Abstract
Prostate cancer is one of the most common malignant tumors in men. Patients with local infiltration and distant metastasis often have a poor prognosis. The present study aimed to investigate the expression and regulatory mechanism of the circular RNA cerebellar degeneration-related protein 1, anti-sense (circCDR1as) in prostate cancer cell lines. MicroRNAs (miRNAs) regulated by circCDR1as and target genes regulated by miRNAs were predicted using bioinformatics software. Prostate cancer cell lines (LNCaP, 22Rv1 and PC-3), a normal prostate epithelial cell line (RWPE-1) and a human embryonic kidney cell line (293T) were cultured. Relative gene expression was detected using reverse transcription PCR. Small interfering RNAs (siRNAs) targeting circCDR1as and X-linked inhibitor of apoptosis protein (XIAP) and miRNA mimics were designed and transfected into the cell lines using Lipofectamine® 3000. Cell invasion was determined using a Transwell assay, the cell proliferation rate was detected using an MTT assay and cell migration was examined using a scratch assay. Relative protein expression was detected using western blotting. Double fluorescent reporter gene vectors and an anti-Ago2 RNA-binding protein immunoprecipitation assay were used to verify binding. Bioinformatics analyses indicated that there was a binding site between miR-641 and circCDR1as and between miR-641 and XIAP. The expression of circCDR1as and XIAP was higher and the expression of miR-641 was lower in the prostate cancer cell lines compared with the normal prostate epithelial cell line. After effectively reducing the expression of circCDR1as and XIAP and increasing the expression of miR-641 in PC-3 cells, the proliferation, invasion and migration of PC-3 cells were effectively inhibited. circCDR1as could bind to miR-641, which targeted the 3′-untranslated region of XIAP. Reducing the expression of circCDR1 promoted the expression of miR-641 and inhibited the expression of XIAP. Overall, the circCDR1as/miR-641/XIAP regulatory axis plays a role in the invasion and migration of the prostate cancer PC-3 cell line.
Keywords: prostate cancer, circCDR1as, miR-641, XIAP, invasion, migration
Introduction
Prostate cancer is one of the most common malignant tumors in men. Historically, the incidence rate of prostate cancer has been lower in China compared with in Western countries (1). However, with lifestyle changes due to economic growth, the incidence rate of prostate cancer has been rapidly increasing in recent years. A high-fat diet and lack of exercise are among the factors that contribute to prostate cancer (2). In addition, patients with local infiltration and distant metastasis often have a poor prognosis (3). Therefore, inhibiting metastasis in prostate cancer has become a major challenge for clinicians.
Circular RNAs (circRNAs) are natural endogenous RNAs that regulate transcriptional and post-transcriptional steps (4). circRNAs are evolutionarily conserved and stable. Although they are considered a novel type of non-coding RNA, numerous studies have shown that some circRNAs can encode proteins (5). circRNAs have tissue- and cell-specific expression patterns (6). circRNA is involved in the occurrence and development of tumors. Its biological functions include acting as a scaffold for protein complexes, regulating gene expression, alternative splicing and participating in the interaction between RNA and protein (7). Cerebellar degeneration-related protein 1, anti-sense (circCDR1as; hsa_circ_0001946) is derived from the antisense transcript of CDR1, which is located on chromosome Xq27.1 (8). circCDR1as is highly expressed in prostate cancer, but its role in prostate cancer is unclear (9).
microRNAs (miRNAs/miRs) are a class of endogenous single-chain non-coding RNAs consisting of 19–25 nucleotides. miRNAs are widely found in eukaryotic cells and play an important role in gene regulation. They bind to the 3′-untranslated regions (3′-UTRs) of target mRNAs to regulate gene expression, and cause either the translation or degradation of the target mRNAs (10,11). miRNA-mediated inhibition of gene expression helps to regulate cell development, proliferation and adhesion (12).
Recent studies have shown that most circRNAs exist in the cytoplasm and their mechanisms of action are mediated by miRNA sponge interactions through binding to miRNAs (4,13). X-linked inhibitor of apoptosis protein (XIAP) is a major member of a newly identified IAP family of proteins. It is the strongest inhibitor of apoptosis in IAP family. It can directly inhibit caspases and regulate apoptosis in multiple ways. XIAP is overexpressed in prostate cancer cell lines, and its expression is closely associated with tumor progression, recurrence, prognosis and chemoresistance (14). Combined with the results of bioinformatics analysis, the present study used approaches to overexpress and suppress circCDR1as to elucidate its molecular role in prostate cancer invasion at the cellular and molecular levels. The current findings provide the scientific basis for the design of strategies to inhibit invasion with circCDR1as as the target.
Materials and methods
Bioinformatics analysis
The relative expression of circCDR1as was analyzed using MiOncoCirc (https://mioncocirc.github.io/), the relative expression of circCDR1as was analyzed in cancer tissues(ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoama; BRCA, breast invasive carcinoma; chol, cholangiocarcinoma; colo, colorectal cancer; esca, esophageal carcinoma; gbm, glioblastoma multiforme; hcc, hepatocellular carcinoma; hnsc, head and neck squamous cell carcinoma; kdny, kidney cancer; lung, lung cancer; maly, malignant lymphoma; nbl, neuroblastoma; ov, ovarian serous cystadenocarcinoma; paad, pancreatic adenocarcinoma; prad, prostate adenocarcinoma; sarc, sarcoma; secr, cecretory cancer; skcm, skin cutaneous melanoma. and the location and regulatory miRNAs of circCDR1as were identified using circBase (http://www.circbase.org/). The target genes regulated by miRNA were analyzed using TargetScan (http://www.targetscan.org/cgi-bin/targetscan/vert_72/view_gene.cgi?rs=ENST00000371199.3&taxid=9606&members=miR-641/3617-5p&showcnc=1&shownc=1&shownc_nc=1&showncf1=1&subset=1).
Cell culture and transfection
LNCaP, 22Rv1, PC-3, RWPE-1 and 293T cell lines were purchased from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 containing 10% newborn bovine serum (Thermo Fisher Scientific, Inc.) at 37°C in 5% CO2. The cells were digested and subcultured with 0.25% trypsin every 2–3 days. Cells in the logarithmic growth period and with a trypan blue rejection rate >95% were used in all experiments.
