to the electrochemical impedance technique, a small AC excitation signal (2–10) mV under a specific frequency (Hz) range, is emitted to the electrode under investigation, and by measuring the in and out-phase current response, the parameters of electrode's resistance and conductivity are estimated. [52]. At high frequency values the electron transfer can be estimated, while the mass transfer is estimated at low frequency values.
EIS is primarily used for the affinity evaluation of label-free electrochemical immunosensors for the direct control of the antibody-antigen binding process, taking place onto the electrode’s surface.
Indicatively, before the measurement the surface of the working electrode is covered (using electrodeposition method) with a highly conductive polymeric film, where the bioreceptors are incorporated too. The detection process takes place by applying a known potential, which causes a current flux and consequently an electron transfer. The electron transfer resistances, at the electrode and at the bulk of analyte interface, change as the target analyte binds onto the bioreceptor. The resistances change, measured with EIS, is proportional to the antigen concentration. In this way the formation of the antibody-antigen bonding layer can be appropriately monitored, without the necessity of using labels [53]. Nevertheless, the main disadvantage of this type of detection is the low regeneration possibility of the working electrode.
The lower cost, the quicker response of assays, and the higher signal to noise ratio, are some of the main advantages of the electrochemical impedance detection. Nevertheless, the main disadvantage of this type of detection is the low regeneration ability of the working electrode [54].
1.3.3. Potentiometric detection
According to the potentiometric method the sensor is consisted of two reference electrodes that are fixed onto a layer, whose potential change is measured when it reacts selectively with the charged ion of the target analyte. The charged ion produced by the aid of an enzyme, which catalyzes a specific reaction, causes the potential difference the membrane presents before and after the charge ion is produced.
The field effect transistors-based biosensors [55] belong to this category; consisted of a biological recognition compartment and a field-effect transistor (semi-conducting material). Between the analyte solution and the semiconductor there is an insulating (chemically and electrically) layer that is usually a polymer onto which the bioreceptors are placed. When the binding procedure (bioreceptor-target analyte) becomes successful, the surface of the electrolyte-insulator layer electrostatic potential changes is recorded, which accordingly cause an electrostatic gating effect to the semiconductor as well as a current difference between the source and the drain electrodes that is also recorded.
1.4. Efficiency characteristics of electrochemical biosensors
The efficiency of an electrochemical biosensor is determined by its: i) accuracy, ii) response time, iii) selectivity, iv) repeatability, v) reproducibility, vi) detection limit, vii) sensitivity, viii) recovery time and ix) linear range [38], characteristics that are explained in the Table 1 .
Table 1.
Determination factors of the efficiency [38].
| Efficiency of an electrochemical biosensor | |
|---|---|
| Accuracy | The amount of uncertainty in a measurement, compared to a reference. The accuracy is important in order to evaluate the correct quantity (usually molarity) of the target-analyte. |
| Response time | The time interval between the moment the analyte gets into contact with the bioreceptor and the moment the electric signal is received. A quick response time is preferable if the sensor is used as a device of point-of-care. |
| Selectivity | The ability of a sensor to preferably analyze the desired analyte in presence of other substances. The high selectivity ensures higher lifetime and accuracy of a sensor. |
| Repeatability | It ensures that an electrochemical sensor will give the same output for over a period. It represents a factor along with the reproducibility that determines a sensor’s reliability. |
| Reproducibility | The ability of an electrochemical sensor to give the same signals under varied measurement conditions. |
| Detection limit | The lowest detection limit is the lowest concentration of the target-analyte that can be tracked with accuracy. |
| Sensitivity | The lowest difference identified by a sensor, after the change of analyte’s concentration. A sensor with very high sensitivity can even detect very small changes of analyte concentration. |
| Recovery time | The minimum time between two successive measurements. Before the second measurement starts, the sensor should return into its initial condition. |
| Linear range | The electric signal received from the sensor should be linearly dependenton the analyte concentration. So, the minimum and maximum concentration that could be analysed by a sensor, are determined. |
2. Emerging transducer electrode materials for viruses’ electrochemical detection
The development of nanotechnology has helped the growth of biotechnology, allowing emerged new possibilities that have greatly improved the efficiency features of biosensors [56]. The adoption of nanomaterials for the transducer electrode structure of electrochemical biosensors enhanced their sensitivity, selectivity, response time, making them more accessible for virus detection.
Specifically, as many researchers support, the transducer electrode, in which an aptamer or an antibody combines with a nanomaterial (hybrid material), transmits a higher signal, and presents increased sensitivity and affinity towards the target analyte, especially when a nanomaterial is used as label [57]. Among the examined nanomaterials, gold nanoparticles [58], [59] are mostly investigated as they are biocompatible, with excellent electrical and optical properties. Moreover, gold nanoparticles improve the immobilization of the bioreceptors onto the electrode transducer, providing a larger surface area, which allow the immobilization of higher number of bioreceptors [57]. Therefore, today the scientists pay more attention to further modify the gold nanoparticles either by exploring different supports, such as multiwall carbon nanotubes, graphene, etc, or adopting other preparation strategies.
In the next sub-section, we discuss indicatively some interesting methods applied the last five years for the fabrication of nanomaterial-based electrochemical MERS-CoV, HCoV and SARS-CoV biosensors. Our attention then is focused to the respective electrochemical biosensors developed for the SARS-CoV-2; we analytically discuss the use of nanomaterials for the fabrication of electrodes and their effect onto biosensor characteristics.
2.1. Electrochemical biosensors for the detection of MERS-CoV, HCoV and SARS-CoV viruses
The use of Au surfaces as electrodes in electrochemical biosensors is not new. On 2005 Abad-Valle et al. [60] developed for the first time a very thin sputtered Au-nanofilm (100 nm thickness) onto a polyimide substrate, as transducer-electrode in a DNA hybridization assay for the SARS virus electrochemical detection. The thickness of the as-prepared electrode allowed researchers to use small reaction volumes, enabling the simultaneous hybridization and sequential detection offering more than twenty assay sites. During the detection, the complementary strand of a specific SARS-DNA sequence was labeled with thiol group and immobilized on the Au surface.
The electrochemical detection is amplified with alkaline phosphatase-labelled streptavidin; while for the expulsion of the undesired molecules from electrode surface, albumin and 1-hexanethiol are tested as blocking agent, as depicted in Fig. 4 A. Moreover, the operational parameters of the biosensor were evaluated, using the following electrochemical methods: i) cyclic voltammetry, ii) differential pulse voltammetry, iii) alternating current and iv) square wave voltammetry. It is remarkable that the lowest detection limit of the biosensor was found to be 6 pM (pM = 10−12 M) [60].
Fig. 4.
(A) Comparison of LSV measurements in 1 μM DNA solution for albumin (a) and 1-hexanethiol (b) blocking agents on electrode surface [60]. (B) The DNA of SARS genome (oligonucleotides sequence) bound on the modified electrode, producing current [61]. (Ci) Ag reduction current recorded by CV. (Cii) Ag ions, contained in the solution, are reduced into metallic silver (Ag) [62]. CVs of methylene blue covalently attached to a single-stranded DNA (Di) and methylene blue molecule (Dii) on bare (dotted line) and nanostructured screen-printed electrodes (solid line) [65]. (Ε) Comparison of CV response between the dsDNA (blue), dsDNA/rAzu (black), and PSD/rAzu (red). In presence of Ag + ions and oligonucleotides molecules (C = 0.5 pM (10−12 M), rAzu: 0.1 × 10−3g/mL) [66]. Reproduced with permission.
Later, in 2007, De la Escosura-Muñiz et al. [61], developed a DNA hybridization electrochemical genosensor for SARS detection that exhibited a lower limit of detection at 15 fM (fM = 10−15 M). The deposition of electroactive Au label complexes onto the modified (with DNA-probe) glassy carbon electrode allowed the electrochemical detection through the Ag electro-reduction reaction, which was catalyzed by the electrodeposited Au complex. As shown in Fig. 4B, the greater is the degree of DNA binding, the greater the amount of the electrodeposited Au on the glassy carbon and thus the greater the Ag reduction current recorded will be. Over the years, it is observed that the Au in combination with the electroactive species instead of enzymes play an important role for the electrochemical genosensors evolvement. However, despite the lower limit of detection that was achieved, there are also several processes that should be further optimized. One of them is the labeling process, which is time consuming, and the process of the binding verification between the metal and the oligonucleotide that should be done through the label (e.g. thiol group).
Moreover, in 2009 Martinez-Paredes et al. [62] suggested DNA hybridization over Au nanoparticles supported onto carbon screen-printed transducer electrodes for SARS virus electrochemical genosensors fabrication. Thus, for the first time Au nanoparticles were investigated along with small reaction volumes. The Au-nanoparticles in combination with the use of screen-printed electrodes, led to reduced interaction time between Au and thiol (label), high sensitivity (1.76 mA/pM) and relatively low detection limit (2.5 pM). At the same time, the Au-nanoparticles managed to retain their electrocatalytic activity, to immobilize the analyte molecules and to amplify the DNA detection ability. This threefold function led to less analysis time and improved stability of the biosensor.
