Abstract
A clinical challenge to nearly every primate facility in North America is chronic idiopathic diarrhea (CID), the pathogenesis of which has yet to be fully elucidated. However, wild macaques appear resistant to CID, a trend that we observed in the free-ranging population of the Caribbean Primate Research Center. The gastrointestinal microbiota has been shown to have a significant role in the pathogenesis of disease and in maintaining normal health and development of the gut. In humans, chronic diarrhea is associated with alteration of the gut microbiota, which has lower bacterial diversity than does the microbiota of healthy humans. The current study was designed to describe and compare the fecal bacterial microbiota of healthy corralled, CID corralled, and healthy, free-ranging macaques. Fresh fecal samples were collected from healthy corralled (HC; n = 30) and CID (n = 27) rhesus macaques and from healthy macaques from our free-ranging colony (HF; n = 43). We excluded macaques that had received antibiotics during the preceding 60 d (90 d for healthy animals). Bacterial DNA was extracted, and the V4 region of the 16S rRNA gene was sequenced and compared with known databases. The relative abundance of Proteobacteria was higher in CID animals than HC animals, but otherwise few differences were found between these 2 groups. HF macaques were differentially enriched with Christensenellaceae and Helicobacter, which are highly associated with a ‘healthy’ gut in humans, as compared to corralled animals, whereas CID animals were enriched with Proteobacteria, which are associated with dysbiosis in other species. These results indicate that environment has a greater influence than health status on the gut microbiota. Furthermore, the current data provided targets for future studies on potential clinical interventions, such as probiotics and fecal transplants.
Abbreviations: CID, chronic idiopathic diarrhea; HC, healthy corralled; HF, healthy free-ranging; IBD, inflammatory bowel disease; PCoA, principal coordinate analysis; SSFS, Sabana Seca Field Station
Chronic idiopathic diarrhea (CID; also called idiopathic chronic diarrhea and chronic enterocolitis) is a clinical challenge that plagues nearly every large primate facility in North America. For example, the Oregon National Primate Center reports that CID comprises nearly 30% of their clinical caseload.20 At the Caribbean Primate Research Center, a review of the medical records database at the Sabana Seca Field Station (SSFS), where animals are housed in large, outdoor corrals, indicates that treatment for diarrhea comprises nearly 50% of the clinical caseload.
Information on CID in wild macaques is sparse, and an exact cause for CID in research macaques has not been identified, despite extensive study. Fecal bacterial culture has yielded mixed results, with no specific pathogen consistently isolated from animals with CID. An increased prevalence of Campylobacter, Shigella, and Yersinia species in animals with chronic diarrhea compared with healthy animals has been reported.59 However, the overall prevalence in diarrheic animals was around 25% for Campylobacter and well below 25% for Shigella and Yersinia.59 Similarly, one study reported that approximately 30% of chronic diarrheic animals had at least one historic bout of diarrhea that was culture positive and 40% culture positive for Campylobacter at the time of necropsy.38 Others have reported that fecal cultures are regularly negative for these and other common gastrointestinal pathogen,28,38 which is consistent with our experience.
The collective, interacting genomes of the symbiotic microorganisms in the gastrointestinal tract are referred to as the gastrointestinal microbiome.34 The microbiome has a significant role in the pathogenesis of disease and contributes to normal health and development of the gut.19,67 In humans, chronic diarrhea due to Clostridium difficile infection is associated with alteration of the gut microbiota (also known as dysbiosis), which has lower bacterial diversity than does the microbiota of healthy humans. This finding led to the successful use of fecal bacterial transplantation to restore the flora to normal.17,39 Similarly, our group identified significant differences in the bacterial microbiota and enrichment of Proteobacteria (a phylum associated with dysbiosis) in diarrheic calves and horses as compared with healthy ones.3,23 We also reported that diarrheic calves had lower relative abundance of genes responsible for metabolism of various nutrients, indicating that nutrient availability can be altered in diarrheic states.21 A better understanding of the organisms present in the gut of healthy and diarrheic macaques may offer new insights into the pathogenesis of this condition, and lead to new approaches to prevent and treat CID in NHP.
The current study was designed to describe and compare the fecal bacterial microbiota of healthy free-ranging, semiwild rhesus macaques (HF group), healthy macaques living in large, outdoor corrals (HC group), and corralled macaques with CID. The composition of the fecal bacterial microbiota from these 3 groups was compared to determine whether differences in bacterial composition are present among the groups. Identification of such changes may provide feasible starting points for studying the role of the intestinal microbiota in the pathophysiology of CID and possible treatment and preventive measures.
Materials and Methods
Animal housing.
The study was conducted at 2 AAALAC-accredited field sites using Indian-origin rhesus macaques (Macaca mulatta). At the SSFS, macaques are socially housed in outdoor corrals. At the Cayo Santiago Field Station, animals are free ranging on a 15.2-ha (37.5 acres) island near Puerto Rico and live in distinct social groups. At both stations, animals are fed Teklad NIB Primate Diet (Envigo, Madison WI) and provided filtered water, although both populations regularly consume standing water that results from the ample rain of Puerto Rico. At SSFS, animals receive additional food enrichment, such as fresh produce, seeds, and popcorn. At Cayo Santiago, free-ranging animals can forage on native foliage found on the island.
Housing and care for all animals are provided in accordance with the standards set forth in the Guide for the Care and Use of Laboratory Animals and the Animal Welfare Act.1,29 All procedures for this study were approved by the Caribbean Primate Research Center IACUC (protocol no. 338300).
Sample size calculation.
The sample size was determined by using a Dirichlet multinomial distribution model.37 With an expected number of 20,000 sequence reads (per animal) available for comparison and an α of 1%, 25 subjects per group were required for a power of 90%.
Animal selection.
HC macaques were selected according to the schedule for upcoming routine semiannual exams. A list of male and female macaques, 2 to 10 y old, from a variety of social groups was created. Medical records were reviewed, and animals were excluded for any of the following criteria: previous episode of diarrhea, recent use (<90 d from sample collection) of antibiotics for any reason, and other clinical comorbidities. Once animals were determined to be eligible for inclusion, samples were collected directly from the rectum during routine annual exams performed under sedation with ketamine (10 to 15 mg/kg IM; Dechra Veterinary Products, Overland Park, KS).
CID macaques were all housed in corrals. Colony animals were screened using our internal database system for a diagnosis of ‘diarrhea, chronic’ and age 2 to 10 y. From the selection of animals, individual medical records were reviewed. Animals diagnosed with CID were considered for inclusion in the study when at least one of the following was met: documented as having diarrhea for ≥ 30 d during the preceding 90 d; multiple episodes of diarrhea unresponsive to nonantimicrobial treatment; permanent removal from the social group due to ongoing need for treatment; or removal from social group at least 3 times for diarrhea treatment within preceding 1 y.20 Once this list of eligible animals was created, it was cross-referenced whenever an animal presented to the clinic for diarrhea, as determined by animal health technician during daily observations. The clinical veterinarian (NC) then conducted a secondary review of the individual medical record to determine final eligibility for inclusion. Animals with CID were excluded from the study if any antimicrobial had been administered within the 60 d period preceding sample collection or if any concurrent health problem was diagnosed by the veterinarian. Different criteria for recent antibiotic use were used for CID and HC groups because it was prohibitively difficult to identify CID animals that had not received antibiotics within 90 d of sample collection. Approximately10 g of fresh fecal samples were taken from the collection pan of animal's cage immediately after observation of a bowel movement, labeled, and stored in a –80° freezer until processing. All samples were collected during May through July 2018.
Samples from HF animals were collected during the annual trapping event during October through December 2018. As part of the ongoing colony management to address overpopulation on HF, a single social group is removed from the island each year. Animals were trapped in feeding corrals, sedated with ketamine (10 to 15 mg/kg IM), and placed in a transport cage overnight with food and water. The next morning, animals were transported to SSFS. Animals were sedated with ketamine (10 to 15 mg/kg IM) and xylazine (100 mg/mL, 0.1 mL/animal IM; Bayer, Shawnee Mission, KS), various morphometric data were collected, and animals were euthanized by intravenous overdose of sodium pentobarbital (390 mg/mL, 1 mL/5 kg; Med-PharmEx, Pomona, CA). Macaques approximately 2 to 10 y old were selected for sample collection when they appeared generally healthy, were in good body condition, and had formed stool. Fecal samples were collected directly from the rectum, labeled, and stored in a –80° freezer until processing.
