Abstract
The let-7 gene encodes a highly conserved microRNA with critical functions integral to cell fate specification and developmental progression in diverse animals. In Caenorhabditis elegans, let-7 is a component of the heterochronic (developmental timing) gene regulatory network, and loss-of-function mutations of let-7 result in lethality during the larval to adult transition due to misregulation of the conserved let-7 target, lin-41. To date, no bilaterian animal lacking let-7 has been characterized. In this study, we identify a cohort of nematode species within the genus Caenorhabditis, closely related to C. elegans, that lack the let-7 microRNA, owing to absence of the let-7 gene. Using Caenorhabditis sulstoni as a representative let-7-lacking species to characterize normal larval development in the absence of let-7, we demonstrate that, except for the lack of let-7, the heterochronic gene network is otherwise functionally conserved. We also report that species lacking let-7 contain a group of divergent let-7 paralogs—also known as the let-7-family of microRNAs—that have apparently assumed the role of targeting the LIN-41 mRNA.
Keywords: let-7 microRNA, Elegans supergroup, Elegans group, Japonica group, Caenorhabditis sulstoni, Caenorhabditis macrosperma, Caenorhabditis elegans
Introduction
MicroRNAs are ∼22 nucleotide (nt) noncoding RNAs that negatively regulate protein expression through base pairing of nts 2–8 of the microRNA (known as the microRNA seed) to complementary sequences in target mRNA 3′ UTRs. Base pairing with nts 9–22 can also contribute to target repression but non-seed base pairing is less constrained than seed pairing. Accordingly, evolutionary conservation of microRNA sequences is generally highest for nts 2–8, and less so for non-seed nts (Bartel 2009; Ambros and Ruvkun 2018).
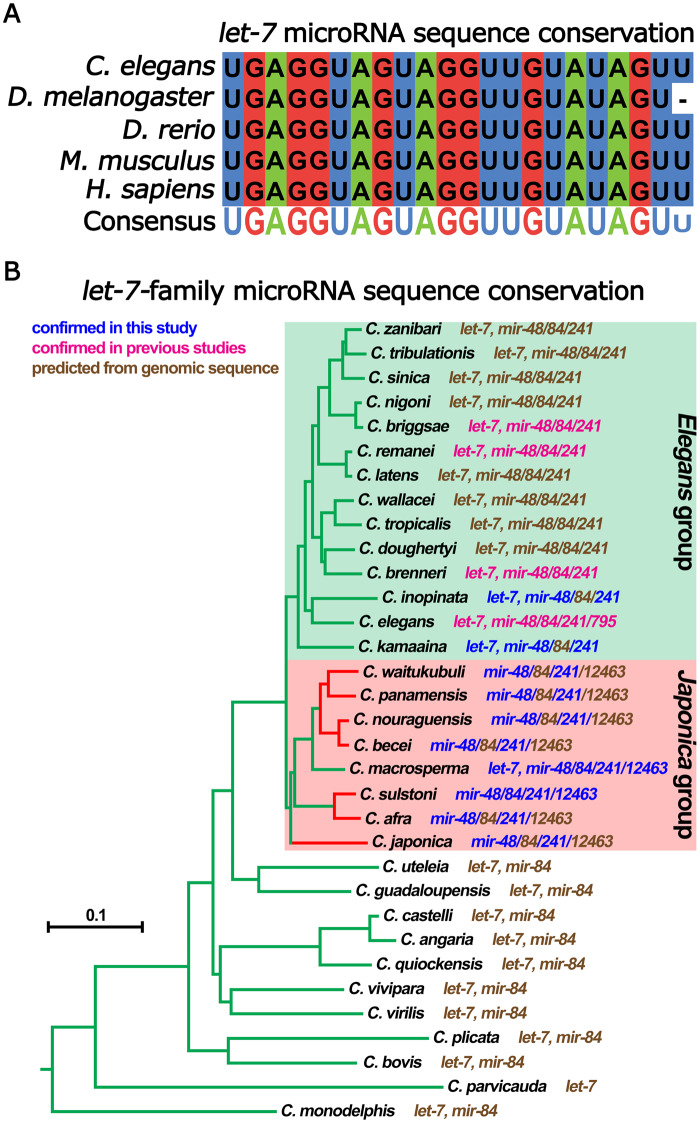
Unlike most microRNAs, the entirety of nts 1–22 of let-7 RNA are highly conserved across bilaterians (Figure 1A) (Pasquinelli et al. 2000). Why let-7 RNA non-seed sequences are so deeply conserved remains a mystery. Alongside the deep conservation of the entire let-7 sequence, let-7 microRNA function is also conserved; across diverse animal phyla, let-7 RNA expression coincides with differentiation and opposes stem cell pluripotency (Pasquinelli et al. 2000; Reinhart et al. 2000; Roush and Slack 2008; Balzeau et al. 2017). Accordingly, in certain contexts let-7 RNA functions as a tumor suppressor through restricting the expression of proteins involved in cell proliferation, growth, and metabolism, among others (Balzeau et al. 2017). We believe that the deep conservation of let-7 sequence holds secrets to important evolutionarily conserved molecular interactions vital to let-7 function, and therefore, a better understanding of let-7 conservation will reveal insights into mechanisms of microRNA function and regulation.
Figure 1.
Conservation of the let-7 microRNA. (A) Sequence conservation of let-7 microRNA. (B) Phylogenetic tree of Caenorhabditis species based on phylogeny from Stevens et al. (2020). For each species, the makeup of their let-7-family microRNAs is indicated. let-7-family microRNAs are predicted from their genomic sequences (brown font), detection in previous studies (magenta font), or detection in this study (blue font). Green branches highlight species that are either predicted or confirmed to have let-7. Red branches highlight species that are either predicted or confirmed to lack let-7. Outlined in the green box are species recovered in the Elegans group. Outlined in the red boxes are species recovered in the Japonica group. Branch lengths are proportional to substitutions per site. Scale bar is shown.
In most bilaterian animal clades, , including mammals and nematodes, the let-7 gene family has been expanded into a number of related microRNAs that share the same seed sequence but differ in their non-seed nts (Roush and Slack 2008). In Caenorhabditis elegans, the let-7-family consists of let-7 microRNA, miR-48, miR-84, miR-241, and miR-795 (Abbott et al. 2005). Despite the presence of multiple let-7-family microRNAs (which could in principle substitute for one another by seed-mediated base pairing to targets), most animals have nevertheless retained the original let-7 microRNA with its non-seed nt sequence conserved (Pasquinelli et al. 2000; Roush and Slack 2008).
The C. elegans heterochronic gene (developmental timing) regulatory network consists of genes that either promote or restrict developmental cell fate progression. Integrated into the heterochronic pathway is the protein coding genes lin-14, lin-28, lin-29, lin-41, lin-46, and hbl-1, as well as the microRNA genes lin-4 and the let-7-family (let-7, mir-48, mir-84, and mir-241). Loss-of-function mutations of genes that restrict cell fate progression, such as lin-14, lin-28, lin-41, or hbl-1, results in precocious development of the hypodermis through the skipping of larval-specific cellular events; whilst loss-of-function mutations of genes that promote cell fate progression, such as lin-29, lin-46, lin-4, or the let-7-family, results in retarded development of the hypodermis through the reiteration of larval-specific cellular divisions (Chalfie et al. 1981; Ambros and Horvitz 1984; Ambros 1989; Fay et al. 1999; Reinhart et al. 2000; Slack et al. 2000; Lin et al. 2003; Pepper et al. 2004; Abbott et al. 2005).
In C. elegans, expression of the lin-4 microRNA increases during the L1 and L2 stages to promote the transition from L1 cell fates to later larval cell fates through its targeted repression of synthesis of the LIN-14 and LIN-28 proteins (which promote L1 and L2 larval stage cell fates) by base-pairing to the 3′ UTRs of lin-14 and lin-28 mRNAs (Chalfie et al. 1981; Ambros and Horvitz 1984; Ambros 1989; Lee et al. 1993; Moss et al. 1997; Lim et al. 2003). Similarly, miR-48, miR-84, and miR-241 (miR-48/84/241), whose expressions increase during the L2 and L3 larval stages to promote transitions to later larval stages by negatively regulating LIN-14, LIN-28, and HBL-1 through base-pairing to their respective 3′ UTRs (Abrahante et al. 2003; Lin et al. 2003; Abbott et al. 2005; Tsialikas et al. 2017).
In C. elegans, let-7 is required for cell cycle exit and differentiation in hypodermal cell lineages at the end of larval development, which is reflected by the dramatic up regulation of mature let-7 microRNA in terminal larval stages (Reinhart et al. 2000). let-7 function and regulation are deeply integrated into the C. elegans heterochronic gene regulatory network; LIN-28 posttranscriptionally restricts let-7 microRNA biogenesis to later larval stages, and let-7 microRNA negatively regulates the evolutionarily conserved pluripotency-promoting genes lin-41 and lin-28 through base-pairing to the 3′ UTRs of the LIN-41 and LIN-28 mRNAs (Reinhart et al. 2000; Slack et al. 2000; Vella et al. 2004; Ding and Großhans 2009; Lehrbach et al. 2009; Van Wynsberghe et al. 2011; Ecsedi et al. 2015; Stefani et al. 2015). The reciprocal direct regulation between let-7 microRNA and LIN-28, and the direct regulation of LIN-41 mRNA by let-7 microRNA, are evolutionarily conserved across vertebrates and invertebrates, suggesting that the unusual conservation of let-7 sequence may be related to these conserved intimate interactions of let-7 microRNA with the LIN-28 and LIN-41 mRNAs and with LIN-28 protein (Reinhart et al. 2000; Slack et al. 2000; Kloosterman et al. 2004; Vella et al. 2004; Schulman et al. 2005; Lin et al. 2007; Heo et al. 2008; Newman et al. 2008; Rybak et al. 2008; Viswanathan et al. 2008; Lehrbach et al. 2009; Nam et al. 2011; Van Wynsberghe et al. 2011; Piskounova et al. 2011; Stratoulias et al. 2014; Ecsedi et al. 2015; Stefani et al. 2015; Balzeau et al. 2017).