The evaluation of the as-fabricated biosensor is achieved by using cyclic voltammetry (Fig. 4Ci), estimating the produced current from the electrochemical oxidation of the deposited metallic Ag, which acted as electroactive species. As seen in Fig. 4Cii, before the detection procedure, the silver ions contained in the solution are reduced to metallic silver (Ag) and deposited next to the enzymatic label AP (alkaline phosphatase), using a reducing agent produced by the 3-indoxyl phosphate substrate. The amount of the deposited Ag was measured, using anodic stripping voltammetry.
Thus, for many scientists the question about the effect of the amount of Ag and other electroactive species arises, as well as of the amount of the immobilized elements, on the efficiency of the biosensor. The answer was provided by Layqah and Eissa [63], who also investigated the maximum concentration of antibodies that could be accepted on Au-nanoparticles and the binding-time required. It should be mentioned that the immunosensor performance is affected by both parameters, in terms of limits of detection and response time.
Initially when the science of nanotechnology was introduced into the biosensors field, managed to make the analysis simpler, quicker, with higher sensitivity, including many other desirable characteristics. However, point-of-care diagnosis, with a real sample, without the need of an electrochemical cell, was a real challenge that should be overcome. So, the robustness for on-site analysis was emerged with the appearance of screen-printed carbon electrodes (SPCEs) combined with nanotechnology. SPCEs was and still remains a leading technology, mainly due to the attractive characteristics offered by carbon material, such as low cost, chemical inertness, low background currents and good conductivity under specific conditions [64].
One of the first carbon material investigated from the scientific community were screen-printed carbon electrodes for the DNA hybridization, during the DNA electrochemical detection. In the case of an ideal immobilization, the probes are oriented towards the immobilized layers, being ready to facilitate the detection procedure.
So, since hybridization process is based on the immobilization of a single stranded DNA (ssDNA) and on the complementary binding of the DNA sequence on the sensing carbon electrode, owning a surface area that can be easily modified, (for example with metal nanoparticles [61], or polymers or other nanomaterials) and thus consisting a very attractive option all over the years. In addition, the subsequent appearance of carbon nanotubes (CNT) and their chemical, mechanical and unique electrical properties, have motivated the scientific community to explore their application in the field of electrochemical biosensors, modifying with them the SPCEs surface.
García-González [65] reported for the first time the fabrication of an electrochemical DNA biosensor based on carbon nanotubes (CNTs) modified Au-SPCEs (commercial) and methylene blue labelled DNA, aiming at the SARS-CoV detection. The use of the CNTs enables the methylene blue utilization as an electrochemical label that is covalently attached to a single-stranded DNA, thus facilitating the higher electron transfer, as seen in Fig. 4Di, compared to methylene blue molecule without DNA-attachment (Fig. 4Dii).
More specifically, the chemistry of the CNTs surface allows the adsorption of such molecules, which in their turn amplify the analytical signal. Following this strategy, it has been managed a seven times higher sensitivity, fact that led to use differential pulse voltammetry and square wave voltammetry, which are very sensitive electrochemical detection techniques and usually applied for the measurement of very low current values. However, comparable to Au transducer electrodes, the sensitivity (0.2864 μA μM−1) was not very high, and the limit of detection remained at the nanoscale (800 nM), indicating that further research is necessary.
Recently, for the simultaneous detection of various CoV viruses, [63]. Au nanoparticles electrodeposited on carbon disposable-microarray transducer electrodes (eight in number), were used for the first time. The electrodeposition procedure was performed using cyclic voltammetry at a specific negative potential range. After the Au-electrodeposition, human corona virus (HCoV) or MERS-CoV antigens (recombinant spike protein S1) were immobilized on Au via a simple and time-consuming incubation procedure. According to the optimal operating conditions of the immunosensor for the successful detection, approximately 20 min of analyte sample incubation was required to take place and a specific concentration of antibodies (~10 μg L−1) should be included in the sample.
However, the main novelty of the as-proposed biosensor [61] derives from the way in which the advantages of the combination between nanotechnology and electrochemical science were exploited. Specifically, by using four electrodes for MERS-CoV detection and two electrodes for HCoV detection, duplicate measurements for each sample were achieved.
The square wave voltammetry technique and ferro/ferricyanide redox-couple system (electroactive substance) were employed [63] in order to evaluate both the antigen binding onto the working electrodes (after being modified with serum albumin, BSA - bovine serum albumin - that prevents undesired molecules to bound onto the electrode) and the investigation of the novel immunosensors operational parameters. When the peak reduction current (derived from the redox reactions) is decreased, the antibody is successfully bound to the immobilized antigen (spike protein S1), blocking the access of the ferro/ferricyanide redox couple to the working electrode. Thus, even after many years of research the Au nanoparticles-based electrochemical biosensors, which are considered better devices as compared to the other types of biosensors, present some limitations, such as the lower limit of detection that depends on the amount of the antigen (consequently the virus antibody) immobilized on the nanomaterials [57], and few others.
Understanding these limitations, few research groups [66], [67], [68], [69] adopted metalloproteins making labels or other linkers unnecessary. Namely, Mohammadniaei et al. [66] immobilized the recombinant azurin protein (r-Azu) on a Au-electrode. The protein had a cross-sectional diameter (the smallest circle through which a molecule can pass) and thus, the as-fabricated electrode acted as an arrayed molecule with selectivity towards the analyte molecule, receiving only one DNA strand [at its N-terminus: the free amine group (–NH2) located at the end of a polypeptide or protein], for the development of electrochemical biosensors for the detection of MERS-CoV and other viruses.
The researchers in order to reduce the possibility of foreign molecules to be anchored onto the Au-electrode, fabricated a series of azurin- single stranded DNA-parallel two double stranded DNA (like a chain), which was repeated all over the Au-surface. This unique strategy of the adoption of two double stranded DNA in a parallel arrangement gave the opportunity for a higher specificity detection of small nucleic acids, such as miRNA or virus DNA. Moreover, at the plasma pH (7.4), the azurin caused a negative charge of the electrode surface, which was responsible for the vertical arrangement of the DNA strands onto the electrode. This vertical position of the DNA strand helped to eliminate the lateral adsorptions of it [66].
The as-suggested electrode was evaluated by measuring the electrical conductivity change, using Ag ion (Ag+) reduction, which played the role of the charge carrier in the case of mismatch between pairs or virus detection. For the Ag+ reduction reaction the cyclic voltammetry technique was chosen. By scanning at negative potential values (reduction), the charge is transferred from the Au electrode to azurin (copper protein), then to DNA that acts as a charge mediator and finally to the Ag+, which is reduced [66], as seen in Fig. 4E. The formation of distinct peak current curves clearly defines that the Ag+ can sufficiently play the role of the redox element that accepts electrons in free-label DNA electrochemical biosensors. Despite the main advantage of the as-suggested biosensor, the optimum detection time was one hour, indicating that further research should be done to reduce the response time.
The above-mentioned limitations, such as the concentration of antibodies, the need for higher sensitivity, the binding of undesired species and the low reproducibility of an electrochemical biosensor, require a more mannered treatment (or functionalization) of the surface of nano catalyst or the exploitation of other nanomaterials than gold nanoparticles to be further investigated.
2.2. Electrochemical biosensors for the detection of SARS-CoV-2
Facing the need of developing tools for rapid SARS-CoV-2 detection, many research groups responded immediately by fabricating and studying a few novel electrochemical platforms based mostly on Au and carbon or graphene nanomaterials. In the current section the most relevant works published in international literature are discussed in terms of the used electrode transducer materials and electrochemical detection techniques. The response time, the lower limit of detection and the electrochemical techniques of biosensor evaluation characteristics are reported and discussed in terms of electrode materials choice.
2.2.1. Au-based SARS-CoV-2 electrochemical biosensors
Table 2 lists the Au-based SARS-CoV-2 electrochemical biosensors, their response time, the used electrochemical techniques for each biosensor evaluation characteristics and their limit of detection. As above-mentioned, antibodies-sensors can play a significant role mainly in identifying the prevalence of a disease, which in turn could help the doctors to decide the best treatment and the scientists to understand the kinetics (evolution) of a virus over time and so providing them more information for developing the appropriate vaccines.
Table 2.