DNA extraction.
Frozen samples were softened (typically approximately 2 h) until they could be reasonably and gently stirred to homogenize in a biosafety cabinet. A commercial DNA extraction kit (E.Z.N.A. Stool DNA Kit, Omega Bio-Tek, Norcross, GA) was used to extract and isolate bacterial DNA from the stool sample according to the manufacturer's protocol. Briefly, 0.2 g of stool was placed in a collection tube, and samples were lysed in a formulated detergent-containing buffer. After a heat–freeze step, proteins, polysaccharides, and cellular debris were precipitated by using a buffer in the kit. A provided reagent and buffer were used to bind DNA and remove contaminants after centrifugation. The supernatant was transferred to a HiBind DNA Mini Column, the column was washed to remove trace contaminants, and the purified DNA was eluted with elution buffer. The buffered DNA was stored in a –20° freezer until sequencing.
Targeted library preparation.
The samples were processed and analyzed by using the ZymoBIOMIHF Targeted Sequencing Service for Microbiome Analysis (Zymo Research, Irvine, CA). Bacterial 16S ribosomal RNA gene targeted sequencing was performed by using the Quick-16S NGS Library Prep Kit (Zymo Research. The bacterial 16S primers amplified the V4 region of the 16S rRNA gene. The sequencing library was prepared by using a library preparation process in which PCR reactions were performed in real-time PCR machines to control cycles and therefore limit PCR chimera formation. The final PCR products were quantified with qPCR fluorescence readings and pooled together to achieve equal molarity. The final pooled library was cleaned with the Select-a-Size DNA Clean and Concentrator (Zymo Research) and then quantified by using TapeStation (Agilent Technologies, Santa Clara, CA) and Qubit (Thermo Fisher Scientific, Waltham, WA).
Control samples.
The ZymoBIOMIHF Microbial Community DNA Standard (Zymo Research) was used as a positive control for each targeted library preparation. Negative controls (i.e., blank extraction control, blank library preparation control) were included to assess the bioburden level due to the wet-lab process.
Sequencing.
The final library was sequenced on Illumina MiSeq with a V4 reagent kit (600 cycles; Zymo Research). The sequencing was performed by spiking with at least 10% φX DNA.
Bioinformatic and statistical analysis.
Unique amplicon sequences were inferred from raw reads, and chimeric sequences were removed by using the DADA2 pipeline.9 Taxonomy assignment was performed by using Uclust from QIIME version 2.0 and the Zymo Research Database, a 16S RNA database that is internally designed and curated, as a reference. Composition visualization, α diversity, β diversity analyses, Bray–Curtis dissimilarity matrices, and weighted and unweighted UniFrac distances were performed by using QIIME version 2.0.10 Vegan function ADONIS was used to perform PERMANOVA to assess differences in β diversity. Taxonomic groups that had significant enrichment among different groups were identified by linear discriminant analysis for effect size by using default settings.58 Those default settings were α parameters for pairwise tests set to 0.05 for both class normality and subclass tests, and the threshold on the logarithmic score of linear discriminate analysis was set to 2.0. Analysis of heatmaps and principal coordinate analysis plots were performed with internal scripts.
Animal weights and ages were checked for normal distribution by using the Shapiro–Wilk goodness-of-fit test within JMP (version 15.1, SAS, Cary, NC). Differences between groups in weight and age were assessed by using the Student t-test and Kruskal–Wallis rank–sum test, respectively. Differences in the relative abundance of taxa and α-diversity parameters between groups were determined by using the Steel–Dwass method for nonparametric comparisons for all pairs (version 15.1, SAS). P values were adjusted for multiple comparisons according to the Benjamini–Hochberg false discovery rate.5
Results
Animal demographics.
A total of 100 samples were collected and sequenced from rhesus macaques at 2 fields sites. Animal weights were normally distributed, whereas animal ages were not. Neither weight nor age differed between groups (Table 1). Significantly more female than male macaques were included in the HF and the CID groups as compared with HC group, but no differences between male and female animals for any parameter, and thus data from both sexes were considered together throughout (data not shown).
Table 1.
Demographics of rhesus macaques from which fecal samples were collected and used in the final analysis. Healthy HF = healthy free-ranging, Healthy SSFS = healthy corralled, CID SSFS = corralled animals with chronic idiopathic diarrhea.
| Group | Median age (y; range) | Weight (kg; mean ± 1 SD) | no. of males | no. of females | Total |
| Healthy free-ranging | 5.5 (0.5–14.5)* | 5.0 ± 2.6 | 17 | 26 | 43 |
| Healthy corralled | 4.2 (2.1–9.5) | 5.9 ± 2.5 | 15 | 15 | 30 |
| CID | 4.1 (1.8–10.3) | 5.7 ± 2.6 | 9 | 18 | 27 |
Rounded to nearest half-year because exact birthdates for the free-ranging population are unknown
Rhesus macaque fecal microbiota: overall assessment.
A total of 15,765,498 raw sequences were obtained, with a range of 57,330 to 327,494 and median of 157,655 sequences per sample. A subsample of 20,000 reads per sample was used to normalize sequence numbers across samples and was considered adequate, as evidenced by greater than 99% coverage for all samples and plateau of rarefaction curves (data not shown).
A total of 17 bacterial phyla were identified; however, only 6 had greater than 1% of the total sequences identified, and Firmicutes and Bacteroidetes together accounted for over 90% of the total sequences (Table 2). Bacteria were identified from 209 genera; however only 11 included more than 1% of the total sequences identified and could be classified to a known genus (Table 3). A total of 30 orders, 18 classes, and 81 families were identified (Tables 4, Table 5 and 6).
Table 2.
Relative abundance of predominant bacterial phyla (overall relative abundance >0.5%) present in the feces of rhesus macaques
| Healthy free- ranging (HF) | Healthy corralled (HC) | CID | HF compared with HC | HF compared with CID | HC compared with CID | HF compared with HC+CID | |
| Phylum | Median (range) | P | |||||
| Firmicutes | 73 | 65 | 61 | 0.004 | 0.003 | 0.511 | <0.001 |
| (53–95) | (44–92) | (4.6–86) | |||||
| Bacteroidetes | 19 | 28 | 29 | <0.001 | 0.009 | 0.983 | <0.001 |
| (3.0–35) | (5.0–50) | (0.7–46) | |||||
| Proteobacteria | 1.3 | 0.9 | 1.7 | 0.212 | 0.320 | 0.007 | not applicable |
| (0–11) | (0.1–5.4) | (0.1–76) | |||||
| Spirochaetae | 0.8 | 0.7 | 1.3 | 0.937 | 0.074 | 0.308 | 0.138 |
| (0.0–3.8) | (0.1–8.1) | (0.0–7.6) | |||||
| Actinobacteria | 0.5 | 0.5 | 0.4 | 0.558 | 0.308 | 0.558 | 0.136 |
| (0.0–3.1) | (0.2–1.3) | (0.1–56) | |||||
| Euryarchaeota | 0.0 | 0.4 | 0.4 | 0.203 | 0.039 | 0.354 | 0.049 |
| (0.0–15) | (0.0–3.3) | (0.0–5.0) | |||||
| Tenericutes | 1.0 | 0.4 | 0.4 | <0.001 | <0.001 | 0.986 | <0.001 |
| (0.0–3.1) | (0.0–1.1) | (0.0–2.0) | |||||
Table 3.
