Abstract
Transcriptomic approaches are increasingly used in reproductive medicine to identify candidate endometrial biomarkers. However, it is known that endometrial progression in the molecular biology of the menstrual cycle is a main factor that could affect the discovery of disorder-related genes. Therefore, the aim of this study was to systematically review current practices for considering the menstrual cycle effect and to demonstrate its bias in the identification of potential biomarkers. From the 35 studies meeting the criteria, 31.43% did not register the menstrual cycle phase. We analysed the menstrual cycle effect in 11 papers (including 12 studies) from Gene Expression Omnibus: three evaluating endometriosis, two evaluating recurrent implantation failure, one evaluating recurrent pregnancy loss, one evaluating uterine fibroids and five control studies, which collected endometrial samples throughout menstrual cycle. An average of 44.2% more genes were identified after removing menstrual cycle bias using linear models. This effect was observed even if studies were balanced in the proportion of samples collected at different endometrial stages or only in the mid-secretory phase. Our bias correction method increased the statistical power by retrieving more candidate genes than per-phase independent analyses. Thanks to this practice, we discovered 544 novel candidate genes for eutopic endometriosis, 158 genes for ectopic ovarian endometriosis and 27 genes for recurrent implantation failure. In conclusion, we demonstrate that menstrual cycle progression masks molecular biomarkers, provides new guidelines to unmask them and proposes a new classification that distinguishes between biomarkers of disorder or/and menstrual cycle progression.
Keywords: gene expression, endometrial pathologies, endometriosis, recurrent implantation failure, recurrent pregnancy loss, uterine fibroids, transcriptomic analysis, confounding variable, menstrual cycle progression, differential expression
Introduction
The human endometrium is hormonally regulated and changes throughout the menstrual cycle (Noyes et al., 1975; Murphy, 2004; Talbi et al., 2006). During most of the menstrual cycle, the endometrium is not receptive to embryonic implantation; it becomes receptive during a period of two to four-five days within the mid-secretory phase known as the window of implantation (Harper, 1992; Wilcox et al., 1999). To aid in assisted reproduction, endometrial dating methods have been exhaustively investigated over the past 40 years, as have potential reliable biomarkers of receptive status, including days from luteinising hormone (LH) peak or exogenous progesterone administration, morphological changes detected by ultrasounds, histological features and endometrial gene expression profiles (Noyes et al., 1975; Díaz-Gimeno et al., 2011; Niederberger et al., 2018; Craciunas et al., 2019).
Suboptimal endometrial receptivity and altered embryo-endometrial dialogue are considered to be responsible for one third of implantation failures in in-vitro fertilisation (IVF) cycles (Somigliana et al., 2018). Uterine disorders are complex, polygenic and multifactorial gynecological alterations affecting endometrial gene expression. Endometrial pathologies such as endometriosis, uterine fibroids and adenomyosis are associated with infertility, and may impact endometrial receptivity (Devlieger et al., 2003; Dunselman et al., 2014; Zepiridis et al., 2016; Devesa-Peiro et al., 2020). Some disorders remain undiagnosed in IVF patients and current treatments are either invasive (e.g. surgical removal) or have low-to-moderate efficiency (Wang et al., 2009; Dunselman et al., 2014; Harada et al., 2016; Zepiridis et al., 2016; Tanbo and Fedorcsak, 2017). Moreover, issues such as recurrent implantation failure or pregnancy loss of endometrial origin remain incompletely understood with neither an efficient diagnosis nor effective infertility treatment (Jauniaux et al., 2006; Diaz-Gimeno et al., 2017; Sebastian-Leon et al., 2018). Therefore, identifying reliable biomarkers of uterine disorders is a priority to understand the molecular bases of the pathology and to improve diagnosis, prognosis and treatment.
With the advent of high-throughput technologies, uterine disorders are evaluated by transcriptomic analysis to identify genes (Burney et al., 2007; Hawkins et al., 2011; Lédée et al., 2011; Tamaresis et al., 2014; Koot et al., 2016; Pathare et al., 2017) that could be useful as diagnostic biomarkers and/or therapeutic targets. Although many genes are reported as potential uterine disorder biomarkers, the results of individual studies overlap poorly (Altmäe et al., 2017; Lessey and Kim, 2017; Miravet-Valenciano et al., 2017; Sebastian-Leon et al., 2018), and reliable biomarkers and potential therapeutic targets that could be clinically meaningful to address suboptimal endometrial receptivity remain elusive. This lack of reproducibility between studies may arise from low sample sizes, sample heterogeneity, undetected confounding variables in gene expression experimental procedures or differing experimental designs and data analysis protocols (Gurevitch et al., 2018; Suhorutshenko et al., 2018; Craciunas et al., 2019; Devesa-Peiro et al., 2020).
Menstrual cycle progression has a profound influence on gene expression (Talbi et al., 2006; Koot et al., 2016; Diaz-Gimeno et al., 2017; Sebastian-Leon et al., 2018; Saare et al., 2019). This effect could mask the discovery of candidate uterine disorder biomarkers whose expression responds to both endometrial progression and alterations in uterine disorders. Consequently, it is unclear whether the observed changes in transcriptomic studies reporting differentially expressed genes (DEGs) reflect variations related to the disorder, to menstrual cycle progression or to both. To address this research gap, we quantified the menstrual cycle effect on biomarkers associated with uterine disorders and described current practices in endometrial transcriptomic analysis. Based on our findings, we propose new guidelines to correct for menstrual cycle bias in biomarker identification.
Materials and methods
Ethics statement
This is a retrospective study using case and control data from multiple studies of endometrial gene expression in women with uterine disorders and women with no endometrial pathology. The raw gene expression data and patient meta-data were downloaded from National Center for Biotechnology Information (NCBI) functional genomics data repository Gene Expression Omnibus (GEO), where data are made available for the scientific community. Following GEO policies, in these repository patient data are anonymised and encrypted, and no additional institutional review board is required for downloading (Edgar, 2002).
Study search and selection
A systematic search and review of individual studies was conducted between October 2016 and January 2019 at the NCBI repository GEO (Edgar, 2002). The search identified experiments involving human transcriptomic case versus control raw data evaluating uterine disorders.
Keywords searched included endometrium, endometriosis, uterine fibroids, recurrent implantation failure (RIF) and recurrent pregnancy loss (RPL), among others (see Supplementary Table SI for a full list of search terms). No restrictions were placed on publication date or language. The final inclusion criteria were that: there was at least one uterine disorder evaluated in the study design; RNA was extracted directly from human endometrial biopsies; information regarding the menstrual cycle at the time of biopsy was available for all samples; sample sizes were greater than three for both case and control groups belonging to the same study; microarray or RNA sequencing data were obtained using Affymetrix, Illumina or Agilent gene expression platforms; and raw gene expression data were made freely available to download from GEO. Studies evaluating endometrial gene expression at different times of the menstrual cycle in women with normal endometrium were retrieved from GEO using the same keywords and criteria.
Pre-processing and exploratory analysis
Pre-processing and exploratory analyses were completed according to the gene expression platform used: raw data were downloaded and pre-processed using the affy v. 1.52.0 R package (Gautier et al., 2004) for studies measuring endometrial gene expression with Affymetrix microarray platforms and the limma v.3.30.13 R package (Ritchie et al., 2015) was used for studies using Agilent or Illumina devices. Normalisation between samples was applied using quantile normalisation (limma R package v.3.30.13); (Ritchie et al., 2015) and annotation from probeset to gene symbol was established with the biomaRt R package v. 2.30.0 (Durinck et al., 2009). For studies evaluating gene expression through RNA-Seq, low-count filtering and normalisation was achieved with the edgeR R package v. 3.16.5 (Robinson et al., 2010).
Exploratory analyses were performed to detect batch effects such as sequencing run or microarray slide. Detected effects were treated using linear models [limma R package v.3.30.13 (Ritchie et al., 2015)]. Afterward, menstrual cycle effect was evaluated through principal component analysis plots drawn with the ggplot2 R package v. 3.2.0 (Wickham, 2016). The proportion of endometrial biopsies collected at different stages of the menstrual cycle were compared between case and control groups using Fisher’s exact test (Fisher, 1922) implemented in the R environment (R Core Team, 2016).
Menstrual cycle effect correction and differential expression analysis
The effect of menstrual cycle progression on endometrial biopsy collection was removed from gene expression data using the removeBatcheffect function based on linear models implemented in the limma R package v.3.30.13 (Ritchie et al., 2015) because this function enables correcting the bias from a known batch effect (e.g., menstrual cycle) while indicating the group differences to be retained in the data (e.g., uterine disorder versus control). Specifically, we used removeBatcheffect for being a slightly safer option than Combat (Espín-Pérez et al., 2018), specifying the menstrual cycle phase of endometrial biopsy collection as the batch to remove, and defining the design matrix in relation to the condition to be preserved (case versus control samples).
Case versus control differential expression analyses were applied (limma R package v.3.30.13 (Ritchie et al., 2015)) with and without removing the menstrual cycle effect; the proportion of differentially expressed genes (false discovery rate; FDR < 0.05) were compared for demonstrating the menstrual cycle bias.
To highlight the advantage of this method and validate its statistical power, an alternative approach was compared evaluating case versus control DEGs within each menstrual cycle phase using the limma R package v.3.30.13 (Ritchie et al., 2015). The proportion of DEGs (FDR < 0.05) obtained was compared with those obtained after menstrual cycle effect correction. All comparisons of DEG proportions were performed with one-sided Fisher’s exact test [adjusted by FDR (Benjamini and Hochberg, 1995)]. The statistical power of both methods was calculated with sizepower R package v.1.52.0 (Qiu et al., 2018).
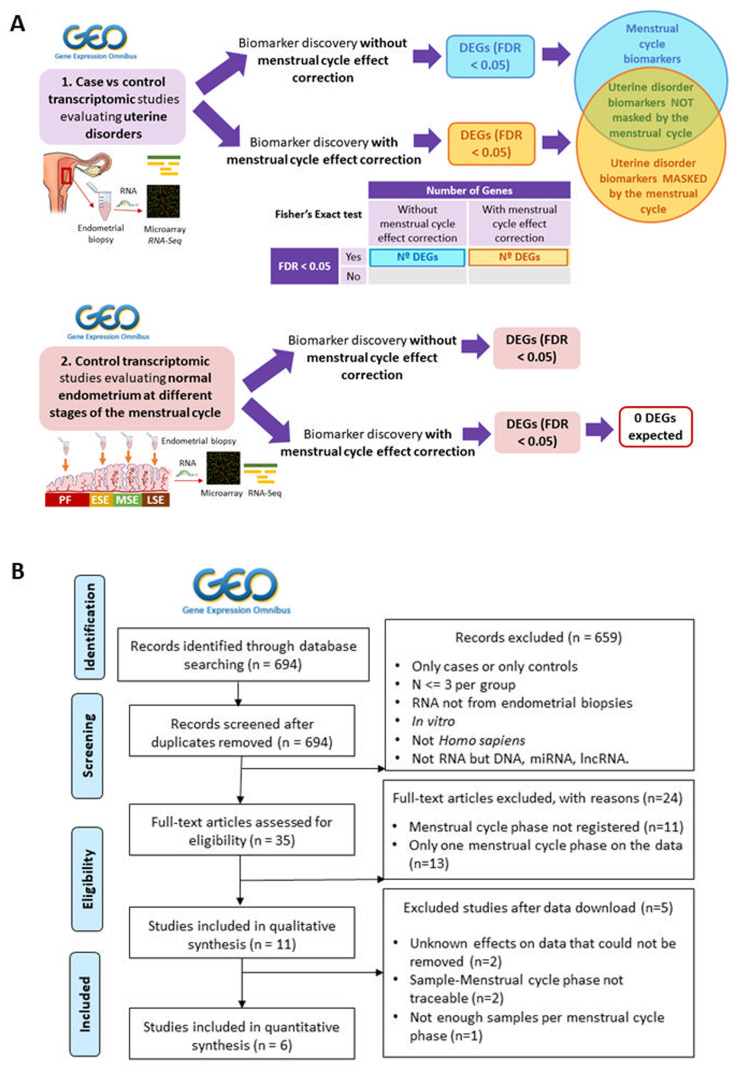
Additionally, to validate the reliability and robustness of the method used to remove the menstrual cycle effect, the aforementioned approaches of differential expression analysis (with and without menstrual cycle correction) were applied to GEO individual studies comparing endometrial gene expression profiles between different menstrual cycle phases. If the method properly removed the menstrual cycle effect from transcriptomic data, the differential expression analysis between endometrial phases after correcting this effect should indicate no DEGs. The study design is detailed in Fig. 1A.
Figure 1.
(A) Study design and flowchart for transcriptomic studies evaluating uterine disorders. Systematic search and review of (i) transcriptomic case versus control studies evaluating uterine disorders and (ii) transcriptomic studies evaluating menstrual cycle progression in control women with normal endometrium were performed at the Gene Expression Omnibus (GEO) database. After filtering and selection, raw data were downloaded and pre-processed. For each included study, a differential expression analysis was applied with and without removing the menstrual cycle effect on the data using linear models. For case versus control studies (i), the proportions of differentially expressed genes (DEGs; FDR < 0.05) obtained under both approaches were compared using a Fisher’s Exact test and distinct types of endometrial biomarkers were established. Control studies evaluating menstrual cycle progression (ii) were used to evaluate the reliability of menstrual cycle effect correction; if the method properly removed the menstrual cycle effect from transcriptomic data, the differential expression analysis between endometrial phases after correcting this effect should indicate no differentially expressed genes. (B) Flowchart of the selection process of case versus control transcriptomic studies evaluating uterine disorders. The selection of suitable individual transcriptomic studies at GEO and the number of individual studies excluded and remaining after each filtering step are shown. GEO, Gene Expression Omnibus. n, number of studies. N, sample size. lncRNA, long noncoding RNA.
Finally, we compared the fold change (FC), P-value, gene expression average, and standard deviation (SD) before and after menstrual cycle effect correction for understanding the aetiology of the potential endometrial biomarkers. All statistical analyses were run under R software v.3.3.2 (2016-10-31) (R Core Team, 2016).
Results
Current practices for transcriptomic studies evaluating the endometrium in uterine disorders
Of the endometrial studies found in GEO (n = 694), 35 studies met the inclusion criteria (Fig. 1B). Of these, 31.43% (11 studies) did not register the menstrual cycle phase at the time of endometrial biopsy collection and 37.14% (13 studies) collected all endometrial samples at only the proliferative or secretory phases, with no further subdivision for secretory samples into early-, mid- or late-secretory endometrial stages (Fig. 1B). Only six papers with seven case versus control transcriptomic studies reporting the time point in which the biopsy was collected were suitable for analysis: two of eutopic endometrium in stages I–IV endometriosis (n = 37, n = 81), one of ectopic endometrium in ovarian endometriosis (n = 14), one of uterine fibroids (n = 43), two of RIF (n = 115, n = 18) and one of RPL (n = 20) (Table IA; detailed filtering steps in Fig. 1B).
Table I.
Characterization of transcriptomic studies evaluating uterine disorders. A) Clinical characterization of participants and study designs.
| GEO ID (Study name) | UD | Diagnostic and Sub-classification of the studied uterine disorder | Cycle type | Cycle phase dating method | N° Samples per cycle phase | Age | BMI |
|---|---|---|---|---|---|---|---|
| GSE6364 (Burney 2007) | EU | Laparoscopy proven, surgically documented and histologically validated. Subtype: Ovaric, peritoneal, rectovaginal. Stage: III–IV (rAF) (The American Fertility Society) | Natural; Regular; 3 months without hormonal treatment | 4 blind histopathologists (Noyes et al., 1975). | PF (n = 11), ESE (n = 9), MSE (n = 17) |
D: 22–44. C: N/A |
N/A |
| GSE23339 (Hawkins 2011) | EC | Surgical pathology reports. Subtype: Ovaric. | Regular; 30 days without hormonal treatment | Last menstrual period confirmed by pathology | PF (n = 12), S (n = 2) |
D : 20–43. C : 39–48 |
N/A |
| EU | Laparoscopy proven. Subtype: Ovaric, peritoneal, pelvic, vaginal, rectovaginal. Stage: I–II (n = 16), III–IV (n = 37) (rAF) (American Society for Reproductive Medicine, 1997) | 3 months without hormonal treatment | 2 pathologists (Noyes et al., 1975), confirmed by serum estradiol and P4 levels and corroborated by 2 independent bioinformatics methods: clustering in unsupervised whole-transcriptome principal component analysis and cycle phase assignment classifier analysis |
PF (n = 34), ESE (n = 15), MSE (n = 32) PF (n = 23), ESE (n = 7), MSE (n = 12), LSE (n = 1). |
E : 20–48. UF : 40–50. C : 23–40. |
N/A | |
| UF | Participants’ operative and pathology reports | N/A | |||||
| RIF |
≥ 3 failed IVF/ICSI or ≥10 good quality transferred embryos without pregnancy after IVF/ICSI. P: Previous implantations: 0–1. Embryo implantations: 3–12. Embryos replaced: 3–1. C: Previous implantations: 1. Embryo implantations: 1–7, 18. Embryos replaced: 1–8, 30. |
Natural. Regular (25–35 days), 30 days without hormonal treatment | Urinary LH ovulation predictor kit | LH + 5 (n = 8), LH + 6 (n = 27), LH + 7 (n = 70), LH + 8 (n = 10). | D: 27–38 C: 26–39 |
D: 19–37 C: 19–53 |
|
| RIF |
≥ 2 IVF cycles/ET with good quality embryos without previous conception (Polanski et al., 2014) |
Controlled ovarian stimulation | Days from first administration of hCG | hCG + 6 (n = 13), hCG + 7 (n = 5) |
D : 27–40 C : 21–30 |
N/A | |
| RPL | ≥ 3 1st trimester losses | Natural. 3 months without hormonal treatment | LH surge | LH + 6 (n = 3), LH + 7 (n = 5), LH + 8 (n = 6), LH + 9 (n = 4), LH + 10 (n = 2). | D: 31–40 C: 31–44 | D: 22–32 C: 18–33 |
| A) Clinical characterisation of participants and study designs. | |||||
|---|---|---|---|---|---|
| GEO ID (Study name) | Ethnicity | N° Cases and Controls (D) (C) | N° Evaluated Genes | Platform | Ref. |
| GSE6364 (Burney 2007) |
D: Caucasian (n = 13), Asian (n = 4), Black (n = 1), Asian Indian (n = 1), unknown (n = 2). C: N/A |
(21) (16) | 19 361 | hgu133plus2 Affymetrix | (Burney et al., 2007) |
| GSE23339 (Hawkins 2011) |
D: Latina (n = 5), Caucasian (n = 2). C: Latina (n = 5), African American (n = 2) |
(7) (7) | 24 613 | Illumina human-6 v2.0 expression beadchip | (Hawkins et al., 2011) |
|
E: Caucasian (n = 38), Asian (n = 4), Black (n = 1), Asian Indian (n = 1), Hispanic (n = 1), unknown (n = 8). UF: Caucasian (n = 11), Asian (n = 1), Black (n = 3). C: Caucasian (n = 19), Asian (n = 4), Black (n = 3), Hispanic (n = 1), unknown (n = 1) |
(53) (28) | 19 361 | hgu133plus2 Affymetrix | (Tamaresis et al., 2014) | |
| (15) (28) | 19 361 | ||||
| N/A | (43) (72) | 21 773 | Agilent G2565BA Scanner | (Koot et al., 2016) | |
| Indian | (10) (8) | 31 426 | Illumina Human HT-12 V4.0 expression beadchip | (Pathare et al., 2017) | |
| N/A | (10) (10) | 21 332 | Illumina HiSeq 2000 | (Lucas et al., 2016) | |
| B) Comparison between case and control groups in relation to the menstrual cycle phase of endometrial biopsy collection. | ||||
|---|---|---|---|---|
| Study | UD | N.° Samples per cycle phase |
Fisher P-value | |
| Cases | Controls | |||
| Burney 2007 |
EU |
PF (n = 6), ESE (n = 6), MSE (n = 9) |
PF (n = 5), ESE (n = 3), MSE (n = 8) |
00.834 |
| Hawkins 2011 | EC | PF (n = 6), S (n = 1) | PF (n = 6), S (n = 1) | 1 |
| Tamaresis 2014 | EU | PF (n = 18), ESE (n = 11), MSE (n = 24). | PF (n = 16), ESE (n = 4), MSE (n = 8) | 0.150 |
| UF | PF (n = 7), ESE (n = 3), MSE (n = 4), LSE (n = 1) | PF (n = 16), ESE (n = 4), MSE (n = 8) | 0.618 | |
| Koot 2016 | RIF | LH + 5 (n = 2), LH + 6 (n = 13), LH + 7 (n = 26), LH + 8 (n = 2) | LH + 5 (n = 6), LH + 6 (n = 14), LH + 7 (n = 44), LH + 8 (n = 8) | 0.398 |
| Pathare 2017 | RIF | hCG + 6 (n = 8), hCG + 7 (n = 2) | hCG + 6 (n = 5), hCG + 7 (n = 3) | 0.608 |
| Lucas et al., 2016 | RPL | LH + 6 (n = 2), LH + 7 (n = 1), LH + 8 (n = 3), LH + 9 (n = 2), LH + 10 (n = 2) | LH + 6 (n = 1), LH + 7 (n = 4), LH + 8 (n = 3), LH + 9 (n = 2) | 0.529 |
(A) Clinical characterisation of participants and study designs. The GEO identifier, study name given in this work, uterine disorder and clinical information about participants including diagnostic method and sub-classification of patients belonging to the case group, cycle type, endometrial dating method, cycle phase in which the endometrial biopsies were collected along with number of samples collected at each menstrual cycle phase, age, BMI, ethnicity and number of samples for both case and control groups are presented for each study. The transcriptomic platform used to measure gene expression and the publication in which data were initially employed are also presented together with the number of evaluated genes. Tamaresis 2014 includes samples from both endometriosis and uterine fibroid patients along with controls. N/A, not available. D, patients belonging to the case the control group. GEO ID, Gene Expression Omnibus identifier. UD, uterine disorder. BMI, body mass index. RIF, recurrent implantation failure. RPL, recurrent pregnancy loss. EU, eutopic endometriosis. EC, ectopic endometriosis. UF, uterine fibroids. rAF, revised American Fertility Society classification system. LH, luteinizing hormone. AMH, anti-müllerian hormone. ET, embryo transfer. FSH, follicle-stimulating hormone. PF, proliferative. ESE, early secretory. MSE, mid-secretory. LSE, late secretory. S, secretory. ICSI, intracytoplasmic sperm injection. IVF, in vitro fertilisation. PCOS, polycystic ovary syndrome. (B) For each study, the number of samples collected at each stage of the menstrual cycle is indicated independently for cases and controls, together with the two-sided Fisher’s exact test P-value obtained after evaluating whether the proportion of samples collected at each endometrial stage significantly differed between groups.
While studies evaluating endometriosis and uterine fibroids mainly used Noyes histopathological criteria and collected biopsies along both the proliferative and secretory phases (Table IA), those evaluating RIF and RPL used days from LH peak or first administration of human chorionic gonadotropin (hCG) for endometrial biopsies collected in the secretory phase (Table IA). For all studies, case and control groups were balanced in terms of the proportion of samples collected at each endometrial stage (P > 0.05; Table IB).
Menstrual cycle effect on transcriptomic studies searching for uterine disorder biomarkers
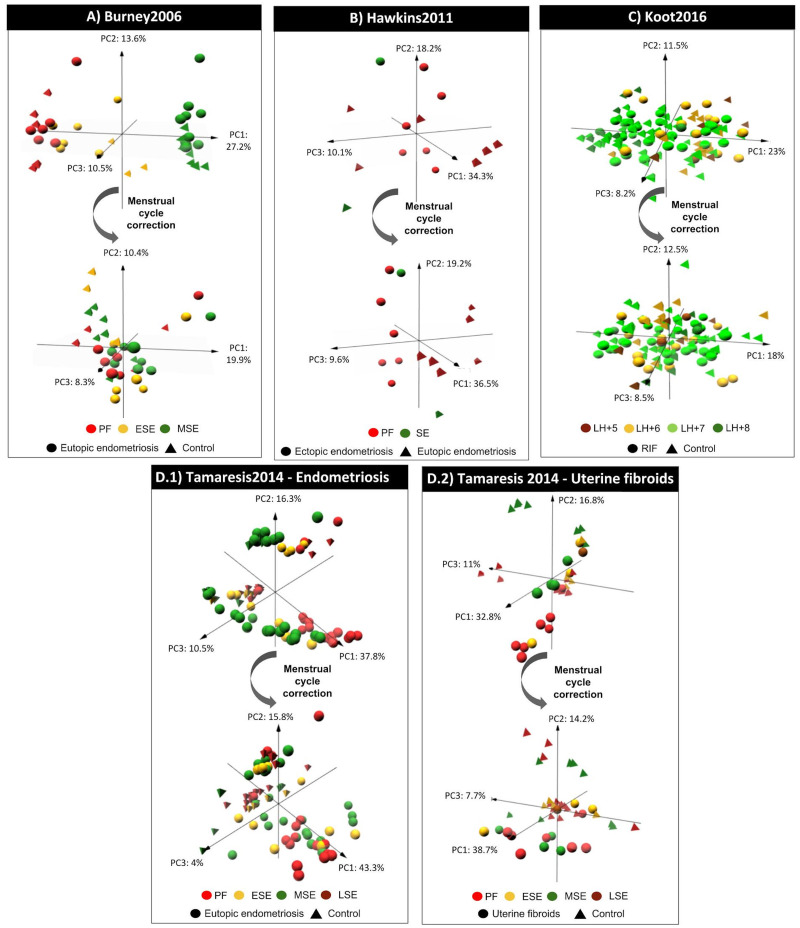
All studies collecting endometrial biopsies at different cycle time points had a menstrual cycle effect on gene expression (see PCAs on Figs 2 and 3).
Figure 2.
Menstrual cycle effect on endometrial gene expression in transcriptomic studies by Burney 2006, Hawkins 2011, Koot 2016 and Tamaresis 2014. For each study (A–D), the result of the principal component analysis is plotted for the first three components before and after applying the menstrual cycle effect correction to evaluate whether the samples are primarily grouped by the menstrual cycle phase of endometrial biopsy collection (colour code) and/or by the uterine disorder (shape code) based on their gene expression profiles. PC, principal component. PF, proliferative. ESE, early secretory. MSE, mid-secretory. SE, secretory. LH, luteinising hormone. RIF, recurrent implantation failure.
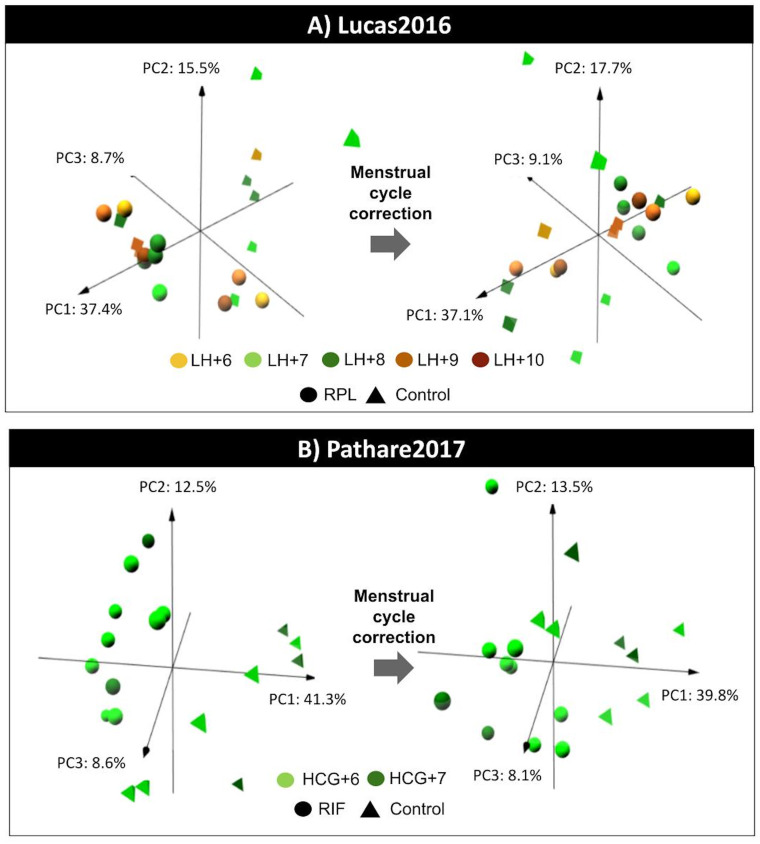
Figure 3.
Menstrual cycle effect on endometrial gene expression for Lucas et al., 2016 and Pathare 2017. For each study (A and B), the result of the principal component analysis is plotted for the first three components before (right) and after (left) applying the menstrual cycle effect correction to evaluate whether the samples are primarily grouped by the menstrual cycle phase of endometrial biopsy collection (colour code) and/or by the uterine disorder (shape code) based on their gene expression profiles. PC, principal component. hCG, human chorionic gonadotrophin. LH, luteinising hormone. RIF, recurrent implantation failure. RPL, recurrent pregnancy loss.
Samples from Burney 2007, Hawkins 2011, Tamaresis 2014 and Koot 2016 were grouped by endometrial phase before menstrual cycle effect correction (Fig. 2). However, samples were mainly grouped by the uterine disorder (Fig. 2) and a significant average of 44.19% more biomarkers were detected (one-sided Fisher’s exact tests FDR ≤ 5.53 × 10–06) when the effect of the menstrual cycle was corrected (Table IIA). These newly revealed uterine disorder biomarkers that were previously masked by the menstrual cycle effect were, in fact, identified despite balanced endometrial cycle stages between case and control groups (Table IB). Masked uterine disorder biomarkers were also detected in Koot 2016, among endometrial biopsies collected at different time points within the mid-secretory phase (LH + 5-LH + 8) (Table IB).
Table II.
Uterine disorder biomarkers after removing the menstrual cycle bias.
A) Differential expression analysis with and without correcting the menstrual cycle effect on endometrial transcriptomic data.
| Study | UD | N° DEGs without menstrual cycle correction | N° DEGs with menstrual cycle correction | % of newly detected disorder biomarkers | Fisher’s test FDR | Menstrual cycle biomarkers | UD biomarkers not masked by the menstrual cycle | UD biomarkers masked by the menstrual cycle |
|---|---|---|---|---|---|---|---|---|
| Burney 2007 | EU | 127 | 812 | 84.36% | 6.6 × 10–16 | 0 | 127 | 685 |
| Hawkins 2011 | EC | 903 | 1205 | 25.06% | 1.92 × 10–11 | 0 | 903 | 302 |
| Tamaresis 2014 | EU | 13 397 | 13 797 | 6.05% | 5.53 × 10–06 | 435 | 12 962 | 835 |
| UF | 9715 | 10 909 | 11.71% | 6.6 × 10–16 | 83 | 9632 | 1277 | |
| Koot 2016 | RIF | 2 | 32 | 93.75% | 5.15 × 10–08 | 0 | 2 | 30 |
| Pathare 2017 | RIF | 2783 | 2492 | −2.29% | 1 | – | – | – |
| Lucas et al., 2016 | RPL | 0 | 0 | 0% | – | – | – | – |
| B) Newly discovered uterine disorder biomarkers previously masked by the menstrual cycle effect. | ||||
|---|---|---|---|---|
| Study | UD | UD biomarkers masked by the menstrual cycle |
||
| Previously reported | Newly discovered | Total | ||
| Burney 2007 | EU | 141 | 544 | 685 |
| Hawkins 2011 | EC | 144 | 158 | 302 |
| Koot 2016 | RIF | 3 | 27 | 30 |
| C) Differential expression analysis (DEA) for each menstrual cycle phase | ||||
|---|---|---|---|---|
| Study | DEA for each menstrual cycle phase |
DEA with menstrual cycle correction |
||
| Menstrual cycle phase (n) | N° DEGs | Statistical Power % | Statistical Power % (n) | |
| Burney 2007 | PF (11) | 0 | 29.74% | 98.7% (37) |
| ESE (9) | 100 | 9.28% | ||
| MSE (17) | 0 | 65.5% | ||
| Hawkins 2011 | PF (12) | 541 | 40.01% | 52.1% (14) |
| S (2) | — | — | ||
| Koot 2016 | LH + 5 (8) | 0 | 3.21% | 100% (115) |
| LH + 6 (27) | 0 | 94.43% | ||
| LH + 7 (70) | 0 | 99.99% | ||
| LH + 8 (10) | 0 | 3.21% | ||
(A) Differential expression analysis with and without correcting the menstrual cycle effect on endometrial transcriptomic data. For each study, this table presents the number of differentially expressed genes (DEGs) obtained with and without menstrual cycle correction, the % of disorder-specific genes newly identified when correcting the menstrual cycle effect, the false discovery rate (FDR) associated with the different proportions of DEGs detected with and without menstrual cycle correction (one-sided Fisher’s exact test, as we expected to identify a higher number of DEGs with menstrual cycle correction), and the number of genes belonging to the three types of endometrial biomarkers identified in the study: biomarkers of the menstrual cycle alone (DEGs only detected without correcting the menstrual cycle effect), biomarkers of the uterine disorder that are masked by the menstrual cycle (DEGs only detected with the menstrual cycle correction) and uterine disorder biomarkers that are not masked by the menstrual cycle (intersected DEGs between both approaches). UD, uterine disorder. EU, eutopic endometriosis. EC, ectopic endometriosis. UF, uterine fibroids. RIF, recurrent implantation failure. RPL, recurrent pregnancy loss. (B) Newly discovered uterine disorder biomarkers unmasked by the menstrual cycle effect correction. From the total number of uterine disorder biomarkers that were unmasked after applying the menstrual cycle effect correction method (DEGs only detected with the menstrual cycle correction, type C biomarkers in Fig. 3), the table shows the number of biomarkers that had been previously associated with the uterine disorder either in the original articles or in the databases Disgenet v.6 (42), Phenopedia v.6.2.3 (43), and/or GeneCards v.4.14.0 (41), together with the number of uterine disorder biomarkers not previously reported and thus newly discovered in this work. Keywords used in database searches: ‘endometriosis’, ‘uterine fibroids OR leiomyoma OR myoma’, and ‘recurrent implantation failure’. (C) Differential expression analysis (DEA) for each menstrual cycle phase. This table presents the number of differentially expressed genes (DEGs) in each study and the statistical power (%) obtained when the analysis was performed for samples collected at each menstrual cycle phase separately. The statistical power (%) of the analysis with menstrual cycle phase correction is also indicated for comparison. For both approaches, sample sizes (n) are indicated between parentheses. PF, proliferative. ESE, early secretory. MSE, mid-secretory. S, secretory. LH, luteinizing hormone.
In contrast, samples from Lucas et al., 2016 and Pathare 2017 were primarily grouped by the disorder rather than by the time point of endometrial biopsy collection before menstrual cycle effect correction; and no more DEGs were identified after correcting the menstrual cycle effect (Table IIA, Fig. 3).
While in Burney 2007, Hawkins 2011 and Koot 2016, the DEGs detected before menstrual cycle correction where included in those identified after the correction was applied. For Tamaresis 2014, there were DEGs detected only before applying the menstrual cycle correction (Table IIA).
A new classification of endometrial transcriptomic biomarkers in uterine disorders
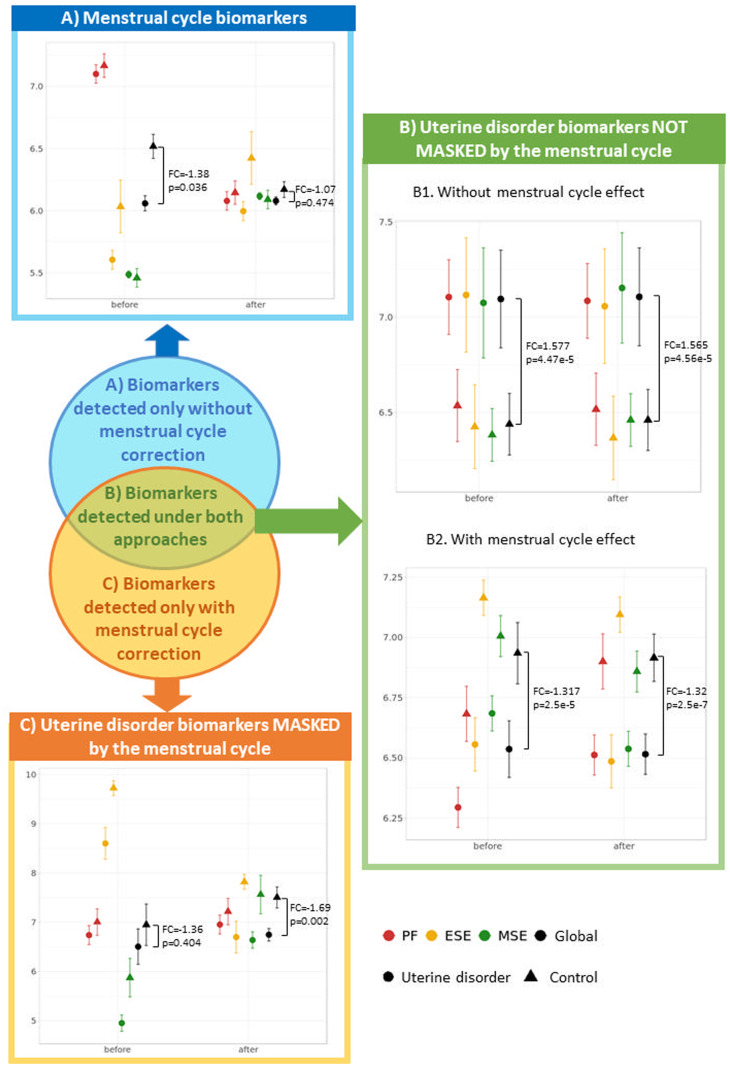
Comparison of the expression profiles of significant genes identified before and after applying the menstrual cycle correction allowed us to detect their aetiology and define a new classification of endometrial biomarkers (Table IIA, Fig. 4).
Figure 4.
A new classification of endometrial biomarkers according to their gene expression profiles. For each type of endometrial biomarker (A–C), the average and standard deviation of an example gene is represented before and after menstrual cycle effect correction separated by the uterine disorder (cases and controls) and the menstrual cycle phase. The global measure of cases and controls is also represented together with the correspondent P-values and fold changes (FC) obtained in the differential expression analyses with and without correcting the menstrual cycle. P-values were used instead of FDR as, for each gene, P-value adjustment for multiple testing is dependent on the P-values obtained in the other genes rather than gene-exclusive. (A) Menstrual cycle biomarkers detected only without correction: before the menstrual cycle effect correction, the differences in gene expression among samples from distinct endometrial phases was high and made this gene globally significant (P = 0.036) without being truly affected by the uterine disorder, as the average gene expression of case and control groups was not different within the proliferative and mid-secretory phases. After correcting for the menstrual cycle, the distance between gene expression patterns in different endometrial phases shortened and became non-significant (P = 0.474). Example gene: PPP2R2C from Tamaresis 2014. (B) Uterine disorder biomarkers not masked by the menstrual cycle and detected both before and after the correction: for B1 biomarkers, samples belonging to different phases had similar average expression both before and after applying the menstrual cycle correction, suggesting that expression is not affected by the menstrual cycle and is significantly distinct between cases and controls regardless of whether the correction is applied. In contrast, B2 biomarkers had different average expression between samples collected at different menstrual cycle phases, but the differences between cases and controls were higher, allowing these genes to be also identified as uterine disorder biomarkers before correcting for the effect of the menstrual cycle. Example genes: MYL10 (B1) and SESN1 (B2) from Burney2006. (C) Uterine disorder biomarkers masked by the menstrual cycle and only detected after the correction: Expression differences between menstrual cycle phases are greater than those between cases and controls, making the depicted gene not significant before menstrual cycle correction (P = 0.404). After correcting for the effect of the menstrual cycle, expression differences between menstrual cycle phases are minimised, reducing the variability within case and control groups and making the gene globally significant (P = 0.002). Example gene: CTNNA2 from Burney2006. DEGs, differentially expressed genes. PF, proliferative. ESE, early secretory. MSE, mid-secretory.
Menstrual cycle biomarkers (Fig. 4A) are genes detected only before menstrual cycle correction, suggesting that they respond to endometrial progression but not to the uterine disorder itself (higher expression differences were observed between endometrial stages than between cases and controls). This type of biomarker was identified by Tamaresis 2014, where a gene is differentially expressed without being truly affected by the uterine disorder (Fig. 4A). After correcting for the menstrual cycle, the distance between gene expression patterns in different endometrial phases shortened and became non-significant.
Uterine pathology biomarkers not masked by the menstrual cycle (Fig. 4B) are genes detected with and without menstrual cycle correction because there was no effect of endometrial progression (Fig. 4.B.1, no expression differences were observed between endometrial stages) or the effect was lower than that of the uterine disorder (Fig. 4.B.2, higher expression differences between cases and controls than between endometrial stages).
Uterine disorder biomarkers masked by the menstrual cycle (Fig. 4C) are genes detected only after menstrual cycle correction because the effect of endometrial progression greatly outweighs that of the uterine disorder (higher expression differences were observed between endometrial phases than between cases and controls). Consequently, these genes remain masked and not significant before menstrual cycle correction and can only be detected as uterine disorder biomarkers after removal of menstrual cycle effect.
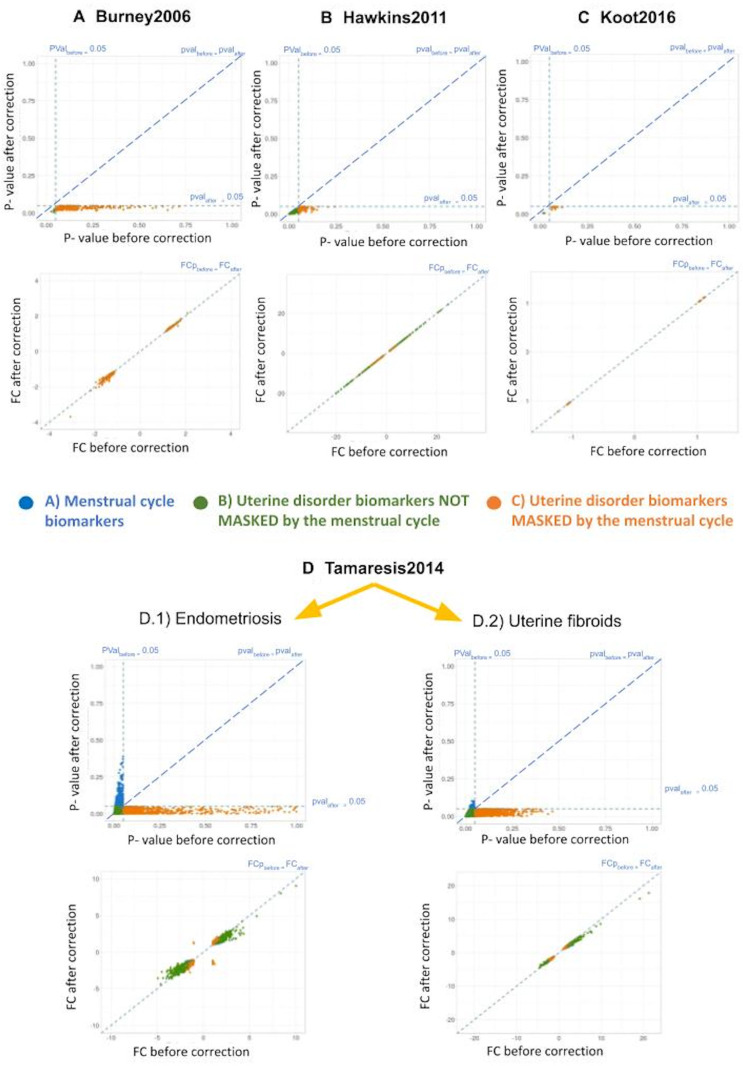
For the three types of endometrial biomarkers, the menstrual cycle effect correction method only affected the variability of gene expression explained by endometrial progression, thus changing the P-value when comparing case and control groups (Fig. 5). In contrast, fold changes between cases and controls did not substantially change in any gene belonging to any type of endometrial biomarker (Fig. 5), indicating that the correction method successfully maintained the expression differences associated with uterine disorders. Consequently, P-value changes were not caused by alteration in case versus control mean expression differences but by the removal of menstrual cycle induced variation in gene expression (Fig. 5).
Figure 5.
Changes in P-value and fold change (FC) due to menstrual cycle effect correction on gene expression. For each study (A–D) and type of biomarker (colour code) shown in Fig. 4, P-values (up) and FCs (down) before and after applying the menstrual cycle effect correction method are plotted against each other. Diagonal dotted lines indicate absence of changes in P-values/FCs before and after applying the correction method. Vertical and horizontal dotted lines for P-values represent P-value = 0.05 before and after correcting for the menstrual cycle effect, respectively.
Discovery of new potential biomarkers
Among the type C uterine disorder biomarkers that remained undetected before correcting the menstrual cycle effect on gene expression (Fig. 4C, Supplementary Table SII) (Yu et al., 2010; Stelzer et al., 2016; Piñero et al., 2020), we discovered 544 new candidate biomarkers of eutopic endometriosis (Burney 2007), 158 of ectopic ovarian endometriosis (Hawkins 2011) and 27 of recurrent implantation failure (Koot 2016) that had not been previously reported (Table IIB and Supplementary Table SII) (Burney et al., 2007; Yu et al., 2010; Hawkins et al., 2011; Koot et al., 2016; Stelzer et al., 2016; Piñero et al., 2020). These new biomarkers presented an expression difference between cases and controls of 12–121% for eutopic endometriosis, 15–359% for ectopic ovarian endometriosis and 2–11% for RIF (Supplementary Table SII).
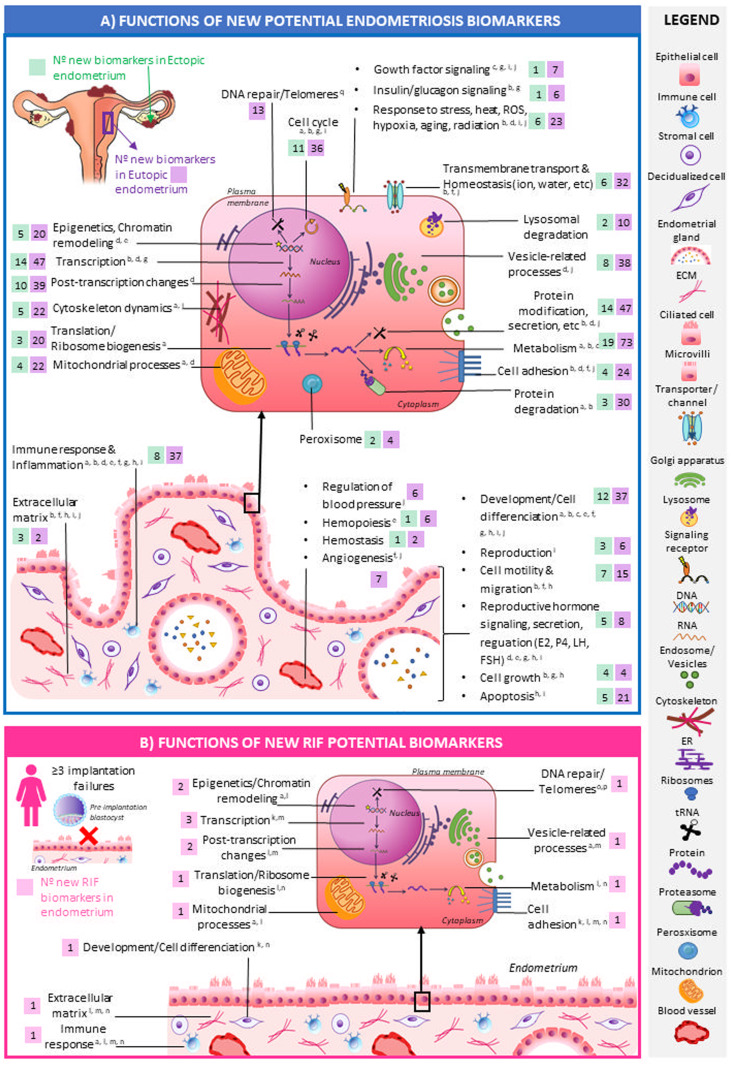
To better understand their functional role in the context of uterine disorder pathophysiology, these new biomarkers were functionally annotated (Fig. 6, Supplementary Table SIII). New candidate endometriosis biomarkers (both ectopic and eutopic) were mainly related to metabolism (19 ectopic and 73 eutopic), transcription regulation processes (14 ectopic and 47 eutopic) and protein-modification processes (14 ectopic and 47 eutopic) (Fig. 6A). In addition, functions widely reported to be involved in endometriosis such as immune response and inflammation (Tomassetti et al., 2006; Burney et al., 2007; Burney and Giudice, 2012; Liu et al., 2015; Patel et al., 2018; Anderson, 2019; Marquardt et al., 2019; Devesa-Peiro et al., 2020) and cell differentiation and development (Tomassetti et al., 2006; Burney et al., 2007; Burney and Giudice, 2012; Crispi et al., 2013; Liu et al., 2015; Patel et al., 2018; Marquardt et al., 2019; Zhang et al., 2019; Devesa-Peiro et al., 2020) were notably annotated to these new potential biomarkers of endometriosis (Fig. 6A). New RIF candidate biomarkers were mostly associated with transcription regulation (3 biomarkers: CHD4, IGHMBP2, ZBTB48), post-transcriptional changes (2 biomarkers: FAM182B and RN7SK), and epigenetics and chromatin remodelling processes (2 biomarkers: CDH4, FAM182B); these functions were previously reported as significantly altered in RIF patients (Cakmak and Taylor, 2011; Koot et al., 2016; Pathare et al., 2017; Devesa-Peiro et al., 2020) (Fig. 6B). Gene names of new candidate biomarkers of ectopic/eutopic endometriosis and RIF belonging to each functional group are listed in Supplementary Table SIII together with literature references supporting the role of each functional group in the context of each uterine disorder.
Figure 6.
Functional roles of the newly discovered potential biomarkers of endometriosis and recurrent implantation failure (A) and recurrent implantation failure (RIF) (B). Functions associated with the new biomarkers of endometriosis (A) (eutopic: n = 544, ectopic: n = 158) and RIF (n = 27) (B) are shown in the endometrial cellular context together with references supporting the role of each function in the pathophysiology of the corresponding disorder. The number of biomarkers of eutopic (A, purple boxes) and ectopic (A, green boxes) endometriosis and RIF (B, pink boxes) annotated to each function are represented. New potential biomarkers belonging to each represented function are fully listed in Supplementary Table SIII. Functional annotations were performed for each new biomarker using Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways (Ogata et al., 1999) and Gene Ontology (GO) (Ashburner et al., 2000) databases. For GO annotation, the three ontologies were used (biological processes, molecular functions, cellular components), only experimental-evidenced GO-gene associations were included, a propagated GO version was used for considering the whole GO-tree structure, and annotated GO terms were filtered by those having more than five and less than 500 associated genes. Obtained GO and KEGG annotated terms were then grouped in broader functional categories, which are represented in this figure. Keywords used in PubMed to search for functions altered in endometriosis and RIF patients included ‘endometriosis’, ‘RIF’, ‘recurrent implantation failure’, ‘function’, ‘pathway’, ‘gene ontology’ and ‘KEGG’. References: a—(Devesa-Peiro et al., 2020); b—(Burney et al., 2007); c—(Crispi et al., 2013); d—(Anderson, 2019); e—(Burney and Giudice, 2012); f—(Liu et al., 2015); g—(Marquardt et al., 2019); h—(Patel et al., 2018); I—(Tomassetti et al., 2006); j—(Zhang et al., 2019); k—(Cakmak and Taylor, 2011); l—(Koot et al., 2016); m—(Pathare et al., 2017); n—(Huang et al., 2017); o—(Long et al., 2016); p—(Hapangama et al., 2008); q—(Attar et al., 2010); r—(von Adamek et al., 2005); s—(Kiyomizu et al., 2006).
Genes obtained from Tamaresis 2014 were not used to report newly discovered biomarkers because their samples were separated into two groups according to an unknown effect that we were not able to associate with any technical, biological or clinical registered variable in the original study. This unknown effect could be responsible for the remarkably large number of DEGs obtained both before and after applying the correction of the menstrual cycle effect (Table IIA). Considering all this, we cannot assess how this unknown effect on gene expression may impact the results.
Comparison of the proposed menstrual cycle effect correction method versus transcriptomic analyses within each menstrual cycle phase for the identification of uterine disorder biomarkers in endometrium
A significantly lower proportion of DEGs (FDR < 0.05) was obtained when analysis was performed independently for each menstrual cycle phase compared to when correcting for the menstrual cycle effect on gene expression (Fisher’s exact test FDR < 2.2 × 10−16; Table IIC). Indeed, significant genes from Burney 2007 and Hawkins 2011 were only detected in the early secretory phase and in the proliferative phase, respectively (Table IIC). As expected, due to the reduction in sample size, the statistical power was lower for the per menstrual cycle phase analyses compared to the menstrual cycle correction method (Table IIC).
Validation of the menstrual cycle effect correction method
To check the robustness and reliability of the method used to remove the menstrual cycle effect on gene expression, we applied the aforementioned approaches of differential expression analysis (with and without menstrual cycle correction) to five independent endometrial transcriptomic studies that compared endometrial gene expression profiles across different menstrual cycle phases (Table IIIA). Three studies used the LH peak for endometrial dating (Bradley 2010, Altmae 2017, Sigurgeirsson 2016), one used histopathological criteria of Noyes et al., 1975 (Talbi 2006), and the other did not report the dating methodology (Kelleher2017). For the five evaluated studies, samples were mainly grouped by the menstrual cycle phase according to principal component analysis (Supplementary Fig. S1). Consequently, we identified significantly DEGs between endometrial phases in all of them before applying the menstrual cycle effect correction (Table IIIB). However, samples were no longer grouped by endometrial phase (Supplementary Fig. S1) and no DEGs were obtained between the distinct endometrial phases after the menstrual cycle effect was removed (Table IIIB), demonstrating that the correction worked as expected.
Table III.
Transcriptomic studies evaluating differences between menstrual cycle phases in women with normal endometrium: Validating the menstrual cycle effect correction method.
A) Clinical characterisation of participants.
| GEO ID (Study Name) | Cycle type | Cycle phase dating method | N° Samples per cycle phase | Age | BMI | Ethnicity | Platform | Ref. |
|---|---|---|---|---|---|---|---|---|
| GSE4888 (Talbi 2006) | Normo-ovulatory. Regular (24–35 days). 3 months since last hormonal treatment | 4 pathologists (Noyes et al., 1975) |
PF (n = 6) ESE (n = 4) MSE (n = 9) LSE (n = 8) |
23–49 | N/A | Caucasian (n = 17), Black (n = 6), Asian (n = 1), Other (n = 2), Unknown (n = 1) | hgu133plus2 Affymetrix | (Talbi et al., 2006) |
| GSE29981 (Bradley 2011) | Regular (glandular epithelium alone) | Days from LH peak: PF (LH-14—LH-1), ESE (LH + 1—LH + 4), MSE (LH + 6 - LH + 7) |
PF (n = 10) ESE (n = 6) MSE (n = 4) |
20–39 | N/A | N/A | hgu133plus2 Affymetrix | N/A |
| GSE98386 (Altmae 2017) | Natural cycle | Days from LH peak |
LH + 2 (n = 20) LH + 8 (n = 20) |
N/A | N/A | Estonia | Illumina HiSeq 2500 | (Altmäe et al., 2017; Rekker et al., 2018; Teder et al., 2018) |
| GSE86491 (Sigurgeirsson 2016) | Regular. 3 months since last hormonal treatment | PF: 6–8 days after the start of the subsequent menstruation. MSE-LSE: LH + 7-LH + 9, Urinary LH ovulation predictor kit. Both confirmed by a gynaecological pathologist through histopathological examination. |
PF (n = 7) MSE-LSE (n = 7) |
24–30 | 19.8–33.2 | N/A | Illumina HiSeq 2500 | (Sigurgeirsson et al., 2016) |
| GSE119209 (Kelleher 2018) | N/A | N/A |
PF (n = 6) MSE (n = 5) |
N/A | N/A | N/A | Illumina HiSeq 2500 | N/A |
| B) Differential expression analysis with and without correcting for menstrual cycle. | |||
|---|---|---|---|
| GEO ID | Comparison | N° DEGs without menstrual cycle correction | N° DEGs with menstrual cycle correction |
| Talbi 2006 | PF vs ESE | 1478 | 0 |
| PF vs MSE | 3130 | 0 | |
| PF vs LSE | 3790 | 0 | |
| ESE vs MSE | 1309 | 0 | |
| ESE vs LSE | 3075 | 0 | |
| MSE vs LSE | 1435 | 0 | |
| ANOVA | 624 | ||
| Bradley 2011 | PF vs ESE | 1559 | 0 |
| PF vs MSE | 1720 | 0 | |
| ESE vs MSE | 35 | 0 | |
| ANOVA | 53 | ||
| Altmae 2017 | ESE vs MSE | 6788 | 0 |
| Sigurgeirsson 2016 | PF vs MSE-LSE | 5959 | 0 |
| Kelleher 2018 | PF vs MSE | 7532 | 0 |
(A) Clinical characterisation of participants. The GEO identifier, study name given in this work, and clinical information about participants including cycle type, endometrial dating method, cycle phase in which the endometrial biopsies were collected along with number of samples for each menstrual cycle phase, age, BMI, and ethnicity are presented for each included study. The transcriptomic platform used to measure gene expression and the publication in which data were initially employed are also presented. Altmae2017 and Sigurgeirsson 2016 have paired samples. N/A, not available. GEO ID, Gene Expression Omnibus GSE identifier. BMI, body mass index. LH, luteinizing hormone. PF, proliferative. ESE, early secretory. MSE, mid-secretory. LSE, late secretory. S, secretory. (B) Differential expression analysis with and without correcting for menstrual cycle. For each study, the number of differentially expressed genes (DEGs) obtained after ANOVA and pairwise comparisons between the distinct menstrual cycle phases is shown, with and without menstrual cycle correction.
Discussion
This study is the first to assess the current practices in identifying transcriptomic biomarkers of uterine disorders in endometrium, especially in relation to the menstrual cycle phase of endometrial biopsy collection. We found that one third of the studies did not report the menstrual cycle phase of the samples, including all of the suitable studies evaluating endometrial adenocarcinoma, leiomyosarcoma and adenomyosis. Other studies (37.14%) collected endometrial biopsies within the same menstrual cycle phase but with no further sub-classification in early-, mid- or late-secretory stages was made. Noyes histopathological criteria (Noyes et al., 1975) is one of the most utilised methods for endometrial dating, even though endometrial transcriptomics is superior in both accuracy and reproducibility (Díaz-Gimeno et al., 2013).
Our description of current practices demonstrated that, although it is widely known that the menstrual cycle progression affects most of the genes expressed in endometrium (Talbi et al., 2006; Koot et al., 2016; Diaz-Gimeno et al., 2017; Sebastian-Leon et al., 2018; Saare et al., 2019), new guidelines are needed for avoiding the menstrual cycle bias in transcriptomic analysis, and a more in-depth registration and consideration of endometrial stage is required.
Menstrual cycle correction enabled identification of an average of 44.19% more uterine disorder biomarkers that would have otherwise remained undiscovered. This phenomenon held true regardless of the endometrial dating method or whether the endometrial biopsies were collected along the entire menstrual cycle (Burney 2007, Hawkins 2011, Tamaresis 2014) or only within the secretory phase (Koot 2016). The highest evidence was shown when the menstrual cycle effect correction was needed even if the study design included samples balanced across the cycle between case and control groups. Our findings suggest that the current practice for avoiding menstrual cycle bias in transcriptomic studies is unable to prevent endometrial progression from masking potential uterine disorder biomarkers. In addition, our results showed biopsies collected at the secretory phase must be further subdivided into early-, mid- and late-secretory stages to correct for menstrual cycle bias.
One of the limitations of this study is that it depends on publicly available datasets. Therefore, the effect of the menstrual cycle on biomarker discovery could only be evaluated in a limited number of uterine disorders. However, the effect of the menstrual cycle was present in all the evaluated conditions; thus, this effect is likely also present in other endometrial pathologies not included here. Recently, Suhorutshenko and colleagues demonstrated that endometrial receptivity biomarkers are biased by the distinct proportions of stromal and epithelial cells within the collected endometrial biopsies (Suhorutshenko et al., 2018). Here, we demonstrate that menstrual cycle progression biases the biomarker search and if the proportion of cell types described by Suhorutshenko and colleagues is inherent to menstrual cycle progression, this produces a cycle-based bias rather than a technical bias in biopsy collection as has been suggested (Suhorutshenko et al., 2018). We also demonstrate that menstrual cycle progression affects the discovery of endometrial transcriptomic biomarkers and call for an assessment of best practices of endometrial transcriptomic analysis.
We propose a novel classification of endometrial biomarkers for gene expression studies evaluating uterine disorders. This new classification identifies biomarkers depending on the aetiology of gene expression changes, distinguishing between menstrual cycle biomarkers, uterine disorder biomarkers not masked by the menstrual cycle (which are sub-classified as genes whose expression is not affected by the menstrual cycle and genes with a menstrual cycle effect but in which the phase-dependent expression changes is less than those explained by the uterine disorder) and uterine disorder biomarkers masked by the menstrual cycle (identified after the menstrual cycle effect is corrected). This latter type of endometrial biomarker is likely to remain undetected under current practices of transcriptomic studies that do not control for menstrual cycle bias. Using this methodology, we unveiled new potential biomarkers: 544 for eutopic endometriosis, 158 for ectopic ovarian endometriosis and 27 for recurrent implantation failure, all of which had not been previously reported in the included studies (Burney et al., 2007; Hawkins et al., 2011; Tamaresis et al., 2014; Koot et al., 2016) or other studies consulted through Disgenet (Piñero et al., 2020), Phenopedia (Yu et al., 2010) and GeneCards (Stelzer et al., 2016) databases. These new candidate biomarkers were involved in functions known to be altered by the corresponding uterine disorder, such as immune response and inflammation, cell differentiation and development in endometriosis (Tomassetti et al., 2006; Burney and Giudice, 2012; Crispi et al., 2013; Tamaresis et al., 2014; Liu et al., 2015; Patel et al., 2018; Anderson, 2019; Marquardt et al., 2019; Zhang et al., 2019; Devesa-Peiro et al., 2020) or epigenetics and transcription and post-transcription regulation in RIF (Cakmak and Taylor, 2011; Koot et al., 2016; Pathare et al., 2017; Devesa-Peiro et al., 2020), highlighting their relevance in the pathophysiology of the disease.
Our method of menstrual cycle correction proved to be robust and reliable, as samples were no longer grouped by the endometrial phase and we did not identify any DEGs between distinct menstrual cycle phases after we applied this correction to studies evaluating menstrual cycle changes in women with normal endometrium. This was consistent regardless of the statistical method employed for differential expression analysis (ANOVA or pairwise comparisons) and/or endometrial dating method. In addition, the correction method maintained the average differences between case and control samples for studies evaluating uterine disorders, demonstrating that the effect of the condition was not removed in the correction process and that the observed changes in the P-values were explained only by the removal of the gene expression variability explained by menstrual cycle progression.
When comparing our approach of uterine disorder biomarker detection with those followed in the included studies, we found that only Koot and colleagues corrected for menstrual cycle effect also using linear models (Koot et al., 2016). From the remaining five studies, Burney and colleagues and Tamaresis and colleagues addressed menstrual cycle bias by dividing samples according to the menstrual cycle phase and performing an independent differential expression analysis at a probeset level (Burney et al., 2007; Tamaresis et al., 2014). Unlike our proposed correction method, this strategy allows identification of phase-specific uterine disorder biomarkers (e.g., genes whose expression only differs significantly between cases and controls in the proliferative phase but not in the secretory phase). Although identifying phase-specific uterine disorder biomarkers is useful in understanding the relationship between the disorder and the menstrual cycle, we demonstrated that this strategy retrieves significantly fewer potential uterine disorder biomarkers compared to menstrual cycle effect correction, as it sacrifices statistical power due to lower sample sizes. In contrast, our proposed correction method identifies uterine disorder biomarkers regardless of menstrual cycle phase during biopsy. Notably, removing the menstrual cycle effect does not impede identifying potential biomarker genes that are responding to both the menstrual cycle and the uterine disorder. In fact, those genes more greatly influenced by the menstrual cycle than by the uterine disorder are the ones that the correction method can unmask.
Considering these findings, we define new guidelines for the detection of reliable uterine disorder biomarkers according to distinct scenarios. If endometrial biopsies are collected at different stages of the menstrual cycle, the menstrual cycle effect must be always corrected in the transcriptomic analysis as endometrial timing is masking genes whose expression is affected by the uterine disorder. In unbalanced studies in which the menstrual cycle stage was not corrected, we expected an additional risk of identifying genes as uterine disorder biomarkers whose expression is indeed dependent on the menstrual cycle and not on the evaluated condition. Therefore, applying the correction for the menstrual cycle in these studies is even more crucial. We observed this in Tamaresis 2014, the only study in which menstrual cycle biomarkers were identified and where the endometriosis samples were mostly secretory and the control samples proliferative. However, this hypothesis needs further confirmation, as this study presented an unknown effect on gene expression.
Single-cell studies are increasingly used to evaluate endometrial gene expression changes throughout the menstrual cycle, increasing our molecular understanding of endometrial function and receptivity acquisition (Wang et al., 2020). Although further studies are needed, we could expect from our results that the menstrual cycle effect will also need to be corrected in single-cell studies aimed to identify biomarkers of uterine disorders on specific cell types and in which samples are collected at different endometrial stages. Although the correction method proposed in this study was previously applied to single-cell studies (Tran et al., 2020), other methodologies have recently arisen to specifically correct known effects in this type of gene expression data (Haghverdi et al., 2018; Tran et al., 2020).
In-depth registration and consideration of endometrial stage is needed in any transcriptomic study to optimise the detection of reliable biomarkers of uterine disorders. Here, we introduce a novel classification of endometrial transcriptomic biomarkers depending on the DEG aetiology and set new guidelines to accurately detect uterine disorder biomarkers through differential expression analysis with high reproducibility and statistical power. Using these methods, we unmasked new endometriosis and RIF potential biomarkers to improve diagnosis, prognosis and treatment. The application of these methods in future research on biomarker discovery of uterine disorders would further contribute to delineating their aetiology and progression, and ultimately leading towards improved treatments and increased pregnancy rates in these patients.
Data availability
The raw gene expression data and patient meta-data associated with the studies that were reprocessed and reanalysed in this work are freely available to download from GEO given their unique identifiers: GSE6364 (Giudice, 2007), GSE23339 (Creighton et al., 2011), GSE51981 (Tamaresis et al., 2014b), GSE58144 (van Hooff et al., 2015), GSE92324 (Pathare et al., 2019), GSE65099 (Brosens et al., 2015), GSE4888 (Talbi et al., 2006b), GSE29981 (Bradley et al., 2011), GSE98386 (Suhorutshenko et al., 2017), GSE86491 (Sigurgeirsson et al., 2016) and GSE119209 (Kelleher et al., 2018). Supplementary Tables SII and SIII of this article are available at the Figshare Repository, https://dx.doi.org/10.6084/m9.figshare.13643147
Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Supplementary Material
Acknowledgements
The authors thank the IVI-RMA IVI Foundation and the University of Valencia for their research support. We also thank the team of Fresh Eyes Editing, LLC., especially Dr. Sheila Cherry, for their professional assistance in manuscript preparation.
Authors’ roles
The original idea of this research was defined by P.D.-G. with the help of A.P. Study design was defined by P.D.-.G with the help of A.D.-P. and P.S.-L. Systematic review and dataset selection and acquisition were performed by A.D.-P. and P.S.-L., supervised by P.D.-G. Transcriptomic analyses were executed by A.D.-P. and P.S.-L. under the supervision of P.D.-G. Biomedical interpretation and implications were provided by P.D.-G. and A.P. with the help of A.D.P.; and functional interpretation was completed by A.D.-P., supervised by P.D.-G. Tables and figures were designed by A.D.-P., P.S.-L. and P.D.-G. Manuscript writing was primarily done by P.D.-G. and A.D.-P., being supervised by all co-authors (P.S.-L. and A.P.).
Funding
Research was supported by the Instituto de Salud Carlos III through a Health Research Project programme (PI19/00537) (Spanish Government) and Spanish Ministry of Economy and Competitiveness through the Miguel Servet programme (CP20/00118) granted to Patricia Diaz-Gimeno, and co-funded by FEDER and by the IVI-RMA IVI Foundation (1706-FIVI-041-PD). A. Devesa-Peiro is supported by the FPU/15/01398 program fellowship from the Ministry of Science, Innovation and Universities (Spanish Government).
Conflict of interest
The authors do not have any competing interests to declare.
This research was partially presented at the 35th Annual Meeting of the European Society of Human Reproduction and Embryology (ESHRE), Vienna, Austria, June 24–26, 2019.
References
- Altmäe S, Koel M, Võsa U, Adler P, Suhorutšenko M, Laisk-Podar T, Kukushkina V, Saare M, Velthut-Meikas A, Krjutškov K. et al. Meta-signature of human endometrial receptivity: a meta-analysis and validation study of transcriptomic biomarkers. Sci Rep 2017;7:10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Society for Reproductive. Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertility and Sterility 1997;67:817–821. 10.1016/S0015-0282(97)81391-X [DOI] [PubMed] [Google Scholar]
- Anderson G. Endometriosis pathoetiology and pathophysiology: roles of vitamin A, estrogen, immunity, adipocytes, gut microbiome and melatonergic pathway on mitochondria regulation. Biomol Concepts 2019;10:133–149. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. et al. Gene ontology: tool for the unification of biology. Nat Genet 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attar R, Cacina C, Sozen S, Attar E, Agachan B.. DNA repair genes in endometriosis. Genet Mol Res 2010;9:629–636. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300. [Google Scholar]
- Bradley G, O'Regan P, Pullen N. Expression data from human endometrium, Gene Expression Omnibus, Accession number GSE29981. 2011.
- Brosens JJ, Chan Y, Lucas ES. Loss of endometrial plasticity in recurrent pregnancy loss (RNA-Seq), Gene Expression Omnibus, Accession number GSE65099. 2015.
- Burney R, Giudice L.. Pathogenesis and Pathophysiology of Endometriosis. Fertil Steril 2012;98: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burney RO, Talbi S, Hamilton AE, Kim CV, Nyegaard M, Nezhat CR, Lessey BA, Giudice LC.. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology 2007;148:3814–3826. [DOI] [PubMed] [Google Scholar]
- Cakmak H, Taylor HS.. Implantation failure: Molecular mechanisms and clinical treatment. Hum Reprod Update 2011;17:242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craciunas L, Gallos I, Chu J, Bourne T, Quenby S, Brosens JJ, Coomarasamy A.. Conventional and modern markers of endometrial receptivity: a systematic review and meta-analysis. Hum Reprod Update 2019;25:202–223. [DOI] [PubMed] [Google Scholar]
- Creighton C, Matzuk M, Hawkins S. Gene expression profiles of endometriosis, Gene Expression Omnibus, Accession number GSE23339. 2006.
- Crispi S, Piccolo MT, D'avino A, Donizetti A, Viceconte R, Spyrou M, Calogero RA, Baldi A, Signorile PG.. Transcriptional profiling of endometriosis tissues identifies genes related to organogenesis defects. J Cell Physiol 2013;228:1927–1934. [DOI] [PubMed] [Google Scholar]
- Devesa-Peiro A, Sebastian-Leon P, Garcia-Garcia F, Arnau V, Aleman A, Pellicer A, Diaz-Gimeno P.. Uterine disorders affecting female fertility: what are the molecular functions altered in endometrium? Fertil Steril 2020;113:1261–1274. [DOI] [PubMed] [Google Scholar]
- Devlieger R, D’Hooghe T, Timmerman D.. Uterine adenomyosis in the infertility clinic. Hum Reprod Update 2003;9:139–147. [DOI] [PubMed] [Google Scholar]
- Díaz-Gimeno P, Horcajadas JA, Martínez-Conejero JA, Esteban FJ, Alamá P, Pellicer A, Simón C.. A genomic diagnostic tool for human endometrial receptivity based on the transcriptomic signature. Fertil Steril 2011;95:50–60.e15. [DOI] [PubMed] [Google Scholar]
- Díaz-Gimeno P, Ruiz-Alonso M, Blesa D, Bosch N, Martínez-Conejero JA, Alamá P, Garrido N, Pellicer A, Simón C.. The accuracy and reproducibility of the endometrial receptivity array is superior to histology as a diagnostic method for endometrial receptivity. Fertil Steril 2013;99:508–517. [DOI] [PubMed] [Google Scholar]
- Diaz-Gimeno P, Ruiz-Alonso M, Sebastian-Leon P, Pellicer A, Valbuena D, Simon C.. Window of implantation transcriptomic stratification reveals different endometrial subsignatures associated with live birth and biochemical pregnancy. Fertil Steril 2017;108:703–710. [DOI] [PubMed] [Google Scholar]
- Dunselman GAJ, Vermeulen N, Becker C, Calhaz-Jorge C, D'Hooghe T, De Bie B, Heikinheimo O, Horne AW, Kiesel L, Nap A. et al. ESHRE guideline: Management of women with endometriosis. Hum Reprod 2014;29:400–412. [DOI] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, Huber W.. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 2009;4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espín-Pérez A, Portier C, Chadeau-Hyam M, Veldhoven K, van Kleinjans J, Kok T. D.. Comparison of statistical methods and the use of quality control samples for batch effect correction in human transcriptome data. PLoS One 2018;13:e0202947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. On the Interpretation of χ2 from Contingency Tables, and the Calculation of P. J R Stat Soc 1922;85:87. [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA.. Affy - Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307–315. [DOI] [PubMed] [Google Scholar]
- Giudice L. Gene Profiling of Endometrium Reveals Progesterone Resistance and Candidate Genetic Loci in Women with Endometriosis, Gene Expression Omnibus, Accession number GSE6364. 2010.
- Gurevitch J, Koricheva J, Nakagawa S, Stewart G.. Meta-analysis and the science of research synthesis. Nature 2018;555:175–182. [DOI] [PubMed] [Google Scholar]
- Haghverdi L, Lun A, Morgan M, Marioni J.. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 2018;36:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapangama DK, Turner MA, Drury JA, Martin-Ruiz C, von ZT, Farquharson RG, Quenby S.. Endometrial telomerase shows specific expression patterns in different types of reproductive failure. Reprod Biomed Online 2008;17:416–424. [DOI] [PubMed] [Google Scholar]
- Harada T, Khine YM, Kaponis A, Nikellis T, Decavalas G, Taniguchi F.. The Impact of Adenomyosis on Women’s Fertility. Obstet Gynecol Surv 2016;71:557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper MJ. The implantation window. Baillieres Clin Obs Gynaecol 1992;6:351–371. [DOI] [PubMed] [Google Scholar]
- Hawkins SM, Creighton CJ, Han DY, Zariff A, Anderson ML, Gunaratne PH, Matzuk MM.. Functional microRNA involved in endometriosis. Mol Endocrinol 2011;25:821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Qin H, Yang Y, Chen X, