Abstract
Non-small cell lung cancer (NSCLC) has high morbidity and mortality rates worldwide, and tumor metastasis is generally associated with poor prognosis. Chemotherapy resistance aggravates the challenges associated with treating NSCLC. Therefore, identifying effective targets and developing therapies based on these findings could bring novel perspectives for patients with metastatic NSCLC. The expression levels of receptor tyrosine-protein kinase erbB-2 (ERBB2) are associated with NSCLC progression. Differential microRNA (miR) expression profiles have been identified in tumors and can be used to identify multiple malignant phenotypes. miR-133a-3p expression is dysregulated in a variety of tumors. However, to the best of our knowledge, the association between miR-133a-3p and the NSCLC pathogenesis process has not been demonstrated yet. The present study revealed a decrease in miR-133a-3p expression in both tissues and cell lines, which was detected using reverse transcription-quantitative (RT-q)PCR, and western blotting and RT-qPCR demonstrated ERBB2 levels were increased at both protein and mRNA levels. Bioinformatics analysis and dual-luciferase reporter assays demonstrated that ERBB2 was a direct target of miR-133a-3p. Furthermore, MTT, wound healing and Transwell assays revealed that overexpression of miR-133a-3p suppressed proliferation, invasion and migration of NSCLC cells, respectively, effects that were inhibited following ERBB2 overexpression. In addition, immunofluorescence assays demonstrated that overexpression of ERBB2 upregulated N-cadherin expression, while E-cadherin expression was downregulated. In conclusion, the present data demonstrated that miR-133a-3p acted as a tumor suppressor by negatively regulating ERBB2 expression. The miR-133a-3p/ERBB2 axis may be a potential target for the diagnosis and treatment of NSCLC in the future.
Keywords: non-small cell lung cancer, microRNA-133a-3p, erb-b2 receptor tyrosine kinase 2, proliferation, invasion, migration
Introduction
Lung cancer is a common malignant tumor with high morbidity and mortality rates (1,2). In 2015, ~158,040 patients succumbed to lung cancer in the United States (3). Non-small cell lung cancer (NSCLC) accounts for 87% of all lung cancers, and 40% of NSCLC cases are diagnosed with tumor metastasis (3). Although targeted therapies, such as gefitinib, are available for patients with gene mutations, including EGFR mutations and anaplastic lymphoma kinase rearrangements, these mutations are not common in NSCLC (4). Therefore, most patients are treated with conventional chemotherapy (5). Systemic chemotherapy can improve survival and alleviate disease-related symptoms. However, a large proportion of patients develop drug resistance after several chemotherapy courses, resulting in tumor relapse, invasion and metastasis (6,7). Therefore, it would be of great clinical value to identify novel, potent targets for the treatment of NSCLC, and to explore their association with invasion and metastasis of NSCLC.
Receptor tyrosine-protein kinase erbB-2 (ERBB2) is a 185 kDa cell membrane receptor encoded by the oncogene erbB-2, which is a member of the EGFR family. ERBB2 is involved in multiple cell events, such as proliferation, differentiation and apoptosis (8,9). ERBB2 is highly expressed in multiple malignant tumors, such as breast cancer, ovarian cancer and hepatocellular carcinoma (8,10–13). Downregulation of ERBB2 suppresses the proliferation and invasion of breast cancer and gastric cancer cells (12,14,15). ERBB2 expression is positively associated with NSCLC, and high expression levels of ERBB2 are associated with resistance to radiotherapy and chemotherapy (16,17). Therefore, ERBB2 may be a potential prognostic biomarker and therapeutic target for NSCLC.
MicroRNAs (miRNAs/miRs) are endogenous short non-coding sequences with a length of 20–24 bases. miRNAs serve a role in multiple biological processes, such as cell proliferation, differentiation and apoptosis, by regulating protein expression at the posttranscriptional level (18,19). miRNAs regulate mRNAs by binding to the 3′ untranslated region (3′UTR) end of mRNA, degrading mRNA or repressing protein translation. miR-133a is highly conserved and widely expressed in various organisms, and can be expressed as the subtypes miR-133a-3p and miR-133a-5p (20). In a variety of cancer types, including gastric cancer, bladder cancer and oral squamous cell carcinoma, miR-133a-3p expression is downregulated (21–23). Yang et al (24) reported that patients with NSCLC with high miR-133a expression in tumor cells had a favorable prognosis. To the best of our knowledge, no reports have demonstrated the association between miR-133a-3p and NSCLC pathogenesis.
The present study aimed to investigate the association between dysregulated miR-133a-3p expression and NSCLC pathogenesis, and to explore the role of miR-133a-3p and ERBB2 in NSCLC carcinogenesis. The expression of miR-133a-3p and ERBB2 in NSCLC tissues and cell lines was detected using reverse transcription-quantitative (RT-q)PCR and western blotting, respectively. The effects of miR-133a-3p on the malignant phenotypes of NSCLC cells were then evaluated by multiple experiments, such as MTT and Transwell assays, and the underlying mechanisms were further investigated. The present data indicate that the miR-133a-3p/ERBB2 axis could be a potential therapeutic target of NSCLC.
Materials and methods
Sample collection
Material was collected from 77 patients (45 males and 32 females, aged 66.58±14.38 years) primarily diagnosed with NSCLC who underwent primary surgery at Zhejiang Hospital (Hangzhou, China) between January 2010 and January 2017. None of the participants received chemotherapy or radiotherapy before surgery and none of the participants was diagnosed other malignant diseases before. The present study was conducted in accordance with the Helsinki Declaration, and all patients or direct relatives consented to participate in the study. Furthermore, the present study was approved by the Ethics Committee of Zhejiang Hospital. After surgery, the tumor and adjacent healthy tissues (≥1-cm from the edge of tumor) were collected and stored in liquid nitrogen (−196°C).
Cell culture and transfection
The normal human lung epithelial cell line BEAS-2B and human NSCLC HCC827 and H1299 cell lines were purchased from the Institute of Shanghai Biochemistry and Cell Biology. BEAS-2B was cultured in BEBM medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (HyClone; Cyvita). HCC827 and H1299 cells were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 1% penicillin/streptomycin (100 U/ml penicillin and 0.1 mg/ml streptomycin; Thermo Fisher Scientific, Inc.). All cells were cultured with 5% CO2 at 37°C.
For transfection, HCC827 and H1299 were collected and seeded in 6-well plates at the density of 1×105/well. After incubated for 24 h, cells treated with 100 nM mimic control (5′-CAGCUGGUUGAAGGGGACCAAA-3′; Shanghai GenePharma Co., Ltd.), 75 nM miR-133a-3p mimic (5′-UUUGGUCCCCUUCAACCAGCUG-3′; Shanghai GenePharma Co., Ltd.), 600 ng/well pcDNAn empty vector negative control (pcDNA-NC; Shanghai GenePharma Co., Ltd.) or 600 ng/well pcDNA-ERBB2 (Shanghai GenePharma Co., Ltd.) using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturers' protocols. The 1 µg nucleic acid fragment and 2 µl Lipofectamine 3000 were mixed with 1 ml RPMI-1640 medium without serum, and then incubated for 15 min at room temperature. Then, the mixture was mixed with 4 ml serum-free RPMI-1640 medium and added into plates. After cultured for 24 h, the culture medium was removed and replaced with RPMI-1640 medium containing 10% FBS. After 24–72 h, cells were used for subsequent experiments.
MTT assay
Following treatment with control mimics, miR-133a-3p mimics or pcDNA-ERBB2, H1299 and HCC827 cells were collected and seeded into 96-well plates at the density of 1×104 cells/well. After culturing for 24, 48 and 72 h, medium was removed and replaced with medium containing MTT (KGA312; Nanjing KeyGen Biotech Co., Ltd.) at a final concentration of 0.5 mg/ml and cells were cultured for another 4 h at 37°C. Finally, the medium was replaced with 200 µl DMSO. After 10 min, the absorbance at 490 nm was measured using a spectrometer (Thermo Fisher Scientific, Inc.).
Western blotting
The protein expression levels of ERBB2 were assessed by western blotting. Following treated with different nucleotide fragments as aforementioned, cells were collected and lysed in RIPA buffer (Nanjing KeyGen Biotech Co., Ltd.). Samples were quantified using a Bicinchoninic Acid Assay kit (Beyotime Institute of Biotechnology). Samples (50 µg per lane) were separated using 10% SDS-PAGE and then transferred onto 0.45-µm PVDF membranes (Merck KGaA). The membranes were blocked with 5% skim milk powder solution, followed by incubation overnight at 4°C with antibodies against ERBB2 (dilution, 1:1,000; 18299-1-AP; ProteinTech Group, Inc.) and GAPDH (dilution, 1:3,000; 10494-1-AP; ProteinTech Group, Inc.). Subsequently, the membranes were incubated with HRP-conjugated Affinipure Rabbit Anti-Goat IgG(H+L) (dilution, 1:5,000; SA00001-4; ProteinTech Group, Inc.) for 2 h at 37°C. Subsequently, bands were detected by incubation with ECL mixture solution (Advansta Inc.), and immediately detected using a Bio-Rad Gel Imaging System (Bio-Rad Laboratories, Inc.). ImageJ software (version 1.8.0; National Institutes of Health) was used to analyze the intensity of the bands.
Reverse transcription-quantitative PCR (RT-qPCR)
After undergoing different treatments, cells were collected and lysed using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) to extract total RNA. RNA was transcribed into cDNA using a cDNA reverse transcription kit (Takara Bio, Inc.). The conditions used for reverse transcription were as follows: 42°C For 15 min, then 5 sec at 85°C and storage at 4°C for further analysis. Subsequently, cDNA samples were amplified using an RT-qPCR (Applied Biosystems; Thermo Fisher Scientific, Inc.) system and SYBR® Green Premix Ex Taq™ (Takara Bio, Inc.) according to the manufacturer's protocols. Thermocycling conditions for qPCR were: Pre-denaturation for 30 sec at 95°C; followed by 40 cycles of 5 sec at 95°C and 30 sec at 60°C; and dissociation at 95°C for 15 sec, 60°C for 30 sec and 95°C for 15 sec. GAPDH and U6 were used as the reference genes to normalize the expression levels of ERBB2 and miR-133a-3p, respectively. mRNA and miRNA expression levels were quantified using the 2−∆∆Cq method (25). The primers used were as follows: ERBB2 forward, 5′-CCAGCCTTCGACAACCTCTATT-3′ and reverse, 5′-TGCCGTAGGTGTCCCTTTG-3′; miR-133a-5p forward, 5′-CTTTAACCATTCTAGCTTTTCCAGGTA−3′ and reverse, 5′-GACTTCGGCTGTGGACAAGATTAG−3′; U6 forward, 5′-CGCTTCGGCAGCACATATACTA-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCA-3′; and GAPDH forward, 5′-AGGTCGGTGTGAACGGATTTG−3′ and reverse, 5′-GGGGTCGTTGATGGCAACA-3′.
Wound healing assay
The present study measured the cell migration ability using a wound healing assay. After seeding into 24-well plates at the density of 3×105 cells per well, H1299 and HCC827 cells were transfected with different nucleotide fragments as aforementioned. After cell reached 80% confluence, scratches were created across the monolayer using a 10-µl pipette tip, followed by washing with cold PBS. An image of each scratch was captured under a light microscope. Subsequently, cells were incubated in RPMI-1640 medium containing 1% FBS (26). After culturing for 24 h at 37°C, images of the scratches were captured again. The migration distance of each group was measured using ImageJ software, and analyzed with the following formula: (W 0 h-W 24 h)/W 0 h ×100%, where W is the wound.
Cell invasion assay
A Transwell assay was performed to detect the cell invasion ability following the overexpression of miR-133a-3p or ERBB2. The Transwell inserts were purchased from Corning, Inc. Prior to the experiment, the upper chambers were covered with 100 µl of a 1:1 mixture of Matrigel and RPMI-1640 medium and incubated at 37°C for 1 h. NSCLC cells were trypsinized, resuspended in serum-free RPMI-1640 medium, and then seeded into the upper chambers (2×104 cells/well), while medium containing 20% FBS was placed in the lower chambers. After culturing at 37°C for 24 h, non-invaded cells in the upper chamber were wiped off using sterile cotton swabs. The invaded cells were fixed using 4% paraformaldehyde at room temperature for 10 min and stained with 0.1% crystal violet solution for 20 min at room temperature. The invaded cells were imaged using an light microscope (magnification, ×400).