Relative abundance of predominant bacterial genera (overall relative abundance >1.0%) present in the feces of rhesus macaques
| Healthy free- ranging (HF) | Healthy corralled (HC) | CID | HF compared with HC | HF compared with CID | HC compared with CID | HF compared with HC+CID | ||
| Genus | Median (range) | P | ||||||
| Lactobacillus | 22 | 20 | 12 | 0.302 | 0.009 | 0.088 | not applicable | |
| (0.0–73) | (1.4–42) | (0.0–41) | ||||||
| Prevotella | 2.4 | 4.5 | 4.0 | 0.002 | 0.165 | 0.685 | 0.008 | |
| (0.4–7.1) | (0.2–12) | (0.0–8) | ||||||
| Streptococcus | 0.2 | 3.4 | 1.3 | <0.001 | <0.001 | 0.099 | not applicable | |
| (0.0–11) | (0.2–17) | (0.1–9.1) | ||||||
| Phascolarctobacterium | 0.8 | 2.6 | 1.5 | 0.001 | 0.321 | 0.070 | not applicable | |
| (0.0–5.5) | (0.3–5.3) | (0.0–8.8) | ||||||
| Alloprevotella | 0.4 | 2.3 | 3.0 | <0.001 | <0.001 | 0.181 | <0.001 | |
| (0.0–2.8) | (0.1–6.2) | (0.0–8.6) | ||||||
| Faecalibacterium | 0.6 | 1.8 | 2.4 | <0.001 | <0.001 | 0.309 | <0.001 | |
| (0.0–5.1) | (0.1–4.9) | (0.1–4.8) | ||||||
| Blautia | 0.9 | 1.1 | 1.0 | 0.274 | 0.622 | 0.654 | 0.209 | |
| (0.1–4.9) | (0.3–4.7) | (0.0–3.1) | ||||||
| Subdoligranulum | 0.9 | 1.4 | 1.6 | 0.045 | 0.016 | 0.436 | 0.002 | |
| (0.1–2.6) | (0.1–4.1) | (0.1–7.7) | ||||||
| Megasphaera | 0.0 | 0.7 | 2.3 | <0.001 | <0.001 | 0.008 | <0.001 | |
| (0.0–0.1) | (0.0–5.7) | (0.1–11) | ||||||
| Roseburia | 0.3 | 1.5 | 1.3 | <0.001 | <0.001 | 0.715 | <0.001 | |
| (0.0–3.0) | (0.2–2.7) | (0.0–3.2) | ||||||
| Ruminococcus | 1.2 | 0.8 | 0.3 | 0.018 | <0.001 | <0.001 | not applicable | |
| (0.0–8.8) | (0.3–2.0) | (0.0–2.0) | ||||||
Table 4.
Relative abundance of predominant bacterial phyla (overall relative abundance >0.5% among 17 classifiable orders) present in the feces of rhesus macaques
| Healthy free- ranging (HF) | Healthy corralled (HC) | CID | HF compared with HC | HC compared with CID | HF compared with CID | HF compared with HC+CID | |||||||
| Class | Min | Max | Median | Min | Max | Median | Min | Max | Median | ||||
| Clostridia | 12 | 66.6 | 42.2 | 24.1 | 60.9 | 32.45 | 2.4 | 56.4 | 32.6 | <0.001 | 0.017 | 0.799 | <0.001 |
| Bacteroidia | 3 | 34.9 | 18.6 | 5 | 49.9 | 29.25 | 0.7 | 46 | 29 | 0.004 | 0.005 | 0.799 | <0.001 |
| Bacilli | 0.2 | 72.9 | 22.9 | 2.4 | 45.7 | 26 | 0.3 | 41.8 | 16.3 | 0.799 | 0.064 | 0.040 | n/a |
| Negativicutes | 0 | 5.8 | 1.9 | 2.3 | 12.3 | 5.3 | 0.2 | 17.4 | 6.7 | <0.001 | <0.001 | 0.050 | n/a |
| Erysipelotrichia | 0.5 | 7.4 | 1.7 | 0.5 | 3.6 | 1.05 | 0.1 | 16 | 1.2 | 0.011 | 0.128 | 0.263 | 0.004 |
| Spirochaetes | 0 | 3.8 | 0.8 | 0.1 | 8.1 | 0.7 | 0 | 7.6 | 1.3 | 0.953 | 0.031 | 0.191 | 0.138 |
| Methanobacteria | 0 | 14.7 | 0 | 0 | 3.3 | 0.35 | 0 | 5 | 0.4 | 0.195 | 0.038 | 0.354 | 0.050 |
| Epsilonproteobacteria | 0 | 3.7 | 0.3 | 0 | 1.3 | 0.1 | 0 | 48.9 | 0.7 | 0.010 | 0.405 | 0.001 | n/a |
| Mollicutes | 0.1 | 6.4 | 0.8 | 0 | 3.7 | 0.65 | 0 | 3.1 | 0.8 | <0.001 | <0.001 | 0.802 | <0.001 |
| Actinobacteria | 0 | 3.1 | 1 | 0 | 1.1 | 0.4 | 0 | 2 | 0.4 | 0.090 | 0.031 | <0.001 | n/a |
| Deltaproteobacteria | 0 | 1.1 | 0 | 0 | 0.1 | 0 | 0 | 55.7 | 0 | 0.493 | 0.411 | 0.987 | 0.366 |
Table 5.
Relative abundance of predominant bacterial phyla (overall relative abundance >0.5% among 27 classifiable orders) present in the feces of rhesus macaques
| Healthy free- ranging (HF) | Healthy corralled (HC) | CID | HF compared with HC | HF compared with CID | HC compared with CID | HF compared with HC+CID | |||||||
| Order | Min | Max | Median | Min | Max | Median | Min | Max | Median | ||||
| Bacteroidales | 3 | 34.9 | 18.6 | 5 | 49.9 | 29.25 | 0.7 | 46 | 29 | <0.001 | 0.020 | 0.998 | <0.001 |
| Methanobacteriales | 0 | 14.7 | 0 | 0 | 3.3 | 0.35 | 0 | 5 | 0.4 | 0.482 | 0.277 | 0.634 | 0.047 |
| Corynebacteriales | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 55.4 | 0 | 1.000 | 0.365 | 0.417 | 0.222 |
| Coriobacteriales | 0 | 2 | 0.4 | 0.2 | 1.3 | 0.5 | 0.1 | 2.3 | 0.5 | 0.582 | 0.993 | 0.993 | 0.357 |
Table 6.
Relative abundance of predominant bacterial phyla (overall relative abundance >0.5% among 69 classifiable families) present in the feces of rhesus macaques
| Healthy free- ranging (HF) | Healthy corralled (HC) | CID | HF compared with HC | HF compared with CID | HC compared with CID | HF compared with HC+CID | ||||||||
| Family | Min | Max | Median | Min | Max | Median | Min | Max | Median | |||||
| Lactobacillaceae | 0 | 72.9 | 22.5 | 1.4 | 42 | 20.3 | 0 | 40.9 | 12 | 0.291 | 0.009 | 0.090 | not applicable | |
| Ruminococcaceae | 5.2 | 47.6 | 24.1 | 10.6 | 41.6 | 15.65 | 1.5 | 28.2 | 14.6 | <0.001 | <0.001 | 0.523 | <0.001 | |
| Prevotellaceae | 1.2 | 26.6 | 9.5 | 4.3 | 43.6 | 25.3 | 0.7 | 36.1 | 25.6 | <0.001 | <0.001 | 0.696 | <0.001 | |
| Lachnospiraceae | 4.6 | 24.8 | 12 | 7.1 | 25.4 | 13.35 | 0.4 | 27.8 | 15.3 | 0.352 | 0.149 | 0.131 | 0.088 | |
| Unclassified Bacteroidales | 0.1 | 14.6 | 3.9 | 0.3 | 12.8 | 2.6 | 0 | 26.7 | 2.3 | 0.040 | 0.026 | 0.808 | 0.003 | |
| Veillonellaceae | 0 | 2.1 | 0.2 | 0.1 | 10.9 | 2.5 | 0.2 | 14.4 | 5.3 | <0.001 | <0.001 | 0.016 | not applicable | |
| Streptococcaceae | 0 | 11.2 | 0.2 | 0.2 | 16.7 | 3.4 | 0.1 | 9.2 | 1.3 | <0.001 | <0.001 | 0.094 | not applicable | |
| Acidaminococcaceae | 0 | 5.5 | 0.8 | 0.3 | 5.3 | 2.55 | 0 | 9.1 | 1.7 | <0.001 | 0.146 | 0.161 | 0.001 | |
| Rikenellaceae | 0.1 | 8.6 | 1.6 | 0 | 10.7 | 0.85 | 0 | 5.3 | 1.1 | 0.017 | 0.006 | 0.630 | 0.004 | |
| Christensenellaceae | 0.1 | 7.4 | 2.9 | 0.1 | 3 | 0.65 | 0 | 1.9 | 0.5 | <0.001 | <0.001 | 0.132 | <0.001 | |
| Erysipelotrichaceae | 0.5 | 7.4 | 1.7 | 0.5 | 3.6 | 1.05 | 0.1 | 16 | 1.2 | 0.010 | 0.139 | 0.262 | 0.004 | |
| Spirochaetaceae | 0 | 3.8 | 0.7 | 0.1 | 8.1 | 0.7 | 0 | 5.7 | 1.3 | 0.874 | 0.024 | 0.189 | 0.098 | |
| Family XIII | 0.2 | 3.2 | 1 | 0.5 | 3.2 | 1 | 0 | 4 | 1.2 | 0.294 | 0.072 | 0.319 | 0.041 | |
| Unclassfied Clostridiales | 0.1 | 5.9 | 1.2 | 0.2 | 3.5 | 0.95 | 0 | 2.1 | 0.5 | 0.467 | 0.001 | 0.001 | not applicable | |
| Campylobacteraceae | 0 | 1.5 | 0 | 0 | 1 | 0 | 0 | 42.5 | 0.7 | 0.610 | <0.001 | <0.001 | not applicable | |
| Unclassified Mollicutes | 0 | 3.1 | 1 | 0 | 1.1 | 0.4 | 0 | 1.9 | 0.3 | <0.001 | <0.001 | 0.729 | <0.001 | |
| Corynebacteriaceae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 55.4 | 0 | <0.001 | 0.141 | 0.228 | 0.222 | |
α and β diversity.