The construction of a lentivirus vector expressing siRNA targeting circCDR1as was carried out by Shanghai GenePharma Co., Ltd. 293T cells in logarithmic growth phase were inoculated at a density of 5×106/ml and cultured at 37°C with 5% CO2. The recombinant si-circCDT1as vector was packaged with 10 ug plasmid and incubated with Lipofectamine® 3000 to form a complex. The cells were treated with DMEM containing 1% FBS 24 h after transfection. After 48 h of transfection, the culture medium was collected. The transfection sequences were as follows: miR-641 Mature sequence (MI0003656), 5′-UGGGUGAAAGGAAGGAAAGACAUAGGAUAGAGUCACCUCUGUCCUCUGUCCUCUACCUAUAGAGGUGACUGUCCUAUGUCUUUCCUUCCUCUUACCCCU-3′; miRNA mimic negative control (scrambled sequences, mimic-NC, Guangzhou Ruibo Biotechnology Co., Ltd.), 5′-GUUACCAUGGAUGUAAU-3′; small interfering (si)-circCDR1as, 5′-GCACCTGTGTCAAGGTCTTTT-3′ and siRNA-NC (scrambled sequence), 5′-CGGGATTGGGATAAGCC-3′. siRNA-NC was used for the transfection of si-XIAP (Guangzhou Ruibo Biotechnology Co., Ltd.). si-XIAP sequences are presented in Table SI and the most effective sequence was used for subsequent studies. In total, 20 pmol of siRNAs, miR-641 mimic and respective controls (cell group, untransfected control cells) were transfected to PC-3 cell line using Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Transfections were performed at room temperature for 6 h.
Reverse transcription-quantitative (RT-q)PCR
Primer 6.0 (Canada primer, inc. www.premierbiosoft.com) was used to design the primers (Table I), which were synthesized by Thermo Fisher Scientific, Inc. LNCaP, 22Rv1, PC-3, and RWPE-1 cells were cultured and treated with trypsin to make a cell suspension. After counting the cells with a cell counting plate, a cell suspension containing ~1.0×106 cells was transferred to an RNase-free centrifuge tube to collect the cells. TRIzol® reagent was used to extract RNA (Invitrogen; Thermo Fisher Scientific, Inc.), and M-MLV reverse transcriptase (Thermo Fisher Scientific, Inc.) was used to reverse transcribed RNA into cDNA. Reverse transcription was performed at 16°C for 30 min, 42°C for 40 min and 85°C for 5 min. cDNA was used as a template for PCR reaction using SYBR Green qPCR supermax ((Takara Biotechnology Co., Ltd.). Under the following conditions: 50°C For 2 min, 95°C for 2 min, 95°C for 15 sec and 60°C for 32 sec for 40 cycles. GAPDH and U6 were used as internal references, and the 2−ΔΔCq method was used quantify the results (15).
Table I.
Primers for genes.
| Gene | Forward, 5′-3′ | Reverse, 5′-3′ |
|---|---|---|
| GAPDH | GTTGGTGGTGCAGGAGGCA | CTCGCTTCGGCAGCACA |
| circCDR1 | GGTTTCCGATGGCACCTG | GGTTTCCGATGGCACCTG |
| miR-641 | AAAGACATAGGATAGAGTCACCT C | GTGCAGGGTCCGAGGT |
| XIAP | CATCCATGGCAGATTATGAAGCA | CTTCACTGGGCTTCCAATCAGTTAG |
| RT-miRNA | TCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGA | AACGCTTCACGAATTTGCGT |
| U6 | GTTCCCATCTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
miR/miRNA, microRNA; XIAP, X-linked inhibitor of apoptosis protein.
MTT assay
There were six experimental groups: Cell group (untransfected cells), siRNA-NC group (negative control sequence + Lipofectamine 3000), miR-641 (miR-641 mimic + Lipofectamine 3000), mimic-NC group (negative control miRNA mimic + Lipofectamine 3000), si-circCDR1as (siRNA circCDR1as + Lipofectamine 3000), si-XIAP (siRNA XIAP + Lipofectamine 3000). PC-3 cells in the logarithmic growth phase were collected in serum-free DMEM/F12 and the cell concentration was adjusted to 1.0×105 cells/ml with dilution with serum-free DMEM/F12. Next, 96-well plates were inoculated with 100 µl/well. After 24, 48 and 72 h, 20 µl MTT solution [5 mg/ml in phosphate-buffered saline (PBS)] was added to each well. The cells were incubated for 4 h at 37°C and the culture supernatant was carefully removed from each well and discarded. Suspended cells were centrifuged (1,000 × g at 37°C for 3 mins) prior to removing the culture medium. Then, 150 µl dimethyl sulfoxide was added to each well, and the formazan crystals were dissolved by shaking for 10 min. The optical absorption value of each well at a wavelength of 490 nm was measured using a plate reader.
Transwell assay
The six aforementioned experimental groups were analyzed. Precooled serum-free medium was used to dilute the membrane matrix (Matrigel; Corning Inc.) at a volume ratio of 1:3, 40 µl of which was added to the precooled Transwell cells and incubated at 37°C for 2 h to coagulate the membrane matrix. Excess liquid was removed from the chamber, and 100 and 600 µl serum-free medium was added to the upper and lower chambers, respectively. The cells were incubated overnight at 37°C. On the second day of cell transfection, 1.0×105 cells were counted use a cell counter (Thermo Fisher Scientific, Inc.), and resuspended in 100 µl serum-free DMEM/F12. The cells were added to the upper chamber of the Transwell chamber and 600 µl DMEM/F12 containing 10% newborn bovine serum was added to the lower chamber. After incubation at 37°C and 5% CO2 for 24 and 48 h, the cells in the upper chamber were removed and wiped with a cotton swab (15). The cells in the lower chamber were observed under an inverted light microscope and images were captured. The cells were stained with crystal violet (37°C for 10 min), which was eluted in 33% acetic acid, and absorbance at a wavelength of 570 nm was measured using a plate reader.
Scratch assay
The aforementioned six experimental groups were analyzed. When the cell fusion rate reached 100%, cells in the logarithmic growth stage were washed three times with PBS after being scratched vertically with a pipette tip. The cells debris were removed and added to the serum-free medium for further culture. At 0 and 48 h after scratching, images were captured under a light microscope and the mean distance between cells was calculated using ImageJ version 3.0 (National Institutes of Health) according the following formula: Healing rate (%)=(1-measured width/0 h mean measured width) ×100 (16).
