Abstract
Nanoparticles are small particles sized 1–100 nm, which have a large surface-to-volume ratio, allowing efficient adsorption of drugs, proteins, and other chemical compounds. Consequently, functionalized nanoparticles have potential diagnostic and therapeutic applications. A variety of nanoparticles have been studied, including those constructed from inorganic materials, biopolymers, and lipids. In this review, we focus on recent work targeting the severe acute respiratory syndrome coronavirus 2 virus that causes coronavirus disease (COVID-19). Understanding the interactions between coronavirus-specific proteins (such as the spike protein and its host cell receptor angiotensin-converting enzyme 2) with different nanoparticles paves the way to the development of new therapeutics and diagnostics that are urgently needed for the fight against COVID-19, and indeed for related future viral threats that may emerge.
Keywords: Nanoparticles, COVID-19, SARS-CoV-2, Proteins, Therapeutics, Diagnostics
Graphical abstract
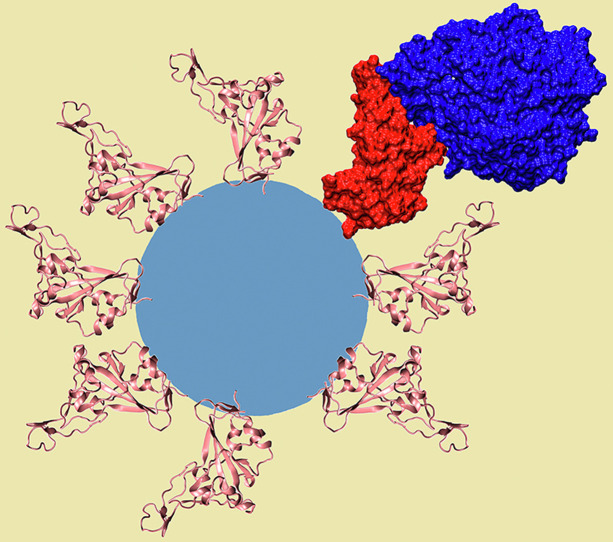
Schematic representation of the severe acute respiratory syndrome coronavirus 2 spike protein receptor-binding domain decorating a nanoparticle. The proteins are shown as a secondary structure colored in pink, whereas one of them is represented as a red surface complexed with the angiotensin-converting enzyme 2 receptor, which is shown in dark blue.
Introduction
Nanoparticles (NPs) are very small materials with a dimension between 1 and 100 nm. Their key physicochemical properties include a high surface area-to-volume ratio, solubility, surface topology/morphology, and controllable aggregation, making them suitable for application in a variety of commercial and domestic sectors, including electronics, catalysis, environment, imaging, energy, automotive, and health care [1]. There are various types of NPs, from inorganic materials, such as gold, silica, graphene, and iron oxide, to organic materials where the main groups include liposomes, micelles, protein/peptides, and dendrimers. They are particularly useful in health care applications, mainly because of their high capacity for adsorbing biomolecules [2].
Pharmaceutical nanotechnology is the development of therapeutic materials and devices at a nanometer scale, and there are several advantages to exploiting NPs in drug delivery. These include but are not limited to (1) improvement in the solubility of certain drugs; (2) controlled, sustained release of drugs for a long-term effect; (3) reduction of the side effects of some drugs; (4) targeting of specific cells; (5) administration routes; and (6) delivery of drugs in a secure manner, so that they are protected from degradation in the body and can effectively reach the target cells intact [3]. NPs can display efficient adsorption of proteins, drug molecules, and a variety of other chemical compounds. Therefore, NPs can carry a varied cargo load [4], making them efficient not only for drug delivery but also diagnostic and therapeutic applications.
In this review, we explore how NPs have been used to develop approaches to tackling COVID-19, focusing on the interactions between NPs and the adsorption of molecules such as proteins and drugs. We start with a brief overview of properties of the NP and their potential antiviral applications. We then review the SARS coronavirus (SARS-CoV-2) that causes COVID-19 and its proteins that are the targets for new technologies before turning to the various types of NPs that can be used as the basis for these technologies. Alternative approaches to treating COVID-19, for example, by repurposing drugs that were previously successful against other viruses, is discussed, followed by an overview of developments in diagnostics. We conclude the review with a summary and forward look as to how understanding the interactions between the different molecules and NPs could be used to rationally design new technologies to help tackle this pandemic and future coronavirus disease.
NP-biomolecule interactions and applications
Physicochemical properties
Selective and targeted delivery of modified NPs could enable specific detection and even destruction of viruses. To ensure this happens efficiently, it is important that the NPs are correctly optimized to ensure maximum efficacy and correct bioavailability, as well as negating any toxic effects, particularly those related to the formation of reactive oxygen species (ROS) [5]. Furthermore, the rate of cellular uptake of the NPs depends on their physicochemical properties and the membrane characteristics at the site of interaction [6].
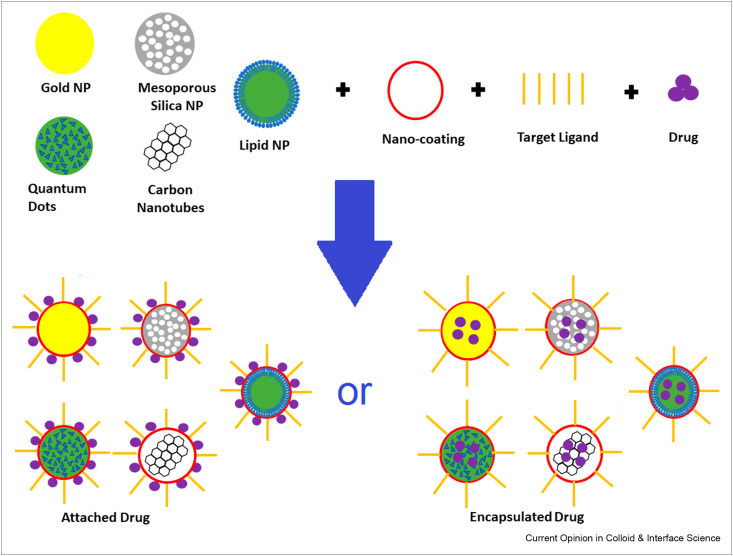
The key properties of NPs (Figure 1 ) make them ideal for a variety of effective systems. They can be porous or even hollow and are often amenable to surface chemistry modification. Proteins adsorbed on NPs normally form a dynamic corona, and protein conformational changes associated with the adsorption influence the overall in vivo bioreactivity [7]. The nature of NPs can influence the folding and unfolding properties of the protein, and by tuning the properties of the NPs, it can open new prospects in producing biologically active molecules. Thus, understanding the properties of the corona is essential [8]. The interactions between NPs and a particular protein can use a noncovalent route, with the solvent having a critical role to facilitate the interaction [8]. Consequently, it is vital to use a solvent in vitro that mediates the same interactions in vivo [9].
Figure 1.
A schematic diagram showing drug loading options in NP targeted drug delivery.
The biodegradation of NPs also requires attention, as uniform biodistribution kinetics and sustained drug release are key elements in the drug design process. Absorption, distribution, metabolism, and excretion are pharmacokinetic features linking directly to the nature and profile of these systems, and it is therefore crucial to account for all these factors when designing a nanoparticulate therapy [10].
