Abstract
The Support vector regression (SVR) was used to investigate quantitative structure–activity relationships (QSAR) of 75 phenolic compounds with Trolox-equivalent antioxidant capacity (TEAC). Geometric structures were optimized at the EF level of the MOPAC software program. Using Pearson correlation coefficient analysis, four molecular descriptors [n(OH), Cosmo Area (CA), Core-Core Repulsion (CCR) and Final Heat of Formation (FHF)] were selected as independent variables. The QSAR model was developed from the training set consisting of 57 compounds and then used the leave-one-out cross-validation (LOOCV) correlation coefficient to evaluate the prediction ability of the QSAR model. Used Artificial neural network (ANN) and multiple linear regression (MLR) for comparing. The RMSE (root mean square error) values of LOOCV in SVR, ANN and MLR models were 0.44, 0.46 and 0.54. The RMSE values of prediction of external 18 compounds were 0.41, 0.39 and 0.54 for SVR, ANN and MLR models, respectively. The obtained result indicated that the SVR models exhibited excellent predicting performance and competent for predicting the TEAC of phenolic compounds.
Subject terms: Biochemistry, Chemical biology, Molecular biology, Molecular medicine, Chemistry
Introduction
Phenolic compounds are natural products and can be extracted easily from many plants1. They show extensive biological activities such as anti-hepatotoxic2, antitumor3, anti-inflammatory4,5 and antioxidant activity6–8. Among them, antioxidant activity depends mainly on the structure9–11, so numerous researcher establish many quantitative structure–activity relationships (QSAR) models to investigated the antioxidant activity of flavonoids and interpret the relationship between phenolic compounds structure and their antioxidant activity12–16, the optimized QSAR model is helpful for researchers to design and synthesize antioxidants. Because of the complex relationship between phenolic compounds structure and antioxidant activity, simple linear models are insufficient to explain the effect of structural parameters on antioxidant activity17,18. Therefore, it is essential to use machine learning algorithms such as multiple linear regression (MLR), artificial neural networks (ANNs) to improve the predictability of QSAR19,20. Djeradi et al. have used Fukui indices and MLR method for prediction antioxidant activity of DPPH test of 24 flavonoids, the square of correlation coefficient (R2) of their model is 0.81621. Cerit et al. have used a multilayer perceptron (MLP) ANN to predict the effect of ferric ion on the antioxidant capacity of phenolic, the average errors of prediction of the training set and validation sets are 8.5 and 10.1%22. Li et al. have used MLP-ANN model to predict the antioxidant activity of polysaccharides in DPPH test and used sensitivity analysis to interpret the effect of the input variables on the target values23. Petar et al. and Fatemi et al. have used ANN and MLP-ANN QSAR models to evaluate the contribution of the quantum mechanical molecular descriptors to the Trolox-equivalent antioxidant capacity (TEAC) in an optimized ANN model19,24. Although the prediction accuracy of ANN is higher than MLR, most of the current ANN methods used to predict antioxidant activity are more like a black box that has overfitting risk and lead to unreliable predictions. Besides, it comprises a single hidden layer with an arbitrary activation function that must be bonded.
In addition to the above algorithm, support vector regression (SVR) is a useful machine learning algorithms that can be used to solve linear and nonlinear problems25, especially for small sample sizes. It has been proved to be suitable for the QSAR analyses of flavonoids26, drug activity prediction and design27. For instance, Minaoui et al. have used support vector regression to investigate the relationship between structure and activity of 38 cyclicurea derivatives, inhibiting HIV protease. In their work, each molecule is described by four descriptors, and the parameters of the SVR model are optimized by grid optimization. Then they compared the R2 and RMSE values of the prediction results of MLR, ANN, and SVR methods. The obtained results show that the SVR model has better qualities and better generalization capabilities than other methods. By evaluating the contribution of the molecular descriptors to the model established by the SVR, they also found that the molar volume and dipole moment parameters of the compounds take the most relevant part in the molecular description and controlling the biological activity of cyclic-urea derivatives28.
In addition to modelling methods, a reliable QSAR models also need to select appropriate variables, the QSAR models usually using topological and quantum mechanical parameters. Density functional theory (DFT) is an accurate but time consuming method for calculating electronic structure parameters29. While the Semi-empirical Hamiltonians method can obtain reliable molecular parameters for building QSAR models in a more time-efficient way30, especially when there is a lack of experience in selecting descriptors.
This study use Semi-empirical Hamiltonians (PM7, MOPAC 2016) to obtain molecular descriptors, then use Pearson correlation coefficient analysis for selecting molecular descriptors, then use the SVR method to develop a QSAR model to predict the antioxidant activity of 75 phenolic compounds. For comparing the prediction ability, ANN and MLR methods are used to build the QSAR models, too.