Western blotting
The cells grouped were the same as aforementioned. The PC3 cells of the different experimental groups were collected, and protein was extracted using a nuclear extraction kit (Thermo Fisher Scientific, Inc.). Protein concentration was measured using the BCA method. Samples of 30 µg protein were loaded per lane and separated using 10% gels and SDS-PAGE and transferred to a PVDF membrane. The PVDF membrane was incubated in a PBS solution containing 5% skimmed milk powder first for 2 h at room temperature and then overnight at 4°C to block non-specific binding sites. The membrane was incubated with the following antibodies: Anti-XIAP (1;500; cat. no. ab229050; Abcam, Inc.), anti-GAPDH (1:500; cat. no. ab9485; Abcam) at room temperature for 3 h, washed three times with 0.1% Tween 20, and incubated at room temperature for 1 h with a horseradish peroxidase-labeled mouse anti-XIAP, anti-GAPDH IgG antibody (1:1,000; cat. no. ab205719; Abcam). The protein bands were analyzed by AlphaEaseFC 4.0 software (Protein Simple) and semi-quantitative quantification was performed.
Luciferase reporter assay
There were five experimental groups: Cell group (untreated cells), NC group [NC sequence + psiCHECK-2-circCDR1as-wild-type (wt), psiCHECK-2-circCDR1as-mutant (mut), psiCHECK-2-XIAP-3′-untranslated region (UTR)-wt or psiCHECK-2-XIAP-3′-UTR-mut], inhibitor-NC group (inhibitor-NC + psiCHECK-2-circCDR1as-wt/psiCHECK-2-circCDR1as-mut or psiCHECK-2-XIAP-3′-UTR-WT/psiCHECK-2-XIAP-3′-UTR-mut), miR-641 group (miR-641 mimic + psiCHECK-2-circCDR1as-wt/psiCHECK-2-circCDR1as-mut or psiCHECK-2-XIAP-3′-UTR-WT/psiCHECK-2-XIAP-3′-UTR-mut) and miR-641 inhibitor group (miR-641 inhibitor + psiCHECK-2-circCDR1as-wt/psiCHECK-2-circCDR1as-mut or psiCHECK-2-XIAP-3′-UTR-wt/psiCHECK-2-XIAP-3′-UTR-mut). psiCHECK-2 vector purchased from Promega Corporation. The genomic DNA of PC-3 cells was extracted as a PCR amplification template, this was performed as aforementioned, and the 3′-UTR sequences of linear circCDR1as and XIAP were amplified with genomic DNA as a template. The PCR amplification products were double digested with restriction enzymes Xho and NotIR, and the amplification fragments were ligated to the psiCHECK-2 vector. DH5a Escherichia coli were transformed with the ligated products. Positive clones were screened using blue and white spot analysis and enzyme digestion identification, and were sent for sequencing and double fluorescence detection (17). Luciferase activity was calculated using the formula: (Firefly Luciferase/Rellina Luciferase) and was measured using a Dual Luciferase Reporter Assay System kit (Promega Corporation) on a Tecan M200 Pro luminescence reader according to the manufacturer's protocol (Beijing KeyuXingye Science and Technology Development Co., Ltd.).
Anti-Ago2 RNA-binding protein immunoprecipitation (RIP) assay
There were three experimental groups: Input group (input), IgG group (IgG antibody) and Ago2 group (anti-Ago2 antibody). Biotin-coupled miRNA and circRNA pull-down assays were performed as previously described (11). Briefly, PC-3 cells were transfected with 20 nM of 5′-end biotinylated circCDR1as (digoxin-5′-CTCAGGATTATCTGGAAGAC-3′) (digoxin; Guangzhou RiboBio Co., Ltd.) for 1 day. Biotin-coupled RNA complexes were pulled down by incubating the cell lysates with MyOne Streptavidin C1 Dynabeads (Invitrogen; Thermo Fisher Scientific, Inc.). The abundance of circCDR1as or miRNA in the bound fractions was evaluated by RT-qPCR analysis (18).
Statistical analysis
Data are shown as the mean ± standard deviation. The experiment repeated three times. SPSS 20.0 (IBM, Corp.) was used to process the data, and one-way ANOVA followed by Bonferroni's correction test was used to analyze the significance of differences between different groups. Unpaired t-tests were used for comparisons between groups. P<0.05 was considered to indicate a statistically significant difference.
Results
circCDR1as is highly expressed in prostate cancer and there are interactions between circCDR1as and miR-641 and between miR-641 and XIAP
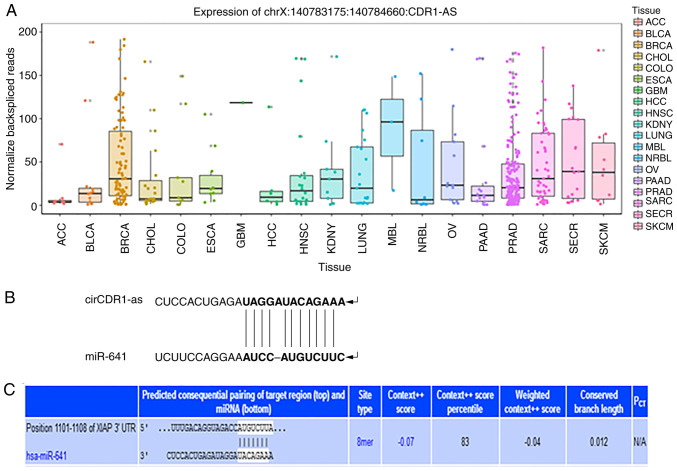
Bioinformatics analysis indicated that the relative expression of circCDR1as was significantly higher in breast cancer, prostate cancer and sarcoma samples compared with other cancer tissues. Binding sites were identified between circCDR1as and miR-641 and between miR-641 and XIAP. These results indicated that there is a regulatory relationship between circCDR1as, miR-641 and XIAP (Fig. 1).
Figure 1.
Bioinformatics software prediction results. (A) Relative expression levels of circCDR1as in cancer tissue. (B) Schematic diagram of the binding site between circCDR1as and miR-641. (C) Schematic diagram of the binding site between miR-641 and XIAP. circCDR1as, circular RNA cerebellar degeneration-related antigen 1, anti-sense; miR, microRNA; XIAP, X-linked inhibitor of apoptosis protein; hsa, homo sapien; ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoama; BRCA, breast invasive carcinoma; chol, cholangiocarcinoma; colo, colorectal cancer; esca, esophageal carcinoma; gbm, glioblastoma multiforme; hcc, hepatocellular carcinoma; hnsc, head and neck squamous cell carcinoma; kdny, kidney cancer; lung, lung cancer; maly, malignant lymphoma; nbl, neuroblastoma; ov, ovarian serous cystadenocarcinoma; paad, pancreatic adenocarcinoma; prad, prostate adenocarcinoma; sarc, sarcoma; secr, cecretory cancer; skcm, skin cutaneous melanoma.
circCDR1as and XIAP expression is high in prostate cancer cell lines and miR-641 expression is low in prostate cancer cell lines
RT-qPCR showed that circCDR1as and XIAP mRNA were highly expressed in prostate cancer cell lines (LNCaP, 22Rv1 and PC-3) compared with normal prostate epithelial cells (RWPE-1), with the highest expression in PC-3 cells. In contrast, miR-641 was expressed at a low level in prostate cancer cells compared with normal prostate epithelial cells. The results suggested that circCDR1as and XIAP may promote the occurrence and development of prostate cancer, and that miR-641 may inhibit the occurrence and development of prostate cancer (Fig. 2).
Figure 2.
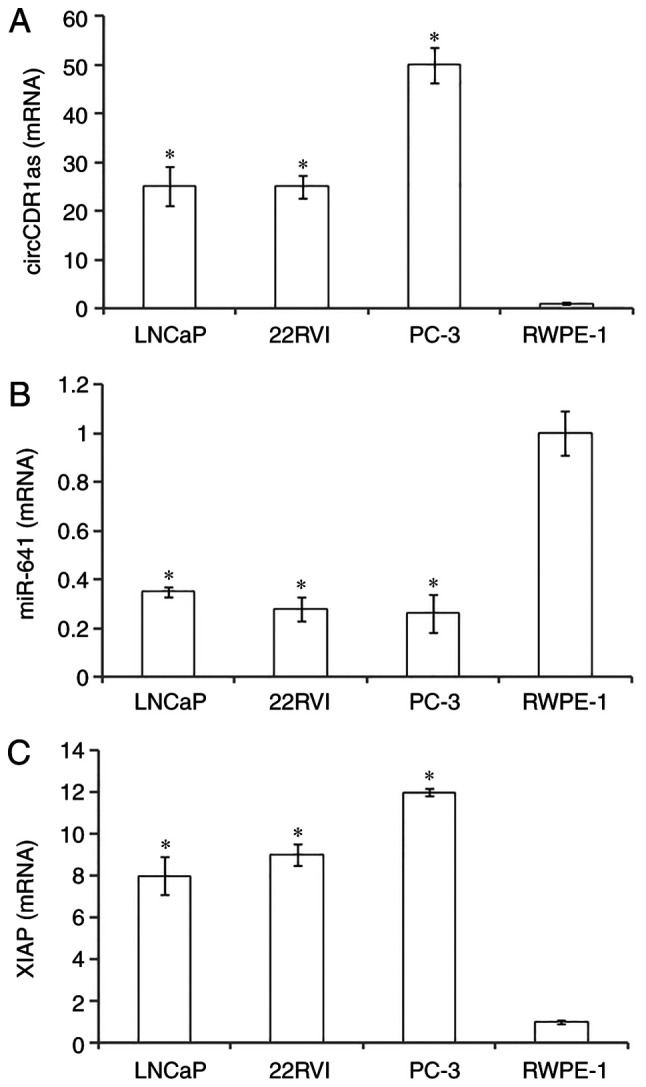
Relative expression of genes detected by reverse transcription-quantitative PCR. Relative expression levels of (A) circCDR1as, (B) miR-641 and (C) XIAP in prostate cancer cell lines and a normal prostate epithelial cell line. *P<0.05 compared with RWPE-1 cells. circCDR1as, circular RNA cerebellar degeneration-related antigen 1, anti-sense; miR, microRNA; XIAP, X-linked inhibitor of apoptosis protein.
Reduction of circCDR1as and XIAP expression and overexpression of miR-641 inhibit the proliferation of PC-3 cells
After the expression levels of circCDR1as and XIAP were suppressed and miR-641 was overexpressed (Fig. S1). MTT assay results showed that si-circCDR1as, si-XIAP and miR-641 mimic inhibited the proliferation of PC-3 cells in a time-dependent manner compared with respective controls (Fig. 3).
Figure 3.
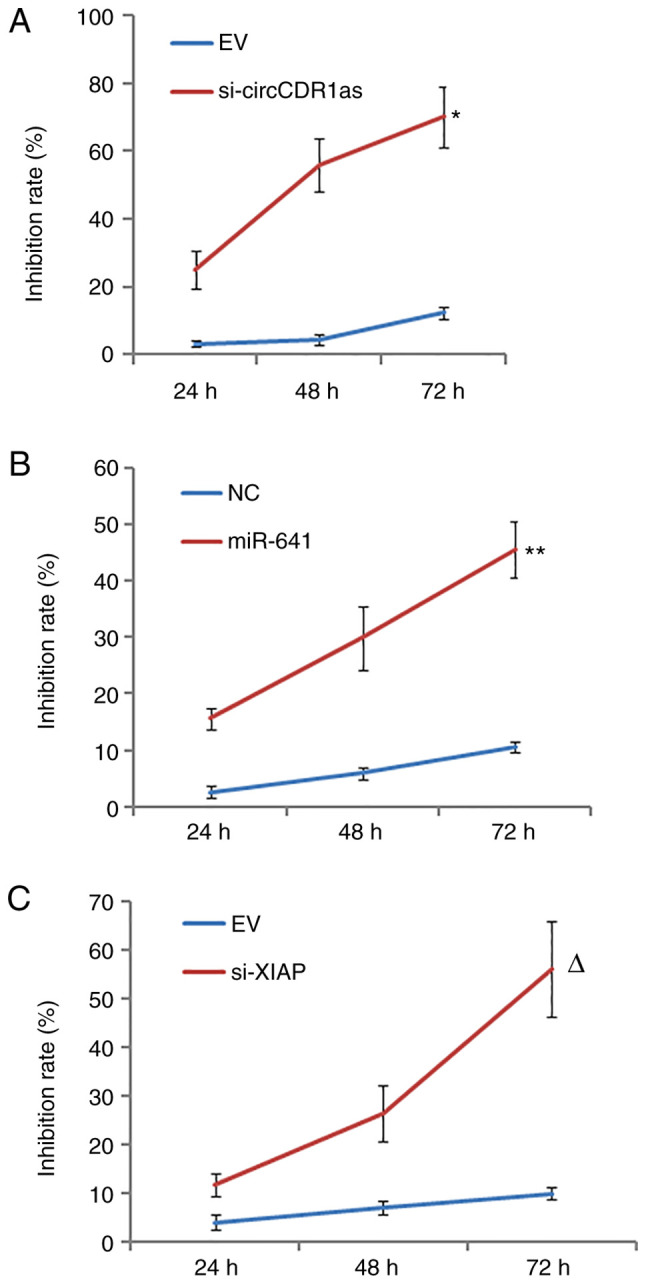
MTT assay of cell proliferation rate. Effect of (A) si-circCDR1as, (B) miR-641 and (C) si-XIAP on PC-3 cell proliferation. *P<0.05 compared with the EV group, **P<0.05 compared with mimic-NC group, ∆P<0.05 compared with the EV group. si, small interfering; circCDR1as, circular RNA cerebellar degeneration-related antigen 1, anti-sense; miR, microRNA; XIAP, X-linked inhibitor of apoptosis protein; NC, negative control.
Reduction of circCDR1as and XIAP expression and overexpression of miR-641 inhibit the invasion and migration of PC-3 cells
After the expression of circCDR1as and XIAP were suppressed and miR-641 was overexpressed, a Transwell assay showed that si-circCDR1as, si-XIAP and miR-641 mimic inhibited the invasion of PC-3 cells (Fig. 4). A scratch assay showed that the migration of PC-3 cells could be inhibited by si-circCDR1as, si-XIAP and miR-641 mimic (Fig. 5). These findings showed that the invasion and migration of PC-3 cells could be inhibited by decreasing the expression of circCDR1as and XIAP and increasing the expression of miR-641.
Figure 4.
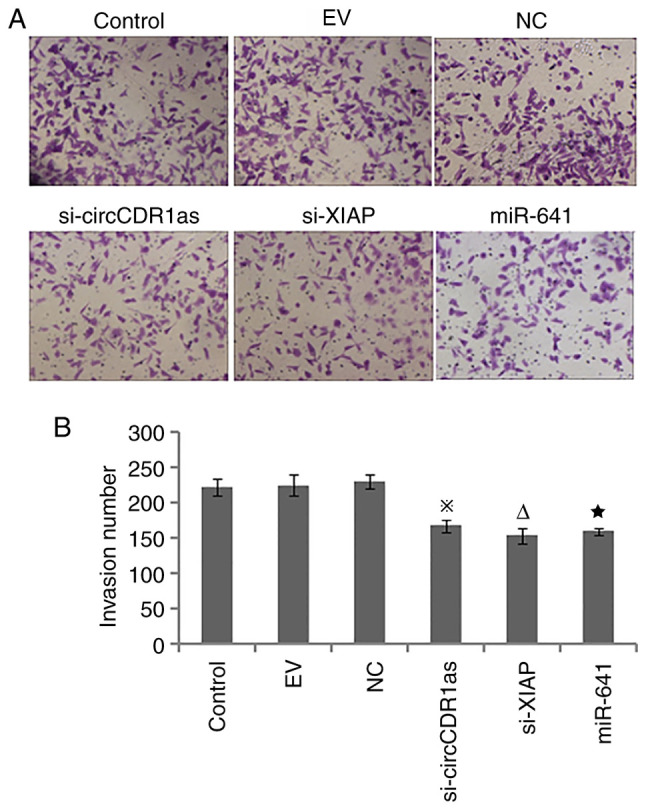
Detection of cell invasion by Transwell assay. (A) Representative images of invasion (magnification, ×100). (B) Invasion number of PC-3 cells. ※P<0.05 compared with the siRNA-NC and control groups. ∆P<0.05 compared with the mimic-NC and control groups. ★P<0.05 compared with the siRNA-NC and control groups. circCDR1as, circular RNA cerebellar degeneration-related antigen 1, anti-sense; miR, microRNA; XIAP, X-linked inhibitor of apoptosis protein; NC, negative control.
Figure 5.
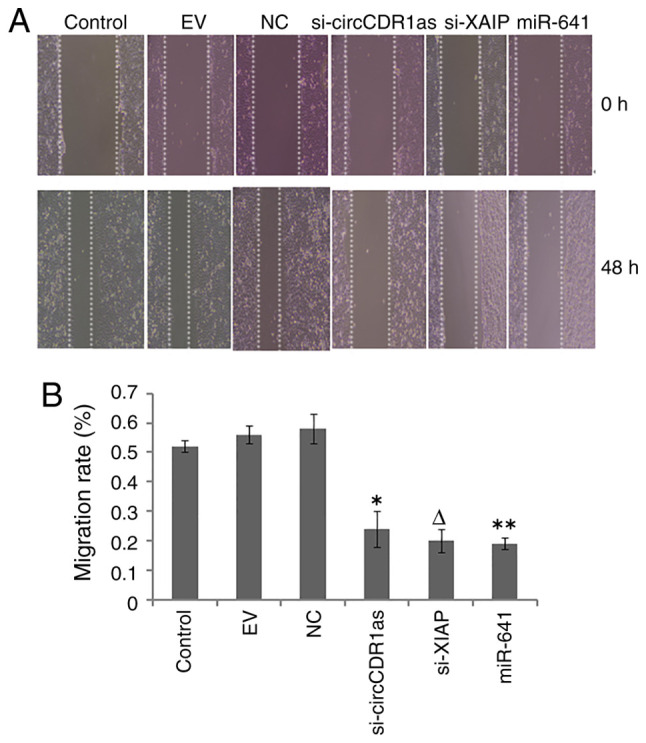
Scratch assay for cell migration. (A) Representative images of cell migration (magnification, ×50). (B) Comparison of PC-3 cell migration. *P<0.05 compared with the siRNA-NC and control groups. **P<0.05 compared with the mimic-NC and control groups. ∆P<0.05 compared with the siRNA-NC and control groups. circCDR1as, circular RNA cerebellar degeneration-related antigen 1, anti-sense; miR, microRNA; XIAP, X-linked inhibitor of apoptosis protein; NC, negative control.
