Abstract
DNA methylation as a process that regulates gene expression is crucial in immune cells biology. Global and gene specific methylation changes have been described in autoimmunity, especially in Systemic Lupus Erythematosus. These changes not only contribute to the understanding of the disease, but also some have been proposed as diagnostic or disease activity biomarkers. The present review compiles the most recent discoveries on this field on each type of immune cells, including specific changes in signalling pathways, genes of interest and its possible applications on diagnosis or treatment.
Keywords: DNA methylation, Systemic Lupus Erythematosus, autoimmunity
Introduction
Systemic Lupus Erythematosus (SLE) is an autoimmune, multi-organic and chronic disease characterised by the presence of auto-antibodies directed to double-stranded DNA and other cellular components [1]. It is 2–4 times more frequent and with greater severity in populations such as African Americans, Hispanics and Asians. It is also nine times more frequent in women than in men, especially in those who are of reproductive age (18–40 years) [2].
It is postulated that multiple factors such as genetic susceptibility and environmental exposure are involved in the pathogenesis of SLE. However, it is striking that genetics alone fails to explain the presence of lupus. The evidence of the discordance between monozygotic twins of which one presents the disease, and the other does not, an observation that suggests that external factors play a substantial role in pathogenesis [3]. Recently, this field of study has been directed towards epigenetic changes such as DNA methylation, histone changes and the presence of some non-coding RNAs; which modulate the expression of susceptible genes in SLE [4].
Epigenetics is in turn dependent on environmental factors, the diet, the use of certain medications, cigarette smoking, etc., and since it is potentially heritable, it represents a pathway that links external conditions with gene expression [5]. Indeed, specific epigenetic modifications have been described in patients with SLE [6], so its influence on the pathophysiology of the disease can play a role.
DNA methylation is the most studied epigenetic change in SLE and consists of the addition of methyl groups (CH3) to cytosine. These are found at specific points called CpG sites that are usually found in the promoter, which leads to repression of the expression of the gene in question. This modification is carried out by DNA methyltransferases (DNMT) [7]. There are several types of DNMT, such as DNMT1 that handles global methylation after replication and DNMT3a and DNMT3b responsible for adding de novo methyl groups and the DNMT3L that does not have genomic action but regulates de novo DNA methylation [8].
One of the most specific pieces of evidence of the association between DNA methylation and SLE is lupus induced by drugs such as procainamide and hydralazine [9]. Hypomethylation is observed in these patients since these drugs are direct inhibitors of DNMT or signalling pathways like the Ras-MAPK-ERK pathway [10]. Another study that supports DNA methylation as an event in the pathophysiology of SLE was carried out by Javierre et al. with 15 pairs of discordant monozygotic twins for Dermatomyositis, Rheumatoid Arthritis and SLE. They found differences in DNA methylation only for discordant couples in SLE, specifically in 49 genes related to immune response, cell proliferation and cytokine production [11].
Regarding the role of environmental exposure, factors such as ultraviolet light and Epstein-Barr virus infection have been described as triggers of SLE [12]. How it is believed that they mediate pathogenesis is precisely through epigenetics, which is known to be sensitive to external stimuli. These findings suggest that events other than genetic predisposition play a role in the onset of the disease. Given the relevance that epigenetic changes could have a role in the pathophysiology of SLE, the existing shreds of evidence of the changes in DNA methylation described for different immune cells will be reviewed in the present work and are summarised in Table 1.
Table 1.
Summary of main changes in DNA methylation by cell.
| Cell type | Methylation change in SLE | Reference |
|---|---|---|
| CD4+ T CELLS | Global Hypomethylation | [13] |
| Hypomethylation of CD9, MMP9 and PDGFRA | [34] | |
| Hypomethylation of CD40L | [38] | |
| Hypomethylation of CHST12 | [39] | |
| Hypomethylation of IL-10 and IL-13 | [41] | |
| Hypomethylation of CREMα | [42] | |
| Hypermethylation of miRNA-142 | [28] | |
| B cells | Hypomethylation of CD5-E1B | [60] |
| Hypomethylation of IFN regulated genes in ARID3aH cells | [61] | |
| Hypomethylation of HRES-1 | [62] | |
| Hypomethylation of IFI44, IFITM1, YBX1 and TAF8 | [57] | |
| Hypermethylation of SOX12, ARFGAP3, CCDC81 and MEG3 | [57] | |
| Monocytes | Differential methylation on interferon related genes | [36] |
| Hypomethylation of IFI44L, LGALS3BP, and DTX3L | [66] | |
| Hypermethylation of B3GALT4 and PRIC285 | [66] | |
| Neutrophils | Hypomethylation of MX1 and IFI44L | [69] |
| Hypomethylation of LINE-1 | [70] | |
| Dendritic cells | Global hypermethylation on patients with severe LN | [72] |
| PBMC | Hypomethylation of IFI44L | [63] |
In this review, we assess current research exploring the link between epigenetic changes and pathogenesis of Systemic Lupus Erythematosus. Many signalling pathways, proteins, transcription factors and non-coding RNAs affect the DNA methylation status of CD4+ T Cells, B cells, monocytes, neutrophils and dendritic cells, which will be reviewed next.
CD4+ T cells
The epigenetic phenomenon described most frequently in CD4+ T cells of patients with lupus is global hypomethylation, being more pronounced in active patients [13]. Many signalling pathways, proteins, transcription factors and non-coding RNAs affect the DNA methylation status of these cells, which will be reviewed next.
