Abstract
Collagen hybridizing peptides (CHPs) have a great potential for use in targeted drug delivery, diagnostics, and regenerative medicine due to their ability to specifically bind to denatured collagens associated with many pathologic conditions. Since peptides generally suffer from poor enzymatic stability, resulting in rapid degradation and elimination in vivo, CHP’s serum stability is a critical parameter that may dictate its pharmacokinetic behavior. Here, we report the serum stability of a series of monomeric CHP derivatives and establish how peptide length, amino acid composition, terminal modification, and linker chemistry influence their availability in serum. We show that monomeric CHPs comprised of the collagen-like Gly-Pro-Hyp motif are resistant to common serum proteinases and that their stability can be further increased by simple N-terminal labeling which negates CHP’s susceptibility to proline-specific exopeptidases. When fluorescent dyes are conjugated to a CHP via maleimide–thiol reaction, the dye can transfer from CHP onto serum proteins (e.g., albumin), resulting in an unexpected drop in signal during serum stability assays and off-target accumulation during in vivo tests. This work is the crucial first step toward understanding the pharmacokinetic behavior of CHPs, which can facilitate the development of CHP-based theranostics.
Keywords: extracellular matrix, collagen mimetic peptide, peptide degradation, proline peptidase pharmacokinetics, theranostics
Graphical Abstract
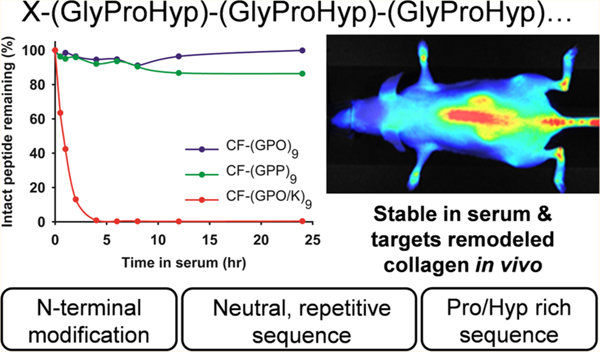
INTRODUCTION
Collagen is the most abundant protein in the human body and is a major component of the extracellular matrix (ECM).1,2 Although collagen degradation and synthesis are delicately coordinated during natural tissue homeostasis, excessive collagen remodeling is a common feature in a variety of injuries and diseases, such as cancer, arthritis, and fibrosis.3 Once cleaved by ECM-specific proteinases, such as matrix metalloproteinases (MMPs), collagen molecules become thermally unstable and spontaneously denature at body temperature.4,5 Such denatured collagen molecules can be present at high concentrations in pathological tissues and can serve as a unique biomarker for diagnosis or targeted drug delivery.6–8 Although there are various types of collagen binding molecules, such as antibodies raised against collagens,6 peptide/protein domains derived from collagen binding proteins,6,9 and peptides selected from phage display,10 most of such molecules target native collagens, not denatured collagen, and are ineffective at detecting degraded collagens associated with pathologic conditions. Therefore, a targeting agent that can specifically distinguish denatured collagen from intact collagen could lead to new diagnostics and therapeutics.
Recently, our group developed a collagen hybridizing peptide (CHP) that binds only to denatured collagen and not to intact collagen.7,8,11 The design of this peptide was based on the triple helix, a unique supersecondary protein structure almost exclusively seen in the family of collagen proteins,1,12,13 where three individual polyproline II-like chains intertwine with one another by forming interchain hydrogen bonds.1,13 The CHP is comprised of repeating units of glycine-proline-hydroxyproline (GPO): a triplet with a strong propensity to fold into the triple helix.12–15 We discovered that a single strand (GPO)9 can hybridize to the denatured collagen chains7,8,11 in a fashion similar to a primer binding to a melted DNA strand during PCR. This peptide and its analogs were previously called collagen mimetic peptides (CMPs) or collagen-like peptides (CLPs),8,16 but we believe CHP is a more descriptive name when it is used to bind to denatured collagen. The CHP sequence inherently self-assembles into a triple helical homotrimer which has no capacity to hybridize with collagen strands.6,17 To address this problem, we developed a caged CHP with a photocleavable nitrobenzyl (NB) group attached to the central glycine [sequence: (GPO)4NBGPO(GPO)4],7 which sterically prevents the monomeric CHP strands from folding into a triple helix; yet, brief UV irradiation which removes the cage group can restore the triple helical folding propensity, allowing CHP-collagen hybridization on demand.7,18 By in vivo injection of the UV activated caged CHP labeled with a near-infrared-fluorophore (NIRF), we were able to image denatured collagens in tissues undergoing normal (e.g., in bone and cartilage) and pathological remodeling (e.g., in cancer xenografts or transgenic mice with Marfan syndrome).7 These results demonstrated an entirely new way to target diseased tissues, and they showcased new translational opportunities for CHP in detecting, staging, and delivering therapeutics to many types of pathologic conditions associated with abnormal ECM remodeling.
Although understanding the pharmacokinetics of the CHPs is essential for their diagnostic and therapeutic applications, we know very little about the stability of CHPs against proteinases in blood. Without any strategic structural modification, peptides generally suffer from short lifetime in the bloodstream because they are easily digested by the proteinases abundant in serum.19 It is well-known that collagen and collagen mimetic peptides, which are in triple helical conformation, are resistant to most proteases4,20,21 except for collagen-specific ones, such as MMPs and cathepsin K. In fact, most of the intravenously injected triple helical peptides are eliminated through glomerular filtration in the kidneys.22 Despite the abundance of literature on the serum stability of triple helical CMPs, there is limited information on the serum stability for nontriple helical, single strand CHP. Our previous in vivo NIRF imaging results17 demonstrated that the binding of CHP in the targeted tissue is stable for days,7 suggesting high serum stability. Therefore, we set off to study the proteolytic stability of monomeric CHP in serum as the first step in assessing its pharmacokinetics. Such a study will also benefit a number of applications based on collagen mimetic peptides in developing tissue scaffolds, drug delivery systems, and hydrogels.23–26 Here we report the serum stability of a series of monomeric CHP derivatives that vary in length, amino acid composition, and terminal modifications as determined by reversed phase high-performance liquid chromatography (RP-HPLC) and mass spectrometry (MS). Our results indicate that the CHPs based on GPO repeats are stable in serum even in monomeric form, and that their stability can be further increased through N-terminal modification. Additionally, we show that when a fluorescent dye is conjugated to CHP via a thioether link (formed by maleimide–thiol reaction), the dye can transfer from CHP to serum proteins (e.g., albumin), giving off-target distribution during in vivo imaging experiments. Finally, we suggest the most ideal CHP structure for targeting and imaging denatured collagens in vivo.
MATERIALS AND METHODS
Peptide Synthesis and Purification.
All peptides were synthesized on rink-amide-MBHA resin purchased from Peptides International (Louisville, KY, USA). Fmoc-protected amino acid residues were purchased from CreoSalus (Louisville, KY, USA). Peptides were synthesized using standard Fmoc-chemistry on solid phase using a Focus XC autosynthesizer from Aapptec (Louisville, KY, USA). The 5(6)-carboxyfluorescein tag, from Sigma-Aldrich (St. Louis, MO, USA), was conjugated manually on resin using HBTU chemistry. Peptides containing the nitrobenzyl (NB) cage group were synthesized as previously described.7 Crude peptides were cleaved from resin using a mixture of 95% trifluoroacetic acid (TFA), 2.5% triisopropylsilane (TIS), and 2.5% water, before purification by an Agilent Prep-Star HPLC instrument (Santa Clara, CA, USA) equipped with an Agilent Zorbax SB C-18 analytical column (Santa Clara, CA, USA). A mixture of water (0.1% TFA) and acetonitrile (0.1% TFA) in a linear gradient (5–50% acetonitrile in 45 min) was used as a mobile phase during HPLC. The molecular weights of the purified peptides were verified using matrix assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS on a Bruker UltrafleXtreme (Billerica, MA, USA) (Table S1). The peptide conformation was studied by Circular Dichroism (CD) on a Jasco J-1500 CD spectrometer (Easton, MD, USA) following a previously reported protocol.7
NIRF-CHP Conjugation Reaction.
Near-infrared fluorescent (NIRF) dyes (IR800CW-maleimide, IR800CW-NHS, IR680RD-maleimide, and IR680RD-NHS) (NHS: N-hydroxysuccinimide) were purchased from LI-COR Biotechnology (Lincoln, NE, USA). For NIRF dye conjugation to peptides, 0.5 mg of dye was dissolved in 50 μL of DMSO, followed by addition to a PBS solution containing approximately 2 mg of Ac-C-Ahx-NB(GPO)9 (for reaction of dye with maleimide) or NH2-Ahx-NB(GPO)9 (for reaction of dye with NHS) (Ahx: aminohexanoic acid) and was allowed to mix overnight at 4 °C. Crude NIRF-conjugated CHPs were purified by HPLC as described above. Excess dithiothreitol was added to the maleimide reaction mixture prior to HPLC injection to breakup disulfide bonds between CHPs.
Serum Stability Study.