Despite the sharing of a common seed sequence and the seemingly redundant potential to regulate the same targets, the let-7-family microRNAs (miR-48/84/241) and let-7 do not function interchangeably. Specifically, miR-48/84/241, as a semi-redundant cohort, primarily regulate lin-14, lin-28, and hbl-1 during L1–3 cell fate transitions, whereas let-7 regulates lin-41 during a larval to adult cell fate switch (Slack et al. 2000; Vella et al. 2004; Abbott et al. 2005; Ecsedi et al. 2015; Tsialikas et al. 2017; Ilbay and Ambros 2019). let-7 loss-of-function (let-7(lf)) mutants display phenotypes distinguishable from those of triply-mutant mir-84(lf); mir-48(lf)mir-241(lf) animals, as a consequence of the stage-specific de-repression of their respective targets—essentially gain-of-function of lin-41, or gain-of-function lin-14/lin-28/hbl-1, respectively (Abbott et al. 2005; Aeschimann et al. 2019). The specificity of let-7 for regulation of lin-41 is thought to be conferred by base pairing of 3’ non-seed sequences of let-7 microRNA to the let-7 complementary sites in the LIN-41 mRNA 3′ UTR (Reinhart et al. 2000; Vella et al. 2004; Ecsedi et al. 2015), suggesting that conservation of let-7 microRNA non-seed sequences could reflect evolutionary pressure to conserve functional distinctions between let-7 microRNA and other let-7-family microRNAs.
The deep evolutionary roots of let-7 in the heterochronic pathway, including the apparent conservation of specific targeting of LIN-41 mRNA by let-7 microRNA suggests that hypothetical evolutionary loss of let-7 could be expected to be accompanied by significant divergence, compared with C. elegans, in the functions of heterochronic genes downstream of let-7, such as lin-41 and lin-29, and/or upstream genes, such as lin-14, lin-28, and hbl-1. Exploration of these questions would require the identification of species closely related to C. elegans that lack let-7.
Here, we identify a faction of Caenorhabditis species within the Japonica group, a sister group to the Elegans group, that lack the let-7gene. As far as we know, this is the first described instance of two sister clades where all known species of one clade have retained let-7, whereas numerous species of the sister clade do not have let-7. We demonstrate that for an exemplary let-7-lacking species, Caenorhabditis sulstoni, the functional architecture of the heterochronic pathway is otherwise conserved compared with C. elegans. Our findings indicate thatLIN-41 mRNA is apparently regulated by the remaining let-7-family microRNAs in most Japonica group species, suggesting that the heterochronic pathway can evolve to re-delegate let-7-family function under certain evolutionary circumstances.
Materials and methods
Nematode methods
All Caenorhabditis species were cultured on nematode growth medium (NGM; Brenner 1974) and fed with Escherichia coli HB101 except for all RNA interference (RNAi) experiments, in which C. elegans and C. sulstoni were fed E. coli HT115. Synchronized populations of developmentally staged worms were obtained using standard methods (Stiernagle 2006). All experiments involving C. elegans, unless otherwise noted, were performed at 20°C. All experiments with the other Caenorhabditis species, unless otherwise noted, were performed at 25°C. A list of strains used in this study is in Supplementary Table S5.
Developmental and phenotypic analyses of C. elegans, C. sulstoni, and Caenorhabditis macrosperma
To characterize the effects of temperature on development, synchronized populations of C. elegans and C. sulstoni were plated at 15°C, 20°C, 25°C, 30°C, 33°C, and 35°C. A synchronized population of C. macrosperma was plated at 25 °C. Except for C. sulstoni animals plated at 15 °C, developing populations plated at each respective temperature were observed every hour until animal development reached the adult stage. Following the initial 48 h of hourly observation, C. sulstoni animals plated at 15 °C were observed every 12 h until animals reached the adult stage. L1 alae, lethargy, cuticular molting, gonad migration, vulva development, adult alae, and oogenesis were used as markers to determine and calibrate developmental stages.
For heterochronic phenotype analyses, larvae were fed with RNAi food (as described below) starting from the L1 stage, and animals of defined developmental stages (as described above) were picked from healthy uncrowded mixed staged cultures and imaged. DIC microscopy was used to image hypodermis and alae. Fluorescence microscopy was used to image GFP-LIN-41.
For quantification of alae formation, the entire length of the animal’s cuticle was observed using DIC microscopy. Alae with one or more discontinuity were scored as incomplete. Any region where alae branched into multiple directions was scored as a branch.
Microscopy
All DIC and fluorescent images were obtained using a ZEISS Imager Z1 equipped with ZEISS Axiocam 503 mono camera, and the ZEN Blue software. Prior to imaging, worms were anesthetized with 0.2 mM levamisole in M9 buffer and mounted on 2% agarose pads. Adobe Photoshop was used to adjust the brightness and contrast of the images to enhance the visualization of the DIC and fluorescent signals. All fluorescent images were taken using the same microscopy settings and a standard exposure time for each larval stage for each reporter (C. elegans GFP-LIN-41 and C. sulstoni GFP-LIN-41). Identical brightness and contrast adjustments were used for each fluorescent image.
Caenorhabditis genomes
All genomes used in this study were provided by the Caenorhabditis Genomes Project (CGP) (http://caenorhabditis.org (last accessed February 4, 2021)).
Syntenic comparisons
Syntenic comparisons were performed using GEvo (https://genomevolution.org/coge/GEvo.pl; Lyons and Freeling 2008) with the following algorithm settings—Alignment Algorithm: (B)LastZ: Large Regions; Word size: 8; Gap start penalty: 300; Gap extend penalty: 30; Chaining: chain and extend; Score threshold: 2000; Mask threshold: 0; Minimum High-scoring Segment Pair (HSP) length for finding overlapped features: 50.
Gene prediction
Genes predictions were performed using the AUGUSTUS web interface (http://bioinf.uni-greifswald.de/augustus/ (last accessed February 4, 2021)) with the default settings (Keller et al. 2011).
Identification of homologous genes
Identification of all homologs was performed using CoGeBlast and the Caenorhabditis Genomes Project Blast webpages (https://genomevolution.org/coge/CoGeBlast.pl (last accessed February 4, 2021); http://blast.caenorhabditis.org/ (last accessed February 4, 2021)).
Sequence alignments
Sequence alignments were performed using Clustal Omega (www.ebi.ac.uk (last accessed February 4, 2021); Madeira et al. 2019) and visualized using Jalview (www.jalview.org (last accessed February 4, 2021); Waterhouse et al. 2009).
RNA hybridization predictions and minimum free energy calculations
RNA hybridization predictions and minimum free energy (MFE) calculations were performed using RNAhybrid (https://bibiserv2.cebitec.uni-bielefeld.de (last accessed February 4, 2021); Rehmsmeier et al. 2004).
lin-41 phylogeny
lin-41 phylogenetic tree was generated from orthology clustering provided by the Caenorhabditis Genomes Project (Stevens 2020) and visualized using iTOL (https://itol.embl.de/ (last accessed February 4, 2021); Letunic and Bork 2019 ). The lin-41 phylogenetic tree was rooted on the outgroup Diploscapter coronatus.
RNA extraction
Populations of animals were collected and flash-frozen in liquid nitrogen, and total RNA was extracted using Qiazol reagen (Qiagen) as described by Mcjunkin and Ambros (2017).
FirePlex microRNA detection
MicroRNAs were quantified from total RNA using FirePlex miRNA assay (Abcam) following the manufacturer’s instructions. Guava easyCyte 8HT (Millipore) was used for analysis.
Small RNA sequencing and let-7-family microRNA identification and normalization
Samples of total RNA were used to generate all small RNA libraries using a QIAseq miRNA Library Kit (Qiagen) following the manufacturer’s instructions. All libraries were sequenced on an Illumina NextSeq 500 sequencer. let-7-family microRNAs were identified by searching for reads that contained the let-7-family microRNA seed sequence “GAGGTAG” at positions 2–8. let-7-family microRNA reads were considered legitimate if the sequence mapped to the genome and if the read was predicted to form a microRNA-like stem-loop secondary structure with adjacent genomic sequence. RNA secondary structure modeling was performed using RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi (last accessed February 4, 2021); Lorenz et al. 2011). In most instances, reads corresponding to microRNA precursors were identified, adding additional credence to the validation of let-7-family microRNAs. microRNAs were quantified by normalizing the read count of a given microRNA to the total reads in that library.
GFP tagging of C. sulstoni LIN-41
Caenorhabditis sulstoni GFP-lin-41 was generated using CRISPR/Cas9 methods adapted from Paix et al. (2014, 2015) and Dokshin et al. (2018). The germlines of young adult females were injected with a mix of CRISPR RNA (crRNA) that targeted the 5′ end of the lin-41 coding sequence and the “co-CRISPR” marker dpy-10, tracrRNA (Supplementary Table S6), a PCR-derived dsDNA HR template, Cas9 protein (Integrated DNA Technologies), and Integrated DNA Technologies duplex buffer (30 mM HEPES, pH 7.5; 100 mM potassium acetate). L4 females were picked from plates where F1 animals exhibited the co-CRISPR phenotype and mated to a single male picked from the same plate, allowed to lay eggs, and then genotyped using PCR. F2s with GFP expression were cloned from F1s that scored positively by PCR genotyping for the desired modification. Single male and single female progeny were then mated, and a homozygous line was selected by GFP expression and PCR genotyping and subjected to Sanger sequencing for validation. The mutant was then thrice backcrossed to wild type. Sequence of the GFP::lin-41 allele generated in this study can be found in Supplementary Table S7.
Bacterial RNAi feeding strain constructions
cDNA from mixed staged C. elegans and C. sulstoni was generated from total RNA using SuperScript IV Reverse Transcriptase (ThermoFisher) and oligoDT following the manufacturer’s instructions. PCR was then used to amplify portions of C. elegans lin-41, C. sulstoni lin-14, C. sulstoni lin-28, C. sulstoni lin-29, C. sulstoni lin-41, C. sulstoni lin-46, C. sulstoni hbl-1, and C. sulstoni unc-22, respectively. Primers (Supplementary Table S6) used for each respective PCR also added KpnI sites to each end of each PCR product except for C. elegans lin-41, which added a HindIII site to one end and a KpnI site to the other end. The PCR products and the T444T vector were digested with KpnI (NEB) restriction enzyme for all C. sulstoni genes and HindIII and KpnI for C. elegans lin-41. The cut T444T vector was then dephosphorylated, and the cut PCR products and the cut/dephosphorylated vector were gel purified, ligated, and transformed into TOP10 chemically competent cells. Purified plasmids were subjected to Sanger sequencing for validation and transformed into chemically competent E. coli HT115 cells. The T444T plasmid was a gift from Tibor Vellai (Addgene plasmid # 113081; http://n2t.net/addgene:113081 (last accessed February 4, 2021); RRID: Addgene_113081).