Antiviral applications
Several inorganic NPs have been explored previously for their applications in drug delivery for viral infections. Gold NPs have a particular advantage in nanovaccines, as they can function as adjuvants (compounds to boost an immune response) in immunization. For example, their use was investigated against influenza A virus to combat mutations, which made the virus resistant to existing antiviral drugs [11]. Silica NPs were investigated as a vaccine platform against human immunodeficiency virus [12], and quantum dots, which have excellent sensing properties, can be used for antiviral therapeutics as well as for detection and diagnosis [13].
Silver NPs have also been investigated for their antiviral activity [14,15]. Antiviral activity against Peste des petits ruminants virus depends on the NP interaction with virion surface, and this interaction impairs viral entry into target cells [14]. These NPs may lead to better antiviral activity when used in conjunction with bronchodilators in the lungs, and this technology could have promising applications in treating COVID-19 patients [15].
Several organic NPs have also been used in pharmaceutical applications, for example, Cyclodextrin NPs, which are cyclic oligosaccharides with a hydrophilic outer surface and a lipophilic central cavity. Garrido et al. [16] suggested the use of cyclodextrins against COVID-19. These NPs maybe particularly helpful because of their physical properties with polar hydroxy groups oriented specifically, allowing increased solubility and decreased toxicity of the associated drug. Furthermore, they are highly biocompatible, meaning they do not generate an immune response. Lipid NPs (LNPs), often used in novel pharmaceutical formulations, are readily integrated into medicines. This is because of their high biocompatibility, low toxicity, ability to cross membranes, and seamless integration with hydrophobic/hydrophilic drugs.
NPs can be readily made with a similar size to the virus and may interact with proteins associated with the SARS-CoV-2 virus, disrupting viral replication and disease prognosis [17]. The use of NPs against SARS-CoV-2 has tremendous potential because of their specific properties, including (1) precise targeting of cellular entry pathways, (2) targeted binding to the viral genome, (3) modulation of viral transcription, (4) triggering the production of ROS, and (5) activation of signaling pathways at a mitochondrial level [18].
Tabish [18] explored the multivalent nature of nanomedicines and how this may be particularly useful in the fight against COVID-19. Multivalent NPs have several advantages over standard monovalent drugs, including a high density of binding sites on each NP, the ability to form multivalent ligand receptor pairs, multifold RNA hybridization, and the transformation of inactive NPs into multivalent conjugates [18]. Multivalency may work against SARS-CoV-2 effectively with cell entry through receptor-mediated endocytosis [19]. Hassanzadeh [20] also suggested the use of multivalent NPs against COVID-19. Given the similarities in the shape of synthetic NPs and SARS-CoV-2, they could be particularly useful for investigation with drug repurposing, enhancing properties of existing drugs and compounds against COVID-19. However, caution is required because SARS-CoV-2 may induce a hyperinflammatory response, driven by a dysregulated macrophage response [21]. Therefore, it is important to look at the properties of any material to ensure that it does not interact negatively in vivo.
SARS-CoV-2
Description of the virus and its function
SARS-CoV-2 is spread predominantly from person to person by droplets generated when an infected person coughs, sneezes, or talks. Infection may also occur by touching contaminated surfaces and then the face without first washing hands, and the fecal-oral route may also be a source of transmission for the virus [22]. The base symptoms include fever, cough, shortness of breath, fatigue, and loss of taste and/or smell. Depending on other factors such as infection level, age, and ethnicity, the symptoms may be extended to include headache, hemoptysis, or diarrhea. This highlights the severity of the virus, which can be fatal [23]. Therefore, the development of a new treatment for this virus is a priority for researchers globally.
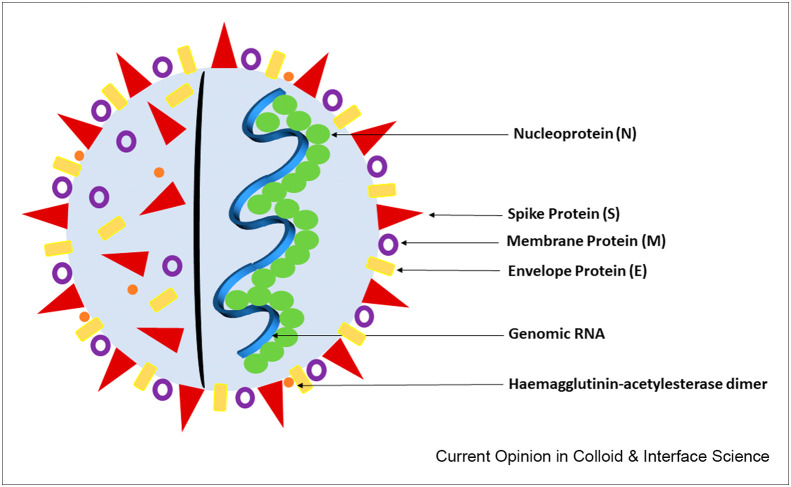
Analysis of the genomic sequence of SARS-CoV-2 [24] shows there are at least six open reading frames (ORFs), which are segments of an RNA molecule that can be translated, allowing the production of four main structural proteins: a spike protein (S), an envelope protein (E), a membrane protein (M), and a nucleocapsid protein (N). There is also the viral hemagglutinin-acetylesterase (HE) glycoprotein receptor, as illustrated in Figure 2 . The M and E proteins are involved in virus morphogenesis and assembly [25]. The N protein guards the RNA inside the M and E proteins, and the S protein is on the outside and the focal point of infection.
Figure 2.
Diagram showing the structural proteins of the SARS-CoV-2 virus.
Potential biomolecular targets
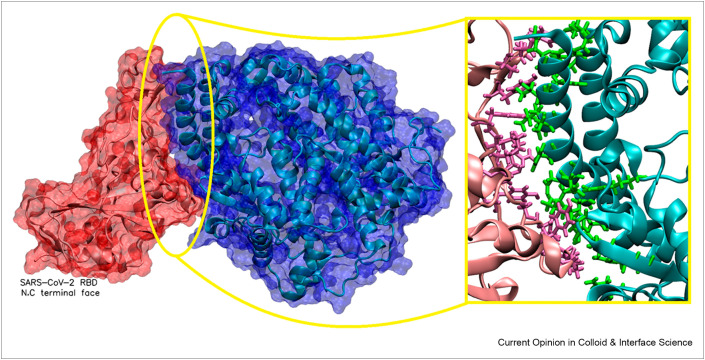
The S protein is an important therapeutic and diagnostic target, as it is responsible for entry into and infiltration of the host cell. It is a homotrimer with two domains, S1 and S2, on each monomer. Analysis of these monomers shows that they are highly glycosylated [26], protecting the protein from the biological environment and allowing evasion from the host immune system. The S1 subunit contains the receptor-binding domain (RBD) that binds to the peptidase domain of angiotensin-converting enzyme 2 (ACE2; Figure 3 ), a cellular receptor expressed on several cell types in human tissues, and this allows entry of SARS-CoV-2 into the cell [27].
Figure 3.