Signalling pathways
The most critical signalling pathway related to DNA methylation is ERK-MAP kinase pathway, depicted in Figure 1. Sawalha et al. showed in a model of transgenic mice that an alteration or inhibition of any of the proteins of the ERK pathway would eventually lead to the downregulation of DNMT. Even more, these mice developed anti dsDNA antibodies [14]. Another signalling protein involved is Protein Kinase C delta (PKCδ), which activates the ERK pathway through phosphorylation. CD4+ T cells of transgenic mice deficient in PKCδ developed hypomethylation of sensitive genes like CD70 and a lupus-like syndrome [15]. Actually, there are evidences that SLE patients have a decreased phosphorylation of PKCδ [16].
Figure 1.
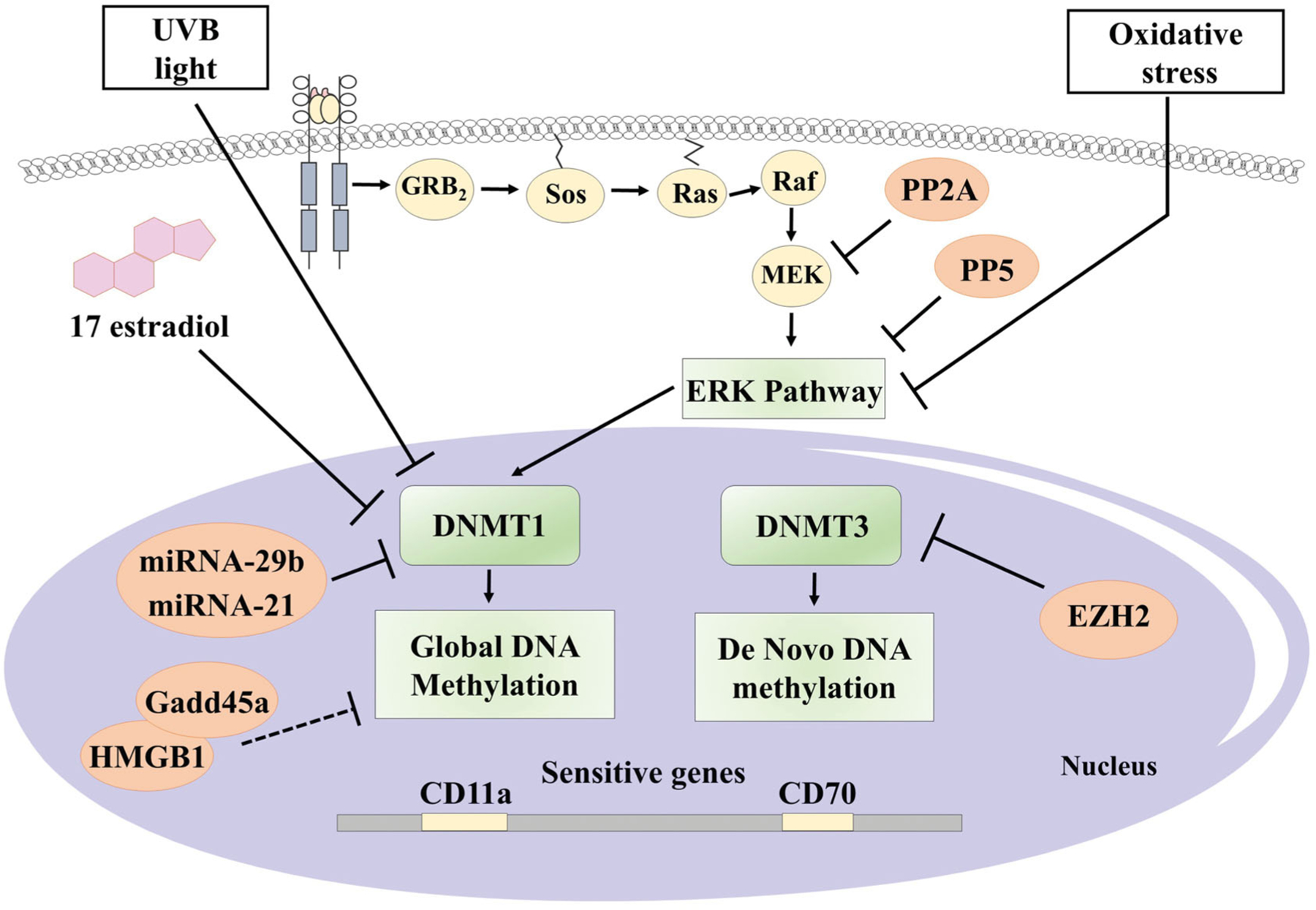
Reported associated factors with DNA methylation changes in CD4+ T cells of patients with SLE. Orange circles represent factors that have been reported with overexpression on patients with SLE. Dashed lines indicate tentative mechanisms. Designed with ⒸMotifolio Drawing Toolkits.
These alterations occur not only in the proteins of the ERK signalling pathway but also in their regulators, such as different types of phosphatases. An increase of the phosphatases activity results in the inhibition of the ERK pathway, whereas suppression of these regulatory phosphatases leads to an easier activation of the ERK pathway. For example, Sunahori et al. described the relation between protein-phosphatase 2 A (PP2A) and the signalling of the ERK pathway. They showed that PP2A specifically inactivates MEK, which in turns inactivates ERK, therefore decreasing DNMT1 activity, finally leading to the hypomethylation of sensitive genes [17]. Indeed, overexpression of PP2A has been reported in T cells of patients with SLE [18].
