Abstract
Phages are versatile agents for delivering a variety of cargo, including nanomaterials, nucleic acids, and small molecules. A potentially important application is treatment of antibiotic-resistant infections. All of these applications require molecular engineering of the phages, including chemical modification and genetic engineering. Phages are remarkably amenable to such engineering. We review some examples, including for controlled phage therapy. We suggest that the ability of phages to support extensive engineering may have evolutionary origins in the billions-year-old 'arms race' between bacteria and phages, which selects for sequences and structures that are robust in the face of rapid evolutionary change. This leads to high tolerance of both naturally evolved mutations and synthetic molecular engineering.
Keywords: Phage, phage therapy, arms race, red queen, robustness
Graphical abstract
Introduction
Phages are naturally evolved nanocarriers for delivering DNA and RNA. Virion production, survival, and attachment to host cells are all under selection to ensure phage propagation. These same features, along with ease of production in bacterial cell culture, make phages highly attractive as delivery vehicles for biomedical applications [1, 2]. As phage therapy returns to the attention of researchers concerned by the growing crisis of antibiotic-resistant infections, so do the associated biosafety concerns. Although phages are incapable of infecting eukaryotic cells, they can alter the properties of a bacterial infection and have public health implications through effects on their bacterial hosts. While, in an ideal situation, phages would lyse their bacterial hosts without other effects, many phages that infect pathogens have problematic behaviors from a clinical perspective. Phages that adopt a temperate rather than strictly lytic life cycle integrate into the host chromosome, enabling them to mediate gene transfer among bacterial cells through transduction. For example, phages of S. aureus transfer antibiotic resistance genes among their hosts [3]. Phages sometimes carry virulence factors, like phage CTXϕ, which encodes and transmits a toxin gene that renders V. cholerae pathogenic to humans [4]. In addition to genetic effects, phage particles can contribute physically to pathological infections, such as phage Pf4, whose virions create liquid crystalline biofilm compartments surrounding P. aeruginosa cells, increasing tolerance to antibiotics [5]. Moreover, Pf4 phage can trigger a maladaptive viral immune response, which suppresses clearance of the bacterial infection [6]. Because of the wide diversity of phages and the fact that the majority of phage sequences are uncharacterized, these negative effects are difficult to anticipate. Such concerns are intrinsic to a biocontrol strategy, namely that the use of a complex, evolving organism to eradicate another creates unpredictability in outcomes. While these risks may be acceptable in extreme circumstances [7], such unpredictability is undesirable in most therapeutic situations [8].
Several strategies can be used to avoid the potential problems, including engineering well-studied phages to have the desired tropism, adapting isolated phage proteins with lytic properties rather than whole phages, and engineering phages that 'self-destruct' through controllable nanomaterials [9]. These strategies raise the interesting issue of how amenable and robust phages are to engineering efforts. In the first section of this article, we review some illustrative examples of ways in which model phages are engineered to be delivery vehicles. These include a controlled version of phage therapy, in which phages would be enhanced by genetic or chemical controls to reduce potential side effects. Then, we reflect on why phages appear to be so tolerant to molecular engineering, a crucial element of any phage-based therapy.
Engineering phages as delivery vehicles
Phages attach to bacterial host cells through receptor-binding proteins (RBPs) present on the virion particles, which have high affinity for a bacterial surface receptor and contribute to host specificity. If the therapeutic target is a bacterial species, RBPs themselves can be used for targeting. However, phages are also considered as delivery vehicles more broadly (e.g., to cancer cells), as the RBPs can be replaced or genetically engineered to bind the desired receptor. For example, a heterologous targeting peptide can be inserted as a fusion to a capsid protein. The targeting peptides might be derived from biological interactions, such as antibodies, or evolved from a library of random peptides by in vitro selection against the desired receptor (phage display). Cargo, such as nucleic acids, nanomaterials, therapeutic drugs, and diagnostic probes, can be attached through physical interaction or chemical conjugation with capsid proteins. Each virion contains hundreds to thousands of copies of capsid proteins, creating a large carrying potential. Solvent-accessible reactive groups, such as amino and carboxyl groups, can be utilized for functionalization [10, 11]. Some examples of phages engineered for cargo delivery are given in Table 1, illustrating a breadth of scaffolds, cargo, loading chemistry, and targeting applications.
Table 1.