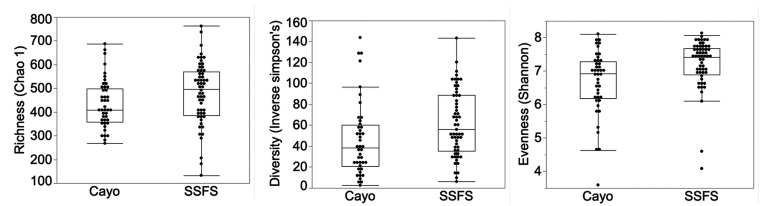
HC and CID animals showed no differences (P > 0.05) in the richness, evenness, or diversity of organisms identified (Chao 1, Shannon, and inverse Simpson analyses, respectively). These groups were therefore combined into a single group for comparison with HF animals. Samples from the combined HC+CID group were more rich, even, and diverse in bacterial organisms than were samples from HF animals (P = 0.034, 0.001, and 0.008, respectively; Figure 1).
Figure 1.
α diversity indices of bacteria observed in the feces of rhesus macaques. (A) Richness. (B) Evenness. (C) diversity.
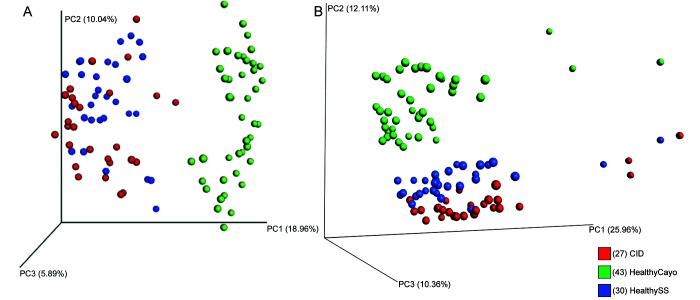
Principal coordinate analysis of the Bray–Curtis distance showed significant differences in the fecal microbiota of the macaques according to housing site but not health status (PERMANOVA P < 0.05), separating along principal component 1 and explaining about 20% of the total variation in the data (Figure 2). Similarly, principal coordinate analysis of unweighted UniFrac distances show clustering of fecal samples by housing site (PERMANOVA P < 0.05), but not by disease status. The samples separate along principal component 1, which explains approximately 26% of the total variation in β diversity (Figure 2).
Figure 2.
Principal coordinate analysis plot with (A) Bray–Curtis and (B) UniFrac unweighted distances, showing clustering of fecal bacterial composition by (A) abundance and (B) abundance and phylogenetic relatedness, respectively. Samples clustered significantly according to housing site but not health status.
Linear discriminant analysis for effect size.
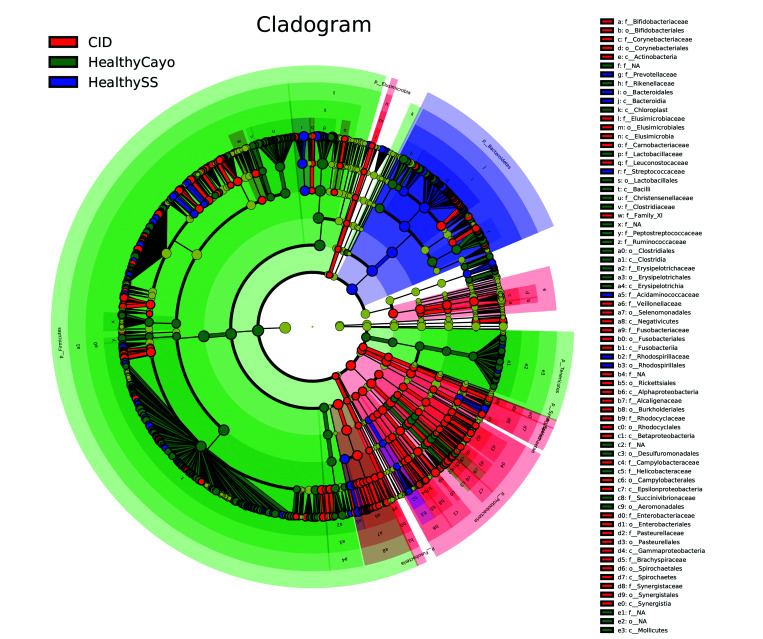
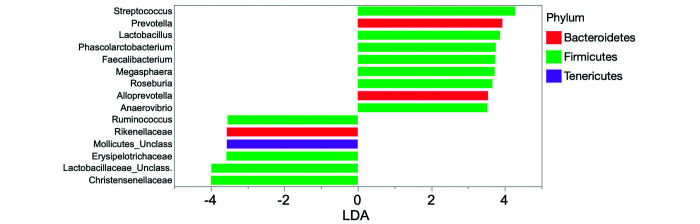
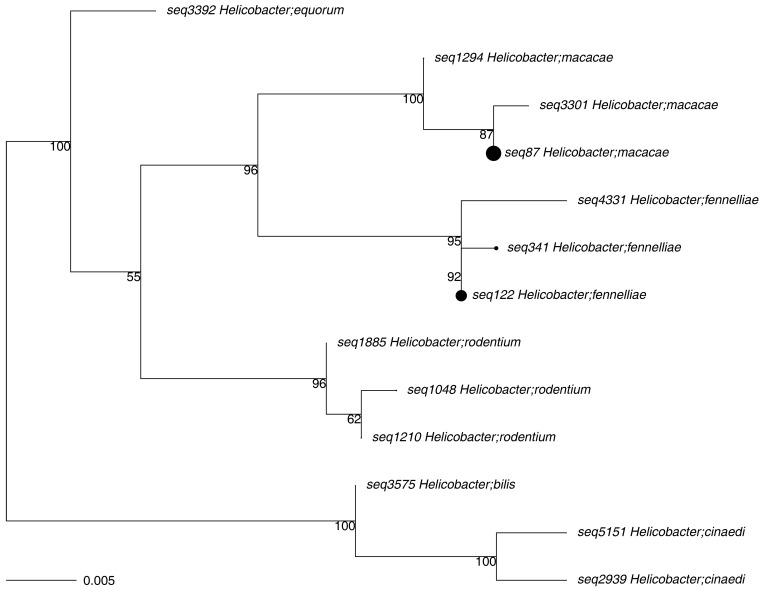
When comparing HC macaques, corralled macaques with CID, and HF macaques, a total of 127, 221, and 236 bacterial taxa (excluding species) were enriched, respectively, among 355 total taxa identified. Of the 3 groups, feces from HF animals had a higher number of bacterial taxa enriched from phylum Firmicutes (173, compared with 68 from HC macaques and 52 from animals with CID). In contrast, macaques with CID had more taxa enriched from phylum Proteobacteria (43, compared with 13 from HF and 5 from HC macaques; Figure 3). Figure 4 shows those taxa found to be enriched in HC compared with HF macaques. Figure 5 highlights the higher number of members of the phylum Proteobacteria, including Campylobacter, in HC compared with CID animals. Those phyla, classes, orders, families, and genera with a linear discriminate analysis score greater than 3.5 among all 3 groups are summarized in Table 7, and selected taxa of clinical interest, Helicobacter, Campylobacter, and Clostridium are shown in Figures 6, Figure 7 and 8. The phylogenetic relationships among Helicobacter species and strains, the only Proteobacteria differentially enriched in HF macaques, are depicted in Figure 9.
Figure 3.
Cladogram created by using linear discriminant analysis for effect size showing differentially bacterial abundant taxa in fecal samples from healthy corralled (blue), chronic diarrheic corralled (red), and healthy semiwild (green) rhesus macaques.