Reducing circCDR1as expression promotes miR-641 expression and inhibits XIAP expression
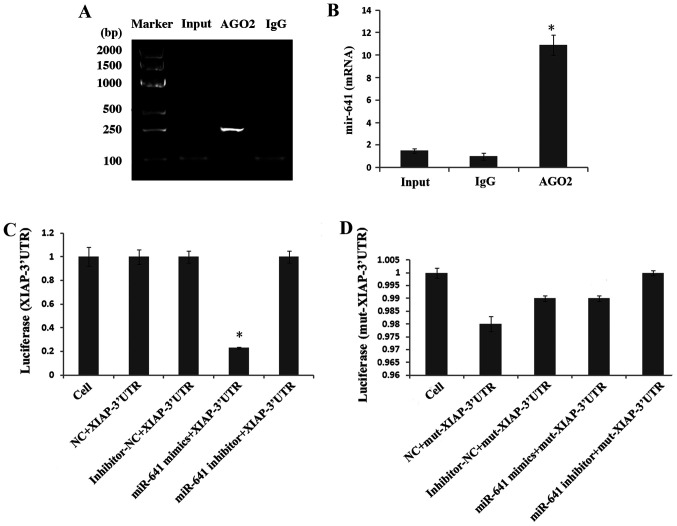
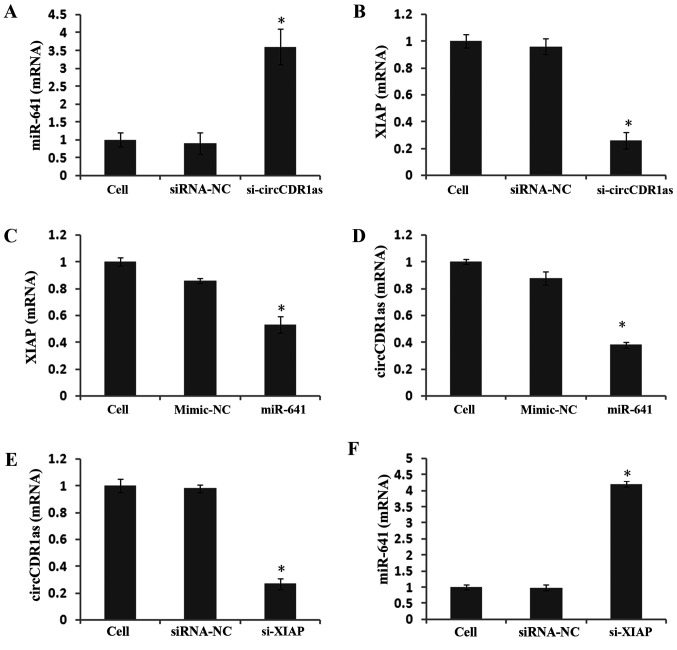
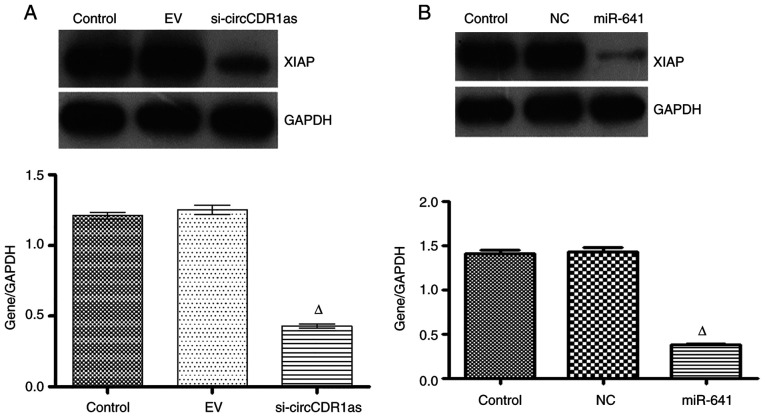
To study the expression and regulatory mechanism of circCDR1as, an anti-Ago2 RIP assay was used to detect binding between circCDR1as and miR-641. The results showed that the relative expression of miR-641 was 11 times higher compared with that of the negative control group in the Ago2 antibody-binding RNA group, indicating binding between circCDR1as and miR-641 (Fig. 6A and B). A double fluorescent reporter gene vector was constructed to confirm the binding site between miR-641 and the 3′-UTR of XIAP. The results showed that the fluorescence activity of 293T cells co-transfected with miR-641 mimic and the 3′-UTR of XIAP was clearly reduced (Fig. 6C), but the fluorescence activity of 293T cells co-transfected with miR-641 mimic and the mut-3′-UTR of XIAP showed no obvious change (Fig. 6D), indicating that miR-641 could bind to the 3′-UTR of XIAP. At 48 h after transfecting PC-3 cells with si-circCDR1as, miR-641 expression was significantly increased (Fig. 7A), whereas XIAP expression was downregulated (Figs. 7B and 8A). At 48 h after transfecting PC-3 cells with miR-641 mimic, the expression of circCDR1as (Fig. 7D) and XIAP was also downregulated (Figs. 7C and 8B). After PC-3 cells were transfected with si-XIAP, circCDR1as expression was downregulated and miR-641 expression was upregulated (Fig. 7E and F, respectively). These results showed that decreasing circCDR1as expression promotes the expression of miR-641 and inhibits the expression of XIAP.
Figure 6.
Detection of interactions between genes. (A) Anti-ago2 RIP electrophoretogram. (B) Anti-Ago2 RIP assay was used to detect binding between circCDR1as and miR-641. (C) Comparison of the fluorescence activity of 293T cells co-transfected with miR-641 mimic and wild-type XIAP-3′-UTR. *P<0.05 compared with the cell, NC, inhibitor-NC and miR-641 inhibitor groups (D) Comparison of the fluorescence activity of 293T cells co-transfected with miR-641 mimic and mut XIAP-3′-UTR. circCDR1as, circular RNA cerebellar degeneration-related antigen 1, anti-sense; miR, microRNA; XIAP, X-linked inhibitor of apoptosis protein; EV, empty vector; NC, negative control; UTR, untranslated region; mut, mutant.