Examples of phages engineered for cargo delivery.
| Phage/Component | Cargo | Loading technique | Targeting | Application | Reference |
|---|---|---|---|---|---|
| M13 | Gold nanorods | Thiolation of g8p by chemical modification to form Au-S bonds | Bacterial receptor-binding proteins displayed on g3p | Photothermal lysis of P. aeruginosa biofilms, controlled phage therapy | [9] |
| fd–tet phage displaying 9-mer peptide | Small interfering RNA (siRNA) | Interaction of positively charged C-terminal end and negatively charged phosphate of siRNA | Tumor-specific peptides isolated from phage library | Delivery to breast cancer cells, leading to specific silencing of the model gene GAPDH | [44] |
| T7 | Fluorescent protein | Displayed as fusion to capsid protein by phage display | Gold-binding-peptide displayed on the tail protein; gold nanoparticles decorated with specific oligonucleotide | Counting miRNA by the naked eye in a Petri dish | [45] |
| M13 | Exogenous proteins | Biotin conjugation to capsid; streptavidin fusion to cargo | Prostate cancer cell-penetrating peptide displayed on g3p | Delivery of functional imaging protein and enzymes | [46] |
| P22 capsid | GdIII-chelating agents | Thiol-maleimide Michael-type addition | Passive targeting | Visualizing murine intravascular system by in vivo magnetic resonance imaging | [47] |
| Lambda | Mammalian expression cassette encoding firefly luciferase | Genetic engineering | An integrin-binding peptide displayed on phage | In vivo lambda phage-mediated gene delivery and expression | [48] |
| MS2 capsid | Capsids radiolabeled with 64Cu | Antibody conjugated to exterior by nitrophenol–amine reaction; cargo loaded by metal chelator maleimide reaction with internal cysteines | Anti-EGFR antibody targeting breast cancer cells | In vivo PET/CT imaging of tumor xenografts in mice | [49] |
Molecular engineering of phages tends to focus on a few model phages that infect E. coli, likely due to a paucity of established cloning techniques for other bacterial organisms and their phages. One of the most extensively investigated nanocarriers for controlled delivery and release is filamentous phages of Inoviridae, rod-shaped viruses including f1, M13 and fd (which are nearly identical to each other) [12, 13]. They are amenable to chemical modification, as the amino groups from lysine residues, the carboxylic acid groups of acidic residues, and the aromatic groups of tyrosine residues of the major coat protein (pVIII) are potentially accessible and reactive. For example, carboxylic acid residues near the N-terminus (Glu2, Asp4, and Asp5) may be modified through EDC chemistry [14, 15]. Alternatively, primary amines of pVIII can be modified (e.g., by N-succinimidyl-S-acetylthiopropionate for thiolation); this approach has fewer reactive sites but might give more evenly distributed modification across the virion surface [9]. While chemical modification is simple, efficient and straightforward, it should be kept in mind that such treatments might also cause crosslinking among virions, denature proteins or alter cargo properties. Whether this occurs in practice must be seen on a case-by-case basis, but phages themselves appear to be remarkably tolerant to chemical modification.
Filamentous phages are also readily genetically engineered using well-established cloning methods [16]. By exchanging the receptor-binding domain of M13 (pIII) for that from a phage that naturally targets a different bacterial host, M13 can be engineered as chimeras to attach to a variety of different hosts, including the pathogens P. aeruginosa and V. cholerae [14, 15]. When coupled to gold nanorods (AuNRs), the chimeric phages can differentially kill specific strains of bacteria, as excitation of the AuNRs by near-infrared light induces surface plasmon resonance, whose energy is released locally as heat [17]. This effect (photothermal ablation) kills both cells and phages, equipping a well-studied phage with a 'self-destruct' strategy and the appropriate tropism to achieve controlled therapy [9]. Thus, use of a model E. coli phage need not limit the target range in certain applications. In addition, while the self-amplifying nature of phages could appear, at first glance, to be an attractive feature for phage therapy, self-replication may actually be undesirable due to the potential for unpredictable dynamics and interactions with the bacterial (or human) host [9]. A model E. coli phage may therefore have the further advantage of preventing replication on non-E. coli hosts. While phages are sometimes said to be very specific, in reality they vary in degree of specificity [18], depending on the level of conservation of the bacterial receptors and other interacting components. For example, phages capable of infecting multiple classes within the phylum Proteobacteria appear to be relatively common among environmental isolates [19], raising the possibility that non-model phages may have unexpected hosts. Such considerations compel development of strategies for controlled phage therapy based on well-studied model phages.
