Abstract
In total, ~25% of familial breast cancer (BC) is attributed to germline mutations of the BRCA1 and BRCA2 genes, while the rest of the cases are included in the BRCAX group. BC is also known to affect men, with a worldwide incidence of 1%. Epigenetic alterations, including DNA methylation, have been rarely studied in male breast cancer (MBC) on a genome-wide level. The aim of the present study was to examine the global DNA methylation profiles of patients with BC to identify differences between familial female breast cancer (FBC) and MBC, and according to BRCA1, BRCA2 or BRCAX mutation status. The genomic DNA of formalin-fixed paraffin-embedded tissues from 17 women and 7 men with BC was subjected to methylated DNA immunoprecipitation and hybridized on human promoter microarrays. The comparison between FBC and MBC revealed 2,846 significant differentially methylated regions corresponding to 2,486 annotated genes. Gene Ontology enrichment analysis revealed molecular function terms, such as the GTPase superfamily genes (particularly the GTPase Rho GAP/GEF and GTPase RAB), and cellular component terms associated with cytoskeletal architecture, such as ‘cytoskeletal part’, ‘keratin filament’ and ‘intermediate filament’. When only FBC was considered, several cancer-associated pathways were among the most enriched KEGG pathways of differentially methylated genes when the BRCA2 group was compared with the BRCAX or BRCA1+BRCAX groups. The comparison between the BRCA1 and BRCA2+BRCAX groups comprised the molecular function term ‘cytoskeletal protein binding’. Finally, the functional annotation of differentially methylated genes between the BRCAX and BRCA1+BRCA2 groups indicated that the most enriched molecular function terms were associated with GTPase activity. In conclusion, to the best of our knowledge, the present study was the first to compare the global DNA methylation profile of familial FBC and MBC. The results may provide useful insights into the epigenomic subtyping of BC and shed light on a possible novel molecular mechanism underlying BC carcinogenesis.
Keywords: female breast cancer, male breast cancer, epigenomics, DNA methylation, methylated DNA immunoprecipitation on chip, Gene Ontology
Introduction
Breast cancer (BC) is a complex and heterogeneous disease and a leading cause of mortality among women (1). Familial BC accounts for 5–7% of all BC cases (2,3). Within this group, only around 25% of patients are carriers of germ line mutations in the two high susceptibility genes for BC, BRCA1 and BRCA2 (4). The patients with familial BC who do not have mutation of these genes fall under the BRCAX category (5).
Tumorigenesis is a multistep process that results from the accumulation of genetic and epigenetic alterations (6). A total of 40–50% of human genes have CpG islands (CGIs) located in or near the promoter and/or first exon. Their methylation level is critical to regulate the expression of these genes and essential for the development and proper functioning of the cell. Alterations of the DNA methylation status are frequently associated with human cancer. To date, several studies have determined the methylation profile of specific genes in familial and sporadic forms of BC and between the patients carrying mutations in BRCA1, BRCA2 or BRCAX (7). Flanagan et al (8) have reported data about the global DNA methylation variations in some cases of female familial BC characterized by a given mutation status. BC may also affect men albeit more rarely than women with an incidence of 1% of all cases of BC diagnosed annually. It has been suggested by numerous studies that male breast cancer (MBC) is different from female breast cancer (FBC), at both the clinical and molecular levels (9–12). In fact, even though MBC treatment follows the same indications as postmenopausal FBC, the clinical and pathological characteristics of MBC do not overlap with those of FBC, which could explain why mortality and survival rates are significantly worse in men, as compared to women (13). At present, the epigenetic alterations in MBC have been the focus of a limited number of studies (14). Among them, some investigated the differences in the DNA methylation level of putative genes between FBC and MBC. Kornegoor et al (15), examined the promoter methylation of 25 cancer-related genes in 108 cases of MBC using methylation specific multiplex ligation dependent probe amplification. These authors concluded that the methylation of promoters was common in MBC and that the high methylation status was correlated with the aggressive phenotype and poor outcome (15). Using the same technique, Vermeulen et al (16) studied the promoter methylation of 25 BC-related genes in situ in pure ductal carcinoma of the male breast. Subsequently, Pinto et al (17) identified different expression patterns in RASSF1A, RARβ and in four selected miRNAs studying the differences between MBC and FBC in a set of 56 familial BC cases. Using methylation-sensitive high resolution techniques, Deb et al (18) tested a panel of 10 genes in 60 men, and concluded that BRCA2-related MBC was characterized by high methylation levels of specific genes and that the average methylation index might be a useful prognostic marker. Finally, in a study by Rizzolo et al (19), the results of promoter methylation analysis of genes involved in signal transduction and hormone signaling in 69 men with BC, showed that variations in methylation patterns were common in BC and might identify specific subgroups based on BRCA1/2 mutation status or certain clinicopathologic features.
The aim of the present study was therefore to study DNA methylation pattern using a novel global approach (MeDip-chip; based on human promoter arrays comprising 4.6 million probes tiled through 25,500 human promoter regions) in familiar BC to identify differences between FBC and MBC, as well as among BRCA1, BRCA2 and BRCAX mutations within FBC. In particular, comparisons in DNA methylation levels were performed based on sex and mutation status and the differentially methylated genes in each comparison class were subjected to functional enrichment analysis. The enriched Gene Ontology terms and molecular pathways could be useful for identifying sex and/or mutation related biological differences with a potential clinical impact.
Materials and methods
Clinical and pathological characteristics of BC patients
The present study involved 24 patients with familial BC, including 7 men and 17 women who underwent surgery between May 1997 and February 2012 in 5 different Italian hospital institutes as indicated in Table I. The group of men was between 45 and 79 years (mean 56±13.5) and the group of women was between 33 and 60 years (mean 44.56±7.39). All patients were previously subjected to genomic DNA sequencing from peripheral blood mononuclear cells as requested by the medical genetic counsellors to identify germline mutations of the BRCA1 and BRCA2 genes. Among the men, one patient carried a mutation of the BRCA2 gene while the remaining patients were wild type for BRCA1 or BRCA2 genes and included in the group BRCAX. Among the group of women, four patients carried BRCA1 gene mutations, three carried BRCA2 gene mutations, and ten were included in the BRCAX group (see Table I for the list of mutations). Ethical approval for the study was obtained from Ethic Committee of Spedali Civili of Brescia (approval no. NP 1439). Written informed consent for research purpose in the fields of genomics and epigenomics was obtained from each patient at the original date of the surgery.
Table I.
Clinicopathological characteristics of patients with breast cancer enrolled in the present study.
| Case number | Sex | Age, years | Mutation | ER | PR | HER2 | Ki67/MIB1 | Molecular subtype predicted | Surgical resection, year | Hospital institutea |
|---|---|---|---|---|---|---|---|---|---|---|
| Case 01 | Female | 41 | BRCA 2 458stop | – | – | – | – | – | 1997 | SC-BS |
| Case 02 | Female | 46 | BRCA X | – | – | – | – | – | 1997 | SC-BS |
| Case 03 | Female | 42 | BRCA 2 T703N | Positive | Positive | Negative | Negative | Luminal A | 2005 | FP-BS |
| Case 04 | Female | 43 | BRCA 1 C64R | Negative | Negative | Negative | Positive | Triple-negative | 2011 | FP-BS |
| Case 05 | Female | 49 | BRCA X | Positive | Positive | – | Negative | Luminal A | 2005 | SC-BS |
| Case 06 | Female | 38 | BRCA 1 C64R | Negative | Negative | Negative | Positive | Triple-negative | 2008 | SA-BS |
| Case 07 | Female | 33 | BRCA 2 Q2960X | Positive | Positive | Positive | Positive | Luminal B | 2011 | FP-BS |
| Case 08 | Female | 51 | BRCA X | Positive | Positive | Negative | Negative | Luminal A | 2005 | SA-BS |
| Case 09 | Female | 60 | BRCA X | Positive | Positive | – | – | Luminal A | 2007 | SA-BS |
| Case 10 | Female | 59 | BRCA X | Positive | Positive | Positive | Negative | Luminal B | 2008 | SA-BS |
| Case 11 | Female | 36 | BRCA 1C64R | Negative | Negative | Negative | Positive | Triple-negative | 2009 | FP-BS |
| Case 12 | Female | 50 | BRCA X | Positive | Positive | Negative | Negative | Luminal A | 2008 | FP-BS |
| Case 13 | Female | 38 | BRCA1 M1652I | Negative | Negative | Negative | Positive | Triple-negative | 2001 | SC-BS |
| Case 14 | Female | 43 | BRCA X | Positive | Positive | Negative | Positive | Luminal A | 2007 | SC-BS |
| Case 15 | Female | 41 | BRCA X | Positive | Positive | Positive | Negative | Luminal B | 2000 | SC-BS |
| Case 16 | Female | 43 | BRCA X | Positive | Positive | Negative | Negative | Luminal A | 2006 | SC-BS |
| Case 17 | Female | 43 | BRCA X | Positive | Positive | Negative | Positive | Luminal A | 2002 | SC-BS |
| Case 18 | Male | 49 | BRCA 2 delA9158FS + 29 stop | Positive | Negative | Positive | Negative | HER2-enriched | 2007 | SC-BS |
| Case 19 | Male | 45 | BRCA X | Positive | Negative | Negative | Positive | Luminal A | 2011 | FP-BS |
| Case 20 | Male | 46 | BRCA X | Positive | Positive | Positive | Positive | Luminal B | 2012 | FP-BS |
| Case 21 | Male | 79 | BRCA X | – | – | – | – | – | 2007 | CR |
| Case 22 | Male | 71 | BRCA X | – | – | – | – | – | 2009 | CR |
| Case 23 | Male | 54 | BRCA X | Negative | Negative | Positive | Negative | HER2-enriched | 2011 | CR |
| Case 24 | Male | 48 | BRCA X | Positive | Positive | Negative | Positive | Luminal A | 2012 | PC |
SC-BS, Spedali Civili of Brescia, Brescia, Italy; FP-BS, Fondazione Poliambulanza, Brescia, Italy; SA-BS, Istituto Clinico Sant'Anna, Brescia, Italy; CR, ASST of Cremona, Hospital of Cremona, Cremona, Italy; PC, Guglielmo da Saliceto Hospital, Piacenza, Italy. -, information not available in the histopathological report; ER, estrogen receptor; MIB1, MIB E3 ubiquitin protein ligase 1; PR, progesterone receptor.