RNAi knockdown of heterochronic genes
RNAi by feeding C. elegans and C. sulstoni with the strains described above was conducted as described in Conte et al. (2015).
Caenorhabditis elegans heterochronic pathway
The C. elegans heterochronic pathway schematic shown in Supplementary Figure S7A was adapted from Resnick et al. (2010).
Data availability
All Caenorhabditis strains are available at the Caenorhabditis Genetics Center (https://cgc.umn.edu (last accessed February 4, 2021)). Reagents used in this study are available upon request. Raw small RNA sequencing data can be found in the NCBI SRA (http://www.ncbi.nlm.nih.gov/sra (last accessed February 4, 2021)) (SUB8541612: accession numbers SAMN16816130, SAMN16816131, SAMN16816132, SAMN16816133, SAMN16816134, SAMN16816135, SAMN16816136, SAMN16816137, SAMN16816138, SAMN16816139, SAMN16816140, SAMN16816141, SAMN16816142, SAMN16816143, SAMN16816144, SAMN16816145, SAMN16816146, SAMN16816147, SAMN16816148, SAMN16816149, SAMN16816150, SAMN16816151, SAMN16816152, SAMN16816153, and SAMN16816154) under BioProject PRJNA678899. Normalized small RNA sequencing data used for Figure 3 and Supplementary Figure S4 can be found in Supplementary Table S8. The exact timing data used for Figure 3, Supplementary Figures S3 and S4 can be found in Supplementary Table S9. The RNAi quantification data used for Figures 4 and 7 can be found in Supplementary Table S10. microRNA data used for the “confirmed in previous study” sections of Figure 1A and Supplementary Table S1 were obtained from Pasquinelli et al. (2000), Reinhart et al. (2000), Lau et al. (2001), Lim et al. (2003), Ruby et al. (2006), de Wit et al. (2009), Shi et al. (2013), and Kozomara et al. (2019).
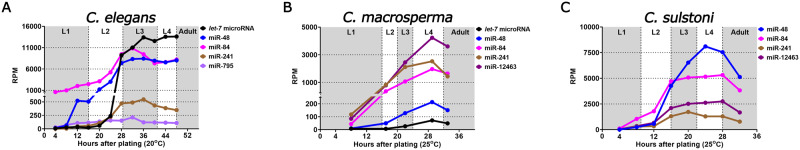
Figure 3.
let-7-family microRNA temporal expression patterns for C. elegans, C. macrosperma, and C. sulstoni. Small RNA sequencing data showing expression of let-7-family microRNAs throughout C. elegans (A), C. macrosperma (B), and C. sulstoni (C) development. RPM refers to reads per million.
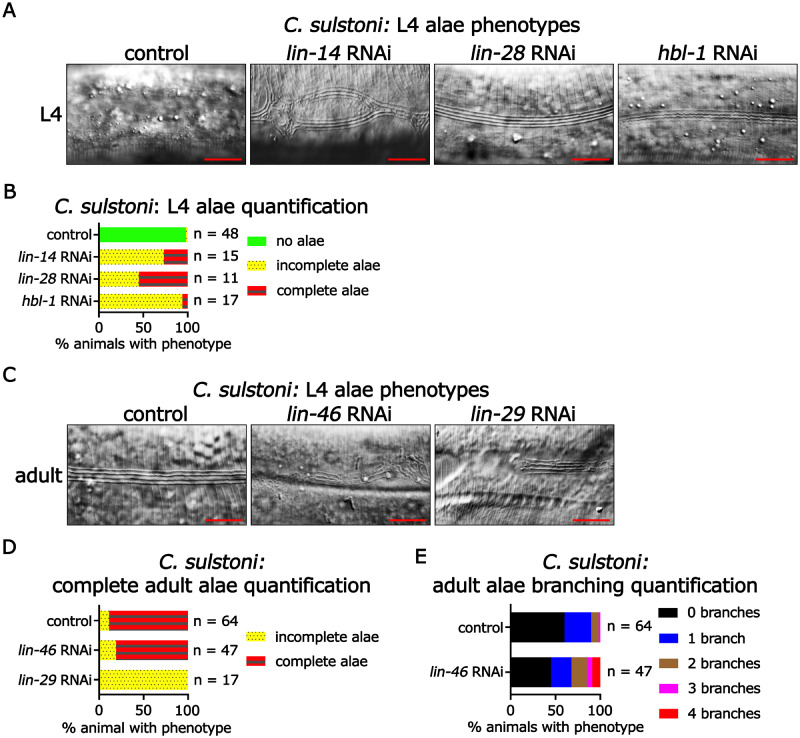
Figure 4.
Heterochronic phenotypes associated with RNAi of C. sulstoni lin-14, lin-28, hbl-1, lin-46, and lin-29. (A) Panels from left to right: representative DIC images of C. sulstoni L4 hypodermis of animals fed control (empty vector), lin-14, lin-28, and hbl-1 RNAi, respectively. (B) Quantification of observed L4 alae phenotypes in (A). Scale bars = 10 µM. (C) Panels from left to right: representative DIC images of C. sulstoni adult hypodermis of animals fed control (empty vector), lin-46, and lin-29 RNAi, respectively. Quantification of complete adult alae (D) and adult alae branching (E) phenotypes observed in (C). Scale bars = 10 µM. Note: the control (empty vector) and lin-46 RNAi animals used for (D) were also used for (E).
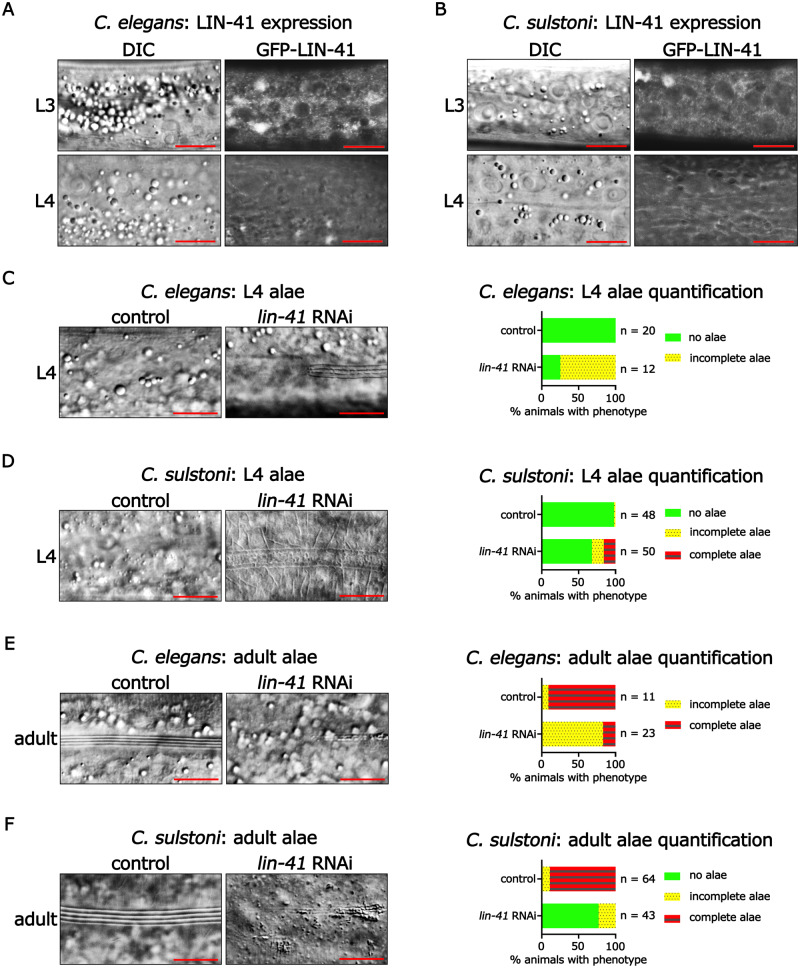
Figure 7.
lin-41 expression and function are conserved between C. elegans and C. sulstoni. (A, B) Representative DIC images (left panels) and endogenously tagged GFP-LIN-41 images (right panels) of hypodermal cells in C. elegans (A) and C. sulstoni (B) L3 (top panels) and L4 animals (bottom panels). Scale bars = 10 µM. Representative DIC images of C. elegans L4 hypodermis (C), C. sulstoni L4 hypodermis (D), C. elegans adult hypodermis (E), and C. sulstoni adult hypodermis (F) of animals fed control (empty vector) (left panels) and lin-41 RNAi (middle panels) and quantification of alae phenotypes (graphs). Scale bars = 10 µM. Note: RNAi experiments used for Figures 4, B and D and 7, D and F were performed together. Therefore, control RNAi data used for Figure 4, B and D were also used for Figure 7, D and F.
Supplementary material is available at https://doi.org/10.25387/g3.13557194.
Results
Most Caenorhabditis species belonging to the Japonica group lack let-7 microRNA
The let-7 microRNA was the first microRNA whose sequence and developmental function in promoting the differentiation of cell fates were shown to be conserved from nematodes to vertebrates (Figure 1A; Pasquinelli et al. 2000; Reinhart et al. 2000; Lin et al. 2007; Caygill and Johnston 2008; Sokol et al. 2008). While studying the evolution of regulatory sequences within the let-7 genomic loci of related nematodes, we discovered that most species of the Japonica group of Caenorhabditis lack the let-7 sequence in their genomic assemblies (Figure 1B and Supplementary Table S1). Interestingly, one exceptional Japonica group species, C. macrosperma, had let-7 sequence in its genomic assembly, which we confirmed using PCR amplification and Sanger sequencing (Supplementary Figure S1).
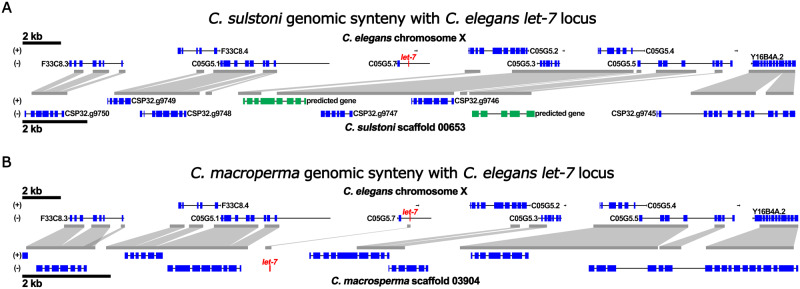
To determine if the lack of let-7 sequence in the genomic assemblies of these Japonica group species could reflect major genomic rearrangements and/or anomalous assembly of genomic sequence, we analyzed the genome sequences surrounding let-7 in C. elegans for potential synteny to corresponding genomic sequences of all Caenorhabditis species predicted to lack the let-7 sequence. We found that the genomic assemblies of all the Caenorhabditis species lacking let-7 sequence contain a region syntenic to the region surrounding the let-7 locus of C. elegans (representative synteny shown in Figure 2A and Supplementary Figure S2A). Except for Caenorhabditis afra, none of the Caenorhabditis species lacking let-7 sequence exhibited any indication that loss of let-7 sequence is associated with genomic rearrangement (Supplementary Figure S3).