Patel et al. also found an increase in the expression of protein phosphatase 5 (PP5) in CD4+ CD28+ T cells from patients with lupus. Then, they analysed the effect of transfection of PP5 to T cells and found a decrease in the expression of DNMT1 and the increased expression of genes already known for their sensitivity to demethylation such as perforin, CD11a, CD70, CD40L, among others [19]. Future studies are needed to know if the effect of PP5 is selective to any of the proteins of the ERK pathway or if this phosphatase regulates the pathway in general. Another controversial protein is the phosphatase DUSP23, which is overexpressed in patients with lupus. However, it is still not known if it has an inhibitory or stimulatory effect of the ERK pathway [20].
Besides signalling pathways, there are also proteins that might regulate DNMTs in several ways. One of them is EZH2, which is part of Polycomb repressive complex 2 (PRC2), as an enzymatic subunit [21]. It was found to be increased in naive CD4+ T cells in lupus patients. Also, it has a function as an epigenetic regulator inducing broad genome methylation changes in T cells [22].
On the other hand, augmentation of “growth arrest and DNA damage–induced 45α” (gadd45A) in CD4+ T cells was associated with DNA hypomethylation changes, as well as overexpression of sensitive genes like CD11a, and CD70 and autoreactivity. Indeed, gadd45A was found to be raised in CD4+ T cells of lupus patients. Also, when gadd45A was forced to underexpression, the result was DNA hypermethylation, down-regulation of CD11a and CD70, as well as a decreased autoreactivity [23]. Another protein that may help gadd45A is HMGB1. Li et al. found that HMGB1 was overexpressed in CD4+ T cells of patients with lupus. Tentative, HMGB1 might directly bind to gadd45A and stimulate its effects on DNA methylation [24]. An additional protein that is believed to affect DNA methylation is MBD4, which appears to have a protective role against demethylation [25]. Indeed, the hypomethylation of a known sensitive gen as CD70 is related to the decreased expression of MBD4 in CD4+ T cells of SLE patients [26].
In addition to phosphatases and other proteins, a regulatory role of the ERK pathway has been described by microRNAs. Although they constitute a separate group of epigenetic changes apart from DNA methylation, some miRNAs specifically regulate either the MAPK pathway or DNMT1. An example of these is miR-29b, described by Qin et al. They found that there is an increase in the expression of miR-29b in CD4+ T cells of patients with lupus. Also, when transferring an inhibitor of miR-29b to CD4+ T cells from patients with SLE, both the expression of DNMT1, as well as the global methylation was increased, with a corresponding decrease in expression of CD11a and CD70 [27]. On the other hand, miRNA-126 inhibited DNMT1 causing hypomethylation of sensitive genes [28].
An even more interesting phenomenon than DNA methylation’s regulation by microRNAs is the up-regulation of these by changes such as associated hypermethylation. As described by Ding et al., hypermethylation of the CGs closest to the start of transcription of the gene for miR-142, is related to a decrease in its expression in CD4+ T cells from patients with lupus. They also showed that miRNA-142 degrades the mRNA of SAP, CD84, and IL-10. The downregulation of this miRNA causes an over-expression of SAP, CD84, and IL-10, leading to both T cell activation and B-cell hyperreactivity invitro. Similarly, this dysregulation of miRNA-142 appears to be specific for SLE, since it was not observed in CD4+ T cells from patients with Psoriasis [29].
Finally, patients with SLE may also have disturbances on the counterpart of DNMTs, the Ten-eleven Translocation proteins (TET). TETs proteins are involved in active demethylation of cytosines [30]. TET are stimulated by an increased concentration of intracellular iron, which is inhibited by the protein BDH2. This protein is decreased in CD4+ cells of SLE patients, because of the inhibitory activity of the overexpressed miRNA-21. This finally leads to the accumulation of iron and overstimulation of TET proteins, leading to DNA hypomethylation and expression of autoreactivity genes [31].
Transcription factors
RFX1 is a transcription factor believed to be able to bind to DNMT1 and recruit it to the promoter region of both CD11a and CD70. RFX1 was found to be downregulated in CD4+ T cells of patients with lupus, and this was associated with an increase in CD11a and CD70, as well as B cell activation and IgG production [32]. Also, RFX1 is regulated by STAT3 pathway and has a direct effect on IL-17A production. Specifically, RFX1 has an inhibitory effect on IL-17A, through epigenetic mechanisms. Then, the down-regulation of RFX1 could lead to a lupus flare due to the increased IL-17A [33], because IL-17 has a proinflammatory effect, inducing the production pro-inflammatory cytokines and chemotaxis [34].
Specific genes
Jeffries et al. performed a Genome-Wide Analysis (GWA) on CD4+ cells of patients with SLE. They found 341 loci with different methylation between healthy and patients with SLE, of which 236 were hypomethylated and 105 hypermethylated. Among the genes reported with hypomethylation were CD9, MMP9, and PDGFRA, and those with hypermethylation were genes for folate and RUNX3 biosynthesis [35]. In another GWA study, they found major hypomethylation in genes associated with IFN type I signalling in all T cells. Even more interesting, interferon genes were also hypomethylated when evaluating specific subpopulations of memory, naive and regulatory T cells [36].
Although the status of DNA methylation of the entire genome of a cell makes it possible to analyse the general picture, the description of the methylation status of specific genes has been associated with phenotypes or clinical manifestations and even specific diagnostic applications of SLE.