Au-based transducer electrode for the SARS-CoV-2 electrochemical detection.
| Material | Electrochemical detection technique | Target analyte Immobilized element | Response time | Limit of detection |
|---|---|---|---|---|
| Au-electrodes on commercial polyethylene terephthalate (PET) platform [70] | Electrochemical impedance spectroscopy (EIS) | SARS-CoV-2 monoclonal antibody CR3022/SARS-CoV-2 spike RBD protein | <5 min | – |
| Au-based onto Si-based transducer [71] | Square-wave voltammetry (SWV) | SARS-CoV-2 cDNA/SARS-CoV-2 antigen | – | Artificial target:0.02 pg |
| Au onto Ti substrate [72] | Amperometry | viral RNA or cDNA/complementary thiolated probes | – | – |
| Eight-channel Au screen-printed electrode assembly [73] | Chrono-potentiometry | SARS-CoV-2 spike S1 protein/SARS-CoV-2 Spike S1 antibody | 3 min | Artificial target:1 pg/L |
| Au nanoparticles/RGO [74] | Electrochemical impedance spectroscopy (EIS) | SARS‐CoV‐2 spike S1 protein and RDB antibodies/SARS-CoV-2 spike S1 & RBD antigens | 11.5sec | Artificial targets: 1) 1 pM for spike S1 protein 2) 0.001 pM for RBD (receptor-binding-domain) antibodies |
| Au@Fe3O4 onto reduced graphene oxide (RGO)/Screen-printed carbon electrode [75] | 1) Differential pulse voltammetry (DPV) | SARS-CoV-2 RNA (without amplification)/SARS-CoV-2 capture probe (CP) label probe (LP), and auxiliary probe (AP) | <10 sec | Artificial target: 3aM = 3 × 10−6 pM Clinical sample: 2 × 102copies/mL |
Au-based electrodes placed on a commercial sensing polyethylene terephthalate (PET) substrate platform (Fig. 5 Ai) were adopted by Rashed et al. [70] in order to develop a capacitive immunosensor for SARS-CoV-2 antibodies detection. Gold (Au) is a safe choice, as among the electrochemical biosensors it is the most investigated material, being explored in combination with various substrate platforms (screen printed electrodes, polymer base platforms, etc). However, the properties of the substrate material affect the efficiency of sensors, while many concerns have been expressed about the reproducibility of the deposition of the sensing layer onto the substrate surface [76].
Fig. 5.
(Ai) A schematic illustration of a label-free platform, able to immobilize COVID-19 antibodies on the Au electrodes, coated with RBD SARS-CoV-2 spike-protein; (Aii) Change of the electrical double layer resistance between the Au surface and the RBD bound on it, when antibodies are detected (samples A,B are infected with the virus, while sample C is not) [70]; (Bi) Au-based SARS-CoV-2 sensor in one chip with three operating modes; (Bii) Comparison of electroanalytical signals between SARS-CoV-2 (2003) cDNA and SARS-CoV2-cDNA [71]; (C) Electrodeposited Au nanoparticles, where thiol is immobilized/self-assembled and operated as probe for bound the RNA/c-RNA of SARS-CoV-2 [72]; (Di, Dii) Change in electric properties of a cell-membrane, when the S1 protein (antigen) of SARS-CoV-2 is bound with the immobilized antibody on the Au electrode surface (screen-printed electrode) [73]. Reproduced with permission.
The PET platform produced by Rashed et al. [70] was label-free and able to immobilize COVID-19 antibodies onto the Au electrodes coated with the RBD (receptor binding domain) SARS-CoV-2 spike protein. The detection tests were implemented using electrochemical impedance spectroscopy (EIS), by measuring the changes of electrode’s conductivity. More precisely, the interface between the Au surface and the RBD SARS-CoV-2 spike protein along with the bound antibodies form an electrical double layer, as depicted in Fig. 5Ai, whose resistance increases when in the analysis sample anti-SARS-CoV-2 monoclonal antibodies are present. According to the reported results [70], it was observed that the thickness of the electrical double layer, its shape and therefore the measured conductivity, depend on the binding events (probe-target binding) and the time-range they occur. The binding events depend on the concentration of virus’ antibodies in the analysis sample, as well as on the probe's concentration on the electrode surface; both parameters affect the frequency under which the EIS measurement is performed. For example, they analyzed three samples in a specific frequency. Two of them (samples A and B) contained different concentrations of the analysis target (SARS-CoV-2 antibodies), while the third one (sample C) contained no analysis target. As seen in Fig. 5Aii, when the binding event occurs, a conduction peak is instantly recorded (in the initial 5–10 s), while in ~ 30 s the conductivity is rapidly reduced. This change of the electrical behavior is attributed to the association and dissociation event of the antibody/target (binding event) that alters the whole conductivity. However, one might wonder to what extent the EIS technique could be sufficiently employed, at the same operational parameters (frequency and mV), under higher or lower concentrations of the under-detection target. It can be concluded that as the process evolves, i.e. as the experimental conditions change (when the antibodies of the sample are reduced), the EIS detection sensitivity is reduced leading to false conclusions. Thus, the optimum parameters of this electrochemical biosensor type should be defined each time.
Similarly, Bajo et al. [71] exploiting the special properties of Si substrate, developed an Au-based SARS-CoV-2 sensor in one chip having three operating modes: i) an electrical heater, ii) a temperature sensor and iii) an electrochemical sensor for detecting the virus targeted nucleic acids (NAs) (Fig. 5Bi). The NAs detection was achieved having methylene blue electroactive mediator, while the sample of interest could be analyzed either under a specific temperature or under a cyclic operation between different temperatures (with a precision of ±1.3 °C). Non-covalent methylene blue (MB+) constitutes a commonly used intercalative redox indicator that is interposed into the DNA base stack [77]. As discussed in section two, the intercalators (or redox-active reporters) are substances that form intermolecular (non-covalent) bonds with DNA, and they are used in the time-consuming chemical cross-linking steps [77] as they are able to be reduced and oxidized, producing electric current when the target of interest bounds to the electrode’s probe [78].
Moreover, Bajo et al. [71], amplified the DNA (repeated copying of a piece of DNA) and measured in real-time the concentration of amplicons (one or more copies of a genetic fragment of DNA), using square wave voltammetry (SWV), performing an indirect measurement of the produced current. When new amplicons are produced during amplification, the methylene blue (MB) is interacting with them, without interacting with the working electrode, therefore the measured current is originated from the redox reactions between MB and amplicons. In order to evaluate the produced electroanalytical signal, for comparison reasons, fragments from another coronavirus (SARS-CoV (2003) cDNA) were used. As shown in Fig. 5 Bii, after 35 PCR cycles the electroanalytical signals for the two viruses differ by about four times, indicating the ability of the sensor to separate the two viruses. For the designed sensor, a LOD of 20 fg (fg = 10−15g) (or 0.02 pg (pg = 10−12 g)) was calculated, in a solution containing 50–150 nM of SARS-CoV-2 cDNA (synthetic).
Square wave voltammetry (SWV) is such a sensitive pulse-voltammetry technique the analysis results of which can be compared with those of spectroscopic and chromatographic techniques. Based on its operational principle, the produced current varies according to both the potential step and the duration of potential application [79]. Moreover, similarly to the cyclic voltammetry technique, when the sweep rate increases the recorded current increases too, with the difference that the peak potential increases accordingly instead of remaining constant, making the SWV more popular among sensitive medical diagnostic analysis.
Furthermore, compared to the other electrochemical detection techniques, SWV requires higher frequency values, thus reducing the analysis time. However, similarly to the EIS technique, the SWV measurement should be optimized in terms of applied frequency, voltage step, amplitude and target analyte concentration. Therefore, in the future the modification of SWV operational parameters under various target analysis concentrations should be examined.
Following the safe choice offered by Au nanoparticles, Tripathy et al. [72] electrodeposited them onto a Ti substrate and suggested the development of an electrochemical label-free COVID-19 transducer. In the last years, it was observed the adoption of titanium substrate as surface support for metallic nanoparticles, and its application as transducer electrode for electrochemical biosensors, due to its reasonable cost, improved mechanical stability and its good conductivity [80].
As depicted in Fig. 5C, after the Au electrodeposition thiol (probe), as a self-assembled element, is immobilized onto the Au nanoparticles. The rest of the surface is covered with a blocking agent in order to block the binding of undesired molecules on the sensing electrode. When the target DNA binds to the probe, the hybridization procedure modifies the charge of the sensing surface. The produced current, which according to the authors can be measured with the amperometry technique, is proportional to the concentration of the bound-target, for a specific probe density bound on the electrode [72]. However, further research about the response time, lower limit of detection and other efficiency characteristics should be explored for the above sensor.
