Abstract
The highly conserved striatin-interacting phosphatase and kinase (STrIPAK) multimeric complex regulates the hippo signaling pathway through phosphatase activity. A recent structure of the core STrIPAK hub reveals how striatins tetramerize to serve as a scaffolding platform for the assembly of an intricate architecture, which is distinct from that of all other protein phosphatase 2A (PP2A) complexes.
As the most common post-translational modification (PTM), protein phosphorylation plays a key role in regulating the majority of cell signaling networks. The PP2A family functions to remove PTM phosphate decorations from phosphoserine and phosphothreonine residues on thousands of substrates. To recognize the vast repertoire of targets and achieve specificity in target selection, the PP2A A–C core dimer recruits a third, regulatory subunit, B, which is separated into structurally distinct B (B55), B′ (B56) and B′′ (PR72/130) classes (Fig. 1a). Striatin/STRN (B′′′) represents the fourth class of PP2A B subunits, which assemble into much larger structures to form the STRIPAK complex that regulates cellular pathways, including the Hippo signaling pathway1. Whereas B, B′ and B′′ PP2A complexes assemble into heterotrimeric holoenzymes, the structure and stoichiometric composition of STRIPAK has remained a mystery.
Fig. 1 |. comparison of PP2a complexes from four major B subunit families.
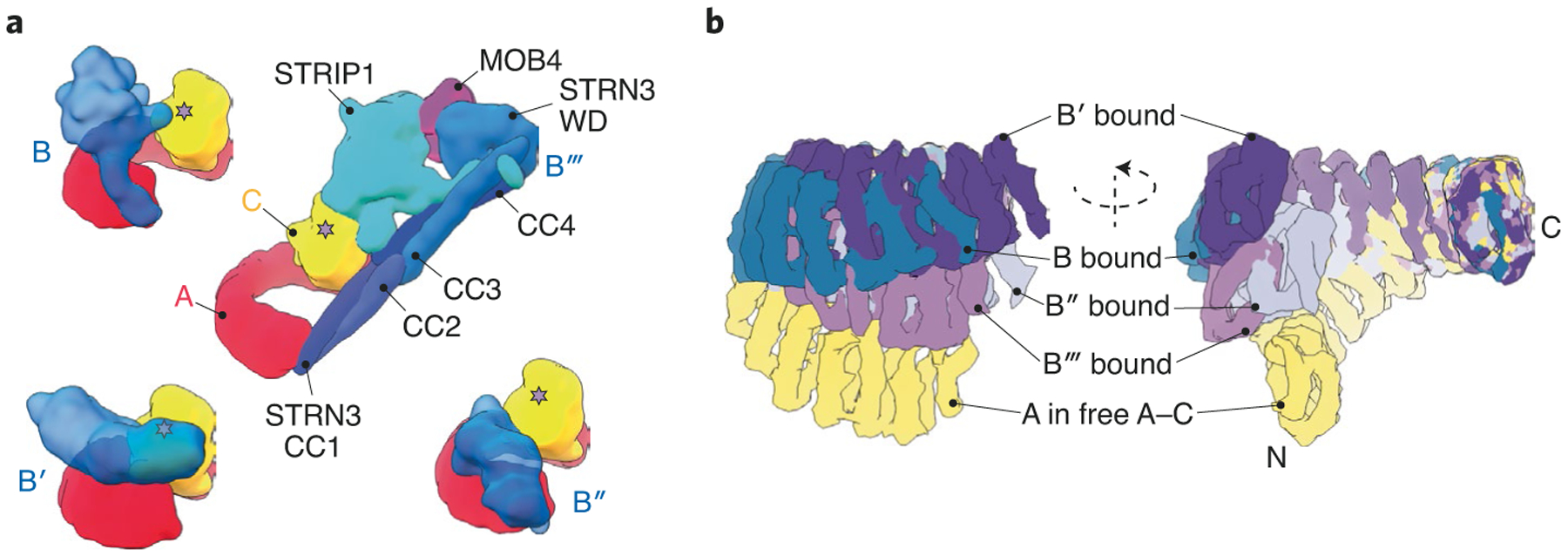
a, representative architectures of PP2A holoenzyme complexes from each family (B, B′, B′′ and B′′′). The catalytic active site is marked by a star on the C subunit. b, Superposition of A subunits at the constant A–C dimeric interface reveals flexibility at the distal N terminus of the scaffolding A subunit for each PP2A family member. PP2A A–C conformations in the context of different B-subunit-containing complexes are color coded and labeled. The overlay of subunits is shown as two different views, rotated around the vertical axis as indicated. Structures and overlays were generated using PDB IDs 3DW8 (B–PP2A), 2IAE (B′–PP2A), 4I5L (B′′–PP2A), 7K36 (B′′′–PP2A) and 2IE4 (A–C dimer).