Regarding the association between clinical manifestations and DNA methylation, Zhao et al. analysed separately patients with specific symptoms, patients were grouped with cutaneous involvement only (S), cutaneous and renal (SK) and finally, cutaneous, renal, articular compromise (KJS) categories. They found 1433 hypermethylated genes and 2287 hypomethylated genes in the first group “S,” 1046 hypermethylated and 3360 hypomethylated for “SK” and 2105 hypermethylated with 3296 hypomethylated in “SKJ” as opposed to healthy controls [37]. This suggests that methylation changes might also be specific to clinical manifestations, which could lead to further research to diagnostic markers and targeted treatment options.
Specific epigenetic changes have also been associated with particular manifestations of SLE. For example, in a study Renauer et al. discovered that in naive CD4+ T cells, patients with malar rash had unique Differentially Methylated Regions (DMR), that differ to those with discoid rash or SLE without cutaneous involvement. It is striking that the three groups of patients with SLE shared the hypomethylated region of the IFI44L gene [38].
Regarding specific genes and their relationship with SLE, in another study, Lu and colleagues explored the methylation of the CD40L gene, which is encoded on the X chromosome and is expressed by T cells for the co-stimulation of the B cell. They found that in women with SLE, unlike healthy women and men with lupus, the promoter of this gene was demethylated in both chromosomes, which contributed to the overexpression of this gene in CD4+ T cells [39].
In addition to the above, the methylation status of some genes has been proposed as a biomarker for both diagnosis and monitoring of the activity of the disease. One of these studies focussed on identifying genes related to kidney damage, finding that hypomethylation of CG sites in the “carbohydrate (chondroitin 4) sulfotransferase 12 gene” (CHST12) has a sensitivity of 85.7% and a specificity of 64.3% to identify patients with lupus nephritis [40].
Cytokines
Cytokines play a critical role in SLE pathophysiology. Recent evidence has shown that epigenetic mechanisms interact and sometimes lead to the characteristic cytokine dysregulation observed in SLE. For example, Hedrich et al. found that STAT3 was related with the hypomethylation of the promoter and subsequent overexpression of IL-10 on T cells, a feature of SLE patients [41]. Another study showed that IL-10 and IL-13 were hypomethylated in CD4+ cells of patients with SLE leading to an increase in its mRNA [42].
One more is the CREMα promoter, which is hypomethylated and with an augmented expression in CD4+ cells of patients with lupus. The mechanism proposed is through the binding of Set1, which inhibits DNMT3a through the increase of H3K4me3 [43]. On the other hand, CREM has two different actions on both IL-17A and IL-2. CREM activates DNMT3a to hypermethylate IL-2 promoter; however, CREM does the contrary effect on IL-17, increasing its expression in naive CD4+ T cells of SLE patients [44].
There are also miRNAs that regulate DNA methylation of some cytokine’s genes. The overexpression of DLK1-Dio3 derived miRNAs were related to the production of pro-inflammatory cytokines like IL-1β, IL-6, and IFN γ. Indeed, DLK1-Dio3 was hypomethylated in MRL/lpr lupus-prone mice [45].
Inducible methylation
Finally, one of the main characteristics of epigenetics is its modulation by different environmental exposures. On this, Wu and colleagues described the effect of UVB light on CD4+ T cells from patients with SLE. They demonstrated that UVB light stimulated the global hypomethylation of DNA in CD4+ T cells, through the inhibition of the catalytic activity of DNMT1, without affecting the expression of the protein [46]. Another environmental factor that can alter DNA Methylation is 17b-estradiol; through its receptor, it mediates the underexpression of DNMT1 in SLE CD4+ T cells [47].
Li et al. explored the role of oxidative stress in DNA methylation, using a treatment with H2O2 or ONOO– as oxidisers. It resulted in the reduction of DNMT-1, as well as the ERK pathway signalling. Closely related to this, PKC phosphorylation was inhibited. All these events finally caused a decrease in DNA methylation and the increased expression of sensitive genes [48]. Even more, Strickland et al. observed that mice developed anti-dsDNA antibody and glomerulonephritis when they received transferred CD4+ T cells that were previously treated with oxidants [49].
SLE treatment drugs and methylation
Current research has found that the drugs used in SLE treatment have epigenetic effects on immune cells. Changes in histones, non-coding RNAs and DNA methylation have been described for common prescriptions such as glucocorticoids and antimalarials [50]. We will review next the changes in DNA methylation described for some of these drugs.
Cyclophosphamide is a cytotoxic drug used in cancer and autoimmunity. Zhang et al. showed that treatment with cyclophosphamide increased DNMT1 protein level, but not its mRNA, in Jurkat T cells. This result in an increase in the percentage of global DNA methylation at day 4 of treatment in vitro [51]. On the other hand, Peters et al. described that Mycophenolate (MPA) produced DNA re-methylation of IFNγ gene of naive T cells but not in memory T cells [52]. However, in another study, Yang et al. did not find differences in global DNA methylation, and either in CD40L specific DNA methylation, after MPA treatment in lupus CD4+ T cells in vitro [53].
Finally, Cribbs et al. investigated the effect of Methotrexate MTX on CD4+ cells and found no differences in the global DNA methylation status. However, they did observe changes in a specific gen. MTX treatment increased FoxP3 expression, through the hypomethylation of its enhancer, which was related to downregulation of DNMT1 [54].