Materials and methods
Methods
Support vector regression (SVR)
As a statistical learning method, SVR uses a kernel function (including the linear kernel function (LKF), the polynomial kernel function (PKF), and the radial basic function (RBF) kernel function) to map the vectors into a higher dimensional feature space. By introducing an alternative loss function and kernel function, SVR can be applied to linear regression of the target variable in this space. For detailed information on the optimal regression function and related Lagrangian expressions, see Refs.20,31.
Leave one out cross-validation (LOOCV)
LOOCV process: first, each sample in the training dataset will be removed, and then use the remaining samples to build a model and predict the target value of the removed sample. In this work, the reliability was evaluated by LOOCV, and used tenfold-cross-validation (tenfold-CV) to search for the optimal kernel function type and corresponding parameters32,33.
Sensitivity analysis (SA)
Sensitivity analysis is often used to obtain the influence degree of variables on the target variable. SA can provide an effective method to characterize the uncertainties between characteristic parameters and models34,35. Based on the straightforward characteristics of SA, it was used in this work to explain the influence of parameters on TEAC.
Model accuracy
To obtain appropriate kernel function and capacity parameter C, insensitive loss function ε and the corresponding parameters gamma g of the kernel function in this computation, the least root mean square error (RMSE) and correlation coefficient R were used as the evaluation criterion20. RMSE is defined as follows:
| 1 |
where n is the number of total samples, ei and pi are the experimental value and the predicted value of sample i, respectively. Generally, the smaller RMSE means the better expected predictive ability.
The prediction power of the training set and test set also validated by statistical parameters of correlation coefficient (Q2)36,37, Q2 is defined as
| 2 |
All the methods calculated on the ExpMiner Software (version 2.1.1.0, Laboratory of Materials Data Mining, Department of Chemistry, College of Sciences, Shanghai University, China).
Data sets
75 phenolic compounds and TEAC values
The antioxidant activity (TEAC values, ABTS·+ assay) of 75 phenolic compounds were obtained from a study by Cai et al.38, The data set was randomly divided into the training set (57 phenolic compounds, ~ 75%) and the testing set (18 phenolic compounds, ~ 25%).
Molecular descriptors
The molecular descriptors of each phenolic compound were calculated by MOPAC software with EF geometry optimization and PM7 Semi-empirical Hamiltonians (MOPAC2016, J.J.P. Stewart, Stewart Computational Chemistry, Colorado Springs, CO, USA).
The name and molecular descriptors of phenolic compounds were given in Table S1.
Results
Descriptor selection and data set
Due to the existence of irrelevant or redundant features redundancy of the parameters, it is necessary to select the parameter most relevant to the target variable, especially when the sample set is small. The purpose of feature selection is to select a variables subset of n features from the set of m obtained variables (n < m) without significantly reducing the predictive ability of the model27. In this work, the total number of calculated molecular descriptors was eight. Used Pearson correlation selection modules to select descriptors (ExpMiner software), then the most significant three descriptors were selected. Since n(OH) is a critical variable and easy to get, added it to the variables. Finally, a total of four descriptors were chosen to construct the QSAR models, the descriptions of descriptors are shown in Table 1.
Table 1.
Molecular descriptors involved in the QSAR models.
| Molecular descriptor | Description |
|---|---|
| n(OH) | Number of OH groups |
| CA | Cosmo area |
| CCR | Core–core repulsion |
| FHF | Final heat of formation |
Grid-search for parameter optimization
In the modeling process, the parameters of the model were selected by grid search method, and the parameters of the lowest RMSE were found with three different kernel functions (RBF, PKF and LKF kernel function), that is, the optimal parameters.
By tenfold cross-validation in the grid-search process, RMSE values were calculated with capacity parameter (C, C = 1–500, step = 10) and e-insensitive loss function parameter (ε, ε = 0.01–0.1, step = 0.01)) with LKF and PKF, C (C = 1–500, step = 10), ε (ε = 0.01–0.1, step = 0.02) and Gamma (g, g = 0.5–1.5, step = 0.1) with RBF kernel function. The minimum RMSE values of RBF, PKF and LKF kernel function were 0.41, 0.45 and 0.50, respectively (see Supporting Information Fig. S1). Hence, the optimal SVR model is SVR-RBF kernel function with C = 121, ε = 0.07, g = 0.6 and the corresponding equation is:
| 3 |
where αi − αi* is the Lagrange coefficient corresponding to the 24 support vectors, the correlation coefficient between the predicted value and the experimental value is 0.967, as shown in Fig. 1.
Figure 1.