DNA isolation from formalin-fixed paraffin-embedded (FFPE) BC tissues
The genomic DNA was extracted from FFPE tissues (10 of 5 µm sections for each patient) using QIAamp DNA FFPE Tissue Kit supplied by Qiagen, Inc.. The genomic DNA was digested using the micrococcal nuclease (New England Biolabs, Inc.), following the manufacturer's specifications, in order to obtain DNA fragments ranging from 200 to 500 bp (labelled input DNA). Agilent Bioanalyzer with the RNA 6000 Nano LabChip Kit was used to check the size, quality and quantity of fragmented DNA.
Methylated DNA immunoprecipitation on chip (MeDip-chip)
The DNA methylome of 24 patients with BC was obtained by MeDip followed by Affymetrix Human Promoter 1.0R Tiling Arrays hybridization (MeDip-chip) using the modified protocol of the Affymetrix chromatin immunoprecipitation assay as previously described (20). The human promoter array is a single array comprising 4.6 million probes tiled through 25,500 human promoter regions. Sequences used in the design of the human promoter arrays were selected from NCBI human genome assembly (BUILD 34). Purified DNA (4 µg), named input DNA, was immunoprecipitated with 10 µl anti-5-MethylCytosine Antibody (cat. no. BI-MECY-0100; Eurogentec) using the MeDip protocol (21), with minor modifications. The antibody-DNA complexes were immunoprecipitated using Dynabeads® Protein G immunoprecipitation kit (Thermo Fisher scientific, Inc.) and the enriched methylated DNA (labelled MeDip DNA) was purified by standard phenol/chloroform procedure and precipitated with isopropanol. A total of 200 ng input or MeDip DNA were amplified using the Affymetrix Chromatin Immunoprecipitation Assay Protocol. Hybridization on Human Promoter 1.0R array was performed using the GeneChip®Hybridization, Wash, and Stain Kit (Thermo Fisher scientific, Inc.) and the GeneChip® 640 hybridization oven. Arrays were washed and stained using the Fluidics Station 450 (Thermo Fisher scientific, Inc.), were scanned with the GeneChip Scanner 3000 7G and raw data were extracted with the GeneChip Operating System (GCOS) software. In total, 2 microarrays were used for each patient (one for MeDip DNA and one for input DNA). Data obtained from MeDip and input DNA microarrays have been deposited in the NCBI Gene 97 Expression Omnibus (GEO) data repository (GSE153636).
Quantification of methylated DNA by qPCR
To verify the effectiveness of the protocol, the input and MeDip DNA of three random patients with BC were amplified by qPCR. The methylated DNA was amplified using specific primers for the H19 gene, which is known to be hypermethylated (H19, chr11: 1,973,061-1,973,234 hg18) and primers for a control region without CpG dinucleotides (CTRL, chr7: 84,768,017-84,768,155 hg18). H19 forward primer: 5′-CGAGTGTGCGTGAGTGTGAG-3′and reverse primer: 5′-GGCGTAATGGAATGCTTGAA-3′. CTRL forward primer: 5′-GAGAGCATTAGGGCAGACAAA-3′ and reverse primer: 5′-GTTCCTCAGACAGCCACATTT-3′. DNA (25 ng) was used for each reaction and assayed in triplicate. qPCR was run with a first step of denaturation at 95°C for 10 min, then 40 cycles at 95°C for 30 sec, 56°C for 30 sec, 72°C for 30 sec. GoTaq qPCR Master Mix with SYBR Green was used (Promega corporation).
The enrichment level for the methylated regions was calculated using the qPCR threshold cycle (Ct) values applied to the following formula previously described (22,23): 2−[(H19me-H19in)-(CTRLme-CTRLin)] where H19me and H19in are the qPCR Ct values obtained for the H19 gene qPCR primers pair using MeDip and Input DNA as template, respectively; CTRLme and CTRLin are the qPCR Ct values obtained for the control region qPCR primers pair using MeDip and Input DNA as a template, respectively. Samples with low Ct value (<10) were considered as non-enriched and were therefore excluded from the study.
Statistical analysis
The raw data of 48. CEL files (24 from input DNA and 24 from MeDip DNA) were imported into Partek Genomics Suite (PGS) software version 6.6, normalized with the RMA algorithm and converted into log2 values. Hierarchical clustering and Principal Component Analysis were performed to verify that input DNA clustered in the same group compared to the MeDip DNA samples confirming the success of the MeDip protocol. For each patient, the DNA methylation was scored as Δ methylation value: The MeDip. CEL files were normalized against input DNA by subtracting the log2 of the signal intensity value of each of the 4.6 million array probes of the input DNA to the corresponding log2 signal intensity values for MeDip. The association between the DNA methylation levels of keratin genes and the pathological characteristics of the patients was evaluated using Fisher's exact test. For each gene, the median Δ methylation value was selected as the cutoff point to classify keratin methylation levels as ‘high’ or ‘low’. One-way ANOVA and Tukey's pairwise comparison tests were used to determine the statistical differences in the mean Δ methylation values among the different molecular subtypes of BC.
Significant differentially methylated regions (DMRs) were obtained using the model-based analysis of tiling arrays (MAT) algorithm (24) using the parameters described in Data S1. The DMRs of patients grouped based on some of their characteristics (Table II) were analyzed by ANOVA. In each pairwise comparison, the positive or negative MAT score indicated hypermethylation or hypomethylation, respectively of a given DMR compared to the other in the pair. The genomic coordinates of the DMRs were calculated based on UCSC human genomic assembly version 18 and PGS was used for the corresponding DMR gene annotation. The DNA methylation levels of gene promoters and gene bodies were calculated considering the DMRs located between-5,000 bp upstream the transcription start site (TSS) and +5,000 bp downstream the end codon of the nearest gene. Since both FBC and MBC cases were analyzed, DMRs located on chromosome Y were not considered. DAVID (25) was used for functional enrichment analysis to identify gene ontology terms and molecular signaling pathways. The P-value cut-offs used for each comparison have been included in Data S1.
Table II.
Comparisons performed and the relative DMRs found.
| Groups compared | Cases considered | Number of DMRs found | Hypomethylated DMRs, % | Hypermethylated DMRs, % | Number of DMRs associated with genes |
|---|---|---|---|---|---|
| 1: FBC vs. MBC (17 cases vs. 7 cases) | All cases | 2,846 | 67.83 | 32.17 | 2,486 |
| 2: FBC vs. MBC (10 cases vs. 6 cases) | BRCAX cases only | 1,242 | 93.56 | 6.44 | 1,102 |
| 3: BRCAX vs. BRCA1/2 (16 cases vs. 8 cases) | All cases | 364 | 46.70 | 53.30 | 357 |
| 4: BRCA1 vs. BRCA2 (4 cases vs. 3 cases) | Female cases only | 802 | 54.74 | 45.26 | 755 |
| 5: BRCA1 vs. BRCAX (4 cases vs. 10 cases) | Female cases only | 484 | 60.54 | 39.46 | 464 |
| 6: BRCA2 vs. BRCAX (3 cases vs. 10 cases) | Female cases only | 673 | 52.01 | 47.99 | 629 |
| 7: BRCA1 vs. BRCA2/X (4 cases vs. 13 cases) | Female cases only | 861 | 43.79 | 56.21 | 819 |
| 8: BRCA2 vs. BRCA1/X (4 cases vs. 14 cases) | Female cases only | 1,380 | 38.19 | 61.81 | 1,251 |
| 9: BRCAX vs. BRCA1/2 (10 cases vs. 7 cases) | Female cases only | 962 | 51.14 | 48.86 | 914 |
DMRs, differentially methylated regions; FBC, female breast cancer; MBC, male breast cancer.
Results
MeDip-chip analysis of BC tissues
The present study was designed to establish the global DNA methylation profile of 24 familial BC cases in order to identify sex-(FBC vs. MBC) and mutation-related differences among women with BRCA1, BRCA2 and BRCAX mutations.