A novel approach based on the combination of the membrane engineering field with the electrochemical analysis field, was proposed by Mavrikou et al. [73] in order to electrochemically detect SARS-CoV-2 S1 protein. They electrodeposited SARS-CoV-2 Spike S1 antibody into kidney cells, as depicted in Fig. 5Di, which then are immobilized onto Au-screen printed carbon electrodes (commercial). As shown in Fig. 5Dii, when the S1 protein (antigen) of SARS-CoV-2 bound onto the immobilized antibody, the potential of the cell-membrane is altered and appropriately measured. For a quick reading of the results, the novel biosensor can also be connected to a smartphone or a tablet. More precisely, the authors [73] achieved a lower detection limit of 1 fg/mL (fg/mL = 10−15 g/mL) and a good selectivity for SARS-CoV-2 antigen, when other virus-associated proteins were also present. Specifically, a semi-linear behavior was observed in the range between 10 fg/mL and 1 μg/mL. However, at concentrations higher than 10 fg/mL the sensor response is decreased. This means that maybe more cells is necessary to be bound onto the Au surface in order to facilitate more targets or independently target-molecules competing each other for the binding sites. The main advantage of this biosensor [73] was the ability to detect the virus antigen within 3 min, without need any prior sample processing. However, the main limitation of the as-suggested electrochemical detection method is the necessity for the cell culture, using special equipment and staff. Moreover, the factors of cost and stability of the cell culture over time are some questions that arise and should be answered with a future research.
Au nanoparticles supported onto reduced graphene oxide (r-GO) were explored by some groups as transducer electrode material. Currently, graphene and its derivatives (graphene oxide or reduced graphene oxide) are considered among the most promising materials for the development of biosensors, not only because of their high conductivity, but also of their very good biocompatibility with the human body [81], [82]. However, graphene sheets own strong π-π and hydrogen interactions that cause its aggregation [83], [84]. Before being applied into a biosensor, the modification of the structure or the surface, as well as the functionalization of graphene is necessary. When water-soluble macrocycles or cyclic oligomers are used as functional groups, a new material is obtained, owing to the properties of graphene (large surface area, conductivity, etc.) and the functional groups (very high molecular recognition and ability) [85].
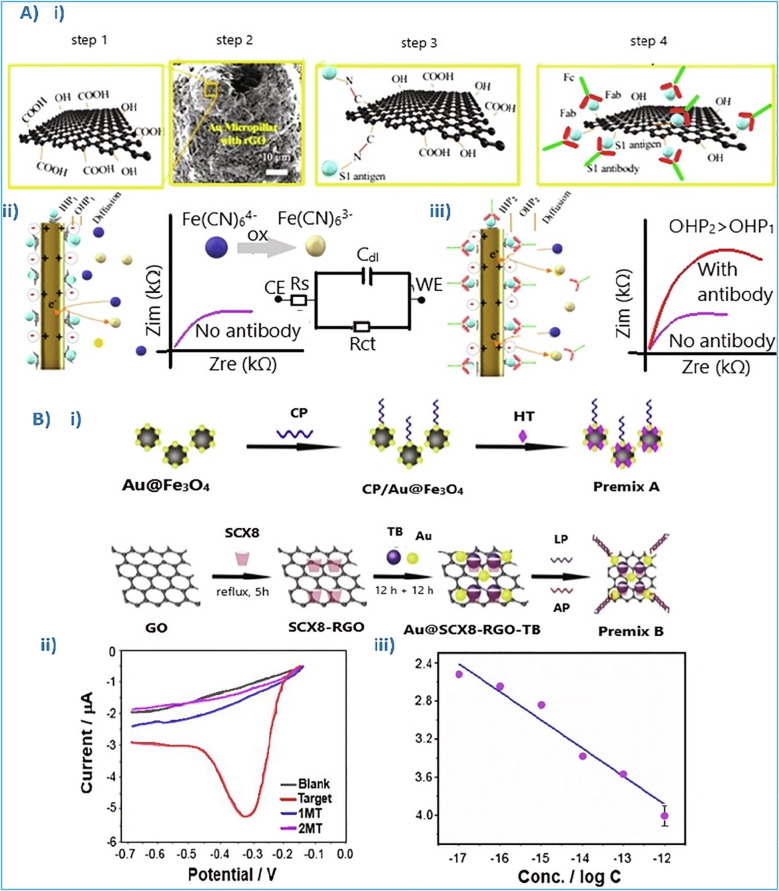
Very recently Ali et al. [74], exploiting the novel aerosol jet 3D-printing method [86], [87], [88] along with the r-GO properties, fabricated 3D Au pillars, being decorated with r-Go nanoflakes (Fig. 6 Ai) and tested as transducer electrode in a novel SARS-CoV-2 capacitive sensor. The functional carboxylate groups of r-GO structure (–COOH and –OH) combined with the structural properties of Au, offer more binding sites for viral antigens, using EDC (1-ethyl-3- (-3-dimethylaminopropyl) carbodiimide hydrochloride) and DCC (N’, N’, Dicyclohexylcarbodiimide) carbodiimide compounds, as schematically depicted in Fig. 6Ai. Thus, increase the as-suggested sensor sensitivity, allowing higher redox current (from the electroactive species). Moreover, the adjusted r-GO to the 3D Au pillars formed a secondary 3D network, facilitating bind of a greater number of antigens and therefore showing higher sensitivity.
Fig. 6.
(Ai) Functionalized r-GO sheets with carboxylated groups, using a drop-casting process, enriching the r-GO surface with –COOH and –OH groups. (Aii) Schematic representation of the electrical double layer (consisted of Helmholtz planes: OHP & IHP), created between the electrode and the electrolyte. (Aiii) After the targeted antibodies bound with electrode’s antigens, the charge capacitance increases with the increase of the double layer width [74]. (Bi) Schematic illustration of the Au nanoparticles treatment. (Bii) Differential pulse voltammetry (DPV) measurements of the as prepared electrode: i) in blank solution (black), ii) in 10−12 M of the target (red), iii) in 10−12 M of 2 different mismatch artificial targets (MT) (blue and purple). (Biii) Linear relationship (current vs logCanalyte), ranging from 10−17 to 10−12 M [75]. Reproduced with permission.
The response time of the biosensor suggested by Ali et al. [74] was only 11.5 sec. In the case of phosphate-buffered saline (PBS), the novel biosensor was able to detect the SARS-CoV-2 protein at concentrations close to 1 fg/mL (fg/mL = 10−15 g/mL), while in the case of clinical transport media, the limit increased at 100 fg/mL. Moreover, the produced signal is possible to be recorded by a smartphone-based user interface. The biosensor successfully detected SARS-CoV-2, with a LOD of 16 pfu/mL in culture media and of 2.42 × 102 copies/mL in clinical samples. The pfu is the plaque-forming unit used in virology to describe the number of virus particles able of forming plaques/colonies, while the copies/mL is the measure of the virus amount in a blood sample. In addition, after eluting the antibody-antigen binding, the as-suggested platform could be used for ten successive screenings, presenting accuracy and reproducibility among the measurements.
The technique of electrochemical impedance spectroscopy was used for the evaluation of the as-fabricated sensor. More specifically, when the analysis sample does not contain any virus molecule, an electrical double layer (Cdl) between the electrode and the electrolyte (buffer saline solution with ferro/ferricyanide) is created, which is consisted of an inner and an outer Helmholtz plane (IHP &OHP). However, when a virus is present in the test sample, by applying a specific potential the targeted antibodies combine with the antigens already bound onto the r-GO nanoflakes, increasing the double layer thickness, thus changing its capacitance, as presented in Fig. 6(Aii & Aiii). The binding event is validated by the current produced from the electrochemical oxidation and the ferro/ferricyanide reaction [74].
Examining how the target concentration affect the impedance signal, the authors [74] concluded that when the binding sites were all occupied the impedance signal was not increased, even if the target analyte concentration increased. This means that there is an impedance signal saturation. This arises questions like whether such sensors would extract real quantitative results when the target’s concentration is higher than the saturation value. In this case, the high concentrations might cause competitive adsorption phenomena between the antibodies on the surface of the antigens and maybe the diffusion phenomena will prevail, causing false charge measurements [54]. All the above parameters affect the behavior of electrochemical biosensor and require further research efforts, when the validation of the stability of such biosensor under various environments is desired.
Zhao et al. [75], exploiting the unique properties of r-GO, designed a portable electrochemical biosensor for the detection of SARS-CoV-2 RNA. More precisely, they functionalized the r-GO surface with p-sulfocalix[8]arene (SCX8) onto which they deposited Au@Fe3O4 nanoparticles. Label probes and auxiliary probes were also adopted.
Αs schematically seen in Fig. 6Bi, before the deposition of Au nanoparticles on the modified r-GO, the deposition of toluidine blue (TB) is preceded. The TB is an electrochemical mediator, being selected because of its high affinity with the SCX8 functional material of the r-GO. The inclusion of SCX8 in the r-GO structure enhances its sensing ability. Among the materials developed to be applied as transducer electrodes for SARS-CoV-2 electrochemical biosensors (Table 2), the above-presented exhibited the lowest response time, as well the lowest detectable concentration limit [75]. More precisely, the as-suggested sensor is based on nucleic acid following the super sandwich-sensing type, which is consisted of: i) a capture probe (CP, is captured on the catalytic surface area, usually an antibody), ii) a target sequence (usually oligonucleotides or DNA or RNA, when the complementary sequence binds with the target sequence, then an electric signal is produced), iii) a label probe (organic dyes, enzymes or redox molecules play the role of the label, which displays a signal when the DNA hybridization is detected) and iv) an auxiliary probe, hexane-1-thiol (HT) (to ensure the analyte molecule capture).