Genetic engineering, including phage display technology, can also be used to deliver cargo such as peptides, vaccines, and small molecules [20, 21]. For example, M13 can display collagen mimetic peptide on pIII along with a streptavidin-binding peptide on pVIII [22], to deliver streptavidin-linked fluorescent agents to the abnormal collagens expressed on human lung adenocarcinoma cells. Furthermore, filamentous phages are highly immunogenic, possibly due to the repeated structure of pVIII proteins and the relatively large dimensions. Therefore, whole phages can be used to enhance the immune response for vaccine delivery, and the vaccine itself may be stabilized against environmental damage by the phage-like particles [23]. Antigens may be expressed through genetic engineering [22] or by bioconjugation to the surface of phage particles [24]. At the same time, recent findings illustrating mechanisms by which another filamentous phage, Pf4, enhances the virulence of its host, P. aeruginosa [5,6], emphasize the importance of implementing multiple levels of control over the phage, such as self-destruction by nanomaterials [15] and cross-species barriers that prevent completion of a replication cycle on the targeted pathogenic species.
Another prominent model phage is MS2, which has very small virions (~27 nm in diameter) with a symmetric icosahedral capsid containing 180 copies of the coat protein [25]. Unlike the ssDNA-containing filamentous phages, MS2 phages contain a single-stranded RNA genome (family Leviviridae), making genetic engineering less straightforward [26]. However, MS2 capsids have 32 pores that allow access to the inside of the capsid [27], enabling chemical modification of the interior as well as exterior surfaces. For example, using orthogonal bioconjugation chemistry, the exterior can be modified with a targeting reagent while the cargo is attached to the interior. In one example, fluorescent dyes were attached to the interior of genome-free MS2 capsids by site-specific alkylation, while the outer surface was decorated with DNA aptamers by a NaIO4-mediated oxidative coupling reaction [28]. The resulting particles could bind to tyrosine kinase receptors on a cancer cell line and were readily endocytosed. For a cytotoxic application, the MS2 interior was modified with porphyrins that generate singlet oxygen upon illumination, such that the capsids could deliver photodynamic therapy to targeted cancer cells [29]. Peptides are commonly used for targeting, such as peptide SP94, which enables selective delivery of a variety of cargo to human hepatocellular carcinoma cells [30]. Since the natural cargo of MS2 phage is single-stranded RNA, it is also a promising nanocarrier for RNA delivery [31, 32]. In one case, engineered MS2 particles activated osteoblast differentiation by delivering mRNA for transcription factors into bone-marrow mesenchymal stem cells [33], illustrating how MS2 can be considered for delivery of a transient, though limited, load for gene therapy.
When delivery of large amounts of genetic cargo is needed, one must turn to larger phages [34]. Among these is lambda phage, whose temperate lifestyle became a classic subject of study during the early days of molecular biology. Lambda phages contain a double-stranded DNA genome of 48,502 bp and have the 'lunar lander' shape characteristic of the large taxon Caudovirales, including an icosahedral head, tail, and tail nanofibers [35]. An interesting potential application is DNA vaccine delivery [36, 37], although whether phages possess sufficient advantages over eukaryotic viruses for gene delivery to mammalian cells is unclear. Nevertheless, gene delivery to prokaryotic cells could be a natural niche. For applications that require peptide display rather than gene transfer, lambda phages are particularly useful for large peptides [38, 39], where expression can be two to three orders of magnitude more efficient than M13-based vectors [40, 41]. This difference stems from the abundance of the coat proteins capable of accommodating large peptides; protein gpD on lambda phage is present in ~420 copies per virion, while g3p on M13 phage is present at ~5 copies per virion (with g8p on M13 being essentially unable to display peptides longer than 16 aa) [42]. For example, lambda phage displaying a HER2/neu peptide on the capsid activated cytotoxic T-lymphocytes against a murine breast cancer model [43]. Thus, lambda phage may be useful as delivery agents for larger cargo including both genes and antigens. Other Caudovirales phages, including T4, T7, and P22, are not reviewed here, but have also been used for display and engineering.
Evolutionary robustness and molecular engineering
Given the examples above, phages appear to be remarkably easy to modify, both by genetic modification such as creation of fusions to capsid proteins, or by chemical modification such as bioconjugation to small molecules or nanoparticles. In our own work [9, 14, 15], we have been struck by the fact that every chimeric capsid protein we created resulted in a functional construct, despite the fact that the proteins had no detectable homology to each other. The fact that virions retain the ability to assemble and attach after quite substantial perturbations, including decoration by both macromolecules and small molecules, points toward high robustness of these activities during engineering. This robustness is quite different from, for example, the challenges encountered when engineering antibodies, another class of biomolecules that is under analogously strong selection for attachment [50, 51].