Figure 4.
Linear discriminate analysis scores of differentially enriched taxa in fecal samples from healthy, semiwild (left panel) and healthy, corralled (right panel) rhesus macaques.
Figure 5.
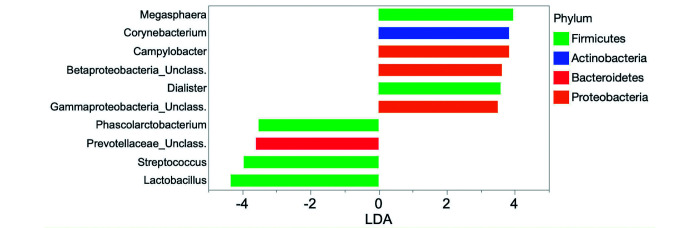
Linear discriminate analysis scores of differentially enriched in fecal samples from healthy (left panel) and chronic diarrheic (right panel) corralled rhesus macaques.
Table 7.
Differentially enriched taxa with a linear discriminate analysis score (LDA) of ≥3.5 when comparing the feces of healthy free-ranging, healthy corralled, and chronic diarrheic rhesus macaques
| Group | Phylum | Taxonomic level | Taxon | LDA |
| Healthy free-ranging | ||||
| Firmicutes | 4.8 | |||
| Class | Bacilli | 4.7 | ||
| Class | Clostridia | 4.6 | ||
| Class | Erysipelotrichia | 3.6 | ||
| Order | Lactobacillales | 4.7 | ||
| Order | Clostridiales | 4.6 | ||
| Order | Erysipelotrichales | 3.6 | ||
| Family | Lactobacillaceae | 4.8 | ||
| Family | Ruminococcaceae | 4.6 | ||
| Family | Christensenellaceae | 4.1 | ||
| Family | Erysipelotrichaceae | 3.6 | ||
| Genus | Lactobacillus | 4.8 | ||
| Genus | Ruminococcus | 3.8 | ||
| Proteobacteria | Family | Helicobacteraceae | 3.5 | |
| Genus | Helicobacter | 3.5 | ||
| Bacteroidetes | Family | Rikenellaceae | 3.9 | |
| Tenericutes | 3.6 | |||
| Mollicutes | 3.6 | |||
| CID | Firmicutes | Class | Negativicutes | 4.5 |
| Order | Selenomonadales | 4.5 | ||
| Family | Veillonellaceae | 4.4 | ||
| Genus | Megasphaera | 4.2 | ||
| Genus | Faecalibacterium | 3.9 | ||
| Genus | Dialister | 3.8 | ||
| Genus | Roseburia | 3.8 | ||
| Genus | Eubacterium | 3.6 | ||
| Genus | Subdoligranulum | 3.5 | ||
| Proteobacteria | 4.1 | |||
| Class | Epsilonproteobacteria | 3.9 | ||
| Class | Betaproteobacteria | 3.6 | ||
| Order | Campylobacterales | 3.9 | ||
| Family | Campylobacteraceae | 3.9 | ||
| Family | Rhodocyclaceae | 3.5 | ||
| Genus | Campylobacter | 3.9 | ||
| Bacteroidetes | Genus | Alloprevotella | 4.1 | |
| Spirochaetes | Spirochaetae | 3.7 | ||
| Class | Spirochaetes | 3.7 | ||
| Order | Spirochaetales | 3.7 | ||
| Actinobacteria | 3.9 | |||
| Order | Corynebacteriales | 3.8 | ||
| Family | Corynebacteriaceae | 3.8 | ||
| Genus | Corynebacterium | 3.8 | ||
| Healthy corralled | Bacteroidetes | 4.7 | ||
| Class | Bacteroidia | 4.7 | ||
| Order | Bacteroidales | 4.7 | ||
| Family | Prevotellaceae | 4.8 | ||
| Genus | Prevotella | 4.0 | ||
| Firmicutes | ||||
| Class | Acidaminococcaceae | 3.7 | ||
| Family | Streptococcaceae | 4.3 | ||
| Genus | Streptococcus | 4.3 | ||
| Genus | Phascolarctobacterium | 3.7 | ||
| Genus | Anaerovibrio | 3.5 |
Figure 6.
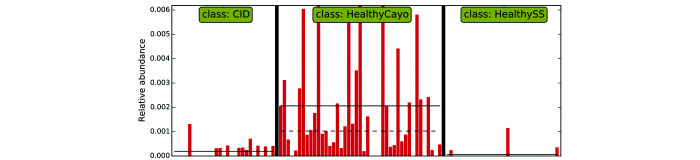
Linear discriminant analysis for effect size indicates that fecal samples from healthy, free-ranging rhesus macaques are differentially enriched with bacteria of the genus Helicobacter.
Figure 7.
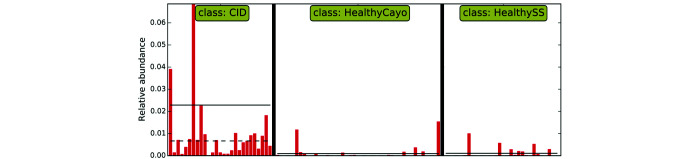
Linear discriminant analysis for effect size indicates that fecal samples from corralled rhesus macaques with CID are differentially enriched with bacteria of the genus Campylobacter.
Figure 8.
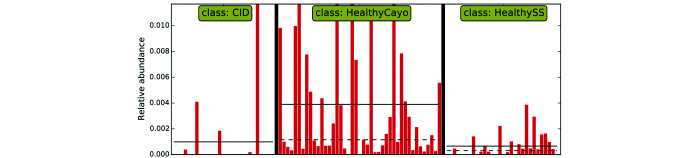
Linear discriminant analysis for effect size indicates that fecal samples from healthy, free-ranging rhesus macaques are differentially enriched with bacteria of the genus Clostridium.
Figure 9.
Phylogenetic tree of Helicobacter species and strains identified in the feces of rhesus macaques. Dot size represents relative contribution to the number of sequences identified.
Discussion
A number of studies20,28,32,38,47,55,57,59 have explored CID in captive macaques, but, to our knowledge, none have specifically characterized and compared the fecal microbiomes of healthy adult and diarrheic animals or of corralled and free-ranging animals by using 16S rRNA gene sequences. The 2 field sites of the Caribbean Primate Research Center offer a unique opportunity to compare macaques in a semiwild state with those in a more traditional large, outdoor-corral facility. Animals at both sites received the same standard monkey chow, had access to purified water, and lived in the same climate and thus offer the most directly comparable populations of free-ranging and corralled NHP that we know of, with one population afflicted with CID and the other unaffected to any significant degree. The HF macaques undergo no veterinary intervention (except for annual health exams, which include tuberculosis testing and vaccination against tetanus) and therefore have never received antibiotics or other treatment that may alter the gut microbiota. In the current study, we showed that rhesus macaques from the same housing site, regardless of disease status, were more similar in fecal bacterial composition than animals of similar health status at different housing sites with different housing modalities. This finding is in agreement with studies in other mammals.15,23
The 2 predominant bacterial phyla regardless of health status or housing modality were Firmicutes and Bacteroidetes, a finding that is consistent across species, including other NHP, humans, mink, rabbits, cows, and mice.15,18,23,36,57,61,68 However, an overall lower relative abundance of Bacteroidetes was found in the HF macaques as compared with corralled animals and as compared with previously published reports in macaques.55,61,72 In addition, taxa from the phylum Bacteroidetes were the most differentially enriched taxa of the HC animals, and were the only nonfirmicute taxa represented. Several studies in mice and humans have shown a negative correlation between obesity and the relative abundance of Bacteroidetes, with leaner subjects showing lower proportions of Bacteroidetes.31,40,41,64 Although we did not account for body condition score and group ages and weights were similar, a cursory evaluation of the relationship between age and weight suggests that the free-ranging animals were leaner than their corralled counterparts. Free-ranging macaques had the highest mean age but the lowest mean weight, whereas HC animals had a lower mean age and highest mean weight. Given the frequent use of rhesus macaques as a model for human obesity and metabolic syndrome,4 additional studies on the relationship between the gut microbiome and body condition in this species are warranted. Reasons for these differences are unclear, but body condition, diet, or water sources may explain, at least in part, the disparities in the abundance of Bacteroidetes between groups.
