Abstract
Objective
The occurrence of osteoarthritis is related to genetic and environmental factors. Among them, the change of chondrocyte gene expression pattern regulated by epigenetic modification is an important participant. This study analyzed the effect of CAMP gene methylation on the level of oxidative stress and inflammation of chondrocytes.
Methods
We analyzed the changes of the transcriptome in the articular cartilage tissue of osteoarthritis (OA) patients from the GSE117999 dataset. The GSE48422 dataset was used to analyze the changes in the methylation level of osteoarthritis cells. Cell Counting Kit-8 (CCK-8) and flow cytometry analysis of short hairpin RNA (shRNA) silencing CAMP gene and 5-μM 5-Aza-2’-Deoxycytidine (AZA) treatment on the proliferation and apoptosis of Human chondrocytes osteoarthritis (HC-OA) cells. The Dichloro-dihydro-fluorescein diacetate (DCFH-DA) assay was used to detect the level of reactive oxygen species (ROS), and the expression level of inflammatory factors was analyzed by Western Blot.
Results
The expression of CAMP in cartilage tissue of OA patients was upregulated, and the level of methylation was downregulated. CAMP was highly expressed in osteoarthritis articular cartilage cells. Silencing CAMP inhibited the proliferation of HC-OA cells and promoted their apoptosis. CAMP gene methylation inhibited ROS levels and tumor necrosis factor-α (TNF-α) expression levels in HC-OA cells, and promoted transforming growth factor beta (TGF-β) expression. CAMP gene methylation inhibited the proliferation of HC-OA cells and promoted their apoptosis.
Conclusion
CAMP gene promoter methylation inhibits ROS levels and inflammation and induces chondrocyte apoptosis.
Keywords: CAMP, Methylation, Osteoarthritis, ROS, Inflammatory factors
Introduction
Osteoarthritis (OA) is a common age-related degenerative disease. Its main pathological feature is articular cartilage degeneration, which is mainly manifested by the loss of articular cartilage and the formation of osteophytes, subchondral bone sclerosis, and synovium inflammation, among others; clinical symptoms such as pain, joint deformity, and dysfunction may appear in the later clinical stage [1–3].
Research in recent years has shown that epigenetic modification plays an important role in the pathogenesis of OA [4, 5]. Epigenetic modification mainly includes DNA methylation, histone modification, chromatin remodeling, and non-coding RNA, among which DNA methylation is closely related to inflammatory diseases [6, 7]. At present, DNA methylation studies of OA mainly focus on articular cartilage, because articular cartilage is the core tissue involved in the disease process [8]. In addition, DNA methylation has become an important regulator of chondrocyte dedifferentiation, severely destroying the results of autologous chondrocyte implantation in patients with cartilage injury [8]. Therefore, it is of great significance to study the DNA methylation profile of chondrocyte dedifferentiation.
At present, the methylation sites or regions identified in OA genome-wide methylation studies are mostly enriched in genes related to immunity or inflammation, cartilage development, transcription factor regulation, and protease activity [9, 10]. Although OA was once considered a non-inflammatory disease, it has been confirmed that inflammation is very important in the occurrence of OA [11].
Tumor necrosis factor-α (TNF-α) is one of the main pro-inflammatory cytokines related to the pathogenesis of OA; it can stimulate the release of matrix metalloenase-1 (MMP-1), MMP-3, and MMP-13 in OA patients, and inhibit the synthesis of proteoglycans and type II collagen [12]. Transforming growth factor beta (TGF-β) is a member of the TGF-β superfamily. It transmits signals to the nucleus through the TGF-β receptor and its intracellular signaling system to play a variety of different physiological regulatory roles [13, 14]. Studies have shown that TGF-β plays an important role in maintaining normal articular cartilage and joint repair [15].
Some researchers have used the demethylating agent AZA for in vitro intervention of OA chondrocytes. For example, Iliopoulos et al. [16] found that AZA could reduce the methylation level of the leptin gene promoter of chondrocytes and increased the expression of leptin mRNA, which then activated MMP-13. Kim et al. [17] reported that when OA articular chondrocytes were co-cultured with 10 μM AZA for 8 days, the methylation level of 6 CpG islands in the Sry-type high-mobility-group box 9 (SOX-9) promoter region of chondrocytes decreased, and the expression level of SOX-9 increased.
The CAMP gene (also known as LL-37) is located on chromosome 3p21.31. The CAMP protein is composed of 37 amino acids in a spiral shape and has a broad-spectrum antibacterial activity. Studies have shown that CAMP has defensive effects, such as regulating inflammation, inducing immune cells to injured or infected sites, binding and neutralizing lipopolysaccharide (LPS), promoting epithelialization, and repairing the damage [18]. Some researchers had found that CAMP affected the expression and distribution of TLR in tissue mast cells [19]. CAMP can be used as a powerful inducer of CCL3 and ROS generation [19]. Research by Li et al. [20] showed that CAMP could promote epithelial and smooth muscle-like differentiation of adipose stem cells through Wnt/β-Catenin and nuclear factor-kappaB (NF-κB) pathways.
In this study, we analyzed CAMP expression and methylation levels in OA chondrocytes through the GSE117999 dataset and GSE48422 dataset, and analyzed the effects of CAMP methylation on chondrocyte oxidative stress and inflammation level in vitro, as well as the impact on chondrocyte proliferation and apoptosis. It is of great significance for further elucidating the mechanism of OA, and it also provides a basis for early diagnosis and treatment of OA.
Materials and methods
Datasets
Both the GSE117999 dataset and GSE48422 dataset [21] can be obtained from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) database. The GSE117999 dataset contains transcript data from OA and non-OA cartilage tissue. The GSE48422 dataset contains gene methylation levels in OA patients and non-OA cartilage tissues.