Peptide stock solutions were prepared in deionized water to a concentration of 1 mM. A reaction mixture (50 μM of peptide) was prepared by adding 50 μL of peptide stock solution to 0.95 mL of diluted serum solution, which was prepared by mixing 700 μL of1 × PBS and 250 μL of mouse serum from EMD Millipore (Billerica, MA, USA). The mixture was then placed in an incubator at 37 °C. CF-(GPP)9 and (GPP)9 were heated to 80 °C for 5 min followed by quenching on ice for 1 min to break up any triple helix formation prior to mixing with the mouse serum. At specified time points (0, 0.5, 1, 2, 4, 6, 8, 12, and 24 h), a 100 μL aliquot of the peptide–serum mixture was added to 200 μL of ice-chilled ethanol followed by vortexing and centrifugation at 13,800g for 2 min at 4 °C to precipitate the serum proteins. For RP-HPLC analysis, 100 μL of the supernatant was added to 900 μL of water with 0.1% TFA followed by injection into a C-18 column at 60 °C. For the mobile phase of RP-HPLC, a mixture of water (0.1% TFA) and acetonitrile (0.1% TFA) in a linear gradient (5–50% acetonitrile in 45 min) was used. UV detection wavelengths were set at 220, 280, 774, and 550 nm for unlabeled peptides, CF-labeled peptides, IR800-labeled peptides, and IR680-labeled peptides, respectively. The level of degradation was determined by the integration of the area under the HPLC peak corresponding to the intact peptide. The peak area for “zero”-minute time point was designated as 100% peptide and each peak area at successive time points was normalized to the zero-minute peak in order to calculate the percent peptide remaining. The test was done in triplicate. From the HPLC, the intact peptide peaks and peaks corresponding to degraded peptide fragments were collected and analyzed using MALDI-TOF MS to determine the location of peptide fragmentation.
Serum Albumin Binding Assay.
The serum stability protocol described above was followed, except the 250 μL of mouse serum was replaced with 250 μL of bovine serum albumin (BSA, Protea Biosciences, Morgantown, WV, USA) in 1 × PBS (40 mg/mL). After precipitation of the BSA by ice-chilled ethanol, the supernatant was analyzed via RP-HPLC to determine the area under the peak corresponding to the intact peptides at each time point. Prior to the precipitation of BSA, the peptide–BSA mixtures were also analyzed by electrospray ionization mass spectrometry (ESI-MS) on a Bruker maXis II with an electron transfer-dissociation system (Billerica, MA, USA) to verify the covalent bonding between the maleimide functionalized dyes and BSA.
In Vivo Near-Infrared Imaging.
All animal studies were performed in compliance with the regulations of the Johns Hopkins Animal Care and Use Committee. As previously described,7,18 a 100 μL PBS solution containing 4 nmol of Ac-C(IR800)-Ahx-NB(GPO)9, IR800-Ahx-NB(GPO)9, Ac-C-(IR680)-Ahx-NB(GPO)9, or IR680-Ahx-NB(GPO)9 was exposed to UV light (365 nm, >25 mW/cm2) for 5 min and immediately injected into a female adult SKH-1 mouse via tail vein. Groups of ≥3 mice were dosed with each compound. Mice were scanned using a LI-COR Pearl Impulse imager at 12, 24, 48, 72, and 96 h post injection. The 710 and 800 nm NIR fluorescence channels were used for IR680 and IR800, respectively. Following the 48 h scan, mice were euthanized and had the skin removed to allow imaging of deep tissues and organs.
RESULTS
Design of the Study.
In predicting the pharmacokinetic behavior of a peptide, it is essential to understand the peptide’s stability against proteinases in the blood, which can be reliably studied in vitro using plasma or serum.19,27–29 To investigate the structural factors that may contribute to the proteolytic stability of CHPs, we synthesized a series of CHPs which vary in length, sequence, and terminal group (Tables 1 and 2). The peptides were incubated at 37 °C in mouse serum for 24 h, and RP-HPLC profiles (representative data in Figure S1) were used to determine the amount of intact peptide at predetermined time points. To obtain reproducible results, the serum was diluted to 25% in PBS, so that the degradation reaction speed was uniformly governed by the serum concentration, and not by the peptide substrate concentrations.29 Low serum concentration also slows down the degradation rates so that their differences between peptides can be easily monitored. It is also easier to isolate digested peptides from serum protein in HPLC when its concentration is low.28 Investigating the serum stability of CHPs with high triple helical propensity (and high Tm), such as (GPO)9 in the monomeric state, is challenging, because such peptides readily self-assemble into triple helices at the digestion temperature (37 °C).7 We decided to use caged CHP to address this problem. As described above, adding a single nitrobenzyl group to the centrally located Gly can completely inhibit the self-trimerization of the entire peptide.7 Therefore, for CHPs with high Tm, the NB caged versions of the CHPs [e.g., NB(GPO)9, Table 1] were used to simulate the stable monomeric condition.
Table 1.
Sequences of Unlabeled CHP Peptidesa
| Peptide | Sequence | Conformation at 37 °C |
|---|---|---|
| (GPO)9 | NH2-GPOGPOGPOGPOGPOGPOGPOGPOGPO | Triple helix |
| NB(GPO)9b | NH2-GPOGPOGPOGPONBGPOGPOGPOGPOGPO | Monomeric |
| S(G9P9O9) | NH2–PGOGPGPOPOGOGOPPGOOPGGOOPPG | Monomeric |
| (GPP)9 | NH2-GPPGPPGPPGPPGPPGPPGPPGPPGPP | Monomeric |
| (GPO)5c | NH2-G▼PO▼G▼POGPOGPOGPO | Monomeric |
C-termini of all sequences: –CONH2.
NBG = N-(2-nitrobenzyl)glycine.
Arrowheads indicate probable cleavage sites.
Table 2.
Sequences of 5(6)-Carboxyfluorescein (CF)-Labeled CHP Peptidesa
| Peptide | Sequence | Conformation at 37 °C |
|---|---|---|
| CF-(GPO)9 | CF-GGG-GPOGPOGPOGPOGPOGPOGPOGPOGPO | Triple helix |
| CF-NB(GPO)9b | CF-GGG-GPOGPOGPOGPONBGPOGPOGPOGPOGPO | Monomeric |
| CF-S(G9P9O9) | CF-GGG-PGOGPGPOPOGOGOPPGOOPGGOOPPG | Monomeric |
| CF-(GPP)9 | CF-GGG -GPPGPPGPPGPPGPPGPPGPPGPPGPP | Monomeric |
| CF-(GPO)5 | CF-GGG-GPOGPOGPOGPOGPO | Monomeric |
| CF-(GPO/K)9c | CF-Ahx-GPOGP▼K▼GPOGPKGPOG▼PKGPOGPKGPO | Monomeric |
C-termini of all sequences: –CONH2.
NBG = N-(2-nitrobenzyl)glycine.
Arrowheads indicate probable cleavage sites.
Serum Stability of CHPs without N-Terminal Modification.
We first investigated the serum stability for a series of CHPs with free N-termini. It is well-known that the triple helical structure of collagen is highly stable against proteolytic enzymes. Although the high serum stability of triple helical collagen mimetic peptides has been validated previously,20–22 the stability of such peptides in monomeric form has not been thoroughly studied. As shown in Figure 1A, (GPO)9, which is in the triple helical structure during the digestion condition, exhibited nearly no degradation in serum during 24 h, which is in agreement with the findings of Koide and co-workers.22 The single strand version of the same peptide, NB(GPO)9, also exhibited high serum stability with approximately 90.8% of intact peptide remaining after 24 h (Figure 1A). In drastic contrast, penetratin, a control peptide that is known to have poor half-life in blood, had only 7.3% left after digestion under the same conditions (Figure S2). These results clearly indicate that the monomeric CHP, NB(GPO)9, has high serum stability and is nearly as stable as the triple helical (GPO)9.
Figure 1.
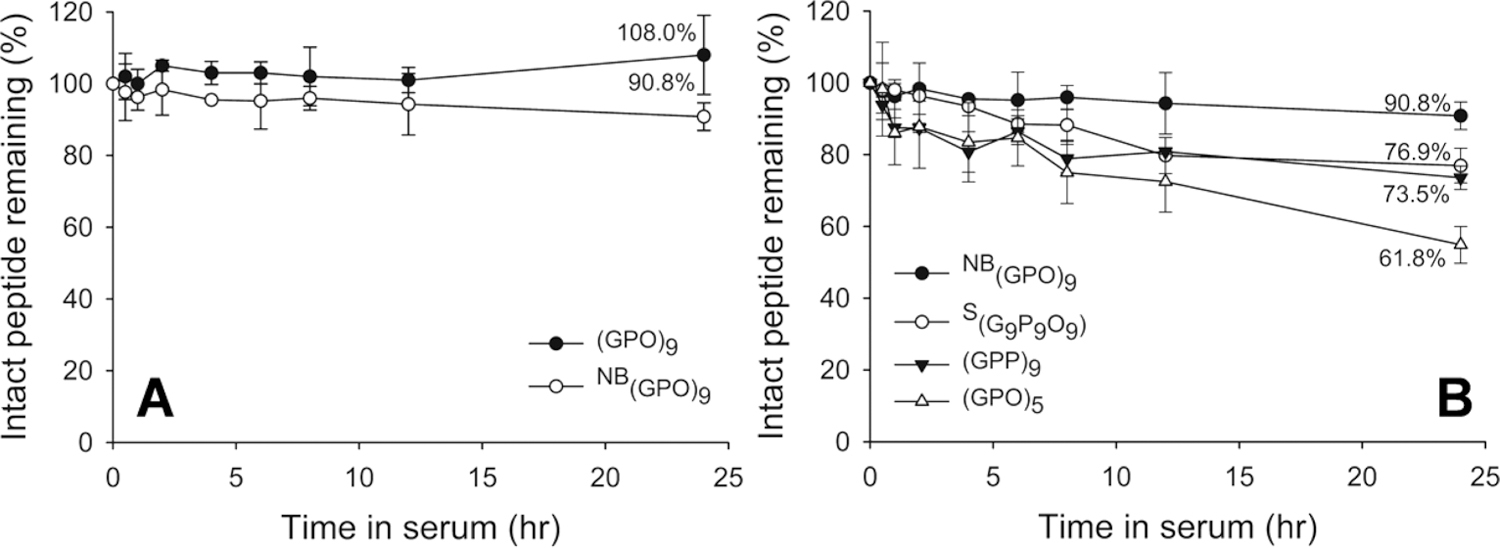
Stability profiles of unlabeled peptides in mouse serum. (A) Serum stability of triple helical (GPO)9 and monomeric NB(GPO)9. Both trimeric and monomeric CHPs exhibit high serum stability. (B) Serum stability of other unlabeled CHPs. (GPO)5, (GPP)9, and the scrambled control peptide S(G9P9O9) all exhibit lower serum stability than NB(GPO)9, with (GPO)5 showing the lowest stability, with 61.8% remaining after 24 h. Amounts of intact peptides were determined from the areas under the HPLC peak after incubation in serum at 37 °C for indicated time intervals. Percentages of intact peptides remaining after 24 h are shown. All tests were run in triplicate.