Proteobacteria were differentially enriched in diarrheic animals. This phylum was the only one that differed in the relative abundance among HC and CID animals. This pattern is expected, given that higher proportions of Proteobacteria are regularly identified in diarrheic animals across species and are considered a hallmark of dysbiosis.21,23,33,44,51,60 However, an unexpected finding was the lack of difference in the relative abundance of Proteobacteria between diarrheic and HF animals. In all other regards, the free-ranging animals would be considered to have a ‘normal’ or ‘healthy’ fecal microbiota, but the relative abundance of Proteobacteria in the HF group was even higher than that of the HC animals. This finding may challenge the hypothesis that, overall, enrichment of Proteobacteria is an indicator of dysbiosis or altered gut microbiota. Rather, it highlights the importance of taking into account gastrointestinal location when making generalizations regarding what determines a dysbiotic state. Certain bacteria may have a commensal or protective role in one region of the gut but cause pathology if unchecked in other regions.
The unexpectedly high proportion of Proteobacteria in the free-ranging group is accounted for by the differential enrichment of the genus Helicobacter and its corresponding family, Helicobacteraceae, the only bacteria from this phylum that was enriched in this group. One study reported that the colon from healthy control rhesus macaques had a superficial mucosa densely populated with epithelium-adherent bacteria, identified as 100% Helicobacter macacae, whereas in animals with CID, these organisms were largely absent.38 Similarly, healthy 8-mo-old rhesus macaques were differentially enriched with and had a significantly higher relative abundance of H. macacae in feces than did their diarrheic counterparts.55 Studies in human diarrheal cases have reported an inverse relationship between H. pylori infection and diarrheal illnesses in children and lower risk of diarrhea of unknown origin and shigellosis in H. pylori-positive adults.14,56 The literature on the relationship between H. pylori infection and inflammatory bowel disease (IBD) in humans, similar in nature to CID of macaques, is diverse. A meta-analysis found a higher prevalence of H. pylori infection in healthy people compared with those with IBD,45 although the authors acknowledged that differing methodologies and possible publication bias may limit the certainty of their findings. Similar results were reported in a systematic review investigating the mechanisms underlying the potential link between H. pylori and IBD.52 Moreover, a systematic review and meta-analysis found that H. pylori infection was indeed negatively associated with IBD, regardless of ethnicity, age, or detection methods.11 Our findings, together with other studies in rhesus macaques39,56 and extrapolation from the human literature, suggest that H. macacae may be important in maintaining homeostasis of the gut or exert a protective role in preventing CID.15,39,56,57
Campylobacter spp. are an important cause of diarrhea in humans and NHP worldwide, with the Center of Disease Control estimating that Campylobacter is the number-one cause of bacterial diarrheal illness in the United States.13 Indeed, multiple studies have identified a higher prevalence of Campylobacter species in bacterial cultures of diarrheic as compared with asymptomatic rhesus macaques.38,59 Similarly, using next-generation sequencing techniques, one study reported that identification of Campylobacter (C. jejuni and C. coli) was strongly associated with diarrhea, and 2 other studies each found that Campylobacter were differentially enriched in relative abundance and gene expression, respectively, in symptomatic as compared with asymptomatic counterparts.47,55,71 However, identification of Campylobacter in as many as 42% of healthy subjects brings into question whether its presence is as a primary or an opportunistic pathogen.32,38,59 Campylobacter spp. in diarrheic subjects overall had a much higher level of virulent gene expression than did Campylobacter spp. of healthy controls, and Campylobacter-specific transcriptomes suggested a closer association with the mucosa in chronic diarrhea than in controls.71 Taken together, differing expression of virulence factors, resulting in a more virulent phenotype, may explain why some animals positive for Campylobacter develop diarrhea whereas others do not.
An alternative explanation for the presence of Campylobacter in diarrheic animals is rooted in its close phylogenetic relationship to Helicobacter, both of which are members of the order Campylobacterales (Helicobacter were previously classified as Campylobacter organisms). As previously discussed, H. macacae may have a role in prevention of CID. We showed that healthy macaques were differentially enriched with Helicobacter and diarrheic animals were differentially enriched with Campylobacter, mirroring results in healthy and diarrheic rhesus infants and in humans with and without IBD.11,55 The inverse relationship between these closely related genera may represent a niche displacement event, with some inciting factor leading to displacement of Helicobacter, either prior to or concurrent with, increased expression of virulence factors by Campylobacter, ultimately leading to overgrowth and adherence of Campylobacter. The importance of this relationship is likely only recently being explored because of the inherent limitations of bacterial culture: Helicobacter is fastidious and requires specific, narrow culture conditions that are unlikely to be achieved unless specifically seeking to do so.6 The use of next-generation sequencing allows the identification of organisms that have been historically difficult to culture. Although Helicobacter is known to be associated with gastric disease and ulcers in humans, macaques, and other mammalian species,6,14,56 it is not generally considered to be an important cause of diarrhea; consequently, previous studies would have been unlikely to have specifically evaluated Helicobacter, such that its prevalence and thus its importance would not be recognized.
Bacteria of the class Clostridia are a diverse group of obligate anaerobes that we found to be enriched in HF rhesus macaques, as was its largest order, Clostridiales. This enrichment was due to a corresponding enrichment of 2 families: Ruminococcaceae and Christensenellaceae. Clostridia, including species of the infamous genus Clostridium, many of which produce highly pathogenic toxins and are associated with serious diarrheal disease in humans,7,12,35,49 are known to digest cellulose and hemicellulose.65,73 The free-ranging animals on the island of Cayo Santiago receive standard monkey chow daily but also are able to forage freely on the island fauna, whereas corralled animals at the SSFS have access to fresh produce (primarily fruits and seeds) only in limited supply as part of their enrichment program, suggesting the important role that diet plays in shaping the gut microbiome. Ruminococcaceae are depleted during chronic diarrhea in humans, horses, and piglets.2,16,26,62,66 Christensenellaceae has recently been recognized as highly important to human health,69 with several studies showing a highly negative correlation between relative abundance of Christensenellaceae and body mass index,8,43,54 serum markers of metabolic disease,25,27,43 and metabolic syndrome.25,42,50 Moreover, Christensenellaceae is one of the most highly heritable and transmittable groups of bacteria in humans, with germ-free mice that receive feces amended with Christensenellaceae gain less weight than recipients of unamended feces.24 Finally, Christensenellaceae was identified as a signature of a healthy gut in a meta-analysis of IBD46 and is consistently depleted in individuals with IBD.22,30,51,53 Taken all together, Ruminococcaceae and Christensenellaceae bacteria represent a reasonable starting point for experimental and interventional studies aimed at using or optimizing the gut microbiome to combat CID, as well as obesity, in research macaques.
We recognize several limitations to the current study. Primarily, fecal samples are limited in their representation of all parts of the gut. In macaques, fecal samples were highly correlated with the microbiota of the large intestinal lumen and mucosa but less correlated with small intestinal luminal and mucosal samples.73 The cited study also found that about 95% of the operational taxonomic units within the large intestinal mucosa and lumen were proportionally similar, such that feces can serve as a proxy for colonic contents.72 In addition, sample collection times differed between the free-ranging and corralled groups, because the trapping schedule for the free-ranging macaques occurred during the fall. In horses, season, supplementary feeding and ambient weather conditions (for example, changes in temperature) are associated with changes in fecal microbiota structure and therefore, the differences in collection times could have impacted our results. However, the community membership and structure of the fecal microbiota of CID and HC and HF macaques were similar. The specific changes we found in CID macaques were similar to those previously reported in macaques and other species.3,23,26,32,38,55 Therefore, the influence of different collection times on the fecal microbiota probably was minimal. A final limitation of our study is that frozen samples were softened in a biosafety cabinet for approximately 2 h before processing. During this period, proteases in the stool could have damaged the DNA, thereby reducing the quality of the data. However, several studies, including by our group, have reported that storage of fecal samples at room temperature or refrigerated at 4 °C for less than 6 h has minimal effect on the results of the microbiota analysis.48,63,70
In summary, we found that, overall, the housing site of rhesus macaques appeared to have a greater effect on the composition of the gut microbiome than did their health status. Our results suggest a possible role for Helicobacter macacae in maintaining gut health, and a potentially important niche relationship between the closely related genera Helicobacter and Campylobacter. In addition, our findings provide a basis for future studies on potential clinical interventions, such as probiotics and fecal transplants, which, if effective, could have a profound effect on animal welfare in research settings.