B cells
Changes in both global methylation and specific genes in B cells have been associated with autoreactivity. Some of the mechanisms involved are similar to those described for T cells, such as alterations in the ERK signalling pathway, with the corresponding inhibition of DNMT1 and demethylation of sensitive genes [55]. Others are so specific that could be identified in B cell subpopulations. For example, Scharer et al. described a characteristic chromatic accessibility pattern in resting Naive B cells of SLE patients [56]. Furthermore, they also described in a later study that global DNA methylation changed on each stage of B cell ontology, as cells advance on the differentiation pathway, global DNA methylation decreases and specific hypomethylated and hypermethylated genes could discriminate among B cell subsets like resting Naive and DN2 cells on SLE patients [57].
Signalling pathways
Two signalling pathways have been associated with DNMT activity in B cells, IL-6 and B Cell Receptor (BCR) signalling. BCR signals through nuclear factor-κB (NF-κB) pathway [58] and IL-6 activates the STAT3 pathway [59], although it is not known the exact mechanisms that these pathways use to activate DNMT. However, Garaud et al. reported that activation through BCR increased the expression of DNMT1. Still, in the presence of IL-6, on the contrary, the expression of DNMT1 is decreased. Both activation by BCR and by IL-6, change the methylation and produce an increase in the expression of the CD5-E1B isoform, which produces the decrease of the expression of the second CD5-E1A isoform. This allows a greater activation of the BCR. Indeed, CD5-E1B presented hypomethylation in B cells of patients with SLE. Therefore, the formation of a positive feedback loop is constituted, since the activation of BCR in the presence of IL-6 increases the expression of CD5-E1B, which in turn allows a greater activation of the BCR and so on. This circuit was stopped by using an antibody to block IL-6 [60]. This study has relevance within the pathophysiology of lupus since an increase in BCR activity has been associated with the generation of B cell autoreactivity [58].
Cytokines
A subgroup of B cells with overexpression of “transcription factor A-T rich interacting domain 3a” (ARID3aH), showed a decreased DNA methylation patterns level of IFN related genes. Although the mechanisms are unknown, ARID3aH might epigenetically regulate several promoters at once, leading eventually to the overexpression of these genes, which is a hallmark in the pathophysiology of SLE. This phenomenon was observed primarily in naive B cells, but not in all B cell subpopulations [61].
Specific genes
Absher et al. reported 166 hypomethylated CGs in B cells, those with the most potent effects were grouped into genes, finding 95 genes with a strong association with lupus. Regarding the functional analysis of these genes, at least 60% of them are involved in signalling through interferon [36].
HRES-1 is another of the involved genes and hypomethylation of HRES-1 was found in B cells of patients with SLE. Also, the ERK pathway and IL-6 affected the DNA methylation of HRES-1, even more, blockage of IL-6 re-established its DNA methylation status [62].
Finally, the methylation status of the gen IFI44L has been proposed as a diagnostic marker. The hypomethylation at site 2 of the promoter of IFI44L in PBMC, had a sensitivity of 93.6% and a specificity of 96.4% for the diagnosis of lupus. This was validated in 2 cohorts of patients with a Chinese origin. However, when it was validated in a cohort of European patients the sensitivity dropped to 82.1% and the specificity to 81.8% [63]. This could be indicative that some of these new biomarkers could work better according to ethnicity. Additionally, IFI44L is regulated by interferon, and estrogens stimulate its expression in B cells [64].
SLE treatment drugs and methylation
An increase of global DNA methylation content after treatment with MTX in T cells, B cells, and monocytes has been reported. Although its consequences are still not known because they did not find any change in the expression of DNMT1, DNMT3A, DNMT3B, GADD45A, TET1, TET2 and TET3 [65].
Monocytes
Specific genes
Epigenetic changes in monocytes from patients with SLE are beginning to be researched. In the study by Absher et al. mentioned above, they explored these changes for the first time. They found 97 CpGs differentially methylated only in monocytes. The most potent effects were grouped in genes, finding 27 genes with a strong association with lupus. Regarding the functional analysis of these genes, at least 54% of these are involved in signalling through interferon [36]. In another study on discordant twins with SLE, specific genes were related to recent flares, hypomethylated genes like IFI44L, LGALS3BP, DTX3L, PARP9 and GGT1; as well as hypermethylated genes like B3GALT4 and PRIC285 were identified [66].
Also, Notley et al. showed that monocyte-derived macrophages, when phagocyte apoptotic CD4+ T cells with hypomethylated DNA, increased its production of pro-inflammatory cytokines like IL-6 [67].
Neutrophils
In recent years, the role of neutrophils in the pathophysiology of SLE has been related since it has been seen that the cells of these patients do NETosis more frequently. This exposes the genetic material against which antibodies are generated [68]. Although there are few studies on epigenetic changes in neutrophils of patients with lupus, some of them will be described below.
Specific genes
In the first of these studies, methylation difference was found in 293 GC sites, of which 68% were hypomethylated and 32% hypermethylated. Within the genes with hypomethylation were IFITM1, IFIT3, DDX60, ISG15 and the two with the largest hypomethylation were MX1 and IFI44L, which are regulated by interferon. As a consequence, the expression of the mentioned genes was increased [69]. On the other hand, Sukapan et al. identified hypomethylation of LINE-1 (Long Interspersed Nuclear Elements) in neutrophils of patients with SLE. The genes with LINE-1 content were correspondingly over-expressed and were related to cellular functions such as apoptosis. Finally, no correlation was reported between methylation patterns of LINE-1 with disease activity measured by SLEDAI [70]. Also, LINE-1 appear to participate during the neutrophil differentiation process [71].
Dendritic cells
Very little is known about epigenetic changes on dendritic cells on SLE. Wardowska et al., described that patients with severe Lupus Nephritis had myeloid dendritic cells with a higher global DNA methylation percentage when compared to healthy controls [72].
Conclusion
Although DNA methylation alterations have been related to autoimmunity and SLE in several studies, most of these have been done in T cells, especially CD4+ lymphocytes. Recently, the epigenetic landscape of other immune cells, such as B cells, neutrophils, monocytes, and even dendritic cells, that also participate in SLE pathogenesis, is begging to be elucidated. Further research on the DNA methylation status and epigenetics of these cells is needed, and this could increase our understanding of SLE disease.
Footnotes
Disclosure statement
This review did not receive financial support and the authors have no potential conflict of interest to declare.
References
- [1].Ghodke-Puranik Y, Niewold TB. Immunogenetics of systemic lupus erythematosus: a comprehensive review. J Autoimmun 2015;64:125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pons-Estel GJ, Alarcón GS, Scofield L, et al. Understanding the epidemiology and progression of systemic lupus erythematosus. Semin Arthritis Rheum 2010;39(4):257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen L, Morris DL, Vyse TJ. Genetic advances in systemic lupus erythematosus: an update. Curr Opin Rheumatol 2017; 29(5):423–433. [DOI] [PubMed] [Google Scholar]
- [4].Patel DR, Richardson BC. Dissecting complex epigenetic alterations in human lupus. Arthritis Res Ther 2012;15(1):201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ospelt C Epigenetic biomarkers in rheumatology – the future? Swiss Med Wkly 2016;146:w14312. Available from: http://doi.emh.ch/smw.2016.14312. [DOI] [PubMed] [Google Scholar]
- [6].Jeffries MA, Sawalha AH. Epigenetics in systemic lupus erythematosus: leading the way for specific therapeutic agents. Int J Clin Rheumatol 2011;6(4):423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen SH, Lv QL, Hu L, et al. DNA methylation alterations in the pathogenesis of lupus: DNA methylation alterations in lupus. Clin Exp Immunol 2017;187(2):185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu C, Ou T, Wu C, et al. Global DNA methylation, DNMT1, and MBD2 in patients with systemic lupus erythematosus. Lupus 2011;20(2):131–136. [DOI] [PubMed] [Google Scholar]
- [9].Deng C, Lu Q, Zhang Z, et al. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum 2003;48(3):746–756. [DOI] [PubMed] [Google Scholar]
- [10].Sun B, Hu L, Luo Z-Y, et al. DNA methylation perspectives in the pathogenesis of autoimmune diseases. Clin Immunol 2016; 164:21–27. [DOI] [PubMed] [Google Scholar]
- [11].Javierre BM, Fernandez AF, Richter J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res 2010;20(2): 170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Correia Azevedo P, Murphy G, Isenberg DA. Pathology of systemic lupus erythematosus: the challenges ahead. Methods Mol Biol 2014;1134:1–16. [DOI] [PubMed] [Google Scholar]
- [13].Wang Z, Chang C, Lu Q. Epigenetics of CD4+ T cells in autoimmune diseases. Curr Opin Rheumatol 2017;29(4):361–368. [DOI] [PubMed] [Google Scholar]
- [14].Sawalha AH, Jeffries M, Webb R, et al. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun 2008;9(4):368–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gorelik G, Sawalha AH, Patel D, et al. T cell PKCd kinase inactivation induces lupus-like autoimmunity in mice. Clin Immunol 2015;158(2):193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gorelik G, Fang JY, Wu A, et al. Impaired T cell protein kinase Cd activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J Immunol 2007;179(8): 5553–5563. [DOI] [PubMed] [Google Scholar]
- [17].Sunahori K, Nagpal K, Hedrich CM, et al. The catalytic subunit of protein phosphatase 2A (PP2Ac) promotes DNA hypomethylation by suppressing the phosphorylated mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase (MEK)/phosphorylated ERK/DNMT1 protein pathway in T-cells from controls and systemic lupus erythematosus patients. J Biol Chem 2013;288(30):21936–21944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sunahori K, Juang Y-T, Kyttaris VC, et al. Promoter hypomethylation results in increased expression of protein phosphatase 2A in T cells from patients with systemic lupus erythematosus. JI 2011;186(7):4508–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Patel D, Gorelik G, Richardson B. Protein phosphatase 5 contributes to the overexpression of epigenetically regulated T-lymphocyte genes in patients with lupus. Lupus (Los Angel) 2016;1:120. [PMC free article] [PubMed] [Google Scholar]
- [20].Balada E, Felip L, Ordi-Ros J, et al. DUSP23 is over-expressed and linked to the expression of DNMTs in CD4+ T cells from systemic lupus erythematosus patients: DUSP23 and lupus. Clin Exp Immunol 2017;187(2):242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011;469(7330):343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tsou P-S, Coit P, Kilian NC, et al. EZH2 modulates the DNA methylome and controls T cell adhesion through junctional adhesion molecule A in lupus patients. Arthritis Rheumatol 2018;70(1):98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li Y, Zhao M, Yin H, et al. Overexpression of the growth arrest and DNA damage-induced 45a gene contributes to autoimmunity by promoting DNA demethylation in lupus T cells. Arthritis Rheum 2010;62(5):1438–1447. [DOI] [PubMed] [Google Scholar]
- [24].Li Y, Huang C, Zhao M, et al. A possible role of HMGB1 in DNA demethylation in CD4+ T cells from patients with systemic lupus erythematosus. Clin Dev Immunol 2013;2013:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sanders MA, Chew E, Flensburg C, et al. MBD4 guards against methylation damage and germ line deficiency predisposes to clonal hematopoiesis and early-onset AML. Blood 2018; 132(14):1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liao W, Li M, Wu H, et al. Down-regulation of MBD4 contributes to hypomethylation and overexpression of CD70 in CD4+ T cells in systemic lupus erythematosus. Clin Epigenetics 2017; 9:104. Available from: 10.1186/s13148-017-0405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Qin H, Zhu X, Liang J, et al. MicroRNA-29b contributes to DNA hypomethylation of CD4+ T cells in systemic lupus erythematosus by indirectly targeting DNA methyltransferase 1. J Dermatol Sci 2013;69(1):61–67. [DOI] [PubMed] [Google Scholar]
- [28].Zhao S, Wang Y, Liang Y, et al. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum 2011;63(5):1376–1386. [DOI] [PubMed] [Google Scholar]
- [29].Ding S, Liang Y, Zhao M, et al. Decreased microRNA-142–3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis Rheum 2012;64(9):2953–2963. [DOI] [PubMed] [Google Scholar]
- [30].Li D, Chen J, Pei D. The battle between TET proteins and DNA methylation for the right cell. Trends Cell Biol 2018; 28(12):973–975. [DOI] [PubMed] [Google Scholar]
- [31].Zhao M, Li M, Gao X, et al. Downregulation of BDH2 modulates iron homeostasis and promotes DNA demethylation in CD4+ T cells of systemic lupus erythematosus. Clin Immunol 2018;187:113–121. [DOI] [PubMed] [Google Scholar]
- [32].Zhao M, Sun Y, Gao F, et al. Epigenetics and SLE: RFX1 downregulation causes CD11a and CD70 overexpression by altering epigenetic modifications in lupus CD4+ T cells. J Autoimmun 2010;35(1):58–69. [DOI] [PubMed] [Google Scholar]
- [33].Zhao M, Tan Y, Peng Q, et al. IL-6/STAT3 pathway induced deficiency of RFX1 contributes to Th17-dependent autoimmune diseases via epigenetic regulation. Nat Commun 2018;9:583. Available from: http://www.nature.com/articles/s41467-018-02890-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Koga T, Ichinose K, Tsokos GC. T cells and IL-17 in lupus nephritis. Clin Immunol 2017;185:95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jeffries M, Dozmorov M, Tang Y, et al. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics 2011;6(5):593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Absher DM, Li X, Waite LL, et al. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. O’Shea J, editor. PLoS Genet 2013;9(8):e1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhao M, Liu S, Luo S, et al. DNA methylation and mRNA and microRNA expression of SLE CD4+ T cells correlate with disease phenotype. J Autoimmun 2014;54:127–136. [DOI] [PubMed] [Google Scholar]
- [38].Renauer P, Coit P, Jeffries MA, et al. DNA methylation patterns in naïve CD4+ T cells identify epigenetic susceptibility loci for malar rash and discoid rash in systemic lupus erythematosus. Lupus Sci Med 2015;2(1):e000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lu Q, Wu A, Tesmer L, et al. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol 2007;179(9):6352–6358. [DOI] [PubMed] [Google Scholar]
- [40].Coit P, Renauer P, Jeffries MA, et al. Renal involvement in lupus is characterized by unique DNA methylation changes in naïve CD4+ T cells. J Autoimmun 2015;61:29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hedrich CM, Rauen T, Apostolidis SA, et al. Stat3 promotes IL-10 expression in lupus T cells through trans-activation and chromatin remodeling. Proc Natl Acad Sci USA 2014;111(37): 13457–13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhao M, Tang J, Gao F, et al. Hypomethylation of IL10 and IL13 promoters in CD4+ T cells of patients with systemic lupus erythematosus. J Biomed Biotechnol 2010;2010:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang Q, Ding S, Zhang H, et al. Increased Set1 binding at the promoter induces aberrant epigenetic alterations and up-regulates cyclic adenosine 5’-monophosphate response element modulator alpha in systemic lupus erythematosus. Clin Epigenetics 2016;8:126. Available from: 10.1186/s13148-016-0294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Rauen T, Hedrich CM, Juang Y-T, et al. cAMP-responsive element modulator (CREM)a protein induces interleukin 17A expression and mediates epigenetic alterations at the interleukin-17A gene locus in patients with systemic lupus erythematosus. J Biol Chem 2011;286(50):43437–43446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dai R, Lu R, Ahmed SA. The upregulation of genomic imprinted DLK1-Dio3 miRNAs in murine lupus is associated with global DNA hypomethylation. Bobé P, editor. PLoS One 2016;11(4):e0153509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wu Z, Li X, Qin H, et al. Ultraviolet B enhances DNA hypomethylation of CD4+ T cells in systemic lupus erythematosus via inhibiting DNMT1 catalytic activity. J Dermatol Sci 2013; 71(3):167–173. [DOI] [PubMed] [Google Scholar]
- [47].Wu Z, Sun Y, Mei X, et al. 17β-oestradiol enhances global DNA hypomethylation in CD4-positive T cells from female patients with lupus, through overexpression of oestrogen receptor-α-mediated downregulation of DNMT1. Clin Exp Dermatol 2014;39(4):525–532. [DOI] [PubMed] [Google Scholar]