Figure 2.
The genomic region containing the let-7 sequence in C. elegans is syntenic to genomic regions in C. sulstoni and C. macrosperma. Synteny of a portion of C. elegans chromosome X containing the let-7 sequence with C. sulstoni scaffold 00653 (A) and with C. macrosperma scaffold 03904 (B). Regions with sequence similarity are outlined in gray. let-7 is shown in red. Annotated genes are shown in blue. Predicted genes are shown in green.
The absence of the let-7 sequence in the genomic assemblies of Japonica group species could reflect incomplete sequence coverage. In such cases, let-7 microRNA could be expressed from DNA that was, for some reason, not detected by genomic sequencing. Therefore, to gather evidence, independent of genomic sequence, for whether or not let-7 microRNA is expressed in Japonica group species, we profiled microRNAs using FirePlex miRNA assays in RNA samples from mixed-stage populations of eight Japonica species available from the Caenorhabditis Genetics Center (CGC), Caenorhabditis waitukubuli, Caenorhabditis panamensis, Caenorhabditis nouraguensis, Caenorhabditis becei, Caenorhabditis macrosperma, Caenorhabditis sulstoni, C. afra, and Caenorhabditis japonica.
For all seven of the Japonica group species that lack let-7 in their genome assemblies as well as C. macrosperma, which has let-7 sequence in its genome assembly (Figures 1B and 2B, Supplementary Figure S1 and Table S1), we failed to detect let-7 microRNA expression by FirePlex assay. In contrast, let-7 microRNA was readily detectable using FirePlex in mixed-staged total RNA samples from species of the closely related Elegans group, Caenorhabditis inopinata, C. elegans, and Caenorhabditis kamaaina (Figure 1B and Supplementary Table S1). These FirePlex data also confirmed the expression of other microRNAs, including lin-4 microRNA and the let-7-family microRNAs miR-48 and miR-241 in all eight Japonica group species, as well as in the three Elegans group species (Figure 1B and Supplementary Table S1).
Caenorhabditis macrosperma expresses let-7
Caenorhabditis macrosperma is the one exceptional Japonica species in our experimental set that does contain let-7 genomic sequence. Despite confirming the presence of the let-7 sequence in the C. macrosperma genomic sequence assembly and its synteny to the C. elegans let-7 genomic locus (Supplementary Figure S1 and Figure 2B; expanded synteny Supplementary Figure S2B), we failed to detect let-7 microRNA expression using the FirePlex assay (see above). The FirePlex assay employs a panel of hybridization probes complementary to C. elegans microRNAs, which allows detection of microRNAs that precisely match the corresponding probe, or in some cases, that differ by a single internal nt. Although the C. macrosperma genomic sequence assembly contains an apparent let-7-5p guide RNA sequence identical to the C. elegans let-7 microRNA, it is possible that let-7 microRNA was not detected by FirePlex in our C. macrosperma RNA samples owing to significant 5′ or 3′ end variation and/or expression levels below the limits of detection by FirePlex. Similarly, other microRNAs that were not detected in our FirePlex data (such as the let-7 microRNA paralog miR-84, which was detected in C. elegans only; Figure 1B and Supplementary Table S1), could have been missed owing to low expression levels and/or significant interspecific variation in their non-seed sequences.
To definitively determine if let-7 microRNA is expressed in C. macrosperma, we performed small RNA sequencing of RNA samples from each larval stage as well as early adult stage for both C. elegans and C. macrosperma (staging times shown in Supplementary Figure S4, A and B). From these data, we confirmed that let-7 microRNA is indeed expressed in C. macrosperma (Figure 1B and Supplementary Table S1). Unlike C. elegans let-7 microRNA, whose expression dramatically increases during the L3 stage and peaks at the L4 stage and accumulates to levels similar to miR-48 and miR-84, C. macrosperma let-7 microRNA undergoes a more gradual and blunted increase in level during C. macrosperma development and never exceeds the level of any other let-7-family microRNA (Figure 3, A and B).
Small RNA sequencing also confirmed the expression of C. macrosperma lin-4 microRNA, as well as the let-7-family microRNAs, miR-48, miR-241, miR-84 (which is one nt shorter and has four internal nt differences from C. elegans miR-84), and a novel let-7-family microRNA, miR-12463 (Figures 1B and 3B, Supplementary Figure S5B and Table S1). Interestingly, the C. macrosperma mir-12463 genomic sequence is located 106 bp downstream of the mir-84 sequence, suggesting that miR-84 and miR-12463 may be produced from a common primary transcript. Similar to the C. elegans let-7-family microRNAs, the developmental expression profiles of miR-48/84/241/12463 in C. macrosperma consist of gradual increases during the L1 to L2 stages and peaks during the L3 and L4 stages (Figure 3B). We note a relatively low expression of miR-48 in C. macrosperma compared with C. elegans miR-48 (Figure 3, A and B).
The temporal expression profile of C. macrosperma lin-4 microRNA was similar to that of C. elegans, consisting of an increase in expression during early larval stages, followed by a broad peak during middle stages, and a decrease around the L4/adult transition (Supplementary Figure S5, A and B).
Robust assignment of C. macrosperma to the Japonica group
Caenorhabditis macrosperma was the only member of the Japonica group analyzed here to contain a let-7 gene in its genome, implying the presence of let-7 in a Japonica ancestor and loss of let-7 during the divergence of these Japonica species. An alternative explanation for the exceptionalism of C. macrosperma as a let-7-containing Japonica species could be an erroneous phylogenic assignment of C. macrosperma. In this regard, C. macrosperma was first recovered as a member the Japonica group based on a comparison of selected gene segments across Caenorhabditis species (Kiontke et al. 2011; Felix et al. 2014). Subsequent genome-wide analyses that included additional Caenorhabditis species again recovered C. macrosperma in the Japonica group with maximal support (a Bayesian posterior probably of 1.0 and a bootstrap value of 100, respectively) (Stevens et al. 2019, 2020), indicating a high likelihood of its correct assignment.
To obtain additional evidence confirming the evolutionary affinity of C. macrosperma with the Japonica group, we examined the phylogeny of the conserved let-7 target gene, lin-41. We reasoned that if the presence of a let-7 gene reflects evolutionary affinity of C. macrosperma with a clade outside Japonica, then such hypothetical divergence from Japonica might be reflected in the gene-specific phylogeny of lin-41, a conserved let-7-specific target. When we constructed a lin-41 gene tree across 33 Caenorhabditis species, we observed a lin-41 phylogeny nearly identical to the species phylogeny (Supplementary Figure S6), confirming the Japonica affinity of C. macrosperma lin-41, and strongly supporting the conclusion that C. macrosperma is an exceptional Japonica group species that has retained let-7.
Further support for the assignment of C. macrosperma to the Japonica group comes from considering the novel let-7-family microRNA, miR-12463, which we initially identified in small RNA sequencing from C. macrosperma. BLAST search analyses of Caenorhabditis species genomes revealed mir-12463 homologs to be present in all eight Japonica group species and in no species outside of the Japonica group (Figure 1B and Supplementary Table S1). This suggests that miR-12463 is a Japonica group specific let-7-family microRNA, and its presence in C. macrosperma supports the view that C. macrosperma is indeed a member of the Japonica group.
The mir-84 loci in the Japonica group are polycistronic with novel let-7-family microRNAs
The identification of mir-12463 within the mir-84 locus of C. macrosperma led us to explore if there are more predicted novel let-7-family microRNAs in the mir-84 loci of other Caenorhabditis species. To do this, we searched the genomic sequence surrounding the mir-84 sequence for the presence of the let-7-family seed sequence (GAGGUAG) in all Caenorhabditis species. We next used in silico RNA folding to predict if the RNA would fold into a stem-loop structure indicative of a microRNA precursor. From this analysis, we concluded that all members of the Japonica group have between one and five extra let-7-family microRNAs in their mir-84 loci. Interestingly, miR-84 is always the 5′ most let-7-family microRNA in these loci, and these additional let-7-family microRNAs are most likely polycistronic with miR-84 as they are no further than 221 bp apart (average of 116 ± 40 bp). We found no evidence of additional let-7-family microRNAs in the mir-84 loci in any Caenorhabditis species outside of the Japonica group (Figure 1B and Supplementary Table S1).
Caenorhabditis sulstoni serves as a representative species lacking let-7
Caenorhabditis sulstoni is a gonochoristic bacteriovore that was isolated from the feces of the east African millipede Archispirostreptus gigas purchased at an insect market in Berlin in spring 2013 (Stevens et al. 2019). Based on its robust and consistent developmental trajectory and ease of experimental manipulation (including sensitivity to RNAi), we adopted C. sulstoni as a representative let-7-lacking species for our experiments. Caenorhabditis sulstoni grows on standard NGM plates seeded with E. coli. Caenorhabditis sulstoni larval development appears largely similar to C. elegans, containing six distinct phases: an embryonic stage, L1 through L4 larval stages, and adult stage. The overall organismal morphology and cellular anatomy of C. sulstoni males and females appear markedly similar to the corresponding sexes of C. elegans. Particularly, the development of the C. sulstoni hypodermal cell lineages, the morphologies of stage-specific cuticles, and the ontogeny of the gonad and vulva are similar to C. elegans.
Like C. elegans, C. sulstoni embryos can be isolated by sodium hypochlorite/sodium hydroxide treatment of populations that include gravid adults. After allowing embryos to hatch in the absence of food, populations of developmentally arrested L1 larvae (L1 diapause) are obtained. Addition of food to a population of L1 diapause larvae triggers synchronous initiation of larval development, thereby enabling the preparation of populations of developing larvae of defined larval stages for biochemical and molecular experiments. Unlike C. elegans, which is typically cultured between 15°C and 25°C, C. sulstoni can be cultured between 20°C and 30°C (Supplementary Figure S4, A and C). Interestingly, at 25°C C. sulstoni develop from the L1 to adulthood 13 h faster than C. elegans, 28 vs 41 h, respectively (Supplementary Figure S4, A and C). Moreover, at 30°C and 33°C (temperatures that do not support C. elegans development), C. sulstoni develops from the L1 stage to adulthood in ∼23 h (Supplementary Figure S4A and C). 15°C and 33°C seem to define the low and high limits of C. sulstoni temperature tolerance under our culture conditions; C. sulstoni can develop from L1 to adulthood at 15°C or 33°C but are sterile when raised at either temperature. Populations of C. sulstoni grown at 15°C become asynchronous and take ∼6 days for the first animals to reach adulthood (Supplementary Figure S4C).