Acknowledgments
The authors thank Natalia Melchor for her help with sample processing, as well as Samuel Bauman, Indya Thompson, and the technical staff at the CPRC for their contributions to sample and metadata collection. This work was funded by the AALAS Grants for Laboratory Animal Science (GLAS) Program. The CPRC currently holds 2 NIH grants supporting operations, labor, and veterinary care: NIH P40 OD012217 and NIHU42OD021458. The authors declare no conflicts of interest.
References
- 1.Animal Welfare Act as Amended. 2008. 7 USC §2131–2156.
- 2.Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH, Wang GP. 2013. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol 51:2884–2892. 10.1128/JCM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arroyo LG, Rossi L, Santos BP, Gomez DE, Surette MG, Costa MC. 2020. Luminal and mucosal microbiota of the cecum and large colon of healthy and diarrheic horses. Animals (Basel) 10:1–11. 10.3390/ani10081403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer SA, Arndt TP, Leslie KE, Pearl DL, Turner PV. 2011. Obesity in rhesus and cynomolgus macaques: a comparative review of the condition and its implications for research. Comp Med 61:514–526. [PMC free article] [PubMed] [Google Scholar]
- 5.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 6.Blanchard TG, Nedrud JG. 2006. Laboratory maintenance of Helicobacter species. Curr Protoc Microbiol 00:8B.1.1–8B.1.13. 10.1002/9780471729259.mc08b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brook I. 2008. Current concepts in the management of Clostridium tetani infection. Expert Rev Anti Infect Ther 6:327–336. 10.1586/14787210.6.3.327. [DOI] [PubMed] [Google Scholar]
- 8.Brooks AW, Priya S, Blekhman R, Bordenstein SR. 2018. Gut microbiota diversity across ethnicities in the United States. PLoS Biol 16:1–24. 10.1371/journal.pbio.2006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: High resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez Peña A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castaño-Rodríguez N, Kaakoush NO, Lee WS, Mitchell HM. 2017. Dual role of Helicobacter and Campylobacter species in IBD: a systematic review and meta-analysis. Gut 66:235–249. 10.1136/gutjnl-2015-310545. [DOI] [PubMed] [Google Scholar]
- 12.Caya JG, Agni R, Miller JE. 2004. Clostridium botulinum and the clinical laboratorian: a detailed review of botulism, including biological warfare ramifications of botulinum toxin. Arch Pathol Lab Med 128:653–662. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. [Internet]. 2020. Campylobacter (Campylobacteriosis). [Cited June 6 2020]. Available at: https://www.cdc.gov/campylobacter/index.html
- 14.Cohen D, Shoham O, Orr N, Muhsen K. 2011. An inverse and independent association between Helicobacter pylori infection and the incidence of shigellosis and other diarrheal diseases. Clin Infect Dis 54:e35–e42. 10.1093/cid/cir916. [DOI] [PubMed] [Google Scholar]
- 15.Compo NR, Gomez DE, Tapscott B, Weese JS, Turner PV. 2018. Fecal bacterial microbiota of Canadian commercial mink (Neovison vison): Yearly, life stage, and seasonal comparisons. PLoS One 13:1–18. 10.1371/journal.pone.0207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa MC, Stampfli HR, Allen-Vercoe E, Weese JS. 2016. Development of faecal microbiota in foals. Equine Vet J 48:681–688. 10.1111/evj.12532. [DOI] [PubMed] [Google Scholar]
- 17.Duplessis CA, You D, Johnson M, Speziale A. 2011. Efficacious outcome employing fecal bacteriotherapy in severe Crohn's colitis complicated by refractory Clostridium difficile infection. Infection 40:469–472. 10.1007/s15010-011-0226-1. [DOI] [PubMed] [Google Scholar]
- 18.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ericsson AC, Davis JW, Spollen W, Bivens N, Givan S, Hagan CE, Hagan CE, McIntosh M, Franklin CL. 2015. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One 10:1–19. 10.1371/journal.pone.0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrecchia CE, Hobbs TR. 2013. Efficacy of oral fecal bacteriotherapy in rhesus macaques (Macaca mulatta) with chronic diarrhea. Comp Med 63:71–75. [PMC free article] [PubMed] [Google Scholar]
- 21.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. 2010. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8:292–300. 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, Gonzalez A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. 2014. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15:382–392. 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez DE, Arroyo LG, Costa MC, Viel L, Weese JS. 2017. Characterization of the fecal bacterial microbiota of healthy and diarrheic dairy calves. J Vet Intern Med 31:928–939. 10.1111/jvim.14695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Y, Wu W, Wu S, Zheng H-M, Li P, Sheng H-F, Chen M-X, Chen Z-H, Ji G-Y, Zheng Z-D-X, Mujagond P, Chen X-J, Rong Z-H, Chen P, Lyu L-Y, Wang X, Xu J-B, Wu C-B, Yu N, Xu Y-J, Yin J, Raes J, Ma W-J, Zhou H-W. 2018. Linking gut microbiota, metabolic syndrome and economic status based on a population-level analysis. Microbiome 6:1–11. 10.1186/s40168-018-0557-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernández M, de Frutos M, Rodríguez-Lázaro D, López-Urrutia L, Quijada NM, Eiros JM. 2019. Fecal microbiota of toxigenic Clostridioides difficile-associated diarrhea. Front Microbiol 9:3331. 10.3389/fmicb.2018.03331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hibberd AA, Yde CC, Ziegler ML, Honoré AH, Saarinen MT, Lahtinen S, Stahl B, Jensen HM, Stenman LK. 2019. Probiotic or synbiotic alters the gut microbiota and metabolism in a randomised controlled trial of weight management in overweight adults. Benef Microbes 10:121–135. 10.3920/BM2018.0028. [DOI] [PubMed] [Google Scholar]
- 28.Howell S, Whiteb D, Ingrama S, Jackson R, Larina J, Morales P, Garcia AP, Hicks C, Hopper K, Wagner J. 2012. A bio-behavioral study of chronic idiopathic colitis in the rhesus macaque (Macaca mulatta). Appl Anim Behav Sci 137:208–220. 10.1016/j.applanim.2012.01.003. [DOI] [Google Scholar]
- 29.Institute for Laboratory Animal Research. 2011. Guide for the care and use of laboratory animals, 8th ed. Washington (DC): National Academies Press. [Google Scholar]
- 30.Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, Spekhorst LM, Alberts R, Franke L, van Dullemen HM, Steege RWFT, Huttenhower C, Dijkstra G, Xavier RJ, Festen EAM, Wijmenga C, Zhernakova A, Weersma RK. 2016. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 67:108–119. 10.1136/gutjnl-2016-312135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ismail NA, Ragab SH, ElBaky AA, Shoeib ARS, Alhosary Y, Fekry D. 2011. Frequency of firmicutes and bacteroidetes in gut microbiota in obese and normal weight Egyptian children and adults. Arch Med Sci 7:501–507. 10.5114/aoms.2011.23418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalashnikova VA, Dzhikidze EK, Stasilevich ZK, Chikobava MG. 2002. Detection of Campylobacter jejuni in healthy monkeys and monkeys with enteric infections by PCR. Bull Exp Biol Med 134:299–300. 10.1023/A:1021528122942. [DOI] [PubMed] [Google Scholar]
- 33.Kang Z, Lu M, Jiang M, Zhou D, Huang D. 2019. Proteobacteria acts as a pathogenic risk-factor for chronic abdominal pain and diarrhea in post-cholecystectomy syndrome patients: a gut microbiome metabolomics study. Med Sci Monit 25:7312–7320. 10.12659/MSM.915984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinross JM, Darzi AW, Nicholson JK. 2011. Gut microbiome-host interactions in health and disease. Genome Med 3:14. 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiu R, Hall LJ. 2018. An update on the human and animal enteric pathogen Clostridium perfringens. Emerg Microbes Infect 7:1–15. 10.1038/s41426-018-0144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kylie J, Weese JS, Turner PV. 2018. Comparison of the fecal microbiota of domestic commercial meat, laboratory, companion, and shelter rabbits (Oryctolagus cuniculi). BMC Vet Res 14:1–15. 10.1186/s12917-018-1464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.La Rosa PS, Brooks JP, Deych E, Boone EL, Edwards DJ, Wang Q, Sodergren E, Weinstock G, Shannon WD. 2012. Hypothesis testing and power calculations for taxonomic-based human microbiome data. PLoS One 7:1–13. 10.1371/journal.pone.0052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laing ST, Merriam D, Shock BC, Mills S, Spinner A, Reader R, Hartigan-O'Connor DJ. 2018. Idiopathic colitis in rhesus macaques is associated with dysbiosis, abundant enterochromaffin cells and altered T-cell cytokine expression. Vet Pathol 55:741–752. 10.1177/0300985818780449. [DOI] [PubMed] [Google Scholar]