Additional CHP derivatives which have low triple helical folding propensity, and are present in monomeric form at 37 °C (Figures 1B, S3 and S4) were also examined for stability under these conditions.7 After 24 h, only 61.8% of (GPO)5 (shorter GPO repeat sequence), 76.9% of S(G9P9O9) [scrambled version of (GPO)9], and 73.5% of (GPP)9 [O → P substituted version of (GPO)9] remained intact in serum, which were markedly lower than NB(GPO)9 (90.8%). The decreased stability of these monomeric CHPs suggests that the long peptide length, the GPO triplet sequence, as well as the presence of hydroxyproline residue are all contributing to the enhanced stability of NB(GPO)9 against serum proteinases.
Serum Stability of CHPs with N-Terminal CF-Modification.
Since N-terminal modification has been reported to improve peptide’s proteolytic resistance by interfering with exopeptidases,27,29 we also investigated the serum stability of CHPs with N-terminal modification. Previously, we used CHPs terminated with 5(6)-carboxyfluorescein (CF) at the N-terminus for detecting denatured collagens in SDS-PAGE and in histological stains,17 as well as for patterning gelatin substrates.30 In this study, the CF-labeled versions of the CHPs discussed above and a CF-labeled control peptide based on the GPO sequence that has K replacing O in every other GPO triplet (Table 2) were assayed. HPLC analysis indicated that monomeric CF-(GPO)5 and CF-NB(GPO)9, as well as triple helical CF-(GPO)9 remained completely intact after 24 h of incubation in serum (Figure 2A). Monomeric CF-NB(GPP)9 (86.3% remaining) and CF-S(G9P9O9) (79.1% remaining) showed moderate degradation after 24 h. The control peptide CF-(GPO/K)9 was quickly degraded with less than half remaining after 1 h of incubation in serum. When the results were compared between N-terminal free CHPs and CF-labeled CHPs (Figure 2B), there was a substantial increase in stability for (GPO)5, going from 38.2% of peptide digested in 24 h to virtually none after N-terminal CF-labeling, while only a small improvement in stabilities was seen for CF-NB(GPO)9 and CF-NB(GPP)9. In contrast, no statistically significant change in stability was found for S(G9P9O9) after N-terminal labeling (p > 0.05), suggesting that the degradation mechanism of this scrambled sequence may be different from other peptides tested.
Figure 2.
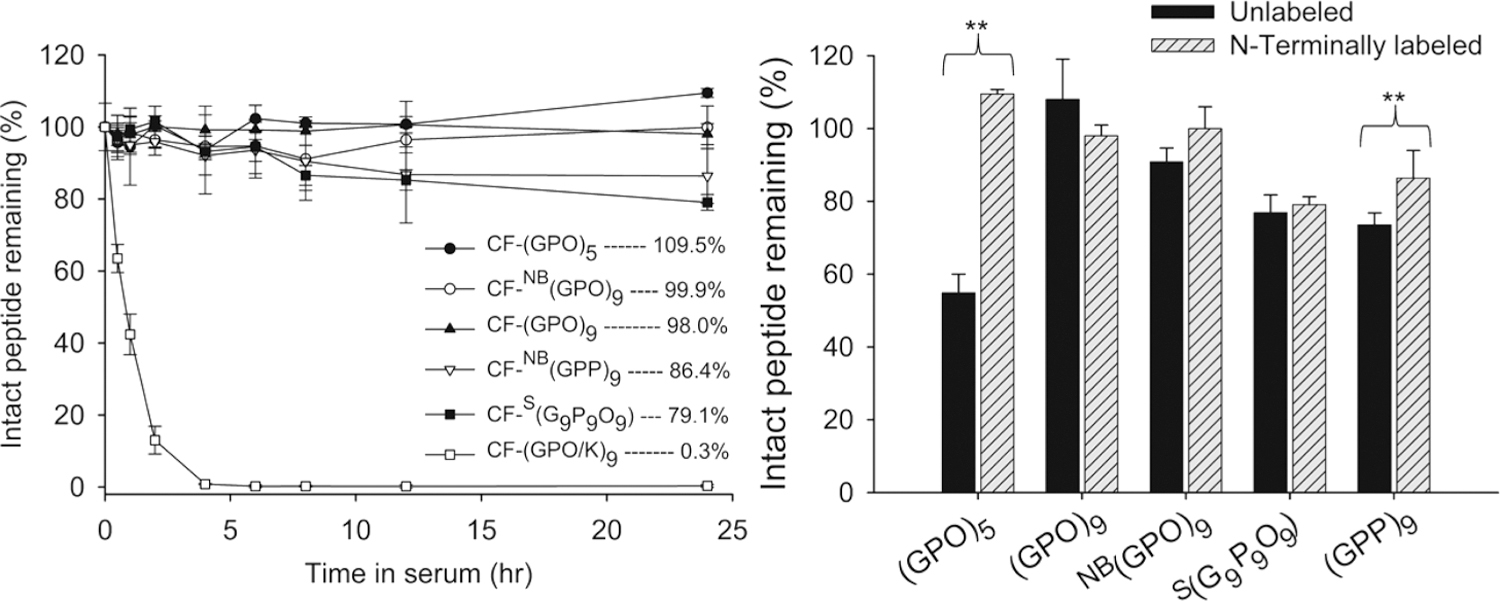
Stability profiles of peptides with N-terminal CF labels in mouse serum (left) and comparison of stability between peptides with and without N-terminal labeling (right). Amounts of intact peptides were determined from the areas under the HPLC peak after incubation in serum at 37 °C for indicated time intervals. All tests were run in triplicate. Error bars represent standard deviation. **p < 0.05 (Student’s t test), and differences in all other pairs are insignificant (p > 0.05).
Serum Stability of NIRF-Labeled CHPs (Amide Link vs Thioether Link).
After studying the structural factors affecting the CHP’s serum stability, we investigated the serum stability of CHPs labeled with near-infrared fluorophores (NIRF) which were used in previously described in vivo studies.7,18 Since commercially available NIRF dyes provide convenient conjugation to CHPs using two different chemistries: (i) N-hydroxy succinimide ester (NHS)-amine reaction resulting in an amide bond between the dye and the CHP, and (ii) maleimide–thiol reaction resulting in a thioether bond, we wanted to see if there are any effects from the two most common linker chemistries. Before NIRF conjugation with either chemistry, the N-terminus of the caged NB(GPO)9 was extended with Ahx spacer to account for the large size of the NIRF (Figure 3A). In making IR800-Ahx-NB(GPO)9, the N-terminal amine of the Ahx linker was directly reacted with IR800 NHS-ester. Ac-C(IR800)-Ahx-NB(GPO)9 was prepared by first adding an extra Cys to the N-terminus of the Ahx, followed by N-terminal acetylation, and reacting the thiol group from the Cys with IR800 maleimide. Serum stability experiments indicated that the stability of the amide-conjugated IR800-Ahx-NB(GPO)9 was high, with 81.2% of intact peptide remaining after 24 h incubation (Figure 3B). In contrast, the thioether-conjugated Ac-C(IR800)-Ahx-NB(GPO)9 exhibited rapid removal from the serum with only 16.7% remaining after 24 h (Figure 3B). This was a surprising result, since this peptide had a similar structure as the CF-NB(GPO)9 except for the spacer and the large fluorophore, both of which were not expected to affect the serum stability of the CHP. In fact, we expected that such structural changes made to the NIRF-labeled CHP would enhance the peptide’s serum stability.
Figure 3.