Dual-luciferase reporter gene assay
Following bioinformatics analysis of the association between miR-133a-3p and the 3′UTR of the ERBB2 transcript (http://www.targetscan.org/vert_72/), the dual-luciferase reporter gene assay was employed using TargetScanHuman 7.2 to verify this prediction. The 3′UTR clones of the ERBB2 transcript, including mutant (MUT) 3′UTRs and wild type (WT) 3′UTRs, were designed and synthesized by Sangon Biotech Co., Ltd. The nucleotide fragments were ligated into the luciferase plasmid (Promega Corporation). H1299 cells were seeded into 24-well plates at the density of 1×105/well, and co-transfected with 300 ng/well plasmid for pmir/ERBB2-WT or pmir/ERBB2-MUT and 50 ng/well miR-133a-3p mimics or miR-negative control (NC) and 100 ng Renilla luciferase plasmid (pRL-TK; Promega Corporation) using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), and H1299 cells transfected with pRL-TK alone were regarded as the negative control, followed by incubation for 48 h at 37°C. Finally, cells were lysed and the luciferase activities in different groups were detected using a Dual-Luciferase Reporter Assay System (Promega Corporation) following the manufacturer's guidance. Firefly luciferase activities were normalized to Renilla luciferase activities.
Immunofluorescence assay
Immunofluorescence assays were carried out to evaluate E-cadherin and N-cadherin expression. A glass slide was placed in a 6-well plate, and 2×105 cells/well were seeded into the plate. After 24 h, the medium was removed and cells were fixed with 4% paraformaldehyde at 4°C for 30 min. Then, cells were blocked with 10% goat serum at room temperature (22±5°C) for 30 min. Subsequently, cells were incubated with anti-N-cadherin (dilution, 1:1,000; 22018-1-AP; ProteinTech Group, Inc.) or anti-E-cadherin (dilution, 1:1,000; 20874-1-AP; ProteinTech Group, Inc.) antibodies overnight at 4°C. Afterwards, cells were incubated with the Fluorescein-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) secondary antibody (dilution, 1:100; SA00003-2; ProteinTech Group, Inc.) at room temperature for 1 h, followed by staining using DAPI at room temperature for 5 min. Finally, images were captured using a fluorescence microscope (magnification, ×400).
Statistical analysis
The data analysis in the present study was conducted using SPSS 23.0 (IBM Corp.) and GraphPad 6.0 software (GraphPad Software, Inc.). Student's t-test was carried out to compare differences between two groups. Statistical differences among three groups were analyzed using one-way ANOVA followed by Duncan's post hoc test (LSR) at the 95% confidence interval (P<0.05 was considered to indicate statistical significance). Statistical differences among four groups were analyzed using one-way ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference. Data were expressed as mean ± standard deviation. Each assay was repeated three times.
Results
Expression levels of miR-133a-3p and ERBB2 are dysregulated in NSCLC tissues and cell lines
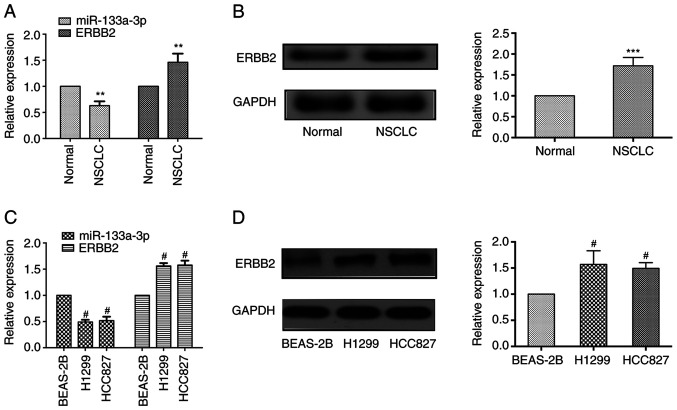
The present study assessed miR133a-3p expression at the mRNA level and ERBB2 expression at the mRNA and protein levels in NSCLC tissues and normal adjacent tissues. As shown in Fig. 1A, RT-qPCR results revealed that miR-133a-3p expression was markedly downregulated in NSCLC tissues compared with adjacent healthy tissues, while the mRNA expression levels of ERBB2 were upregulated in cancer tissues. Western blotting demonstrated that the protein expression levels of ERBB2 were elevated in NSCLC tissues (Fig. 1B).
Figure 1.
miR-133a-3p and ERBB2 expression are dysregulated in both NSCLC tissues and cell lines. (A) miR-133a-3p expression was downregulated in cancer tissues at the mRNA level, whereas ERBB2 expression was upregulated in cancer tissues compared with in normal tissues. (B) ERBB2 protein expression was increased in NSCLC tissues. (C) miR-133a-3p expression was downregulated in NSCLC cell lines (H1299 and HCC827) at the mRNA level, while ERBB2 expression was upregulated. (D) ERBB2 expression was increased in NSCLC cell lines at the protein level. **P<0.01 and ***P<0.001 vs. normal. #P<0.05 vs. BEAS-2B. ERBB2, erb-b2 receptor tyrosine kinase 2; miR-133a-3p, microRNA-133a-3p; NSCLC, non-small cell lung cancer.
In addition, the present study evaluated the expression levels of these molecules in NSCLC cells. Consistent with the results identified in tissue samples, lower miR-133a-3p expression was observed in NSCLC cells (H1299 and HCC827 cell lines), while ERBB2 expression was upregulated both at the mRNA and protein levels compared with that in the normal pulmonary epithelial cells (BEAS-2B; Fig. 1C and D). These results indicated that miR-133a-3p expression was decreased, while ERBB2 expression was increased in NSCLC tissues and cells.