The as-fabricated biosensor [75] was tested using laboratory and clinical samples. Its detection ability was estimated by employing the technique of differential pulse voltammetry (DPV); once the analysis sample contains the virus (target) a certain current is produced (Fig. 6Bii). As Fig. 6Biii shows, the current is linearly proportional to the concentration of the analyte (current vs logCanalyte), ranging from 10−17 to 10−12 M (mol/L) and exhibiting a lower limit of detection (LOD) close to 3 aM (aM = 10−18 mol/L). Moreover, its response time is <10sec, being one of the fastest SARS sensors proposed. It is also remarkable that no method was used to amplify the signal detection. However, in the case of clinical samples analysis, the 200 copies/mL was the lowest detection limit measured, achieving an accuracy of 100%. Moreover, the ability of the biosensor to be connected directly to a smartphone, with no need of nucleic acid amplification and reverse transcription, makes it more accessible as a diagnostic tool.
Thus, some of the as-developed sensors for SARS-CoV-2 detection are based on Au-working electrodes, as Au material consists a safe choice [89]. For the preparation of these electrodes four different strategies were adopted: i) the first one, in which Au nanoparticles are deposited on different electrode substrate (Ti, Si or other) that offers better characteristics, ii) the second one, in which the Au nanoparticles are modified with a second element, e.g. r-GO nanoflakes that increase the Au-nanoparticles binding capacity and sensitivity, iii) the third one, in which the Au-modified nanoparticles are deposited on a nano-support (r-GO) that offers more sites for increasing the sensitivity of host–guest recognition electrochemical biosensors and iv) the fourth one, in which the membrane cells are used as mediators for the virus detection.
Recently, some research groups focused their attention on the development of pure carbon or graphene-based electrodes for application in SARS-CoV-2 electrochemical biosensors, as it is below discussed.
2.2.2. Carbon and graphene-based SARS-CoV-2 electrochemical biosensors
The application of carbon or graphene-based nanomaterials, especially in the field of electrochemical biosensors, has become more intense the last decade [90], [91], [92], [93], [94], since it is found that the nano-structuring of the transducer-electrode improves by far all their operation and efficiency features. For example, Chin et al. [95], modifying the screen printed electrodes with carbon nanoparticles, they noticed that the charge transfer kinetics was significantly enhanced, and so the current response of the biosensor was also improved by 63%.
Very recently Mahari et al. [96], modified the screen-printed electrodes surface with carbon nanoparticles, fabricating a highly sensitive SARS-CoV-2 electrochemical platform (denoted as eCovSens). For comparison reasons they also fabricated a home-made biosensor equipped with a Au-based fluorine doped tin-oxide (FTO) electrode. The detection procedure of the as-proposed platform was based on the conductivity changes, caused by the binding events between the nCovid-19 monoclonal antibody (nCovid-19Ab) and the nCovid-19 antigen (nCovid-19 Ag) immobilized on the electrode. Using the technique of differential pulse voltammetry, they found that both biosensors exhibited high sensitivity in the Covid-19Ag concentration range between 10−15 M - 10−6 M. However, the low detection limit of eCovSens was calculated at 10 fM (fM = 10−15 M), while the Au-FTO one was estimated at 120 fM [96]. Taking into consideration the high sensitivity, the ability to be portable, the quick response time (10–30 s), as well as the low cost of the eCovSens, one might state that carbon-based electrochemical biosensors can be strong competitors of the Au-based ones.
Fabiani et al. [97] also modified the surface of graphite screen-printed electrodes with carbon magnetic beads, fabricating a SARS-CoV-2 electrochemical immunoassay. Since the magnetic beads have been set as biocompatible materials [98], their combination with carbon black is another biosensor configuration, exploited increasingly. The magnetic beads (placed under the working electrode) are operating as the support of the immunological chain and they are able to: i) accept higher antibody loading due to the high surface to volume ratio, ii) be moved to a specific volume of analyte and washed to remove interfering species, iii) increase the selectivity of the working electrode towards the bioreceptor and iv) improve its sensitivity [99].
The S and N proteins of the SARS-CoV-2 were used as target analytes, bonded with the immobilized-antibodies on the carbon surface. For achieving the optimum bonding between the SARS-CoV-2 protein and the antibody, the latter was labeled with alkaline phosphate enzyme (1-naphthyl phosphate enzymatic substrate). The use of carbon is enhanced the sensitivity of the screen-printed electrode, while the use of magnetic beads improved its selectivity.
In Fig. 7 A (i & ii), the differential pulse voltammetry results from the infected saliva sample analysis are displayed. The sensor is more sensitive, towards S-protein (Fig. 7Ai) detection than to N-protein (Fig. 7Aii). This difference was attributed to the fact that in SARS-CoV-2 cell the concentration of S-protein is three-times lower than that of N-protein [97]. In addition, the LOD was estimated to be 19 ng/mL (ng = 10−9g/mL) and 8 ng/mL for N and S-protein, respectively (Table 3 ). However, the suggested biosensor required a relatively high analysis time close to 30 min, which should be ameliorated with further research.
Fig. 7.
Voltammogram of: (Ai) S-protein detection and (Aii) P-protein detection [97]; (Bi) TiO2 nanotubes functionalized by Co (Co-TNTs), as electrode material for a SARS-CoV-2 biosensor; (Bii) Linear response for concentration range between 14 nM and 1400 nM and a detection response time of ~ 30 sec[102]; (Ci) Schematic illustration of –NH2 groups of the SARS-CoV-2 proteins or antibodies attached to 1-Pyrenebutyric acid (PBA); (Cii) Current response of real samples detection of the N-proteins, S1-IgG, S1-IgM and CRP [103]; (Di) FET SARS-CoV-2 biosensor; (Dii) For laboratory sample the lower limit of detection of SARS protein, was recorded at 1 fg/mL and 100 fg/mL [104]. Reproduced with permission.
Table 3.
Carbon or graphene-based transducer electrode for the SARS-CoV-2 electrochemical detection.
| Material | Electrochemical detection techniques | Target analyte/Immobilized element | Response time | Limit of detection |
|---|---|---|---|---|
| Carbon/screen-printed carbon electrode [96] | Differential pulse voltammetry | nCovid-19 Ag/nCovid-19 monoclonal Ab | 10 ~ 30 sec | Artificial target: 0.01 pM |
| Carbon magnetic beads/screen printed electrode [97] | Linear sweep voltammetry | SARS-CoV-2, S, N proteins | 30 min | Real sample: 19 × 106 pg/L for S, N protein Artificial target: 8 × 106 pg/Lfor S, N protein |
| Co– functionalized TiO2 nanotubes [102] |
Amperometry (bias voltage: −0.8 V) |
S-RBD/none | 30 sec | Artificial target: 14 × 103 pM |
| Laser-engraved graphene [103] | Amperometric technique | SARS-CoV-2 Biomolecules (N-protein, S1-IgM, S1IgG, and C-reactive protein)/blood and saliva | 10 min | 1) 0.1 × 1012 pg/L and 0.5 × 109pg/L for N-protein in artificial and real sample 2) 20 × 1012 pg/L and 0.2 × 1012 pg/L for S1-IgG in artificial and real sample 3) 20 × 1012 pg/L and 0.6 × 1012 pg/L for S1-IgM in artificial and real sample 4) 10 × 1012 pg/L and 0.1 × 1012pg/L for C-reactive protein in artificial and real sample |
| Graphene-sheets coated/field-effect transistor [104] | – | 1) SARS-CoV-2 virus’ Genome/SARS-CoV-2 spike protein | – | 1) 2.42 × 102 copies/mL (in real samples) 2) 100 pg/L (in artificial targets) |
The modification of carbon (or graphene) structure or the alteration of its surface composition strongly improves the properties of carbon (or graphene), motivating more and more research groups to explore them in many versions [98]. Surface functionalization, which is the most common strategy, can be succeeded either by physical interactions or chemical conjugations [100], [101].
The functionality of a nano-material increases the ratio of electrode surface area to volume, and consequently the electrochemical reaction rate [14], [95]. The titania nanotubes functionalization with Co (Fig. 7Bi), adopted by Vadlamani et al. [102], was the key element for ensuring: i) the stability and selectivity of the novel SARS-CoV-2 biosensor and ii) the successful immobilization of SARS-CoV-2 spike protein molecules. As the authors observed, the functionalization of the catalyst improved three main characteristics of the sensor: i) its selectivity towards the S-Receptor Binding Domain (SRBD) protein, ii) its sensitivity, showing a linear response in the concentration range between 14 nM and 1400 nM and iii) its response time (30 sec) (Fig. 7Bii).