- 39.Lee WJ, Lattimer LDN, Stephen S, Borum ML, Doman DB. 2015. Fecal microbiota transplantation: A review of emerging indications beyond relapsing Clostridium difficile toxin colitis. Gastroenterol Hepatol (N Y 11:24–32. [PMC free article] [PubMed] [Google Scholar]
- 40.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102:11070–11075. 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023. 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 42.Lim MY, You HJ, Yoon HC, Kwon B, Lee JY, Lee S. 2017. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut 66:1031–1038. [DOI] [PubMed] [Google Scholar]
- 43.López-Contreras BE, Morán-Ramos S, Villarruel-Vázquez R, Macías-Kauffer L, Villamil-Ramírez H, León-Mimila H, Vega-Badillo P, Sánchez-Muñoz J, Llanos-Moreno F, Canizalez-Román LE, del Río-Navarro A, Ibarra-González I, Vela-Amieva M, Villarreal-Molina T, Ochoa-Leyva A, Aguilar-Salinas CA, Canizales-Quinteros S. 2017. Composition of gut microbiota in obese and normal-weight Mexican school-age children and its association with metabolic traits. Pediatr Obes 13:381–388. 10.1111/ijpo.12262. [DOI] [PubMed] [Google Scholar]
- 44.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:119–129. 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Luther J, Dave M, Higgins PD, Kao JY. 2010. Association between Helicobacter pylori infection and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Inflamm Bowel Dis 16:1077–1084. 10.1002/ibd.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mancabelli L, Milani C, Lugli GA, Turroni F, Cocconi D, van Sinderen D, Ventura M. 2017. Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol Ecol 93:1093. 10.1093/femsec/fix153. [DOI] [PubMed] [Google Scholar]
- 47.McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, Lozupone CA, Hamady M, Knight R, Bushman FD. 2008. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog 4:1–12. 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muiños-Bühl A, González-Recio O, Muñoz M, Óvilo C, García-Casco J, Fernández AI. 2018. Evaluating protocols for porcine faecal microbiome recollection, storage and DNA extraction: from the farm to the lab. Curr Microbiol 75:651–657. 10.1007/s00284-017-1429-1. [DOI] [PubMed] [Google Scholar]
- 49.Mylonakis E, Ryan ET, Calderwood SB. 2001. Clostridium difficile–associated diarrhea: a review. Arch Intern Med 161:525–533. 10.1001/archinte.161.4.525. [DOI] [PubMed] [Google Scholar]
- 50.Org E, Blum Y, Kasela S, Mehrabian M, Kuusisto J, Kangas AJ. 2017. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol 18:1–14. 10.1186/s13059-017-1194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, Giannoukos G, Ciulla D, Tabbaa D, Ingram J, Schauer DB, Ward DV, Korzenik JR, Xavier RJ, Bousvaros A, Alm EJ. 2012. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One 7:1–12. 10.1371/journal.pone.0039242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papamichael K, Konstantopoulos P, Mantzaris GJ. 2014. Helicobacter pylori infection and inflammatory bowel disease: is there a link? World J Gastroenterol 20:6374–6385. 10.3748/wjg.v20.i21.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pascal V, Pozuelo M, Borruel N, Casellas F, Campos D, Santiago A, Martinez X, Varela E, Sarrabayrouse G, Machiels K, Vermeire S, Sokol H, Guarner F, Manichanh C. 2017. A microbial signature for Crohn's disease. Gut 66:813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peters BA, Shapiro JA, Church TR, Miller G, Trinh-Shevrin C, Yuen E. 2018. A taxonomic signature of obesity in a large study of American adults. Sci Rep 8:1–13. 10.1038/s41598-018-28126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rhoades N, Barr T, Hendrickson S, Prongay K, Haertel AJ, Gill L, Garzel L, Whiteson K, Slifka M, Messaoudi I. 2019. Maturation of the infant rhesus macaque gut microbiome and its role in the development of diarrheal disease. Genome Biol 20:1–16. 10.1186/s13059-019-1789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rothenbacher D, Blaser MJ, Bode G, Brenner H. 2000. Inverse relationship between gastric colonization of Helicobacter pylori and diarrheal illnesses in children: results of a population-based cross-sectional study. J Infect Dis 182:1446–1449. 10.1086/315887. [DOI] [PubMed] [Google Scholar]
- 57.Seekatz AM, Panda A, Rasko DA, Toapanta FR, Eloe-Fadrosh EA, Khan AQ, Liu Z, Shipley ST, Detolla LJ, Sztein MB, Fraser CM. 2013. Differential response of the cynomolgus macaque gut microbiota to Shigella infection. PLoS One 8:1–13. 10.1371/journal.pone.0064212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:1–18. 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sestak K, Merritt CK, Borda J, Saylor E, Schwamberger SR, Cogswell F, Didier ES, Didier PJ, Plauche G, Bohm RP, Aye PP, Alexa P, Ward RL, Lackner AA. 2003. Infectious agent and immune response characteristics of chronic enterocolitis in captive rhesus macaques. Infect Immun 71:4079–4086. 10.1128/IAI.71.7.4079-4086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suchodolski JS, Markel ME, Garcia-Mazcorro JF, Unterer S, Heilmann RM, Dowd SE, Kachroo P, Ivanov I, Minamoto Y, Dillman EM, Steiner JM, Cook AK, Toresson L. 2012. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One 7:1–13. 10.1371/journal.pone.0051907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun B, Wang X, Bernstein S, Huffman MA, Dong-Po X, Zhiyuan G, Rui C, Sheeran LK, Wagner RS, Jinhua L. 2016. Marked variation between winter and spring gut microbiota in free-ranging Tibetan Macaques (Macaca thibetana). Sci Rep 6:1–8. 10.1038/srep26035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun J, Du L, Li X, Zhong H, Ding Y, Liu Z, Ge L. 2019. Identification of the core bacteria in rectums of diarrheic and non-diarrheic piglets. Sci Rep 9:1–10. 10.1038/s41598-019-55328-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tal M, Verbrugghe A, Gomez DE, Chau C, Weese JS. 2017. The effect of storage at ambient temperature on the feline fecal microbiota. BMC Vet Res 13:1–8. 10.1186/s12917-017-1188-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Mitchell L, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2008. A core gut microbiome in obese and lean twins. Nature 457:480–484. 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Dyke MI, McCarthy AJ. 2002. Molecular biological detection and characterization of Clostridium populations in municipal landfill sites. Appl Environ Microbiol 68:2049–2053. 10.1128/AEM.68.4.2049-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. 2015. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 65:57–62. 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Velasquez-Manoff M. 2015. Gut microbiome: the peacekeepers. Nature 518:S3–11. 10.1038/518S3a. [DOI] [PubMed] [Google Scholar]
- 68.Wang J, Lang T, Shen J, Dai J, Tian L, Wang X. 2019. Core gut bacteria analysis of healthy mice. Front Microbiol 10:1–14. 10.3389/fmicb.2019.00887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waters JL, Ley RE. 2019. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol 17:1–11. 10.1186/s12915-019-0699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weese JS, Jalali M. 2014. Evaluation of the impact of refrigeration on next generation sequencing-based assessment of the canine and feline fecal microbiota. BMC Vet Res 10:1–6. 10.1186/s12917-014-0230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Westreich ST, Ardeshir A, Alkan Z, Kable ME, Korf I, Lemay DG. 2019. Fecal metatranscriptomics of macaques with idiopathic chronic diarrhea reveals altered mucin degradation and fucose utilization. Microbiome 7:1–17. 10.1186/s40168-019-0664-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yasuda K, Oh K, Ren B, Tickle TL, Franzosa EA, Wachtman LM, Miller AD, Westmoreland SV, Mansfield KG, Vallender EJ, Miller GM, Rowlett JK, Gevers D, Huttenhower C, Morgan XC. 2015. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 17:385–391. 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu L, Wua Q, Daia J, Zhang S, Weia F. 2011. Evidence of cellulose metabolism by the giant panda gut microbiome. Proc Natl Acad Sci USA 108:17714–17719. 10.1073/pnas.1017956108. [DOI] [PMC free article] [PubMed] [Google Scholar]