Summary
Measurements of cellular tRNA abundance are hampered by pervasive blocks to cDNA synthesis at modified nucleosides and the extensive similarity among tRNA genes. We overcome these limitations with modification-induced misincorporation tRNA sequencing (mim-tRNAseq), which combines a workflow for full-length cDNA library construction from endogenously modified tRNA with a comprehensive and user-friendly computational analysis toolkit. Our method accurately captures tRNA abundance and modification status in yeast, fly, and human cells and is applicable to any organism with a known genome. We applied mim-tRNAseq to discover a dramatic heterogeneity of tRNA isodecoder pools among diverse human cell lines and a surprising interdependence of modifications at distinct sites within the same tRNA transcript.
Keywords: translation regulation, transfer RNA, tRNA quantitation, RNA modification
Graphical abstract

Highlights
-
•
mim-tRNAseq overcomes experimental and computational hurdles to tRNA quantitation
-
•
mim-tRNAseq includes a comprehensive computational toolkit for tRNA read analysis
-
•
tRNA abundance, aminoacylation, and modification status quantified in one reaction
-
•
mim-tRNAseq reveals an interdependence of modifications at distinct tRNA positions
Quantification of tRNAs by sequencing has been hampered by premature RT termination at modified sites and the extensive sequence similarity among tRNA genes. Behrens et al. overcome these two key challenges with modification-induced misincorporation tRNA sequencing (mim-tRNAseq), which combines an efficient library construction protocol with a comprehensive, user-friendly computational toolkit.
Introduction
Transfer RNAs (tRNAs) are short, abundant molecules required for translating genetic information into protein sequences. The composition of cellular tRNA pools is critical for efficient mRNA decoding and proteome integrity. tRNA expression is dynamically regulated in different tissues and during development (Dittmar et al., 2006; Ishimura et al., 2014; Kutter et al., 2011; Schmitt et al., 2014), and defective tRNA biogenesis is linked to neurological disorders and cancer (Kirchner and Ignatova, 2015).
Nevertheless, the regulation of tRNA levels and its physiological significance remain under-appreciated because of a lack of accurate, high-resolution methods for tRNA quantitation. A major challenge is posed by the stable structure and pervasive Watson-Crick face modifications, which block reverse transcriptase (RT) (Motorin et al., 2007). Library generation workflows without a strategy for overcoming RT blocks yield mostly short reads due to premature RT stops at modified sites, as for instance in quantitative mature tRNA sequencing (QuantM-tRNAseq) (Pinkard et al., 2020). Hybridization-based approaches can circumvent the need for cDNA synthesis, but they can only distinguish tRNAs differing by at least eight nucleotides (Dittmar et al., 2006). This limitation is problematic given the extensive sequence similarity among tRNA transcripts, which can differ by a single nucleotide even if they read different codons (Chan and Lowe, 2016). Strategies to overcome structure- and modification-induced RT barriers have included tRNA fragmentation (Arimbasseri et al., 2015; Gogakos et al., 2017; Karaca et al., 2014), the use of a thermostable template-switching RT in thermostable group II intron RTsequencing (TGIRT-seq and DM-tRNAseq) (Katibah et al., 2014; Qin et al., 2016; Zheng et al., 2015), and enzymatic removal of some base methylations in AlkB-facilitated RNA methylation sequencing (ARM-seq) and DM-tRNAseq (Cozen et al., 2015; Zheng et al., 2015).
Although these methods have improved tRNA representation in sequencing libraries, several limitations remain. First, all of these methods relieve only a fraction of RT blocks, which can bias recovery toward tRNA subsets with few modified sites or those that are better substrates for demethylation in vitro. Second, removing modifications eliminates information about their presence and stoichiometry, which could be inferred from signatures of RT stops and misincorporations (Clark et al., 2016; Ebhardt et al., 2009; Hauenschild et al., 2015; Katibah et al., 2014; Li et al., 2017; Motorin et al., 2007; Qin et al., 2016; Ryvkin et al., 2013; Safra et al., 2017; Zheng et al., 2015). RNA modification profiling based solely on misincorporation signatures would be advantageous, as RT stops can also arise from RNA degradation or structure. Conditions that enable readthrough of Watson-Crick face modified sites while abrogating stops, however, have not been described for any RT so far (Werner et al., 2020). A variant of the HIV-1 RT with improved readthrough of N1-methyladenosine (m1A) was recently derived by protein evolution (Zhou et al., 2019), but whether this enzyme can also overcome any of the other types of RT-blocking tRNA modifications is unknown.
The computational analysis of tRNA sequencing data also presents significant challenges that are often overlooked. The number of predicted tRNA anticodon families in different genomes ranges from 33 in M. hominis to 57 in humans, with many tRNAs encoded by multiple gene copies. In eukaryotes, there is also considerable sequence variation among tRNAs with identical anticodons, which becomes more pronounced with increasing organismal complexity (Goodenbour and Pan, 2006). While the 41 tRNA anticodon families in budding yeast constitute 54 distinct tRNA transcripts, ∼400 unique tRNA molecules can be potentially produced in human cells (Chan and Lowe, 2016). Some of these can have tissue-specific functions even in the presence of closely related isodecoders (tRNAs that share an anticodon but differ in sequence elsewhere; Ishimura et al., 2014).
The exceptional degree of tRNA sequence similarity can undermine alignment accuracy, particularly for short reads resulting from premature RT stops (Pinkard et al., 2020) or tRNA fragmentation (Arimbasseri et al., 2015; Gogakos et al., 2017). The problem is compounded by multiple mismatches between tRNA-derived reads and the genomic reference that arise from RT misincorporation during modification readthrough. Current alignment approaches allow mismatches at any position of a read (Arimbasseri et al., 2015; Gogakos et al., 2017; Hoffmann et al., 2018; Katibah et al., 2014; Pinkard et al., 2020; Qin et al., 2016; Zheng et al., 2015), which can decrease mapping accuracy for nearly identical tRNAs. The total number of mismatches is also limited in some approaches, which can eliminate reads from highly modified tRNAs. Computational tool choice can thus substantially affect measurements of tRNA abundance and modification.
Here, we present a novel workflow that overcomes the experimental and computational hurdles to quantitative tRNA profiling through modification-induced misincorporation tRNA sequencing (mim-tRNAseq). We combine a sensitive method for cDNA library construction from endogenously modified tRNAs with a new computational framework for read alignment, data analysis, and visualization. By identifying conditions that enable efficient RT readthrough of modified sites, we achieve uniform sequence coverage of tRNA pools from yeast, fly, and human cells while retaining modification signatures. In parallel, we developed a comprehensive and user-friendly computational toolkit, which yields measurements of tRNA abundance, charging fractions, and modification profiles with unprecedented accuracy and resolution. mim-tRNAseq identified a wide variation in tRNA isodecoder abundance among different human cell lines and an interdependence among tRNA modifications at distinct sites. As our workflow is sensitive, robust, and applicable to any organism with a known genome, we anticipate it will help shed new light on previously intractable aspects of tRNA biology.
Design
Efficient sequencing library generation from native eukaryotic tRNA pools
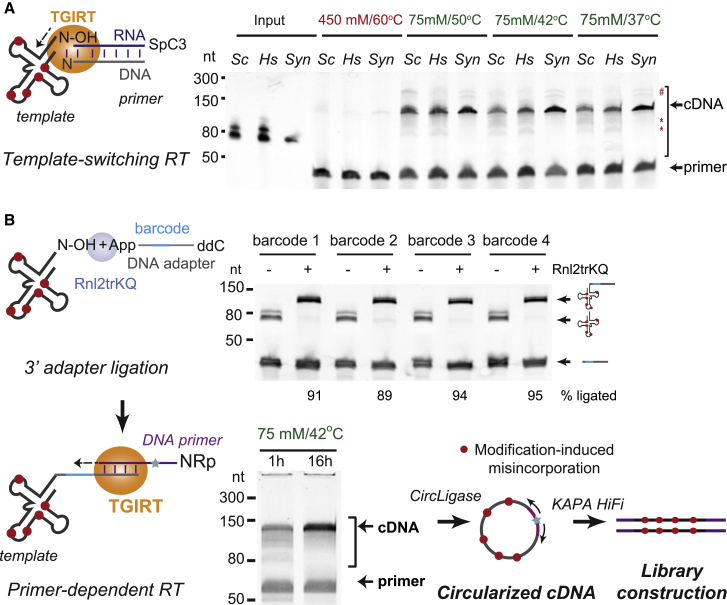
To develop a method for high-resolution tRNA quantitation, we focused on improving the efficiency of full-length cDNA synthesis from endogenously modified tRNAs by TGIRT. This enzyme can attach adapter sequences to RNA by template switching (Mohr et al., 2013), which circumvents potential hindrances to 3′ adapter ligation and RT posed by tRNA structure (Katibah et al., 2014; Qin et al., 2016; Zheng et al., 2015). TGIRT can also read through a subset of Watson-Crick face modifications more efficiently than other commercial RTs (Li et al., 2017), albeit with reduced fidelity (Katibah et al., 2014; Qin et al., 2016; Zheng et al., 2015). Despite these advantages, RT stops at modified sites in tRNA are still pervasive in TGIRT-mediated reactions (Clark et al., 2016; Zheng et al., 2015), and cDNA yield is extremely low (Zhao et al., 2018; Zheng et al., 2015).
As TGIRT is active in a wide range of conditions (Mohr et al., 2013), we asked whether its efficiency on tRNA templates can be further improved. To test this, we first purified tRNA pools from S. cerevisiae and human K562 cells by gel size selection of 60–100 nt RNAs from total RNA. We then used these, along with a synthetic unmodified E. coli tRNA-Lys-UUU, in template-switching TGIRT reactions. The cDNA yield from all templates was minimal under conditions previously used for tRNA sequencing (450 mM salt, 60°C; Katibah et al., 2014; Qin et al., 2016; Zheng et al., 2015) but dramatically improved at lower temperatures and salt concentration (Figure 1A). Although a considerable fraction of cDNAs we obtained were full length, some RT stops still occurred, and larger products potentially derived from two tRNA molecules linked by template switching were also present (Figure 1A). To circumvent these issues and the known sequence bias of TGIRT during template switching (Xu et al., 2019), we introduced DNA adapters at the 3′ end of tRNA with T4 RNA ligase 2. We reasoned that the stable structure of mature tRNAs would not pose a challenge, as their 3′ ends contain the stretch of at least two unpaired nucleotides that is required for efficient 3' adapter ligation (Zhuang et al., 2012). To further minimize potential bias and enable sample pooling prior to RT, we designed four barcoded adapters with limited potential to co-fold with tRNA and confirmed that they can be ligated to size-selected yeast tRNA pools with 89%–95% efficiency (Figure 1B). Pooled adapter-containing tRNA samples were then subjected to primer-dependent RT with TGIRT in a low-salt buffer at 42°C. Strikingly, we found that extending the reaction time eliminated nearly all premature RT stops on endogenously modified yeast and human tRNAs (Figure 1B) without compromising template integrity (Figure S1A). The primer for cDNA synthesis contained a 5′ RN dinucleotide to ensure efficient cDNA circularization (Heyer et al., 2015; McGlincy and Ingolia, 2017) prior to PCR amplification with KAPA HiFi DNA Polymerase, which exhibits minimal bias for fragment length or GC content (Quail et al., 2011). This optimization enabled us to construct Illumina sequencing libraries starting from as little as 50 ng of endogenously modified tRNA with only five or six PCR cycles, minimizing sample input requirements and amplification bias.
Figure 1.
An optimized workflow for full-length cDNA library construction from eukaryotic tRNA pools
(A) Schematic of template-switching TGIRT reactions primed by an RNA/DNA duplex with a single-nucleotide 3′ overhang and a gel image of cDNA products from endogenously modified tRNA pools from S. cerevisiae (Sc), K562 cells (Hs), or a synthetic unmodified tRNA (Syn) at different reaction temperatures and salt concentration. Red, reaction conditions previously used for tRNA library construction; asterisks, premature stops to cDNA synthesis; hash, potential products from end-to-end linkage of tRNAs.
(B) Schematic of the mim-tRNAseq library generation workflow. Top gel image: 3′ adapter ligation reactions with four barcoded adapters. Ligation efficiency was measured by normalizing input tRNA band intensity to that in reactions from which Rnl2trKQ was omitted. Bottom gel image: comparison of cDNA yield in short (1 h) or extended (16 h) primer-dependent TGIRT RT on a mix of adapter-ligated tRNA pools from S. cerevisiae and human K562 and HEK293T cells.
See also Figure S1 and STAR methods.