Now, Jeong et al.2 have used cryo-EM to solve the structure of the STRIPAK core complex, which consists of eight protein constituents (Fig. 1a). The completion of the structures of PP2A complexes with representatives of all four major B subunit classes provides new insights into PP2A biology. For example, the flexibility of the PP2A A subunit is critical in regulating B subunit binding and thus substrate specificity, as demonstrated by the wide spectrum of conformations adopted by the A subunit across the PP2A complex structures solved (Fig. 1b). Pathological mutations within the A subunit compromise this flexibility to prevent or enhance interactions with different regulatory subunits, promoting tumorigenesis3,4. Similarly, small molecules that promote the assembly of specific PP2A complexes while preventing the formation of others inhibit cancer progression5–7. The structure of STRIPAK will undoubtedly aid in the further characterization of the PP2A phosphatome network and in the development of novel phosphatase-based therapies.
Instead of incorporating a single prototypical regulatory B subunit, as observed in other PP2A heterotrimers, the new structure reveals that the PP2A A–C heterodimer in STRIPAK assembles with four STRN3 proteins along with the regulatory proteins STRIP1 and MOB4 (Fig. 1a). The N-terminal coiled-coil (CC) domains of STRN3 proteins coalesce to form a homotetramer that serves as an assembly hub for the other proteins. In contrast to the CC domain, which is highly conserved among STRN proteins, the tryptophan-aspartate (WD)-repeat domain exhibits higher sequence variability. Since only one of the four WD domains is structurally involved in the STRIPAK core, it is possible that the other three behave as dynamic ‘tentacles’ extruding away from the assembly to engage substrates and reel them in for catalysis. Whether different STRN proteins can form heterotetramers through interactions between conserved CC regions remains to be determined. Since WD domains can specifically recognize phosphorylation status8, the sequence variations in different WD repeats would expand the number of interaction partners of a single STRIPAK complex to include diverse targets that are recognized by different STRN proteins.
STRIP1 forms the central hub of the STRIPAK core complex and interacts with PP2A C, STRN3 and MOB4. Interestingly, Jeong et al. observed an inositol hexaphosphate (IP6) molecule bound at the center of STRIP1 in STRIPAK. IP6 is a metabolite that senses the cellular phosphate balance to regulate diverse signaling pathways9. Mechanistically, binding of IP6 stabilizes several proteins that catalyze other PTMs, including ubiquitin ligases and acetyltransferases10,11. In this context, it seems that IP6 signaling plays a similar role in regulating phosphatase activity by stabilizing STRIP1, which is a prerequisite for STRIPAK assembly. STRIP1 interacts with the PP2A substrate and Hippo signaling initiator, MST2 (ref.12). As such, STRIP1 appears to play a similar role to that of a canonical B subunit, regulating the assembly of STRIPAK and selectively recruiting substrates to the active site of the PP2A C subunit.
It remains unclear which proteins within the STRIPAK complex are responsible for recognizing and recruiting substrates for PP2A-catalyzed dephosphorylation. Despite the presence of the STK25–CCM3 regulatory module in the sample interrogated by Jeong et al., only weak density that probably corresponded to this module was seen in the cryo-EM map. This supports the notion that STK25–CCM3 assists in binding MST1/2 kinases and that both proteins dissociate from the STRIPAK core immediately after the phosphatase-mediated deactivation of MST1/2. It will be important to identify the conserved sequence or structural motif required for recognition of the multiple STRIPAK substrates identified in mass spectrometry studies13–15, as has been done for the B56 family of PP2A phosphatases16–18. Also, in addition to defining the role of IP6 signaling in STRIP1 stability and STRIPAK assembly, it will be equally important to define the role of other PTMs in STRIPAK function. A priority will be to determine whether carboxymethylation of the PP2A C subunit has any effect on STRIPAK complex assembly, as it does for so many other PP2A complexes19. Interestingly, a minor population of STRIPAK core dimers was observed by Jeong et al. While this dimer was suggested to be an artifact of in vitro reconstitution, it is possible that the STRN CC domains that appear to mediate this dimerization similarly contribute to functional interactions in the cell. For example, an analogous CC interaction might be responsible for directing STRIPAK to specific subcellular locations, potentially through association with cytoskeleton microtubules20. Indeed, a component of the Drosophila STRIPAK complex has been shown to associate with microtubules to regulate neuronal morphogenesis21.