Overexpression of miR-133a-3p inhibits NSCLC cell proliferation, migration and invasion
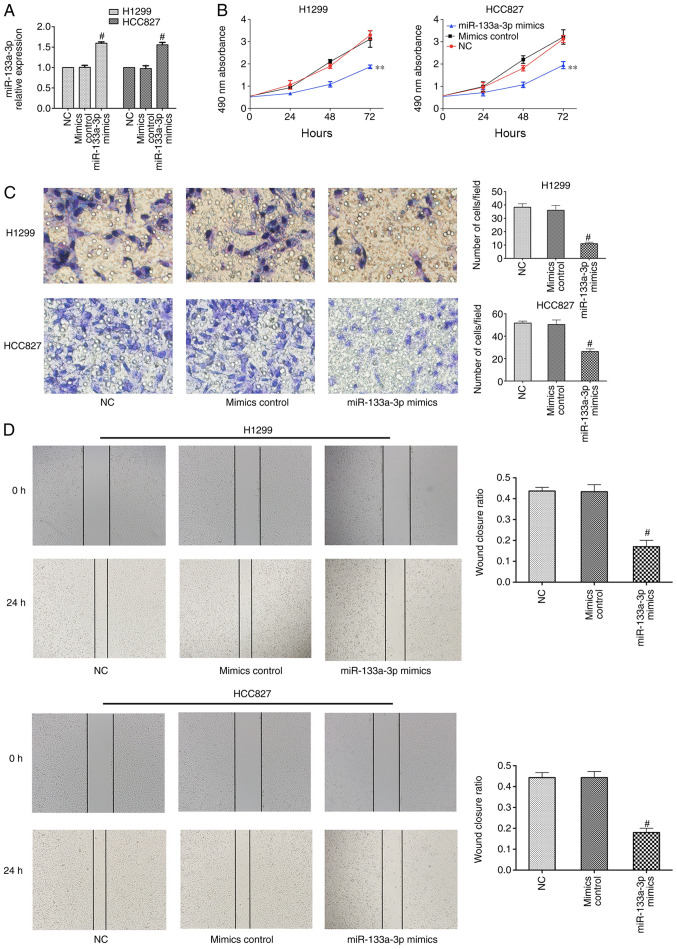
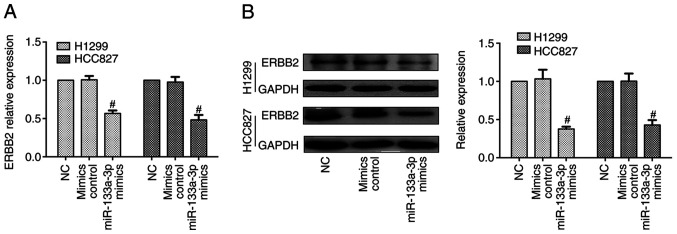
To determine the biological role of miR-133a-3p, H1299 and HCC827 cells were transfected with control or miR-133a-3p mimic. RT-qPCR was performed to evaluate transfection efficiency, which demonstrated that miR-133a-3p expression was significantly increased after transfection with miR-133a-3p mimic (Fig. 2A). An MTT assay was used to assess the effect of miR-133a-3p on NSCLC cell proliferation. As shown in Fig. 2B, overexpression of miR-133a-3p led to a suppression of proliferation in NSCLC cells. The Transwell assay indicated that overexpression of miR133a-3p inhibited the invasion abilities of these cells (Fig. 2C). In addition, the wound healing assay indicated the inhibitory effects of miR-133a-3p on migration (Fig. 2D). Therefore, the present results demonstrated that overexpression of miR-133a-3p inhibited the proliferation, migration and invasion of NSCLC cells. Furthermore, RT-qPCR and western blotting data indicated that the mRNA and protein expression levels of ERBB2 were decreased in the miR-133a-3p mimics group, compared with mimics control groups (Fig. 3).
Figure 2.
Overexpression of miR-133a-3p inhibits non-small cell lung cancer cell proliferation, migration and invasion. (A) miR-133a-3p mimics transfection successfully increased the miR-133a-3p levels in H1299 and HCC827 cells. (B) Proliferation was suppressed in cells transfected with miR-133a-3p mimics. (C) Invasion abilities were suppressed in cells transfected with miR-133a-3p mimics. Magnification, ×400. (D) Migration abilities were inhibited in cells following overexpression of miR-133a-3p. Magnification, ×400. **P<0.01 vs. NC. #P<0.05 vs. NC. miR-133a-3p, microRNA-133a-3p; NC, negative control.
Figure 3.
Overexpression of miR-133a-3p suppresses ERBB2 expression. (A) ERBB2 expression was decreased in non-small cell lung cancer cell lines (H1299 and HCC827) at the mRNA level following miR-133a-3p mimics treatment. (B) ERBB2 protein expression was decreased after miR-133a-3p mimic transfection. #P<0.05 vs. NC. ERBB2, erb-b2 receptor tyrosine kinase 2; miR-133a-3p, microRNA-133a-3p; NC, negative control.
Overall, upregulation of ERBB2 and decreased expression levels of miR-133a-3p were observed in NSCLC tissues and cell lines. Downregulation of ERBB2 expression was observed in cells transfected with miR-133a-3p mimic, indicating that ERBB2 may be an indirect or direct target of miR-133a-3p.
miR-133a-3p directly targets the 3′UTR of the ERBB2 transcript in NSCLC cells
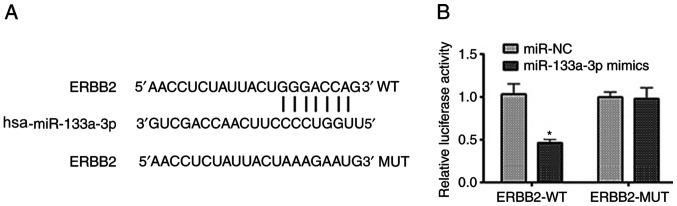
As aforementioned, a negative association between ERBB2 and miR-133-3p expression was noted. To further analyze this interaction, bioinformatics analysis was performed using TargetScan (Fig. 4A). This suggested that the 3′UTR of the ERBB2 transcript is a potential target of miR-133a-3p. A dual-luciferase reporter assay was performed to verify this prediction. As shown in Fig. 4B, miR-133a-3p overexpression markedly suppressed the luciferase activity in cells transfected with pmir/ERBB2-WT, while the luciferase activity was not changed in cells transfected with pmir/ERBB2-MUT. Therefore, these data provide evidence that miR-133a-3p directly targets the 3′UTR of the ERBB2 transcript in NSCLC cells.