A comprehensive computational framework for tRNA sequencing data analysis
We reasoned that the increase in full-length cDNA reads would reduce alignment ambiguity. However, given TGIRT’s low fidelity at modified sites, we expected many tRNA-derived reads to contain multiple mismatches to the reference genome. Another source of mismatches are non-templated nucleotides added to 3′ cDNA ends by TGIRT and other RTs (Chen and Patton, 2001; Mohr et al., 2013). Such read extensions are penalized by most algorithms but can be recognized and dynamically processed (“soft-clipped”) by some aligners. We therefore asked how two short-read aligners commonly used for tRNA analysis—Bowtie (Langmead et al., 2009) and Bowtie 2 (Langmead and Salzberg, 2012)—would perform on a tRNA sequencing dataset from human HEK293T cells obtained with our improved library construction protocol (Figure 1B).
We first generated a non-redundant reference of 420 mature tRNA transcripts from 596 curated nuclear- and mitochondrial-encoded tRNA genes retrieved from GtRNAdb and mitotRNAdb (Chan and Lowe, 2016; Jühling et al., 2009; Figure 2A; STAR methods). Alignment was performed with Bowtie or Bowtie 2 with parameters previously used for tRNA sequencing analysis (Clark et al., 2016; Cozen et al., 2015; Katibah et al., 2014; Qin et al., 2016; Zheng et al., 2015). Bowtie end-to-end alignment allows a maximum of three mismatches to the reference at any position. Its inability to distinguish modification-induced misincorporations from other mismatches can lead to data loss for highly modified tRNAs or misalignment for highly similar tRNAs. Indeed, only 25% of reads from our HEK293T tRNA library aligned with Bowtie, with a third of those mapping to multiple tRNA references (Figure 2A). Trimming a fixed number of nucleotides from 5′ read ends prior to alignment, which can remove non-templated nucleotides, expectedly improved mapping rates (Figure S1B). The variable length of non-templated additions, however, makes such a trimming approach imprecise, and many trimmed reads still failed to align or were multi-mapped (Figure S1B).
Figure 2.
The mim-tRNAseq computational pipeline: a comprehensive framework for tRNA sequencing data analysis
(A) Bowtie and Bowtie 2 alignment strategies and mapping statistics for a tRNA library from HEK293T cells constructed with the mim-tRNAseq workflow (n = 1).
(B) Outline of the mim-tRNAseq computational pipeline.
(C) Alignment statistics of HEK293T data (as in A; n = 1) using the mim-tRNAseq pipeline.
(D) Uniquely aligned read proportions for inosine 34 (I34)- and uridine 34 (U34)-containing Ser and Pro tRNA isoacceptors using the three alignment strategies on a HEK293T dataset generated as in Figure 1B.
(E) Distribution of uniquely aligned reads among tRNA isotypes in published datasets and mim-tRNAseq from HEK293-derived cell lines (hydro-tRNAseq and QuantM-tRNAseq: HEK293 T-Rex Flp-IN; DM-tRNAseq control or AlkB-treated [+AlkB] and mim-tRNAseq library construction: HEK293T). Proportions were obtained from published counts per tRNA (“publ”) or after re-analysis of the datasets with the mim-tRNAseq pipeline (“new”). tRNA families that carry the same amino acid (isotypes) are sorted by the number of RT barriers annotated in MODOMICS (decreasing from top to bottom; grayscale, isotypes without MODOMICS annotation).
See also Figure S1 and STAR methods.
In contrast, Bowtie 2’s lack of mismatch restrictions and ability to soft-clip read ends make it seem more suited for tRNA read mapping. High mismatch tolerance, however, compounds the problem of misalignment: while Bowtie 2 increased alignment rates of our HEK293T-derived dataset to 82%, most mapped reads (85%) could not be assigned to a single reference (Figure 2A). Multi-mapping rates were similarly high when human QuantM-tRNAseq data were aligned using Bowtie 2 with the published settings (Pinkard et al., 2020) (85%; Figure S1C). These high rates of data loss indicate that standard read alignment approaches are poorly suited to the complexity of tRNA sequencing data, with consequences for the accuracy of all downstream analyses.
Given these limitations, we reasoned that an accurate tRNA read analysis workflow requires solutions to two main challenges: alignment bias against reads with modification-induced misincorporations and multi-mapping of reads from nearly identical tRNAs. To tackle the first issue, we took advantage of the comprehensive annotation of tRNA modifications in MODOMICS (Boccaletto et al., 2018) and used these data to enable position-specific mismatch tolerance during alignment (Figure 2B, top panel). To achieve this, we chose GSNAP, an aligner designed for detecting complex variants in sequencing reads (Wu and Nacu, 2010). Unlike most other algorithms, GSNAP considers alignments to a reference and an alternative allele equally in SNP-tolerant alignment mode while also effectively soft-clipping read ends. To address multi-mapping, we devised a strategy to cluster reference sequences by a sequence identity (ID) threshold. Given that many reads still map to multiple references with the commonly used strategy of clustering only completely identical tRNA genes (ID = 1; Figure 2A) (Clark et al., 2016; Hoffmann et al., 2018; Zheng et al., 2015), we reasoned that alignment ambiguity could be decreased by lowering the sequence ID threshold. To maintain isoacceptor resolution, we chose to only cluster tRNA transcripts that share an anticodon regardless of sequence ID.
On the basis of these premises, we developed a new computational workflow to suit the intricacies of tRNA sequencing data (Figure 2B; STAR methods). To generate an alignment reference, mature tRNA transcript sequences are matched to MODOMICS to index known modified sites and clustered by anticodon according to sequence ID. Reads are aligned to the resulting indexed reference using GSNAP in SNP-tolerant mode. Unannotated potentially modified sites are detected by a mismatch rate of >10% and included in an updated index, followed by re-alignment of all reads with a more stringent tolerance to mismatches outside of modified sites to further boost alignment accuracy. To restore single-transcript resolution for subsequent analyses, we developed a deconvolution algorithm that assigns cluster-aligned reads to unique tRNA species (Figure 2B, middle panel; STAR methods). For this, each cluster is assessed for single-nucleotide differences that distinguish unique tRNA sequences, on the basis of which each read is separated from the cluster “parent” and assigned to an individual transcript. Analysis of coverage, 3′-CCA, differential tRNA abundance, and modification profiling is then performed after read deconvolution (Figure 2B, bottom panel). The entire computational framework for tRNA read alignment, analysis, and visualization is packaged in an open-source tool with a command-line interface and a broad set of customizable parameters.
This computational workflow dramatically improved both the efficiency and accuracy of tRNA read alignment. Both clustering and SNP tolerance at modified sites prevented data loss for defined tRNA subsets. A cluster ID of 0.95 maximized unique transcript resolution and minimized multi-mapping for human tRNAs (Figure S1D), yielding 86% uniquely mapped and only 2.5% ambiguously aligned reads (Figure 2C). Multi-mapping rates were 5-fold higher when only completely identical tRNA transcripts were clustered, resulting in data loss for selected tRNAs (e.g., tRNA-Asn-GTT-2 and tRNA-Pro-AGG-1; Figures S1D and S1E). Aligning without SNP tolerance had similar effects, particularly for transcripts with inosine at position 34 (I34), which is encoded as an A but yields a G in cDNA libraries. The number of reads mapping to tRNA-Val-AAC, for example, increased by 300-fold in SNP-tolerant mode, and virtually all of these contained a G34 (Figures S1F and S1G). This high mismatch rate at I34 also presented obvious challenges for Bowtie and Bowtie 2. Almost no reads mapped to the I34-containing tRNA-Ser-AGA and tRNA-Pro-AGG with these algorithms, while many were assigned to tRNA-Ser-UGA and tRNA-Pro-UGG instead (Figure 2D). The same dramatic under-representation of tRNA-Ser-AGA and tRNA-Pro-AGG was evident in published counts for QuantM-tRNAseq libraries, which were generated by Bowtie 2 local alignment (Figure S1H). In contrast, our computational workflow yielded a more balanced representation of these four tRNA species for both mim-tRNAseq (Figure 2D) and QuantM-tRNAseq libraries (Figure S1H). The choice of read alignment parameters can thus yield very different tRNA abundance estimates.
Results
The mim-tRNAseq workflow alleviates tRNA sequencing bias
To benchmark our workflow, we used mim-tRNAseq to analyze HEK293T tRNAs and compared our results with those published for the same cell type with DM-tRNAseq (Zheng et al., 2015) and from the closely related HEK293 T-Rex Flp-IN line (Lin et al., 2014) obtained with hydro-tRNAseq (Gogakos et al., 2017) or QuantM-tRNAseq (Pinkard et al., 2020). To distinguish experimental from computational differences, we also re-analyzed the published datasets using our computational pipeline (Figure 2B). Reads from tRNA isotypes with a single known barrier to RT (Boccaletto et al., 2018) were substantially overrepresented in DM-tRNAseq (tRNA-Val, 19%–21%) and hydro-tRNAseq (tRNA-Gly, 30%) compared with our dataset (∼6%). In QuantM-tRNAseq, tRNA-Arg constituted 16% of published tRNA counts versus 3.5% in hydro-tRNAseq, 7%–9% in DM-tRNAseq, and 9% in our dataset. This isotype over-representation persisted regardless of alignment algorithm (Figure 2E, “publ” versus “new”), suggesting that it originated during library construction. In contrast, tRNA-Tyr, which has five known RT-blocking modifications, constituted ∼4% of mapped reads in our dataset versus only 1% for published hydro-tRNAseq and DM-tRNAseq counts and 0.3% for QuantM-tRNAseq. This under-representation was largely relieved when DM-tRNAseq and QuantM-tRNAseq datasets were re-analyzed with our computational pipeline (Figure 2E). Thus, mim-tRNAseq recovers highly modified tRNAs more efficiently than current methods through a combination of advances in library construction and data analysis.
mim-tRNAseq improves tRNA coverage and abundance estimates
We extended our analysis to single-cell and multicellular eukaryotes by preparing mim-tRNAseq libraries from exponentially growing S. cerevisiae and S. pombe, as well as D. melanogaster BG3-c2 cells and human induced pluripotent stem cells (hiPSCs) with a normal karyotype. We determined the optimal cluster ID threshold as 0.90 for budding yeast and 0.95 for fission yeast, Drosophila, and human tRNA pools (Figures S1D and S2A). These settings yielded between 85% and 91% of uniquely mapped reads (Figure 3A), with a median of 65%–83% full-length ones (Figures 3B and 3C). In contrast, unique alignment rates were lower for datasets from DM-tRNAseq and QuantM-tRNAseq and for libraries we generated with the standard TGIRT protocol (Figure S2B). tRNA coverage in those datasets also had substantial 3′ end bias, consistent with RT stops at modified sites (Figures S2C–S2E). Accordingly, unique tRNA transcripts were represented by a median of <11% and 6% full-length reads in DM-tRNAseq and QuantM-tRNAseq, respectively (Figures S2F–S2H).
Figure 3.
mim-tRNAseq improves quantitative analysis of tRNA pools in cells from diverse eukaryotes
(A) Alignment statistics for mim-tRNAseq datasets from the indicated cell types. Bars and labels indicate average values, dots show individual sample values (n = 2).
(B) Metagene analysis of scaled sequence coverage across nuclear-encoded tRNA isotypes ordered per sample by differences between 3′ and 5′ coverage (decreasing order from top to bottom; n = 1). y axis values normalized to the second-to-last bin from the 3′ end. Each x axis bin represents 4% of tRNA length. Indicated are major known barriers to RT.
(C) Boxplots of full-length fraction per tRNA transcript in datasets from (B) (center line and label: median; box limits: upper and lower quartiles; whiskers: 1.5 × interquartile range).
(D) Correlation plots of unique tRNA gene copy number and corresponding proportion of uniquely aligned tRNA reads in single replicates (same samples as in B) from S. cerevisiae, S. pombe, and D. melanogaster BG3-c2 cells and hiPSCs. Solid blue lines: linear regression model; shaded gray: 95% confidence interval (CI).
See also Figures S2 and S3.
Most reads in mim-tRNAseq datasets mapped to cytosolic tRNA, with mitochondrial tRNA fractions ranging from 0.5% in budding yeast to 3% in hiPSCs (Figure S2I). Importantly, nearly all mapped reads (>96%) spanned the post-transcriptionally added 3′-CCA stretch (Figures S3A–S3D), indicating that they originate from mature tRNA. This was not due to bias toward A-ending RNA species, as our workflow accurately captured the 3:1 ratio of two synthetic E. coli tRNA-Lys-UUU tRNAs with either 3′-CCA or 3′-CC spiked in prior to library construction. cDNA circularization also did not introduce appreciable length bias, as tRNA coverage after alignment mirrored initial cDNA size (Figures 3B and 1B). Moreover, circularization sequence context is very similar for all cDNAs, as most have a stretch of one to three non-templated Ts at their 5′ ends, corresponding to non-templated A added to cDNA 3′ ends by TGIRT (Figures S3E and S3F), which were effectively soft-clipped during GSNAP alignment. Indeed, nucleotide frequencies downstream of non-templated nucleotides were highly similar to those obtained by aligning the 5′ ends of predicted tRNA transcripts (Figures S3E and S3F).
We asked whether these experimental and computational advances would enable more accurate tRNA quantitation. We first sought to compare our measurements of absolute tRNA abundance with data obtained with an orthogonal, hybridization-based approach. Absolute RNA quantitation by, for example, Northern blotting or arrays requires highly specific probes and careful comparisons of signal in serial sample dilutions to calibration curves with known target amounts. The design of specific probes for tRNAs, however, is extremely challenging: even with full-length probes, a difference of at least 8 nt is required to avoid cross-hybridization (Dittmar et al., 2004, 2006). Probe design is particularly problematic for human tRNA pools, which can contain >400 tRNA species from 57 anticodon families. As the major tRNA transcript for each anticodon family can differ among cell types (Ishimura et al., 2014), probe selection can unduly influence measurement accuracy. In contrast, the 41 anticodon families of S. cerevisiae consist of only 54 tRNA species, and most major anticodon variants differ sufficiently in sequence to be distinguished by hybridization. We therefore compared fluorescence intensity measurements for 39 of the 41 budding yeast anticodon families obtained by direct hybridization to a tRNA microarray (Tuller et al., 2010) to the fraction of reads mapping to those anticodon families in mim-tRNAseq datasets. This comparison yielded Pearson’s r = 0.75 (p = 3.8 × 10−8), corroborating the quantitative nature of mim-tRNAseq (Figure S3G).
The main regulatory elements for tRNA transcription are intrinsic and overlap with conserved structural regions of mature tRNAs, and it remains unclear how selective tRNA gene expression is achieved in metazoans (Ishimura et al., 2014; Kutter et al., 2011; Schmitt et al., 2014). In rapidly growing yeast cells, however, nearly all tRNA loci are transcribed (Harismendy et al., 2003; Roberts et al., 2003). tRNA gene copy number thus positively correlates with the abundance of tRNA anticodon families during exponential growth measured by hybridization (R2 = 0.47 in microscale thermophoresis and R2 = 0.60 in tRNA microarray; Jacob et al., 2019; Tuller et al., 2010). We leveraged the superior resolution of mim-tRNAseq to probe this relationship at the level of individual tRNA transcripts (Figure 3D). We obtained the highest correlation between gene copy number and tRNA abundance reported so far (adjusted R2 = 0.92 for S. cerevisiae and R2 = 0.91 for S. pombe, p < 3.71 × 10−30), further underscoring the quantitative nature of mim-tRNAseq. This correlation decreased substantially for a S. cerevisiae library from budding yeast generated by template switching in otherwise identical RT conditions (R2 = 0.61; Figures S3H and S3I), consistent with 3′ sequence preferences of TGIRT in this setup (Xu et al., 2019). An even more drastic reduction was seen in a S. cerevisiae library generated with Superscript III (R2 = 0.31), which displayed substantial 3′ end coverage bias despite high rates of unique read alignment (Figures S3H, S3J, and S3K).
The correlation between gene copy number and tRNA abundance was also lower in Drosophila BG3-c2 cells (adjusted R2 = 0.79) and hiPSCs (adjusted R2 = 0.62). The values were similar regardless of whether we used copy numbers for all predicted human tRNA genes or only the high-confidence tRNA gene set (Figure S3L). These findings are consistent with differential tRNA gene use in distinct cell types (Dittmar et al., 2006; Ishimura et al., 2014; Kutter et al., 2011; Schmitt et al., 2014) and highlight that mechanisms beyond gene copy number shape metazoan tRNA pools.
mim-tRNAseq captures differences in tRNA abundance and aminoacylation
To establish whether mim-tRNAseq can accurately detect differences in tRNA abundance, we first compared the tRNA pools of karyotypically normal hiPSCs with those in two aneuploid human cell lines (K562 and HEK293T). Of the 368 cytosolic tRNA species resolved quantitatively by mim-tRNAseq, 205 were undetectable in one or more cell lines (≤0.005% of tRNA-mapped reads). Remarkably, more than half of the detectable tRNAs were differentially expressed, some by up to 3 orders of magnitude (adjusted p < 0.05; Figure 4A; Table S1). In contrast, the relative levels of tRNAs with a given anticodon differed by only up to 1.7-fold among the three cell lines (Figure 4B; Table S1). Of the 47 tRNA anticodon families passing our detection threshold, 11 differed in abundance between HEK293T cells and hiPSCs, and 21 differed in abundance between K562 cells and hiPSCs (Figure 4B; Table S1). Each cell line exhibited a distinct pattern of tRNA expression, with differences being more pronounced for low-abundance transcripts (Figure 4C; base mean expression given by line plot in rightmost panel). These data suggest that different cell types can converge on similar anticodon pools via distinct tRNA transcript subsets, possibly through the relatively stable expression of major tRNA isodecoders (Kutter et al., 2011).
Figure 4.
mim-tRNAseq accurately captures differential tRNA expression and aminoacylation with single-transcript resolution
(A) Differential expression analysis of unique tRNA transcripts in HEK293T and K562 relative to hiPSCs. Axes represent log-transformed normalized read counts from DESeq2, with significant down- and upregulation in hiPSCs indicated with closed orange and green triangles, respectively (false discovery rate [FDR]-adjusted one-sided Wald test p ≤ 0.01, n = 2).
(B) Differential expression analysis as in (A) for counts per tRNA anticodon family.
(C) Left panel: hierarchically clustered expression heatmap showing scaled z score of normalized unique transcript counts in HEK293T, K562, and hiPSCs (n = 2). Middle panels: differential expression for HEK293T and K562 relative to iPSCs (values, log2 fold changes; bar plots, numbers of up- and downregulated genes in green and orange, respectively). Right panel: base mean normalized per tRNA transcript across all samples.
(D) Northern blot analysis of tRNA-Arg-UCU-4 and tRNA-Gly-CCC-2 in HEK293T, K562, and hiPSCs (n = 2, matched samples to those used for mim-tRNAseq). Band intensities were quantified by densitometry and normalized to the mean value for HEK293T.
(E) Relative abundance of tRNA-Arg-UCU-4 and tRNA-Gly-CCC-2 in HEK293T, K562, and hiPSCs measured by mim-tRNAseq (C) or northern blotting (D), normalized to the mean value for HEK293T (n = 2, matched samples).
(F) tRNA charging analysis in wild-type and trm7Δ S. cerevisiae. Charged tRNA are represented by proportion of reads with 3′-CCA ends (light green, in %). Light green bars and tRNA-Phe-GAA labels, average charged tRNA fractions (% CCA; n = 3).
We validated the changes in relative abundance by Northern blotting for two tRNA species: tRNA-Arg-UCU-4 and tRNA-Gly-CCC-2, which differ sufficiently from their isodecoders to avoid probe cross-hybridization and represent tRNAs with low and high abundance. tRNA-Arg-UCU-4 and its mouse ortholog are highly expressed in the central nervous system and are also present at low levels in HEK293T cells (Ishimura et al., 2014; Torres et al., 2019). mim-tRNAseq detected 6- to 8-fold lower levels of tRNA-Arg-UCU-4 in K562 and hiPSCs versus HEK293T (Table S1), and a similar 5- to 10-fold decrease was observed by Northern blotting (Figures 4D and 4E). Differential abundance estimates by mim-tRNAseq and Northern blotting were also highly concordant for the abundant tRNA-Gly-CCC-2 (∼1% of tRNA-mapped reads; Figures 4D and 4E).
We then confirmed the ability of mim-tRNAseq to accurately measure tRNA aminoacylation. Charged tRNAs have periodate-resistant 3′ ends and can be quantified as a fraction of tRNAs with 3′-CCA versus 3′-CC following oxidation and β-elimination (Evans et al., 2017). We compared mim-tRNAseq data from oxidized tRNA of wild-type (WT) yeast and a trm7Δ strain, which has a tRNA-Phe-GAA charging defect (Han et al., 2018). This defect was evident by a 2.5-fold decrease in 3′-CCA proportions for both tRNA-Phe-GAA isodecoders in tRNA pools from trm7Δ cells in the absence of other changes in aminoacylation status (Figure 4F). Thus, mim-tRNAseq enables the sensitive and accurate quantitation of differences in tRNA abundance or charging.
Improved readthrough facilitates the discovery and annotation of Watson-Crick face tRNA modifications
Mismatches to reference and/or premature RT stop signatures are frequently used to detect Watson-Crick face RNA modifications (Clark et al., 2016; Ebhardt et al., 2009; Hauenschild et al., 2015; Katibah et al., 2014; Li et al., 2017; Motorin et al., 2007; Qin et al., 2016; Ryvkin et al., 2013; Safra et al., 2017; Zheng et al., 2015), but their analysis is prone to both experimental and computational artifacts (Sas-Chen and Schwartz, 2019). As tRNA-derived reads are particularly misalignment-prone with standard algorithms (Figures 2A and 2D), this could affect the accuracy of modification calling.
In contrast, mim-tRNAseq abrogated nearly all RT stops and yielded reproducibly high levels of mismatches coinciding with frequently modified tRNA positions (Figure 5A). We quantified the extent of readthrough at annotated Watson-Crick face tRNA modifications by calculating the proportion of aligned reads extending past a given position. We then took the minimum value in a 3-nt window centered around it to avoid readthrough overestimation. The median readthrough values we obtained with this approach were ∼100% at the most common RT barriers in tRNA such as m1A, N1-methylguanosine (m1G), N2,N2-dimethylguanosine (m22G), and N3-methylcytosine (m3C), as well as bulkier modifications such as wybutosine (yW) and other wyosine derivatives (Figure 5B). All 162 annotated Watson-Crick face modifications in tRNA from budding yeast (100%) and 232 of the 250 annotated ones in human tRNA (93%) had a readthrough efficiency of >80% (Table S3). This is due to both experimental and computational advances, as readthrough was much lower in libraries generated with standard TGIRT conditions or in DM-tRNAseq (Figures S4A and S4B). There was a notably large variation in bypass of the same modification type in different tRNAs in libraries made with Superscript IV (Figure S4C).
Figure 5.
Near-complete modification readthrough in mim-tRNAseq datasets enables modification discovery and annotation
(A) Average proportion of stops (red) and misincorporation rates (blue) per nucleotide for all tRNA unique transcripts (n = 2) in S. cerevisiae, S. pombe, and D. melanogaster BG3-c2 cells and hiPSCs. x axis, canonical tRNA position at major sites with known RT barriers.
(B) RT readthrough per annotated modification aggregated for cytosolic and mitochondrial tRNA from the four species.
(C) Boxplots of misincorporation signatures for annotated modified sites as in (B) (center line: median; box limits: upper and lower quartiles; whiskers: 1.5 × interquartile range). Signatures stratified by upstream context (rows) and modification type (columns); proportion per nucleotide scaled to total misincorporation at this site.
(D) Boxplot of misincorporation signature at G37 of tRNA-Phe-GAA from WT and trm7Δ S. cerevisiae (n = 3).
(E) Modified site discovery by mim-tRNAseq (“new”) compared with misincorporation-inducing modified sites previously annotated in MODOMICS (“annot.”). Labels indicate percentage of newly detected sites relative to annotated ones.
The only RT blocks remaining in mim-tRNAseq datasets were at rare hypermodified positions. These include 2-methylthio-derivatives of A37 (ms2t6A/ms2i6A in human cytosolic tRNA-Lys-UUU and 3-4 mitochondrial tRNAs in Drosophila and human cells) and rare stretches of two modified sites (m22G26/27 and 20/20a N3-[3-amino-3-carboxypropyl]-uridines [acp3U]; Figures 5B, S2I, S4D, and S4E). These few remaining RT stops do not affect tRNA quantitation, as the cDNA fragments derived from them are sufficiently long (39–56 nt) for unambiguous read alignment with our pipeline.
We then examined whether different modifications are marked by specific signatures of nucleotide misincorporation. This can depend on the processivity and fidelity of an RT, the reaction conditions, and the sequence context of the modified site (Hauenschild et al., 2015; Katibah et al., 2014; Li et al., 2017; Qin et al., 2016; Safra et al., 2017). Signature analysis is especially challenging when RT stops are pervasive, as mismatches at read ends stemming from non-templated nucleotide addition during RT may manifest as misincorporation and lead to spurious modification calls (Sas-Chen and Schwartz, 2019). As mim-tRNAseq enables near-complete modification readthrough (Figure 5B), we examined misincorporation patterns at annotated sites as a function of modification type and sequence context. We found distinct and highly reproducible misincorporation signatures at specific modifications (Figure 5C). The ones at m1G, m22G, and m3C were largely independent of sequence context, whereas those at m1A and acp3U were influenced by the upstream template nucleotide (Figure 5C). We also observed distinct signatures for wyosine derivatives, inosine and N1-methylinosine (m1I), where the tRNA sequence space is not sufficiently large to explore the impact of sequence context. In contrast, misincorporation signatures of Superscript IV were much less specific for distinct modifications, with a high prevalence of T mismatches regardless of modification type (Figure S4F). A recent comparison of 13 RTs found a similar lack of distinguishable signatures for m1G and m22G (Werner et al., 2020).
To validate the specificity of these signatures, we compared misincorporation patterns at G37 in tRNA-Phe-GAA from WT and trm7Δ yeast (Figure 5D). The conversion of m1G37 to yW in this tRNA requires 2′-O-methylation of C32 and G34 by Trm7 (Guy et al., 2012). Accordingly, the misincorporation signature at G37 in tRNA-Phe-GAA from trm7Δ cells was distinct from that in WT (Figure 5D) and nearly identical with that of m1G in our aggregate analysis (Figure 5C).
This remarkable consistency enables the use of misincorporation signatures not only for mapping RNA modifications but also for predicting their identity. We therefore probed our datasets from S. cerevisiae, S. pombe, and Drosophila BG3-c2 cells and human cells for misincorporation-inducing modifications not annotated in MODOMICS. Such sites were identified by a mismatch frequency of >10% and the presence of a distinct misincorporation signature to limit spurious modification calls due to genomic misannotation or SNPs. Modification type was then predicted by combining information on the canonical tRNA position, nucleotide type, and misincorporation signature in comparison with known sites (Figure 5C). Performing this analysis with single-transcript resolution revealed many uncatalogued modifications (Figure 5E; Table S2), including 30 sites in S. cerevisiae and 358 sites in human tRNAs, despite comprehensive existing annotation. Discovery rates were higher in poorly annotated species such as S. pombe and D. melanogaster. Our predictions generally agreed with prior annotation of modified sites based on RT stops and/or misincorporations (Table S2), with some important differences. First, we identified one m1G9 site, two m22G26 sites, and seven m1A58 sites in tRNAs from S. pombe, which had not been detected by hydro-tRNAseq (Arimbasseri et al., 2015). Second, we found no detectable misincorporation at G37 in human tRNA-Pro-AGG or C47d in human tRNA-Ser-AGA and tRNA-Ser-CGA, although these positions have been annotated as m1G37 and m3C47d, respectively (Arimbasseri et al., 2016; Clark et al., 2016). These differences likely result from our workflow’s improved resolution of nearly identical tRNAs, as human tRNA-Pro-UGG and tRNA-Ser-UGA contain m1G37 and m3C47d, respectively (Figures 2D and S5C). These data demonstrate that mim-tRNAseq can map potentially modified tRNA sites and predict modification type with high sensitivity and specificity.
Accurate quantitation of RNA modification stoichiometry on the basis of misincorporation rates
Proportions of RT stops and/or misincorporations are widely used to estimate RNA modification levels (Arimbasseri et al., 2015; Clark et al., 2016; Gogakos et al., 2017; Ryvkin et al., 2013), but whether such measurements are quantitative is unknown. Misincorporation rates at individual modified positions in mim-tRNAseq datasets varied remarkably across tRNA species (Figure 6A) despite efficient readthrough (Figures 5B and S5). To test whether this variation reflects modification stoichiometry, we sequenced endogenously modified tRNA from WT and mutant yeast lacking m1G9 (trm10Δ) (Jackman et al., 2003) or m22G26 (trm1Δ) (Ellis et al., 1986) pooled in defined ratios prior to library construction. Misincorporations were predictably absent at G9 or G26 sites in samples from the knockout strains. Strikingly, their rates had a near-perfect linear correlation to initial pooling ratios in mixed samples (R2 = 0.97; Figures 6B and 6C). Mismatch proportions in mim-tRNAseq datasets thus accurately reflect the stoichiometry of m1G and m22G26, and possibly all other misincorporation-inducing modified tRNA bases. Calibration curves with endogenously modified tRNAs are not feasible for all misincorporation-inducing modifications (Figure 5B), however, as some of them are essential for cell viability (Anderson et al., 1998; Gerber and Keller, 1999).
Figure 6.
Misincorporation rates in mim-tRNAseq reflect modification stoichiometry
(A) Global heatmap of average misincorporation proportions in S. cerevisiae per unique tRNA transcript with coverage above 2,000 reads (n = 2; top bar graph, mean misincorporation per position; right bar graph, number of sites per transcript with detectable misincorporation signatures in ≥10% of reads spanning that position).
(B) Relative misincorporation proportions at G9 in samples from wild-type (WT) S. cerevisiae and trm10Δ (lacking m1G9) or mixes thereof (filtered for clusters with ≥10% misincorporation in WT and scaled to WT proportion; solid blue line, linear regression model; shaded gray, 95% CI).
(C) Analysis as in (B) but for misincorporation at G26 in samples from WT S. cerevisiae or trm1Δ (lacking m22G26).
(D) Misincorporation proportions per canonical nucleotide position and identity (aggregated per species; e2, second nucleotide of variable loop; n = 2).
(E) Significant changes in misincorporation rates in trm1Δ relative to WT S. cerevisiae (FDR-adjusted chi-square p ≤ 0.01, log2 fold change ≥ 0.5, n = 1).
See also Figures S5 and S6 and Table S3.
These findings enabled us to profile modified tRNA fractions with single-transcript resolution in cells from four eukaryotic species. Misincorporation rates were ∼100% at all instances of I34 and of wyosine derivatives at position 37, suggesting these modifications are present in stoichiometric levels (Figures 6D and S6A). We observed a similar trend for m22G26, with a clear separation between a majority of fully modified tRNAs and a very small number of transcripts with 10%–30% misincorporation. In contrast, the modified fractions of m1G, m3C, and m1A varied substantially among individual tRNAs independently of sequence context (Figures 6D, S6A, and S6B). Instances of very high misincorporation were detectable for all three modifications (m1A, 100%; m3C, 94%; m1G, 88%), indicating that mim-tRNAseq can capture high stoichiometry at these sites if it is present (Figure 6D; Table S3). However, some tRNAs seem to contain these modifications at sub-stoichiometric levels. Sub-stoichiometric m3C32 and m1G37 are consistent with the regulatory rather than structural roles of modifications within the tRNA anticodon loop. The stoichiometry of m1G37 measured by mim-tRNAseq ranged from 14% to 80% in tRNAs from the four eukaryotic species (Table S3). In bacteria, m1G37 in tRNA-Pro-UGG and tRNA-Pro-GGG aids in reading frame maintenance (Gamper et al., 2015; Maehigashi et al., 2014). Eukaryotic cells, however, lack tRNA-Pro-GGG because of toxicity from its high miscoding capacity (Pernod et al., 2020). A recent study estimated m1G37 stoichiometry in bacterial tRNA-Pro-UGG by primer extension at 68% in E. coli and 73% in Salmonella enterica (Masuda et al., 2019). Our workflow estimated m1G37 stoichiometry at 53% in yeast tRNA-Pro-UGG and 72% for tRNA-Leu-UAA (Table S3). Gel-based primer extension assays with AMV RT, which is blocked by m1G, were consistent with these measurements (Figures S5D and S5E), providing an orthogonal validation of mim-tRNAseq modification stoichiometry estimates.
In contrast to the regulatory roles of anticodon loop modifications, m1A58 is important for the maturation and stability of initiator tRNA-Met in yeast (Anderson et al., 1998) and may play a similar role in other eukaryotic tRNA species. A sequence comparison of budding yeast tRNAs with high or low m1A58 levels revealed no notable differences, however, indicating that sequence alone is unlikely to be a major determinant of modification stoichiometry at this position (Figure S6C).
To examine whether the stoichiometry of misincorporation-inducing tRNA modifications differs in distinct cell types or states, we calculated log odds ratios of misincorporation proportions across all tRNA positions (see STAR methods). There were very few statistically significant changes when comparing mim-tRNAseq datasets from hiPSCs and HEK293T or K562 cells (Figures S6D and S6E), suggesting that most tRNAs are modified to a similar extent in these cell lines. A comparison of datasets from WT and trm10Δ or trm1Δ yeast, however, revealed the striking precision of our approach in detecting transcripts with large reductions in m1G9 or m22G26 (Figures S6F and S6G). Unexpectedly, in trm1Δ yeast cells that lack m22G26, there were also differences in modification levels at other tRNA sites. These included a 3- to 6.5-fold increase in m1G9 levels in four tRNAs (tRNA-Lys-CUU-1, tRNA-Thr-AGU-1, tRNA-Arg-CCU-1, and tRNA-Asn-GUU-1) and a 2-fold decrease in m3C32 of tRNA-Ser-UGA-1 (Figures 6E and S6G). m1G9 levels in tRNA-Lys-CUU-1 and tRNA-Thr-AGU-1 also increase upon Trm10 overexpression in yeast (Swinehart et al., 2013). Sequence comparisons between tRNAs with increased versus unchanged m1G9 levels in trm1Δ cells indicate that a U7:A66 pair rather than G7:C66 pair may be linked to m1G9 hypermethylation in the absence of m22G26 (Figure S6H). These findings reveal an interdependence between Watson-Crick face modifications at distinct tRNA sites and suggest that their stoichiometry is determined by structural features.
Discussion
The abundance, charging, and modification status of individual tRNA species can differ in distinct cellular environments. Measuring these properties on a global scale, however, has not been feasible because of technical limitations. No library construction method so far has allowed the efficient reverse transcription of these highly modified RNAs, while the lack of computational tools suited to the complexity of tRNA sequencing data has been another major methodological gap.
We describe conditions that permit near-complete tRNA modification readthrough by TGIRT, dramatically improving cDNA yield and the fraction of full-length products from tRNA templates. All but one rare tRNA modification roadblock are resolved by mim-tRNAseq, which alleviates the bias of existing tRNA quantitation methods toward low-modified tRNAs species. Our library construction protocol circumvents the need to purify enzymes for modification removal (Zheng et al., 2015) or RT (Zhou et al., 2019), which can introduce unwanted variation. We also describe multiple conceptual advances in the analysis of tRNA sequencing data, including the use of modification annotations, which permits position-specific mismatch tolerance during read alignment. Collectively, these advances enable the efficient and accurate mapping and analysis of tRNA-derived reads with single-transcript resolution.
One poignant example of the substantial improvements in our computational workflow concerns tRNAs with I34, which is essential for wobble pairing during decoding. Inosines are interpreted as cytosines during RT, resulting in the stoichiometric presence of G in sequencing reads. When using Bowtie or Bowtie 2 to align tRNA datasets from human cells, we found that reads with G34 were frequently mapped to nearly identical tRNA isoacceptors with U34. Such misalignment can have wide-ranging implications, as it would not only skew abundance estimates but can also lead to spurious conclusions about tRNA modification status and stoichiometry. These findings highlight the importance of both sensitivity and accuracy of read alignment in the context of analyzing tRNA transcriptomes.
The robust misincorporation signatures deposited by TGIRT reveal the location, type, and stoichiometry of Watson-Crick face base modifications in tRNA. Calibration measurements of observed versus expected modified fractions in existing approaches for sequencing-based modification analysis are either lacking (Ryvkin et al., 2013; Zheng et al., 2015) or display a non-linear relationship (Zhou et al., 2019), likely because of persistent RT stops. In contrast, mim-tRNAseq enables efficient readthrough of almost all tRNA modifications, while modification ID is also discernible by highly specific misincorporation patterns. Improved readthrough permits accurate measurements of modification stoichiometry from misincorporation rates alone, evident from calibration curves with near-perfect linear regression for m1G and m22G (R2 = 0.97). Performing this calibration with mixtures of endogenously modified tRNA pools shows that our entire workflow is free of bias toward low-modified tRNAs.
Remarkably, we find that although some tRNA positions are almost always fully modified (e.g., m22G26 and I34), others are sub-stoichiometric in some tRNA species. This is in line with a model in which some modifications are deposited because of overlapping substrate specificities in RNA modification enzymes (Phizicky and Alfonzo, 2010). Indeed, methylation at G9 in some yeast tRNAs is enhanced when they lack m22G26, while methylation of C32 is decreased, suggesting that a tRNA conformational change upon m22G26 loss (Steinberg and Cedergren, 1995) might change the affinity of other modification enzymes for individual tRNAs.
In summary, mim-tRNAseq is a sensitive and accurate start-to-finish technique for quantitation of tRNA abundance and charging, which also reports on the presence and stoichiometry of misincorporation-inducing RNA modifications. The robust library construction workflow and the easy-to-use and freely available computational toolkit make mim-tRNAseq broadly applicable for studying key aspects of tRNA biology in a range of organisms and cell types. Our experimental workflow can also be implemented for the discovery and quantitation of modified sites in other RNA species.
Limitations
mim-tRNAseq currently reports on the presence and stoichiometry of those Watson-Crick face tRNA modifications that elicit robust misincorporation during RT with TGIRT. Various protocols for chemical treatment of the “RT-silent” modifications (e.g., pseudouridine, N5-methylcytosine, N7--methylguanosine) have been developed to enable their detection via misincorporation (Motorin and Helm, 2019). Combining them with mim-tRNAseq can expand the modification range detectable in a single sequencing reaction. Our stoichiometry measurements for m1G and m22G26 were validated with mixtures of endogenously modified tRNA pools from WT and modification-deficient strains, but such validation is not feasible for modifications essential for cell viability. Finally, mim-tRNAseq requires low starting material but is not compatible with single-cell tRNA profiling.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Commercial reagents | ||
| mTeSR1 | STEMCELL Technologies | Cat# 85850 |
| Micro Bio-Spin P30 columns, RNase-free | BioRad | Cat# 7326251 |
| Glycogen | Ambion | Cat# AM9510 |
| T4 Polynucleotide Kinase | New England Biolabs | Cat# M0201L |
| T4 RNA ligase 2 (truncated KQ) | New England Biolabs | Cat# M0373L |
| SUPERase In | Ambion | Cat# AM2694 |
| TGIRT | InGex | Cat# TGIRT50 |
| Superscript III | Invitrogen | Cat# 18080044 |
| AMV RT | Promega | Cat# M9004 |
| CircLigase ssDNA ligase | Lucigen | Cat# CL4115K |
| KAPA HiFi DNA Polymerase | Roche | Cat# KK2102 |
| DNA Clean&Concentrator-5 PCR purification kit | Zymo Research | Cat# D4013 |
| Immobilon NY+ | Millipore | Cat# INYC00010 |
| Deposited data | ||
| Raw and analyzed sequencing data | This paper | GEO: GSE152621 |
| DM-tRNAseq raw data for H. sapiens HEK293T | Zheng et al., 2015 | GEO: GSE66550 |
| Hydro-tRNAseq raw data for H. sapiens HEK293 T-Rex Flp-IN | Gogakos et al., 2017 | GEO: GSE95683 |
| QuantM-tRNAseq raw data for H. sapiens HEK293 T-Rex Flp-IN | Pinkard et al., 2020 | GEO: GSE141436 |
| Experimental models: cell lines | ||
| D. melanogaster BG3-c2 cells | P. Becker, LMU | N/A |
| HEK293T cells | O. Griesbeck, MPIN | N/A |
| HPSI0214i-kucg_2 cells | Kilpinen et al., 2017; ECACC | Cat# 77650065 |
| Experimental models: organisms/strains | ||
| S. cerevisiae: strain BY4741 | Euroscarf | N/A |
| S. cerevisiae: strain BY4741 trm1Δ::kanMX | Euroscarf | N/A |
| S. cerevisiae: strain BY4741 trm7Δ::kanMX | Euroscarf | N/A |
| S. cerevisiae: strain BY4741 trm10Δ::kanMX | Euroscarf | N/A |
| S. pombe: strain ED668 h+ | S. Braun, LMU | N/A |
| Oligonucleotides | ||
| RNA sequences, primers for library construction, and probes for primer extension and Northern blotting, see Table S4 | This paper | N/A |
| Software and algorithms | ||
| mim-tRNAseq v0.2.5.6 | This paper | https://github.com/nedialkova-lab/mim-tRNAseq |
| Bowtie v1.2.2 | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/index.shtml |
| Bowtie2 v2.3.3.1 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| GSNAP v2019-02-26 | Wu and Nacu, 2010 | http://research-pub.gene.com/gmap/ |
| Samtools v1.11 | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Bedtools v2.29.2 | Quinlan and Hall, 2010 | https://bedtools.readthedocs.io/en/latest/ |
| BLAST+ v2.9.0 | Camacho et al., 2009 | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastDocs&DOC_TYPE=Download |
| Infernal v1.1.2 | Nawrocki and Eddy, 2013 | http://eddylab.org/infernal/ |
| usearch v10.0.240_i86linux32 | Edgar, 2010 | https://www.drive5.com/usearch/ |
| R/DESeq2 v1.26.0 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| R/ComplexHeatmap v2.2.0 | Gu et al., 2016 | https://www.bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html |
| Python/Biopython v1.70 | Cock et al., 2009 | https://biopython.org/ |
| Other | ||
| Detailed protocol for mim-tRNAseq library construction | This paper | Methods S1 |
Resource availability
Lead contact
Please direct any requests for further information or reagents to the Lead Contact, Danny Nedialkova (nedialkova@biochem.mpg.de).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The accession number for the sequencing data reported in this paper is GEO: GSE152621. The mim-tRNAseq computational pipeline is available under a GNU public License v3 at https://github.com/nedialkova-lab/mim-tRNAseq. A package description and installation guide are available at https://mim-trnaseq.readthedocs.io/en/latest/.
Experimental model and subject details
Cell lines and strains
S. cerevisiae cells (BY4741 wild-type, trm7Δ, trm1Δ and trm10Δ) were grown in yeast extract-peptone-dextrose (YPD) medium. S. pombe cells (ED668 h+, ade6-M216 ura4-D18 leu1-32) were cultured in yeast extract with supplements (YES). Overnight cultures were diluted to an optical density 600 (OD600) of 0.05, grown at 30°C at 250 revolutions per minute, and harvested at OD600 = 0.5 by rapid filtration and snap-freezing in liquid nitrogen. D. melanogaster BG3-c2 cells were cultured at 26°C in Schneider’s Drosophila Medium (GIBCO) supplemented with 10% fetal calf serum, 1% penicillin/streptomycin, and 10 μg/ml human insulin. HEK293T cells were grown at 37°C and 5% CO2 in DMEM supplemented with 10% fetal bovine serum (Sigma Aldrich). The HPSI0214i-kucg_2 human induced pluripotent stem cell line (obtained from HipSci; Kilpinen et al., 2017) was cultured at 37°C and 5% CO2 in mTeSR1 (STEMCELL Technologies). K562 cells were grown at 37°C and 5% CO2 in RPMI 1640 supplemented with 10% fetal calf serum and 2mM L-Glutamine.
Method details
RNA isolation
RNA from Drosophila BG3-c2, HEK293T, and human iPS cells was isolated with Trizol (Sigma Aldrich) according to the manufacturer’s instructions. For total RNA isolation from yeast, frozen cells were resuspended in 100 mM sodium acetate pH = 4.5, 10 mM EDTA pH = 8.0, 1% SDS (1 mL per 50 OD600 units). An equal volume of hot acid phenol (pH = 4.3) was added, and the cell suspension was vortexed vigorously followed by incubation at 65°C for 5 min (S. cerevisiae) or 45 min (S. pombe) with intermittent mixing. After addition of 1/10 volume 1-Bromo-3-chloropropane (BCP, Sigma Aldrich), samples were centrifuged at 10,000 x g for 5 min and the aqueous phase was transferred to a new tube. Following an additional round of hot acid phenol/BCP and a round of BCP only extraction, RNA was precipitated from the aqueous phase by the addition of 3 volumes of 100% ethanol. Pellets were washed in 80% ethanol, briefly air-dried, and resuspended in RNase-free water. For RNA isolation from yeast under conditions that preserve tRNA charging, frozen cells were resuspended in ice-cold 100 mM sodium acetate pH = 4.5, 10 mM EDTA pH = 8.0. One volume of cold acid phenol (pH = 4.3) was added and cells were lysed with 500 μm-diameter glass beads by three rounds of vortexing for 45 s with a 1-min incubation on ice in between. One-tenth volume of BCP was then added and the samples were centrifuged at 10,000 x g/4°C for 5 min, followed by a second round of cold phenol-BCP and one round of BCP-only extraction. RNA was ethanol-precipitated from the aqueous phase and pellets were washed in 80% ethanol containing 50 mM sodium acetate, pH = 4.5, briefly air-dried, and resuspended in 50 mM sodium acetate pH = 4.5, 1 mM EDTA pH = 8.0. RNA concentration was determined with NanoDrop and samples were frozen at −80°C in single-use aliquots.
RNA oxidation and β-elimination
To measure tRNA charging levels, RNA oxidation and β-elimination were performed as described (Evans et al., 2017) with minor modifications. 25 μg of total RNA were resuspended in 10 mM sodium acetate pH 4.5 and oxidized by the addition of freshly prepared NaIO4 to a final concentration of 50 mM in a 58 μL volume for 30 min at 22°C. The reaction was quenched by addition of 6 μL 1 M glucose for 5 min at 22°C. RNA was purified with Micro Bio Spin P30 columns (BioRad) followed by two rounds of ethanol precipitation in the presence of 0.3M sodium acetate pH = 4.5. Pellets were resuspended in 20 μL RNase-free water and β-elimination was performed by addition of 30 μl 100 mM sodium borate pH = 9.5 (freshly prepared) for 90 min at 45°C. RNA was recovered with Micro Bio Spin P30 columns followed by ethanol precipitation, resuspended in RNase-free water, quantified on a NanoDrop, and stored at −80°C in single-use aliquots.
tRNA purification by gel size selection
Two synthetic RNA standards corresponding to E.coli tRNA-Lys-UUU with intact 3′-CCA (5′-GGGUCGUUAGCUCAGUUGGUAGAGCAGUUGACUUUUAAUCAAUUGGUCGCAGGUUCGAAUCCUGCACGACCCACCA-3′) or a 3′-CC (5′-GGGUCGUUAGCUCAGUUGGUAGAGCAGUUGACUUUUAAUCAAUUGGUCGCAGGUUCGAAUCCUGCACGACCCACC-3′) were added to total RNA in a 3:1 molar ratio at 0.06 pmol/μg, followed by incubation at 37°C in 50 mM Tris-HCl pH = 9.0 to deacylate tRNAs. Deacylation was omitted for samples subjected to oxidation and β-elimination. Total RNA was subsequently dephosphorylated with 10 U of T4 PNK (NEB) at 37°C for 30 min and purified by ethanol precipitation in 0.3M sodium acetate pH = 4.5 with 25 μg glycogen (Ambion) as a carrier. RNA was resolved on a denaturing 10% polyacrylamide/7M urea/1XTBE gel alongside Low Range ssRNA marker (NEB) and visualized with SYBR Gold. Species migrating at the size range of mature tRNAs (60 – 100 nt) were excised and gel slices were crushed with disposable pestles. Low-retention tubes and tips (Biotix, Axygen) were used for all subsequent steps of sequencing library construction to maximize nucleic acid recovery. Following addition of 400 μl gel elution buffer (0.3M sodium acetate pH = 4.5, 0.25% SDS, 1mM EDTA pH = 8.0), the gel slurry was incubated at 65°C for 10 min, snap-frozen on dry ice, and thawed at 65°C for 5 min. RNA was eluted overnight at room temperature with continuous mixing. Gel pieces were removed with Costar Spin-X centrifuge tube filters and RNA was recovered from the flow-through by ethanol precipitation in the presence of 25 μg of glycogen. This protocol typically recovers 5%–10% of total RNA in the 60 – 100 nt fraction, consistent with estimates of tRNA proportions in cells (Warner, 1999).
3′ adapter ligation
50 to 200 ng of gel-purified tRNA was ligated to one of four adapters with distinct barcodes:
I1: 5′-pGATATCGTCAAGATCGGAAGAGCACACGTCTGAA/ddC/-3′;
I2: 5′-pGATAGCTACAAGATCGGAAGAGCACACGTCTGAA/ddC/-3′;
I3: 5′-pGATGCATACAAGATCGGAAGAGCACACGTCTGAA/ddC/-3′;
I4: 5′-pGATTCTAGCAAGATCGGAAGAGCACACGTCTGAA/ddC/-3′ (barcodes italicised; underlined sequence complementary to RT primer). The adapters are blocked by the 3′ chain terminator dideoxycytidine to prevent concatemer formation, and 5′- phosphorylated to enable pre-adenylation by Mth RNA ligase prior to ligation (McGlincy and Ingolia, 2017). Ligation was performed for 3 hours at 25°C in a 20-μl reaction volume containing pre-adenylated adapter and RNA substrate in a 4:1 molar ratio, 1x T4 RNA Ligase Reaction Buffer, 200 U of T4 RNA ligase 2 (truncated KQ; NEB), 25% PEG 8000, and 10 U SUPERase In (Ambion). Ligation products were separated from excess adapter on denaturing 10% polyacrylamide/7M urea/1XTBE gels. Bands migrating at 95-125 nt were excised and ligation products were recovered from crushed gel slices.
Reverse transcription
All reactions contained 125 nM primer, 125 nM template and 500 nM TGIRT (InGex) or 200 U Superscript III (Invitrogen). To prime reverse transcription in template-switching reactions, a synthetic RNA/DNA duplex with a single-nucleotide 3′ overhang was generated by annealing an RNA oligonucleotide (5′-GAGCACACGUCUGAACUCCACUCUUUCCCUACACGACGCUCUUCCGAUCU-3′) to a DNA oligonucleotide (5′-pRAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGGAGTTCAGACGTGTGCTCN-3′). The DNA oligonucleotide contained a phosphorylated A/G at its 5′ end, which is a preferred substrate for CircLigase used in subsequent cDNA circularization (Heyer et al., 2015; McGlincy and Ingolia, 2017). For primer-dependent reverse transcription reactions, adapter-ligated tRNA and RT primer (5′-pRNAGATCGGAAGAGCGTCGTGTAGGGAAAGAG/iSp18/GTGACTGGAGTTCAGACGTGTGCTC-3′; underlined sequence complementary to 3′ adapter, 5′-RN to ameliorate potential biases during circularization) were mixed in MAXYMum Recovery PCR Tubes (Axygen), denatured at 82°C for 2 min and annealed at 25°C for 5 min in a Thermocycler. TGIRT reactions were assembled in a 20-μl final volume by combining template and primer with 10 U SUPERase In, 5 mM DTT (from a freshly made 100 mM stock) and manufacturer-recommended TGIRT buffer (20 mM Tris-HCl pH = 7.6, 450 mM NaCl, 5 mM MgCl2) or low salt buffer (50 mM Tris-HCl pH = 8.3, 75 mM KCl, 3 mM MgCl2). After TGIRT addition, samples were pre-incubated at reaction temperature for 10 min (primer-dependent reactions) or 22°C for 30 min (template-switching reactions), initiated by addition of dNTPs to a final concentration of 1.25 mM, and incubated in a Thermocycler for 1 hour or 16 hours. For Superscript III RT, template and primer were denatured at 75°C for 5 min and chilled on ice, and reverse transcription was performed in the presence of 1X First-Strand Buffer, 5 mM DTT, 0.5 mM dNTPs, 10 U SUPERase In, and 200 U Superscript III (Invitrogen) at 57°C for 60 min.
Template RNA was subsequently hydrolyzed by the addition of 1 μl 5M NaOH and incubation at 95°C for 3 min and reaction products were separated from unextended primer on denaturing 10% polyacrylamide/7M urea/1XTBE gels. Gels were stained with SYBR Gold, the region between 60 and 150 nt was excised and cDNA was eluted from crushed gel slices in 400 μl 10 mM Tris-HCl pH = 8.0, 1 mM EDTA at 70°C/2000 rpm for 1 hour in a Thermoblock, followed by ethanol precipitation in 0.3M sodium acetate pH = 5.5 in the presence of 25 μg glycogen.
cDNA circularization and library construction PCR
Purified cDNA was circularized with CircLigase ssDNA ligase (Lucigen) in 1X reaction buffer supplemented with 1 mM ATP, 50 mM MgCl2, and 1M betaine for 3 hours at 60°C, followed by enzyme inactivation for 10 min at 80°C. One-fifth of circularized cDNA was directly used for library construction PCR with a common forward (5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCT∗C-3′) and unique indexed reverse primers (5′-CAAGCAGAAGACGGCATACGAGATNNNNNNGTGACTGGAGTTCAGACGTGT∗G-3′, asterisks denote a phosphorothioate bond and NNNNNN corresponds to the reverse complement of an Illumina index sequence). Amplification was performed with KAPA HiFi DNA Polymerase (Roche) in 1X GC buffer with initial denaturation at 95°C for 3 min, followed by five to six cycles of 98°C for 20 s, 62°C for 30 s, 72°C for 30 s at a ramp rate of 3°C/sec. PCR products were purified with DNA Clean&Concentrator 5 (Zymo Research) and resolved on 8% polyacrylamide/1XTBE gels alongside pBR322 DNA-MspI Digest (NEB). The 130-220 bp region of each lane was excised and DNA was eluted from crushed gel slices in 400 μl water with continuous mixing at room temperature overnight. After ethanol precipitation in 0.3M sodium acetate pH = 5.5 and 25 μg glycogen, libraries were dissolved in 10 μl 10 mM Tris-HCl pH = 8.0, quantified with the Qubit dsDNA HS kit, and sequenced for 150 cycles on an Illumina NextSeq platform.
Northern blotting
Two micrograms of total RNA were resolved on denaturing 10% polyacrylamide/7M urea/1XTBE gels. RNA was transferred to Immobilon NY+ membranes (Millipore) in 1XTBE for 40 min at 4mA/cm2 on a TransBlot Turbo semi-dry blotting apparatus (Bio-Rad) and crosslinked at 0.04 J in a Stratalinker UV crosslinker. Membranes were incubated at 80°C for one hour and pre-hybridized in 20 mM Na2HPO4 pH = 7.2, 5xSSC, 7% SDS, 2x Denhardt, 40 μg/ml sheared salmon sperm DNA at 55°C for 4 hours. The buffer was exchanged and 10 pmol 5′-end 32P-labeled probe (Arg-UCU-4: 5′-CGGAACCTCTGGATTAGAAGTCCAGCGCGCTCGTCC-3′; Gly-CCC-2: 5′-CGGGTCGCAAGAATGGGAATCTTGCATGATAC-3′) was added, followed by hybridization at 55°C overnight. Membranes were washed three times in 25 mM Na2HPO4 pH = 7.5, 3xSSC, 5% SDS, 10x Denhardt, once in 1xSSC, 10% SDS, and exposed to PhosphorImager screens, which were subsequently scanned on a Typhoon FLA 9000 (GE Healthcare). Band intensity was quantified with ImageQuant (GE Healthcare).
Primer extension analysis of m1G37
The extent of RT arrest at m1G37 in tRNA-Leu-UAA and tRNA-Pro-UGG from S. cerevisiae was quantified via primer extension with AMV RT, an enzyme with low processivity at this modification (Werner et al., 2020). The primers were designed to enable a 4-nucleotide extension to m1G37 (tRNA-Leu-UAA: 5′-CGCGGACAACCGTCCAAC-3′; tRNA-Pro-UGG: 5′-TGAACCCAGGGCCTCT-3′) and 5'-end-labeled with γ-32P-ATP. 3 μg of total RNA from exponentially growing yeast cells was mixed with 1 pmol end-labeled primer and incubated at 95°C for 3 min followed by slow cooling to 37°C. RT reactions were assembled by adding 15 U AMV RT (Promega), 0.5 mM dNTPs, 20 U SUPERase In (Ambion) and 1X AMV RT buffer in a 5-μl volume. Following incubation at 37°C for 45 min, reactions were stopped by addition of 5 μl 2X RNA loading dye (47.5% Formamide, 0.01% SDS, 0.01% bromophenol blue, 0.005% Xylene Cyanol, 0.5 mM EDTA), boiled at 95°C for 5 min, and resolved on a denaturing 15% PAA/7M urea/1X TBE gel. The gel was exposed at −80°C to a PhosphorImager screen, which was scanned on a Typhoon FLA 9000 (GE Healthcare). Band intensity was quantified with ImageQuant (GE Healthcare).
Quantification and statistical analysis
Read preprocessing
Sequencing libraries were demultiplexed using cutadapt v2.5 (Martin, 2011) and a fasta file (barcodes.fa) of the first 10 nt for the four different 3′ adapters (see 3′ adapter ligation above). Indels in the alignment to the adapter sequence were disabled with --no-indels. Following demultiplexing, reads were further trimmed to remove the two 5′-RN nucleotides introduced by circularization from the RT primer with -u 2. In both processing steps, reads shorter than 10 nt were discarded using -m 10. Example commands for demultiplexing and 5′ nucleotide trimming:
cutadapt --no-indels -a file:barcodes.fa -m 10 -o mix1_{name}_trim.fastq.gz mix.fastq.gz
cutadapt -j 40 -m 10 -u 2 -o mix1_barcode1_trimFinal.fastq.gz mix1_barcode1_trim.fastq.gz
Modification indexing and clustering
mim-tRNAseq uses modification data from MODOMICS (Boccaletto et al., 2018) to guide accurate alignment of short reads from tRNAs. A prepackaged set of data is available for S. cerevisiae, S. pombe, C. elegans, D. melanogaster, M. musculus, H. sapiens and E. coli, and can be specified with the --species parameter. For other organisms, mim-tRNAseq requires a fasta file of predicted genomic tRNA sequences (-t) and a tRNAscan-SE “out” file containing information about tRNA introns (-o), both of which should be obtained from GtRNAdb (Chan and Lowe, 2016) or from running tRNAscan-SE (Lowe and Chan, 2016) on the genome of interest. Lastly, a user-generated sample input file is required which contains two tab-separated columns specifying the path to trimmed tRNA-seq reads in fastq format, and the experimental condition of each fastq file. Additionally, a mitochondrial tRNA fasta reference is supplied with the prepackaged data inputs listed above, or may be supplied (-m) for custom genomes as a fasta file obtained from mitotRNAdb (Jühling et al., 2009). mim-tRNAseq automatically removes nuclear-encoded mitochondrial tRNAs (nmt-tRNAs) and tRNA species with undetermined anticodons (where applicable), generates mature, processed tRNA sequences (with appended 3′-CCA if necessary, 5′-G for tRNA-His, and spliced introns), and fetches species-matched MODOMICS entries accordingly. Transcript sequences are then matched to MODOMICS entries using BLAST in order to index all known instances of residues modified at the Watson-Crick face within each tRNA. An additional modifications file for modifications reported in the literature but not yet added to MODOMICS may be supplied and is automatically processed by the pipeline (e.g., I34 annotation; Arimbasseri et al., 2015; Torres et al., 2015). tRNA clustering is enabled with the --cluster parameter, which utilizes the usearch --cluster_fast algorithm (Edgar, 2010) to cluster tRNA sequences by a user-defined sequence identity threshold (customizable with --cluster-id). Regardless of the chosen threshold, only tRNAs sharing an anticodon are clustered to maintain isoacceptor resolution in cases where tRNA transcripts differ by a single nucleotide in the anticodon. The clusters are re-centered based on the number of identical sequences, and this is used to re-cluster and improve the selection of a representative centroid/parent sequence for each cluster (https://www.drive5.com/usearch/manual7/recenter.html). Polymorphisms between cluster members are recorded, and mismatches at these sites during alignment are tolerated, but they are not included in misincorporation analysis for modified sites. Since inosine is interpreted as a G during reverse transcription, annotated inosines are changed to G in tRNA reference sequences.
Read alignment and modification discovery
After clustering, reads are aligned using GSNAP to the representative centroid cluster sequences of mature tRNA transcripts. By enabling SNP-tolerant alignment with --snp-tolerance, the indexed modified sites are treated as pseudo-SNPs to allow modification-induced mismatches at these sites in a sequence- and position-specific manner. Soft-clipping during alignment in combination with the GSNAP parameter --ignore-trim-in-filtering = 1 ensures that non-templated nucleotide extensions are not counted as mismatches during alignment. Mismatch tolerance outside of indexed SNPs is controlled using the --max-mismatches parameter, where an integer of allowed mismatches per read can be provided, or a relative mismatch fraction of read length between 0.0 and 0.1 can be supplied (default 0.1). If --remap is specified, then misincorporation analysis is performed and new, unannotated modifications are called where --misinc-thresh (total misincorporation proportion at a residue; default is 0.1 or 10%) and --min-cov (minimum total coverage for a cluster) regulate the calling of new modifications, which exclude mismatch sites between cluster members appearing as misincorporations in this analysis. The existing SNP index is then updated with these new sites, and realignment of all reads is performed with a mismatch tolerance set using --remap-mismatches. New potential inosine sites are classified for position 34 where a reference A nucleotide is misincorporated with a G in 95% or more total misincorporation events. Both --remap and --max-mismatches are extremely useful for detecting unknown modifications in poorly annotated tRNAs, subsequently allowing more accurate and efficient read alignment, which improves the results of all downstream analyses. Users should consider a low mismatch tolerance during remap to avoid inaccuracy resulting from lenient alignment parameters. We recommend a relative mismatch fraction of 0.075 during remapping (--remap-mismatches 0.075). Only uniquely mapped reads are retained for post-alignment analyses.