The increment of the sensitivity was attributed to the as-formed Co(OH)2 bonded with TiO2 on the titania nanotubes (TNTs) surface. During the detection, the OH-functional groups of the protein are electrochemically oxidized to O−, which in turn interact with Co2+ ions of the Co-TNTs. Titania nanotube arrays are already reported as an effective material for application in electrochemical biosensors [105]. Their good sensing ability is mainly attributed to their special morphology that offers high area to volume ratio, more pathways for charge transfer and good adhesion to the substrate [106]. Moreover, their surface functionalization and the addition of defects, seem to create more oxygen vacancy sites, which act as adsorption sites for the protein or the enzyme of the viruses cells [107].
Graphene alteration by engraving before functionalization its surface with laser, is suggested as an effective strategy for producing high-sensitive biosensors [108], [109]. Applying the laser-engraving strategy, graphene can be produced from polymer films, avoiding the chemical routes that require high temperature processing [110], thus permitting low cost mass production and better properties.
Recently, Torrente-Rodriguez et al. [103] fabricated a wireless, portable SARS-CoV-2 electrochemical biosensor, consisted of four laser-engraved graphene working electrodes on polyamide substrate. For accepting the analyte molecule, graphene surface was functionalized and modified, using 1-pyrenebutyric acid (PBA), as the functional group (linker). The π-electrons and the carboxylic groups of PBA form hydrophobic or non-covalent interactions (π-stacking) with the aromatic rings of graphene (electrostatic bonds). Then, the –NH2 groups of the SARS-CoV-2 proteins or antibodies are attached to the PBA, while BSA protein is also introduced, hindering other molecules to be adsorbed onto the surface (Fig. 7Ci).
Contrary to the common functionalization, in which the functional groups directly bind the sp2 carbon atom by modifying its edges and by introducing defects, pyrene can provide a non-covalent functionalization, assuring that the graphene structure and consequently its electronic structure will not be disturbed when the required receptor is attaching on its surface. When disruptions appear, the receptor and consequently the detection ability of the biosensor will be partially deactivated [111].
The step-by-step functionalization of graphene was initially evaluated by applying differential pulse voltammetry and electrochemical impedance, under open circuit conditions. It was observed that after each modification step the peak current was decreased, while the resistance increased, proving the successful functionalization. The performance of the biosensor platform [103] was then evaluated, using the amperometric technique and by increasing the analyte concentration and recording the current response. The detected target-analytes were: i) proteins (N-protein and C-reactive protein), existing in the virus genome and ii) specific immunoglobulins (S1-IgG and S1-IgM), produced from the immune response of the infected patient.
The specific lower limit of detection, recorded for each one of the analytes, is listed in Table 3. More precisely, the as-fabricated biosensor was evaluated using electrochemical impedance spectroscopy (EIS) and differential pulse voltammetry (DPV). Fig. 7Cii shows the current response of the biosensor being tested with real samples, containing: N-proteins, S1-IgG, S1-IgM and C-reactive protein (CRP). Furthermore, the lower detection limits, in blood and saliva respectively, were as follows: i) 0.1 g/L and 0.5 mg/L for N-protein, ii) 20 g/L and 0.2 g/L for S1-IgG, iii) 20 g/L and 0.6 g/L for S1-IgM, and iv) of 10 g/L and 0.1 g/L for C-reactive protein. Additionally, the as-proposed sensor [103] showed a quick response and accuracy in a time period of 10 min (Table 3).
Graphene, as a two-dimensional material, is able to adsorb the analyte molecule without any pretreatment, however, running the risk to be easily contaminated [112]. Thus, in order to overcome this challenge, different strategies for changing graphene’s properties to the desired ones, including surface modification as well as functionalization, have been proposed [113].
Another type of biosensors for the detection of viruses is the field effect transistors (FET), in which graphene was adopted the last few years as the active material [113], [114], [115]. Recently, a promising electrochemical FET SARS-CoV-2 biosensor, applying graphene sheets as sensing material, was fabricated by Seo et al. [104]. The operation principle of the as-fabricated biosensor is based on the detection of SARS-CoV-2 spike protein bound on its antibody, which can be immobilized on graphene sheets through a probe linker (1-pyrenebutyric acid N-hydroxysuccinimide ester, denoted as PBASE) (Fig. 7Di). The PBASE inclusion in the graphene sheets caused its chemical functionalization, creating π-π bonds between them. The operational parameters of the suggested biosensor were studied amperometrically over laboratory cultured SARS-CoV-2 and over real patient samples. The sensor's LOD for SARS protein, was recorded at 1 fg/mL and 100 fg/mL for laboratory and for real samples, respectively (Fig. 7Dii). While the LOD for the SARS-CoV-2 biosensor was 1.6 × 10 pfu/mL in laboratory medium and 2.42 × 102 copies/mL in real samples [116].
3. Concluding remarks
Looking for the most effective COVID-19 detection ways, electrochemical biosensors could be a very promising one for: i) accurate, ii) low cost, iii) high sensitivity, iv) quick and v) portable detection, acting as supplementary to the current diagnostic tools for controlling the COVID disease expansion. In this report we present a detailed review on the up-to-date developed SARS-CoV-2 electrochemical biosensors, focusing on their classification based on the transducer electrode material.
Currently, two main groups of materials have been thoroughly explored as transducer electrodes: i) the Au-based and ii) the carbon or graphene-based ones. Both present faster response time (few seconds) along with higher accuracy than the current detection methods. They display high sensitivity values, reaching even the aM scale (=10−18M), while their specificity is also comparable to the current detection methods and they can be portable and miniaturized.
Comparing the two groups, we concluded that the carbon or graphene-based electrodes can compete the Au-based ones, as they have similar or even better operational characteristics, also offering the advantage of lower cost. The use of nanomaterials into electrochemical biosensors can improve their characteristics by far. However, their synthesis methods, the combination of the different nanomaterials, their nanostructure (nanoparticles, nanotubes, sheets, functionalized or not), their size and the way they are used in electrochemical biosensors (labels, support, or auxiliary elements for signal enhancement) significantly affect their characteristics.
Therefore, in the current review we recognized that in the case of the Au-based electrodes, Au was mainly used in the form of nanoparticles onto alternative supports (polymer based or other) or supported onto reduced graphene oxide, before being deposited onto the basic platform. The inclusion of the r-GO to the Au nanoparticles significantly ameliorates SARS-CoV-2 sensor characteristics as it mainly expands the detection area, on which the virus binds.
In the case of carbon or graphene-based electrodes, the surface functionalization constitutes the main strategy followed. Especially graphene and its derivatives, which are considered the most promising materials, do not contain chemically reactive functional groups that could help immobilizing analyte biomolecules. Thus, we observe its surface or structure alteration by: i) doping graphene with another(bio)element, or ii) creating structure defects, or iii) being used as they are to modify screen printed carbon electrodes.