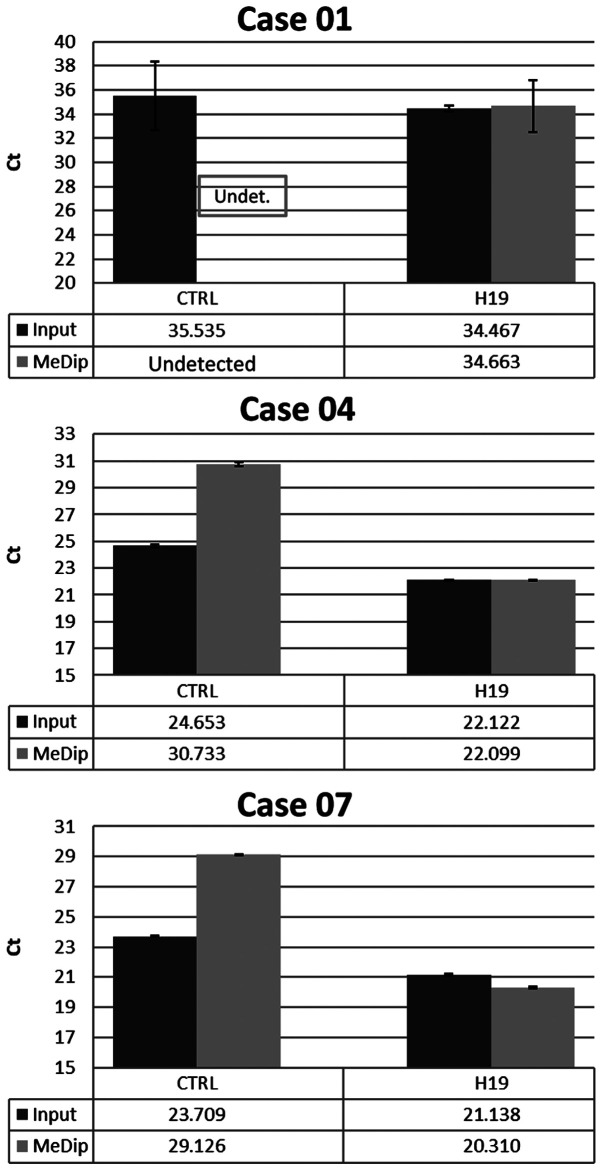
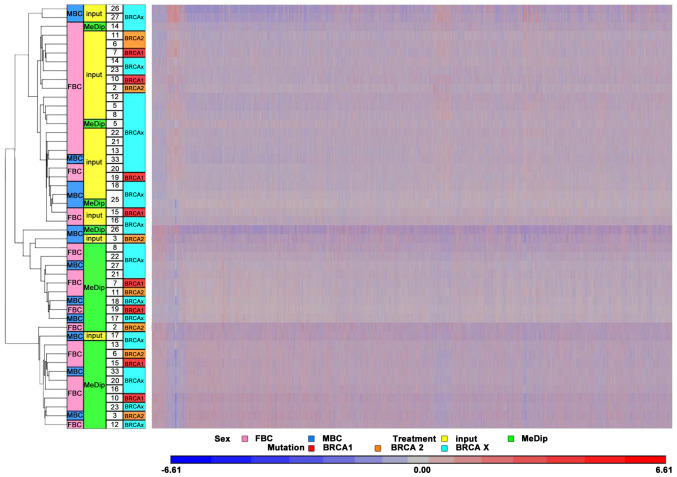
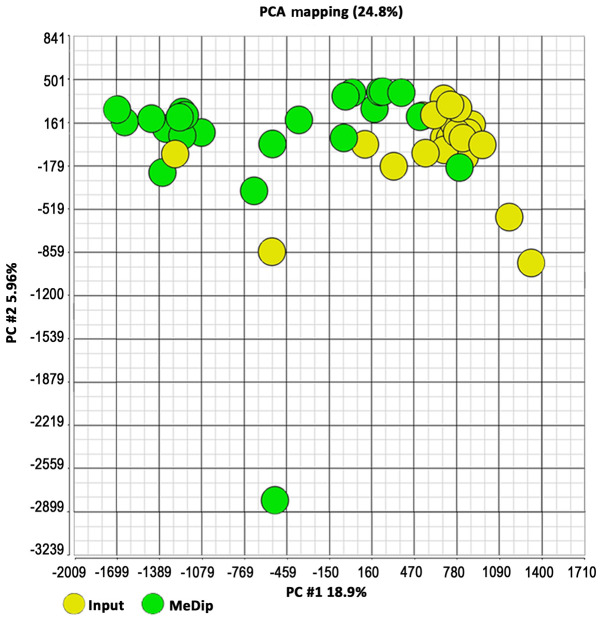
In detail, the DNA methylome was examined in 17 FBC (4 with BRCA1 mutations, 3 with BRCA2 mutations and 10 with BRCAX condition) and 7 MBC (1 with BRCA2 and 6 with BRCAX conditions) (Table I). MeDip was performed followed by hybridization on Affymetrix Promoter 1.0 Tiling arrays to identify genomic regions that were either hypo- or hypermethylated when compared to the same regions in patients from different groups. To quantify the enriched methylated DNA following the MeDip process, a DNA region containing the highly methylated H19 gene and a genomic DNA region without any CpG dinucleotides named CTRL were amplified by qPCR in randomly selected BC cases. The results showed that the average Ct values for H19 in input DNA and MeDip DNA were similar. On the contrary, the average Ct values for CTRL were higher in MeDip DNA samples as compared with DNA input samples. These results indicated the loss of the unmethylated DNA in the MeDip phase and thus an enrichment of methylated DNA (Fig. 1). To further assess the quality of the MeDip-chip protocol, the hierarchical clustering of the raw data was generated using Partek Genomic Suite (PGS) software. The heat map showed two robust clusters: One cluster including MeDip DNA (green) and one including input DNA (yellow; Fig. 2). The same clusters were obtained by Principal Component Analysis (Fig. 3).
Figure 1.
Evaluation of MeDip enrichment. The Ct values of H19 gene and CTRL regions were evaluated by qPCR in MeDip DNA and input DNA fractions from 3 randomly selected breast cancer cases. The enrichment levels for the methylated regions were calculated by the qPCR threshold cycle using the formula 2−[(H19me-H19in)-(CTRLme-CTRLin)], where, in case of an undetected Ct value, a value of 40 was used. Each sample was tested in triplicate. Error bars represent Ct standard error. Ct, threshold cycle; qPCR, quantitative PCR; MeDip, methylated DNA immunoprecipitation; Undet., undetermined; CTRL, control.
Figure 2.
Hierarchical clustering of the Human Promoter array methylation data. Hierarchical clustering of microarray methylation data of input and MeDip DNA from the examined BC cases. Two arrays per BC case (1 input DNA and 1 MeDip DNA) were performed. BC, breast cancer; FBC, female BC; MBC, male BC; MeDip, methylated DNA immunoprecipitation.
Figure 3.
PCA of the Human Promoter array methylation data. PCA of microarray methylation data of input (yellow) and MeDip (green) DNA from the examined BC cases. Two arrays per BC case (1 input DNA and 1 MeDip DNA) were performed. BC, breast cancer; MeDip, methylated DNA immunoprecipitation; PCA, principal component analysis.
Identification of DMRs in FBC as compared to MBC and between groups with different BRCA mutation statuses
The DNA methylation profiles of the 24 BC cases were achieved by subtracting the mean signal obtained from the Input array from the matching MeDip array for each probe. A total of nine pairwise comparisons were performed according to sex and mutation status (Table II). The number of the significant differentially methylated regions (DMRs) were obtained by Partek Genomics Suite using ANOVA and MAT algorithms with specific parameters (see Materials and methods).
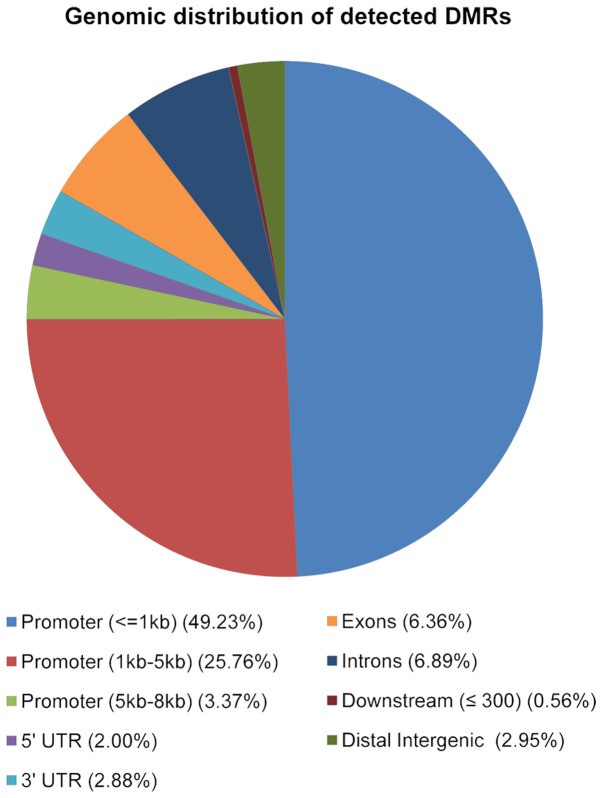
A list of DMRs for each pairwise comparison was obtained (nine lists in total) and their associated genes were identified. The lists of genes associated with the 20 most significant DMRs are described in Table SI A-I and Figs. S1–S9. All genes associated with the DMRs of the nine comparisons considered were catalogued using the online repository of HGNC (HUGO gene nomenclature committee) in order to obtain an overview of the gene classes involved (Table SIIA-I) and were analyzed in relation to their genomic location. As shown in Fig. 4, the majority of 2,846 DMRs were located in promoter regions (49% were located in the 1kb region upstream the TSS; 26% in the region 1-5kb upstream the TSS and 3% in the region between 5-8kb upstream the TSS), 7% in introns, 6% in exons, 3% in the 3′UTR and, 2% in the 5′UTR (Fig. 4).
Figure 4.
Genomic distribution of identified DMRs. Pie chart showing the percentage of total DMRs according to their functional genomic distribution. AnnotatePeak (ChIPSeeker) was used to assign the DMRs according to their genomic positions, considering the promoter regions (in 1 kb region upstream from TSS-in the region between 1 kb and 5 kb upstream from TSS-in the region between 5 kb and 8 kb upstream from TSS), 3′ and 5′UTR regions, exons, introns, the downstream regions defined as the downstream of gene end, and distal intergenic regions. DMRs, differentially methylated regions; TSS, transcriptional start sites; UTR, untranslated region.
Sex-related DNA methylation differences between FBC and MBC
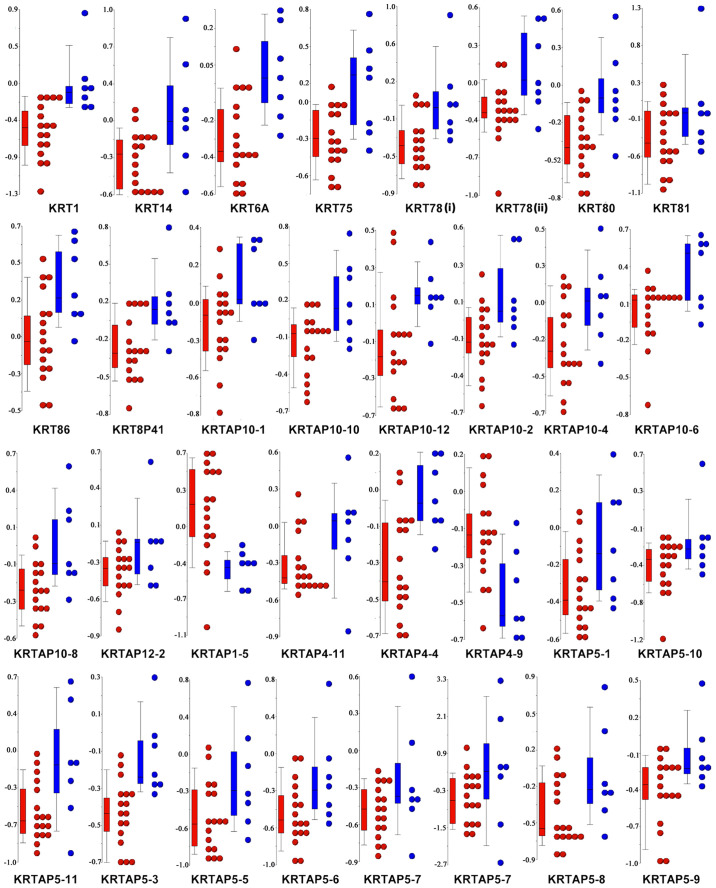
The methylome of FBC (n=17) was compared with that of MBC (n=7) and 2,846 DMRs associated with 2,486 annotated genes were identified. Functional enrichment analysis performed by DAVID identified nine enriched GO terms (Table III). The most significant enriched terms were the GO Cellular Component (CC) concerning the structure of the cytoskeleton, including ‘GO:0045095: Keratin filament’, ‘GO:0005882: Intermediate filament’ and ‘GO:0044430: Cytoskeletal part’. As indicated by the negative MAT score (Table IV) and the Δ methylation value for each patient (Fig. 5), almost all the genes associated with the ‘GO:0045095: Keratin filament’ term were significantly hypomethylated in FBC, as compared with MBC. This result prompted us to combine the methylation level of keratin (KRT) genes with the pathological characteristics patients with BC. Of note, the DNA methylation levels of KRT14, KRT81, and KRT86 were significantly associated with the progesterone receptor status, and KRT75 was found to be differentially methylated among the BC molecular subtypes (Table SIII). No correlations were found between the methylation level of the remaining KRTs and the other pathological characteristics, including estrogen receptor (ER), HER2, and Ki67 status.