Except for the lack of let-7, the heterochronic pathway is functionally conserved in C. sulstoni
In C. elegans, let-7 functions as a significant component of the heterochronic pathway by ensuring proper developmental cell fate progression, particularly during the larval to adult cell fate transition (Supplementary Figure S6A; Reinhart et al. 2000). Because this hallmark microRNA of the heterochronic pathway is lacking in C. sulstoni, we sought to determine the status of other major components of the heterochronic pathway. As mentioned in the Introduction section, the protein coding genes lin-14, lin-28, lin-46, hbl-1, lin-41, and lin-29 as well as the microRNA genes lin-4, mir-48/84/241, and let-7 are critical components of the heterochronic pathway in C. elegans (Supplementary Figure S7A). In C. elegans, loss of lin-14, lin-28, hbl-1, or lin-41 results in precocious development and the subsequent early formation of adult-specific structures including adult lateral alae (Ambros and Horvitz 1984; Fay et al. 1999; Slack et al. 2000; Abrahante et al. 2003; Lin et al. 2003). In contrast, loss of lin-4, lin-46, mir-48/84/241, or lin-29 result in retarded development, characterized by incomplete formation of adult-specific structures including adult lateral alae (Chalfie et al. 1981; Ambros 1989; Ambros and Horvitz 1984; Pepper et al. 2004; Abbott et al. 2005).
To assess the conservation of heterochronic pathway gene function between C. elegans and C. sulstoni, we first characterized the larva-to-adult cell fate transition in the hypodermis of C. sulstoni. During the L4-adult transition in C. elegans, hypodermal seam cells, which consist of a longitudinal string of cells of either side of the animal, finish dividing, fuse with each other to form a lateral line syncytium, and produce the adult-specific cuticular structure called lateral alae, which signifies the terminal differentiation of the seam cells (Sulston and Horvitz 1977; Ambros and Horvitz 1984). Similar to C. elegans, in C. sulstoni we observed seam cell fusion and the formation for adult alae during the L4-adult transition (Figure 4, A–D).
From our FirePlex microRNA profiling, we had already confirmed the expression of lin-4 microRNA and the let-7-family microRNAs miR-48 and miR-241 in C. sulstoni (Figure 1B and Supplementary Table S1). We sought to determine if these microRNAs are integrated into the C. sulstoni heterochronic network through their targeted repression of the LIN-14, LIN-28, and HBL-1 mRNA 3′ UTRs as they are in C. elegans. Because 3′ UTRs are not annotated in C. sulstoni, we used the C. elegans 3′ UTRs as a framework and examined sequences downstream of each respective gene’s stop codon for lin-4 and let-7-family microRNAs complementary sites (Supplementary Table S2). For the C. sulstoni lin-14, lin-28, and hbl-1 3′ UTR regions, predicted sites for lin-4 and let-7-family microRNAs were identified, indicating conservation of the targeting of these heterochronic genes by lin-4 and let-7-family microRNAs between C. elegans and C. sulstoni (Supplementary Table S2 and Figure S7B).
As mentioned previously, the FirePlex assay we used to detect lin-4 microRNA and miR-48/241 in C. sulstoni employs a panel of hybridization probes complementary to C. elegans microRNAs, which does not allow for the detection of divergent orthologs of these microRNAs . This would include C. sulstoni miR-84, whichis predicted to be one nt shorter and have three internal nt differences compared with C. elegans miR-84.
To assay for miR-84 and to potentially identify novel let-7-family microRNAs in C. sulstoni, and to determine their temporal expression patterns during development, we performed small RNA sequencing of RNA samples from each larval stage as well as the early adult stage of C. sulstoni. From these data, we confirmed the expression of C. sulstoni lin-4 microRNA, miR-48, miR-84, miR-241, and miR-12463, and found that their developmental dynamics in C. sulstoni are similar to C. elegans and C. macrosperma (Figure 3, A–C, Supplementary Figure S5, A–C and Table S1).
To assess the potential functional conservation of protein coding components of the heterochronic pathway, we used RNAi to knock down heterochronic gene homologs in C. sulstoni. Previous studies reported that loss-of-function mutations of lin-14, lin-28, or hbl-1 in C. elegans result in precocious alae formation (Ambros and Horvitz 1984; Ambros 1989; Fay et al. 1999; Abrahante et al. 2003; Lin et al. 2003). Similarly, in C. sulstoni RNAi of lin-14, lin-28, or hbl-1 caused precocious phenotypes (Figure 4, A and B) like those previously reported for C. elegans.
Previous studies also reported that lin-46(lf) results in a mild retarded phenotype manifesting as minor gaps and branches in adult alae (Pepper et al. 2004), and lin-29(lf) results in a more severe retarded phenotype manifesting as significant gaps in, or complete absence of adult alae (Ambros and Horvitz 1984; Ambros 1989). RNAi of lin-46 or lin-29 in C. sulstoni resulted in retarded phenotypes (Figure 4, C–E) like those previously reported for C. elegans lin-46(lf) or lin-29(lf) mutants.
The similarities in expression of lin-4 and the let-7-family, the conservation of complementary sites in the 3′ UTRs of LIN-14, LIN-28, and HBL-1 mRNAs, and the RNAi knock down phenotypes for lin-14, lin-28, lin-46, hbl-1, and lin-29 indicate that the heterochronic pathway is largely conserved between C. sulstoni and C. elegans.
Temporal regulation and function of LIN-41 are largely conserved in C. sulstoni
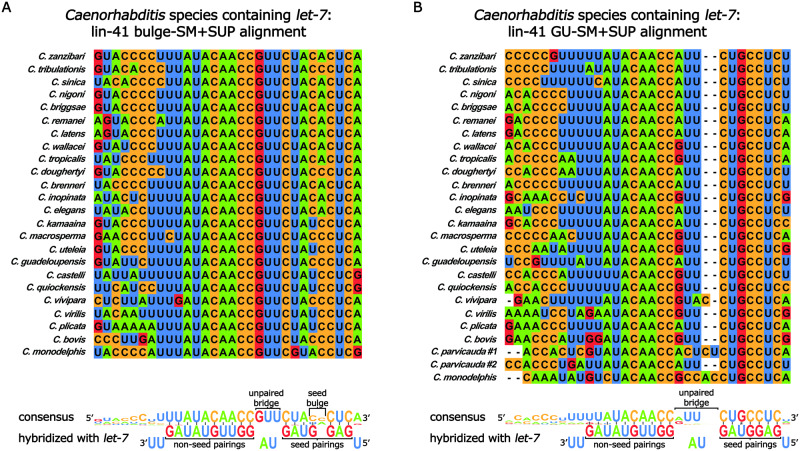
MicroRNA families are groups of microRNAs that share an identical seed sequence (nts 2–8) but differ in their non-seed sequence (Roush and Slack 2008). In principle, members of the same microRNA family can regulate the same target via seed pairing, but at the same time, differences in non-seed sequences can allow family member specificity of targeting through base-pairing of non-seed nts (Moore et al. 2015; Broughton et al. 2016; Brancati and Großhans 2018). In C. elegans, the let-7-family of microRNAs consists of let-7 microRNA, miR-48, miR-84, miR-241, and miR-795 (Supplementary Table S1; Abbott et al. 2005; Roush and Slack 2008). Interestingly, C. elegans let-7 microRNA targets the LIN-41mRNA 3′ UTR via two sites that are “weakly” complementary to the let-7-family microRNA seed sequence (hereafter referred to as SM for “seed match”), both of which are supplemented with significant non-seed pairings to let-7 microRNA. This base pairing configuration (hereafter referred to as SM+SUP) is thought to confer specificity for regulation of LIN-41 mRNA by let-7 microRNA, to the exclusion of the other let-7-family microRNAs. Interestingly, the “weak” seed of each SM is distinct from the other: pairing of let-7 microRNA to the first SM results in a bulged target adenine in the seed helix between g4 and g5 (hereafter referred to as the “bulge-SM”; Figure 5A); for the second SM, the seed helix contains a G-U wobble base pair at g5 (hereafter referred to as the “GU-SM”; Figure 5B; Reinhart et al. 2000; Vella et al. 2004; Ecsedi et al. 2015).
Figure 5.
The let-7 SM+SUP sites in the 3′ UTR region of lin-41 are highly conserved in Caenorhabditis species that contain let-7. Sequence alignment of the bulge-SM+SUP site (A) and the GU-SM+SUP (B) in the 3’ UTR region of lin-41 in Caenorhabditis species containing let-7. Shown at the bottom of each panel is the respective consensus sequence predicted hybridization with let-7 microRNA. Watson–Crick base pairing is shown with a solid line between the paired bases. The G-U base pair in the seed is shown with an asterisk between the paired bases.
To gauge the phylogenetic conservation of the let-7 microRNA SM+SUP configurations in the lin-41 3′ UTR regions of other related species, we aligned predicted lin-41 SM+SUP sequences for all available Caenorhabditis species genomes. To identify potential let-7-family microRNA base paring SM+SUP sequences, we searched downstream of the stop codon of lin-41 homologs for sequences complementary to the let-7 microRNA seed sequence, including bulges and G-U pairings. With the exception of the species lacking let-7, all Caenorhabditis species have two “weak” SM+SUPs in their lin-41 3′ UTR regions, and apart from C. parvicauda, which has two GU-SM+SUPs, all let-7-containing Caenorhabditis species have one bulge-SM+SUP positioned 5′ of one GU-SM+SUP (Figure 5, A and B, Supplementary Tables S3 and S4). Moreover, the SM+SUPs are always in relatively close proximity to each other, with the 3′ most base-paired nt of the bulge-SM being no further than 39 nts away from the 5′ most base-paired nt of the GU-SM (Supplementary Tables S3 and S4). These results indicate that there is strong selective pressure to maintain a strict let-7-specific regulation of lin-41 in Caenorhabditis species that express let-7 microRNA.