- [48].Li Y, Gorelik G, Strickland FM, et al. Oxidative stress, T Cell DNA methylation, and lupus: oxidative stress and lupus. Arthritis Rheumatol 2014;66(6):1574–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Strickland FM, Li Y, Johnson K, et al. CD4+ T cells epigenetically modified by oxidative stress cause lupus-like autoimmunity in mice. J Autoimmun 2015;62:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Guo Y, Sawalha AH, Lu Q. Epigenetics in the treatment of systemic lupus erythematosus: potential clinical application. Clin Immunol 2014;155(1):79–90. [DOI] [PubMed] [Google Scholar]
- [51].Zhang J, Yuan B, Zhang F, et al. Cyclophosphamide perturbs cytosine methylation in jurkat-T cells through LSD1-mediated stabilization of DNMT1 protein. Chem Res Toxicol 2011; 24(11):2040–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Peters FS, Peeters AMA, Hofland LJ, et al. Interferon-gamma DNA methylation is affected by mycophenolic acid but not by tacrolimus after T-cell activation. Front Immunol 2017;8:822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yang Y, Tang Q, Zhao M, et al. The effect of mycophenolic acid on epigenetic modifications in lupus CD4+ T cells. Clin Immunol 2015;158(1):67–76. [DOI] [PubMed] [Google Scholar]
- [54].Cribbs AP, Kennedy A, Penn H, et al. Methotrexate restores regulatory T cell function through demethylation of the FoxP3 upstream enhancer in patients with rheumatoid arthritis. Arthritis Rheumatol 2015;67(5):1182–1192. [DOI] [PubMed] [Google Scholar]
- [55].Renaudineau Y, Garaud S, Le Dantec C, et al. Autoreactive B cells and epigenetics. Clin Rev Allergy Immunol 2010;39(1): 85–94. [DOI] [PubMed] [Google Scholar]
- [56].Scharer CD, Blalock EL, Barwick BG, et al. ATAC-seq on bio-banked specimens defines a unique chromatin accessibility structure in naïve SLE B cells. Sci Rep 2016;6:27030. Available from: http://www.nature.com/articles/srep27030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Scharer CD, Blalock EL, Mi T, et al. Epigenetic programming underpins B cell dysfunction in human SLE. Nat Immunol 2019;20(8):1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rawlings DJ, Metzler G, Wray-Dutra M, et al. Altered B cell signalling in autoimmunity. Nat Rev Immunol 2017;17(7): 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Mihara M, Hashizume M, Yoshida H, et al. IL-6/IL-6 receptor system and its role in physiological and pathological conditions. Clin Sci 2012;122(4):143–159. [DOI] [PubMed] [Google Scholar]
- [60].Garaud S, Le Dantec C, Jousse-Joulin S, et al. IL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylation. J Immunol 2009;182(9):5623–5632. [DOI] [PubMed] [Google Scholar]
- [61].Ward JM, Ratliff ML, Dozmorov MG, et al. Human effector B lymphocytes express ARID3a and secrete interferon alpha. J Autoimmun 2016;75:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Fali T, Le Dantec C, Thabet Y, et al. DNA methylation modulates HRES1/p28 expression in B cells from patients with lupus. Autoimmunity 2014;47(4):265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhao M, Zhou Y, Zhu B, et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann Rheum Dis 2016;75(11):1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Fan H, Zhao G, Ren D, et al. Gender differences of B cell signature related to estrogen-induced IFI44L/BAFF in systemic lupus erythematosus. Immunol Lett 2017;181:71–78. [DOI] [PubMed] [Google Scholar]
- [65].de Andres MC, Perez-Pampin E, Calaza M, et al. Assessment of global DNA methylation in peripheral blood cell subpopulations of early rheumatoid arthritis before and after methotrexate. Arthritis Res Ther 2015;17:233. Available from: http://arthritis-research.com/content/17/1/233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ulff-Møller CJ, Asmar F, Liu Y, et al. Twin DNA methylation profiling reveals flare-dependent interferon signature and B cell promoter hypermethylation in systemic lupus erythematosus. Arthritis Rheumatol 2018;70(6):878–890. [DOI] [PubMed] [Google Scholar]
- [67].Notley CA, Jordan CK, McGovern JL, et al. DNA methylation governs the dynamic regulation of inflammation by apoptotic cells during efferocytosis. Sci Rep 2017;7:42204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sawalha AH. Neutrophils in systemic lupus erythematosus. In: Tsokos GC, editor. Systemic lupus erythematosus Amsterdam: Academic Press; 2016. p. 127–130. Available from: http://link-inghub.elsevier.com/retrieve/pii/B9780128019177000152. [Google Scholar]
- [69].Coit P, Yalavarthi S, Ognenovski M, et al. Epigenome profiling reveals significant DNA demethylation of interferon signature genes in lupus neutrophils. J Autoimmun 2015;58:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sukapan P, Promnarate P, Avihingsanon Y, et al. Types of DNA methylation status of the interspersed repetitive sequences for LINE-1, Alu, HERV-E and HERV-K in the neutrophils from systemic lupus erythematosus patients and healthy controls. J Hum Genet 2014;59(4):178–188. [DOI] [PubMed] [Google Scholar]
- [71].Zhu Y, Gong K, Denholtz M, et al. Comprehensive characterization of neutrophil genome topology. Genes Dev 2017;31(2): 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wardowska A, Komorniczak M, Bułło-Piontecka B, et al. Transcriptomic and epigenetic alterations in dendritic cells correspond with chronic kidney disease in lupus nephritis. Front Immunol 2019;10:2026. Available from: 10.3389/fimmu.2019.02026/full. [DOI] [PMC free article] [PubMed] [Google Scholar]