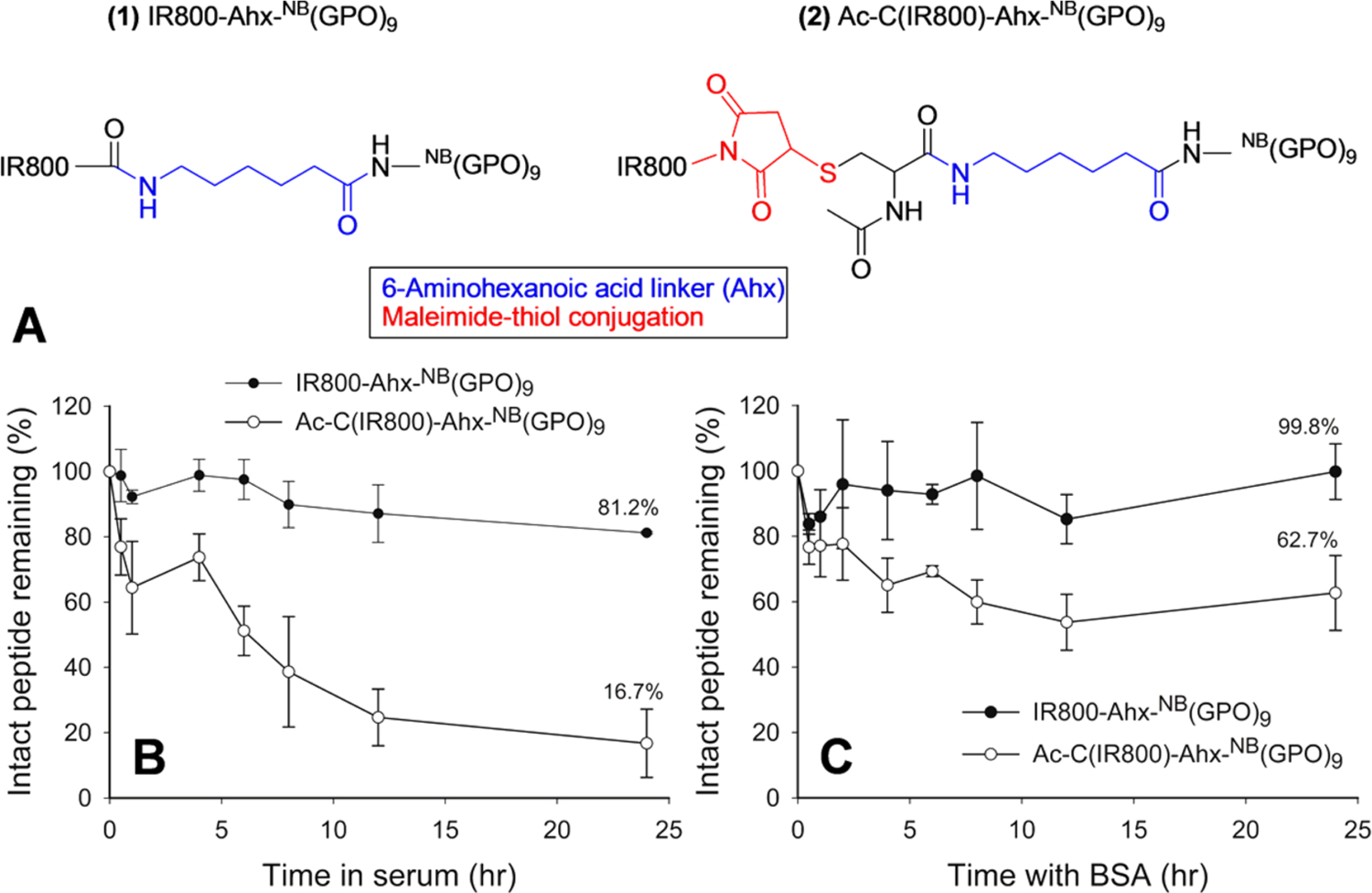
Serum stability of IR800-labeled CHPs. (A) The two different linker chemistries used to conjugate IR800-Ahx-NB(GPO)9 and Ac-C(IR800)-Ahx-NB(GPO)9. (B) Stability profiles of IR800-Ahx-NB(GPO)9 and Ac-C(IR800)-Ahx-NB(GPO)9 in mouse serum. (C) Percentages of intact IR800-Ahx-NB(GPO)9 and Ac-C(IR800)-Ahx-NB(GPO)9 remaining after incubation with BSA without serum. All tests were run in triplicate.
During the serum stability assay of Ac-C(IR800)-Ahx-NB(GPO)9, we noted a green color in the protein pellets during the serum protein separation step (Figure S5). This observation prompted us to suspect that the decrease of intact Ac-C(IR800)-Ahx-NB(GPO)9 in serum during the stability test was not due to peptide degradation but due to IR800 dye transferring to serum proteins, via the retro-Michael reaction.31,32 To test this hypothesis, we incubated both IR800-labeled CHPs in pure BSA solutions for 24 h without serum, precipitated out the BSA, and quantified the NIRF-CHP remaining in the solutions using the absorbance of IR800 dye at 774 nm (see Methods). Even without any proteolytic activity, only 62.7% of Ac-C(IR800)-Ahx-NB(GPO)9 remained intact after 24 h (Figure 3C), while 100% of IR800-Ahx-NB(GPO)9 remained intact, which could only be explained by an interaction between Ac-C(IR800)-Ahx-NB(GPO)9 and BSA. When ESI-MS was used to determine the mass of the BSA after incubation with Ac-C(IR800)-Ahx-NB(GPO)9, a mass corresponding to an IR800-maleimide-conjugated BSA molecule was clearly detected, whereas no such peak was found in the sample incubated with IR800-Ahx-NB(GPO)9 (Figure 4). The results indicate that although NB(GPO)9 has high serum stability, the dyes conjugated onto CHP via maleimide chemistry can transfer to BSA (and possibly other serum proteins) by an exchange reaction. Such transfer reactions have been reported by others.32–35
Figure 4.
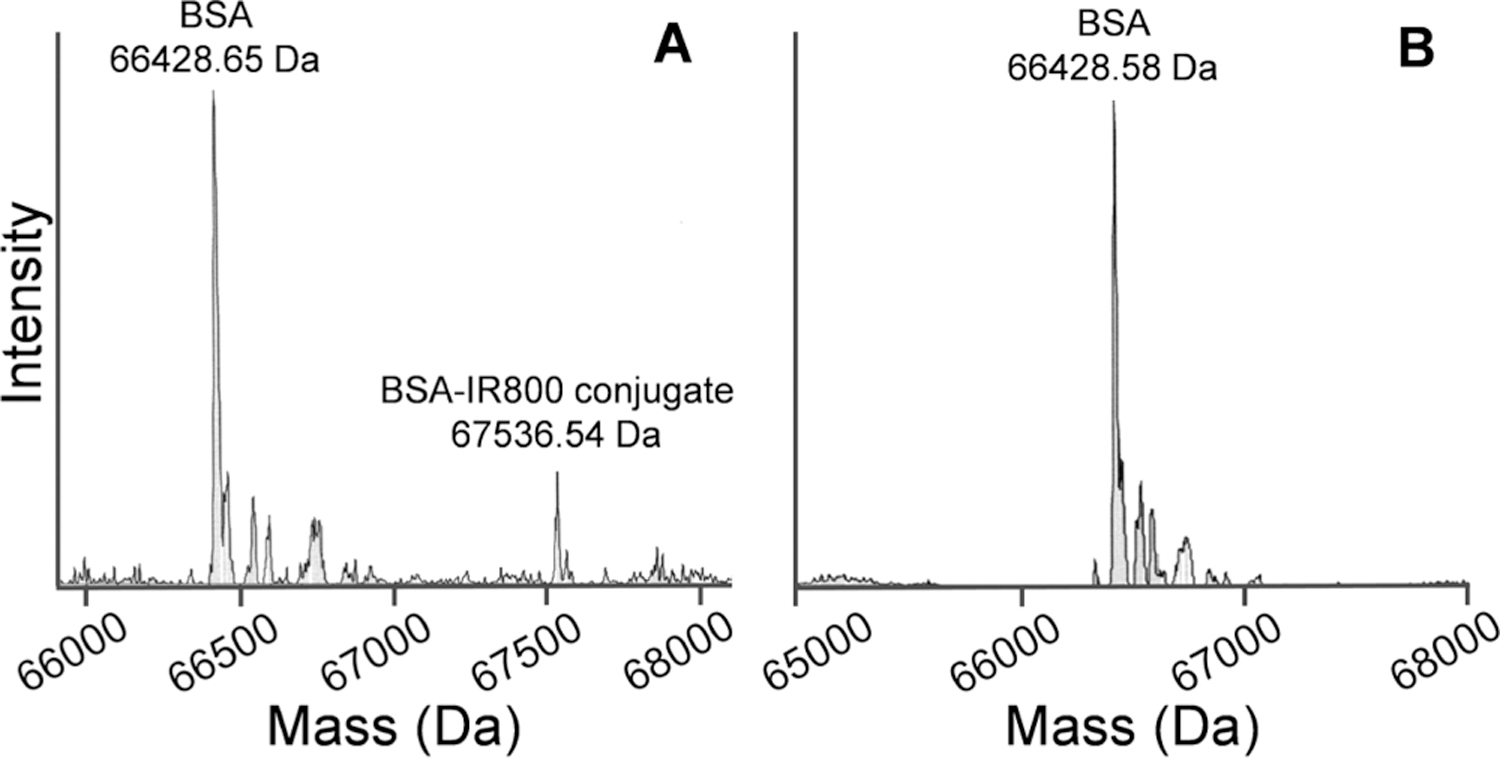
ESI-MS spectra of Ac-C(IR800)-Ahx-NB(GPO)9 (A) and IR800-Ahx-NB(GPO)9 (B) after 24 h incubation with BSA, clearly showing the presence of the BSA-IR800 conjugate only in Ac-C(IR800)-Ahx-NB(GPO)9.
We conducted another set of serum stability and dye transfer validation experiments using the same CHP conjugated with a different type of NIRF, the IR680. The results were similar to those of the IR800 case above in that the IR680-Ahx-NB(GPO)9 exhibited higher serum stability than Ac-C(IR680)-NB(GPO)9 (96.6% vs 71.2%, respectively), with evidence of maleimide–thiol mediated dye transfer reaction to BSA (Figures S6 and S7). However, the difference in stability between the two linker chemistries was noticeably less than that of the IR800 conjugated CHPs. The thioether linked Ac-C(IR680)-Ahx-NB(GPO)9 appeared to be more stable than Ac-C-(IR800)-Ahx-NB(GPO)9 in serum (71.2% vs 16.7% content remaining intact after 24 h incubation), and the extent of IR680 dye transfer to BSA was significantly lower than the IR800 dye transfer (Figures 3C and S6), suggesting that the structure of the dye affected the dye transfer reactions.
In Vivo NIR Fluorescence Imaging.
To examine the effects of the linker chemistries on the in vivo behaviors of the NIRF-labeled CHPs, we compared the distributions of the single strand Ac-C(IR800)-Ahx-(GPO)9 and IR800-Ahx-(GPO)9 in normal mice. The NB cage group on their precursors was removed by UV exposure immediately prior to intravenous injection to allow the peptide to bind to denatured collagen strands during circulation.7,18 Both peptides showed clear uptake in the skeleton, especially in the spine, mandibles, and joints (knees, ankles, and wrists) (Figure 5), which was the result of high levels of collagen turnover in the skeletal system during normal bone renewal, as reported before.7,18 The amide-linked IR800-Ahx-(GPO)9 exhibited some uptake in kidneys and minimal uptake in liver, which indicated that renal clearance is the primary elimination pathway for such a small and chemically inert peptide.22,36 In comparison, the thioether-linked Ac-C(IR800)-Ahx-(GPO)9 showed significantly higher uptake in kidneys and liver (Figure 5). These off-target accumulations are likely the results of the lower stability of the thioether linker chemistry, which we noted in the in vitro serum stability test (Figures 3 and 4). When the IR800-maleimide dyes covalently transfer from CHPs onto the serum proteins (e.g., albumin),37 their biological fate was altered. We believe that, due to the large size, the dye-conjugated serum proteins were not able to be filtered by the glomeruli (renal accumulation) and, instead, were metabolized in the liver, which led to the high NIRF signals observed in kidneys and liver. The in vivo behaviors of both CHP conjugates labeled with IR680 via the two linker chemistries showed similar biodistribution patterns, with high skeletal and low liver uptake (Figure S8), suggesting that the dye transfer reaction was not significant for the IR680. This result is also in agreement with the in vitro serum stability assay, where the maleimide–thiol mediated dye transfer reaction took place at a much higher level for IR800 than for IR680 (Figures 3 and S6). These results underscore the importance of the dye structures as related to the interaction of CHP–dye conjugates with serum proteins and their distribution in vivo.
Figure 5.
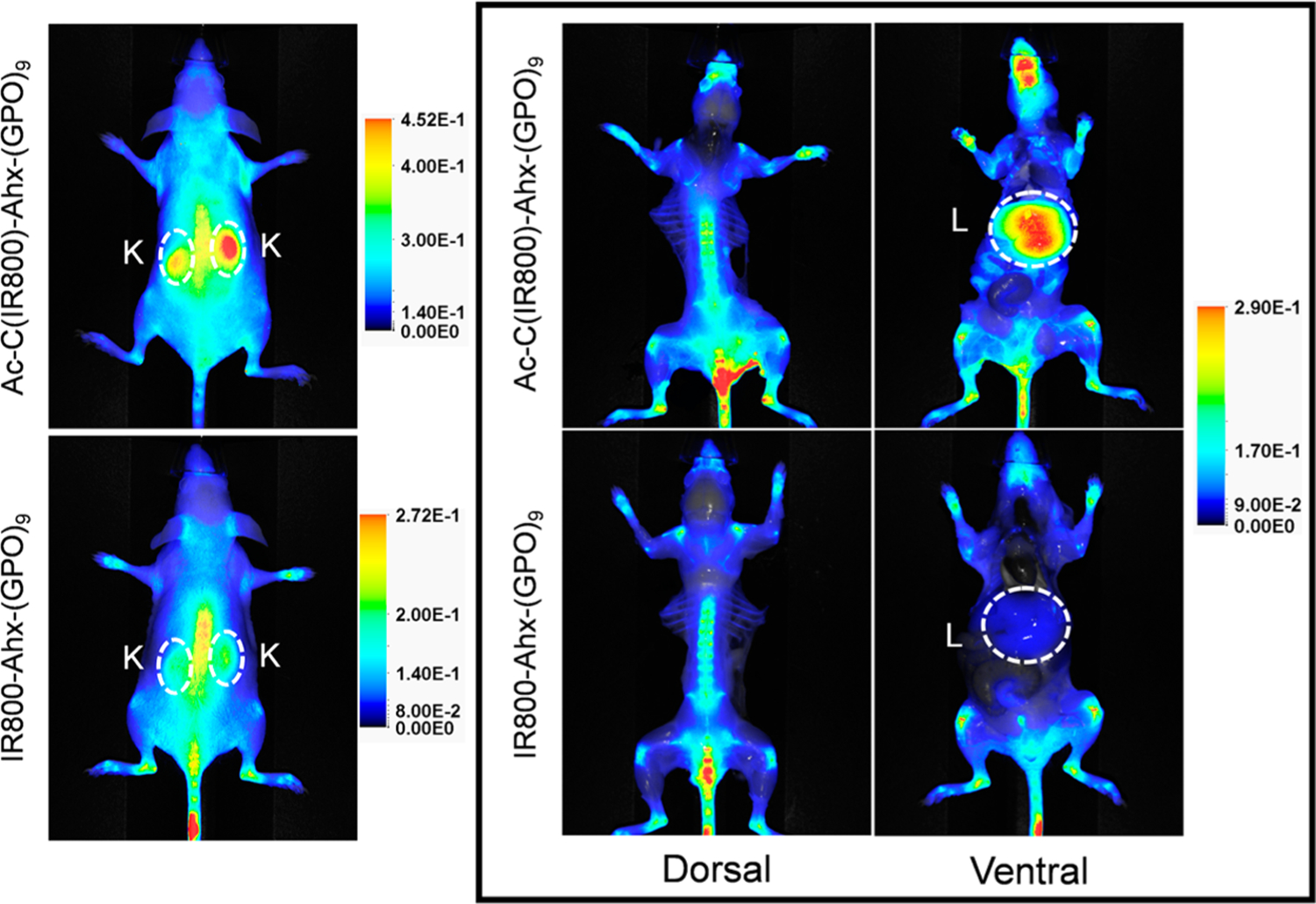
NIR fluorescence imaging showing the in vivo distribution of Ac-C(IR800)-Ahx-(GPO)9 and IR800-Ahx-(GPO)9 in mice 48 h post tail vein injection. Both conjugates showed similar uptake in bones and joints due to the presence of denatured collagen produced during normal remodeling, while only Ac-C(IR800)-Ahx-(GPO)9 showed high intensity signals in the kidneys and liver. The kidneys (K) and livers (L) are highlighted by dashed circles. The images in the box were taken after skin removal for clear display of signals from the skeleton and internal organs, and their fluorescence intensities were adjusted to the same scale for direct comparison. All in vivo experiments were performed three times with similar results.
DISCUSSION
There are two main methods used for determining a proteolytic degradation profile of a compound in biological fluids: blotting analysis and direct stability assays. Blotting analysis is useful for investigating the action of a specific peptidase, while stability assays can provide a wide range of information, including the type of peptidase, enzyme specificity, and the degradation profile for a specific peptide sequence.28 It has been well demonstrated that the in vivo stability of peptides, which could help understand the peptide’s pharmacokinetics, can be reliably modeled in vitro using either plasma or serum.19,27,28
Our study confirmed that the triple helical collagen mimetic peptide, (GPO)9, is highly stable in serum and that the single strand CHPs based on G–P-P/O repeats are generally resistant to proteolytic degradation when compared to peptide comprised of a non-G–P-P/O sequence. The Gly, Pro, and Hyp are hydrophilic and neutral amino acids which are all considered to be inert amino acids with respect to protein interactions. Typically, charged, hydrophobic, or aromatic amino acids occupy the proteolytic location because they can interact with specific residues for cleavage site recognition. For example, thrombin and factor X, which are abundant in blood, specifically cleave at the carboxyl side of Arg, and hydrophobic and aromatic residues (such as Ala, Phe, Ile, Leu, and Val) are generally found near the cleaved bond.38,39 It is the absence of such charged or hydrophobic residues that makes the CHP resistant to common serum proteases. This fact is most clearly reflected in our result where a peptide that was virtually indigestible became highly susceptible to digestion (completely digested in 5 h) after only four Hyp → Lys substitutions [Figure 2A, CF-(GPO/K)9]. When analyzing the serum stability of CF-(GPO/K)9, we found HPLC peaks corresponding to peptide fragments resulting from specific proteolytic digestion. The most probable cleavage sites were validated by MALDI-TOF MS analysis (Figure S9) and are shown as arrowheads between the amino acids in Table 2. The cleavage sites of CF-(GPO/K)9 are all located at or in close proximity to the Lys residues, suggesting the involvement of this charged residue in peptide bond cleavage.
Although CHPs based on GPP and GPO triplets are resistant to common proteases, they are susceptible to a family of proteases specialized for hydrolyzing proline containing peptides. This family contains both endo- and exopeptidases, and their mechanisms have been fully documented in the literature (Figure 6). Some of the well-known proteases in thisfamily are prolyl endopeptidase (PE), which can hydrolyze the bond on the carboxyl side of proline within the interior of a peptide,40–43 dipeptidyl peptidases II and IV (DPPs), which are exopeptidases that can cleave at N-terminal proline residues in blocks of two to three amino acids,41,44,45 and finally amino peptidase P (APP) and proline iminopeptidase (PIP), both of which can release a single N-terminal amino acid by cleaving either the amino side or carboxyl side of Pro, respectively.41,46
Figure 6.