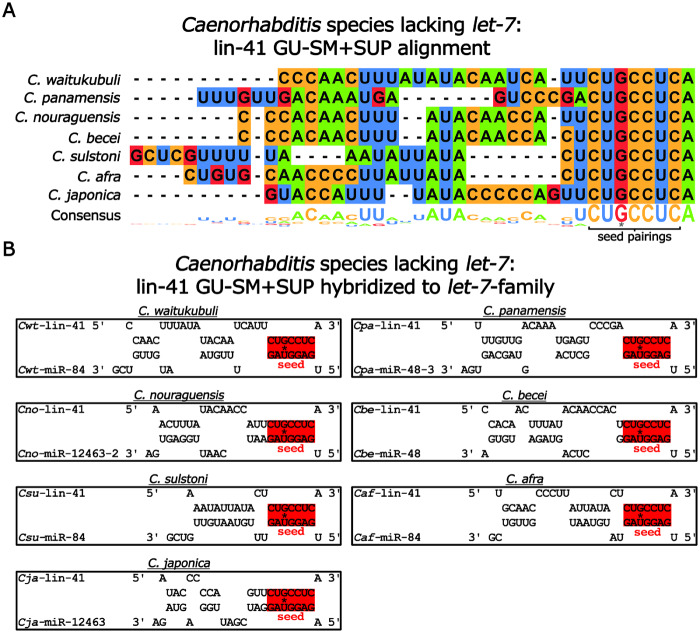
One possible explanation for how the absence of let-7 is accommodated in Japonica species is that one or more of the other let-7-family microRNAs may have adopted the role of regulating lin-41. To test this possibility, we examined the lin-41 3′ UTR regions of let-7-lacking genomes for sequences complementary to the remaining let-7-family microRNAs. Remarkably, in all seven of these genomes, we observed conservation of the GU-SM, indicating conservation of the regulation of LIN-41 mRNA by one or more let-7-family microRNAs, despite the absence of let-7 itself (Figure 6, A and B and Supplementary Table S4). Interestingly, we failed to observe any significant conservation of lin-41 3′ UTR region sequences adjacent to the GU-SM, arguing against any conservation of supplemental non-seed pairing (SUP) by a particular let-7-family microRNA (Figure 6, A and B and Supplementary Table S4).
Figure 6.
The SM +SUP site in the 3′ UTR region of lin-41 are divergent amongst Caenorhabditis species lacking let-7. (A) Sequence alignment of the GU-SM+SUP in the 3′ UTR region of lin-41 in Caenorhabditis species lacking let-7. The G-U base pair in the seed pairings is shown with an asterisk. (B) Predicted base pairing of let-7-family microRNAs with the GU-SM+SUP in the lin-41 3′ UTR region in Caenorhabditis species lacking let-7. Shown are the predicted base pairing of the let-7-family microRNA with the most favorable hybridization (lowest MFE) for each respective species. The G-U base pair in the seed pairings is shown with an asterisk.
Owing to the lack of conservation of supplemental non-seed matching sequence in the lin-41 3′ UTR regions of Caenorhabditis species lacking let-7, we hypothesized that one or more let-7-family microRNAs could regulate LIN-41 mRNA via GU-SM+SUP base-pairing pattern, but the particular let-7-family microRNAs with the best match toLIN-41 mRNA could vary for each species. We presumed that for a let-7-family microRNA to assume a similar role as let-7 microRNA in regulating LIN-41 mRNA, the MFE between that let-7-family microRNA and the LIN-41mRNA GU-SM+SUP should be similar to that for the interaction of let-7 microRNA with LIN-41 mRNA in let-7-containing species. To test this, we calculated the MFE of base pairing for each let-7-lacking species’ predicted let-7-family microRNAs to their respective lin-41 GU-SM+SUP and compared it with the MFEs of the let-7-family microRNAs hybridized to the bulge-SM+SUP as well as GU-SM+SUP in all let-7-containing species. In all let-7-containing species, the lowest (most favorable) MFE pairing of a let-7-family microRNA to the lin-41 bulge-SM+SUP and GU-SM+SUP was for the interaction with let-7 microRNA (average MFE of −27.2 ± 0.5 and −29.2 ± 1.2 kcal/mol, respectfully; Supplementary Figure S8, A and B, Tables S3 and S4). In contrast, in let-7-lacking species, the let-7-family microRNA that had the lowest MFE pairing to the lin-41 GU-SM+SUP varied between species and was higher (less favorable) than the MFE of let-7 microRNA with either the bulge-SM+SUP or GU-SM+SUP in species that contain let-7 (average MFE −21.5 ± 2.1 kcal/mol; Supplementary Figure S8B and Table S4).
LIN-41 is an RNA binding protein that, in C. elegans, forms distinct foci in the cytoplasm particularly around the periphery of the nucleus (Figure 7A;Spike et al. 2014). In the hypodermis of C. elegans, LIN-41 is expressed during the L1 through L3 stages and is undetectable in the L4 and adult stages due to its translational repression by let-7 microRNA (Figure 7A;Slack et al. 2000). The relatively weaker predicted MFE of the interaction between let-7-family microRNAs and lin-41 3′ UTR regions in species lacking let-7 compared with let-7 microRNA and lin-41 3′ UTR regions in species containing let-7 suggests that the let-7-family could have a less prominent role in down regulation of LIN-41 mRNA in species lacking let-7 than let-7 microRNA does in species containing let-7. To determine whether LIN-41 protein is down regulated in a representative let-7-lacking species as observed in C. elegans, we used CRISPR to GFP-tag the N-terminus of endogenous LIN-41 in C. sulstoni. We observed a nearly identical expression pattern in C. sulstoni to what we observed in C. elegans: hypodermal LIN-41 is expressed in the L1 through L3 stages and is downregulated in the L4 and adult stages (Figure 7, A and B). This down regulation of LIN-41 is indicative of prominent temporal regulation similar to what is observed in C. elegans and could reflect the action of one or more let-7-family microRNAs.
In C. elegans, lin-41(lf) animals precociously produce adult alae during the L4 stage (Slack et al. 2000). To determine if LIN-41 could be functionally conserved in species that lack let-7, we used RNAi to knock down lin-41 in C. sulstoni. Similar to C. elegans, when lin-41 was knocked down in C. sulstoni we observed L4 animals with adult alae. However, precocious alae occurred in a lower percentage of animals than previously reported for lin-41(lf) in C. elegans, and generally the precocious adult alae were of rather indistinct morphology compared with bona fide adult alae (Figure 7, D and E; Reinhart et al. 2000; Slack et al. 2000; Banerjee et al. 2005; Nolde et al. 2007). The relatively low penetrance of the precocious alae phenotype could possibly reflect poor RNAi knock down of lin-41. To test this, we knocked down lin-41 in our GFP-tagged strain and confirmed efficient knockdown by an absence of detectable GFP-LIN-41 fluorescence in hypodermal cells. Moreover, animals in which lin-41 was knocked down were sterile, a hallmark of lin-41 loss-of-function for its germline function in C. elegans (data not shown; Slack et al. 2000).
To our surprise, RNAi of lin-41 in C. sulstoni caused not only precocious and morphologically abnormal alae at the L4 stage, but also caused a highly penetrant alae formation phenotype in adult stage animals, where alae were often absent, incomplete, and/or morphologically abnormal (Figure 7F). This sort of adult stage phenotype had not been reported previously for lin-41(lf) in C. elegans. The previous studies of lin-41(lf) in C. elegans employed partial loss-of-function mutations and not the high-efficiency RNAi expression vector that we used to knock down lin-41 in C. sulstoni (T444T vector; Sturm et al. 2018). We hypothesized that the functions of lin-41 could be conserved between C. elegans and C. sulstoni, and the discrepancies in the phenotypes previously reported and observed here could be due to the partial function of the lin-41 alleles used in previous studies. To test this hypothesis, we knocked down lin-41 in C. elegans using the new, high-efficiency vector (T444T), and we observed similar phenotypes observed in C. sulstoni: L4 larvae with adult alae and incomplete/weak alae in adults (Figure 7, C–F). Interestingly, the L4 alae phenotypes observed in C. sulstoni were still less penetrant and weaker in appearance than what we observed in C. elegans (Figure 7, C and D). Moreover, the alae phenotypes observed in C. elegans adults appeared weaker than what we observed in C. sulstoni (Figure 7, E and F). Aside from these differences, our results indicate that the temporal patterning and function of LIN-41 are largely conserved between C. elegans and C. sulstoni.
Discussion
let-7 appears to be indispensable across diverse bilaterian phyla, indicating deep and pervasive evolutionary constraints on maintaining the entire 22-nt sequence of let-7 microRNA. Moreover, let-7’s function in promoting cellular differentiation and repression of pluripotency are conserved as well, which could reflect some degree of conservation of orthologous targets. Chiefly among these are lin-41 (TRIM71 in mammals) and lin-28 (LIN28 in mammals) both of which encode pluripotency promoting RNA binding proteins whose expression is directly repressed by let-7 microRNA in invertebrates and vertebrates alike (Slack et al. 2000; Kloosterman et al. 2004; Schulman et al. 2005; Lin et al. 2007; Vella et al. 2004; Ding and Großhans 2009; Ecsedi et al. 2015). Consequently, loss-of-function of let-7 results in dysregulation of developmental progression (Reinhart et al. 2000; Kloosterman et al. 2004; Sokol et al. 2008; Caygill and Johnston 2008) and can lead to disease such as cancer (Balzeau et al. 2017). Thus, let-7 is an important regulator of animal development and tissue homeostasis.
In this study, we characterized for the first time a cohort of animal species lacking the let-7 microRNA sequence in their genomes. Using microRNA profiling, we confirmed the absence of mature let-7 microRNA expression, while also confirming the conserved expression of the let-7-family microRNAs miR-48, miR-84, and miR-241. We also found that, except for the absence of let-7, the heterochronic pathway, into which let-7 is deeply integrated in C. elegans, appears otherwise functionally conserved in a representative let-7-lacking species, C. sulstoni. Finally, we provide evidence that lin-41, a let-7-specific target in C. elegans, is regulated by other members of the let-7-family of microRNAs in Caenorhabditis species that lack let-7.
Our finding that C. macrosperma appears to be an exceptional member of the Japonica group that contains let-7 raises interesting questions. Based on the current phylogenetic tree of Caenorhabditis species, for C. macrosperma to retain let-7, whilst the remaining members of the Japonica group lack let-7, three possibilities are suggested: (1) C. macrosperma does not belong in the Japonica group, (2) C. macrosperma is correctly assigned to the Japonica group, and gained the let-7 sequence after it was lost in a Japonica ancestor, or (3) C. macrosperma is correctly assigned to Japonica, and let-7 was lost at least three times during the evolution of the Japonica group.