Table III.
Enriched GO terms found by Database for Annotation, Visualization and Integrated Discovery when all female breast cancer cases were compared with all male breast cancer cases.
| Category | Term | Genes | Count | P-value | Benjamini |
|---|---|---|---|---|---|
| GO: CC | GO:0045095 Keratin filament | KRT1, KRT14, KRT6A, KRT75, KRT78, KRT80, KRT81, KRT86, KRT8P41, KRTAP10-1, KRTAP10-10, KRTAP10-12, KRTAP10-2, KRTAP10-4, KRTAP10-6, KRTAP10-8, KRTAP12-2, KRTAP1-5, KRTAP4-11, KRTAP4-4, KRTAP4-9, KRTAP5-1, KRTAP5-10, KRTAP5-11, KRTAP5-3, KRTAP5-5, KRTAP5-6, KRTAP5-7, KRTAP5-8, KRTAP5-9 | 30 | 0.000000264 | 0.0000156 |
| GO: CC | GO:0044430 Cytoskeletal part | ADORA2A, AKT1, ARHGAP32, ATM, B9D2, BMX, CAMK2N1, CAMSAP1, CAPZB, CC2D2A, CCDC85B, CCIN, CDH23, CEP135, CEP170B, CEP72, CETN2, CHRM1, CLASP2, CNN3, CNTLN, DFNB31, DLG4, DLGAP2, DNAH14, DNAH2, DNAI2, DNAL4, DYNLT1, EML1, EML4, EML6, EVI5, FILIP1L, GAS7, GAS8, HAUS1, HAUS7, HAUS8, HIPK2, HOOK3, IFFO1, JAKMIP1, KATNAL1, KIF13B, KIF16B, KIF21A, KIF24, KIF25, KIF2A, KIFC2, KLC3, KLC4, KRT1, KRT14, KRT15, KRT16P2, KRT17, KRT20, KRT6A, KRT75, KRT78, KRT80, KRT81, KRT86, KRT8P41, KRTAP10-1, KRTAP10-10, KRTAP10-12, KRTAP10-2, KRTAP10-4, KRTAP10-6, KRTAP10-8, KRTAP12-2, KRTAP1-5, KRTAP4-11, KRTAP4-4, KRTAP4-9, KRTAP5-1, KRTAP5-10, KRTAP5-11, KRTAP5-3, KRTAP5-5, KRTAP5-6, KRTAP5-7, KRTAP5-8, KRTAP5-9, LLGL1, LMNB2, MAP1A, MAPT, MC1R, MCPH1, MED12, MYBPH, MYH14, MYH2, MYH6, MYH7, MYH7B, MYL12A, MYL12B, MYL3, MYLPF, MYO15A, MYO1G, MYO7B, MYO9B, NAV1, NDRG2, NEFH, NIN, NTRK2, NUP62, PDE4D, PDE4DIP, PDLIM7, PIN4, PPP1R9A, PPP1R9B, RAB3GAP2, RAB3IP, RANBP1, RHOU, RMDN2, RNF19A, SEPT11, SEPT8, SEPT9, SHANK3, SIRT2, SMEK1, SPAG6, SPDL1, SPTB, SPTBN5, SSNA1, SVIL, SYNM, SYNPO, TACC1, TACC2, TBCD, TNNT3, TPM4, TPX2, TRIM55, TTLL5, TTLL7, TUBA3E, TUBB1, TUBB3, TUBB8, UBXN6 | 153 | 0.0000111 | 0.0032717 |
| GO: CC | GO:0005882 Intermediate filament | ADORA2A, DLGAP2, IFFO1, KRT1, KRT14, KRT15, KRT16P2, KRT17, KRT20, KRT6A, KRT75, KRT78, KRT80, KRT81, KRT86, KRT8P41, KRTAP10-1, KRTAP10-10, KRTAP10-12, KRTAP10-2, KRTAP10-4, KRTAP10-6, KRTAP10-8, KRTAP12-2, KRTAP1-5, KRTAP4-11, KRTAP4-4, KRTAP4-9, KRTAP5-1, KRTAP5-10, KRTAP5-11, KRTAP5-3, KRTAP5-5, KRTAP5-6, KRTAP5-7, KRTAP5-8, KRTAP5-9, LMNB2, NEFH, SYNM | 40 | 0.0000301 | 0.0044294 |
| GO: CC | GO:0045111 Intermediate filament cytoskeleton | ADORA2A, DLGAP2, IFFO1, KRT1, KRT14, KRT15, KRT16P2, KRT17, KRT20, KRT6A, KRT75, KRT78, KRT80, KRT81, KRT86, KRT8P41, KRTAP10-1, KRTAP10-10, KRTAP10-12, KRTAP10-2, KRTAP10-4, KRTAP10-6, KRTAP10-8, KRTAP12-2, KRTAP1-5, KRTAP4-11, KRTAP4-4, KRTAP4-9, KRTAP5-1, KRTAP5-10, KRTAP5-11, KRTAP5-3, KRTAP5-5, KRTAP5-6, KRTAP5-7, KRTAP5-8, KRTAP5-9, LMNB2, MACF1, NEFH, SYNM | 41 | 0.0000228 | 0.0044762 |
| GO: MF | GO:0008047 Enzyme activator activity | ABR, ACAP2, AGAP3, AGAP6, AGAP9, AHSA2, AIFM3, ANGPT4, ANKRD27, APOA2, APOA5, ARAP1, ARAP3, ARFGAP3, ARHGAP11A, ARHGAP19, ARHGAP19-SLIT1, ARHGAP23, ARHGAP32, ARHGAP35, ARHGAP40, BCRP2, CDK5R2, CHM, CTAGE4, CTAGE5, DEPDC1B, DLC1, DOCK2, EVI5, FAM13B, FN1, FZR1, GAPVD1, GDI2, GHRL, GMIP, GPSM3, GRTP1, IGFBP3, IQGAP2, MALT1, MMP15, MMP16, MYO9B, NRG3, PPP1R12B, PRKAG2, PRR5-ARHGAP8, RANBP1, RASA3, RASA4, RASA4CP, RGS12, RGS3, RGS5, RGS6, SEC14L2, SH3BP1, SRGAP1, TBC1D10B, TBC1D16, TBC1D19, TBC1D2, TBC1D22A, TBC1D25, TBC1D29, TBC1D3B, TBC1D3F, TBC1D8B, TBC1D9B, TIAM2, USP6 | 69 | 0.0000779 | 0.0052097 |
| GO: MF | GO:0005096 GTPase activator activity | ABR, ACAP2, AGAP3, AGAP6, AGAP9, ANKRD27, ARAP1, ARAP3, ARFGAP3, ARHGAP11A, ARHGAP19, ARHGAP19-SLIT1, ARHGAP23, ARHGAP32, ARHGAP35, ARHGAP40, BCRP2, CHM, DEPDC1B, DLC1, DOCK2, EVI5, FAM13B, GAPVD1, GDI2, GMIP, GPSM3, GRTP1, IQGAP2, MYO9B, PRR5-ARHGAP8, RANBP1, RASA3, RASA4, RASA4CP, RGS12, RGS3, RGS5, RGS6, SH3BP1, SRGAP1, TBC1D10B, TBC1D16, TBC1D19, TBC1D2, TBC1D22A, TBC1D25, TBC1D29, TBC1D3B, TBC1D3F, TBC1D3G, TBC1D8B, TBC1D9B, TIAM2, USP6 | 51 | 0.0000459 | 0.0061401 |
| GO: CC | GO:0005856 Cytoskeleton | ABL2, ACTB, ADORA2A, AFAP1, AKT1, ANK1, ARAP3, ARHGAP32, ARHGAP35, ATM, B9D2, BMX, CAMK2N1, CAMSAP1, CAPZB, CC2D2A, CCDC85B, CCIN, CDH23, CDK5, CEP135, CEP170B, CEP72, CETN2, CHRM1, CLASP2, CNFN, CNN2, CNN3, CNTLN, CORO2B, CYLD, DAPK1, DFNB31, DLG4, DLGAP2, DMD, DMTN, DNAH14, DNAH2, DNAI2, DNAL4, DOCK2, DPYSL2, DYNLT1, EDA, EML1, EML4, EML6, EPB41L1, EPPK1, ESPN, EVI5, FAM65B, FARP2, FGD5, FHL2, FILIP1L, FRMD1, FRMD7, GAS7, GAS8, HAUS1, HAUS7, HAUS8, HINT1, HIPK2, HOOK3, HRNR, IFFO1, IQGAP2, JAKMIP1, KALRN, KATNAL1, KIF13B, KIF16B, KIF21A, KIF24, KIF25, KIF2A, KIFC2, KLC3, KLC4, KRIT1, KRT1, KRT14, KRT15, KRT16P2, KRT17, KRT20, KRT6A, KRT75, KRT78, KRT80, KRT81, KRT86, KRT8P41, KRTAP10-1, KRTAP10-10, KRTAP10-12, KRTAP10-2, KRTAP10-4, KRTAP10-6, KRTAP10-8, KRTAP12-2, KRTAP1-5, KRTAP4-11, KRTAP4-4, KRTAP4-9, KRTAP5-1, KRTAP5-10, KRTAP5-11, KRTAP5-3, KRTAP5-5, KRTAP5-6, KRTAP5-7, KRTAP5-8, KRTAP5-9, LLGL1, LMNB2, MACF1, MAP1A, MAP3K1, MAP6D1, MAPT, MC1R, MCPH1, MED12, MICAL3, MYBPH, MYH14, MYH2, MYH6, MYH7, MYH7B, MYL12A, MYL12B, MYL3, MYLPF, MYO15A, MYO1G, MYO7B, MYO9B, NAV1, NDRG2, NEFH, NF2, NIN, NTRK2, NUP62, NXF2B, PDE4D, PDE4DIP, PDLIM7, PIN4, PNP, POTEJ, PPP1R9A, PPP1R9B, RAB3GAP2, RAB3IP, RANBP1, RDX, RHOU, RMDN2, RNF19A, SEPT11, SEPT8, SEPT9, SGCA, SGCE, SHANK3, SIRT2, SLC4A1, SMEK1, SPAG6, SPDL1, SPRR2B, SPTB, SPTBN5, SSNA1, STRBP, SVIL, SYNE2, SYNM, SYNPO, TACC1, TACC2, TBCD, TGM1, TNNT3, TPM4, TPX2, TRADD, TRIM55, TTLL5, TTLL7, TUBA3E, TUBB1, TUBB3, TUBB8, TWF1, UBXN6, ZNF174 | 203 | 0.000145 | 0.0170134 |
| GO: MF | GO:0060589 Nucleoside-triphosphatase regulator activity | ABR, ACAP2, AGAP3, AGAP6, AGAP9, AHSA2, ANKRD27, ARAP1, ARAP3, ARFGAP3, ARHGAP11A, ARHGAP19, ARHGAP19-SLIT1, ARHGAP23, ARHGAP32, ARHGAP35, ARHGAP40, ARHGEF10, ARHGEF25, ARHGEF5, BCRP2, CHM, DEPDC1B, DLC1, DOCK2, DOCK3, DOCK8, EVI5, FAM13B, FARP2, FGD5, GAPVD1, GDI2, GMIP, GPSM3, GRTP1, IQGAP2, ITSN2, KALRN, KNDC1, KRIT1, MINK1, MYO9B, PLEKHG1, PLEKHG4B, PLEKHG7, PRR5-ARHGAP8, PSD4, RAB3IP, RANBP1, RAPGEF3, RASA3, RASA4, RASA4CP, RASGRP2, RGL2, RGS12, RGS3, RGS5, RGS6, RIMS2, RPH3AL, SH3BP1, SRGAP1, SYTL2, SYTL3, TBC1D10B, TBC1D16, TBC1D19, TBC1D2, TBC1D22A, TBC1D25, TBC1D29, TBC1D3B, TBC1D3F, TBC1D3G, TBC1D8B, TBC1D9B, TIAM1, TIAM2, TNK2, USP6 | 78 | 0.000568 | 0.0188541 |
| GO: MF | GO:0030695 GTPase regulator activity | ABR, ACAP2, AGAP3, AGAP6, AGAP9, ANKRD27, ARAP1, ARAP3, ARFGAP3, ARHGAP11A, ARHGAP19, ARHGAP19-SLIT1, ARHGAP23, ARHGAP32, ARHGAP35, ARHGAP40, ARHGEF10, ARHGEF25, ARHGEF5, BCRP2, CHM, DEPDC1B, DLC1, DOCK2, DOCK3, DOCK8, EVI5, FAM13B, FARP2, FGD5, GAPVD1, GDI2, GMIP, GPSM3, GRTP1, IQGAP2, ITSN2, KALRN, KNDC1, KRIT1, MINK1, MYO9B, PLEKHG1, PLEKHG4B, PLEKHG7, PRR5-ARHGAP8, PSD4, RAB3IP, RANBP1, RAPGEF3, RASA3, RASA4, RASA4CP, RASGRP2, RGL2, RGS12, RGS3, RGS5, RGS6, RIMS2, RPH3AL, SH3BP1, SRGAP1, SYTL2, SYTL3, TBC1D10B, TBC1D16, TBC1D19, TBC1D2, TBC1D22A, TBC1D25, TBC1D29, TBC1D3B, TBC1D3F, TBC1D3G, TBC1D8B, TBC1D9B, TIAM1, TIAM2, TNK2, USP6 | 77 | 0.000467 | 0.0206520 |
GO, Gene Ontology; CC, cellular component; MF, molecular function.
Table IV.
Genes corresponding to the DMRs associated with GO term ‘GO: 0045095: Keratin filament’ when all female breast cancer cases were compared with all male breast cancer cases.
| Gene symbol | P-value | MAT score | Chromosome | Region start | Region end | DMR length, bp | Probes in region | DMR position |
|---|---|---|---|---|---|---|---|---|
| KRT1 | 1.42×10−5 | −5.317 | chr12 | 51362805 | 51365485 | 2,681 | 48 | Upstream TSS |
| KRT14 | 8.51×10−5 | −4.142 | chr17 | 36996508 | 36998231 | 1,724 | 42 | Promoter |
| KRT6A | 2.84×10−5 | −4.458 | chr12 | 51172288 | 51173829 | 1,542 | 43 | Promoter |
| KRT75 | 8.51×10−5 | −4.166 | chr12 | 51117744 | 51119993 | 2,250 | 56 | Upstream TSS |
| KRT78 | 8.51×10−5 | −4.104 | chr12 | 51516089 | 51517502 | 1,414 | 24 | Downstream CDS end |
| KRT78 | 7.09×10−5 | −4.282 | chr12 | 51518324 | 51520224 | 1,901 | 53 | Promoter |
| KRT80 | 8.51×10−5 | −4.101 | chr12 | 50873925 | 50876076 | 2,152 | 57 | Upstream TSS |
| KRT81 | 8.51×10−5 | −4.123 | chr12 | 50969856 | 50971294 | 1,439 | 39 | Exon |
| KRT86 | 8.51×10−5 | −4.229 | chr12 | 50981621 | 50983635 | 2,015 | 55 | Promoter |
| KRT8P41 | 1.42×10−5 | −5.508 | chr11 | 9071904 | 9074578 | 2,675 | 76 | Exon |
| KRTAP10-1 | 8.51×10−5 | −4.109 | chr21 | 44782903 | 44784728 | 1,826 | 47 | Exon |
| KRTAP10-10 | 8.51×10−5 | −4.208 | chr21 | 44880449 | 44882191 | 1,743 | 44 | Promoter |
| KRTAP10-12 | 7.09×10−5 | −4.255 | chr21 | 44941119 | 44942642 | 1,524 | 38 | Exon |
| KRTAP10-2 | 8.51×10−5 | −4.224 | chr21 | 44795043 | 44796629 | 1,587 | 43 | Promoter |
| KRTAP10-4 | 7.09×10−5 | −4.273 | chr21 | 44817595 | 44819882 | 2,288 | 63 | Exon |
| KRTAP10-6 | 8.51×10−5 | −4.226 | chr21 | 44835328 | 44836747 | 1,420 | 40 | Promoter |
| KRTAP10-8 | 1.42×10−5 | −6.221 | chr21 | 44855891 | 44858376 | 2,486 | 66 | Exon |
| KRTAP12-2 | 8.51×10−5 | −4.093 | chr21 | 44910070 | 44911658 | 1,589 | 45 | Exon |
| KRTAP1-5 | 5.68×10−5 | 4.200 | chr17 | 36436999 | 36438048 | 1,050 | 29 | Upstream TSS |
| KRTAP4-11 | 8.51×10−5 | −4.239 | chr17 | 36526929 | 36528798 | 1,870 | 48 | Exon |
| KRTAP4-4 | 8.51×10−5 | −4.142 | chr17 | 36569481 | 36571301 | 1,821 | 49 | Promoter |
| KRTAP4-9 | 2.84×10−5 | 4.518 | chr17 | 36513351 | 36515335 | 1,985 | 50 | Promoter |
| KRTAP5-1 | 1.42×10−5 | −5.387 | chr11 | 1561553 | 1564020 | 2,468 | 54 | Exon |
| KRTAP5-10 | 8.51×10−5 | −4.180 | chr11 | 70953946 | 70955693 | 1,748 | 45 | Exon |
| KRTAP5-11 | 8.51×10−5 | −4.225 | chr11 | 70970806 | 70972121 | 1,316 | 33 | Promoter |
| KRTAP5-3 | 7.09×10−5 | −4.258 | chr11 | 1584784 | 1586547 | 1,764 | 46 | Exon |
| KRTAP5-5 | 8.51×10−5 | −4.222 | chr11 | 1607571 | 1609218 | 1,648 | 39 | Exon |
| KRTAP5-6 | 1.42×10−5 | −5.126 | chr11 | 1674136 | 1675916 | 1,781 | 38 | Exon |
| KRTAP5-7 | 8.51×10−5 | −4.239 | chr11 | 70915825 | 70917331 | 1,507 | 38 | Exon |
| KRTAP5-7 | 1.42×10−5 | −7.019 | chr11 | 70909034 | 70912320 | 3,287 | 74 | Upstream TSS |
| KRTAP5-8 | 1.42×10−5 | −5.033 | chr11 | 70926237 | 70927983 | 1,747 | 48 | Exon |
| KRTAP5-9 | 8.51×10−5 | −4.115 | chr11 | 70936504 | 70937899 | 1,396 | 35 | Promoter |
CDS, coding sequence; DMRs, differentially methylated regions; GO, Gene Ontology; MAT, model-based analysis of tiling arrays; TSS, transcriptional start sites.
Figure 5.
Methylation levels of keratin filament genes in FBC compared with MBC. Dot plots were generated by Partek Genomics Suite and represent the DMRs associated with GO term ‘GO: 0045095: Keratin filament’ in FBC compared with MBC. Each graph refers to a specific KRT gene and KRTAP genes. Within a graph a point represents the Δ methylation value of a DMR for a given breast cancer case. FBC cases are shown in red and MBC cases are shown in blue. A total of two DMRs were detected for the KRT78 gene, indicated as (i) and (ii). The numbers on the y-axis are Δ methylation values. DMR, differentially methylated region; FBC, female breast cancer; GO, Gene Ontology; KRT, keratin; KRTAP, keratin-associated protein; MBC, male breast cancer.
Among the most significantly enriched terms, we also found the GO Molecular Function (MF) term concerning GTPase superfamily ‘GO:0005096: GTPase activator activity’ (Table III). Numerous differentially methylated genes belonged to the five RAS GTPase families. In particular, 4 genes from the ARF family were generally implicated in vesicular transport, 9 genes from the RAB family were mainly involved in membrane trafficking, 2 genes from the RAN family were associated with nuclear transport, 6 genes from the RAS family were implicated in cellular proliferation and 25 genes from the RHO family were involved in cytoskeletal dynamics and morphology (Table V). These results pointed towards a different methylation profile of RAS GTPases genes in FBC, as compared with MBC.