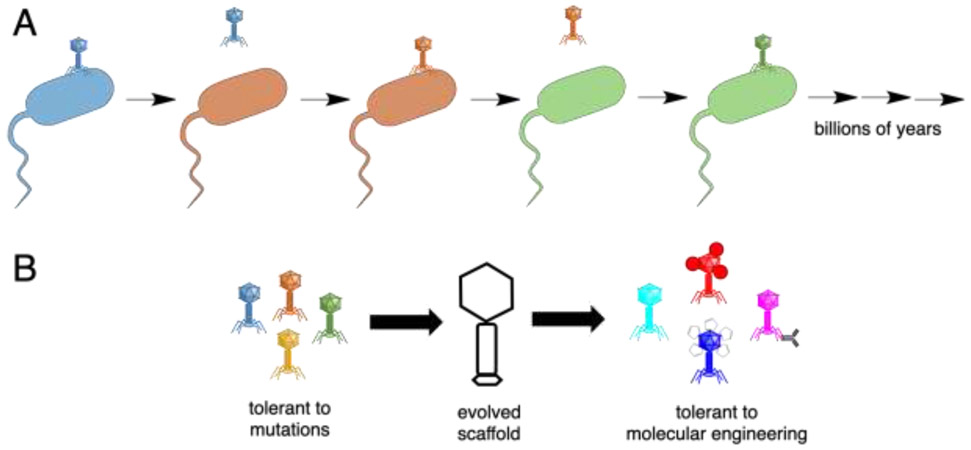
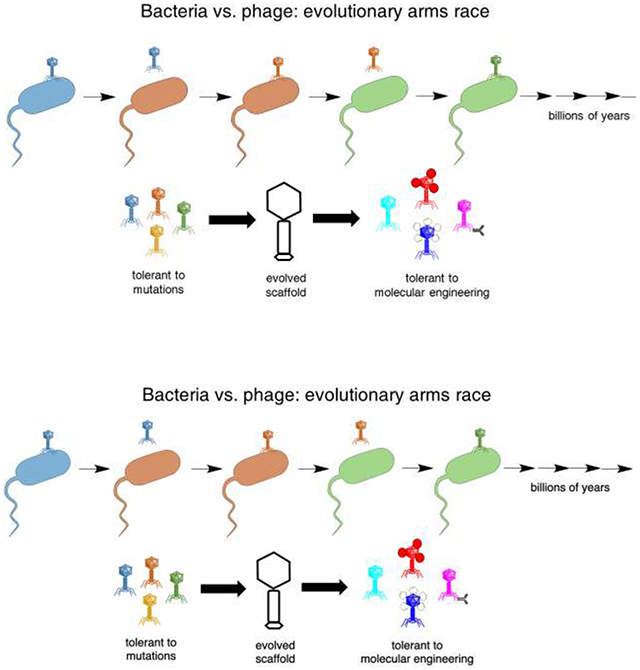
What might drive such robustness to engineering? Robustness itself is a trait under selection. In RNA viruses, which exhibit very high mutation rates (nearly one mutation per generation), strains selected for environmental robustness, such as tolerance of high temperatures, also exhibit increased tolerance of mutations, i.e., increased evolvability [52, 53]. At a molecular level, protein folds that are more environmentally stable appear to be also more stable in the face of genetic changes. Phages are under direct selection for environmental stability, as they must survive the extracellular environment and its thermal, chemical, and physical fluctuations. On top of this, phage proteins are under further selection to tolerate mutations. As bacteria evolve resistance to phages, and phages evolve to counter resistance, the 'evolutionary arms race' between phage and bacteria drives positive selection resulting in rapid evolution and divergence (Figure 1). However, the proteins must fold into similar structures to maintain the virion, despite the rapid pace of evolutionary change. Thus, the protein scaffolds must have evolved to be tolerant to mutations. Yet genetic mutations are only one type of chemical modification. These selective pressures appear to have also led to tolerance of chemical modifications more generally, such as those used for loading cargo, including fairly imprecise, widespread modifications, and to fusion proteins and conjugation to large structures such as nanoparticles. Selection for general adaptability to novel conditions has also been observed in viral adaptation to rapid environmental changes [54]. Thus, natural selection appears to have driven phages to exhibit traits that are particularly favorable for molecular engineering.
Figure 1.
The evolutionary arms race between bacteria and phage drives a rapid evolutionary pace (A). The resulting phage scaffolds were presumably selected for tolerance to mutations. A correlated trait is tolerance to other chemical modifications, such as those used in molecular engineering (B).