The evidence strongly suggests that C. macrosperma is indeed correctly placed in the Japonica clade. Previous publications have routinely recovered C. macrosperma as a member of Japonica group with the most recent publications being a highly extensive, genome-wide studies with high statistical likelihood of C. macrosperma’s correct recovery (Kiontke et al. 2011; Felix et al. 2014; Stevens et al. 2019, 2020). Our results further strengthen this argument. We found that in C. macrosperma, the protein sequence of the let-7-specific target lin-41 was most similar to lin-41 protein sequence in other Japonica group species, and homologs of the novel let-7-family microRNA mir-12463 that we identified in C. macrosperma can only be found in Japonica group species. Put together, these suggest that C. macrosperma’s recovery in the Japonica group is correct.
We can imagine two possible ways by which C. macrosperma could have gained let-7, (1) in a spontaneous manner, possibly by random sequence drift of a let-7-family paralog, or (2) through gene transfer from another Caenorhabditis. The first scenario is unlikely because the let-7 genomic region in C. macrosperma is syntenic to the let-7 region in C. elegans, arguing against spontaneous gain as no other Japonica group member has a let-7-family paralog in that syntenic region. The second scenario also seems unlikely, as there are no known examples of horizontal gene transfer between Caenorhabditis species. Moreover, the presence in C. macrosperma of lin-41 bulge-SM+SUP and GU-SM+SUP sites matching let-7 that are nearly identical to the lin-41 bulge-SM+SUP and GU-SM+SUP sites in every other let-7-containing Caenorhabditis species strongly suggests that C. macrosperma inherited let-7, and its targeting of lin-41, from a common ancestor of the other Japonica species, and that let-7 was lost during the evolution of other Japonica species.
A parsimonious model for loss of let-7 in the Japonica clade suggests at least three independent loss events—once in the common ancestor to C. waitukubuli, C. panamensis, C. nouraguensis, and C. becei, a second loss in a common ancestor to C. sulstoni and C. afra, and a third loss in C. japonica. Multiple independent losses of let-7 in the Japonica group, together with the dramatically lower expression level of let-7 microRNA in C. macrosperma compared with the relatively robust expression of let-7 microRNA in C. elegans, suggests that let-7 became relatively dispensable in a common ancestor of these Japonica species. We suggest that a hallmark of the evolution of the Japonica group may be a reduced dependency on let-7 for functions that are otherwise critical in most other Caenorhabditis species.
The extensive synteny of the let-7 region of C. elegans with C. sulstoni suggests that loss of let-7 was not associated with dramatic genome rearrangements in the region. Only one gene neighboring let-7 in C. elegans, C05G5.7, also appears to have been fully lost in C. sulstoni. The loss of this gene might suggest a functional relationship between let-7 and C05G5.7, perhaps even that loss of C05G5.7 could functionally compensate for loss of let-7. Although C05G5.7 is transcribed on the same primary transcript as let-7 microRNA and serves as a negative regulator of let-7 microRNA processing, nevertheless C05G5.7 appears not to be conserved outside of C. elegans, and loss off C05G5.7 does not suppress let-7 loss-of-function (Nelson and Ambros 2019).
A key component of the heterochronic network in C. elegans is the RNA binding protein LIN-41. In C. elegans, loss of let-7 is lethal, primarily due to the de-repression of lin-41 (Ecsedi et al. 2015; Aeschimann et al. 2019). During wild type C. elegans development, LIN-41 levels decrease during the mid-to-late-L4 stage due to a sharp increase in let-7 microRNA expression and subsequent let-7 microRNA-mediated inhibition of LIN-41 translation. let-7 microRNA interacts with the 3′ UTR of LIN-41 mRNA via base pairing to two non-canonical SM sites—one with a bulge adenosine of the LIN-41 mRNA in the seed helix between the g4 and g5 and the other with a G-U wobble base pair at the g5—combined with extensive SUP. In Caenorhabditis species that contain let-7, both SM+SUPs are highly conserved. In the set of Caenorhabditis species that lack let-7, the lin-41 3′ UTR contains only the GU-SM, with no extensive conservation of non-seed pairings.
To be sure, the presence of a let-7-family SM in the 3’ UTR regions of lin-41 in species lacking let-7 indicates that lin-41 is likely regulated by one or more of the remaining let-7-family microRNAs. However, our observations suggest that let-7-family mediated regulation of lin-41 in these species may be relatively less critical compared with let-7-containing species, where the extensive conservation of two SM+SUP sites indicates that both sites are necessary for robust regulation of LIN-41 mRNA by let-7 microRNA. The lin-41 3′ UTR regions of Caenorhabditis species lacking let-7 contain just one let-7-family SM site of relatively weak predicted binding (higher MFE) and do not exhibit evidence of conserved non-seed pairing.
In species lacking let-7, the particular let-7-family microRNA(s) predicted to regulate lin-41 via SM+SUP pairing varied between species. miR-84 was most favorable in three species (C. waitukubuli, C. sulstoni, and C. afra), miR-48 was most favorable in two species (C. panamensis and C. becei), and miR-12463 was the most favorable in two species (C. nouraguensis and C. japonica; Figure 6B and Supplementary Table S4). Moreover, in some species that lack let-7, the let-7-family microRNA with the most favorable MFE was not much better than the next best MFE. For example, in C. panamensis, miR-48 was the most favorable with an MFE of −25.8 kcal/mol, whereas miR-84 was the next best with an MFE of −25.2 kcal/mol. This variability in which let-7-family microRNA is most favorable in combination with the relatively close MFEs of multiple let-7-family microRNAs within a species suggests that the regulation of LIN-41mRNA by the let-7-family microRNAs is less constrained, in terms of ortholog specificity, in let-7-lacking species than in let-7-containing species.
We found that despite the lack of let-7 in C. sulstoni the temporal expression pattern and function of LIN-41 protein is largely conserved; LIN-41 levels decrease during the L4 stage of C. sulstoni, as is the case in C. elegans, and knockdown of lin-41 results in similar heterochronic phenotypes in C. sulstoni and C. elegans. This indicates a form a compensatory evolution wherein LIN-41 in C. sulstoni has evolved to be regulated in a different way than LIN-41 in C. elegans to achieve the same functional outcome. Although our findings indicate that one or more let-7-family microRNA(s) may stand in for the absent let-7 to developmentally regulate C. sulstoni lin-41, we cannot rule out contributions from transcriptional or other posttranscriptional mechanisms.
Indeed, this instance of compensatory evolution would not be the first example of a regulatory gene and/or pathway being replaced by a related gene/pathway. One example of gene “handoff” is the mosquito Anopheles stephensi, where the gene paired, which is required for the formation of alternating body segments in many insect species (most classically in Drosophila melanogaster), was lost and functionally replaced with a related gene, gooseberry (Jarvela et al. 2020). Another example is the variation in the usage of Wnt signaling as a regulator of vulva induction in nematodes—Wnt signaling is the primary inducer of vulva formation in Pristionchus pacificus but plays a relatively minor role in C. elegans vulva formation (which primarily relies on EGF signaling; Sternberg 2005; Tian et al. 2008).
In this study, we have identified a cohort of related Caenorhabditis species within the Japonica group that lack the (otherwise) highly conserved let-7 microRNA. Our findings that one member of the Japonica group, C. macrosperma, has retained let-7, together with the pattern of species affinities within the Japonica group, suggests that let-7 was lost at least three times during the evolution of the Japonica clade. We do not currently know what allowed for the loss of let-7 in these species. However, it appears that in the Japonica group, let-7 was seemingly released from its evolutionary-entrenched regulatory relationship with lin-41, thereby allowing for the regulation of lin-41 to be taken over by the let-7-family. Further study is required to determine the nature of evolutionary factors that could cause an otherwise deeply conserved microRNA such as let-7 to become dispensable.
Acknowledgments
We thank the members of the Ambros and the Mello laboratories for helpful discussions and comments on this project, especially Ye Duan for help with small RNA sequencing. We thank Lewis Stevens and Mark Blaxter for help with the Caenorhabditis phylogeny and lin-41 gene tree. We thank the Caenorhabditis Genomes project for providing the Caenorhabditis genomic sequences and annotations.
Conceptualization: C.N., V.A.; methodology: C.N., V.A.; formal analysis: C.N., V.A.; investigation: C.N.; resources: v.A.; data curation: C.N.; writing—original draft: C.N.; writing—review and editing: C.N., V.A.; supervision: V.A.; project administration: V.A.; funding acquisition: V.A.
Funding
This research was supported by funding from National Institutes of Health grants R01GM088365, R01GM034028, and R35GM131741 (V.A.). Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). Resources for the genomic analyses were provided by CoGe, which is funded by the National Science Foundation (IOS-1444490).
Conflicts of interest: None declared.