Read deconvolution
This process aims to recapitulate the single-transcript resolution of --cluster-id 1 (see above), but with the alignment accuracy and decreased multi-mapping achieved at lower --cluster-id values. The deconvolution algorithm first searches each cluster of tRNA reference sequences for single-nucleotide differences that distinguish among cluster members. For this, each nucleotide in a reference sequence is assessed for uniqueness at that position when compared to all other reference sequences in the cluster. If a nucleotide is unique in position and identity for a specific tRNA reference in the cluster, it is catalogued. Then, after alignment, each read is assessed for mismatches to the cluster parent to which it was aligned. These are then scanned individually to find potential matches to the previously catalogued set for the cluster which can distinguish unique tRNA references. Based on the presence and identity of a unique distinguishing mismatch, a read is then be assigned to a specific tRNA reference within a cluster. Depending on the organism and/or cluster ID threshold, unique distinguishing mismatches may not always be present for all tRNA references in a cluster. Reads without distinguishing mismatches remain assigned to the cluster parent, which is then marked as not fully deconvolved. Using this algorithm, uniquely aligned reads are assigned to individual tRNA sequences in the reference (where possible) before any of the downstream analyses detailed below. For differential expression analyses of reads summed per tRNA anticodon, read deconvolution is not necessary and therefore not performed.
Modification, RT stop, readthrough and 3′ CCA end analyses
Following read deconvolution, all other mismatched positions for the read are extracted from alignment records in bam files, and converted into positions relative to the unique transcript to which the read was assigned (or the cluster parent if definitive assignment is not possible). The identity of the misincoporated nucleotide is recorded to enable signature analysis, and the counts of mismatches for each of the four nucleotides for all reads with the misincorporation are normalized relative to total read coverage at that position. Stops during reverse transcription are extracted from the alignment start position of each read relative to the reference (5′ read ends correspond to cDNA 3′ ends during RT) and normalized to total read counts for the unique tRNA. Similarly, readthrough for each position is calculated as the fraction of reads that stop at a position relative to read coverage at each position (as opposed to stop proportions which are normalized to total tRNA read coverage). This value is then subtracted from one to estimate the proportion of reads per position that extend beyond that site, and the minimum value in a 3-nucleotide window centered around the modification is recorded. Using a 3-nucleotide window ensures that potential variance in the position at which the RT stalls due to the modification is accounted for. Taking the minimum value of readthrough for these 3 nucleotides reduces the likelihood of readthrough overestimation. Misincorporation, stop data, and readthrough per unique tRNA sequence, per position are output as tab-separated files, and global heatmaps showing misincorporation and stop proportions across all unique tRNA sequences are plotted per experimental condition. Misincorporation signatures are also plotted for well-known conserved modified tRNA sites (9, 20, 26, 32, 34, 37 and 58) separated by upstream and downstream sequence context to assess potential factors influencing misincorporation signatures. Lastly, the dinucleotide at the 3′ ends of reads is quantified, so long as the read aligns to the conserved 3′-CCA tail of the reference. Proportions of transcripts with absent 3′ tails, 3′-C, 3′-CC and 3′-CCA are calculated per unique tRNA sequence and plotted pairwise between conditions for quantitation and comparison of functional tRNA pools, or tRNA charging fractions in periodate oxidation experiments.
Post-alignment analyses
The cluster deconvolution algorithm allows coverage analysis, novel modification discovery and read counting for tRNA quantitation to be done at the level of unique tRNA sequences. Coverage is calculated as the depth of reads at all positions across a tRNA sequence and plotted using custom R scripts. Cytosolic tRNAs with low read coverage can be filtered at the coverage analysis step by supplying a minimum coverage threshold to --min-cov. Unique tRNA sequences filtered out here are excluded from all downstream analyses, except differential expression analysis by DESeq2 (Love et al., 2014) where all unique tRNA sequences are included. Normalized coverage (read fraction relative to library size) is plotted per sample in 25 bins across gene length in a metagene analysis. Normalized coverage is also scaled relative to the second last bin to account for potential differences in 3′ CCA intactness. Read counts per unique tRNA sequence are summed to calculate read counts per isoacceptor family (all tRNAs sharing an anticodon). These counts are subsequently used by a DESeq2 pipeline for count transformations, sample distance analysis using distance matrix heatmaps, PCA plots, and differential expression analysis at the level of isoacceptor families and unique tRNA transcripts (only for completely resolved clusters). In the case that only one experimental condition is supplied, or if there are no replicates for one or more conditions, differential expression analysis is not performed on these samples, but a normalized counts table is still produced for investigations into tRNA abundance.
Data analysis with the mim-tRNAseq package
The following parameters were used for the analysis of mim-tRNAseq generated sequencing datasets (see mimseq --help or https://mim-trnaseq.readthedocs.io/en/latest/intro.html for full explanations of parameters; package version v0.2.5.6):
S. cerevisiae: --cluster --cluster-id 0.90 --snp-tolerance --min-cov 2000 --max-mismatches 0.1 --control-condition Exp --cca-analysis --remap --remap-mismatches 0.075
S. pombe: --cluster --cluster-id 0.95 --snp-tolerance --min-cov 2000 --max-mismatches 0.1 --control-condition Exp --cca-analysis --remap --remap-mismatches 0.075
D. melanogaster: --cluster --cluster-id 0.95 --snp-tolerance --min-cov 2000 --max-mismatches 0.1 --control-condition bg3 --cca-analysis --remap --remap-mismatches 0.075
H. sapiens: --snp-tolerance --cluster --cluster-id 0.95 --min-cov 2000 --max-mismatches 0.1 --control-condition kiPS --cca-analysis --remap --remap-mismatches 0.075
tRNA read alignment with Bowtie and Bowtie 2
To test previously used alignment strategies as in DM-tRNAseq (Zheng et al., 2015) or ARM-seq (Cozen et al., 2015), a non-redundant set of reference human tRNA transcripts was created by fetching the full set of 610 predicted tRNA genes for human genome hg19 from GtRNAdb (Chan and Lowe, 2016) and the 22 mitochondrially encoded human tRNA genes from mitotRNAdb (Jühling et al., 2009). Following intron removal and addition of 3′ CCA (for nuclear-encoded transcripts) and 5′-G (for tRNA-His), a curated set of 596 genes (excluding anticodon/isotype mismatch and nuclear-encoded mitochondrial tRNAs) were collapsed into 420 unique sequences. Corresponding Bowtie and Bowtie 2 indices were built from this set of references. Bowtie alignment was performed with a maximum of 3 allowed mismatches per read (-v 3), filtering for uniquely aligning reads (-m 1) and ensuring the best alignment from the best stratum (i.e., reads with the least number of mismatches) were reported (--best --strata). Bowtie 2 alignments were performed in very sensitive local mode (--very-sensitive --local) and up to 100 alignments per read were allowed (-k 100). Read quality scores were ignored for alignment score and mismatch penalty calculation (--ignore-quals) with increased penalties for ambiguous characters (“N”) in reference or read (--np 5). Output alignments in SAM format were reordered to match read order in input fastq file (--reorder). The alignment commands for both algorithms are given below:
bowtie -v 3 -m 1 --best --strata --threads 40 -S
bowtie2 --local -x -k 100 --very-sensitive --ignore-quals --np 5 --reorder -p 40 -U
QuantM-tRNAseq data for HEK293 T-Rex Flp-IN cells downloaded from the NCBI Gene Expression Omnibus repository was adapter-trimmed and analyzed with Bowtie 2 as described in (Pinkard et al., 2020):
bowtie2 --local --score-min G,1,8 -D 20 -R 3 -N 1 -L 10 -i S,1,0.5
Sequence logo analysis
Alignment files for uniquely aligned reads from human HEK293T and S. cerevisiae cells were utilized to generate frequency plots of untemplated nucleotide additions by TGIRT, and 5′ sequence logos in each sample. Briefly, CIGAR strings for each unique alignment were assessed for GSNAP soft-clipped nucleotides representing untemplated additions. The number of additions per read were recorded and plotted as frequency histograms. Since a total of 3 additions or less were present in > 90% of reads analyzed, we generated sequence logos using the Python package Logomaker (Tareen and Kinney, 2020) for these reads using soft-clipped residues and the first 10 nucleotides after them. For the logo representing all cataloged tRNA genes, we used mature tRNA transcript sequences from each genome present in GtRNAdb, and generated a multiple sequence alignment of these using Infernal (Nawrocki and Eddy, 2013). A sequence logo was then generated from the first 11 nucleotides of each aligned tRNA transcript (in order to include G-1 for tRNA-His, plus 10 additional nucleotides as in the uniquely aligned read logo above).
Differential modification analysis
To test for global differential modification between two conditions, first, misincorporation proportion and coverage data generated by mim-tRNAseq were used to calculate absolute counts of modified and unmodified bases per position for each resolved tRNA transcript. Then, log odds ratios (logOR) were calculated for each position, x, as follows:
where Ma and Mb are the counts of modified nucleotides at position x in condition a and b, and Ua and Ub are the counts of unmodified nucleotides at position x in condition a and b, respectively. Significance for each logOR was determined with chi-square tests using the respective modified and unmodified nucleotide counts for each condition in a two-dimensional contingency table for the Pearson’s chi-square test. Correction for multiple testing was performed with the FDR method. Following significance tests, logOR values were filtered for FDR-adjusted p values ≤ 0.01, absolute log2 fold-changes ≥ 1, and total misincorporation at the given position of 10% or more in at least one of the conditions to ensure only sites with high-confidence misincorporation levels are kept. The resulting logOR were used in generating heatmaps for individual contrasts between cell types or experimental conditions.
Acknowledgments
We are grateful to K. Strasser, F. Kliem, and M. Moschref for assistance with library construction; the Max Planck Institute of Biochemistry Core Facility for high-throughput sequencing; S. Braun, P. Becker, and O. Griesbeck for reagents; T. Wu for help with GSNAP; M. Colomé-Tatché and J. Gagneur for data analysis advice; A. Andersen (Life Science Editors) for manuscript feedback; F. Bonneau and E. Conti for sharing resources; and M. Helm for insightful suggestions. D.D.N. acknowledges the Wellcome Trust Sanger Institute as the source of the HPSI0214i-kucg_2 hiPSC line, which was generated under the Human Induced Pluripotent Stem Cell Initiative supported by the Wellcome Trust (WT098051) and the National Institute for Health Research (NIHR)/Wellcome Trust Clinical Research Facility, and acknowledges Life Science Technologies Corporation as the provider of Cytotune. A.B. and G.R. were supported by the International Max Planck Research School for Molecular Life Sciences. This work was funded by the Max Planck Society and the European Research Council under the European Union’s Horizon 2020 Research and Innovation Programme (ERC Starting Grant No. 803825-TransTempoFold to D.D.N.)
Author contributions
Conceptualization, D.D.N.; Methodology, A.B. and D.D.N.; Investigation, D.D.N. and G.R.; Software, A.B.; Formal Analysis, A.B. and D.D.N.; Visualization, A.B. and D.D.N.; Writing – Original Draft, A.B. and D.D.N.; Writing – Review & Editing, A.B., G.R., and D.D.N.; Supervision and Funding Acquisition, D.D.N.
Declaration of interests
The authors declare no competing interests.
Published: February 12, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.molcel.2021.01.028.