Table V.
Ras GTPase superfamily genes identified to be differentially methylated when female breast cancer cases were compared with male breast cancer cases.
| Gene symbol | Ras subfamily | P-value | MAT score | Chromosome | Region start at: | Region end at: | DMR length, bp | Probes in region | DMR position |
|---|---|---|---|---|---|---|---|---|---|
| ARF1 | Arf | 5.68×10−5 | 4.105 | chr1 | 226336193 | 226338188 | 1,996 | 52 | Promoter |
| ARF4 | Arf | 7.09×10−5 | 4.004 | chr3 | 57557804 | 57559228 | 1,425 | 40 | Promoter |
| ARFGAP3 | Arf | 8.51×10−5 | −4.234 | chr22 | 41520156 | 41522358 | 2,203 | 40 | Downstream CDS end |
| ARL16 | Arf | 2.84×10−5 | −4.533 | chr17 | 77258588 | 77260369 | 1,782 | 34 | Promoter |
| RAB24 | Rab | 7.09×10−5 | −4.266 | chr5 | 176661091 | 176662461 | 1,371 | 38 | Exon |
| RAB26 | Rab | 8.51×10−5 | −4.113 | chr16 | 2141352 | 2142806 | 1,455 | 26 | Exon |
| RAB36 | Rab | 8.51×10−5 | −4.182 | chr22 | 21838547 | 21839857 | 1,311 | 33 | Downstream CDS end |
| RAB3GAP2 | Rab | 2.84×10−5 | 4.379 | chr1 | 218510144 | 218511588 | 1,445 | 41 | Intron |
| RAB3IP | Rab | 5.68×10−5 | 4.103 | chr12 | 68492976 | 68494498 | 1,523 | 42 | Exon |
| RAB4A | Rab | 8.51×10−5 | −4.155 | chr1 | 227474954 | 227476365 | 1,412 | 32 | Intron |
| RAB5A | Rab | 4.26×10−5 | 4.266 | chr3 | 19963331 | 19965486 | 2,156 | 57 | Promoter |
| RAB7L1 | Rab | 7.09×10−5 | −4.254 | chr1 | 204009073 | 204011081 | 2,009 | 48 | Exon |
| RABGGTA | Rab | 4.26×10−5 | −4.383 | chr14 | 23805254 | 23806759 | 1,506 | 42 | Exon |
| RANBP1 | Ran | 8.51×10−5 | −4.095 | chr22 | 18495324 | 18496829 | 1,506 | 30 | Downstream CDS end |
| RANBP1 | Ran | 8.51×10−5 | −4.156 | chr22 | 18491321 | 18493103 | 1,783 | 50 | Exon |
| RANBP3L | Ran | 5.68×10−5 | 4.066 | chr5 | 36282283 | 36283445 | 1,163 | 21 | Downstream CDS end |
| RANBP3L | Ran | 2.84×10−5 | 4.341 | chr5 | 36336981 | 36338826 | 1,846 | 52 | Promoter |
| HRAS | Ras | 1.42×10−5 | −5.077 | chr11 | 520023 | 522094 | 2,072 | 59 | Downstream CDS end |
| RAP1A | Ras | 1.42×10−5 | −5.056 | chr1 | 111991970 | 111993933 | 1,964 | 51 | Intron |
| RASD1 | Ras | 5.68×10−5 | 4.214 | chr17 | 17338882 | 17341351 | 2,470 | 65 | Promoter |
| RASD2 | Ras | 5.68×10−5 | 4.104 | chr22 | 34265450 | 34266847 | 1,398 | 25 | Upstream TSS |
| RASGRP2 | Ras | 7.09×10−5 | −4.299 | chr11 | 64247878 | 64250375 | 2,498 | 68 | Downstream CDS end |
| RASL10A | Ras | 8.51×10−5 | −4.092 | chr22 | 28041625 | 28043157 | 1,533 | 38 | Promoter |
| RHOU | Rho | 8.51×10−5 | −4.149 | chr1 | 226844408 | 226845837 | 1,430 | 33 | Upstream TSS |
| ARHGAP11A | Rho-GAP | 2.84×10−5 | 4.353 | chr15 | 30718918 | 30720431 | 1,514 | 32 | Promoter |
| ARHGAP19 | Rho-GAP | 5.68×10−5 | 4.068 | chr10 | 99019196 | 99020955 | 1,760 | 41 | Intron |
| ARHGAP19-SLIT1 | Rho-GAP | 8.51×10−5 | −4.172 | chr10 | 98938793 | 98940326 | 1,534 | 33 | Intron |
| ARHGAP23 | Rho-GAP | 8.51×10−5 | −4.181 | chr17 | 33872823 | 33874371 | 1,549 | 43 | Exon |
| ARHGAP32 | Rho-GAP | 5.68×10−5 | 4.202 | chr11 | 128444021 | 128445428 | 1,408 | 32 | Intron |
| ARHGAP32 | Rho-GAP | 5.68×10−5 | 4.150 | chr11 | 128438912 | 128440364 | 1,453 | 36 | Exon |
| ARHGAP35 | Rho-GAP | 8.51×10−5 | −4.156 | chr19 | 52193356 | 52195197 | 1,842 | 50 | Exon |
| ARHGAP40 | Rho-GAP | 8.51×10−5 | −4.197 | chr20 | 36676834 | 36678364 | 1,531 | 43 | Intron |
| DLC1 | Rho-GAP | 7.09×10−5 | 4.005 | chr8 | 13414150 | 13415703 | 1,554 | 33 | Intron |
| FAM13B | Rho-GAP | 5.68×10−5 | 4.186 | chr5 | 137395434 | 137396944 | 1,511 | 41 | Promoter |
| GMIP | Rho-GAP | 5.68×10−5 | −4.377 | chr19 | 19605567 | 19607497 | 1,931 | 49 | Exon |
| SH3BP1 | Rho-GAP | 8.51×10−5 | −4.153 | chr22 | 36367680 | 36369434 | 1,755 | 38 | Exon |
| SRGAP1 | Rho-GAP | 5.68×10−5 | −4.352 | chr12 | 62522064 | 62523557 | 1,494 | 13 | Upstream TSS |
| ABR | Rho-GEF | 8.51×10−5 | −4.159 | chr17 | 961466 | 963170 | 1,705 | 42 | Intron |
| ARHGEF10 | Rho-GEF | 1.42×10−5 | −7.052 | chr8 | 1773070 | 1777363 | 4,294 | 103 | Intron |
| ARHGEF25 | Rho-GEF | 8.51×10−5 | −4.212 | chr12 | 56293862 | 56295869 | 2,008 | 55 | Exon |
| ARHGEF34P | Rho-GEF | 1.42×10−5 | −6.205 | chr7 | 143612540 | 143615423 | 2,884 | 80 | Promoter |
| ARHGEF35 | Rho-GEF | 8.51×10−5 | −4.117 | chr7 | 143518580 | 143520964 | 2,385 | 46 | Intron |
| ARHGEF5 | Rho-GEF | 8.51×10−5 | −4.199 | chr7 | 143683885 | 143685539 | 1,655 | 46 | Intron |
| ARHGEF5 | Rho-GEF | 1.42×10−5 | −5.067 | chr7 | 143692916 | 143695114 | 2,199 | 61 | Exon |
| ARHGEF5 | Rho-GEF | 1.42×10−5 | −5.142 | chr7 | 143690467 | 143692900 | 2,434 | 67 | Exon |
| FARP2 | Rho-GEF | 7.09×10−5 | 4.051 | chr2 | 241943420 | 241944877 | 1,458 | 25 | Promoter |
| FGD5 | Rho-GEF | 8.51×10−5 | −4.214 | chr3 | 14835085 | 14836481 | 1,397 | 39 | Promoter |
| ITSN2 | Rho-GEF | 2.84×10−5 | 4.387 | chr2 | 24334440 | 24336066 | 1,627 | 44 | Exon |
| KALRN | Rho-GEF | 8.51×10−5 | −4.138 | chr3 | 125292634 | 125294221 | 1,588 | 44 | Upstream TSS |
| PLEKHG1 | Rho-GEF | 8.51×10−5 | −4.136 | chr6 | 151185066 | 151186484 | 1,419 | 19 | Exon |
| TIAM1 | Rho-GEF | 7.09×10−5 | 4.004 | chr21 | 31851782 | 31853729 | 1,948 | 53 | Promoter |
| TIAM2 | Rho-GEF | 8.51×10−5 | −4.169 | chr6 | 155580243 | 155582141 | 1,899 | 48 | Intron |
CDS, coding sequence; DMRs, differentially methylated regions; MAT, model-based analysis of tiling arrays; TSS, transcriptional start sites.
We further performed the comparison between FBC (n=10) and MBC (n=6) with BRCAX mutation condition. A total of 1,242 DMRs corresponding to 1,102 genes were reported. A total of 8 GO terms generally associated with RAS GTPase superfamily, particularly RHO-GAP and RHO-GEF proteins and RAB GTPase activity, were identified using the DAVID database, (Table SIVA) as already observed when all cases were considered.
Mutation-related DNA methylation differences in FBC among patients with BRCA1, BRCA2 and BRCAX mutations
Initially, we compared the DNA methylation profile of BRCAX patients (n=16) was compared with that of those with BRCA1 and BRCA2 mutations (n=8), irrespective of their sex. A total of 364 DMRs were reported; however, no significant GO terms associated with these genes were identified by DAVID analysis. Therefore, to limit the intrinsic heterogeneity within the groups, the next comparisons were restricted to women. The mutation classes were compared as follows:
a) BRCA1 vs. BRCA2 (n=4; n=3)
Following the comparison between women with BRCA1 and those with BRCA2 mutations, 802 DMRs associated with 755 genes were reported and the term ‘GO:0019787: Small conjugating protein ligase activity’ was found by DAVID analysis to be highly represented (Table SIVB) Despite the limited number of cases, this result may indicate a different modulation of the ubiquitination pathway between BRCA1 and BRCA2 FBC.
b) BRCA1 vs. BRCAX (n=4; n=10)
Following the comparison between patients with BRCA1 and those with BRCAX conditions, 484 DMRs associated with 464 genes were found and the term ‘GO:0051240: Positive regulation of multicellular organismal process’ (Table SIVC) was identified by DAVID analysis. This term is very broad making it challenging to establish the biological differences between these patient groups. Of note, BRCAX patients were heterogeneous and belonged to different molecular subtypes. On the contrary, patients with BRCA1 mutations were more homogeneous and belonged to the triple-negative molecular subtype (Table I).
c) BRCA2 vs. BRCAX (n=3; n=10)
Following the comparison between patients with BRCA2 mutations and those with a BRCAX condition, 673 DMRs corresponding to 629 genes were reported. Following DAVID analysis, 2 significant GO terms and 1 KEGG pathway (Table SIVD) were identified. These terms were ‘GO:0008047: Enzyme activator activity’, ‘GO: 0060589: Nucleoside-triphosphatase regulator activity’ and ‘hsa05220: Chronic myeloid leukemia’. Some genes found differentially methylated and included in the ‘chronic myeloid leukemia’ KEGG pathway, such as CDKN1B (p27) and PIK3R1, were also found frequently mutated in a very large study on BC (26).
d) BRCA1 vs. BRCA2/BRCAX (n=4; n=13)
Following the comparison between patients with BRCA1 and those with BRCA2/BRCAX mutations, 861 DMRs corresponding to 819 genes were reported. Following DAVID analysis, the term ‘GO:0008092: Cytoskeletal protein binding’ was identified (Table SIVE). Certain genes included in the term are known to interact with microfilaments, microtubules and intermediate filaments. This result indicated that these groups of patients have a different DNA methylation profile of the cytoskeleton-related genes when compared against each other and this probably influences the architecture of the cytoskeleton.
e) BRCA2 vs. BRCA1/BRCAX (n=3; n=14)
Following the comparison between patients with BRCA2 and those with BRCA1/BRCAX mutations, found 1,380 DMRs corresponding to 1,251 genes were reported. Following DAVID analysis, 7 enriched KEGG pathways were identified, most of which were associated with cancer (Table SIVF). Of note, PIK3CA and PIK3R1 were found to be frequently mutated in a very large study on BC (26).
However, it is difficult to make conclusions about the importance of these results in discriminating BRCA2 from BRCA1/BRCAX patients, since BRCA2 group consisted of a limited number of cases (n=3).
f) BRCAX vs. BRCA1/BRCA2 (n=10; n=7)
Following the comparison between BRCAX patients and those with BRCA1/BRCA2 mutations, 962 DMRs corresponding to 914 genes were reported. Following DAVID analysis, 3 enriched GO MF terms associated with GTPase regulatory activity were identified (Table SIVG). These results indicated that the DNA methylation levels of the GTPase genes may be a discriminatory factor between BRCA1/BRCA2 and BRCAX cases.
Discussion
Aberrant DNA methylation is an important and frequent event extensively studied in cancer, including BC. Published data have revealed that such epigenetics modifications are directly associated with tumor onset and progression. To the best of our knowledge, a comprehensive global DNA methylation study exploring differences in the methylome of female and male BC has not been performed by using Affymetrix human promoter arrays. This-omic platform allowed the quantification of the methylation levels of the CpG islands located in 25,500 human promoter regions. The DNA methylation profiles of 24 patients with familial BC were studied. Following the comparison between FBC and MBC, 2,486 significant differentially methylated genes were identified. The enrichment analysis suggested that most of the genes encompassed processes associated with the cytoskeleton composition and architecture such as ‘keratin filament’, ‘intermediate filament’ and ‘cytoskeletal part’. Of note, almost all genes included in the GO term: ‘Keratin filament’ were hypomethylated in FBC, as compared with MBC, suggesting their probable over-expression in the former, as compared to the latter. In particular, the hypomethylation of the cytokeratin genes KRT6A and KRT14 was observed in FBC, as compared with MBC. Keratins are considered to be immunohistochemical diagnostic tumor markers and several studies have provided evidence on active keratin involvement in cancer cell invasion and metastasis, as well as treatment responsiveness (27). The overexpression of these genes has been found to be positively correlated with a high tumor grade in BC and the expression of KRT6A and KRT14 to be frequently associated with basal molecular subtype (28).
Following the comparison between the FBC and MCB methylome, several differentially methylated genes that belonged to the RAS GTPase superfamily and whose role in cancer is well documented, were identified. The same results were obtained by limiting the comparison to FBC and MBC patients with BRCAX condition. The RAS GTPase superfamily is composed of 5 families: RHO, RAS, RAB, ARF and RAN; all these families were represented in our findings with numerous RHO genes found to be differentially methylated. Consequently, a different regulation of the expression of proteins in the RHO pathways in male and female BC may occur affecting cell migration and invasion. In fact, the RHO GTPase family plays an important role in cytoskeleton rearrangements and is a key regulator of processes involved in cellular adhesion, migration, proliferation, survival, differentiation and malignant transformation. The RHO family includes RHO-GEF and RHO-GAP proteins that are often deregulated not only in BC but also in several other tumor types (29).
With regard to other RAS GTPase families, RAB genes were found to be both hyper- and hypo-methylated when comparing FBC and MBC methylomes, which could likely down- or upregulate gene expression, respectively. More specifically, 12 members of the RAB-GAP were found to be differentially methylated in FBC, as compared with MBC. The RAB proteins are involved in a wide range of functions including the trafficking between Golgi and endosomes, phagocytosis and the assembly of adherent junctions and mitochondrial dynamics. Certain studies have reported the involvement of RAB GTPases in different types of cancer (30,31) included BC. In a study by Callari et al (32) the global gene expression of 53 FBC was compared to that of 37 MBC by microarray technology and the dysregulation of members of the RAS GTPase superfamily was observed, in line with the present results. Transcriptional alteration of genes belonging to all 5 families was also identify in that study. The authors hypothesized that there was a sex-related modulation of the cytoskeleton organization in BC cells that influenced the cancer invasion process. In combination, the present results suggested that genes involved in cytoskeleton dynamics such as keratins and genes of the RAS GTPase superfamily, have different DNA methylation levels in FBC, as compared to MBC. According to Callari et al (32), we hypothesized that the expression dysregulation, likely determined by the variations of DNA methylation, may influence these cancer aggressive properties, in which the cytoskeleton plays an essential role (i.e. adhesion, migration and invasion).
To identify novel patterns of DNA methylation specific to the different mutations (BRCA1, BRCA2 or BRCAX) only FBC cases were used, since that was the largest group in our cohort of patients. Below, the salient results found in the different comparison classes are discussed.
Following the comparison between patients with BRCA1 and BRCA2 mutations, variations were observed in the DNA methylation of genes involved in the ubiquitination pathway. BRCA1 works with BARD1 to catalyze the transfer of ubiquitin onto protein substrates. The RING domain contained in the N-terminal region of both BRCA1 and BARD1 is responsible for dimerization and ubiquitin ligase activity, which is required for its tumor suppressor function (33). Moreover, the missense mutations in the BRCA1 RING domain were identified in families with a high risk for BC. Therefore, the BRCA1 mutation could alter the ubiquitination pathway and in turn affects the DNA methylation level of the genes belonging to this pathway. The methylome comparison was performed on a limited number of cases; a larger number of patients may confirm this hypothesis.
Following the comparison between patients with BRCA1 and those with BRCA2 or BRCAX mutations, a hypo/hyper-methylation of genes encoding cytoskeletal binding proteins was observed, which may suggest a different expression modulation of genes involved in cytoskeletal dynamics.
The patients with BRCA1 enrolled in the present study had triple-negative/basal-like tumors while the BRCA2/BRCAX cases had luminal A/B tumors. The basal-like tumors originate from normal mammary myoepithelial cells and express genes associated with the normal myoepithelium such as high molecular weight cytokeratins (CK5/6, CK14 and CK17). Conversely, luminal A/B tumors originate from luminal cells of the breast duct and lobule and they express genes associated with luminal cells such as ER, low molecular weight cytokeratins (CK7, CK8, CK18 and CK19), as well as PGR, GATA3, BCL2 and other ER-induced genes (34). In this context, the different methylation pattern of the cytoskeletal binding proteins may be associated with the expression of distinct cytoskeleton-related proteins in tumors with distinct molecular sub-types.
Following the comparison between patients with BRCAX and those with BRCA1 or BRCA2 mutations, variations in DNA methylation were identified among genes associated with the GTPase regulatory activity. The results may help to obtain a better understanding of the biology of BRCAX groups, as compared to BRCA1/BRCA2 groups. Pending confirmation by a study with a larger number of cases, the evaluation of the GTPase regulatory activity-related genes methylation levels could characterize and distinguish these groups of patients. In conclusion, in the current study for the first time, the global DNA methylation was profiled in FBC and MBC patients with a positive family history by using the Affymetrix human promoter array platform. With regards to MBC, only Johansson et al (35) assessed genome-wide DNA methylation profiles using Illumina 450K Infinium methylation arrays and compared them with the transcriptional subgroups of MBC, luminal M1 and M2. They identified two epitypes through unsupervised clustering (ME1 and ME2) associated with the two transcriptional subgroups and the DNA methylation data underscored the heterogeneity of MBC, suggesting it should not be defined using the conventional criteria applied to FBC. The present data reported different DNA methylation levels of GTPase-related genes (RHO-GAP, RHO-GEF and RAB GTPase) and keratin-related genes, which are important components of cytoskeleton, between FBC and MBC. These results may help elucidate an aspect of the molecular differences between male and female BC. The comparisons of DNA methylation profiles among women with BRCA1, BRCA2 or BRCAX mutations led to several observations and conclusions. In patients with BRCA1 or BRCA2 mutations there may be a different modulation of the ubiquitination pathway. Different DNA methylation levels of genes crucial for cancer pathways were identified in patients with BRCA2, as compared to those with BRCAX or BRCA1/BRCAX mutations. Different DNA methylation levels of genes involved in cytoskeleton architecture were identified in patients with BRCA1, as compared to those with BRCA2/BRCAX mutations; this was consistent with the fact that BRCA1 tumors frequently exhibit a basal-like molecular subtype, while BRCA2/X tumors exhibit a luminal molecular subtype. Finally, different DNA methylation levels in certain GTPase genes were observed in BRCAX patients, as compared to BRCA1/2 cases; these results may help better identify and distinguish groups of patients carrying these mutations in the future. In a next prospective study we will increase the number of patients (especially cases of familial MBC) to carry out the analysis of the methylome. We will collect fresh BC biopsy specimens for gene expression analysis in order to correlate the most relevant hyper/hypo-methylated genes to their expression levels.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Marialuisa Crosatti (University of Leicester, Leicester, UK) for the linguistic revision of the manuscript, as well as Professor Marina Colombi and Professor Massimo Gennarelli, who were responsible for the Affymetrix platform at the Department of Molecular and Translational Medicine, Division of Biology and Genetics (University of Brescia, Brescia, Italy).