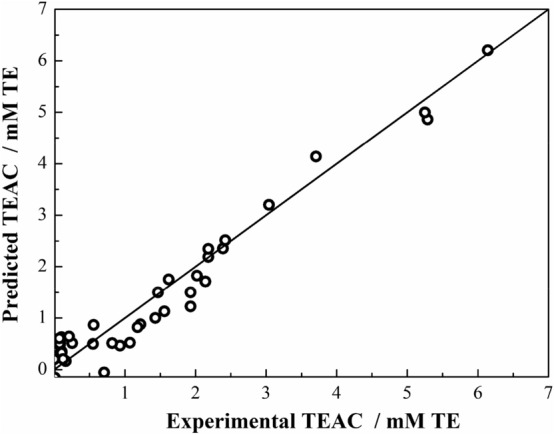
Correlation of experimental and predicted activity of agonists using Eq. (3).
LOOCV result of SVR-QSAR model
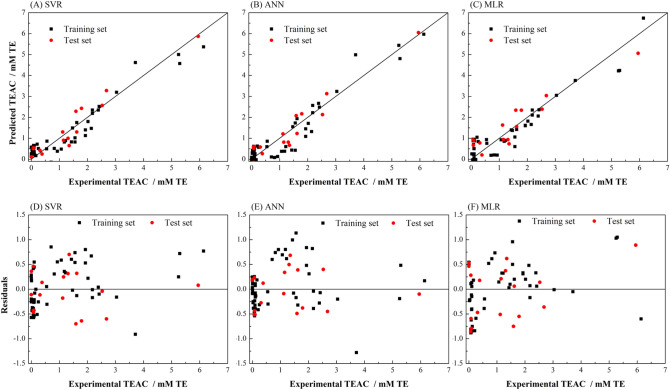
LOOCV was used to verify the reliability of the predictive ability of the QSAR Model. The same parameters were used to model with SVR, ANN and MLR to predict the TEAC values of 57 phenolic compounds (training set), then used the LOOCV method to examine their respective generalization capabilities (Fig. 2). The experimental values, predicted values of the training set and the test set are given in Table 2. The correlation coefficient (R2) between the predicted TEAC values and the experimental TEAC values of LOOCV are 0.904, 0.897 and 0.856 in SVR, ANN and MLR models. The results of Q2 obtained by the three modelling methods are similar to those of R2 (Table 3). The RMSE value of prediction of the test set in SVR is slightly higher than that of ANN, but the SVR model has the lowest predict RMSE of LOOCV, it is suggested that the generalization ability of SVR was superior to ANN and MLR in this work. From the results of residual, SVR is relatively stable in the whole data range, but the residuals of ANN and MLR are larger when the TEAC values are near 1.5 and 0.
Figure 2.
Experimental vs predicted activities of TEAC in LOOCV test and external test set of (A) SVR (using RBF kernel), (B) ANN, and (C) MLR. The plot of predicted residuals vs experimental values of TEAC of (D) SVR, (E) ANN and (F) MLR.
Table 2.
Predicted TEAC with different methods using LOOCV (No. 1–57) and test set (No. 58–75).
| No. | Experimental TEAC/mM TE | Predicted TEAC/mM TE | ||
|---|---|---|---|---|
| SVR | ANN | MLR | ||
| 1 | 1.56 | 0.821 | 0.425 | 0.603 |
| 2 | 0.93 | 0.379 | 0.129 | 0.197 |
| 3 | 0.82 | 0.513 | 0.08 | 0.204 |
| 4 | 0.007 | 0.259 | 0.04 | − 0.508 |
| 5 | 1.22 | 0.89 | 0.603 | 0.897 |
| 6 | 0.037 | 0.41 | 0.422 | 0.25 |
| 7 | 0.025 | 0.456 | 0.298 | 0.256 |
| 8 | 0.028 | 0.462 | 0.281 | 0.251 |
| 9 | 0.092 | 0.359 | 0.414 | 0.209 |
| 10 | 0.005 | 0.261 | − 0.165 | − 0.518 |
| 11 | 5.29 | 4.57 | 4.808 | 4.24 |
| 12 | 3.71 | 4.62 | 4.99 | 3.76 |
| 13 | 3.04 | 3.20 | 3.24 | 3.05 |
| 14 | 2.39 | 2.35 | 2.67 | 2.06 |
| 15 | 2.02 | 1.80 | 1.71 | 1.82 |
| 16 | 2.18 | 2.19 | 2.22 | 2.11 |
| 17 | 1.56 | 1.02 | 1.73 | 1.06 |
| 18 | 0.707 | − 0.145 | 0.107 | 0.186 |
| 19 | 2.42 | 2.52 | 2.49 | 2.36 |
| 20 | 1.93 | 1.40 | 1.45 | 1.60 |
| 21 | 1.43 | 0.826 | 0.435 | 1.40 |
| 22 | 2.18 | 2.35 | 2.57 | 2.35 |
| 23 | 0.081 | 0.608 | 0.619 | 0.948 |
| 24 | 1.47 | 1.49 | 1.55 | 1.37 |
| 25 | 0.083 | 0.656 | 0.318 | 0.705 |
| 26 | 0.003 | − 0.195 | 0.09 | − 0.536 |
| 27 | 0.098 | 0.358 | 0.195 | 0.509 |