Cell culture
HC-OA cells were purchased from Cell Applications (San Diego, CA, USA), and Human Chondrocytes-articular (HC-A) cells were purchased from ScienCell (San Diego, CA, USA). HC-OA cells and HC-A cells were cultured, supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO, USA), 1% penicillin-streptomycin (ThermoFisher Scientific, Waltham, MA) Chondrocyte growth medium (PromoCell, Heidelberg, Germany) at 37 °C in a humidified incubator containing 5% CO2.
Cell transfection and AZA treatment
In order to study the effect of CAMP on the proliferation and apoptosis of chondrocytes, we used small hairpin RNA to silence CAMP gene expression (sh-CAMP) and no template control (sh-NC) and no transfection group (Control) as controls. According to the manufacturer’s instructions, HC-OA cells (5 × 105 cells/well) were transfected with Lipofectamine 3000 (Life Technologies, Gaithersburg, MD, USA) and incubated in a medium containing 10% FBS for 48 h. The detailed process of AZA processing has been previously reported [22]. Quantitative reverse transcription-PCR (qRT-PCR) and western blot were used to detect transfection efficiency.
Cell Counting Kit-8 assay
CCK-8 assay was used to detect HC-OA cell proliferation ability. HC-OA cells were seeded in 96-well plates (1.0 × 104 cells/well), incubated at 37 °C for 0, 24, 48, and 72 h, and then added to CCK-8 (Beyotime, Haimen, China) and incubated for 2 h. A microplate reader (BIOTEK, VT, USA) was used to detect the optical density (OD) value at 450 nm.
ROS level detection
The cultured HC-OA cells were washed with 1 × phosphate-buffered solution (PBS), and the Reactive Oxygen Species Assay Kit (Sigma-Aldrich, Minneapolis, USA) was used to detect ROS levels according to the supplier’s instructions. Three replicate wells in each group, a microplate reader (BIOTEK, VT, USA) was used to detect the absorbance at 485/535 nm.
qRT-PCR
The total RNA in HC-OA cell was extracted by TRIzol reagent (Invitrogen, Carlsbad, CA). The concentration of the extracted RNA was determined by NanoDrop 2000 (Thermo Fisher Scientific). Reverse transcription kit (Takara, Dalian, China) was used to reverse transcribe the extracted total RNA into cDNA. QRT-PCR was performed on an Applied Biosystems StepOnePlus Real-time PCR system (Applied Biosystems, Foster City, CA, USA) using SYBR Green Real-time PCR Master Mix (Toyobo, Osaka, Japan) according to the manufacturer’s protocol. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was served as endogenous control. The CAMP mRNA primer sequences are 5’-GAA GAC CCA AAG GAA TGG CC-3’ (forward) and 5’-TCA GAG CCC AGA AGC CTG AG-3’ (reverse). GAPDH primer sequences: 5’-GAA GGT GAA GGT CGG AGT C-3’ (forward), 5’-GAA GAT GGT GAT GGG ATT TC-3’ (reverse). The relative expression of CAMP mRNA was calculated using the 2−∆∆Ct method.
Western blot
HC-OA cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (Beyotime, Shanghai, China) for half an hour and centrifuged at 17,000 × g for 45 min at 4 °C. Bicinchoninic acid (BCA) protein assay kit (Beyotime, Shanghai, China) was used to detect protein concentration. Then sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate the proteins, and proteins were transferred to polyvinylidene fluoride (PVDF) membrane, and then used Tween–Tris-buffered saline (TTBS) containing 5% skim milk to seal the PVDF membrane at room temperature. Then, anti-CAMP (#ab80895, 1:1000, Abcam, Cambridge, UK) and anti-GAPDH (#ab181602, 1:1000, Abcam, Cambridge, UK) were incubated with PVDF membrane overnight at 4 °C. The membrane was then washed 3 times with TTBS and incubated with a secondary antibody conjugated with horseradish peroxidase at room temperature for 2 h. The blot was visualized with an enhanced chemiluminescence (ECL) kit (Santa Cruz Biotechnology) and scanned by ChemImager 5500 V2.03 software. The relative integrated density value (IDV) was calculated using GAPDH as an internal control.
CpG methyltransferase treatment and luciferase reporter assay
The pCpGfree-CAMP (− 828/ − 1) plasma was treated by M.SssI or M.HpaII methyltransferase, and then the methylation was confirmed by restriction enzyme HpaII. The HC-OA cells were seeded in 6-well plates. When the cells were cultured to 60–90% confluence, methylated or unmethylated plasmids were transfected into HC-OA cells by Lipofectamine 3000 (Invitrogen, California, USA) according to the manufacturer’s recommendations. Forty-eight hours after transfection, the medium was collected, and 20 μl aliquots were added into the 96-well plate. A 100 μl Quanti-Luc™ (InvivoGen, San Diego, CA, USA) was injected into each well and read the luciferase activity.
Statistical analysis
GraphPad Prism 8.0 (GraphPad Software, San Diego, CA) was used for statistical analysis of the data in this study. Two-tailed Student’s t test and analysis of variance (ANOVA) were used to the difference between groups. p < 0.05 was considered to have statistical significance.
Results
The expression of CAMP in cartilage tissue of OA patients was upregulated, and the level of methylation was downregulated
The analysis of the GSE117999 dataset shows that CAMP was upregulated in the articular cartilage of patients with OA (Table 1, Fig. 1a). The analysis of the GSE48422 dataset [21] showed that the level of CAMP methylation was downregulated (Table 2, Fig. 1b).
Table 1.