Interaction between the ACE2 receptor (blue) and the S protein RBD (red). Inset shows key interacting residues between the ACE2 receptor. The crystal structure was obtained from the Protein Databank (PDB entry 6M0J [38]). The crystal structure was viewed and analyzed using VMD (Visual Molecular Dynamics 1.9.1).
On cell entry, two ORFs, 1a and 1b, translate to two polypeptides (1a and 1ab), and this further encodes two proteases, the main protease (Mpro), also known as the chymotrypsin-like cysteine protease (3CLpro), and papain-like protease (PLpro) [28]. These represent significant drug targets because inhibition of these will stop the production of proteins that are critical to viral transcription and replication [29, 30, 31].
The S1 subunit allows entry of the virus into the host cell, and inhibition of this will block the protein from interacting with the ACE2 receptor [32]. For example, immunoadhesins have been investigated for their interactions with the S protein through MD simulations [33]. Another potential target for therapeutics development is the transmembrane protease serine 2 (TMPRSS2) found on host cells [34]. It cleaves (primes) the S protein into its subunits to enable cell entry, and inhibition of this process may prevent the initial entry of the virus.
High-density lipoproteins (HDLs) are particles consisting of several proteins, which transport all fat molecules around the body. HDL-scavenger receptor B type 1 (SR-B1) is a cell surface HDL receptor, which has been shown to facilitate ACE2-dependent entry of SARS-CoV-2 and further enhance uptake and increase the rate of virus entry [35]. Wei et al. [35] suggested that blockage of the cholesterol-binding site on the S1 subunit or treatment with SR-B1 antagonists inhibits HDL-enhanced SARS-CoV-2 infection. Therefore, SR-B1 could also potentially be a target for therapeutic designs. Patel et al. have also suggested HE as a target [36] to inhibit the virus invasion mechanism.
The residues responsible for the interaction between the S protein and the ACE2 receptor have been investigated by Veeramachaneni et al. [37]. This information is important for designing any medicine because the residues required for interaction with the target should remain free to bind to the therapeutic molecule to allow effective inhibition. Their analysis has identified the key residues that interact with the ACE2 receptor (see Figure 3).
NP-biomolecular systems for COVID-19
Inorganic NPs
The potential of NPs for the treatment of COVID-19 is promising because of their various properties. Iron oxide NPs, which have previously been investigated for their antiviral activity, were simulated for their interaction with the RBD of the S1 subunit [39]. It was found that a model Fe3O4 NP forms a stable complex with the protein, interacting through several hydrophobic interactions primarily with residues Leu455, Ser494, and Phe497. Therefore, these NPs, which are currently an approved treatment for anemia, could be repurposed to treat COVID-19 [39].
Carbon nanotubes (CNTs) have a large load capacity and good bioavailability, allowing for easy interaction with biological barriers in the body [40]. The electrical and thermal properties of these materials could be used to develop a CNT functionalized complex, raising the local cellular temperature using a photodynamic thermal effect and treating COVID-19 by inhibiting viral replication [41]. The binding of the S protein to biomedically relevant surfaces has been examined computationally, and it was found that the RBD of the S protein interacts with negatively charged silica surfaces so that the epitope (part of the antigen molecule, RBD in this instance, to which an antibody binds) is exposed. A model gold surface has also shown good interaction with the protein [42]. The use of charged or hydrophobic surfaces in developing therapies may therefore be significant as they show good adsorption [42].
Organic NPs
As researchers globally are working to develop an immediate treatment for this new virus, the development of effective vaccines is also vital. One approach for messenger mRNA (mRNA) vaccines comprises mRNA (encoding a specific protein) encapsulated in organic NPs, most commonly LNPs. Once LNP conjugates reach the host cell, the cell machinery follows the encapsulated mRNA instructions and produces the target protein, which is then displayed on the cell surface and can eventually trigger an immune response [43].
The obvious target for the SARS-CoV-2 virus is the S protein, and an example of mRNA-based vaccine has been developed by BioNTech in collaboration with Pfizer. It has been approved by the US Food and Drug Administration (FDA), the United Kingdom Medicines and Healthcare products Regulatory Agency (MHRA), and the European Medicines Agency (EMA), demonstrating an estimated efficacy of 95% [44,45]. Another mRNA-based vaccine was developed by Moderna, a US-based biotech firm [46]. Phase 3 clinical trial demonstrated that the vaccine has 94.1% efficacy in preventing COVID-19 [47]. At the time of writing, this has been approved by the FDA for emergency use and by the MHRA and EMA.
Self-amplifying RNA (saRNA) is a newer type of RNA vaccine, which contains a viral replication enzyme (replicase), allowing it to amplify [48]. The saRNA enters the host cell, translates the replicase, making a negative copy of the mRNA. The mRNA strand is used by the replicase to synthesize more saRNA while simultaneously binding to a subgenomic promoter in the negative strand. This synthesizes subgenomic mRNA at a 10-fold greater concentration than genomic RNA, encoding the viral antigen more effectively and making a more efficient vaccine.
McKay et al. investigated the vaccine potential of an saRNA molecule encoding the S protein, encapsulated within LNPs [49]. A high concentration of SARS-CoV-2-specific antibody titers in mice was observed. When compared with the results from a natural immune response in recovered COVID-19 human patients, the vaccine resulted in much higher antibody titers, which were able to neutralize both a pseudo-type and wild-type SARS-CoV-2 virus. Furthermore, there was no observation of antibody-dependent enhancement [49], which could result in enhanced respiratory disease and acute lung injury after respiratory virus infection. This is a common concern when developing antibody-dependent vaccines, which could reverse amplify the infection [50].
Administration routes
NPs can open up a variety of administration routes beyond injection. For example, liposomes can be designed for ingestion, protecting the drug from the acidic environment of the digestive tract to release it into the tissue of the gut wall [51]. In addition, liposomes have been used to protect sensitive materials such as mRNA encoding SARS-CoV-2 spike protein, and this technology was adapted in SARS-CoV-2 vaccines developed by Pfizer and Moderna [44, 45∗∗, 46, 47].
For COVID-19, nasal administration would seem to be an attractive proposition. As the virus primarily enters by breathing in particles, providing protection at the site of infection would appear beneficial. One existing flu vaccine, FluMist (https://www.flumistquadrivalent.com/), is sprayed into the patient's nose where the weakened virus induces mucosal immunity represented by IgA antibodies, as well as systemic immunity of the IgG antibodies [52]. This means that the immunized patient has two layers of defense against the virus and reduced likelihood of being able to carry and transmit the virus. Nanoparticulate systems could similarly be administered through inhalation or nasal spray, providing an attractive administration route with potential for greater protection for the patient and more feasible storage conditions for health care providers.
Potential new approaches
Repurposing of existing drugs
Drug repurposing represents the concept of implementing an investigational drug for new uses beyond the original intention [53]. Repurposing drugs for COVID-19 is an attractive approach, given the need to explore all the available options to immediately reduce mortality rates. This approach allows avoidance of the financial, resource, and time implications associated with the novel drug discovery process, and researchers and pharmaceutical companies are increasingly relying on drug repurposing.