There remain questions about the inhibitory role of STRIPAK in the Hippo signaling pathway (Fig. 2). In addition to multiple types of B subunits in the PP2A family, both A and C subunits are expressed as two different isoforms (Aα or Aβ and Cα or Cβ, respectively). It is interesting that the main contacts between Aα and STRN3 in the STRIPAK structure involve the extreme N-terminal region of Aα. This region exhibits the largest sequence divergence when comparing Aα and Aβ proteins. Specifically, the Aβ isoform possesses an additional 12 amino acid residues that are predicted to form an α-helix. The location of this helix begs the question of how it might affect the association of Aβ–C PP2A dimers with other STRIPAK components. While the Aα isoform is the major scaffolding subunit in most tissues, the Aβ isoform may be an even more potent tumor suppressor by targeting different signaling pathways22. Interestingly, PP2A Aβ is expressed at a higher level than Aα during development23, which would correlate well with the need for growth during oogenesis and embryogenesis, both of which are regulated by Hippo signaling. As the role of STRIPAK complexes in cell signaling continues to be investigated, the structure provided by Jeong et al. adds to the mounting evidence that STRIPAK PP2A complexes are mechanistically distinct, both functionally and now structurally, from other canonical B subunit–containing PP2A holoenzymes.
Fig. 2 |. Mammalian Hippo signaling pathway.
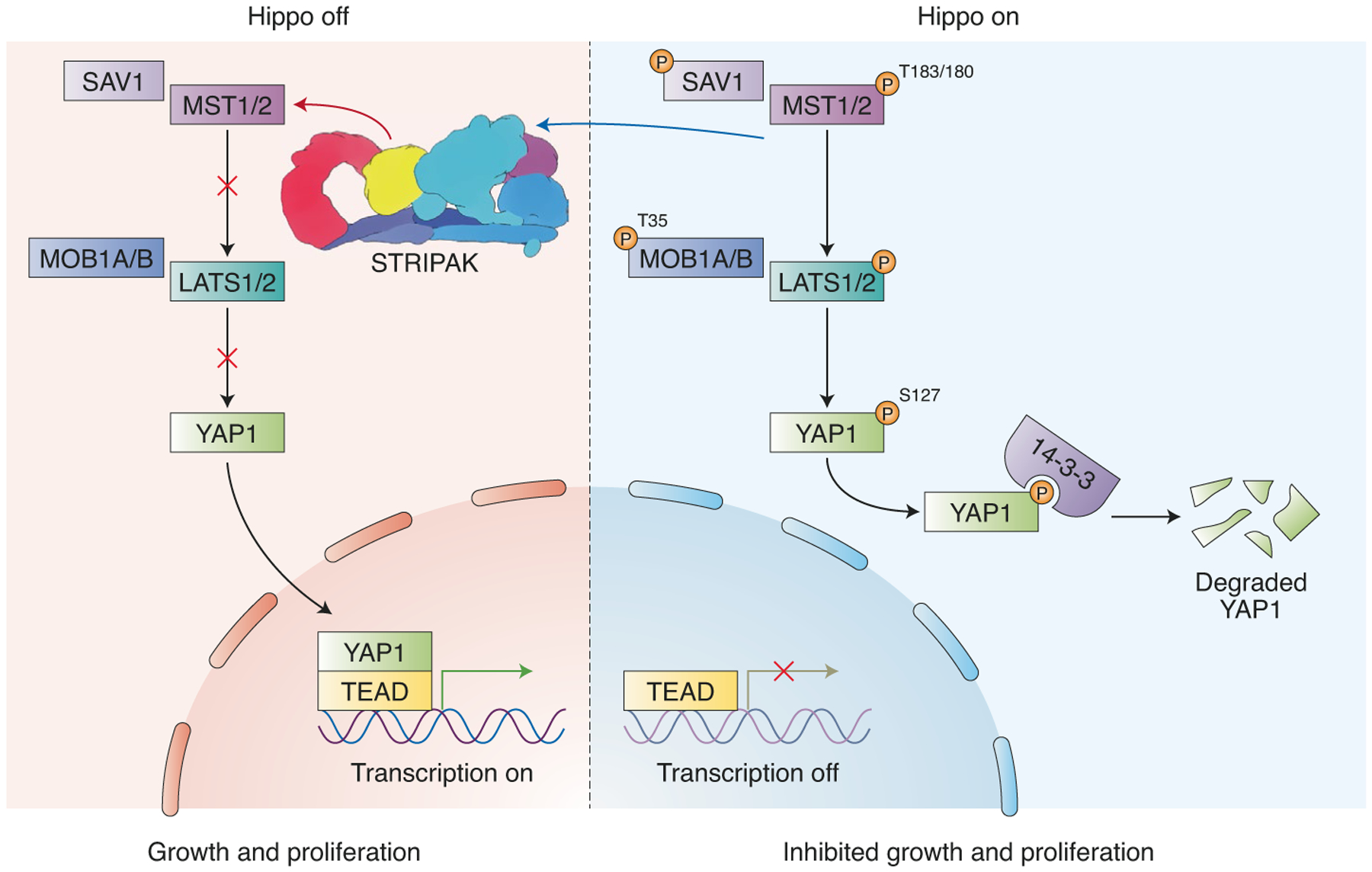
During prosurvival signaling, STrIPAK inhibits the hippo signaling pathway by dephosphorylating and inactivating MST1/2. This allows unphosphorylated YAP1 to transcribe pro-growth proteins. In the absence of STrIPAK, hippo signaling proceeds, allowing phosphorylated MST1/2 to activate LATS1/2–MOB1, which phosphorylates YAP1. 14-3-3 binds to phosphorylated YAP1, leading to its degradation (image generated using Biorender).
Footnotes
Competing interests
W.H., D.L. and D.J.T. provide consultation services to Rappta Therapeutics. D.J.T. has ownership interests in Rappta Therapeutics.
References
- 1.Hwang J & Pallas DC Int. J. Biochem. Cell Biol 10.1016/j.biocel.2013.11.021 (2014). [DOI] [PMC free article] [PubMed]
- 2.Jeong B-C et al. Nat. Struct. Mol. Biol 10.1038/s41594-021-00564-y (2021). [DOI] [PMC free article] [PubMed]
- 3.Taylor SE et al. Cancer Res 10.1158/0008-5472.CAN-19-0218 (2019). [DOI]
- 4.O’Connor CM et al. Oncogene 39, 703–717 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leonard D et al. Cell 181, 688–701.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morita K et al. Cell 181, 702–715.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Tang Y et al. Cancer Cell 10.1016/j.ccell.2020.05.019 (2020). [DOI]
- 8.Hao B, Oehlmann S, Sowa ME, Harper JW & Pavletich NP Mol. Cell 10.1016/j.molcel.2007.02.022 (2007). [DOI] [PubMed]
- 9.Shears SB Adv. Biol. Regul 10.1016/j.jbior.2014.09.015 (2015). [DOI] [PMC free article] [PubMed]
- 10.Gottlieb L & Marmorstein R Structure 26, 925–935.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan X et al. Nature 446, 640–645 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Tang Y et al. Cell Discov 10.1038/s41421-018-0077-3 (2019). [DOI]
- 13.Couzens AL et al. Sci. Signal 10.1126/scisignal.2004712 (2013). [DOI]
- 14.Kwon Y et al. Science 10.1126/science.1243971 (2013). [DOI]
- 15.Wang W et al. Mol. Cell. Proteomics 10.1074/mcp.M113.030049 (2014). [DOI]
- 16.Hertz EPT et al. Mol. Cell 63, 686–695 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Bajaj R, Bollen M, Peti W & Page R Structure 24, 2174–2181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu C-G et al. Cell Discov 3, 17027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogris E, Gibson DM & Pallas DC Oncogene 15, 911–917 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Ma M et al. Cell 10.1016/j.cell.2019.09.030 (2019). [DOI]
- 21.Sakuma C, Okumura M, Umehara T, Miura M & Chihara T Sci. Rep 10.1038/srep17769 (2015). [DOI] [PMC free article] [PubMed]
- 22.Sablina AA et al. Cell 129, 969–982 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosch M et al. Eur. J. Biochem 10.1111/j.1432-1033.1995.1037g.x (1995). [DOI]