Figure 4.
miR-133a-3p directly targets the 3′ untranslated region of the ERBB2 transcript in non-small cell lung cancer cells. (A) Bioinformatics analysis of miR-133a-3p and ERBB2. (B) Co-transfection of miR-133a-3p and pmir/ERBB2-WT markedly suppressed the luciferase activity. *P<0.05 vs. miR-NC. ERBB2, erb-b2 receptor tyrosine kinase 2; miR-133a-3p, microRNA-133a-3p; NC, negative control; WT, wild type; MUT, mutant.
miR-133a-3p inhibits the proliferation, invasion and migration of NSCLC cells by negatively regulating ERBB2 expression
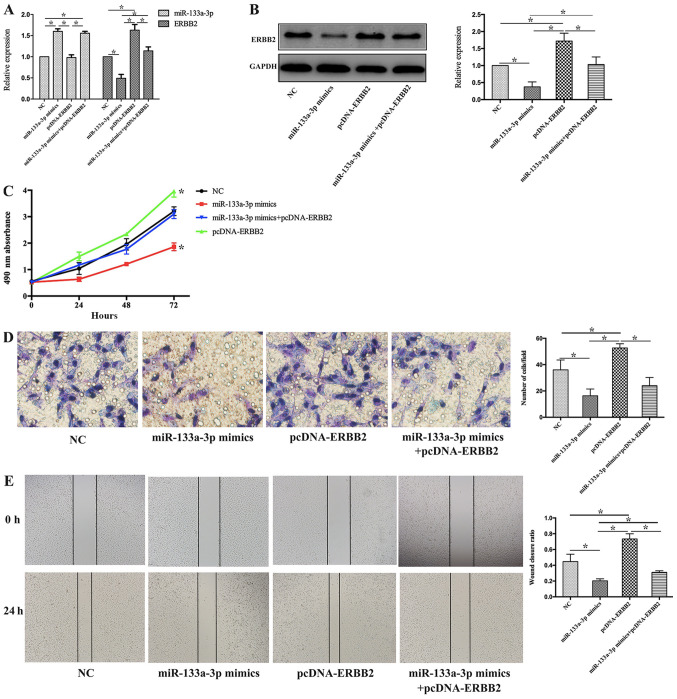
To further explore the biological functions of the miR-133a-3p/ERBB2 axis in NSCLC progression, H1299 cells were treated with miR-133a-3p mimic, pcDNA-ERBB2 or miR-133a-3p mimic + pcDNA-ERBB2. pcDNA-NC and pcDNA-ERBB2 were transfected into H1299 cells, and pcDNA-NC did not affect the expression levels of ERBB2 in H1299 cells (Fig. S1).
RT-qPCR and western blotting were performed to detect miR-133a-3p and ERBB2 expression in each treatment group. As shown in Fig. 5A, miR-133a-3p expression was increased in both the miR-133a-3p mimic and miR-133a-3p mimic + pcDNA-ERBB2 groups but was not significantly altered in the pcDNA-ERBB2 group compared with the NC group. Furthermore, ERBB2 expression was increased in the pcDNA-ERBB2 group, while it was decreased in the miR-133a-3p mimics group, compared with non-treated (NC) group. Compared with the pcDNA-ERBB2 group, ERBB2 expression was suppressed in the miR-133a-3p mimic + pcDNA-ERBB2 group (Fig. 5B).
Figure 5.
miR-133a-3p inhibits the proliferation, invasion and migration of non-small cell lung cancer cells by negatively regulating ERBB2. (A) At the mRNA level, the expression levels of miR-133a-3p were increased in the miR-133a-3p mimic and miR-133a-3p mimic + pcDNA-ERBB2 groups. ERBB2 expression was increased in the pcDNA-ERBB2 group, while it was decreased in cells transfected with miR-133a-3p mimic. (B) ERBB2 protein expression was suppressed in cells transected with miR-133a-3p mimic. (C) MTT assay data demonstrated that upregulated miR-133a-3p and ERBB2 in combination preserved the H1299 cell proliferation activity compared with cells transfected with pcDNA-ERBB2 alone. (D) The invasion ability was attenuated in H1299 cells transfected with miR-133a-3p mimic and pcDNA-ERBB2 in combination compared with cells transfected with pcDNA-ERBB2 alone. Magnification, ×400. (E) Wound healing assays suggested that co-transfection with miR-133a-3p mimic and pcDNA-ERBB2 decreased the wound closure ratio in H1299 cells compared with transfection with pcDNA-ERBB2 alone. Magnification, ×400. *P<0.05 vs. NC. ERBB2, erb-b2 receptor tyrosine kinase 2; miR-133a-3p, microRNA-133a-3p; NC, negative control.
MTT, Transwell and wound healing assays were performed to demonstrate the potential role of the miR-133a-3p/ERBB2 axis in NSCLC cell phenotypes. As shown in Fig. 5C, the MTT data demonstrated that overexpression of miR-133a-3p and ERBB2 in combination preserved the proliferation activity of H1299 cells, which was suppressed in miR-133a-3p mimics group. Compared with pcDNA-ERBB2 group, the proliferation of H1299 cells in miR-133a-3p mimics+pcDNA-ERBB2 group decreased. In addition, the invasive ability was attenuated in H1299 cells transfected with both miR-133a-3p mimics and pcDNA-ERBB2 (miR-133a-3p mimics+pcDNA-ERBB2 group), compared with pcDNA-ERBB2 group (Fig. 5D). Finally, the wound healing assay suggested that the co-transfection of miR-133a-3p mimic and pcDNA-ERBB2 decreased the wound closure ratio in H1299 cells compared with that in cells in the pcDNA-ERBB2 group (Fig. 5E). These results suggest that miR-133a-3p inhibits cell proliferation, invasion and migration of NSCLC cells by negatively regulating ERBB2 expression.