Among the applied detection techniques, electrochemical impedance spectroscopy, amperometry and differential pulse voltammetry are the most used ones. Meanwhile, by applying the amperometric technique, there is a concern about the ‘real’ electric current response of the sensor, when operating in environments with high virus concentrations, as diffusion phenomena may prevail. Electrochemical impedance spectroscopy, square wave voltammetry and differential pulse voltammetry are more sensitive and reliable detection techniques, especially for the very low concentration values of the target analyte. However, for acquiring the ‘real’ charge, the optimal operation conditions are set each time (Hz or voltage step, or scan rate, etc) according to the virus concentration. Therefore, exploiting the potential of nanomaterials over time, by applying emerging synthesis strategies (such as functionalization) for their development, we are optimistic that the above challenge concerning the detection techniques will be overcome and soon the first commercial SARS-CoV-2 electrochemical biosensors will be emerged.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan X., Yang C., He Q., Chen J., Yu D., Li J., Zhai S., Qin Z., Du K., Chu Z., Qin P. ACS Infect. Dis. 2020;6:1998–2016. doi: 10.1021/acsinfecdis.0c00365. [DOI] [PubMed] [Google Scholar]
- 3.FDA: U.S Food & Drug, A Closer Look at Coronavirus Disease 2019 (COVID-19) Diagnostic Testing. 2020 [Google Scholar]
- 4.Ghaffari A., Meurant R., Ardakani A. Diagnostics. 2020;10(7):453. doi: 10.3390/diagnostics10070453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 6.Watson J., Richter A., Deeks J. BMJ. 2020;370 doi: 10.1136/bmj.m3325. [DOI] [PubMed] [Google Scholar]
- 7.Nagura-Ikeda M., Imai K., Tabata S., Miyoshi K., Murahara N., Mizuno T., Horiuchi M., Kato K., Imoto Y., Iwata M. J. Clin. Microbiol. 2020;58(9):e01438–20. doi: 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brouzgou A., Yan L.L., Song S.Q., Tsiakaras P. Appl. Catal. B. 2014;147:481–489. [Google Scholar]
- 9.Brouzgou A., Tsiakaras P. Top. Catal. 2015;58:1311–1327. [Google Scholar]
- 10.Yan L., Brouzgou A., Meng Y., Xiao M., Tsiakaras P., Song S. Appl. Catal. B. 2014;150:268–274. [Google Scholar]
- 11.Brouzgou A., Song S., Tsiakaras P. Appl. Catal. B. 2014;158–159:209–216. [Google Scholar]
- 12.Song S., Wang K., Yan L., Brouzgou A., Zhang Y., Wang Y., Tsiakaras P. Appl. Catal. B. 2015;176–177:233–239. [Google Scholar]
- 13.Brouzgou A., Lo Vecchio C., Baglio V., Aricò A.S., Liang Z.X., Demin A., Tsiakaras P. J. Electroanal. Chem. 2019;855 [Google Scholar]
- 14.Brouzgou A., Gorbova E., Wang Y., Jing S., Seretis A., Liang Z., Tsiakaras P. Ionics. 2019;25:6061–6070. [Google Scholar]
- 15.Bodur O.C., Özkan E.H., Çolak Ö., Arslan H., Sarı N., Dişli A., Arslan F. J. Mol. Struct. 2021;1223 [Google Scholar]
- 16.Koyappayil A., Lee M.-H. Sensors. 2021;21:89. [Google Scholar]
- 17.Ji T., Liu Z., Wang G., Guo X., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahshid S.S., Flynn S.E., Mahshid S. Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miren R.D.E., Loanda R.C., Robert J.F. Electrochem. Commun. 2020;116:106762. doi: 10.1016/j.elecom.2020.106762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan M., Hasan M., Hossain S., Ahommed M., Daizy M. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asif M., Ajmal M., Ashraf G., Muhammad N., Aziz A., Iftikhar T., Wang J., Liu H. Curr. Opin. Electrochem. 2020;23:174–184. doi: 10.1016/j.coelec.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava M., Srivastava N., Mishra P., Malhotra B.D. Sci. Total Environ. 2021;754 doi: 10.1016/j.scitotenv.2020.142363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bukkitgar S.D., Shetti N.P., Aminabhavi T.M. Chem. Eng. J. 2020 doi: 10.1016/j.cej.2020.127575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kudr J., Michalek P., Ilieva L., Adam V., Zitka O. TrAC Trends Anal. Chem. 2021;136 doi: 10.1016/j.trac.2021.116192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotru S., Klimuntowski M., Ridha H., Uddin Z., Askhar A.A., Singh G., Howlader M.M.R. TrAC Trends Anal. Chem. 2021;136 doi: 10.1016/j.trac.2021.116198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peixoto A.C., Silva A.F. In: Bioinspired Materials for Medical Applications. Rodrigues L., Mota M., editors. Woodhead Publishing; 2017. pp. 297–329. Elsevier Ltd. [DOI] [Google Scholar]
- 27.Schöning M.J., Poghossian A. Springer; Germany: 2018. Label-free biosensing: advanced materials, devices and applications. [Google Scholar]
- 28.Acquah C., Danquah M.K., Yon J.L.S., Sidhu A., Ongkudon C.M. Anal. Chim. Acta. 2015;888:10–18. doi: 10.1016/j.aca.2015.05.050. [DOI] [PubMed] [Google Scholar]
- 29.Walter J., Urmann K., Modrejewski J., Scheper T. BioNanoMat. 2017;18:20160012. [Google Scholar]
- 30.Daniels J.S., Pourmand N. Electroanal.: Int. J. Devoted Fundam. Pract. Aspects Electroanal. 2007;19:1239–1257. [Google Scholar]
- 31.Podder A., Lee H.J., Kim B.H. Bull. Chem. Soc. Jpn. 2021;94(3):1010–1035. [Google Scholar]
- 32.Seo H.B., Gu M.B. J. Biol. Eng. 2017;11:11. doi: 10.1186/s13036-017-0054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verdecchia P., Cavallini C., Spanevello A., Angeli F. Eur. J. Int. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sampson T. World Patent Inf. 2003;25:123–129. [Google Scholar]
- 37.Kawamura A., Miyata T. In: Biomaterials Nanoarchitectonics. Mitsuhiro E., editor. Elsevier; Japan: 2016. pp. 157–176. [Google Scholar]
- 38.Ronkainen N.J., Halsall H.B., Heineman W.R. Chem. Soc. Rev. 2010;39:1747–1763. doi: 10.1039/b714449k. [DOI] [PubMed] [Google Scholar]
- 39.Ronkainen-Matsuno N.J., Thomas J.H., Halsall H.B., Heineman W.R. TrAC Trends Anal. Chem. 2002;21:213–225. [Google Scholar]
- 40.Ruiz-Valdepeñas Montiel V.C., Povedano E., Vargas E., Torrente-Rodríguez R.M., Pedrero M., Reviejo A.J., Campuzano S., Pingarrón J.M. ACS Sensors. 2018;3:211–221. doi: 10.1021/acssensors.7b00869. [DOI] [PubMed] [Google Scholar]
- 41.Karastogianni S., Girousi S. Chem. Pap. 2015;69:202–210. [Google Scholar]
- 42.Kokkinos C. Nanomaterials. 2019;9:1361. doi: 10.3390/nano9101361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartlett P.N. John Wiley & Sons; United Kingdom: 2008. Bioelectrochemistry: fundamentals, experimental techniques and applications. [Google Scholar]
- 44.Arrigan D. Fundamentals Experimental Techniques and Applications. Springer; Germany: 2010. Bioelectrochemistry. [Google Scholar]
- 45.Pan M., Gu Y., Yun Y., Li M., Jin X., Wang S. Sensors. 2017;17:1041. doi: 10.3390/s17051041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lazcka O., Del Campo F.J., Munoz F.X. Biosens. Bioelectron. 2007;22:1205–1217. doi: 10.1016/j.bios.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y., Xu H., Zhang J., Li G. Sensors. 2008;8:2043–2081. doi: 10.3390/s8042043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erden P.E., Kılıç E. Talanta. 2013;107:312–323. doi: 10.1016/j.talanta.2013.01.043. [DOI] [PubMed] [Google Scholar]
- 49.Ding S.-N., Shan D., Xue H.-G., Cosnier S. Bioelectrochemistry. 2010;79:218–222. doi: 10.1016/j.bioelechem.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 50.Ensafi A.A. Elsevier Science; Iran: 2019. Electrochemical Biosensors. [Google Scholar]
- 51.Thakur H., Kaur N., Sabherwal P., Sareen D., Prabhakar N. Microchim. Acta. 2017;184:1915–1922. [Google Scholar]
- 52.M.E. Orazem, B. Tribollet, New Jersey, 2008.