Repurposing brings several other advantages because it can lower the risk of failure as the drug has already been evaluated for its toxicity profile. In addition, it can save additional time, as many of the drugs have already undergone preclinical and safety assessments. Moreover, the drugs have already undergone trials, so they may be able to accelerate Phases 1 and 2 and progress to large-scale Phase 3 trials. Furthermore, drug repurposing experiments do not always need major laboratory work, and any required work can often be performed in silico. The identification of suitable effective drugs is an exciting prospect, and further combination with NPs may enhance their biocompatibility and physicochemical properties. Despite the aforementioned advantages, repurposing a drug must be approached with caution, as some drugs can cause polypharmacological side effects, and intellectual property issues may arise [53].
As discussed earlier, the ACE2 receptor, expressed on many cell types, is key to the initial cellular entry by SARS-CoV-2. Therefore, Khelfaoui [54] used molecular docking combined with MD simulations to study drugs similar in structure to chloroquine and hydroxychloroquine, which are both approved medicines, aiming to block the ACE2 receptor. The studies were performed using two structures, the ACE2 receptor and SARS-CoV-2 bound to the ACE2 receptor, and the results showed that ramipril, lisinopril, and delapril, ACE2 receptor inhibitors currently used to treat hypertension, could bind with the ACE2 receptor better than hydroxychloroquine. Drugs that have been investigated for repurposing against key proteins associated with the SARS-CoV-2 virus are summarized in Table 1 . These could then be used in isolation or conjugated to NPs to enhance their properties.
Table 1.
A summary of FDA-approved and other antiviral drugs that have been investigated for repurposing against key proteins involved in the replication of SARS-CoV-2.
| Drug(s) | Existing use | SARS-CoV-2 target protein | Binding residues |
|---|---|---|---|
| Paritaprevir/simeprevir [55] | Hepatitis C virus | Mpro | His41/Cys145 |
| Remdesivir [56] | Ebola virus | RdRp | Ser759, Asp760, Asp761 |
| Hydroxychloroquine [57,58] | Malaria, rheumatoid arthritis, and lupus | Mpro | His41/Cys145 |
| Pyronaridine [59] | Antimalarial agent | Mpro | His41/Cys145 |
| Epirubicin, saquinavir [60, 61, 62] | Chemotherapy, HIV/AIDS | Mpro | His41/Cys145 |
| Mitoxantrone, leucovorin, birinapant, dynasore [63] | Chemotherapy, rectal cancer, breast cancer, perturbs endocytosis | Mpro | His41/Cys145, Glu166 |
| Noscapine ligand 23B [64] | Chemotherapeutic agent | Mpro | Arg40, Tyr54, Cys85, Phe181, Arg188, Glu55, Met82 and Asn84 |
| Lopinavir-ritonavir, tipranavir, raltegravir [65,66] | HIV/AIDS, HIV, HIV/AIDS | Mpro | His41/Cys145 |
| TMB607, TMC310911 [67] | HIV-1 protease inhibitor, HIV/AIDS | Mpro | His41/Cys145 |
| Atazanavir, darunavir [62] | HIV/AIDS | Mpro | His41/Cys145 |
Application of natural compounds
Natural compounds have long been studied for their application in treating disease and have a wide range of diversity in their chemical structures. Their use with drug delivery systems and other technologies might accelerate their exploitation [68]. Han [69] studied peptide inhibitors against the SARS-CoV-2 RBD. The inhibitors were based on the protease domain of ACE2 receptor, and it was shown through MD simulation that the peptides are stable when bound to the RBD, blocking the virus from attaching to the actual ACE2 receptor expressed in human cells, thereby having the potential to stop infection. Of the four inhibitors studied, the work identified high stability with 3, which retained their secondary structures and therefore their fits to the RBD.
In a separate study, Chen et al. [70] looked at the prospect of using polysaccharides in developing treatments for COVID-19. These compounds have several advantages, including low toxicity and good biocompatibility, and they are potential targets for the development of antiviral treatments. This is because they may interfere with the viral pathways by blocking the positive charge on the host cell surface to prevent viral entry [71]. For example, chitosan NPs were investigated against the hepatitis C virus [72]. The applications of natural compounds against COVID-19 are summarized in Table 2 . The versatility of natural compounds may allow for easier interaction with NPs compared with pre-existing drugs.
Table 2.
A summary of natural compounds that have been studied against COVID-19.
| Natural Compound(s) | Origin | Target | Key residues |
|---|---|---|---|
| Oridonin [36] | Compound from the Naturally Occurring Plant-Based Anti-cancer Compound-Activity-Target (NPACT) Database | HE | The114, Thr159, Leu161, Ala176, Arg177, Tyr184, Phe211, Leu212, Ser213, Asn214, Leu267 |
| Epigallocatechin gallate, epicatechin gallate, gallocatechin-3-gallate [73] | Green tea polyphenols | Mpro | His41/Cys145 |
| Peonidin 3-O-glucoside, kaempferol 3-Ob–rutinoside, 4-(3,4-dihydroxyphenyl)-7-methoxy-5-[(6-O-b-D-xylopyranosyl-b-D-glucopyranosyl)oxy]-2H-1-benzopyran-2-one, quercetin-3-D-xyloside, and quercetin 3-O-a-L-arabinopyranoside [74] | Plant-based compounds from the Sigma–Aldrich chemical library | Mpro | His41/Cys145, Leu141, Asn142, Ser144, His163, Glu166 |
| Procyanidin-a [75] | Flavonoid from plants | ACE2, Mpro | Ser44, Ser47, Asp350, Asp382, Tyr385, Arg393, Asn394, His401, Phe40, Phe390 |
| Melatonin [76] | Natural hormone | Mpro | His41/Cys145 |
| C1 and C2 [77] | Natural compounds from Curcuma Ionga L. | Mpro | His41/Cys145, Thr190, Thr25, Glu166, Thr45, Cys44, Ser46, Cys145, Pro168, Met165 |
| Hesperidin, sesamin [78] | Natural herbal medicines | Mpro | His41/Cys145 |
| Theaflavin di-gallate [62,66] | Plant-derived natural drug | Mpro | His41/cys145 |
| Azurin, peptides p18 and p28 [79] | Blue copper bacterial protein produced by Pseudomonas aeruginosa | S protein, Mpro, and PLpro | N-terminal region |
| Human intestinal defensin 5 [80] | Innate defense mechanism | ACE2 | Asp30 and Lys31 |
| NPRL-334 [81] | Natural compound from the Natural Products Research Laboratories (NPRL) library | Mpro | His41/Cys145, His3304, Met3428, Pro3431, Gln3452, Glu3429 |
| TCM 57025, TCM 3495, TCM 20111, TCM 31007 and TCM 5376 [30] | Traditional Chinese medicine database | N7-MTase | Asn306, Arg310, Trp385, Asn388 |
| Luteolin [82] | Flavonoid in honeysuckle | Mpro | His41/Cys145, Gln189, Leu4, Asn142, Thr26. Met49, Val3 |
Promising synthetic chemicals
The drug repurposing approach can also be used to analyze synthetic chemical compounds that might prove to be effective antivirals. This can be achieved by screening a database of small molecules against viral drug targets to identify molecules with possible antiviral activity or by developing chemical compounds inhouse. Promising synthetic chemicals that have been investigated against COVID-19 are summarized in Table 3 .