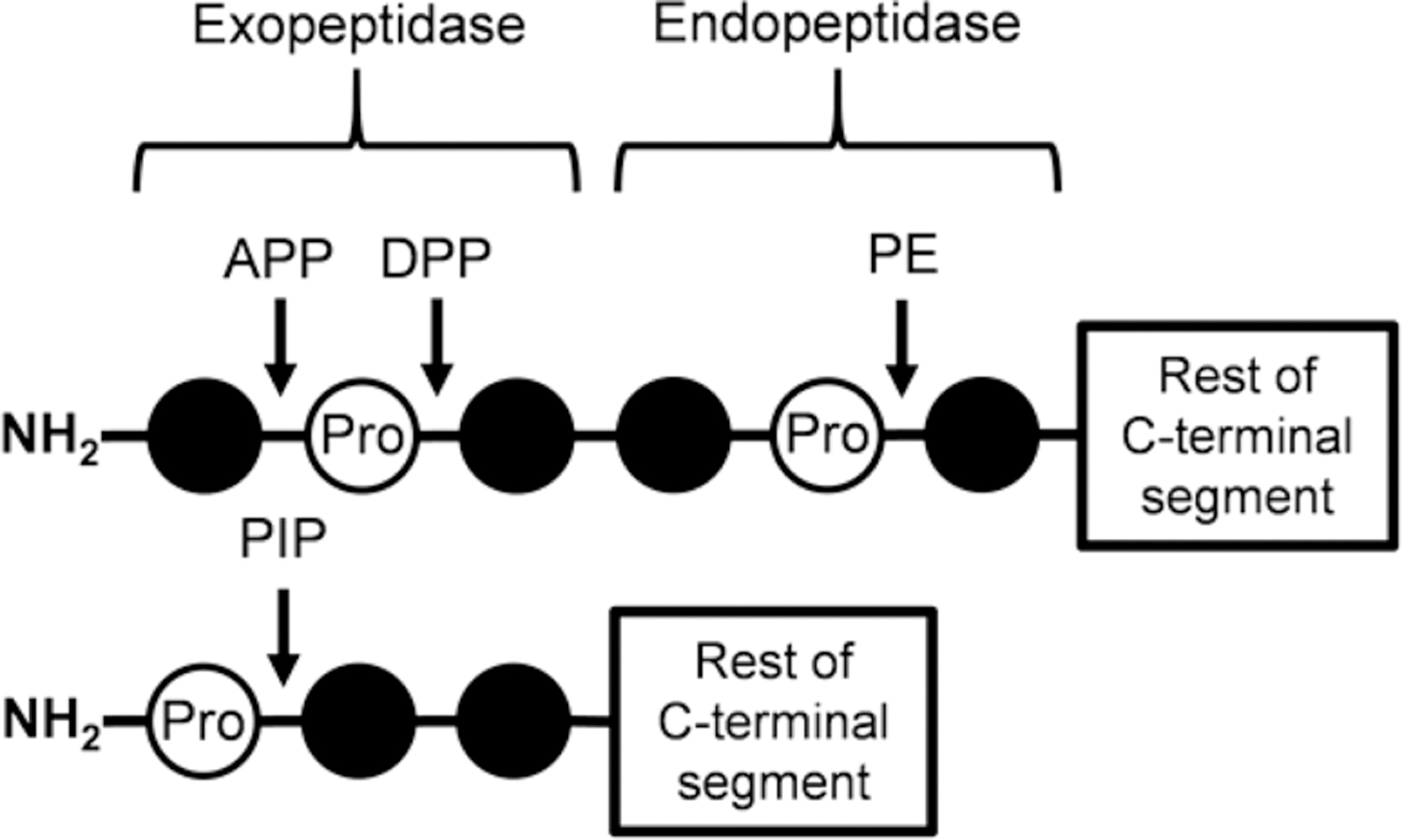
Schematic of cleavage positions for proline-specific peptidases. This shows the general cleavage sites for proline-specific peptidases on a peptide with a free N-terminus. DPPs have the highest cleavage efficiency when removing dipeptides from sequences as shown; however, they can also cleave tri- and tetrapeptides on the carboxyl side of proline residues at a reduced efficiency. PIP prefers to cleave a single proline residue off sequences but can cleave di- and tripeptides on the carboxyl side of prolines at reduced efficiency as well. APP is responsible for the removal of a single residue on the amino side of proline. APP: aminopeptidase P, DPP: dipeptidyl peptidase, PE: prolyl endopeptidase, PIP: proline iminopeptidase.
We were unable to find all the cleaved fragments by HPLC and MALDI-TOF MS; however, by comparing the known mechanisms for these proline-specific proteases and the results of our serum stability test, we were able to identify three structural features critical for the serum stability of CHP comprised of GPO and GPP sequences: (i) capping the N-terminus of the peptide with fluorophores or another moiety protects CHP from exopeptidase digestion, (ii) long peptide length protects the CHP against endopeptidase digestion, and (iii) the presence of hydroxyproline reduces endopeptidase activity.
DPPs, APP, and PIP can only hydrolyze peptides with free N-termini.41,44 Therefore, capping N-termini with CF or CF-Gly3 can increase the serum stability of CHPs. This effect is most clearly seen in the case for (GPO)5, where DPP as well as APP and PIP can digest the peptide from the N-terminus (Figure 1). Using MALDI-TOF MS, we identified three distinct digested products resulting from (i) single APP action [Table 1, first arrowhead from the left in (GPO)5], (ii) single DPP action (second arrowhead from the left), and (iii) DPP action followed by APP digestion (third arrowhead). After the CF labeling we saw a dramatic increase in stability, since these exopeptidases were no longer able to digest the peptide (Figure 7). The stabilization effect resulting from N-capping was also observed for NB(GPO)9 as well as (GPP)9.
Figure 7.
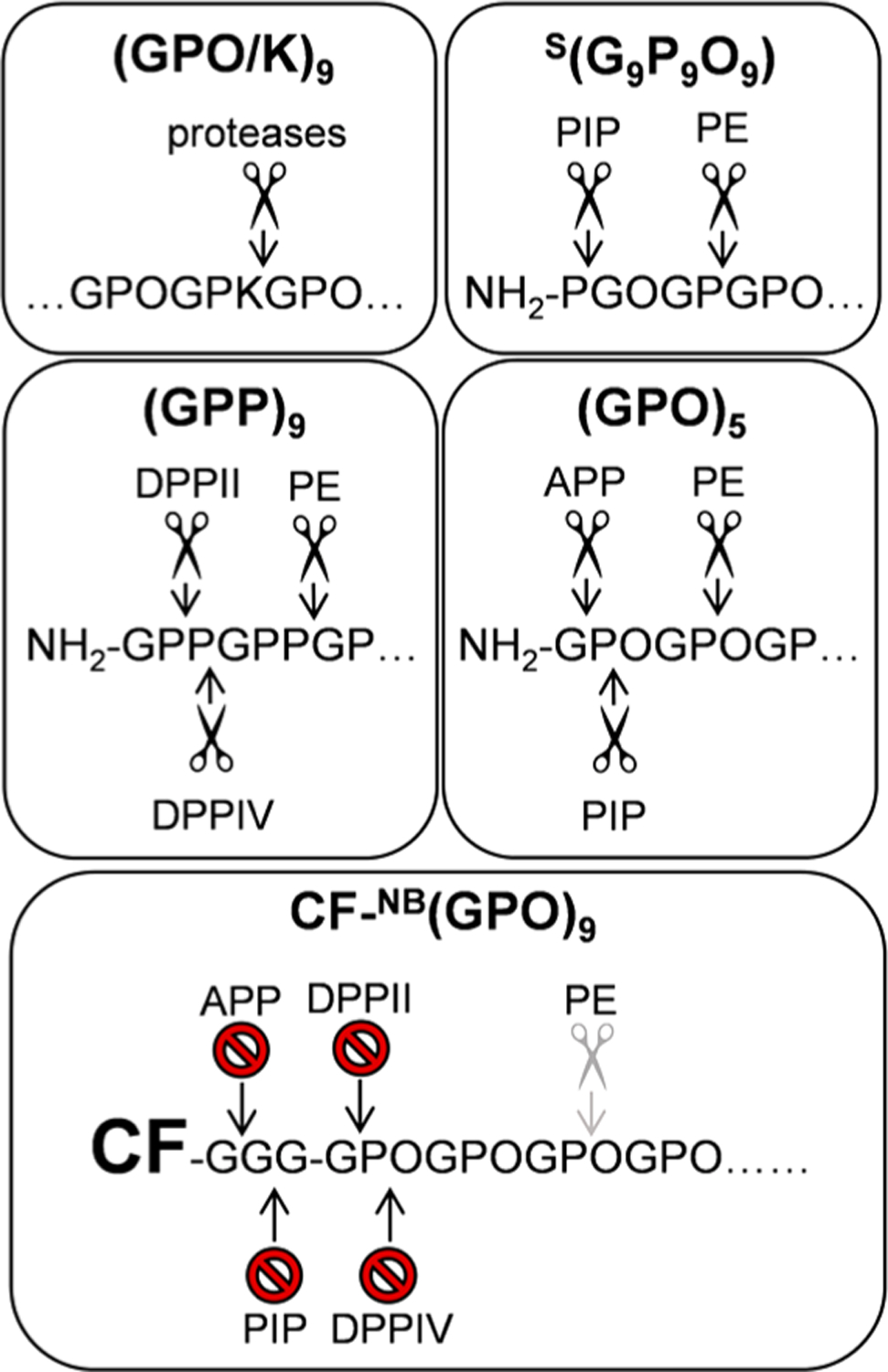
Schematic showing how the CHPs studied here can be digested by proline-specific peptidase. The top four panels highlight the major potential sites of cleavage for general or proline-specific peptidases on (GPO/K)9, S(G9P9O9), (GPP)9, and (GPO)5. The bottom panel shows the inactivation of the cleavage sites by N-terminal modification of (GPO)9. PE may still cleave the monomeric (GPO)9, but its efficiency is significantly reduced (as indicated by gray scissors), possibly due to the presence of O and the increased peptide size. APP: aminopeptidase P, DPPII: dipeptidyl peptidase II, DPPIV: dipeptidyl peptidase IV, PE: prolyl endopeptidase, PIP: proline iminopeptidase.