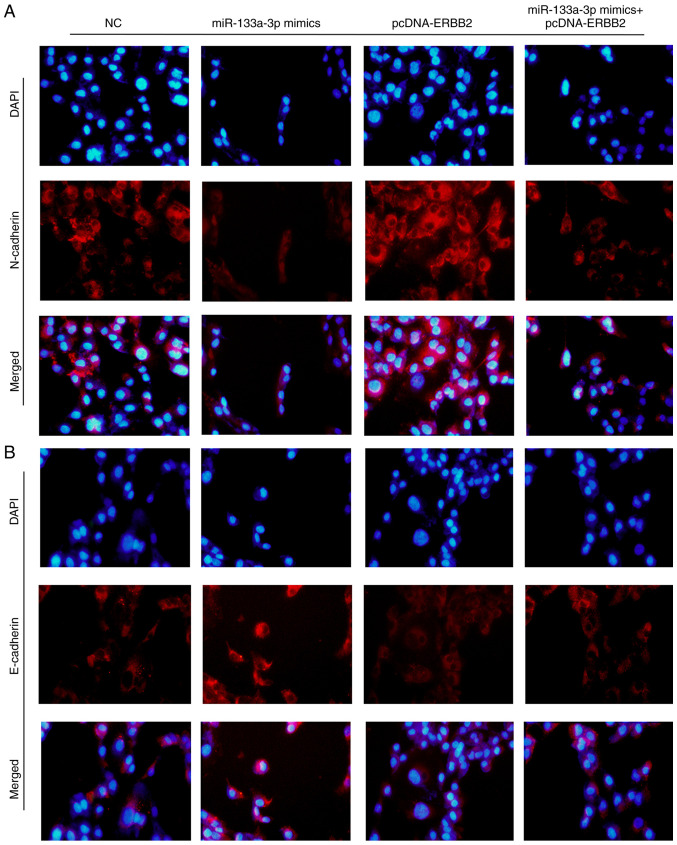
Overexpression of miR-133a-3p regulates the expression levels of epithelial-mesenchymal transition (EMT) biomarkers
E-cadherin and N-cadherin are two important biomarkers of the EMT process. The present study detected E-cadherin and N-cadherin expression using an immunofluorescence assay following transfection with miR-133a-3p mimics, pcDNA-ERBB2 or miR-133a-3p mimics + pcDNA-ERBB2. As shown in Fig. 6A, N-cadherin expression was decreased in the miR-133a-3p mimics group but increased in the pcDNA-ERBB2 group. Consistently, E-cadherin exhibited the opposite trends (Fig. 6B). These results indicate that the overexpression of miR-133a-3p suppresses the EMT process in NSCLC cells.
Figure 6.
Overexpression of miR-133a-3p regulates the expression levels of epithelial-mesenchymal transition biomarkers. (A) N-cadherin expression was decreased in the miR-133a-3p group, while it was increased in the pcDNA-ERBB2 group. Overexpression of miR-133a-3p and ERBB2 rescued the expression levels of N-cadherin. (B) E-cadherin expression was increased in the miR-133a-3p group, whereas it was decreased in the pcDNA-ERBB2 group. Overexpression of miR-133a-3p and ERBB2 rescued the expression levels of E-cadherin. Magnification, ×400. ERBB2, erb-b2 receptor tyrosine kinase 2; miR-133a-3p, microRNA-133a-3p; NC, negative control.
Discussion
At present, surgery and chemotherapy are the mainstream treatment modalities for NSCLC (27). However, the outcome of these treatments is unsatisfactory, since postoperative recurrence and therapy resistance are common in patients with NSCLC (28). With the development of molecular biology, targeted therapy has become available for malignant tumors (29). However, its efficacy remains limited, since most patients eventually develop drug resistance. Therefore, there is still an urgent need for the identification of novel therapeutic targets. In the last decade, miRNAs have become a hot topic in cancer diagnosis and treatment (18). miRNAs are a type of short, non-coding RNA molecules that serve critical regulatory roles in multiple cellular events. Increasing evidence has suggested that miRNA dysregulation is closely associated with the pathogenesis and progression of a variety of diseases, in particular cancer types (30), for example miR-133a-3p expression is downregulated in gastric cancer (21). With the increased understanding of miRNAs, the feasibility of using miRNAs as therapeutic targets is supported (31,32).
Previous studies have suggested a tumor-suppressive role of miR-133a-3p in multiple malignant tumors, including gastric, breast and bladder cancer (21,22,33). Low expression levels of miR-133a-3p are associated with poor prognosis and faster progression (22). Shi et al (33) reported that downregulation of miR-133a-3p promotes the proliferation and invasion of breast cancer cells. Li et al (34) suggested that miR-133a-3p promotes autophagy in gastric cancer cells by negatively regulating forehead box P3. The present study identified decreased miR-133a-3p expression in NSCLC cancer tissues and cell lines. A series of cellular and molecular experiments were performed to identify its biological function in NSCLC pathogenesis. The present results indicated that overexpression of miR-133a-3p effectively suppressed the cell proliferation and invasion of NSCLC cells.
To further assess the molecular mechanisms underlying the tumor-suppressive effect of miR-133a-3p, bioinformatics analysis was performed using TargetScan 7.2. Among the potential target genes, the present study focused on ERBB2, a well-known oncogene in NSCLC (35). ERBB2 is a member of the EGFR family, which is involved in a series of biological events in malignant diseases (36). It has been suggested that ERBB2 could trigger several cellular processes, including cell proliferation, survival and differentiation (8,37). Spencer et al (38) provided evidence that the ERBB2 receptor assists the binding of p130Cas to CRK, and thus, promotes the invasion and migration of breast cancer cells. Furthermore, ERBB2 expression is upregulated in various malignancies, and ERBB2-targeted therapy has become an important component of therapeutic strategies in a number of cancer types (12). In the present study, the luciferase assay supported the notion that miR-133a-3p could target the 3′UTR of ERBB2. Furthermore, a pcDNA-ERBB2 vector was employed to increase ERBB2 expression, and this reversed the effects of the miR-133a-3p mimic on cell proliferation, invasion and migration. Therefore, the tumor-suppressive effect of miR-133a-3p may be mediated, at least in part, by inhibition of ERBB2 expression.