Funding Statement
The present study was supported by funding from Lega Italiana per la Lotta contro i Tumori (LILT) Roma-Brescia (grant no. 2762012), Comitato Nazionale Universitario (CNU)-Brescia, the Italian Ministry of University and Research (FFRB grant), the University of Brescia (local grants). EA was supported by a postdoctoral fellowship from Associazione Davide Rodella Onlus, Brescia. IG was supported by a Postdoctoral fellowship from-Ricerchiamo Onlus-, Brescia, Italy.
Funding
The present study was supported by funding from Lega Italiana per la Lotta contro i Tumori (LILT) Roma-Brescia (grant no. 2762012), Comitato Nazionale Universitario (CNU)-Brescia, the Italian Ministry of University and Research (FFRB grant), the University of Brescia (local grants). EA was supported by a postdoctoral fellowship from Associazione Davide Rodella Onlus, Brescia. IG was supported by a Postdoctoral fellowship from-Ricerchiamo Onlus-, Brescia, Italy.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus repository (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153636).
Authors' contributions
EA, AS and GDP conceived the project. EA performed the experiments, interpreted the results and generated the figures. AS and EA wrote the manuscript. IG contributed to the plotting of the data and their interpretation, as well as the revision of the manuscript. AS and GDP assessed and confirmed the authenticity of all raw data. GDP and MC made their intellectual contribution in the interpretation of the results and the critical revision of the manuscript. The medical doctors, EM and PC, contributed as geneticists; the medical doctors AC, PI, FZ, PLC and CTP provided the formalin-fixed paraffin-embedded tissues sections from patients with breast cancer; all medical doctors contributed to the acquisition of clinical data. Furthermore, EM, PC, AC, PI, FZ, PLC and CTP made substantive contributions to analysis and interpretation of experimental data. All co-authors critically revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Spedali Civili of Brescia (approval no. NP 1439; Brescia, Italy). Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J, Cardoso F. Breast cancer. Nat Rev Dis Prim. 2019;5:66. doi: 10.1038/s41572-019-0111-2. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi H, Ohno S, Sasaki Y, Matsuura M. Hereditary breast and ovarian cancer susceptibility genes (Review) Oncol Rep. 2013;30:1019–1029. doi: 10.3892/or.2013.2541. [DOI] [PubMed] [Google Scholar]
- 3.Marchina E, Fontana MG, Speziani M, Salvi A, Ricca G, Di Lorenzo D, Gervasi M, Caimi L, Barlati S. BRCA1 and BRCA2 genetic test in high risk patients and families: Counselling and management. Oncol Rep. 2010;24:1661–1667. doi: 10.3892/or_00001031. [DOI] [PubMed] [Google Scholar]
- 4.Wang YA, Jian JW, Hung CF, Peng HP, Yang CF, Cheng HCS, Yang AS. Germline breast cancer susceptibility gene mutations and breast cancer outcomes. BMC Cancer. 2018;18:315. doi: 10.1186/s12885-018-4229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melchor L, Benítez J. The complex genetic landscape of familial breast cancer. Hum Genet. 2013;132:845–863. doi: 10.1007/s00439-013-1299-y. [DOI] [PubMed] [Google Scholar]
- 6.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 7.Pinto R, Summa S, Pilato B, Tommasi S. DNA methylation and miRNAs regulation in hereditary breast cancer: Epigenetic changes, players in transcriptional and post-transcriptional regulation in hereditary breast cancer. Curr Mol Med. 2014;14:45–57. doi: 10.2174/1566524013666131203101405. [DOI] [PubMed] [Google Scholar]
- 8.Flanagan J, Kugler S, Waddell N, Johnstone CN, Marsh A, Henderson S, Simpson P, da Silva L, kConFab Investigators. Khanna K, et al. DNA methylome of familial breast cancer identifies distinct profiles defined by mutation status. Breast Cancer Res. 2010;86:420–33. doi: 10.1016/j.ajhg.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gucalp A, Traina TA, Eisner JR, Parker JS, Selitsky SR, Park BH, Elias AD, Baskin-Bey ES, Cardoso F. Male breast cancer: A disease distinct from female breast cancer. Breast Cancer Res Treat. 2019;173:37–48. doi: 10.1007/s10549-018-4921-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornegoor R, van Diest PJ, Buerger H, Korsching E. Tracing differences between male and female breast cancer: Both diseases own a different biology. Histopathology. 2015;67:888–897. doi: 10.1111/his.12727. [DOI] [PubMed] [Google Scholar]
- 11.Moncini S, Salvi A, Zuccotti P, Viero G, Quattrone A, Barlati S, De Petro G, Venturin M, Riva P. The role of miR-103 and miR-107 in regulation of CDK5R1 expression and in cellular migration. PLoS One. 2011;6:e20038. doi: 10.1371/journal.pone.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson I, Ringnér M, Hedenfalk I. The landscape of candidate driver genes differs between male and female breast cancer. PLoS One. 2013;8:e78299. doi: 10.1371/journal.pone.0078299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rizzolo P, Silvestri V, Tommasi S, Pinto R, Danza K, Falchetti M, Gulino M, Frati P, Ottini L. Male breast cancer: Genetics, epigenetics, and ethical aspects. Ann Oncol. 2013;24(Suppl 8):viii75–viii82. doi: 10.1093/annonc/mdt316. [DOI] [PubMed] [Google Scholar]
- 14.André S, Nunes SP, Silva F, Henrique R, Félix A, Jerónimo C. Analysis of epigenetic alterations in homologous recombination dna repair genes in male breast cancer. Int J Mol Sci. 2020;21:2715. doi: 10.3390/ijms21082715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kornegoor R, Moelans CB, Verschuur-Maes AH, Hogenes MCH, de Bruin PC, Oudejans JJ, van Diest PJ. Promoter hypermethylation in male breast cancer: Analysis by multiplex ligation-dependent probe amplification. Breast Cancer Res. 2012;14:R101. doi: 10.1186/bcr3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vermeulen MA, van Deurzen CHM, Doebar SC, de Leng WWJ, Martens JWM, van Diest PJ, Moelans CB. Promoter hypermethylation in ductal carcinoma in situ of the male breast. Endocr Relat Cancer. 2019;26:575–584. doi: 10.1530/ERC-18-0485. [DOI] [PubMed] [Google Scholar]
- 17.Pinto R, Pilato B, Ottini L, Lambo R, Simone G, Paradiso A, Tommasi S. Different methylation and MicroRNA expression pattern in male and female familial breast cancer. J Cell Physiol. 2013;228:1264–1269. doi: 10.1002/jcp.24281. [DOI] [PubMed] [Google Scholar]
- 18.Deb S, Gorringe KL, Pang JM, Byrne DJ, Takano EA, kConFab Investigators. Dobrovic A, Fox SB. BRCA2 carriers with male breast cancer show elevated tumour methylation. BMC Cancer. 2017;17:641. doi: 10.1186/s12885-017-3632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizzolo P, Silvestri V, Valentini V, Zelli V, Zanna I, Masala G, Bianchi S, Palli D, Ottini L. Gene-specific methylation profiles in BRCA-mutation positive and BRCA-mutation negative male breast cancers. Oncotarget. 2018;9:19783–19792. doi: 10.18632/oncotarget.24856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abeni E, Salvi A, Marchina E, Traversa M, Arici B, De Petro G. Sorafenib induces variations of the DNA methylome in HA22T/VGH human hepatocellular carcinoma-derived cells. Int J Oncol. 2017;51:128–144. doi: 10.3892/ijo.2017.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omura N, Li CP, Li A, Hong SM, Walter K, Jimeno A, Hidalgo M, Goggins M. Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol Ther. 2008;7:1146–1156. doi: 10.4161/cbt.7.7.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling G, Waxman DJ. DNase I digestion of isolated nulcei for genome-wide mapping of DNase hypersensitivity sites in chromatin. Methods Mol Biol. 2013;977:21–33. doi: 10.1007/978-1-62703-284-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Network, corp-author. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karantza V. Keratins in health and cancer: More than mere epithelial cell markers. Oncogene. 2011;30:127–138. doi: 10.1038/onc.2010.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao MM, Chan SK, Yu AM, Lam CC, Tsang JY, Lui PC, Law BK, Tan PH, Tse GM. Keratin expression in breast cancers. Virchows Arch. 2012;461:313–322. doi: 10.1007/s00428-012-1289-9. [DOI] [PubMed] [Google Scholar]
- 29.Wertheimer E, Gutierrez-Uzquiza A, Rosemblit C, Lopez-Haber C, Sosa MS, Kazanietz MG. Rac signaling in breast cancer: A tale of GEFs and GAPs. Cell Signal. 2012;24:353–362. doi: 10.1016/j.cellsig.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramani D, Alahari SK. Integrin-mediated function of Rab GTPases in cancer progression. Mol Cancer. 2010;9:312. doi: 10.1186/1476-4598-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishibashi K, Kanno E, Itoh T, Fukuda M. Identification and characterization of a novel Tre-2/Bub2/Cdc16 (TBC) protein that possesses Rab3A-GAP activity. Genes Cells. 2009;14:41–52. doi: 10.1111/j.1365-2443.2008.01251.x. [DOI] [PubMed] [Google Scholar]
- 32.Callari M, Cappelletti V, De Cecco L, Musella V, Miodini P, Veneroni S, Gariboldi M, Pierotti MA, Daidone MG. Gene expression analysis reveals a different transcriptomic landscape in female and male breast cancer. Breast Cancer Res Treat. 2011;127:601–610. doi: 10.1007/s10549-010-1015-8. [DOI] [PubMed] [Google Scholar]
- 33.Wu W, Koike A, Takeshita T, Ohta T. The ubiquitin E3 ligase activity of BRCA1 and its biological functions. Cell Div. 2008;3:1. doi: 10.1186/1747-1028-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertucci F, Finetti P, Birnbaum D. Basal breast cancer: A complex and deadly molecular subtype. Curr Mol Med. 2011;12:96–110. doi: 10.2174/156652412798376134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansson I, Lauss M, Holm K, Staaf J, Nilsson C, Fjällskog ML, Ringnér M, Hedenfalk I. Genome methylation patterns in male breast cancer-Identification of an epitype with hypermethylation of polycomb target genes. Mol Oncol. 2015;9:1565–1579. doi: 10.1016/j.molonc.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus repository (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153636).