Due to a small, 4 Å opening in the catalytic site,40,47 PE hydrolyzes shorter sequences faster than longer ones,48 and cannot cleave peptides over 30 amino acids long.41 The fact that no signs of degradation were seen for CF-NB(GPO)9 (and other N-protected peptides), which is over 30 amino acids long, may have been due to the long-length of these peptides.
The presence of hydroxyproline residues in the GPO triplet seems to have significantly decreased the cleaving efficiency of PE. It was reported that the Pro → Hyp substitution in a peptide substrate greatly reduced the efficiency of PE digestion.41,49 This reduced efficiency is very likely the reason for (i) the increased stability of NB(GPO)9 in comparison to (GPP)9 in both CF-free or CF-labeled peptides, and (ii) the extremely high stability of CF-(GPO)5, which would otherwise be easily degraded by PE, since it is shorter than 30 amino acids. It is unclear why CF-S(G9P9O9) exhibited low serum stability despite being over 30 amino acids and N-protected. We speculate that there may be other proteases, particularly an endopeptidase which is not specific to proline, that is playing a major role in digesting this peptide.
The availability of intact CHP–dye conjugates in serum depends not only on resistance to proteinases but also on the chemistry used to link the peptide and the dye molecules. We found that the rapid decrease of serum concentration of Ac-C(IR800)-Ahx-NB(GPO)9 was not due to peptide degradation but due to the unexpected transfer of the IR800-maleimide group from CHP to serum proteins through a thiol-exchange reaction (Figures 3 and 4). As reported in recent years, this retro-Michael side reaction occurs readily in biological environments where excess thiol groups are present32–35,37 and is a major drawback of the popular thiol-maleimide chemistry, particularly in the preparation of antibody–drug conjugates (ADC). Similar to our study, a drug–maleimide moiety was reported to release from an ADC and covalently conjugate to serum albumin.32 Visual observation (Figure S5) showed that this retro-Michael reaction happens rapidly in serum. The NIRF-CHP linked by an amide bond, IR800-Ahx-NB(GPO)9, was significantly more stable than the thioether linked version in serum (Figure 3), resulting in more efficient targeting of skeletal tissues and less off-target distribution in vivo (Figure 5). These results highlight that the NHS-mediated conjugation is more reliable and should be favored in preparation of future CHP theranostics.
We also discovered that even if a CHP is made of the same thioether linker chemistry, Ac-C(IR680)-Ahx-NB(GPO)9 has a significantly lower level of retro-Michael reaction with albumin and consequently exhibits less off-target distribution in vivo when compared to Ac-C(IR800)-Ahx-NB(GPO)9 (Figures 3 and S6). Since the two molecules are identical except for the dyes, the results suggest that the dye structure is affecting the dye transfer reaction during the serum stability test and the in vivo bone targeting experiment. Others have shown that the charge state of a NIR cyanine dye50 as well as the position of a linker within the dye molecule51 impact the in vivo optical properties and pharmacokinetics of the dye-labeled molecules. The effect of dye structure could be more significant in the case of our NIRF-CHP conjugates, since the CHP part of the conjugate is highly inert toward nonspecific interactions. The IR680 and IR800 dyes are different in backbone structure, linker position, net charge, and charge distribution (Figure S10), all of which could have resulted in varying degrees of nonspecific interactions between the dye and serum proteins that led to different levels of retro-Michael reaction and in vivo distribution.
CONCLUSION
We assessed the serum stability of multiple CHP derivatives and established how length, amino acid composition, N-terminal labeling, and linker chemistry contributed to the overall availability of monomeric CHPs in serum over time. We demonstrated that, similar to triple-helical CMPs, monomeric CHPs comprised of repeating Gly-Pro-Hyp triplets are highly resistant to enzymatic degradation by many serum proteases and maintain elevated levels of stability despite their small size. The neutral and hydrophilic peptide sequence deters recognition by common serum enzymes and prevents nonspecific binding to other biomolecules, thereby extending CHP’s availability in serum. Although CHPs avoid recognition by common serum enzymes, the specific class of enzymes known as proline-specific peptidases can act on CHPs with free N-termini, since a majority of them are exopeptidases. Therefore, CHP stability was increased by N-terminal modification with fluorescent dyes which capped the free N-terminus. As reported previously, the dye-conjugated CHPs can target denatured collagens in the bones and cartilage of mice in vivo under normal conditions due to the high remodeling activity of the skeletal system.7,11 Our in vivo experiments indicated that although skeletal targeting behavior was observed in all NIRF-conjugated CHPs, there were subtle differences in biodistribution due to differences in the linker chemistry and the structure of the dye molecules. Specifically, Ac-C(IR800)-Ahx-NB(GPO)9 exhibited a noticeable off target signal, particularly in the liver, which is believed to be caused by transfer of IR800 dye from CHP to serum proteins via thiol exchange reaction as supported by the emergence of albumin–dye conjugates during the serum stability assay. Based on all of our tests, we conclude that the most ideal NIRF-CHP for imaging denatured collagen in vivo is IR680-Ahx-(GPO)9. CHP’s offer high serum stability, which is uncommon for conventional peptides that suffer from poor stability in vivo.19,28,52 CHP’s serum stability provides a strong foundation for transforming this peptide into an in vivo delivery vehicle for theranostic agents in managing a vast number of disease states that are characterized by excessive collagen remodeling, such as arthritis, cancer, and fibrosis.6
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by the NIAMS/NIH (R01-AR060484 and R21-AR065124) and DOD (W81XWH-12-0555) awarded to S.M.Y., NCI/NIH (CA134675) and DOD (W81XWH-12-0555) awarded to M.G.P., and by the Nano Institute of Utah: Nanotechnology Fellowship (University of Utah) awarded to L.L.B. The authors thank Krishna Parsawar and the Health Sciences Center Core Facilities for help with ESI-MS and MALDI-TOF MS.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.molpharmaceut.7b00009.
Representative raw HPLC profiles; serum stability profile of penetratin; CD spectra and thermal unfolding curves of (GPO)5 and (GPP)9; additional data of stability and BSA assays; photographs of protein pellets and ESI-MS of BSA after incubation with NIRF-CHPs; NIRF in vivo imaging of Ac-C(IR680)-Ahx-(GPO)9 and IR680-Ahx-(GPO)9 IR800/680 dye structures; mass of all peptides verified by MALDI-TOF MS (PDF)
The authors declare the following competing financial interest(s): Y.L. and S.M.Y. are cofounders of 3Helix, which commercializes collagen hybridizing peptides.