Conclusion
While phages have important advantages as delivery vehicles, especially when targeting bacterial cells (e.g., controlled phage therapy), important challenges remain. Circulation times may be short as the phages are cleared by the reticuloendothelial system, although PEGylation of the phage particles and genetic engineering can extend these times [55, 56]. Certain phages may have negative consequences for the human host, such as by inappropriate stimulation of an antiviral response [6] or through release of endotoxins [57], or may lead to undesired gene exchange among bacteria, as discussed above. Controlled phage therapy is needed to prevent such consequences, either by directly managing dosing [9], or engineering a lysis-deficient phage [58-60] or a lysogenic phage that expresses lethal but non-lytic proteins [61]. There is reason for optimism that these challenges can be met by molecular engineering, given the extraordinarily robust and tolerant architecture of phages resulting from billions of years of an evolutionary arms race. While phage scaffolds may be tolerant to engineering, genetic engineering can be technically quite challenging with phages and is itself a rapidly growing area of research [62, 63]. An increasing body of work demonstrates the versatility of phages for carrying diverse cargos while targeting can be engineered essentially orthogonally. Continuing successes in engineering phages paves the way for realizing the long-held dream of safely utilizing the exquisitely honed mechanisms phages possess to 'hunt' their bacterial hosts.
Highlights.
Phages can target delivery of cargo, including nanomaterials, drugs, and genes
Molecular engineering is essential to applications such as controlled phage therapy
The phage-bacteria arms race causes rapid evolution and tolerance of mutations
Phages are therefore remarkably tolerant of chemical modification in general
Acknowledgments:
This work was supported by the National Institutes of Health (DP2 GM123457-01) and the Camille Dreyfus Teacher-Scholar Awards Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Nothing declared.
References
- 1.Westwater C, Kasman LM, Schofield DA, Werner PA, Dolan JW, Schmidt MG, and Norris JS (2003). Use of Genetically Engineered Phage To Deliver Antimicrobial Agents to Bacteria: an Alternative Therapy for Treatment of Bacterial Infections. Antimicrobial Agents and Chemotherapy 47, 1301–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dąbrowska K (2019). Phage therapy: What factors shape phage pharmacokinetics and bioavailability? Systematic and critical review. Medicinal Research Reviews 39, 2000–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haaber J, Leisner JJ, Cohn MT, Catalan-Moreno A, Nielsen JB, Westh H, Penades JR, and Ingmer H (2016). Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nature communications 7, 13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waldor MK, and Mekalanos JJ (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914. [DOI] [PubMed] [Google Scholar]
- 5.Tarafder AK, von Kugelgen A, Mellul AJ, Schulze U, Aarts D, and Bharat TAM (2020). Phage liquid crystalline droplets form occlusive sheaths that encapsulate and protect infectious rod-shaped bacteria. Proc Natl Acad Sci U S A 117, 4724–4731.• • Filamentous phages are associated with the biofilms of the prominent lung and wound pathogen P. aeruginosa. This work shows that the virions increase antibiotic resistance by physically encasing P. aeruginosa cells in a protective sheath. This illustrates how some phages enhance bacterial fitness, a concern for phage therapy that must be addressed through molecular engineering.
- 6.Sweere JM, Van Belleghem JD, Ishak H, Bach MS, Popescu M, Sunkari V, Kaber G, Manasherob R, Suh GA, Cao X, et al. (2019). Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 363, eaat9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearson DE, and Callaway RM (2005). Indirect nontarget effects of host-specific biological control agents: Implications for biological control. Biological Control 35, 288–298. [Google Scholar]
- 8.Hesse S, and Adhya S (2019). Phage Therapy in the Twenty-First Century: Facing the Decline of the Antibiotic Era; Is It Finally Time for the Age of the Phage? Annual Review of Microbiology 73, 155–174.• This authoritative review focuses attention on recent advances and practical challenges of phage therapy.
- 9.Peng H, Borg RE, Dow LP, Pruitt BL, and Chen IA (2020). Controlled phage therapy by photothermal ablation of specific bacterial species using gold nanorods targeted by chimeric phages. Proceedings of the National Academy of Sciences 117, 1951–1961.• • M13 was engineered to attach to gold nanorods and to target P. aeruginosa. Activation of the nanorods treated P. aeruginosa biofilms through intense localized heating. By using the M13 scaffold and destroying phages after use, this strategy represents a controllable version of phage therapy.