Table 3.
Summary of promising synthetic chemical compounds.
| Chemical(s) | Origin | Target | Key residues |
|---|---|---|---|
| IH-009 and IH-027 [83] | Inhouse chemicals | PLpro | Pro247, Pro248 |
| Neohesperidin [84] | Selleckchem database | TMPRSS2 | Arg55, Gly97, Asn51 |
| Ligand F2679-0163, Ligand F6355-0442, Ligand 8250 [85] | Life Chemicals Library, Asinex database | Mpro | Leu141, Glu166, Thr190, Gln192, Gly143, Ser144, His41/Cys145 |
| ZINC20601870, ZINC00793735 [86] | ZINC database | Mpro | His41/Cys145, Hie163, Hie41, Met49, Hie164, Glu166, Met165, Thr26, Gly143, Asn142, Leu141, Gln189 |
| α-ketoamide 13b ligand [87, 88, 89] | Inhouse molecule | Mpro | His41/Cys145 |
| ZINC64606047, ZINC05296775 | ZINC database | TMPRSS2 | His296, Asp345, Ser441, Asp435, Ser460, Gly462 |
Other material applications
Nanobiosensor technology has a potential to enhance testing, giving rapid and accurate detection of viruses. This technology works on the premise that the biomolecule of interest selectively binds to the target conjugated to a detector, producing a sensing signal that can be digitally interpreted [90]. Although limited studies have been reported so far, this technology has the potential to offer a better and alternative approach to existing polymerase chain reaction (PCR) testing that is used to diagnose COVID-19.
A dual-functional plasmonic photothermal (PPT) biosensor, combining localized surface plasmon resonance (LSPR) with a PPT effect, can detect viral proteins. Qiu et al. [91] integrated the technologies on a two-dimensional gold nano-island chip, finding that the sensitivity and reliability of the sensor were enhanced when the angle of incidence of the illuminating light was changed. This is because the plasmonic resonances of the two technologies are excited at different wavelengths, giving a real-time and label-free detection of viral sequences from SARS-CoV-2, including RdRp, ORF1ab, and E genes. Furthermore, the in situ PPT enhancement on the chip improved the specificity of genomic detection, meaning similar sequences of RdRp genes from SARS-CoV (previous pandemic between 2002 and 2004) and SARS-CoV-2 can be accurately distinguished. This dual-functional LSPR sensor represents a simple and rapid diagnostic tool, which could improve the accuracy of SARS-CoV-2 testing in clinical diagnosis settings. In addition, it can help or even replace existing PCR tests, which often need several days to obtain results, may return false results, and need professional staff to perform the assay and interpret the results [92].
Lanthanides, a series of rare earth elements, possess unique physical and electronic features, giving rise to properties, such as long luminescence lifetimes and other optical characteristics. Chen et al. [93] investigated lanthanide-doped NPs with a lateral flow immunoassay (LFIA) as a biosensor to detect anti-SARS-CoV-2 IgG antibodies in human sera. The LFIA also included mouse antihuman IgG and rabbit IgG. A nitrocellulose membrane was used as the template to mount a recombinant phosphoprotein of SARS-CoV-2 to confine the IgG. Nineteen samples tested previously with reverse transcription PCR were then retested with the LFIA, which was found to detect anti-SARS-CoV-2 IgG in ∼10 min. Therefore, the LFIA can allow positive identification of SARS-CoV-2 in potential cases and be effectively used to monitor COVID-19 progression and patient responses to treatment.
Biosensor technology is generally promising; however, there are many challenges to overcome, emphasizing why the technology still needs comprehensive research to develop a high-quality sensor for point-of-care diagnostics. These challenges include reproducibility, surface preparation and immobilization conditions, incubation time and temperature, type of biological fluid used, and sample loading. Furthermore, insufficient selectivity and specificity of many of these tests means they are currently unreliable. These factors may restrict the effective use of this technology for overall SARS-CoV-2 detection [94].
Conclusions
This review has primarily focused on the applications of NPs and their interactions with relevant SARS-CoV-2 proteins, as well as suggestions on how NPs may be used to combat COVID-19. Furthermore, existing drugs that may be repurposed against COVID-19 and natural and synthetic compounds that might be enhanced in conjunction with NPs have also been included. Little is currently known about NP-based drug delivery systems for SARS-CoV-2, and a thorough understanding of the pathogenesis of this novel coronavirus is required to aid the development of effective agents. A collaborative global effort is required to find treatments, and the over-arching aim should be to develop antivirals based on previous work, as not only will this save time, but it is likely to work. Further enhancement of these through combination with NPs may well allow effective application of the drug.
As SARS-CoV-2 is a recently identified virus, any attempts to tackle this should be complemented with in silico studies to optimize the NP–drug interaction. Computer simulations have allowed effective interpretation of experimental data [95], for example, the widely used carrier protein bovine serum albumin adsorbing to a silica surface. Simulation has also previously facilitated the development of a new model NP-based vaccine using gonadotrophin-releasing hormone 1 with silica NPs [96]. Computer simulation is currently being used widely to aid efforts against the COVID-19 pandemic, be that in exploring the repurposing of existing drugs [56,58,63,66,67], or the development of new systems with natural compounds [66,79,87]. In our view, this approach will help design and deliver new therapies and diagnostics not only to fight COVID-19 but future viral threats that may emerge.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Acknowledgments
Moahmmed A.H. Farouq is supported by BBSRC UK grant number BB/T508792/1.
This review comes from a themed issue on Hot Topic: COVID-19
Edited by Reinhard Miller and Libero Liggieri
References
- 1.Khan I., Saeed K., Khan I. Nanoparticles: properties, applications and toxicities. Arabian Journal of Chemistry. 2019;12:908–931. [Google Scholar]
- 2.Navya P.N., Daima H.K. Rational engineering of physicochemical properties of nanos for biomedical applications with nanotoxicological perspectives. Nano Convergence. 2016;3:1. doi: 10.1186/s40580-016-0064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gelperina S., Kisich K., Iseman M.D., Heifets L. The potential advantages of nanoparticle drug delivery systems in chemotherapy of tuberculosis. Am J Respir Crit Care Med. 2005;172:1487–1490. doi: 10.1164/rccm.200504-613PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murthy S.K. Nanoparticles in modern medicine: state of the art and future challenges. Int J Nanomed. 2007;2:129–141. [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Z., Li Q., Wang J., Yu Y., Wang Y., Zhou Q., et al. Reactive oxygen species-related nanoparticle toxicity in the biomedical ffield. Nanoscale Research Letters. 2020;15:115. doi: 10.1186/s11671-020-03344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabourian P., Yazdani G., Ashraf S.S., Frounchi M., Mashayekhan S., Kiani S., et al. Effect of physico-chemical properties of nanoparticles on their iintracellular uptake. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21218019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh J.Y., Kim H.S., Palanikumar L., Go E.M., Jana B., Park S.A., et al. Cloaking nanoparticles with protein corona shield for targeted drug delivery. Nat Commun. 2018;9:4548. doi: 10.1038/s41467-018-06979-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Treuel L., Nienhaus G.U. Toward a molecular understanding of nanoparticle-protein interactions. Biophysical reviews. 2012;4:137–147. doi: 10.1007/s12551-012-0072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bresme F., Lehle H., Oettel M. Solvent-mediated interactions between nanoparticles at fluid interfaces. J Chem Phys. 2009;130:214711. doi: 10.1063/1.3148890. [DOI] [PubMed] [Google Scholar]
- 10.Patil V.M., Singhal S., Masand N. A systematic review on use of aminoquinolines for the therapeutic management of COVID-19: efficacy, safety and clinical trials. Life Sci. 2020;254:117775. doi: 10.1016/j.lfs.2020.117775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J., Yeom M., Lee T., Kim H.-O., Na W., Kang A., et al. Porous gold nanoparticles for attenuating infectivity of influenza A virus. J Nanobiotechnol. 2020;18:54. doi: 10.1186/s12951-020-00611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thalhauser S., Peterhoff D., Wagner R., Breunig M. Presentation of HIV-1 envelope trimers on the surface of silica nanoparticles. J Pharmaceut Sci. 2020;109:911–921. doi: 10.1016/j.xphs.2019.10.059. [DOI] [PubMed] [Google Scholar]
- 13.Kostarelos K. Nanoscale nights of COVID-19. Nat Nanotechnol. 2020;15:343–344. doi: 10.1038/s41565-020-0687-4. [DOI] [PubMed] [Google Scholar]
- 14.El-Mohamady R.S., Ghattas T.A., Zawrah M.F., Abd El-Hafeiz Y.G.M. Inhibitory effect of silver nanoparticles on bovine herpesvirus-1. International Journal of Veterinary Science and Medicine. 2018;6:296–300. doi: 10.1016/j.ijvsm.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subhasish S. Silver nanoparticles with bronchodilators through nebulisation to treat Covid 19 patients. Journal of Current Medical Research and Opinion. 2020;3:449–450. [Google Scholar]
- Garrido P.F., Calvelo M., Blanco-González A., Veleiro U., Suárez F., Conde D., et al. The lord of the NanoRings: cyclodextrins and the battle against SARS-CoV-2. Int J Pharm. 2020;588:119689. doi: 10.1016/j.ijpharm.2020.119689. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article looks at the various properties of cyclodextrins, which make these nanoparticles suitable for a variety of applications in treating COVID-19. This includes acting as encapsulating agents for other drugs used to treat COVID-19, as adjuvants to stabilize molecules involved in infection and use as adjuvants in vaccines.
- 17.Sportelli M.C., Izzi M., Kukushkina E.A., Hossain S.I., Picca R.A., Ditaranto N., et al. Can nanotechnology and materials sscience help the fight against SARS-CoV-2? Nanomaterials. 2020;10 doi: 10.3390/nano10040802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabish T.A., Hamblin M.R. Multivalent nanomedicines to treat COVID-19: a slow train coming. Nano Today. 2020;35:100962. doi: 10.1016/j.nantod.2020.100962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao H., Shi W., Freund L.B. Mechanics of receptor-mediated endocytosis. Proc Natl Acad Sci USA. 2005;102:9469. doi: 10.1073/pnas.0503879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassanzadeh P. Nanotheranostics against COVID-19: from multivalent to immune-targeted materials. J Contr Release. 2020;328:112–126. doi: 10.1016/j.jconrel.2020.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S.V., Hemalatha M., Kopperi H., Ranjith I., Kumar A.K. SARS-CoV-2 in environmental perspective: occurrence, persistence, surveillance, inactivation and challenges. Chem Eng J. 2021;405:126893. doi: 10.1016/j.cej.2020.126893. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is an interesting article because it discusses the transmission routes of SARS-CoV-2. As well as the common transmission route by droplets, this article suggests the fecal-oral route as a possible source of SARS-CoV-2 transmission and suggests strategies to combat the enteric virus.
- 23.Hanley B., Naresh K.N., Roufosse C., Nicholson A.G., Weir J., Cooke G.S., et al. Histopathological findings and viral tropism in UK patients with severe fatal COVID-19: a post-mortem study. The Lancet Microbe. 2020;1:e245–e253. doi: 10.1016/S2666-5247(20)30115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat Med. 2020 Apr;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam S.D., Bordin N., Waman V.P., Scholes H.M., Ashford P., Sen N., et al. SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals. Sci Rep. 2020;10:16471. doi: 10.1038/s41598-020-71936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020 doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gui M., Liu X., Guo D., Zhang Z., Yin C.-C., Chen Y., et al. Electron microscopy studies of the coronavirus ribonucleoprotein complex. Protein & Cell. 2017;8 doi: 10.1007/s13238-016-0352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selvaraj C., Dinesh D.C., Panwar U., Abhirami R., Boura E., Singh S.K. Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19. J Biomol Struct Dyn. 2020 Jun 22:1–12. doi: 10.1080/07391102.2020.1778535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grottesi A., Bešker N., Emerson A., Manelfi C., Beccari A.R., Frigerio F., et al. Computational studies of SARS-CoV-2 3CLpro: insights from MD simulations. Int J Mol Sci. 2020;21:5346. doi: 10.3390/ijms21155346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nayak S.K. Inhibition of S-protein RBD and hACE2 interaction for control of SARSCoV-2 infection (COVID-19) Mini Rev Med Chem. 2020 doi: 10.2174/1389557520666201117111259. [DOI] [PubMed] [Google Scholar]
- Bernardi A., Huang Y., Harris B., Xiong Y., Nandi S., McDonald K.A., et al. Development and simulation of fully glycosylated molecular models of ACE2-Fc fusion proteins and their interaction with the SARS-CoV-2 spike protein binding domain. PloS One. 2020;15 doi: 10.1371/journal.pone.0237295. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article looks at the development and potential use of ACE2-Fc fusion proteins and their use against SARS-CoV-2. This structure could effectively compete with the actual ACE2 receptor, preventing viral entry and infection.
- 34.Mollica V., Rizzo A., Massari F. The pivotal role of TMPRSS2 in coronavirus disease 2019 and prostate cancer. Future Oncol. 2020;16:2029–2033. doi: 10.2217/fon-2020-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Wan L., Yan Q., Wang X., Zhang J., Yang X., et al. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat Metab. 2020 doi: 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]; This article suggests the HDL-scavenger receptor B type 1 (SR-B1) facilitates and enhances the ACE2 dependent entry of SARS-CoV-2. The S1 subunit binds to the cholesterol, and the blockage of the binding site with a monoclonal antibody or treatment with an SR-B1 antagonist could represent a new and interesting target for therapeutics.
- 36.Patel C.N., Kumar S.P., Pandya H.A., Rawal R.M. Identification of potential inhibitors of coronavirus hemagglutinin-esterase using molecular docking, molecular dynamics simulation and binding free energy calculation. Mol Divers. 2020:1–13. doi: 10.1007/s11030-020-10135-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veeramachaneni G.K., Thunuguntla V.B.S.C., Bobbillapati J., Bondili J.S. Structural and simulation analysis of hotspot residues interactions of SARS-CoV 2 with human ACE2 receptor. J Biomol Struct Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1773318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 39.Abo-zeid Y., Ismail N.S.M., McLean G.R., Hamdy N.M. A molecular docking study repurposes FDA approved iron oxide nanoparticles to treat and control COVID-19 infection. Eur J Pharmaceut Sci. 2020;153:105465. doi: 10.1016/j.ejps.2020.105465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehra N.K., Mishra V., Jain N.K. A review of ligand tethered surface engineered carbon nanotubes. Biomaterials. 2014;35:1267–1283. doi: 10.1016/j.biomaterials.2013.10.032. [DOI] [PubMed] [Google Scholar]
- 41.Yang J. Inhibition of SARS-CoV-2 replication by acidizing and RNA llyase-modified carbon nanotubes combined with photodynamic thermal effect. Journal of Exploratory Research in Pharmacology. 2020;5:18–23. [Google Scholar]
- Cerofolini L., Fragai M., Luchinat C., Ravera E. Orientation of immobilized antigens on common surfaces by a simple computational model: exposition of SARS-CoV-2 Spike protein RBD epitopes. Biophys Chem. 2020;265:106441. doi: 10.1016/j.bpc.2020.106441. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article looks at the immobilization of a protein complex of the receptor-binding domain of SARS-CoV-2 S protein to several surfaces computationally. This is interesting as the findings can be used to direct experimental efforts in developing nano therapy.
- 43.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines — a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahase E. Covid-19: Pfizer and BioNTech submit vaccine for US authorisation. BMJ. 2020;371:m4552. doi: 10.1136/bmj.m4552. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article basically contains the safety and efficacy data for the BioNTech/Pfizer vaccine. This is an interesting article because it is the first COVID-19 vaccine incorporating LNPs to prove its efficacy and provides 95% protection in two doses.
- 46.Wang F., Kream R.M., Stefano G.B. An evidence based perspective on mRNA-SARS-CoV-2 vaccine development. Med Sci Mon Int Med J Exp Clin Res : international medical journal of experimental and clinical research. 2020;26 doi: 10.12659/MSM.924700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahase E. Covid-19: Moderna vaccine is nearly 95% effective, trial involving high risk and elderly people shows. BMJ. 2020;371:m4471. [Google Scholar]
- 48.Ballesteros-Briones M.C., Silva-Pilipich N., Herrador-Cañete G., Vanrell L., Smerdou C. A new generation of vaccines based on alphavirus self-amplifying RNA. Current opinion in virology. 2020;44:145–153. doi: 10.1016/j.coviro.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat Commun. 2020;11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., et al. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- 51.Daraee H., Etemadi A., Kouhi M., Alimirzalu S., Akbarzadeh A. Application of liposomes in medicine and drug delivery. Artificial Cells, Nanomedicine, and Biotechnology. 2016;44:381–391. doi: 10.3109/21691401.2014.953633. [DOI] [PubMed] [Google Scholar]
- 52.Mantis N.J., Rol N., Corthésy B. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 54.Khelfaoui H., Harkati D., Saleh B.A. Molecular docking, molecular dynamics simulations and reactivity, studies on approved drugs library targeting ACE2 and SARS-CoV-2 binding with ACE2. J Biomol Struct Dyn. 2020:1–17. doi: 10.1080/07391102.2020.1803967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alamri M.A., Tahir ul Qamar M., Mirza M.U., Bhadane R., Alqahtani S.M., Muneer I., et al. Pharmacoinformatics and molecular dynamics simulation studies reveal potential covalent and FDA-approved inhibitors of SARS-CoV-2 main protease 3CLpro. J Biomol Struct Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1782768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koulgi S., Jani V., Uppuladinne M.V.N., Sonavane U., Joshi R. Remdesivir-bound and ligand-free simulations reveal the probable mechanism of inhibiting the RNA dependent RNA polymerase of severe acute respiratory syndrome coronavirus 2. RSC Adv. 2020;10:26792–26803. doi: 10.1039/d0ra04743k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukherjee S., Dasgupta S., Adhikary T., Adhikari U., Panja S.S. Structural insight to hydroxychloroquine-3C-like proteinase complexation from SARS-CoV-2: inhibitor modelling study through molecular docking and MD-simulation study. J Biomol Struct Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1804458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Procacci P., Macchiagodena M., Pagliai M., Guarnieri G., Iannone F. Interaction of hydroxychloroquine with SARS-CoV2 functional proteins using all-atoms non-equilibrium alchemical simulations. Chem Commun. 2020;56:8854–8856. doi: 10.1039/d0cc03558k. [DOI] [PubMed] [Google Scholar]
- 59.Hosseini F.S., Amanlou M. Anti-HCV and anti-malaria agent, potential candidates to repurpose for coronavirus infection: virtual screening, molecular docking, and molecular dynamics simulation study. Life Sci. 2020;258:118205. doi: 10.1016/j.lfs.2020.118205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khan M.A., Mahmud S., Alam A.S.M.R.U., Rahman M.E., Ahmed F., Rahmatullah M. Comparative molecular investigation of the potential inhibitors against SARS-CoV-2 main protease: a molecular docking study. J Biomol Struct Dyn. 2020:1–7. doi: 10.1080/07391102.2020.1796813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khan A., Ali S.S., Khan M.T., Saleem S., Ali A., Suleman M., et al. Combined drug repurposing and virtual screening strategies with molecular dynamics simulation identified potent inhibitors for SARS-CoV-2 main protease (3CLpro) J Biomol Struct Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1779128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.C S, S DK, Ragunathan V., Tiwari P., A S, P BD Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease. J Biomol Struct Dyn. 2020:1–27. doi: 10.1080/07391102.2020.1815584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lokhande K.B., Doiphode S., Vyas R., Swamy K.V. Molecular docking and simulation studies on SARS-CoV-2 Mpro reveals mitoxantrone, leucovorin, birinapant, and dynasore as potent drugs against COVID-19. J Biomol Struct Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1805019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar D., Kumari K., Jayaraj A., Kumar V., Kumar R.V., Dass S.K., et al. Understanding the binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MD simulations at different temperatures. J Biomol Struct Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1752310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar Y., Singh H., Patel C.N. In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing. Journal of Infection and Public Health. 2020;13:1210–1223. doi: 10.1016/j.jiph.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peele K.A., Potla Durthi C., Srihansa T., Krupanidhi S., Ayyagari V.S., Babu D.J., et al. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: a computational study. Informatics in Medicine Unlocked. 2020;19:100345. doi: 10.1016/j.imu.2020.100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ancy I., Sivanandam M., Kumaradhas P. Possibility of HIV-1 protease inhibitors-clinical trial drugs as repurposed drugs for SARS-CoV-2 main protease: a molecular docking, molecular dynamics and binding free energy simulation study. J Biomol Struct Dyn. 2020:1–8. doi: 10.1080/07391102.2020.1786459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Obeid M.A., Al Qaraghuli M.M., Alsaadi M., Alzahrani A.R., Niwasabutra K., Ferro V.A. Delivering natural products and biotherapeutics to improve drug efficacy. Ther Deliv. 2017;8:947–956. doi: 10.4155/tde-2017-0060. [DOI] [PubMed] [Google Scholar]
- Han Y., Král P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article looks at peptide inhibitors to be used against the RBD of SARS-CoV-2, preventing virus action. The inhibitors were formed by a bundle of alpha helices found in the RBD; these were extracted from the ACE-2 receptor.
- 70.Chen X., Han W., Wang G., Zhao X. Application prospect of polysaccharides in the development of anti-novel coronavirus drugs and vaccines. Int J Biol Macromol. 2020;164:331–343. doi: 10.1016/j.ijbiomac.2020.07.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen L., Huang G. The antiviral activity of polysaccharides and their derivatives. Int J Biol Macromol. 2018;115:77–82. doi: 10.1016/j.ijbiomac.2018.04.056. [DOI] [PubMed] [Google Scholar]
- 72.Loutfy S.A., Elberry M.H., Farroh K.Y., Mohamed H.T., Mohamed A.A., Mohamed E.B., et al. Antiviral activity of chitosan nanoparticles encapsulating curcumin against hepatitis C virus genotype 4a in human hepatoma cell llines. Int J Nanomed. 2020;15:2699–2715. doi: 10.2147/IJN.S241702. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors – an in silico docking and molecular dynamics simulation study. J Biomol Struct Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1779818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Majumder R., Mandal M. Screening of plant-based natural compounds as a potential COVID-19 main protease inhibitor: an in silico docking and molecular dynamics simulation approach. J Biomol Struct Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1817787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maroli N., Bhasuran B., Natarajan J., Kolandaivel P. The potential role of procyanidin as a therapeutic agent against SARS-CoV-2: a text mining, molecular docking and molecular dynamics simulation approach. J Biomol Struct Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1823887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Al-Zaqri N., Pooventhiran T., Alsalme A., Warad I., John A.M., Thomas R. Structural and physico-chemical evaluation of melatonin and its solution-state excited properties, with emphasis on its binding with novel coronavirus proteins. J Mol Liq. 2020;318:114082. doi: 10.1016/j.molliq.2020.114082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gupta S., Singh A.K., Kushwaha P.P., Prajapati K.S., Shuaib M., Senapati S., et al. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J Biomol Struct Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1776157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kodchakorn K., Poovorawan Y., Suwannakarn K., Kongtawelert P. Molecular modelling investigation for drugs and nutraceuticals against protease of SARS-CoV-2. J Mol Graph Model. 2020;101:107717. doi: 10.1016/j.jmgm.2020.107717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sasidharan S., Selvaraj C., Singh S.K., Dubey V.K., Kumar S., Fialho A.M., et al. Bacterial protein azurin and derived peptides as potential anti-SARS-CoV-2 agents: insights from molecular docking and molecular dynamics simulations. J Biomol Struct Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1787864. [DOI] [PubMed] [Google Scholar]
- Wang C., Wang S., Li D., Wei D.-Q., Zhao J., Wang J. Human iintestinal defensin 5 inhibits SARS-CoV-2 invasion by cloaking ACE2. Gastroenterology. 2020;159:1145–1147. doi: 10.1053/j.gastro.2020.05.015. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article suggests the human intestinal defensin 5 (HD5), an antibiotic enteric peptide, can effectively mask key interaction sites on the ACE2 receptor protein. These sites, key amino acid residues, are important for the S protein to bind and allow viral entry and infection.
- 81.Yang J.-S., Chiang J.-H., Tsai S.C., Hsu Y.-M., Bau D.-T., Lee K.-H., et al. In silico de Novo Curcuminoid derivatives from the compound library of Natural Products Research lLaboratories inhibit COVID-19 3CLpro activity. Natural Product Communications. 2020;15 [Google Scholar]
- 82.Yu R., Chen L., Lan R., Shen R., Li P. Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int J Antimicrob Agents. 2020;56:106012. doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amin S.A., Ghosh K., Gayen S., Jha T. Chemical-informatics approach to COVID-19 drug discovery: Monte Carlo based QSAR, virtual screening and molecular docking study of some in-house molecules as papain-like protease (PLpro) inhibitors. J Biomol Struct Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1780946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chikhale R.V., Gupta V.K., Eldesoky G.E., Wabaidur S.M., Patil S.A., Islam M.A. Identification of potential anti-TMPRSS2 natural products through homology modelling, virtual screening and molecular dynamics simulation studies. J Biomol Struct Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1784289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yadav R., Imran M., Dhamija P., Chaurasia D.K., Handu S. Virtual screening, ADMET prediction and dynamics simulation of potential compounds targeting the main protease of SARS-CoV-2. J Biomol Struct Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1796812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kumar D., Kumari K., Vishvakarma V.K., Jayaraj A., Kumar D., Ramappa V.K., et al. Promising inhibitors of main protease of novel corona virus to prevent the spread of COVID-19 using docking and molecular dynamics simulation. J Biomol Struct Dyn. 2020:1–15. doi: 10.1080/07391102.2020.1779131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liang J., Pitsillou E., Karagiannis C., Darmawan K.K., Ng K., Hung A., et al. Interaction of the prototypical α-ketoamide inhibitor with the SARS-CoV-2 main protease active site in silico: molecular dynamic simulations highlight the stability of the ligand-protein complex. Comput Biol Chem. 2020;87:107292. doi: 10.1016/j.compbiolchem.2020.107292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sk M.F., Roy R., Jonniya N.A., Poddar S., Kar P. Elucidating biophysical basis of binding of inhibitors to SARS-CoV-2 main protease by using molecular dynamics simulations and free energy calculations. J Biomol Struct Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1768149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bellan L.M., Wu D., Langer R.S. Current trends in nanobiosensor technology. Wiley interdisciplinary reviews Nanomedicine and nanobiotechnology. 2011;3:229–246. doi: 10.1002/wnan.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]; This study looks at the development of a dual-functional plasmonic photothermal biosensor, which can detect viral sequences from SARS-CoV-2. The sensor gave a label-free, real-time detection of RdRp, ORF1ab, and E genes. This represents a promising simple, yet rapid diagnostic tool.
- 92.Liu H.Y., Hopping G.C., Vaidyanathan U., Ronquillo Y.C., Hoopes P.C., Moshirfar M. Polymerase chain reaction and its application in the diagnosis of infectious keratitis. Med Hypothesis, Discov Innovation Ophthalmol J. 2019;8:152–155. [PMC free article] [PubMed] [Google Scholar]
- 93.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., et al. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 94.Demeke Teklemariam A., Samaddar M., Alharbi M.G., Al-Hindi R.R., Bhunia A.K. Biosensor and molecular-based methods for the detection of human coronaviruses: a review. Mol Cell Probes. 2020;54:101662. doi: 10.1016/j.mcp.2020.101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kubiak-Ossowska K., Tokarczyk K., Jachimska B., Mulheran P.A. Bovine serum albumin adsorption at a silica surface explored by simulation and experiment. J Phys Chem B. 2017;121:3975–3986. doi: 10.1021/acs.jpcb.7b01637. [DOI] [PubMed] [Google Scholar]
- 96.Connell D.J., Gebril A., Khan M.A.H., Patwardhan S.V., Kubiak-Ossowska K., Ferro V.A., et al. Rationalising drug delivery using nanoparticles: a combined simulation and immunology study of GnRH adsorbed to silica nanoparticles. Sci Rep. 2018;8:17115. doi: 10.1038/s41598-018-35143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]