Top 15 upregulated and downregulated gene in arthritic samples from patients with and without osteoarthritis ranked by logFC after GEO2R analysis
| GENE_SYMBOL | Adj. p value | logFC | Up/Down |
|---|---|---|---|
| CRB1 | 1.52E–04 | 1.01580843 | Up |
| ZSCAN12 | 1.89E–03 | 1.0271285 | Up |
| LEO1 | 1.21E–04 | 1.03342688 | Up |
| ESR1 | 4.03E–04 | 1.04997982 | Up |
| FAM20A | 2.36E–03 | 1.05411929 | Up |
| ZNF354B | 2.06E–04 | 1.05630409 | Up |
| CCR10 | 1.15E–03 | 1.06099425 | Up |
| INCENP | 1.21E–04 | 1.18121011 | Up |
| CAMP | 3.24E–03 | 1.23685287 | Up |
| THAP5 | 1.31E–05 | 1.27811954 | Up |
| SMNDC1 | 8.17E–04 | 1.29092563 | Up |
| HOXC8 | 9.69E–05 | 1.30931826 | Up |
| LCE2B | 3.73E–04 | 1.34166775 | Up |
| FAM90A1 | 2.87E–03 | 1.40391379 | Up |
| QRFPR | 4.59E–04 | 1.57552192 | Up |
| SLC38A6 | 1.41E–03 | − 1.64465134 | Down |
| PCDHB10 | 6.24E–04 | − 1.43430506 | Down |
| HLA-DRB4 | 1.21E–04 | − 1.4061153 | Down |
| TOLLIP | 8.09E–04 | − 1.31133046 | Down |
| HLA-DQA2 | 9.20E–04 | − 1.29212829 | Down |
| TP53TG3D | 1.99E–03 | − 1.25442713 | Down |
| CHST1 | 2.18E–03 | − 1.1748763 | Down |
| ANO8 | 2.34E–03 | − 1.16784996 | Down |
| TRIM78P | 2.18E–03 | − 1.10578964 | Down |
| ADHFE1 | 1.66E–03 | − 1.07972333 | Down |
| DUT | 3.48E–03 | − 1.07647613 | Down |
| SS18L1 | 2.18E–03 | − 1.07511907 | Down |
| FRMD8 | 1.72E–04 | − 1.06645789 | Down |
| FAM184B | 2.18E–03 | − 1.06047291 | Down |
| TMEM259 | 9.41E–04 | − 1.05326043 | Down |
FC, fold change
Fig. 1.

Transcriptome and methylation level detection in OA patients. a Analysis of the relative expression level of CAMP mRNA in the GSE117999 dataset (ncontrol = 10, nOA = 10). b The methylation level of CAMP gene in the GSE48422 dataset (ncontrol = 4, nOA = 4)
Table 2.
Top 20 hypomethylated genes in arthritic and non-arthritic knee cartilage samples ranked by logFC after GEO2R analysis
| GENE_SYMBOL | p value | logFC | Up/Down |
|---|---|---|---|
| GUCY2D | 0.00902988 | 4.21533489 | Up |
| GPR23 | 0.01044718 | 4.10888772 | Up |
| LMBR1L | 0.0050466 | 4.06301737 | Up |
| MOBP | 0.02433125 | 3.81893625 | Up |
| RASSF8 | 0.00004736 | 3.71186833 | Up |
| SLC6A5 | 0.02136651 | 3.49741335 | Up |
| ING4 | 0.00371036 | 3.40976791 | Up |
| CDK5 | 0.01690306 | 3.39845271 | Up |
| hsa-mir-29b-2 | 0.00810529 | 3.39799728 | Up |
| PITX1 | 0.03355549 | 3.31112165 | Up |
| TLE6 | 0.02379696 | 3.31057234 | Up |
| IL22 | 0.02971686 | 3.29164562 | Up |
| CDIPT | 0.02299476 | 3.24065781 | Up |
| FLJ10986 | 0.0034974 | 3.21953344 | Up |
| KIAA1970 | 0.02280297 | 3.1642642 | Up |
| GDF7 | 0.01267419 | 3.14401884 | Up |
| CAB39L | 0.00421281 | 3.13792295 | Up |
| CHAF1B | 0.00197712 | 3.13636909 | Up |
| C15orf2 | 0.01644456 | 3.07462778 | Up |
| TMPRSS6 | 0.04829902 | 3.06043658 | Up |
| NDUFV3 | 0.01618486 | − 3.45593987 | Down |
| CINP | 0.01970577 | − 3.40575442 | Down |
| ACOX3 | 0.0123907 | − 3.09168387 | Down |
| KLK15 | 0.00245482 | − 3.01407804 | Down |
| RP13-360B22.2 | 0.04529023 | − 2.9067811 | Down |
| C10orf120 | 0.04175249 | − 2.85198326 | Down |
| C10orf26 | 0.03356837 | − 2.83509207 | Down |
| ATP6AP2 | 0.00771311 | − 2.81820993 | Down |
| MED28 | 0.01218592 | − 2.77282343 | Down |
| ZDHHC16 | 0.0367154 | − 2.75390711 | Down |
| LOC57228 | 0.03643365 | − 2.71534123 | Down |
| C15orf5 | 0.00278977 | − 2.67864013 | Down |
| ALDH1A3 | 0.03431337 | − 2.65248752 | Down |
| DNMT2 | 0.03566104 | − 2.64961214 | Down |
| MGC41945 | 0.02997186 | − 2.64282551 | Down |
| ASF1B | 0.0193366 | − 2.62752981 | Down |
| CAMP | 0.01873345 | − 2.62260485 | Down |
| ZNF44 | 0.01006365 | − 2.56469726 | Down |
| IDS | 0.00377758 | − 2.56033268 | Down |
| TCN2 | 0.01701305 | − 2.53741851 | Down |
FC, fold change
CAMP was highly expressed in osteoarthritis articular cartilage cells
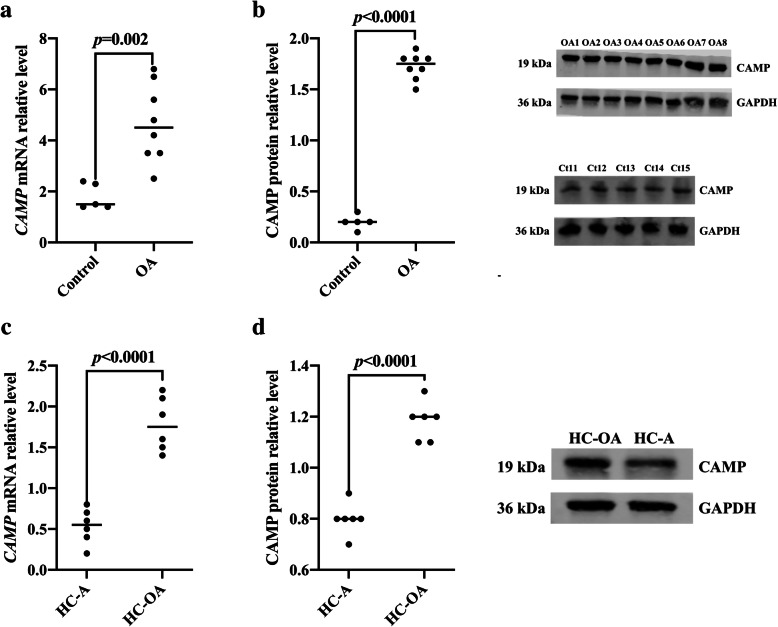
We analyzed the expression levels of CAMP in the cartilage tissue of 8 cases of OA patients undergoing total knee arthroplasty and 5 cases of trauma requiring high amputation (excluding OA) (Control). The results showed that the expression levels of CAMP mRNA and CAMP protein in the articular cartilage tissue of KOA patients were significantly higher than those of the control group (p = 0.002, p < 0.0001, Fig. 2a, b). Our analysis in Human Chondrocytes Osteoarthritis (HC-OA) cells and Human Chondrocytes-articular (HC-A) cells showed that the CAMP mRNA level and CAMP protein level in HC-OA cells were significantly higher than those in HC-A cells (p < 0.001, p < 0.0001, Fig. 2c, d).
Fig. 2.
CAMP was highly expressed in osteoarthritis articular cartilage cells. a The expression level of CAMP mRNA in articular cartilage tissues of OA patients and control group were analyzed by qRT-PCR. b The expression level of CAMP protein in articular cartilage tissues of OA patients and control group were analyzed by western blot. c CAMP mRNA expression levels in Human Chondrocytes Osteoarthritis (HC-OA) cells and Human Chondrocytes-articular (HC-A) cells were analyzed by qRT-PCR. d CAMP protein expression levels in HC-OA cells and HC-A cells were detected by western blot
Silencing CAMP gene inhibited the proliferation of osteoarthritis articular cartilage cells and promoted their apoptosis
In order to study the role of CAMP gene in the proliferation and apoptosis of chondrocytes in OA patients, we used small hairpin RNA to silence CAMP gene (sh-CAMP) and no template control (sh-NC) and no transfection group (Control) as control. QRT-PCR and western blot results confirmed that CAMP gene was successfully knocked out in HC-OA cells (p < 0.001, p < 0.01; Fig. 3a, b). CCK-8 assay results showed that compared with the Control, the proliferation ability of HC-OA cells was significantly inhibited after sh-CAMP transfection (p < 0.01, Fig. 3c). Flow cytometry detection of cell apoptosis showed that the apoptotic rate of HC-OA cells after sh-CAMP transfection was significantly higher than that of the Control (p < 0.01, Fig. 3d).
Fig. 3.
Silencing CAMP gene inhibited the proliferation of osteoarthritis articular cartilage cells and promoted their apoptosis. a The expression level of CAMP mRNA in Control, sh-NC, and sh-CAMP-transfected HC-OA cells was detected by qRT-PCR. b the expression level of CAMP protein in Control, sh-NC, and sh-CAMP-transfected HC-OA cell was detected by western blot. c The cell proliferation ability of Control, sh-NC, and sh-CAMP-transfected HC-OA cells was detected by CCK-8. d The apoptotic ability of Control, sh-NC, and sh-CAMP-transfected HC-OA cells was detected by flow cytometry. **p < 0.01, ***p < 0.001, ****p < 0.0001, compare with Control
CAMP gene methylation inhibited ROS levels and inflammatory response levels in osteoarthritis articular cartilage cells
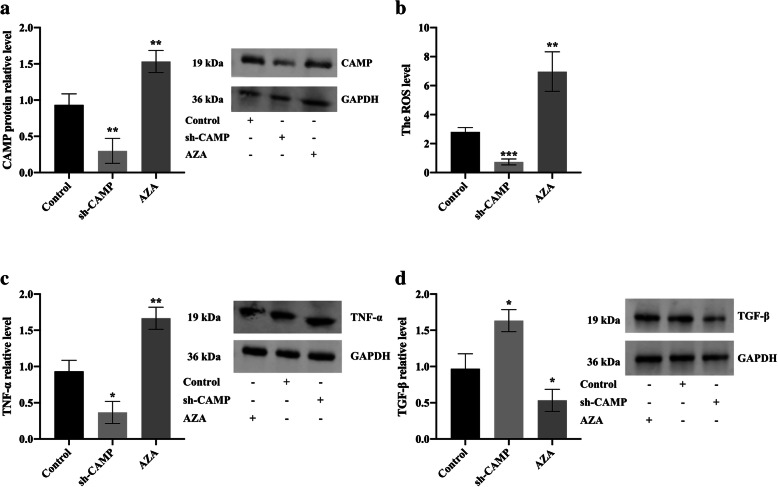
In order to further study the effect of CAMP methylation on the level of oxidative stress and inflammatory response levels in HC-OA cells, we set up 3 groups, namely Control, sh-CAMP-transfected HC-OA cell group (sh-CAMP), and 5-μM AZA treatment group. First, western blot results showed that compared with the Control, the CAMP protein expression in HC-OA cells was significantly downregulated after sh-CAMP transfection, and CAMP expression in HC-OA cells was significantly increased after AZA treatment (Fig. 4a). After transfection with sh-CAMP, the level of ROS in HC-OA cells decreased significantly, while the level of ROS in HC-OA cells increased after AZA treatment (Fig. 4b). Western blot results showed that compared with Control, TNF-α levels in HC-OA cells were significantly decreased after sh-CAMP transfection, and TNF-α levels were significantly increased after AZA treatment (Fig. 4c). The level of TGF-β in HC-OA cells was significantly increased after sh-CAMP transfection, and AZA treatment downregulated the expression of TGF-β (Fig. 4d). These results showed that CAMP gene methylation downregulated the expression of CAMP in HC-OA cells and inhibited the level of oxidative stress and inflammatory response levels.
Fig. 4.
CAMP gene methylation inhibited ROS levels and inflammatory response levels in osteoarthritis articular cartilage cells. a The CAMP protein expression level in Control, sh-CAMP-transfected HC-OA cells, and 5-μM AZA-treated HC-OA cells were analyzed by western blot. b The comparison of the ROS levels in Control, sh-CAMP-transfected HC-OA cells, and 5-μM AZA-treated HC-OA cells. c The TNF-α protein expression levels in Control, sh-CAMP-transfected HC-OA cells, and 5-μM AZA-treated HC-OA cells were analyzed by western blot. d The TGF-β protein expression levels in Control, sh-CAMP-transfected HC-OA cells, and 5-μM AZA-treated HC-OA cells were analyzed by western blot. *p < 0.05, **p < 0.01, ***p < 0.001, compared with Control
CAMP gene methylation inhibited the proliferation of osteoarthritis articular cartilage cells and promoted their apoptosis
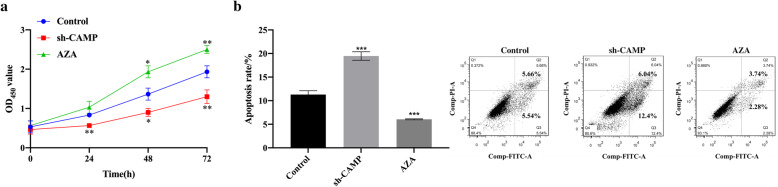
In order to further study the effect of CAMP gene methylation on the proliferation and apoptosis of HC-OA cells, we set up 3 groups, namely Control, sh-CAMP, and AZA. Western blot analysis showed that, compared with Control, the CAMP protein in the sh-CAMP transfection group was significantly downregulated, and the CAMP protein expression level was significantly increased after AZA treatment (Fig. 4a). CCK-8 assay results showed that compared with Control, the proliferation ability of HC-OA cells in the sh-CAMP transfection group was inhibited, and the proliferation ability of HC-OA cells was significantly increased after AZA treatment (Fig. 5a). The results of flow cytometry showed that compared with Control, the apoptotic rate of HC-OA cells in the sh-CAMP transfection group was significantly increased, and the apoptotic rate of HC-OA cells was significantly decreased after AZA treatment (Fig. 5b). These results showed that CAMP gene methylation inhibited the proliferation of osteoarthritis articular cartilage cells and promoted their apoptosis.
Fig. 5.
CAMP gene methylation inhibited the proliferation of osteoarthritis articular cartilage cells and promoted their apoptosis. a The cell proliferation ability of Control, sh-CAMP-transfected HC-OA cells, and 5-μM AZA-treated HC-OA cells were detected by CCK-8. b The apoptotic rate of Control, sh-CAMP-transfected HC-OA cells and 5-μM AZA-treated HC-OA cells were detected by flow cytometry. *p < 0.05, **p < 0.01, ***p < 0.001, compared with Control
DNA methylation directly downregulated the activity of CAMP gene promoter
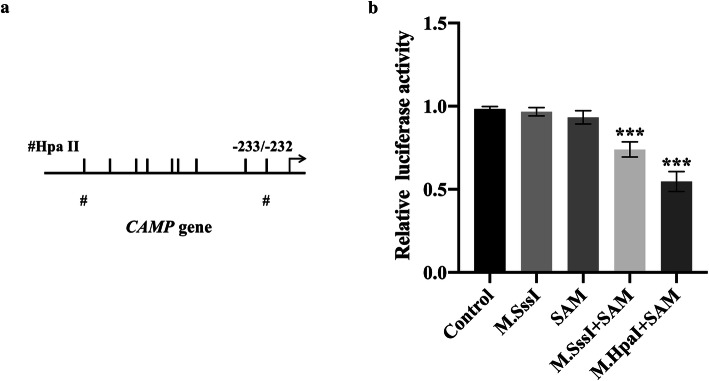
In order to further verify whether DNA methylation can downregulate the activity of the CAMP gene promoter, a pCpGfree-basic-Lucia construct without CpG dinucleotides was used for luciferase reporter assay. S-adenosylmethionine (SAM) is used as the methyl donor for the enzymatic reaction, and pCpGfree-CAMP (− 828/ − 1) was treated with M.SssI and M.HpaII methyltransferase. Untreated, M.SssI-treated or SAM-treated plasmids were used as controls. Then the methylated or unmethylated plasmid was transfected into the HC-OA cells, and the luciferase activity was measured. The results showed that higher promoter activity was detected in HC-OA cells transfected with the unmethylated pCpGfree-CAMP (− 828/ − 1) construct. Compared with the unmethylated construct, when the pCpGfree-CAMP (− 828/ − 1) construct treated with M.SssI was transfected into HC-OA cells, the promoter activity was significantly reduced (Fig. 6). In addition, HC-OA cells transfected with the pCpGfree-CAMP (− 828/ − 1) construct were only methylated by M.HpaII methyltransferase treatment and also showed a similar downregulation of promoter activity (Fig. 6). This indicates that the CpG site of M.HpaII in the CAMP promoter is important for promoter activity regulation, and the scattered methylation on the CpG site may not be able to effectively reduce the activity of the gene promoter. We can also know that the CpG site (− 233/ − 232) closest to the main transcription start site of CAMP is important for transcription regulation. These results were consistent with the results of AZA treatment inducing CAMP expression (Figs. 4 and 5). In summary, these results indicated that DNA methylation directly downregulated the activity of the CAMP promoter in HC-OA cells.
Fig. 6.
Direct downregulation of CAMP gene promoter activity by DNA methylation. a A schematic illustration of the CAMP gene promoter. The major transcriptional start site is indicated by the arrow at position +1. Upper tick marks indicate individual CpG sites in CAMP gene promoter. The symbols of “#” represent HpaII sites. b Luciferase activities of unmethylated and methylated CAMP gene promoter constructs in HC-OA cells. SAM, S-adenosylmethionine. ***p < 0.001, compared with Control
Discussion
In this study, we extracted and analyzed the differentially expressed genes and methylation levels in chondrocytes from public databases, and found that CAMP (also known as LL-37) was upregulated in the articular cartilage of patients with OA, the methylation level of CAMP gene was downregulated. We found that CAMP protein expression was upregulated in articular cartilage tissue of OA patients and HC-OA cells. After silencing the expression of the CAMP gene, we found that the proliferation of HC-OA cells was inhibited, accompanied by a significant increase in the apoptotic rate. After demethylation, the level of ROS and inflammation in HC-OA cells increased significantly, the cell proliferation ability was significantly enhanced, and the apoptosis rate was significantly reduced. Therefore, we speculated that CAMP gene promoter methylation induced chondrocyte apoptosis by inhibiting ROS levels and inflammatory response levels.
Current research evidence shows that risk factors for OA include genetics, inflammation, environmental factors, age, stress stimulation, etc. [23, 24]. In recent years, more and more studies have found that epigenetic modification plays an important role in the pathogenesis of OA. Epigenetic modification works by regulating gene transcription or post-transcriptional regulation, including DNA methylation, histone modification, chromosome remodeling, and non-coding RNAs (ncRNAs) [6, 25, 26]. Usually, DNA methylation occurs in cytosine, guanine, and nucleotides (CpG), which is mainly manifested as the phenomenon of conversion of cytosine to 5-methylcytosine [27, 28]. The research on DNA methylation function mainly focuses on the transcription start site of genes [29]. Methylation of the promoter region can inhibit gene expression; on the one hand, methylation of the CpG site directly interferes with the binding of transcription factors to DNA in the regulatory region; on the other hand, methylated DNA and methylated CpG binding region proteins such as MeCP2 specifically bind to form a complex, which restricts the passage of transcription factors to their binding sites, thereby inhibiting gene expression [30–32].
In this study, we found that the expression level of CAMP protein increased significantly after AZA treatment. The reason might be that the methylation level of the promoter region of CAMP gene decreased after AZA treatment, and the binding efficiency of CAMP gene transcription factors increased; as a result, the expression level of CAMP increased significantly. At present, researchers have tried to use AZA for in vitro intervention of OA chondrocytes. For example, Alvarez-Garcia et al. [33] treated TC28 cells with AZA and the expression of atonal bHLH transcription factor 8 (ATOH8) and T-box transcription factor 4 gene (TBX4) increased significantly. In addition, some researchers found that the DNA methylation level of the inducible nitric oxide synthase (iNOS) enhancer-5.8 kb CpG site decreased after AZA treatment, and the iNOS expression level increased; the methylation of the iNOS enhancer could inhibit the cell cycle process by downregulating NF-κB level and reduce the pro-inflammatory response, which had important therapeutic significance for OA [34].
In this study, we found that CAMP was highly expressed in articular cartilage cells of OA patients. Silencing the expression of CAMP gene inhibited the proliferation of osteoarthritis articular cartilage cells and promoted their apoptosis, suggesting that CAMP may play an important role in the pathogenesis of osteoarthritis. Yu et al. [35] showed that CAMP inhibited inflammation and promoted bone formation of bone marrow stromal cells (BMSC) through purinergic receptor P2X 7 (P2RX7) and mitogen-activated protein kinase (MAPK) signaling pathway. Further, in vitro studies had found that CAMP gene methylation inhibited the level of ROS and TNF-α in chondrocytes, promoted the level of TGF-β expression, inhibited the proliferation of chondrocytes, and promoted their apoptosis. The results of Kuensaen et al. [36] found that high levels of CAMP promote the expression of downstream pro-inflammatory cytokines (especially IL17A), which was related to the pathogenesis of inflammatory arthritis. Hu et al. [37] found that CAMP regulated the production of inflammatory cytokines such as TNF-α and inhibited cell apoptosis. The results of this study suggested that the expression of CAMP gene and methylation in promoter region might be involved in the occurrence of osteoarthritis from the level of oxidative stress and the expression of inflammatory factors.
There are some limitations in this study. First of all, the treatment of the articular cartilage tissue samples used for analysis is unclear, and whether there are drugs that affect the expression of inflammatory factors in tissue is unclear. Secondly, the expression and methylation of CAMP gene may lead to the occurrence of OA through other signaling pathways, which was not confirmed in this study. In addition, the results of this study need to be further confirmed in vivo.
Conclusion
In this study, we found that CAMP gene promoter methylation induces chondrocyte apoptosis by inhibiting ROS levels and inflammatory response levels, which is a potential target for OA therapy.
Acknowledgements
Not applicable
Authors’ contributions
YW contributed to the experiment design, manuscript draft, and data analysis. GW and YL contributed to the experiment implementation, manuscript draft and data analysis. GY, TY, and LH designed and performed the experiments. GW and YW analyzed the data. GW and YL wrote the manuscript. The authors read and approved the final manuscript.
Funding
This work was supported by National Natural Science Foundation of China (No. 81960409, No. 81760403), Yunnan Province Clinical Center for Bone and Joint Diseases (No. ZX2019-03-04), Yunnan Province Medical Leaders Talent Project (No.L-201601), Expert workstation project of Shiyi Chen [No. 2018IC102], and Joint special fund project of applied basic research of Kunming Medical University, Department of Science and Technology of Yunnan province (No. 202001AY070001-043).
Availability of data and materials
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Guoliang Wang and Yanlin Li contributed equally to this work.
Contributor Information
Guoliang Wang, Email: 200301144@163.com.
Yanlin Li, Email: 852387873@qq.com.
Guang Yang, Email: 1447673730@qq.com.
Tengyun Yang, Email: 806991922@qq.com.
Lu He, Email: 809754223@qq.com.
Yang Wang, Email: 358556760@qq.com.
References
- 1.Glyn-Jones S, Palmer AJR, Agricola R, Price AJ, Vincent TL, Weinans H, Carr AJ. Osteoarthritis. Lancet. 2015;386(9991):376–387. doi: 10.1016/S0140-6736(14)60802-3. [DOI] [PubMed] [Google Scholar]
- 2.Mandl LA. Osteoarthritis year in review 2018: clinical. Osteoarthr Cartil. 2019;27(3):359–364. doi: 10.1016/j.joca.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Abramoff B, Caldera FE. Osteoarthritis: pathology, diagnosis, and treatment options. Med Clin North Am. 2020;104(2):293–311. doi: 10.1016/j.mcna.2019.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Shen J, Abu-Amer Y, O’Keefe RJ, McAlinden A. Inflammation and epigenetic regulation in osteoarthritis. Connect Tissue Res. 2017;58(1):49–63. doi: 10.1080/03008207.2016.1208655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim H, Kang D, Cho Y, Kim JH. Epigenetic regulation of chondrocyte catabolism and anabolism in osteoarthritis. Mol Cell. 2015;38(8):677–684. doi: 10.14348/molcells.2015.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Hong T, Wang S, Mo J, Tian T, Zhou X. Epigenetic modification of nucleic acids: from basic studies to medical applications. Chem Soc Rev. 2017;46(10):2844–2872. doi: 10.1039/C6CS00599C. [DOI] [PubMed] [Google Scholar]
- 7.Liberman N, Wang SY, Greer EL. Transgenerational epigenetic inheritance: from phenomena to molecular mechanisms. Curr Opin Neurobiol. 2019;59:189–206. doi: 10.1016/j.conb.2019.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan L, Liang Y, Ma B, Wang D, Liu W, Huang J, Xiong J, Peng L, Chen J, Zhu W, Wang D. DNA methylation profiling in chondrocyte dedifferentiation in vitro. J Cell Physiol. 2017;232(7):1708–1716. doi: 10.1002/jcp.25486. [DOI] [PubMed] [Google Scholar]
- 9.Jeffries MA, Donica M, Baker LW, Stevenson ME, Annan AC, Humphrey MB, James JA, Sawalha AH. Genome-wide DNA methylation study identifies significant epigenomic changes in osteoarthritic cartilage. Arthritis Rheum. 2014;66(10):2804–2815. doi: 10.1002/art.38762. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Yu Y, Hao J, Wen Y, Han J, Hou W, Liu R, Zhao B, He A, Li P, Fan Q, Wu C, Wang S, Wang X, Ning Y, Guo X, Zhang F. Genome-wide DNA methylation profiling of articular cartilage reveals significant epigenetic alterations in Kashin-Beck disease and osteoarthritis. Osteoarthr Cartil. 2017;25(12):2127–2133. doi: 10.1016/j.joca.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Sokolove J, Lepus CM. Role of inflammation in the pathogenesis of osteoarthritis: latest findings and interpretations. Ther Adv Musculoskelet Dis. 2013;5(2):77–94. doi: 10.1177/1759720X12467868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 2011;7(1):33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- 13.Morikawa M, Derynck R, Miyazono K. TGF-beta and the TGF-beta family: context-dependent roles in cell and tissue physiology. Cold Spring Harb Perspect Biol. 2016;8(5). [DOI] [PMC free article] [PubMed]
- 14.Vander Ark A, Cao J, Li X. TGF-beta receptors: in and beyond TGF-beta signaling. Cell Signal. 2018;52:112–120. doi: 10.1016/j.cellsig.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Zhen G, Wen C, Jia X, Li Y, Crane JL, Mears SC, Askin FB, Frassica FJ, Chang W, Yao J, Carrino JA, Cosgarea A, Artemov D, Chen Q, Zhao Z, Zhou X, Riley L, Sponseller P, Wan M, Lu WW, Cao X. Inhibition of TGF-beta signaling in mesenchymal stem cells of subchondral bone attenuates osteoarthritis. Nat Med. 2013;19(6):704–712. doi: 10.1038/nm.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iliopoulos D, Malizos KN, Tsezou A. Epigenetic regulation of leptin affects MMP-13 expression in osteoarthritic chondrocytes: possible molecular target for osteoarthritis therapeutic intervention. Ann Rheum Dis. 2007;66(12):1616–1621. doi: 10.1136/ard.2007.069377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim KI, Park YS, Im GI. Changes in the epigenetic status of the SOX-9 promoter in human osteoarthritic cartilage. J Bone Miner Res. 2013;28(5):1050–1060. doi: 10.1002/jbmr.1843. [DOI] [PubMed] [Google Scholar]