EMT is an essential event in tumor carcinogenesis and progression. As a member of the EGFR family, ERBB2 has been demonstrated to promote EMT in previous study (39). The present study detected the typical EMT biomarkers E-cadherin and N-cadherin and demonstrated that overexpression of miR-133a-3p markedly suppressed the EMT process in NSCLC cells, while overexpression of ERBB2 ameliorated this effect. Therefore, the present findings suggest that miR-133a-3p may suppress EMT by targeting ERBB2.
There were some limitations to the present study. Commonly in wound healing assays non-FBS medium should be used; however, 1% FBS was used in the present study to prevent cell starving. Also, the present study only provided in vitro data, therefore further in vivo experiments, such as tumorigenesis assay, should be performed in future. Besides, further analysis of whether miR-133a-3p can regulate other signaling pathways impacting the phenotypes of NSCLC are needed.
In conclusion, the present results indicate that miR-133a-3p acts as a tumor-suppressive gene via inhibition of the proliferation, invasion and EMT of NSCLC cells. In particular, the inhibitory effects of miR-133a-3p on ERBB2 indicated that miR-133a-3p may be a potent antitumor target for NSCLC treatment.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
The majority of experiments and manuscript were finished by YX. YX performed statistical analysis and interpreted the data. LZ contribute to bioinformatics analysis and dual luciferase reporter gene experiment. LX and XZ designed the research, collected the tissues and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was conducted in accordance with the Helsinki Declaration guidelines, and written consent to participation in the present study were obtained from all patients or their direct relatives. Furthermore, the present study was approved by The Ethics Committee of Zhejiang Hospital (Hangzhou, China; approval no. 20100103).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Duffy SW, Field JK. Mortality reduction with low-dose CT screening for lung cancer. N Engl J Med. 2020;382:572–573. doi: 10.1056/NEJMe1916361. [DOI] [PubMed] [Google Scholar]
- 2.Qi M, Dai D, Liu J, Li Z, Liang P, Wang Y, Cheng L, Zhan Y, An Z, Song Y, et al. AIM2 promotes the development of non-small cell lung cancer by modulating mitochondrial dynamics. Oncogene. 2020;39:2707–2723. doi: 10.1038/s41388-020-1176-9. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Qian G, Zong D, Fan S, Owonikoko TK, Ramalingam SS, Sun SY. Overcoming acquired resistance of epidermal growth factor receptor-mutant non-small cell lung cancer cells to osimertinib by combining osimertinib with the histone deacetylase inhibitor panobinostat (LBH589) Cancer. 2020;126:2024–2033. doi: 10.1002/cncr.32744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgensztern D, Ng SH, Gao F, Govindan R. Trends in stage distribution for patients with non-small cell lung cancer: A national cancer database survey. J Thorac Oncol. 2010;5:29–33. doi: 10.1097/JTO.0b013e3181c5920c. [DOI] [PubMed] [Google Scholar]
- 6.Du L, Morgensztern D. Chemotherapy for advanced-stage non-small cell lung cancer. Cancer J. 2015;21:366–370. doi: 10.1097/PPO.0000000000000141. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Q, Zhou L, Guan Y, Cheng Y, Han X. BENC-511, a novel PI3K inhibitor, suppresses metastasis of non-small cell lung cancer cells by modulating β-catenin/ZEB1 regulatory loop. Chem Biol Interact. 2018;294:18–27. doi: 10.1016/j.cbi.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Yang ZY, Yang L, Xu CW, Wang XJ, Lei L. An insertion mutation of ERBB2 enhances breast cancer cell growth and confers resistance to lapatinib through AKT signaling pathway. Biol Open. 2020;9:bio047662. doi: 10.1242/bio.047662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honkoop H, de Bakker DE, Aharonov A, Kruse F, Shakked A, Nguyen PD, de Heus C, Garric L, Muraro MJ, Shoffner A, et al. Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. Elife. 2019;8:e50163. doi: 10.7554/eLife.50163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albrecht T, Rausch M, Rössler S, Albrecht M, Braun JD, Geissler V, Mehrabi A, Vogel MN, Pathil-Warth A, Mechtersheimer G, et al. HER2 gene (ERBB2) amplification is a rare event in non-liver-fluke associated cholangiocarcinogenesis. BMC Cancer. 2019;19:1191. doi: 10.1186/s12885-019-6320-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menderes G, Bonazzoli E, Bellone S, Black J, Altwerger G, Masserdotti A, Pettinella F, Zammataro L, Buza N, Hui P, et al. SYD985, a novel duocarmycin-based HER2-targeting antibody-drug conjugate, shows promising antitumor activity in epithelial ovarian carcinoma with HER2/Neu expression. Gynecol Oncol. 2017;146:179–186. doi: 10.1016/j.ygyno.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed S, Sami A, Xiang J. HER2-directed therapy: Current treatment options for HER2-positive breast cancer. Breast Cancer. 2015;22:101–116. doi: 10.1007/s12282-015-0587-x. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Jia L, Ding Y. Upregulation of miR-375 inhibits human liver cancer cell growth by modulating cell proliferation and apoptosis via targeting ErbB2. Oncol Lett. 2018;16:3319–3326. doi: 10.3892/ol.2018.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rimawi MF, Schiff R, Osborne CK. Targeting HER2 for the treatment of breast cancer. Annu Rev Med. 2015;66:111–128. doi: 10.1146/annurev-med-042513-015127. [DOI] [PubMed] [Google Scholar]