- 10.Mohan K, and Weiss GA (2016). Chemically Modifying Viruses for Diverse Applications. ACS Chem Biol 11, 1167–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernard JML, and Francis MB (2014). Chemical strategies for the covalent modification of filamentous phage. Frontiers in microbiology 5, 734–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sunderland KS, Yang M, and Mao C (2017). Phage-Enabled Nanomedicine: From Probes to Therapeutics in Precision Medicine. Angew Chem Int Ed Engl 56, 1964–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ju Z, and Sun W (2017). Drug delivery vectors based on filamentous bacteriophages and phage-mimetic nanoparticles. Drug Deliv 24, 1898–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng H, Borg RE, Nguyen ABN, and Chen IA (2020). Chimeric Phage Nanoparticles for Rapid Characterization of Bacterial Pathogens: Detection in Complex Biological Samples and Determination of Antibiotic Sensitivity. Acs Sensors 5, 1491–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng H, and Chen IA (2019). Rapid Colorimetric Detection of Bacterial Species through the Capture of Gold Nanoparticles by Chimeric Phages. Acs Nano 13, 1244–1252.• Chimeric phages were engineered to bind to a variety of different bacteria and thiolated to bind to gold nanoparticles. Despite lack of homology among the phages, all chimeras designed were successful without further engineering, illustrating the robustness of phages to genetic and chemical perturbations.
- 16.Sambrook J (2001). Molecular cloning : a laboratory manual, (Third edition. Cold Spring Harbor, N.Y. : Cold Spring Harbor Laboratory Press, [2001] ©2001). [Google Scholar]
- 17.Murphy CJ, Chang H-H, Falagan-Lotsch P, Gole MT, Hofmann DM, Hoang KNL, McClain SM, Meyer SM, Turner JG, Unnikrishnan M, et al. (2019). Virus-Sized Gold Nanorods: Plasmonic Particles for Biology. Accounts of Chemical Research 52, 2124–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koskella B, and Meaden S (2013). Understanding bacteriophage specificity in natural microbial communities. Viruses 5, 806–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen EC, Schrader HS, Rieland B, Thompson TL, Lee KW, Nickerson KW, and Kokjohn TA (1998). Prevalence of Broad-Host-Range Lytic Bacteriophages of Sphaerotilus natans, Escherichia coli, andPseudomonas aeruginosa. Appl Environ Microbiol 64, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh D, Kohli AG, Moser F, Endy D, and Belcher AM (2012). Refactored M13 Bacteriophage as a Platform for Tumor Cell Imaging and Drug Delivery. ACS Synthetic Biology 1, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ngweniform P, Abbineni G, Cao BR, and Mao CB (2009). Self-Assembly of Drug-Loaded Liposomes on Genetically Engineered Target-Recognizing M13 Phage: A Novel Nanocarrier for Targeted Drug Delivery. Small 5, 1963–1969. [DOI] [PubMed] [Google Scholar]
- 22.Bazan J, Całkosiński I, and Gamian A (2012). Phage display--a powerful technique for immunotherapy: 1. Introduction and potential of therapeutic applications. Hum Vaccin Immunother, 8, 1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark JR, and March JB (2006). Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends Biotechnol 24, 212–218. [DOI] [PubMed] [Google Scholar]
- 24.van Houten NE, Zwick MB, Menendez A, and Scott JK (2006). Filamentous phage as an immunogenic carrier to elicit focused antibody responses against a synthetic peptide. Vaccine 24, 4188–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anobom CD, Albuquerque SC, Albernaz FP, Oliveira AC, Silva JL, Peabody DS, Valente AP, and Almeida FCL (2003). Structural studies of MS2 bacteriophage virus particle disassembly by nuclear magnetic resonance relaxation measurements. Biophys J 84, 3894–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Meerten D, Olsthoorn RCL, van Duin J, and Verhaert RMD (2001). Peptide display on live MS2 phage: restrictions at the RNA genome level. J Gen Virol 82, 1797–1805. [DOI] [PubMed] [Google Scholar]
- 27.Dedeo MT, Finley DT, and Francis MB (2011). Chapter 8 - Viral Capsids as Self-Assembling Templates for New Materials. In Progress in Molecular Biology and Translational Science, Volume 103, Howorka S, ed. (Academic Press; ), pp. 353–392. [DOI] [PubMed] [Google Scholar]
- 28.Tong GJ, Hsiao SC, Carrico ZM, and Francis MB (2009). Viral Capsid DNA Aptamer Conjugates as Multivalent Cell-Targeting Vehicles. Journal of the American Chemical Society 131, 11174–11178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephanopoulos N, Tong GJ, Hsiao SC, and Francis MB (2010). Dual-Surface Modified Virus Capsids for Targeted Delivery of Photodynamic Agents to Cancer Cells. Acs Nano 4, 6014–6020. [DOI] [PubMed] [Google Scholar]
- 30.Ashley CE, Carnes EC, Phillips GK, Durfee PN, Buley MD, Lino CA, Padilla DP, Phillips B, Carter MB, Willman CL, et al. (2011). Cell-Specific Delivery of Diverse Cargos by Bacteriophage MS2 Virus-like Particles. Acs Nano 5, 5729–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan Y, Zhang Y, Jia TT, Zhang K, Li JM, and Wang LN (2012). Development of a microRNA delivery system based on bacteriophage MS2 virus-like particles. Febs J 279, 1198–1208. [DOI] [PubMed] [Google Scholar]
- 32.Galaway FA, and Stockley PG (2013). MS2 Viruslike Particles: A Robust, Semisynthetic Targeted Drug Delivery Platform. Molecular Pharmaceutics 10, 59–68. [DOI] [PubMed] [Google Scholar]
- 33.Prel A, Caval V, Gayon R, Ravassard P, Duthoit C, Payen E, Maouche-Chretien L, Creneguy A, Nguyen TH, Martin N, et al. (2015). Highly efficient in vitro and in vivo delivery of functional RNAs using new versatile MS2-chimeric retrovirus-like particles. Molecular Therapy - Methods & Clinical Development 2, 15039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casjens SR, and Hendrix RW (2015). Bacteriophage lambda: Early pioneer and still relevant. Virology 479-480, 310–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu R, and Kaiser AD (1968). Structure and base sequence in the cohesive ends of bacteriophage lambda DNA. J Mol Biol 35, 523–537. [DOI] [PubMed] [Google Scholar]
- 36.Jepson CD, and March JB (2004). Bacteriophage lambda is a highly stable DNA vaccine delivery vehicle. Vaccine 22, 2413–2419. [DOI] [PubMed] [Google Scholar]
- 37.Ghaemi A, Soleimanjahi H, Gill P, Hassan Z, Jahromi SRM, and Roohvand F (2010). Recombinant λ-phage nanobioparticles for tumor therapy in mice models. Genetic Vaccines and Therapy 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicastro J, Sheldon K, and Slavcev RA (2014). Bacteriophage lambda display systems: developments and applications. Appl Microbiol Biot 98, 2853–2866. [DOI] [PubMed] [Google Scholar]
- 39.Pavoni E, Vaccaro P, D’Alessio V, De Santis R, and Minenkova O (2013). Simultaneous display of two large proteins on the head and tail of bacteriophage lambda. BMC Biotechnology 13, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malik P, Terry TD, Gowda LR, Langara A, Petukhov SA, Symmons MF, Welsh LC, Marvin DA, and Perham RN (1996). Role of Capsid Structure and Membrane Protein Processing in Determining the Size and Copy Number of Peptides Displayed on the Major Coat Protein of Filamentous Bacteriophage. J Mol Biol 260, 9–21. [DOI] [PubMed] [Google Scholar]
- 41.Gupta A, Onda M, Pastan I, Adhya S, and Chaudhary VK (2003). High-density Functional Display of Proteins on Bacteriophage Lambda. J Mol Biol 334, 241–254. [DOI] [PubMed] [Google Scholar]
- 42.Iannolo G, Minenkova O, Petruzzelli R, and Cesareni G (1995). Modifying filamentous phage capsid: limits in the size of the major capsid protein. J Mol Biol 248, 835–844. [DOI] [PubMed] [Google Scholar]
- 43.Razazan A, Nicastro J, Slavcev R, Barati N, Arab A, Mosaffa F, Jaafari MR, and Behravan J (2019). Lambda bacteriophage nanoparticles displaying GP2, a HER2/neu derived peptide, induce prophylactic and therapeutic activities against TUBO tumor model in mice. Scientific Reports 9, 2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bedi D, Gillespie JW, Petrenko VA, Ebner A, Leitner M, Hinterdorfer P, and Petrenko VA (2013). Targeted Delivery of siRNA into Breast Cancer Cells via Phage Fusion Proteins. Molecular Pharmaceutics 10, 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou X, Cao P, Zhu Y, Lu W, Gu N, and Mao C (2015). Phage-mediated counting by the naked eye of miRNA molecules at attomolar concentrations in a Petri dish. Nature Materials 14, 1058–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DePorter SM, and McNaughton BR (2014). Engineered M13 Bacteriophage Nanocarriers for Intracellular Delivery of Exogenous Proteins to Human Prostate Cancer Cells. Bioconjugate Chemistry 25, 1620–1625. [DOI] [PubMed] [Google Scholar]
- 47.Min J, Jung H, Shin H-H, Cho G, Cho H, and Kang S (2013). Implementation of P22 Viral Capsids As Intravascular Magnetic Resonance T1 Contrast Conjugates via Site-Selective Attachment of Gd(III)-Chelating Agents. Biomacromolecules 14, 2332–2339. [DOI] [PubMed] [Google Scholar]
- 48.Lankes HA, Zanghi CN, Santos K, Capella C, Duke CMP, and Dewhurst S (2007). In vivo gene delivery and expression by bacteriophage lambda vectors. Journal of applied microbiology 102, 1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aanei IL, ElSohly AM, Farkas ME, Netirojjanakul C, Regan M, Taylor Murphy S, O’Neil JP, Seo Y, and Francis MB (2016). Biodistribution of Antibody-MS2 Viral Capsid Conjugates in Breast Cancer Models. Molecular Pharmaceutics 13, 3764–3772. [DOI] [PubMed] [Google Scholar]
- 50.Rader C (2014). Chemically programmed antibodies. Trends Biotechnol 32, 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tiller KE, and Tessier PM (2015). Advances in Antibody Design. Annu Rev Biomed Eng 17, 191–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogbunugafor CB, Pease JB, and Turner PE (2010). On the possible role of robustness in the evolution of infectious diseases. Chaos 20, 026108–026108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Domingo-Calap P, Pereira-Gómez M, and Sanjuán R (2010). Selection for thermostability can lead to the emergence of mutational robustness in an RNA virus. J Evol Biol 23, 2453–2460. [DOI] [PubMed] [Google Scholar]
- 54.Gloria-Soria A, Mendiola SY, Morley VJ, Alto BW, and Turner PE (2020). Prior evolution in stochastic versus constant temperatures affects RNA virus evolvability at a thermal extreme. Ecol Evol 10, 5440–5450.• Vesicular stomatitis virus was previously evolved under elevated but constant temperature, or under randomly fluctuating temperature. The strains evolved under fluctuating conditions were later able to adapt to a novel environment, while those evolved under constant conditions were not. This illustrates that adaptability as an evolved trait can be generalized to novel conditions.
- 55.Kim K-P, Cha J-D, Jang E-H, Klumpp J, Hagens S, Hardt W-D, Lee K-Y, and Loessner MJ (2008). PEGylation of bacteriophages increases blood circulation time and reduces T-helper type 1 immune response. Microb Biotechnol 1, 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merril CR, Biswas B, Carlton R, Jensen NC, Creed GJ, Zullo S, and Adhya S (1996). Long-circulating bacteriophage as antibacterial agents. Proceedings of the National Academy of Sciences 93, 3188–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viertel TM, Ritter K, and Horz H-P (2014). Viruses versus bacteria—novel approaches to phage therapy as a tool against multidrug-resistant pathogens. Journal of Antimicrobial Chemotherapy 69, 2326–2336. [DOI] [PubMed] [Google Scholar]
- 58.Hagens S, Habel A, von Ahsen U, von Gabain A, and Bläsi U (2004). Therapy of experimental pseudomonas infections with a nonreplicating genetically modified phage. Antimicrob Agents Chemother 48, 3817–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsuda T, Freeman TA, Hilbert DW, Duff M, Fuortes M, Stapleton PP, and Daly JM (2005). Lysis-deficient bacteriophage therapy decreases endotoxin and inflammatory mediator release and improves survival in a murine peritonitis model. Surgery 137, 639–646. [DOI] [PubMed] [Google Scholar]
- 60.Paul VD, Sundarrajan S, Rajagopalan SS, Hariharan S, Kempashanaiah N, Padmanabhan S, Sriram B, and Ramachandran J (2011). Lysis-deficient phages as novel therapeutic agents for controlling bacterial infection. Bmc Microbiol 11, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hagens S, and Bläsi U (2003). Genetically modified filamentous phage as bactericidal agents: a pilot study. Letters in Applied Microbiology 37, 318–323. [DOI] [PubMed] [Google Scholar]
- 62.Pires DP, Cleto S, Sillankorva S, Azeredo J, and Lu TK (2016). Genetically Engineered Phages: a Review of Advances over the Last Decade. Microbiology and Molecular Biology Reviews 80, 523–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yehl K, Lemire S, Yang AC, Ando H, Mimee M, Torres MT, de la Fuente-Nunez C, and Lu TK (2019). Engineering Phage Host-Range and Suppressing Bacterial Resistance through Phage Tail Fiber Mutagenesis. Cell 179, 459–469.e459. [DOI] [PMC free article] [PubMed] [Google Scholar]