Literature cited
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, et al. 2005. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 9:403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, et al. 2003. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 4:625–637. [DOI] [PubMed] [Google Scholar]
- Aeschimann F, Neagu A, Rausch M, Grosshans H.. 2019. let-7 coordinates the transition to adulthood through a single primary and four secondary targets. Life Sci Alliance. 2:e201900335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. 1989. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 57:49–57. [DOI] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR.. 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 226:409–416. [DOI] [PubMed] [Google Scholar]
- Ambros V, Ruvkun G.. 2018. Recent molecular genetic explorations of Caenorhabditis elegans MicroRNAs. Genetics. 209:651–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzeau J, Menezes MR, Cao S, Hagan JP.. 2017. The LIN28/let-7 pathway in cancer. Front Genet. 8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee D, Kwok A, Lin SY, Slack FJ.. 2005. Developmental timing in C. elegans is regulated by kin-20 and tim-1, homologs of core circadian clock genes. Dev Cell. 8:287–295. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancati G, Großhans H.. 2018. An interplay of miRNA abundance and target site architecture determines miRNA activity and specificity. Nucleic Acids Res. 46:3259–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton JP, Lovci MT, Huang JL, Yeo GW, Pasquinelli AE.. 2016. Pairing beyond the seed supports MicroRNA targeting specificity. Mol Cell. 64:320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill EE, Johnston LA.. 2008. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol. 18:943–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Horvitz HR, Sulston JE.. 1981. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 24:59–69. [DOI] [PubMed] [Google Scholar]
- Conte D Jr, Macneil LT, Walhout AJM, Mello CC.. 2015. RNA interference in Caenorhabditis elegans. Curr Protoc Mol Biol. 109:26.3.1–26.3.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Linsen SE, Cuppen E, Berezikov E.. 2009. Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res. 19:2064–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding XC, Großhans H.. 2009. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. Embo J. 28:213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokshin GA, Ghanta KS, Piscopo KM, Mello CC.. 2018. Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics. 210:781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecsedi M, Rausch M, Großhans H.. 2015. The let-7 microRNA directs vulval development through a single target. Dev Cell. 32:335–344. [DOI] [PubMed] [Google Scholar]
- Fay DS, Stanley HM, Han M, Wood WB.. 1999. A Caenorhabditis elegans homologue of hunchback is required for late stages of development but not early embryonic patterning. Dev Biol. 205:240–253. [DOI] [PubMed] [Google Scholar]
- Felix MA, Braendle C, Cutter AD.. 2014. A streamlined system for species diagnosis in Caenorhabditis (Nematoda: Rhabditidae) with name designations for 15 distinct biological species. PLoS One. 9:e94723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, et al. 2008. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 32:276–284. [DOI] [PubMed] [Google Scholar]
- Ilbay O, Ambros V.. 2019. Regulation of nuclear-cytoplasmic partitioning by the lin-28-lin-46 pathway reinforces microRNA repression of HBL-1 to confer robust cell-fate progression in C. elegans. Development. 146:dev183111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvela AMC, Trelstad CS, Pick L.. 2020. Regulatory gene function handoff allows essential gene loss in mosquitoes. Commun Biol. 3:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller O, Kollmar M, Stanke M, Waack S.. 2011. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics. 27:757–763. [DOI] [PubMed] [Google Scholar]
- Kiontke KC, Felix MA, Ailion M, Rockman MV, Braendle C, et al. 2011. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol. 11:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH.. 2004. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 32:6284–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M, Griffiths-Jones S.. 2019. miRBase: from microRNA sequences to function. Nucleic Acids Res. 47:D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP.. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 294:858–862. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V.. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 75:843–854. [DOI] [PubMed] [Google Scholar]
- Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, et al. 2009. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 16:1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47:W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, et al. 2003. The microRNAs of Caenorhabditis elegans. Genes Dev. 17:991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, et al. 2003. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 4:639–650. [DOI] [PubMed] [Google Scholar]
- Lin YC, Hsieh LC, Kuo MW, Yu J, Kuo HH, et al. 2007. Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development. Mol Biol Evol. 24:2525–2534. [DOI] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, et al. 2011. ViennaRNA package 2.0. Algorithms Mol Biol. 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, Freeling M.. 2008. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 53:661–673. [DOI] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47:W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcjunkin K, Ambros V.. 2017. A microRNA family exerts maternal control on sex determination in C. elegans. Genes Dev. 31:422–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Scheel TK, Luna JM, Park CY, Fak JJ, et al. 2015. miRNA-target chimeras reveal miRNA 3’-end pairing as a major determinant of Argonaute target specificity. Nat Commun. 6:8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V.. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 88:637–646. [DOI] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P.. 2011. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 147:1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C, Ambros V.. 2019. Trans-splicing of the C. elegans let-7 primary transcript developmentally regulates let-7 microRNA biogenesis and let-7 family microRNA activity. Development. 146:dev172031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM.. 2008. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 14:1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolde MJ, Saka N, Reinert KL, Slack FJ.. 2007. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3'UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev Biol. 305:551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, Seydoux G.. 2015. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics. 201:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Wang Y, Smith HE, Lee CY, Calidas D, et al. 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics. 198:1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, et al. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 408:86–89. [DOI] [PubMed] [Google Scholar]
- Pepper AS, Mccane JE, Kemper K, Yeung DA, Lee RC, et al. 2004. The C. elegans heterochronic gene lin-46 affects developmental timing at two larval stages and encodes a relative of the scaffolding protein gephyrin. Development. 131:2049–2059. [DOI] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, Lapierre RJ, Pothoulakis C, et al. 2011. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 147:1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R.. 2004. Fast and effective prediction of microRNA/target duplexes. RNA. 10:1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, et al. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 403:901–906. [DOI] [PubMed] [Google Scholar]
- Resnick TD, Mcculloch KA, Rougvie AE.. 2010. miRNAs give worms the time of their lives: small RNAs and temporal control in Caenorhabditis elegans. Dev Dyn. 239:1477–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S, Slack FJ.. 2008. The let-7 family of microRNAs. Trends Cell Biol. 18:505–516. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, et al. 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 127:1193–1207. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, et al. 2008. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 10:987–993. [DOI] [PubMed] [Google Scholar]
- Schulman BR, Esquela-Kerscher A, Slack FJ.. 2005. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn. 234:1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Montgomery TA, Qi Y, Ruvkun G.. 2013. High-throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes. Genome Res. 23:497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, et al. 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 5:659–669. [DOI] [PubMed] [Google Scholar]
- Sokol NS, Xu P, Jan YN, Ambros V.. 2008. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 22:1591–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike CA, Coetzee D, Eichten C, Wang X, Hansen D, et al. 2014. The TRIM-NHL protein LIN-41 and the OMA RNA-binding proteins antagonistically control the prophase-to-metaphase transition and growth of Caenorhabditis elegans oocytes. Genetics. 198:1535–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Chen X, Zhao H, Slack FJ.. 2015. A novel mechanism of LIN-28 regulation of let-7 microRNA expression revealed by in vivo HITS-CLIP in C. elegans. RNA. 21:985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg PW. 2005. Vulval development. WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.6.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L. 2020. CGP orthology clustering. Zenodo. doi:10.5281/zenodo.4068211. [Google Scholar]
- Stevens L, Felix MA, Beltran T, Braendle C, Caurcel C, et al. 2019. Comparative genomics of 10 new Caenorhabditis species. Evol Lett. 3:217–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L, Rooke S, Falzon LC, Machuka EM, Momanyi K, et al. 2020. The Genome of Caenorhabditis bovis. Curr Biol. 30:1023–1031.e4. [DOI] [PubMed] [Google Scholar]
- Stiernagle T. 2006. Maintenance of C. elegans. WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.101.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratoulias V, Heino TI, Michon F.. 2014. Lin-28 regulates oogenesis and muscle formation in Drosophila melanogaster. PLoS One. 9:e101141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm Á, Saskői É, Tibor K, Weinhardt N, Vellai T.. 2018. Highly efficient RNAi and Cas9-based auto-cloning systems for C. elegans research. Nucleic Acids Res. 46:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR.. 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 56:110–156. [DOI] [PubMed] [Google Scholar]
- Tian H, Schlager B, Xiao H, Sommer RJ.. 2008. Wnt signaling induces vulva development in the nematode Pristionchus pacificus. Curr Biol. 18:142–146. [DOI] [PubMed] [Google Scholar]
- Tsialikas J, Romens MA, Abbott A, Moss EG.. 2017. Stage-specific timing of the microRNA regulation of lin-28 by the heterochronic gene lin-14 in Caenorhabditis elegans. Genetics. 205:251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, et al. 2011. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol. 18:302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ.. 2004. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3’UTR. Genes Dev. 18:132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI.. 2008. Selective blockade of microRNA processing by Lin28. Science. 320:97–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ.. 2009. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All Caenorhabditis strains are available at the Caenorhabditis Genetics Center (https://cgc.umn.edu (last accessed February 4, 2021)). Reagents used in this study are available upon request. Raw small RNA sequencing data can be found in the NCBI SRA (http://www.ncbi.nlm.nih.gov/sra (last accessed February 4, 2021)) (SUB8541612: accession numbers SAMN16816130, SAMN16816131, SAMN16816132, SAMN16816133, SAMN16816134, SAMN16816135, SAMN16816136, SAMN16816137, SAMN16816138, SAMN16816139, SAMN16816140, SAMN16816141, SAMN16816142, SAMN16816143, SAMN16816144, SAMN16816145, SAMN16816146, SAMN16816147, SAMN16816148, SAMN16816149, SAMN16816150, SAMN16816151, SAMN16816152, SAMN16816153, and SAMN16816154) under BioProject PRJNA678899. Normalized small RNA sequencing data used for Figure 3 and Supplementary Figure S4 can be found in Supplementary Table S8. The exact timing data used for Figure 3, Supplementary Figures S3 and S4 can be found in Supplementary Table S9. The RNAi quantification data used for Figures 4 and 7 can be found in Supplementary Table S10. microRNA data used for the “confirmed in previous study” sections of Figure 1A and Supplementary Table S1 were obtained from Pasquinelli et al. (2000), Reinhart et al. (2000), Lau et al. (2001), Lim et al. (2003), Ruby et al. (2006), de Wit et al. (2009), Shi et al. (2013), and Kozomara et al. (2019).
Figure 3.
let-7-family microRNA temporal expression patterns for C. elegans, C. macrosperma, and C. sulstoni. Small RNA sequencing data showing expression of let-7-family microRNAs throughout C. elegans (A), C. macrosperma (B), and C. sulstoni (C) development. RPM refers to reads per million.
Figure 4.
Heterochronic phenotypes associated with RNAi of C. sulstoni lin-14, lin-28, hbl-1, lin-46, and lin-29. (A) Panels from left to right: representative DIC images of C. sulstoni L4 hypodermis of animals fed control (empty vector), lin-14, lin-28, and hbl-1 RNAi, respectively. (B) Quantification of observed L4 alae phenotypes in (A). Scale bars = 10 µM. (C) Panels from left to right: representative DIC images of C. sulstoni adult hypodermis of animals fed control (empty vector), lin-46, and lin-29 RNAi, respectively. Quantification of complete adult alae (D) and adult alae branching (E) phenotypes observed in (C). Scale bars = 10 µM. Note: the control (empty vector) and lin-46 RNAi animals used for (D) were also used for (E).
Figure 7.
lin-41 expression and function are conserved between C. elegans and C. sulstoni. (A, B) Representative DIC images (left panels) and endogenously tagged GFP-LIN-41 images (right panels) of hypodermal cells in C. elegans (A) and C. sulstoni (B) L3 (top panels) and L4 animals (bottom panels). Scale bars = 10 µM. Representative DIC images of C. elegans L4 hypodermis (C), C. sulstoni L4 hypodermis (D), C. elegans adult hypodermis (E), and C. sulstoni adult hypodermis (F) of animals fed control (empty vector) (left panels) and lin-41 RNAi (middle panels) and quantification of alae phenotypes (graphs). Scale bars = 10 µM. Note: RNAi experiments used for Figures 4, B and D and 7, D and F were performed together. Therefore, control RNAi data used for Figure 4, B and D were also used for Figure 7, D and F.
Supplementary material is available at https://doi.org/10.25387/g3.13557194.