- 15.Xiang Z, Huang X, Wang J, Zhang J, Ji J, Yan R, Zhu Z, Cai W, Yu Y. Cross-database analysis reveals sensitive biomarkers for combined therapy for ERBB2+ gastric cancer. Front Pharmacol. 2018;9:861. doi: 10.3389/fphar.2018.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, Li S, Ma R, Sharma A, Bai S, Dun B, Cao H, Jing C, She J, Feng J. Tumor profiling of co-regulated receptor tyrosine kinase and chemoresistant genes reveal different targeting options for lung and gastroesophageal cancers. Am J Transl Res. 2016;8:5729–5740. [PMC free article] [PubMed] [Google Scholar]
- 17.Liang CH, Shiu LY, Chang LC, Sheu HM, Kuo KW. Solamargine upregulation of Fas, downregulation of HER2, and enhancement of cytotoxicity using epirubicin in NSCLC cells. Mol Nutr Food Res. 2007;51:999–1005. doi: 10.1002/mnfr.200700044. [DOI] [PubMed] [Google Scholar]
- 18.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olejniczak M, Kotowska-Zimmer A, Krzyzosiak W. Stress-induced changes in miRNA biogenesis and functioning. Cell Mol Life Sci. 2018;75:177–191. doi: 10.1007/s00018-017-2591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohanian M, Humphreys DT, Anderson E, Preiss T, Fatkin D. A heterozygous variant in the human cardiac miR-133 gene, MIR133A2, alters miRNA duplex processing and strand abundance. BMC Genet. 2013;14:18. doi: 10.1186/1471-2156-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Li Z, Xuan Z, Xu P, Wang W, Chen Z, Wang S, Sun G, Xu J, Xu Z. Novel role of miR-133a-3p in repressing gastric cancer growth and metastasis via blocking autophagy-mediated glutaminolysis. J Exp Clin Cancer Res. 2018;37:320. doi: 10.1186/s13046-018-0993-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Gao L, Li SH, Tian YX, Zhu QQ, Chen G, Pang YY, Hu XH. Role of downregulated miR-133a-3p expression in bladder cancer: A bioinformatics study. Onco Targets Ther. 2017;10:3667–3683. doi: 10.2147/OTT.S137433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He B, Lin X, Tian F, Yu W, Qiao B. miR-133a-3p inhibits oral squamous cell carcinoma (OSCC) proliferation and invasion by suppressing COL1A1. J Cell Biochem. 2018;119:338–346. doi: 10.1002/jcb.26182. [DOI] [PubMed] [Google Scholar]
- 24.Yang ZQ, Wu CA, Cheng YX. Prognostic value of microRNA-133a expression and its clinicopathologic significance in non-small cell lung cancer: A comprehensive study based on meta-analysis and the TCGA database. Oncol Res Treat. 2018;41:762–768. doi: 10.1159/000492343. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Tian L, Chen M, He Q, Yan Q, Zhai C. MicroRNA-199a-5p suppresses cell proliferation, migration and invasion by targeting ITGA3 in colorectal cancer. Mol Med Rep. 2020;22:2307–2317. doi: 10.3892/mmr.2020.11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamerith G, Kocher F, Rudzki J, Pircher A. ASCO 2018 NSCLC highlights-combination therapy is key. Memo. 2018;11:266–271. doi: 10.1007/s12254-018-0444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian Y, Sun C, Zhang L, Pan Y. Clinical significance of miRNA-106a in non-small cell lung cancer patients who received cisplatin combined with gemcitabine chemotherapy. Cancer Biol Med. 2018;15:157–164. doi: 10.20892/j.issn.2095-3941.2017.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y, Liu C, Muyldermans S. Nanobody-based delivery systems for diagnosis and targeted tumor therapy. Front Immunol. 2017;8:1442. doi: 10.3389/fimmu.2017.01442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGuire A, Brown JA, Kerin MJ. Metastatic breast cancer: The potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015;34:145–155. doi: 10.1007/s10555-015-9551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin VY, Chu KM. miRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20:10432–10439. doi: 10.3748/wjg.v20.i30.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi RU, Prieto-Vila M, Kohama I, Ochiya T. Development of miRNA-based therapeutic approaches for cancer patients. Cancer Sci. 2019;110:1140–1147. doi: 10.1111/cas.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi W, Tang T, Li X, Deng S, Li R, Wang Y, Wang Y, Xia T, Zhang Y, Zen K, et al. Methylation-mediated silencing of miR-133a-3p promotes breast cancer cell migration and stemness via miR-133a-3p/MAML1/DNMT3A positive feedback loop. J Exp Clin Cancer Res. 2019;38:429. doi: 10.1186/s13046-019-1400-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li JP, Zhang HM, Liu MJ, Xiang Y, Li H, Huang F, Li HH, Dai ZT, Gu CJ, Liao XH, Zhang TC. miR-133a-3p/FOXP3 axis regulates cell proliferation and autophagy in gastric cancer. J Cell Biochem. 2020;121:3392–3405. doi: 10.1002/jcb.29613. [DOI] [PubMed] [Google Scholar]
- 35.Byeon S, Lee B, Park WY, Choi YL, Jung HA, Sun JM, Ahn JS, Ahn MJ, Park K, Lee SH. Benefit of targeted DNA sequencing in advanced non-small-cell lung cancer patients without EGFR and ALK alterations on conventional tests. Clin Lung Cancer. 2020;21:e182–e190. doi: 10.1016/j.cllc.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 36.Liberelle M, Jonckheere N, Melnyk P, Van Seuningen I, Lebègue N. EGF-containing membrane-bound mucins: A hidden ErbB2 targeting pathway? J Med Chem. 2020;63:5074–5088. doi: 10.1021/acs.jmedchem.9b02001. [DOI] [PubMed] [Google Scholar]
- 37.Voelker R. Another targeted therapy for ERBB2-positive breast cancer. JAMA. 2020;323:408. doi: 10.1001/jama.2020.0222. [DOI] [PubMed] [Google Scholar]
- 38.Spencer KS, Graus-Porta D, Leng J, Hynes NE, Klemke RL. ErbB2 is necessary for induction of carcinoma cell invasion by ErbB family receptor tyrosine kinases. J Cell Biol. 2000;148:385–397. doi: 10.1083/jcb.148.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung DH, Bae YJ, Kim JH, Shin YK, Jeung HC. HER2 regulates cancer stem cell activities via the Wnt signaling pathway in gastric cancer cells. Oncology. 2019;97:311–318. doi: 10.1159/000502845. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.