REFERENCES
- (1).Shoulders MD; Raines RT Collagen structure and stability. Annu. Rev. Biochem 2009, 78, 929–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Brinckmann J Collagens at a glance. In Collagen: Primer in Structure, Processing and Assembly; Brinckmann J, Notbohm H, Müller PK, Eds.; Springer Berlin Heidelberg: Berlin, Heidelberg, 2005; pp 1–6. [Google Scholar]
- (3).Bonnans C; Chou J; Werb Z Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol 2014, 15 (12), 786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Chung L; Dinakarpandian D; Yoshida N; Lauer-Fields JL; Fields GB; Visse R; Nagase H Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. EMBO J 2004, 23 (15), 3020–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Rosenblum G; van den Steen PE; Cohen SR; Bitler A; Brand DD; Opdenakker G; Sagi I Direct visualization of protease action on collagen triple helical structure. PLoS One 2010, 5 (6), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Wahyudi H; Reynolds AA; Li Y; Owen SC; Yu SM Targeting collagen for diagnostic imaging and therapeutic delivery. J. Controlled Release 2016, 240, 323 10.1016/j.jconrel.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Li Y; Foss CA; Summerfield DD; Doyle JJ; Torok CM; Dietz HC; Pomper MG; Yu SM Targeting collagen strands by photo-triggered triple-helix hybridization. Proc. Natl. Acad. Sci. U. S. A 2012, 109 (37), 14767–14772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Yu SM; Li Y; Kim D Collagen mimetic peptides: progress towards functional applications. Soft Matter 2011, 7 (18), 7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Liang H; Li X; Chen B; Wang B; Zhao Y; Zhuang Y; Shen H; Zhang Z; Dai J A collagen-binding EGFR single-chain Fv antibody fragment for the targeted cancer therapy. J. Controlled Release 2015, 209, 101–109. [DOI] [PubMed] [Google Scholar]
- (10).Helms B. a.; Reulen S. W. a; Nijhuis S; De Graaf-Heuvelmans PTHM; Merkx M; Meijer EW High-affinity peptide-based collagen targeting using synthetic phage mimics: from phage display to dendrimer display. J. Am. Chem. Soc 2009, 131 (33), 11683–11685. [DOI] [PubMed] [Google Scholar]
- (11).Li Y; Yu SM Targeting and mimicking collagens via triple helical peptide assembly. Curr. Opin. Chem. Biol 2013, 17 (6), 968–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Engel J; Bachinger HP Structure, stability and folding of the collagen triple helix. Top. Curr. Chem 2005, 247, 7–33. [Google Scholar]
- (13).Brodsky B; Thiagarajan G; Madhan B; Kar K Triple-helical peptides: An approach to collagen conformation, stability, and self-association. Biopolymers 2008, 89 (5), 345–353. [DOI] [PubMed] [Google Scholar]
- (14).Boudko S; Frank S; Kammerer RA; Stetefeld J; Schulthess T; Landwehr R; Lustig A; hinger HP; Engel J Nucleation and propagation of the collagen triple helix in single-chain and trimerized peptides: transition from third to first order kinetics. J. Mol. Biol 2002, 317 (3), 459–470. [DOI] [PubMed] [Google Scholar]
- (15).Persikov AV; Ramshaw JAM; Kirkpatrick A; Brodsky B Amino acid propensities for the collagen triple-helix. Biochemistry 2000, 39 (48), 14960–14967. [DOI] [PubMed] [Google Scholar]
- (16).Luo T; Kiick KL Collagen-like peptides and peptide-polymer conjugates in the design of assembled materials. Eur. Polym. J 2013, 49 (10), 2998–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Li Y; Ho D; Meng H; Chan TR; An B; Yu H; Brodsky B; Jun AS; Michael Yu S Direct detection of collagenous proteins by fluorescently labeled collagen mimetic peptides. Bioconjugate Chem 2013, 24 (1), 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Li Y; Foss CA; Pomper MG; Yu SM Imaging denatured collagen strands in vivo and ex vivo via photo-triggered hybridization of caged collagen mimetic peptides. J. Visualized Exp 2014, 83. 10.3791/51052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Otvos L; Wade JD Current challenges in peptide-based drug discovery. Front. Chem 2014, 2 (62), 8–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Shinde A; Feher KM; Hu C; Slowinska K Peptide internalization enabled by folding: triple helical cell-penetrating peptides. J. Pept. Sci 2015, 21 (2), 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Yamazaki CM; Nakase I; Endo H; Kishimoto S; Mashiyama Y; Masuda R; Futaki S; Koide T Collagen-like cell-penetrating peptides. Angew. Chem., Int. Ed 2013, 52 (21), 5497–5500. [DOI] [PubMed] [Google Scholar]
- (22).Yasui H; Yamazaki CM; Nose H; Awada C; Takao T; Koide T Potential of collagen-like triple helical peptides as drug carriers: their in vivo distribution, metabolism, and excretion profiles in rodents. Biopolymers 2013, 100 (6), 705–713. [DOI] [PubMed] [Google Scholar]
- (23).Urello MA; Kiick KL; Sullivan MO Integration of growth factor gene delivery with collagen-triggered wound repair cascades using collagen-mimetic peptides. Bioeng. Transl. Med 2016, 1 (2), 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Chattopadhyay S; Murphy CJ; McAnulty JF; Raines RT Peptides that anneal to natural collagen in vitro and ex vivo. Org. Biomol. Chem 2012, 10, 5892–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Rubert Perez CM; Rank LA; Chmielewski J Tuning the thermosensitive properties of hybrid collagen peptide-polymer hydrogels. Chem. Commun 2014, 50 (60), 8174–8176. [DOI] [PubMed] [Google Scholar]
- (26).Lee HJ; Lee J-S; Chansakul T; Yu C; Elisseeff JH; Yu SM Collagen mimetic peptide-conjugated photopolymerizable PEG hydrogel. Biomaterials 2006, 27 (30), 5268–5276. [DOI] [PubMed] [Google Scholar]
- (27).Nguyen LT; Chau JK; Perry N. a.; de Boer L; Zaat S. a J.; Vogel HJ Serum stabilities of short tryptophan- and arginine-rich antimicrobial peptide analogs. PLoS One 2010, 5 (9), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Powell MF; Grey H; Gaeta F; Sette A; Colon S Peptide stability in drug development: a comparison of peptide reactivity in different biological media. J. Pharm. Sci 1992, 81 (8), 731–735. [DOI] [PubMed] [Google Scholar]
- (29).Jenssen H; Aspmo SI Serum Stability of Peptides. In Peptide-Based Drug Design; Otvos L, Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, 2008; Vol. 494, pp 177–186. [DOI] [PubMed] [Google Scholar]
- (30).Li Y; San BH; Kessler JL; Kim JH; Xu Q; Hanes J; Yu SM Non-covalent photo-patterning of gelatin matrices using caged collagen mimetic peptides. Macromol. Biosci 2015, 15 (1), 52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Tumey LN; Charati M; He T; Sousa E; Ma D; Han X; Clark T; Casavant J; Loganzo F; Barletta F; et al. Mild method for succinimide hydrolysis on ADCs: Impact on ADC potency, stability, exposure, and efficacy. Bioconjugate Chem 2014, 25 (10), 1871–1880. [DOI] [PubMed] [Google Scholar]
- (32).Alley SC; Benjamin DR; Jeffrey SC; Okeley NM; Meyer DL; Sanderson RJ; Senter PD Contribution of linker stability to the activities of anticancer immunoconjugates. Bioconjugate Chem 2008, 19 (3), 759–765. [DOI] [PubMed] [Google Scholar]
- (33).Fontaine SD; Reid R; Robinson L; Ashley GW; Santi DV Long-term stabilization of maleimide-thiol conjugates. Bioconjugate Chem 2015, 26 (1), 145–152. [DOI] [PubMed] [Google Scholar]
- (34).Baldwin AD; Kiick KL Tunable degradation of maleimidethiol-adducts in reducing environments. Bioconjugate Chem 2011, 22 (10), 1946–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Lyon RP; Setter JR; Bovee TD; Doronina SO; Hunter JH; Anderson ME; Balasubramanian CL; Duniho SM; Leiske CI; Li F; et al. Self-hydrolyzing maleimides improve the stability and pharmacological properties of antibody-drug conjugates. Nat. Biotechnol 2014, 32 (10), 1059–1062. [DOI] [PubMed] [Google Scholar]
- (36).Koide T; Yamamoto N; Taira KB; Yasui H Fecal excretion of orally administered collagen-like peptides in rats: contribution of the triple-helical conformation to their stability. Biol. Pharm. Bull 2016, 39 (1), 135–137. [DOI] [PubMed] [Google Scholar]
- (37).Ponte JF; Sun X; Yoder NC; Fishkin N; Laleau R; Coccia J; Lanieri L; Bogalhas M; Wang L; Wilhelm SD; et al. Understanding how the stability of the thiol-maleimide linkage impacts the pharmacokinetics of lysine-linked antibody-maytansinoid conjugates. Bioconjugate Chem 2016, 27 (7), 1588–1598. [DOI] [PubMed] [Google Scholar]
- (38).Keil B Specificity of Proteolysis; Springer-Verlag: Berlin Heidelberg, 1992. [Google Scholar]
- (39).Fujikawa K; Titani K; Davie EW Activation of bovine factor X (Stuart factor): conversion of factor Xa-alpha to factor Xa-beta. Proc. Natl. Acad. Sci. U. S. A 1975, 72 (9), 3359–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Gass J; Khosla C Prolyl endopeptidases. Cell. Mol. Life Sci 2007, 64 (3), 345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Cunningham DF; O’Connor B Proline specific peptidases. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol 1997, 1343 (2), 160–186. [DOI] [PubMed] [Google Scholar]
- (42).Koida M; Walter R Post-proline cleaving enzyme. J. Biol. Chem 1976, 251 (23), 7593–7599. [PubMed] [Google Scholar]
- (43).Yoshimoto T; Fischl M; Orlowski RC; Walter R Post-proline cleaving enzyme and post-proline dipeptidyl aminopeptidase. J. Biol. Chem 1978, 253 (10), 3708–3716. [PubMed] [Google Scholar]
- (44).Maes MB; Scharpé S; De Meester I Dipeptidyl peptidase II (DPPII), a review. Clin. Chim. Acta 2007, 380 (1–2), 31–49. [DOI] [PubMed] [Google Scholar]
- (45).Polgár L The prolyl oligopeptidase family. Cell. Mol. Life Sci 2002, 59 (2), 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Taylor A Aminopeptidases: structure and function. FASEB J 1993, 7 (2), 290–298. [DOI] [PubMed] [Google Scholar]
- (47).Li M; Chen C; Davies DR; Chiu TK Induced-fit mechanism for prolyl endopeptidase. J. Biol. Chem 2010, 285 (28), 21487–21495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Fülöp V; Böcskei Z; Polgár L Prolyl oligopeptidase: an unusual β-propeller domain regulates proteolysis. Cell 1998, 94 (2), 161–170. [DOI] [PubMed] [Google Scholar]
- (49).Nomura K Specificity of prolyl endopeptidase. FEBS Lett 1986, 209 (2), 235–237. [DOI] [PubMed] [Google Scholar]
- (50).Sato K; Gorka AP; Nagaya T; Michie MS; Nani RR; Nakamura Y; Coble VL; Vasalatiy OV; Swenson RE; Choyke PL; et al. Role of fluorophore charge on the in vivo optical imaging properties of near-infrared cyanine dye/monoclonal antibody conjugates. Bioconjugate Chem 2016, 27 (2), 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Sato K; Nagaya T; Nakamura Y; Harada T; Nani RR; Shaum JB; Gorka AP; Kim I; Paik CH; Choyke PL; et al. Impact of C4′-O-alkyl linker on in vivo pharmacokinetics of near-infrared cyanine/monoclonal antibody conjugates. Mol. Pharmaceutics 2015, 12 (9), 3303–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Powell MF; Stewart T; Otvos LJ; Urge L; Gaeta FCA; Sette A; Arrhenius T; Thomson D; Soda K; Colon SM Peptide stability in drug development II. Effect of single amino acid substitution and glycosylation on peptide reactivity in human serum. Pharm. Res 1993, 10 (9), 1268–1273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