Figure 7.
Relative expression of genes detected by RT-qPCR. RT-qPCR detection of the relative expression of (A) miR-641 and (B) XIAP in PC-3 cells after transfection with si-circCDR1as for 48 h. RT-qPCR detection of the relative expression of (C) circCDR1as and (D) XIAP in PC-3 cells after transfection with miR-641 for 48 h. RT-qPCR detection of the relative expression of (E) circCDR1as and (F) miR-641 in PC-3 cells after transfection with si-XIAP for 48 h. *P<0.05 compared with the control and mimic-NC groups. RT-q, reverse transcription-quantitative; circCDR1as, circular RNA cerebellar degeneration-related antigen 1, anti-sense; miR, microRNA; XIAP, X-linked inhibitor of apoptosis protein; NC, negative control; si, small interfering.
Figure 8.
Relative protein expression of genes detected by western blotting assay. (A) Western blotting of the relative protein expression of XIAP in PC-3 cells after transfection with (A) si-circCDR1as and (B) miR-641 for 48 h. ∆P<0.05 compared with the control or siRNA-NC groups. circCDR1as, circular RNA cerebellar degeneration-related antigen 1, anti-sense; miR, microRNA; XIAP, X-linked inhibitor of apoptosis protein; NC, negative control.
Discussion
circRNAs are generated by reverse splicing of pre-mRNAs, which differs from the production of traditional linear RNAs. In the process of reverse splicing, the upstream 5′ splicing receptor is added to the downstream 3′ splicing donor in the opposite direction, and the binding of 3′ and 5′ phosphodiester bonds at the splicing site results in circRNA molecules forming rings (4,18). circRNA expression is related to a variety of diseases and tumor progression. Due to their wide expression, unique stable structure, and tissue-specific expression, circRNAs play important physiological functions in different biological processes (19). In the present study, circCDR1as was highly expressed in prostate cancer cell lines (LNCaP, 22Rv1, and PC-3), and its expression was reduced in order to inhibit the proliferation and invasion of PC-3 cells. Taken together with the results of bioinformatics analysis, these results suggest that circCDR1as may be a potential diagnostic marker for prostate cancer.
circRNAs exist in the cytoplasm and that their mechanisms of action are mediated mostly by miRNA sponge interactions via binding to miRNAs (8–10). miR-641 exerts an antitumor effect, and overexpression of miR-641 can inhibit the proliferation and migration of lung cancer cells (20). XIAP expression elevated in prostate cancer tissues (21). XIAP is a therapeutic target to induce apoptosis in prostate cancer cells because XIAP plays an important role in human prostate carcinogenesis (22,23).
In the present study, the relative expression of circCDR1as and XIAP was higher in prostate cancer cell lines compared with in normal prostate epithelial cells (RWPE-1), whereas miR-641 expression was higher in prostate epithelial cells compared with in prostate cancer cell lines. After effectively reducing the expression of circCDR1as and XIAP and increasing the expression of miR-641, the proliferation, invasion and migration of PC-3 cells were effectively inhibited. circCDR1as could bind to miR-641, which targets the 3′-UTR of XIAP. The current results showed that the regulatory axis of circCDR1as/miR-641/XIAP affected the proliferation and invasion of PC-3 cells.
The role of circCDR1as has been investigated in previous studies. After knocking down the expression of circCDR1as in hepatocellular carcinoma cells with high circCDR1as expression, the expression of miR-7 was upregulated, the expression of G1/S-specific cyclin E1 and phosphatidylinositol-4,-5-bisphosphate 3-kinase catalytic subunit δ genes are downregulated, and tumor differentiation and invasion are inhibited (24). circCDR1as also upregulates N-cadherin and inhibits E-cadherin to promote the epithelial-mesenchymal transition via miR-7 for osteosarcoma migration (25). Silencing circCDR1as may inhibit the expression of proteasome activating factor γ by removing the competitive inhibitory effect on miR-7, thereby enhancing the sensitivity of drug-resistant breast cancer cells (26). Silencing circCDR1as increases the expression of miR-135b-5p and decreases the expression of hypoxia-inducible factor 1-α inhibitor, thereby increasing the proliferative capacity of ovarian cancer cells (27). An in-depth study on the expression and regulatory mechanism of circCDR1as in tumors may indicate the direction for the targeted therapy of tumors.
In summary, the present study is the first to demonstrate that the circCDR1as/miR-641/XIAP regulatory axis affects the invasion and migration of the prostate cancer PC-3 cell line. These findings may provide a new direction for targeted gene therapy of prostate cancer and lay a theoretical foundation for elucidating the molecular mechanism of prostate cancer. The present study also has some limitations. For example, the expression of circCDR1as in blood was not detected. In future it should be determined whether the circCDR1as/miR-641/XIAP regulatory axis affects the proliferation and metastasis of tumors in animals.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- circCDR1as
circular RNA cerebellar degeneration-related antigen 1, anti-sense
- DMEM
Dulbecco's modified Eagle's medium
- miRNA
microRNA
- NC
negative control
- RIP
RNA-binding protein immunoprecipitation
- RT
reverse transcription
- siRNA
small interfering RNA
- UTR
untranslated region
- XIAP
X-linked inhibitor of apoptosis protein
Funding Statement
This work was supported by grants from the Medical and Health Science and Technology Project of Panyu District, Guangzhou (grant nos. 2017-Z04-18, 2018-Z04-59 and 2019-Z4-02), and the Guangzhou Health and Family Planning Commission Program (grant nos. 20181A011118, 20192A011027 and 20191A011119).
Funding
This work was supported by grants from the Medical and Health Science and Technology Project of Panyu District, Guangzhou (grant nos. 2017-Z04-18, 2018-Z04-59 and 2019-Z4-02), and the Guangzhou Health and Family Planning Commission Program (grant nos. 20181A011118, 20192A011027 and 20191A011119).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YLN and YLZ contributed to the design of the present study, developed the methodology, analyzed the results and wrote the manuscript. YLZ collected the bioinformatics data. JHH performed the experiments. YLZ and KL confirmed the authenticity of all raw data. CGX contributed to the design of the study and critically revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was conducted with the written informed consent of all patients and was approved by The Ethics Committee of the Affiliated Hospital of Guizhou Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Minimally invasive group of urology male genitourinary cancer professional committee of China anti cancer association, corp-author. Consensus of Chinese prostate cancer surgical treatment experts. Zhejiang Med. 2018;40:217–220. [Google Scholar]
- 2.Welch HG, Albertsen PC. Reconsidering prostate cancer mortality-the Future of PSA screening. N Engl J Med. 2020;382:1557–1563. doi: 10.1056/NEJMms1914228. [DOI] [PubMed] [Google Scholar]
- 3.He JH, Zhang JZ, Han ZP, Wang L, Lv YB, Li YG. Reciprocal regulation of PCGEM1 and miR-145 promote proliferation of LNCaP prostate cancer cells. J Exp Clin Canc Res. 2014;33:72. doi: 10.1186/s13046-014-0072-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang S, Li X, Zheng H, Si X, Li B, Wei G, Li C, Chen Y, Chen Y, Liao W, et al. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139:2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18:4. doi: 10.1186/s12943-018-0933-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y, Liang W, Hu C, Liu Y, Li J, et al. Genome-wide lncRNA microarray profiling identifies novel circulating lncRNAs for detection of gastric cancer. Theranostics. 2017;7:213–227. doi: 10.7150/thno.16044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang M, Ma L, Liu Y, He Y, Li G, An X, Cao B. CircRNA-006258 sponge-adsorbs miR-574-5p to regulate cell growth and milk synthesis via EVI5L in goat mammary epithelial cells. Genes (Basel) 2020;11:718. doi: 10.3390/genes11070718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al. The landscape of circular RNA in cancer. Cell. 2019;176:869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Do DN, Dudemaine PL, Fomenky BE, Ibeagha-Awemu EM. Integration of miRNA weighted gene co-expression network and miRNA-mRNA co-expression analyses reveals potential regulatory functions of miRNAs in calf rumen development. Genomics. 2019;111:849–859. doi: 10.1016/j.ygeno.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, Zhou B, Liu M, Liu Y, Gao R. miR-126-5p restoration promotes cell apoptosis in cervical cancer by targeting Bcl2l2. Oncol Res. 2017;25:463–470. doi: 10.3727/096504016X14685034103879. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Sheng Y, Hu R, Zhang Y, Luo W. MicroRNA-4317 predicts the prognosis of breast cancer and inhibits tumor cell proliferation, migration, and invasion. Clin Exp Med. 2020;20:417–425. doi: 10.1007/s10238-020-00625-4. [DOI] [PubMed] [Google Scholar]
- 13.Liu T, Ye P, Ye Y, Lu S, Han B. Circular RNA hsa_circRNA_002178 silencing retards breast cancer progression via microRNA-328-3p-mediated inhibition of COL1A1. J Cell Mol Med. 2020;24:2189. doi: 10.1111/jcmm.14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrera-Vázquez OS, Cancio-Lonches C, Hernández-González O, Chávez-Munguia B, Villegas-Sepúlveda N, Gutiérrez-Escolano AL. The feline calicivirus leader of the capsid protein causes survivin and XIAP downregulation and apoptosis. Virology. 2019;527:146–158. doi: 10.1016/j.virol.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.He JH, Han ZP, Zhou JB, Chen WM, Lv YB, He ML, Li YG. MiR-145 affected the circular RNA expression in prostate cancer LNCaP cells. J Cell Biochem. 2018;119:9168–9177. doi: 10.1002/jcb.27181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He JH, Han ZP, Zou MX, He ML, Li YG, Zheng L. CDX2/mir-145-5p/SENP1 pathways affect LNCaP cells invasion and migration. Front Oncol. 2019;9:477. doi: 10.3389/fonc.2019.00477. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Jiang XM, Li ZL, Li JL, Xu Y, Leng KM, Cui YF, Sun DJ. A novel prognostic biomarker for cholangiocarcinoma: circRNA Cdr1as. Eur Rev Med Pharmacol Sci. 2018;22:365–371. doi: 10.26355/eurrev_201801_14182. [DOI] [PubMed] [Google Scholar]
- 19.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 20.Kong Q, Shu N, Li J, Xu N. miR-641 functions as a tumor suppressor by targeting MDM2 in human lung cancer. Oncol Res. 2018;26:735–741. doi: 10.3727/096504017X15021536183490. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Devi GR. XIAP as target for therapeutic apoptosis in prostate cancer. Drug News Perspect. 2004;17:127–134. doi: 10.1358/dnp.2004.17.2.829046. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Li M, Xia S. Expression and clinical significance of antiapoptosis gene XIAP in prostate cancer. Zhonghua Nan Ke Xue. 2004;10:832–835. (In Chinese) [PubMed] [Google Scholar]
- 23.Berezovskaya O, Schimmer AD, Glinskii AB, Pinilla C, Hoffman RM, Reed JC, Glinsky GV. Increased expression of apoptosis inhibitor protein XIAP contributes to anoikis resistance of circulating human prostate cancer metastasis precursor cells. Cancer Res. 2005;65:2378–2386. doi: 10.1158/0008-5472.CAN-04-2649. [DOI] [PubMed] [Google Scholar]
- 24.Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11:e0158347. doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Xu B, Yang T, Wang Z, Zhang Y, Liu S, Shen M. CircRNA CDR1as/miR-7 signals promote tumor growth of osteosarcoma with a potential therapeutic and diagnostic value. Cancer Manag Res. 2018;10:4871–4880. doi: 10.2147/CMAR.S178213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Yang X, Wang X, Gu J, Zhou D, Wang Y, Yin B, Guo J, Zhou M. Silencing CDR1as enhances the sensitivity of breast cancer cells to drug resistance by acting as a miR-7 sponge to down-regulate REGγ. J Cell Mol Med. 2019;23:4921–4932. doi: 10.1111/jcmm.14305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Mao M, Jiang J, Zhu D, Li P. Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. Onco Targets Ther. 2019;12:3869–3879. doi: 10.2147/OTT.S207938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.