Supplemental information
References
- Anderson J., Phan L., Cuesta R., Carlson B.A., Pak M., Asano K., Björk G.R., Tamame M., Hinnebusch A.G. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri A.G., Blewett N.H., Iben J.R., Lamichhane T.N., Cherkasova V., Hafner M., Maraia R.J. RNA Polymerase III output is functionally linked to tRNA dimethyl-G26 modification. PLoS Genet. 2015;11:e1005671. doi: 10.1371/journal.pgen.1005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri A.G., Iben J., Wei F.Y., Rijal K., Tomizawa K., Hafner M., Maraia R.J. Evolving specificity of tRNA 3-methyl-cytidine-32 (m3C32) modification: a subset of tRNAsSer requires N6-isopentenylation of A37. RNA. 2016;22:1400–1410. doi: 10.1261/rna.056259.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crécy-Lagard V., Ross R., Limbach P.A., Kotter A. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P.P., Lowe T.M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44(D1):D184–D189. doi: 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Patton J.T. Reverse transcriptase adds nontemplated nucleotides to cDNAs during 5′-RACE and primer extension. Biotechniques. 2001;30:574–580. doi: 10.2144/01303rr02. 582. [DOI] [PubMed] [Google Scholar]
- Clark W.C., Evans M.E., Dominissini D., Zheng G., Pan T. tRNA base methylation identification and quantification via high-throughput sequencing. RNA. 2016;22:1771–1784. doi: 10.1261/rna.056531.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., de Hoon M.J.L. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozen A.E., Quartley E., Holmes A.D., Hrabeta-Robinson E., Phizicky E.M., Lowe T.M. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods. 2015;12:879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar K.A., Mobley E.M., Radek A.J., Pan T. Exploring the regulation of tRNA distribution on the genomic scale. J. Mol. Biol. 2004;337:31–47. doi: 10.1016/j.jmb.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Dittmar K.A., Goodenbour J.M., Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221–e229. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt H.A., Tsang H.H., Dai D.C., Liu Y., Bostan B., Fahlman R.P. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res. 2009;37:2461–2470. doi: 10.1093/nar/gkp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Ellis S.R., Morales M.J., Li J.M., Hopper A.K., Martin N.C. Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J. Biol. Chem. 1986;261:9703–9709. [PubMed] [Google Scholar]
- Evans M.E., Clark W.C., Zheng G., Pan T. Determination of tRNA aminoacylation levels by high-throughput sequencing. Nucleic Acids Res. 2017;45:e133. doi: 10.1093/nar/gkx514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper H.B., Masuda I., Frenkel-Morgenstern M., Hou Y.-M. Maintenance of protein synthesis reading frame by EF-P and m(1)G37-tRNA. Nat. Commun. 2015;6:7226. doi: 10.1038/ncomms8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A.P., Keller W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science. 1999;286:1146–1149. doi: 10.1126/science.286.5442.1146. [DOI] [PubMed] [Google Scholar]
- Gogakos T., Brown M., Garzia A., Meyer C., Hafner M., Tuschl T. Characterizing expression and processing of precursor and mature human tRNAs by hydro-tRNAseq and PAR-CLIP. Cell Rep. 2017;20:1463–1475. doi: 10.1016/j.celrep.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenbour J.M., Pan T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006;34:6137–6146. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Guy M.P., Podyma B.M., Preston M.A., Shaheen H.H., Krivos K.L., Limbach P.A., Hopper A.K., Phizicky E.M. Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA. 2012;18:1921–1933. doi: 10.1261/rna.035287.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Guy M.P., Kon Y., Phizicky E.M. Lack of 2′-O-methylation in the tRNA anticodon loop of two phylogenetically distant yeast species activates the general amino acid control pathway. PLoS Genet. 2018;14:e1007288. doi: 10.1371/journal.pgen.1007288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O., Gendrel C.-G., Soularue P., Gidrol X., Sentenac A., Werner M., Lefebvre O. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauenschild R., Tserovski L., Schmid K., Thüring K., Winz M.-L., Sharma S., Entian K.-D., Wacheul L., Lafontaine D.L.J., Anderson J. The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res. 2015;43:9950–9964. doi: 10.1093/nar/gkv895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer E.E., Ozadam H., Ricci E.P., Cenik C., Moore M.J. An optimized kit-free method for making strand-specific deep sequencing libraries from RNA fragments. Nucleic Acids Res. 2015;43:e2. doi: 10.1093/nar/gku1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A., Fallmann J., Vilardo E., Mörl M., Stadler P.F., Amman F. Accurate mapping of tRNA reads. Bioinformatics. 2018;34:1116–1124. doi: 10.1093/bioinformatics/btx756. [DOI] [PubMed] [Google Scholar]
- Ishimura R., Nagy G., Dotu I., Zhou H., Yang X.-L., Schimmel P., Senju S., Nishimura Y., Chuang J.H., Ackerman S.L. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman J.E., Montagne R.K., Malik H.S., Phizicky E.M. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003;9:574–585. doi: 10.1261/rna.5070303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob D., Thüring K., Galliot A., Marchand V., Galvanin A., Ciftci A., Scharmann K., Stock M., Roignant J.-Y., Leidel S.A. Absolute quantification of noncoding RNA by microscale thermophoresis. Angew. Chem. Int. Ed. Engl. 2019;58:9565–9569. doi: 10.1002/anie.201814377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaca E., Weitzer S., Pehlivan D., Shiraishi H., Gogakos T., Hanada T., Jhangiani S.N., Wiszniewski W., Withers M., Campbell I.M., Baylor Hopkins Center for Mendelian Genomics Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell. 2014;157:636–650. doi: 10.1016/j.cell.2014.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katibah G.E., Qin Y., Sidote D.J., Yao J., Lambowitz A.M., Collins K. Broad and adaptable RNA structure recognition by the human interferon-induced tetratricopeptide repeat protein IFIT5. Proc. Natl. Acad. Sci. U S A. 2014;111:12025–12030. doi: 10.1073/pnas.1412842111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H., Gonçalves A., Leha A., Afzal V., Alasoo K., Ashford S., Bala S., Bensaddek D., Casale F.P., Culley O.J. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370–375. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner S., Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015;16:98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- Kutter C., Brown G.D., Gonçalves A., Wilson M.D., Watt S., Brazma A., White R.J., Odom D.T. Pol III binding in six mammals shows conservation among amino acid isotypes despite divergence among tRNA genes. Nat. Genet. 2011;43:948–955. doi: 10.1038/ng.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Xiong X., Zhang M., Wang K., Chen Y., Zhou J., Mao Y., Lv J., Yi D., Chen X.-W. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell. 2017;68:993–1005.e9. doi: 10.1016/j.molcel.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-C., Boone M., Meuris L., Lemmens I., Van Roy N., Soete A., Reumers J., Moisse M., Plaisance S., Drmanac R. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat. Commun. 2014;5:4767. doi: 10.1038/ncomms5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M., Chan P.P. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44(W1):W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehigashi T., Dunkle J.A., Miles S.J., Dunham C.M. Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops. Proc. Natl. Acad. Sci. U S A. 2014;111:12740–12745. doi: 10.1073/pnas.1409436111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M.M.E. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. [Google Scholar]
- Masuda I., Matsubara R., Christian T., Rojas E.R., Yadavalli S.S., Zhang L., Goulian M., Foster L.J., Huang K.C., Hou Y.-M. tRNA methylation is a global determinant of bacterial multi-drug resistance. Cell Syst. 2019;8:302–314.e8. doi: 10.1016/j.cels.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy N.J., Ingolia N.T. Transcriptome-wide measurement of translation by ribosome profiling. Methods. 2017;126:112–129. doi: 10.1016/j.ymeth.2017.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S., Ghanem E., Smith W., Sheeter D., Qin Y., King O., Polioudakis D., Iyer V.R., Hunicke-Smith S., Swamy S. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA. 2013;19:958–970. doi: 10.1261/rna.039743.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y., Helm M. Methods for RNA modification mapping using deep sequencing: established and new emerging technologies. Genes (Basel) 2019;10:35. doi: 10.3390/genes10010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y., Muller S., Behm-Ansmant I., Branlant C. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 2007;425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- Nawrocki E.P., Eddy S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernod K., Schaeffer L., Chicher J., Hok E., Rick C., Geslain R., Eriani G., Westhof E., Ryckelynck M., Martin F. The nature of the purine at position 34 in tRNAs of 4-codon boxes is correlated with nucleotides at positions 32 and 38 to maintain decoding fidelity. Nucleic Acids Res. 2020;48:6170–6183. doi: 10.1093/nar/gkaa221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky E.M., Alfonzo J.D. Do all modifications benefit all tRNAs? FEBS Lett. 2010;584:265–271. doi: 10.1016/j.febslet.2009.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkard O., McFarland S., Sweet T., Coller J. Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat. Commun. 2020;11:4104–4115. doi: 10.1038/s41467-020-17879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Yao J., Wu D.C., Nottingham R.M., Mohr S., Hunicke-Smith S., Lambowitz A.M. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA. 2016;22:111–128. doi: 10.1261/rna.054809.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail M.A., Otto T.D., Gu Y., Harris S.R., Skelly T.F., McQuillan J.A., Swerdlow H.P., Oyola S.O. Optimal enzymes for amplifying sequencing libraries. Nat. Methods. 2011;9:10–11. doi: 10.1038/nmeth.1814. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D.N., Stewart A.J., Huff J.T., Cairns B.R. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl. Acad. Sci. U S A. 2003;100:14695–14700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryvkin P., Leung Y.Y., Silverman I.M., Childress M., Valladares O., Dragomir I., Gregory B.D., Wang L.S. HAMR: high-throughput annotation of modified ribonucleotides. RNA. 2013;19:1684–1692. doi: 10.1261/rna.036806.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M., Sas-Chen A., Nir R., Winkler R., Nachshon A., Bar-Yaacov D., Erlacher M., Rossmanith W., Stern-Ginossar N., Schwartz S. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017;551:251–255. doi: 10.1038/nature24456. [DOI] [PubMed] [Google Scholar]
- Sas-Chen A., Schwartz S. Misincorporation signatures for detecting modifications in mRNA: Not as simple as it sounds. Methods. 2019;156:53–59. doi: 10.1016/j.ymeth.2018.10.011. [DOI] [PubMed] [Google Scholar]
- Schmitt B.M., Rudolph K.L.M., Karagianni P., Fonseca N.A., White R.J., Talianidis I., Odom D.T., Marioni J.C., Kutter C. High-resolution mapping of transcriptional dynamics across tissue development reveals a stable mRNA-tRNA interface. Genome Res. 2014;24:1797–1807. doi: 10.1101/gr.176784.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S., Cedergren R. A correlation between N2-dimethylguanosine presence and alternate tRNA conformers. RNA. 1995;1:886–891. [PMC free article] [PubMed] [Google Scholar]
- Swinehart W.E., Henderson J.C., Jackman J.E. Unexpected expansion of tRNA substrate recognition by the yeast m1G9 methyltransferase Trm10. RNA. 2013;19:1137–1146. doi: 10.1261/rna.039651.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tareen A., Kinney J.B. Logomaker: beautiful sequence logos in Python. Bioinformatics. 2020;36:2272–2274. doi: 10.1093/bioinformatics/btz921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A.G., Piñeyro D., Rodríguez-Escribà M., Camacho N., Reina O., Saint-Léger A., Filonava L., Batlle E., Ribas de Pouplana L. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 2015;43:5145–5157. doi: 10.1093/nar/gkv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A.G., Reina O., Stephan-Otto Attolini C., Ribas de Pouplana L. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc. Natl. Acad. Sci. U S A. 2019;116:8451–8456. doi: 10.1073/pnas.1821120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Werner S., Schmidt L., Marchand V., Kemmer T., Falschlunger C., Sednev M.V., Bec G., Ennifar E., Höbartner C., Micura R. Machine learning of reverse transcription signatures of variegated polymerases allows mapping and discrimination of methylated purines in limited transcriptomes. Nucleic Acids Res. 2020;48:3734–3746. doi: 10.1093/nar/gkaa113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.D., Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Yao J., Wu D.C., Lambowitz A.M. Improved TGIRT-seq methods for comprehensive transcriptome profiling with decreased adapter dimer formation and bias correction. Sci. Rep. 2019;9:7953. doi: 10.1038/s41598-019-44457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Liu F., Pyle A.M. An ultraprocessive, accurate reverse transcriptase encoded by a metazoan group II intron. RNA. 2018;24:183–195. doi: 10.1261/rna.063479.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Qin Y., Clark W.C., Dai Q., Yi C., He C., Lambowitz A.M., Pan T. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods. 2015;12:835–837. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Rauch S., Dai Q., Cui X., Zhang Z., Nachtergaele S., Sepich C., He C., Dickinson B.C. Evolution of a reverse transcriptase to map N1-methyladenosine in human messenger RNA. Nat. Methods. 2019;16:1281–1288. doi: 10.1038/s41592-019-0550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang F., Fuchs R.T., Sun Z., Zheng Y., Robb G.B. Structural bias in T4 RNA ligase-mediated 3′-adapter ligation. Nucleic Acids Res. 2012;40:e54. doi: 10.1093/nar/gkr1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the sequencing data reported in this paper is GEO: GSE152621. The mim-tRNAseq computational pipeline is available under a GNU public License v3 at https://github.com/nedialkova-lab/mim-tRNAseq. A package description and installation guide are available at https://mim-trnaseq.readthedocs.io/en/latest/.