- 18.Nagaoka I, Tamura H, Reich J. Therapeutic potential of cathelicidin peptide LL-37, an antimicrobial agent, in a murine sepsis model. Int J Mol Sci. 2020;21(17). [DOI] [PMC free article] [PubMed]
- 19.Agier J, et al. Cathelicidin LL-37 affects surface and intracellular toll-like receptor expression in tissue mast cells. J Immunol Res. 2018;2018:7357162. doi: 10.1155/2018/7357162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Shan Z, Yang B, Yang D, Men C, Cui Y, Wu J. Cathelicidin LL37 Promotes epithelial and smooth-muscle-like differentiation of adipose-derived stem cells through the Wnt/beta-catenin and NF-kappaB pathways. Biochemistry (Mosc) 2017;82(11):1336–1345. doi: 10.1134/S0006297917110116. [DOI] [PubMed] [Google Scholar]
- 21.Moazedi-Fuerst FC, Hofner M, Gruber G, Weinhaeusel A, Stradner MH, Angerer H, Peischler D, Lohberger B, Glehr M, Leithner A, Sonntagbauer M, Graninger WB. Epigenetic differences in human cartilage between mild and severe OA. J Orthop Res. 2014;32(12):1636–1645. doi: 10.1002/jor.22722. [DOI] [PubMed] [Google Scholar]
- 22.Reynard LN, Bui C, Syddall CM, Loughlin J. CpG methylation regulates allelic expression of GDF5 by modulating binding of SP1 and SP3 repressor proteins to the osteoarthritis susceptibility SNP rs143383. Hum Genet. 2014;133(8):1059–1073. doi: 10.1007/s00439-014-1447-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Neill TW, McCabe PS, McBeth J. Update on the epidemiology, risk factors and disease outcomes of osteoarthritis. Best Pract Res Clin Rheumatol. 2018;32(2):312–326. doi: 10.1016/j.berh.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Scheler F, Verwiebe R. Therapeutic use of diuretics and digitalis glycosides in renal failure. Verh Dtsch Ges Inn Med. 1989;95:252–260. doi: 10.1007/978-3-642-83864-4_44. [DOI] [PubMed] [Google Scholar]
- 25.Skvortsova K, Iovino N, Bogdanovic O. Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol. 2018;19(12):774–790. doi: 10.1038/s41580-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 26.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39(2):157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 27.Mersakova S, et al. Methylation of CADM1 and MAL together with HPV status in cytological cervical specimens serves an important role in the progression of cervical intraepithelial neoplasia. Oncol Lett. 2018;16(6):7166–7174. doi: 10.3892/ol.2018.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu K, Xu C, Lei M, Yang A, Loppnau P, Hughes TR, Min J. Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA. J Biol Chem. 2018;293(19):7344–7354. doi: 10.1074/jbc.RA118.001785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 30.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clemens AW, Wu DY, Moore JR, Christian DL, Zhao G, Gabel HW. MeCP2 Represses enhancers through chromosome topology-associated DNA methylation. Mol Cell. 2020;77(2):279–293. doi: 10.1016/j.molcel.2019.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinde B, Wu DY, Greenberg ME, Gabel HW. DNA methylation in the gene body influences MeCP2-mediated gene repression. Proc Natl Acad Sci U S A. 2016;113(52):15114–15119. doi: 10.1073/pnas.1618737114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alvarez-Garcia O, Fisch KM, Wineinger NE, Akagi R, Saito M, Sasho T, Su AI, Lotz MK. Increased DNA methylation and reduced expression of transcription factors in human osteoarthritis cartilage. Arthritis Rheum. 2016;68(8):1876–1886. doi: 10.1002/art.39643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Andres MC, Takahashi A, Oreffo RO. Demethylation of an NF-kappaB enhancer element orchestrates iNOS induction in osteoarthritis and is associated with altered chondrocyte cell cycle. Osteoarthr Cartil. 2016;24(11):1951–1960. doi: 10.1016/j.joca.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Yu X, Quan J, Long W, Chen H, Wang R, Guo J, Lin X, Mai S. LL-37 inhibits LPS-induced inflammation and stimulates the osteogenic differentiation of BMSCs via P2X7 receptor and MAPK signaling pathway. Exp Cell Res. 2018;372(2):178–187. doi: 10.1016/j.yexcr.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 36.Kuensaen C, Chomdej S, Kongdang P, Sirikaew N, Jaitham R, Thonghoi S, Ongchai S. LL-37 alone and in combination with IL17A enhances proinflammatory cytokine expression in parallel with hyaluronan metabolism in human synovial sarcoma cell line SW982-A step toward understanding the development of inflammatory arthritis. PLoS One. 2019;14(7):e0218736. doi: 10.1371/journal.pone.0218736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Z, Murakami T, Suzuki K, Tamura H, Reich J, Kuwahara-Arai K, Iba T, Nagaoka I. Antimicrobial cathelicidin peptide LL-37 inhibits the pyroptosis of macrophages and improves the survival of polybacterial septic mice. Int Immunol. 2016;28(5):245–253. doi: 10.1093/intimm/dxv113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.