- 53.Xu M., Yadavalli V.K. ACS Sensors. 2019;4:1040–1047. doi: 10.1021/acssensors.9b00230. [DOI] [PubMed] [Google Scholar]
- 54.Bertok T., Lorencova L., Chocholova E., Jane E., Vikartovska A., Kasak P., Tkac J. ChemElectroChem. 2019;6:989–1003. [Google Scholar]
- 55.Vu C.-A., Chen W.-Y. Sensors (Basel) 2019;19:4214. doi: 10.3390/s19194214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aishwaryadev B., Swagata M., Mastrangelo C.H. Phys. Chem. Phys. 2021 [Google Scholar]
- 57.Lee T., Ahn J.-H., Park S.Y., Kim G.-H., Kim J., Kim T.-H., Nam I., Park C., Lee M.-H. Micromachines. 2018;9:651. doi: 10.3390/mi9120651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antiochia R. Microchim. Acta. 2020;187:639. doi: 10.1007/s00604-020-04615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yanez-Sedeno P., Pingarron J. Anal. Bioanal. Chem. 2005;382:884–886. doi: 10.1007/s00216-005-3221-5. [DOI] [PubMed] [Google Scholar]
- 60.Abad-Valle P., Fernández-Abedul M.T., Costa-García A. Biosens. Bioelectron. 2005;20:2251–2260. doi: 10.1016/j.bios.2004.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De la Escosura-Muñiz A., González-García M.B., Costa-García A. Biosens. Bioelectron. 2007;22:1048–1054. doi: 10.1016/j.bios.2006.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martínez-Paredes G., González-García M.B., Costa-García A. Electroanalysis. 2009;21:379–385. [Google Scholar]
- 63.Layqah L.A., Eissa S. Microchim. Acta. 2019;186:224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamanaka K., M. d. C. Vestergaard and E. Tamiya Sensors (Basel) 2016;16:1761. doi: 10.3390/s16101761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.García-González R., Costa-García A., Fernández-Abedul M.T. Sens. Actuators, B. 2014;191:784–790. doi: 10.1016/j.snb.2013.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mohammadniaei M., Lee T., Yoon J., Lee D., Choi J.-W. Biosens. Bioelectron. 2017;98:292–298. doi: 10.1016/j.bios.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mohammadniaei M., Park C., Min J., Sohn H., Lee T. Biomimetic Medical Materials. Springer; 2018. pp. 263–296. [Google Scholar]
- 68.Min J., Lee T., Oh S.-M., Kim H., Choi J.-W. Biotechnol. Bioprocess Eng. 2010;15:30–39. [Google Scholar]
- 69.Fourmond V., Léger C. Practical Approaches to Biological Inorganic Chemistry. Elsevier; 2020. pp. 325–373. [Google Scholar]
- 70.Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N., Valdez J., Flynn J., Williams S.J. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nunez-Bajo E., Kasimatis M., Cotur Y., Asfour T., Collins A., Tanriverdi U., Grell M., Kaisti M., Senesi G., Stevenson K., Güder F. bioRxiv. 2020 doi: 10.1101/2020.03.23.002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tripathy S., Singh S.G. Trans. Indian Nat. Acad. Eng. 2020;5:205–209. doi: 10.1007/s41403-020-00103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S. Sensors. 2020;20:3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ali M.A., Hu C., Jahan S., Yuan B., Saleh M.S., Ju E., Gao S.J., Panat R. Adv. Mater. 2020:2006647. doi: 10.1002/adma.202006647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao H., Liu F., Xie W., Zhou T.-C., OuYang J., Jin L., Li H., Zhao C.-Y., Zhang L., Wei J., Zhang Y.-P., Li C.-P. Sens. Actuators, B. 2021;327 doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tavella F., Ampelli C., Leonardi S.G., Neri G. Sensors. 2018;18:3566. doi: 10.3390/s18103566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Furst A., Hill M.G., Barton J.K. Polyhedron. 2014;84:150–159. doi: 10.1016/j.poly.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ge C., Li Y., Yin J.-J., Liu Y., Wang L., Zhao Y., Chen C. NPG Asia Mater. 2012;4:e32. [Google Scholar]
- 79.Simões F.R., Xavier M.G. William Andrew Publishing; 2017. Nanoscience and its Applications; pp. 55–178. [DOI] [Google Scholar]
- 80.Li X., Liu J., Ji X., Jiang J., Ding R., Hu Y., Hu A., Huang X. Sens. Actuators, B. 2010;147:241–247. [Google Scholar]
- 81.Vermisoglou E., Panáček D., Jayaramulu K., Pykal M., Frébort I., Kolář M., Hajdúch M., Zbořil R., Otyepka M. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu Z. Nano-Micro Lett. 2017;9:25. doi: 10.1007/s40820-017-0128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen X., Gibson C.T., Britton J., Eggers P.K., Wahid M.H., Raston C.L. Chem. Commun. 2015;51:2399–2402. doi: 10.1039/c4cc09368b. [DOI] [PubMed] [Google Scholar]
- 84.Marcano D.C., Kosynkin D.V., Berlin J.M., Sinitskii A., Sun Z., Slesarev A., Alemany L.B., Lu W., Tour J.M. ACS Nano. 2010;4:4806–4814. doi: 10.1021/nn1006368. [DOI] [PubMed] [Google Scholar]
- 85.Yang L., Xie X., Cai L., Ran X., Li Y., Yin T., Zhao H., Li C.-P. Biosens. Bioelectron. 2016;82:146–154. doi: 10.1016/j.bios.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 86.Kwon S.N., Kim S.W., Kim I.G., Hong Y.K., Na S.I. Adv. Mater. Technol.gies. 2019;4:1800500. [Google Scholar]
- 87.Zhao D., Liu T., Zhang M., Liang R., Wang B. Smart Mater. Struct. 2012;21 [Google Scholar]
- 88.Wilkinson N., Smith M., Kay R., Harris R. Int. J. Adv. Manuf. Technol. 2019;105:4599–4619. [Google Scholar]
- 89.Sotiropoulou S., Gavalas V., Vamvakaki V., Chaniotakis N.A. Biosens. Bioelectron. 2003;18:211–215. doi: 10.1016/s0956-5663(02)00183-5. [DOI] [PubMed] [Google Scholar]
- 90.Yu X., Zhang W., Zhang P., Su Z. Biosens. Bioelectron. 2017;89:72–84. doi: 10.1016/j.bios.2016.01.081. [DOI] [PubMed] [Google Scholar]
- 91.Shao Y., Wang J., Wu H., Liu J., Aksay I.A., Lin Y. Electroanal.: Int. J. Devoted Fundam. Pract. Aspects Electroanal. 2010;22:1027–1036. [Google Scholar]
- 92.Suvarnaphaet P., Pechprasarn S. Sensors. 2017;17:2161. doi: 10.3390/s17102161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Novodchuk I., Bajcsy M., Yavuz M. Carbon. 2020;172:431–453. [Google Scholar]
- 94.Ognjanović M., Stanković V., Knežević S., Antić B., Vranješ-Djurić S., Stanković D.M. Microchem. J. 2020;158 [Google Scholar]
- 95.Chin S.F., Lim L.S., Pang S.C., Sum M.S.H., Perera D. Microchim. Acta. 2017;184:491–497. [Google Scholar]
- 96.Mahari S., Roberts A., Shahdeo D., Gandhi S. bioRxiv. 2020 doi: 10.1101/2020.04.24.059204. [DOI] [Google Scholar]
- 97.Fabiani L., Saroglia M., Galatà G., De Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reverté L., Prieto-Simón B., Campàs M. Anal. Chim. Acta. 2016;908:8–21. doi: 10.1016/j.aca.2015.11.050. [DOI] [PubMed] [Google Scholar]
- 99.Pedrero M., Campuzano S., Pingarrón J.M. Electroanalysis. 2012;24:470–482. doi: 10.1002/elan.201100528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jeykumari D.R.S., Kalaivani R., Narayanan S.S. Nano-Micro Letters. 2012;4:220–227. [Google Scholar]
- 101.Xu W., Kang Y., Jiao L., Wu Y., Yan H., Li J., Gu W., Song W., Zhu C. Nano-Micro Letters. 2020;12:184. doi: 10.1007/s40820-020-00520-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vadlamani B.S., Uppal T., Verma S.C., Misra M. Sensors. 2020;20:5871. doi: 10.3390/s20205871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. Matter. 2020;3:1–18. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 105.Lee H.-C., Zhang L.-F., Lin J.-L., Chin Y.-L., Sun T.-P. Sensors. 2013;13:14161–14174. doi: 10.3390/s131014161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bhattacharyya D., Smith Y.R., Misra M., Mohanty S.K. Mater. Res. Express. 2015;2 [Google Scholar]
- 107.Xiao P., Zhang Y., Cao G. Sens. Actuat., B. 2011;155:159–164. [Google Scholar]
- 108.Tehrani F., Bavarian B. Sci. Rep. 2016;6:27975. doi: 10.1038/srep27975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang L., Su J., Song Y., Ye R. Nano-Micro Lett. 2020;12:157. doi: 10.1007/s40820-020-00496-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lin J., Peng Z., Liu Y., Ruiz-Zepeda F., Ye R., Samuel E.L.G., Yacaman M.J., Yakobson B.I., Tour J.M. Nat. Commun. 2014;5:5714. doi: 10.1038/ncomms6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Karachevtsev V.A., Stepanian S.G., Glamazda A.Y., Karachevtsev M.V., Eremenko V.V., Lytvyn O.S., Adamowicz L. J. Phys. Chem. C. 2011;115:21072–21082. [Google Scholar]
- 112.Ambrosi A., Chua C.K., Latiff N.M., Loo A.H., Wong C.H.A., Eng A.Y.S., Bonanni A., Pumera M. Chem. Soc. Rev. 2016;45:2458–2493. doi: 10.1039/c6cs00136j. [DOI] [PubMed] [Google Scholar]
- 113.Cordaro A., Neri G., Sciortino M.T., Scala A., Piperno A. Nanomaterials. 2020;10:1014. doi: 10.3390/nano10061014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Roberts A., Chauhan N., Islam S., Mahari S., Ghawri B., Gandham R.K., Majumdar S.S., Ghosh A., Gandhi S. Sci. Rep. 2020;10:14546. doi: 10.1038/s41598-020-71591-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ono T., Kanai Y., Ohno Y., Maehashi K., Inoue K., Matsumoto K. Carbon Related Materials: Commemoration for Nobel Laureate Professor Suzuki. Springer; Singapore: 2020. pp. 91–101. [Google Scholar]
- 116.Jung Y.J., Park G.-S., Moon J.H., Ku K., Beak S.-H., Kim S., Park E.C., Park D., Lee J.-H., Byeon C.W., Lee J.J., Maeng J.-S., Kim S.J., Kim S.I., Kim B.-T., Lee M.J., Kim H.G. Am. Chem. Soc.: Infectious Diseas. 2020;6:2513–2523. doi: 10.1021/acsinfecdis.0c00464. [DOI] [PubMed] [Google Scholar]