| 28 | 0.104 | 0.358 | − 0.006 | 0.503 |
| 29 | 0.000 | − 0.284 | − 0.246 | − 0.544 |
| 30 | 0.101 | 0.642 | 0.491 | 0.947 |
| 31 | 0.072 | − 0.344 | − 0.182 | − 0.04 |
| 32 | 0.005 | − 0.217 | − 0.026 | − 0.537 |
| 33 | 5.25 | 5.00 | 5.44 | 4.22 |
| 34 | 6.14 | 5.37 | 5.97 | 6.74 |
| 35 | 2.14 | 1.47 | 1.32 | 1.66 |
| 36 | 1.62 | 1.75 | 1.94 | 1.42 |
| 37 | 0.558 | 0.863 | 0.858 | 0.756 |
| 38 | 0.002 | 0.574 | − 0.03 | − 0.503 |
| 39 | 1.18 | 0.821 | 0.378 | 0.854 |
| 40 | 0.164 | 0.166 | − 0.023 | − 0.018 |
| 41 | 0.001 | 0.2 | − 0.021 | − 0.506 |
| 42 | 0.253 | 0.509 | 0.566 | 0.841 |
| 43 | 0.104 | − 0.443 | 0.182 | 0.048 |
| 44 | 0.209 | 0.719 | 0.624 | 1.048 |
| 45 | 1.93 | 1.13 | 1.09 | 1.62 |
| 46 | 1.07 | 0.483 | 0.376 | 0.925 |
| 47 | 0.548 | 0.496 | 0.6 | 0.937 |
| 48 | 0.076 | 0.486 | 0.587 | 0.884 |
| 49 | 0.069 | 0.501 | 0.582 | 0.928 |
| 50 | 0.068 | 0.511 | 0.528 | 0.886 |
| 51 | 0.077 | 0.465 | 0.537 | 0.905 |
| 52 | 0.076 | 0.507 | 0.517 | 0.955 |
| 53 | 0.072 | 0.606 | 0.512 | 0.953 |
| 54 | 0.105 | − 0.341 | 0.031 | − 0.029 |
| 55 | 0.009 | − 0.162 | − 0.098 | − 0.525 |
| 56 | 0.105 | 0.318 | 0.263 | 0.254 |
| 57 | 0.124 | 0.204 | 0.259 | 0.876 |
| 58 | 1.31 | 0.995 | 0.812 | 0.938 |
| 59 | 1.15 | 0.903 | 0.812 | 0.94 |
| 60 | 5.95 | 5.87 | 6.05 | 5.06 |
| 61 | 2.68 | 3.28 | 3.13 | 3.04 |
| 62 | 1.59 | 2.29 | 2.08 | 2.34 |
| 63 | 1.12 | 1.30 | 1.21 | 1.63 |
| 64 | 1.79 | 2.43 | 2.17 | 2.34 |
| 65 | 0.001 | 0.110 | − 0.179 | − 0.463 |
| 66 | 0.097 | 0.524 | 0.608 | 0.889 |
| 67 | 0.077 | 0.543 | 0.511 | 0.671 |
| 68 | 2.53 | 2.57 | 2.13 | 2.39 |
| 69 | 1.35 | 0.649 | 0.666 | 0.732 |
| 70 | 0.383 | 0.246 | 0.261 | 0.204 |
| 71 | 0.003 | − 0.357 | − 0.191 | − 0.527 |
| 72 | 0.308 | 0.423 | 0.582 | 0.778 |
| 73 | 1.62 | 1.30 | 1.23 | 1.56 |
| 74 | 0.068 | 0.501 | 0.581 | 0.924 |
| 75 | 0.073 | − 0.388 | − 0.158 | − 0.208 |
Table 3.
RMSE and the squared correlation coefficient (R2 and Q2) of antioxidant activity prediction in LOOCV and test set of three models (SVR, ANN and MLR).
| SVR | ANN | MLR | |
|---|---|---|---|
| LOOCV | |||
| RMSE | 0.440 | 0.464 | 0.539 |
| R2 | 0.904 | 0.897 | 0.856 |
| Q2 | 0.903 | 0.892 | 0.855 |
| Test set | |||
| RMSE | 0.410 | 0.386 | 0.536 |
| R2 | 0.925 | 0.931 | 0.861 |
| Q2 | 0.917 | 0.927 | 0.859 |
Sensitivity analysis (SA) of SVR-QSAR model
Sensitivity analysis was used for analysis the correlation of molecular descriptors with TEAC, From Fig. 3, it can be suggested that the value of TEAC increased with the increase of n(OH) and CA, decreased with the increase of CCA and FHF. Further analysis showed that the order of the descriptors' influence on TEAC in descending is n(OH) > CA > FHF > CCR.
Figure 3.
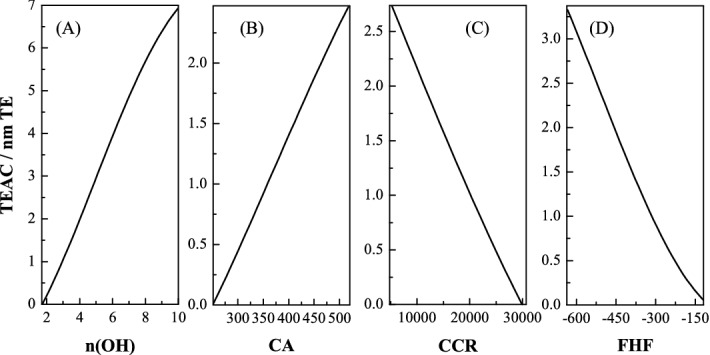
TEAC vs n(OH), CA, FHF and CCR by SA.
Discussion
The QSAR model based on SVR
In LOOCV test, SVR is superior to ANN and MLR. In the test set, the prediction ability of SVR is better than that of MLR, and is basically equal to that of ANN. From the result of residual error, SVR also shows good stability of prediction ability. However, the selection of kernel function and the optimization of parameters in SVR modelling were more time-consuming than ANN and MLR. There may be other more suitable parameters outside the scope of the gridding parameter selection. However, SVR is still a kind of regression method with higher accuracy, and it can be used for the establishment and analysis of QSAR models. In the future, further algorithm optimization can be carried out to shorten the kernel function selection and grid parameter selection process.
The relationship between TEAC and molecular descriptors
Sensitivity analysis in the SVR-QSAR model had shown that four characteristic parameters significantly affect the TEAC of phenolic compounds (Fig. 3).
Based on the hydrogen transfer mechanisms in the antioxidant process, an increase in hydroxyl groups means more hydrogen atoms that can be transferred, thereby increasing the TEAC value39. Core-Core Repulsion is relevant to molecular size, the shorter bond length means the larger CCR value. Some studies have shown that changes in CCR value affect the rate of intermolecular reactions40,41. In this work, the lower CCR value was beneficial to increased antioxidant activity of phenolic compounds. As for the Final heat of formation value, which reflects the stability of the molecule, a more stable molecule lead to lower antioxidant activity. The effect of Cosmo Area on TEAC is opposite to that of Core-Core Repulsion, large Cosmo Area lead to better antioxidant activity.
Compare the previous similar research based on the DFT parameters (minimum bond dissociation enthalpy (BDE(min)), HOMO and LUMO energies of the neutral species, ionization potential (IP), and dipole moment of the neutral species)42,43. This work reveals the potential modelling and prediction capabilities of the model use parameters obtained by Semi-empirical Hamiltonians, which is more time-efficient.
Applicability domain analysis
If a QSAR model is to be used for screening new compounds, the domain of application of this QSAR model must be defined28. The leverage hi of a compound can be used for judging the compound is in the domain or not, which is defined as follows:
| 4 |
where xi is the descriptor vector of the considered compound and x is the descriptor matrix derived from the training set. The superscript T refers to the transpose of the matrix/vector. The warning leverage h* is fixed at 3(p + 1)/n, where n is the number of training compounds and p is the number of model parameters. In this model, the value of h* is 0.263. A leverage greater than the warning leverage h* means that the predicted response may not be reliable.
The plot of leverage and standard residuals for the SVR-QSAR model is shown in Fig. 4. As shown in the Williams plot (Fig. 4), hi values of all the compounds in the training and test sets are lower than the warning value (h* = 0.263). The training set has great representativeness, and none of the compounds is particularly influential in the model space. The standardized residual of compound number 12 was slightly larger than three standard deviation units (3 s), which may be due to its different antioxidant activity mechanism.
Figure 4.
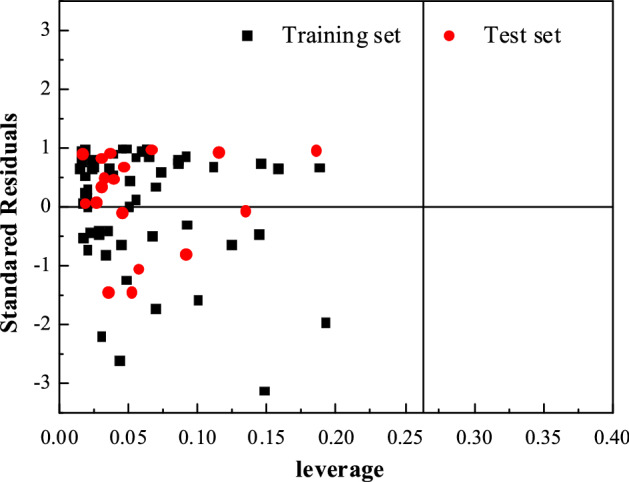
The plot of standardized residuals vs leverage, with a warning leverage of h* = 0.263.
Conclusions
In this study, an SVR-QSAR model of 75 phenolic compounds TEAC values was developed. The Pearson correlation coefficient method was employed in the parameter selection process in QSAR model development. Satisfactory prediction results were obtained using four parameters calculated by Semi-empirical Hamiltonians PM7. Although the SVR-QSAR model shows good stability of prediction ability, the SVR still has some shortcomings, such as selecting kernel function and the optimization of modeling parameters were more time-consuming than ANN and MLR. There may be other more suitable parameters outside the scope of the gridding parameter selection. Continuous optimization algorithms can be used in the future to reduce the time-consuming of the SVR-modelling process. Gupta et al. have done a series of work in this field44–47, they proposed a new unconstrained convex minimization problem formulation equivalent to the Lagrangian dual of the 2-norm twin support vector regression (TSVR), using the proposed formulation on synthetic and real-world datasets demonstrates a significant increase in learning speed with better accuracy in performance in accordance with the classical support vector regression and twin support vector regression47. Therefore, in order to obtain a better and faster SVR model in the subsequent work, it is necessary to continuously optimize the algorithm.
Supplementary Information
Acknowledgements
Many thanks to Prof. Lu Wen-Cong at the Department of Chemistry, College of Sciences, Shanghai University (Shanghai, China) for their excellent ExpMiner software.
Author contributions
Y.S. designed, experimented, statistically analyzed and wrote the manuscript.
Funding
This research was funded by the Natural Science Foundation of China Research, Grant Number 22063006, and the Foundation of Baotou Teachers’ College for High-Level Talents Introduction Grant Number 01108022/023.
Competing interests
The author declares no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88341-1.
References
- 1.Burda S, Oleszek W. Antioxidant and antiradical activities of flavonoids. J. Agric. Food. Chem. 2001;49(6):2774–2779. doi: 10.1021/jf001413m. [DOI] [PubMed] [Google Scholar]
- 2.Soicke H, Leng-Peschlow E. Characterisation of flavonoids from Baccharis trimera and their antihepatotoxic properties. Planta. Med. 1987;53(1):37–39. doi: 10.1055/s-2006-962613. [DOI] [PubMed] [Google Scholar]
- 3.Deschner EE, Ruperto J, Wong G, Newmark HL. Quercetin and rutin as inhibitors of azoxymethanol-induced colonic neoplasia. Carcinogenesis. 1991;12(7):1193–1196. doi: 10.1093/carcin/12.7.1193. [DOI] [PubMed] [Google Scholar]
- 4.Landolfi R, Mower RL, Steiner M. Modification of platelet function and arachidonic acid metabolism by bioflavonoids. Structure-activity relations. Biochem. Pharmacol. 1984;33(9):1525–1530. doi: 10.1016/0006-2952(84)90423-4. [DOI] [PubMed] [Google Scholar]
- 5.Carvalho JC, Ferreira LP, da Silva Santos L, Corrêa MJ, de Oliveira CLM, Bastos JK, Sarti SJ. Anti-inflammatory activity of flavone and some of its derivates from Virola Michelli Heckel. J. Ethnopharmacol. 1999;64(2):173–177. doi: 10.1016/S0378-8741(98)00109-3. [DOI] [PubMed] [Google Scholar]
- 6.Wang MY, Ma ZL, He CL, Yuan XY. The Antioxidant activities of flavonoids in jerusalem artichoke (Helianthus Tuberosus L.) leaves and their quantitative analysis. Nat. Prod. Res. 2020;20:1–5. doi: 10.1080/14786419.2020.1839464. [DOI] [PubMed] [Google Scholar]
- 7.Zeng Y, Song J, Zhang M, Wang H, Zhang Y, Suo H. Comparison of in vitro and in vivo antioxidant activities of six flavonoids with similar structures. Antioxidants. 2020;9(8):732–746. doi: 10.3390/antiox9080732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao X, Chen R, Shi Y, Zhang X, Tian C, Xia D. Antioxidant and anti-inflammatory activities of six flavonoids from Smilax Glabra Roxb. Molecules. 2020;25(22):5295–5318. doi: 10.3390/molecules25225295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nenadis N, Wang LF, Tsimidou M, Zhang HY. Estimation of scavenging activity of phenolic compounds using the ABTS(*+) Assay. J. Agric. Food. Chem. 2004;52(15):4669–4674. doi: 10.1021/jf0400056. [DOI] [PubMed] [Google Scholar]
- 10.Borgohain R, Handique JG, Guha AK, Pratihar S. A Theoretical study on antioxidant activity of ferulic acid and its ester derivatives. J. Theor. Comput. Chem. 2016;15(4):1650028–1650046. doi: 10.1142/S0219633616500280. [DOI] [Google Scholar]
- 11.Villaño D, Fernández-Pachón MS, Troncoso AM, García-Parrilla MC. Comparison of antioxidant activity of wine phenolic compounds and metabolites in vitro. Anal. Chim. Acta. 2005;538(1):391–398. doi: 10.1016/j.aca.2005.02.016. [DOI] [Google Scholar]
- 12.Heim KE, Tagliaferro AR, Bobilya DJ. Flavonoid antioxidants: chemistry: Metabolism and structure-activity relationships. J. Nutr. Biochem. 2002;13(10):572–584. doi: 10.1016/S0955-2863(02)00208-5. [DOI] [PubMed] [Google Scholar]
- 13.Rice-Evans CA, Miller NJ, Paganga G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free. Radic. Biol. Med. 1996;20(7):933–956. doi: 10.1016/0891-5849(95)02227-9. [DOI] [PubMed] [Google Scholar]
- 14.Seyoum A, Asres K, El-Fiky FK. Structure-radical scavenging activity relationships of flavonoids. Phytochemistry. 2006;67(18):2058–2070. doi: 10.1016/j.phytochem.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Rackova L, Firakova S, Kostalova D, Stefek M, Sturdik E, Majekova M. Oxidation of liposomal membrane suppressed by flavonoids: Quantitative structure-activity relationship. Bioorg. Med. Chem. 2005;13(23):6477–6484. doi: 10.1016/j.bmc.2005.07.047. [DOI] [PubMed] [Google Scholar]
- 16.Farkas O, Jakus J, Héberger K. Quantitative structure-antioxidant activity relationships of flavonoid compounds. Molecules. 2004;9(12):1079–1088. doi: 10.3390/91201079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katritzky AR, Kuanar M, Slavov S, Hall CD, Karelson M, Kahn I, Dobchev DA. Quantitative correlation of physical and chemical properties with chemical structure: Utility for prediction. Chem. Rev. 2010;110(10):5714–5789. doi: 10.1021/cr900238d. [DOI] [PubMed] [Google Scholar]
- 18.Fernández M, Caballero J, Helguera EA, González MP, González MP. Quantitative structure-activity relationship to predict differential inhibition of aldose reductase by flavonoid compounds. Bioorg. Med. Chem. 2005;13:3269–3277. doi: 10.1016/j.bmc.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 19.Žuvela P, David J, Wong MW. Interpretation of ANN-based QSAR models for prediction of antioxidant activity of flavonoids. J. Comput. Chem. 2018;39(16):953–963. doi: 10.1002/jcc.25168. [DOI] [PubMed] [Google Scholar]
- 20.Niu B, Lu WC, Yang SS, Cai YD, Li GZ. Support vector machine for SAR/QSAR of phenethyl-amines. Acta. Pharmacol. Sin. 2007;28(7):1075–1086. doi: 10.1111/j.1745-7254.2007.00573.x. [DOI] [PubMed] [Google Scholar]
- 21.Djeradi H, Rahmouni A, Cheriti A. Antioxidant activity of flavonoids: A QSAR modeling using fukui indices descriptors. J. Mol. Model. 2014;20(10):2476–2485. doi: 10.1007/s00894-014-2476-1. [DOI] [PubMed] [Google Scholar]
- 22.Inci CAY, Serap C, Omca D, Muhammed KU, Demirkol A. Estimation of antioxidant activity of foods using artificial neural networks. J. Food. Nutr. Res. 2017;56(2):138–148. [Google Scholar]
- 23.Li Z, Nie K, Wang Z, Luo D. Quantitative structure activity relationship models for the antioxidant activity of polysaccharides. PLoS ONE. 2016;11(9):e0163536. doi: 10.1371/journal.pone.0163536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fatemi MH, Rostami EG. Prediction of the radical scavenging activities of some antioxidant from their molecular structure. Ind. Eng. Chem. Res. 2013;52(28):9525–9531. doi: 10.1021/ie4001426. [DOI] [Google Scholar]
- 25.Mei H, Zhou Y, Liang G, Li ZL. Support vector machine applied in QSAR modelling. Chin. Sci. Bull. 2005;50(20):2291–2296. doi: 10.1007/BF03183737. [DOI] [Google Scholar]
- 26.Huang M, Wei Y, Wang J, Zhang Y. Support vector regression-guided unravelling: Antioxidant capacity and quantitative structure-activity relationship predict reduction and promotion effects of flavonoids on acrylamide formation. Sci. Rep. 2016;6(1):32368–32382. doi: 10.1038/srep32368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, Li M, Su Q, Wu M, Gu T, Lu W. QSAR studies on pyrrolidine amides derivatives as DPP-IV inhibitors for type 2 diabetes. Med. Chem. Res. 2013;22(11):5274–5283. doi: 10.1007/s00044-013-0527-2. [DOI] [Google Scholar]
- 28.Darnag R, Minaoui B, Fakir M. QSAR models for prediction study of HIV protease inhibitors using support vector machines, neural networks and multiple linear regression. Arab. J. Chem. 2017;10(1):S600–S608. doi: 10.1016/j.arabjc.2012.10.021. [DOI] [Google Scholar]
- 29.Hegde G, Bowen RC. Machine-learned approximations to density functional theory hamiltonians. Sci. Rep. 2017;7:42669–42680. doi: 10.1038/srep42669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cojocaru C, Airinei A, Fifere N. Molecular structure and modeling studies of azobenzene derivatives containing maleimide groups. Springerplus. 2013;2(1):586–605. doi: 10.1186/2193-1801-2-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunn, S. R. Support Vector Machines for Classification and Regression. Department of Electronics and Computer Science, University of Southampton. May 14. Report No.: ISIS-1-98 (1998).
- 32.Chou KC, Zhang CT. Prediction of protein structural classes. Crit. Rev. Biochem. Mol. Biol. 1995;30(4):275–349. doi: 10.3109/10409239509083488. [DOI] [PubMed] [Google Scholar]
- 33.Schulerud H, Albregtsen F. Many are called, but few are chosen feature selection and error estimation in high dimensional spaces. Comput. Methods. Programs. Biomed. 2004;73(2):91–99. doi: 10.1016/S0169-2607(03)00018-X. [DOI] [PubMed] [Google Scholar]
- 34.Errico RM, Vukicevic T. Sensitivity analysis using an adjoint of the PSU-NCAR mesoseale model. Mon. Weather. Rev. 1992;120(8):1644–1660. doi: 10.1175/1520-0493(1992)120<1644:SAUAAO>2.0.CO;2. [DOI] [Google Scholar]
- 35.Cacuci DG. Sensitivity theory for nonlinear systems. I. Nonlinear functional analysis approach. J. Math. Phys. 1981;22(12):2794–2802. doi: 10.1063/1.525186. [DOI] [Google Scholar]
- 36.Eriksson L, Jaworska J, Worth AP, Cronin MT, McDowell RM, Gramatica P. Methods for reliability and uncertainty assessment and for applicability evaluations of classification- and regression-based QSARs. Environ. Health Perspect. 2003;111(10):1361–1375. doi: 10.1289/ehp.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gramatica P. Principles of QSAR models validation: Internal and external. QSAR Comb. Sci. 2007;26(5):694–701. doi: 10.1002/qsar.200610151. [DOI] [Google Scholar]
- 38.Cai YZ, Mei S, Jie X, Luo Q, Corke H. Structure-radical scavenging activity relationships of phenolic compounds from traditional Chinese medicinal plants. Life. Sci. 2006;78(25):2872–2888. doi: 10.1016/j.lfs.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Huang D, Ou B, Prior RL. The chemistry behind antioxidant capacity assays. J. Agric. Food. Chem. 2005;53(6):1841–1856. doi: 10.1021/jf030723c. [DOI] [PubMed] [Google Scholar]
- 40.Long X, Niu J. Estimation of gas-phase reaction rate constants of alkylnaphthalenes with chlorine, hydroxyl and nitrate radicals. Chemosphere. 2007;67(10):2028–2034. doi: 10.1016/j.chemosphere.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 41.Eddy NO, Momoh-Yahaya H, Oguzie EE. Theoretical and experimental studies on the corrosion inhibition potentials of some purines for aluminum in 0.1 m HCl. J. Adv. Res. 2015;6(2):203–217. doi: 10.1016/j.jare.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amić D, Davidović-Amić D, Beslo D, Rastija V, Lucić B, Trinajstić N. SAR and QSAR of the antioxidant activity of flavonoids. Curr. Med. Chem. 2007;14(7):827–845. doi: 10.2174/092986707780090954. [DOI] [PubMed] [Google Scholar]
- 43.Tafazoli S, Wright JS, O'Brien PJ. Prooxidant and antioxidant activity of vitamin E analogues and troglitazone. Chem. Res. Toxicol. 2005;18(10):1567–1574. doi: 10.1021/tx0500575. [DOI] [PubMed] [Google Scholar]
- 44.Gupta D. Training primal K-nearest neighbor based weighted twin support vector regression via unconstrained convex minimization. Appl. Intell. 2017;47:962–991. doi: 10.1007/s10489-017-0913-4. [DOI] [Google Scholar]
- 45.Balasundaram S, Gupta D. Training Lagrangain twin support vector regression via unconstrained convex minimization. Knowl. Based Syst. 2014;59:85–96. doi: 10.1016/j.knosys.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 46.Balasundaram S, Gupta D. On optimization based extreme learning machine in primal for regression and classification by functional iterative method. Int. J. Mach. Learn. Cyber. 2016;7:707–728. doi: 10.1007/s13042-014-0283-8. [DOI] [Google Scholar]
- 47.Balasundaram S, Gupta D, Kapil S. Lagrangain support vector regression via unconstrained convex minimization. Neural. Netw. 2014;51:67–79. doi: 10.1016/j.neunet.2013.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.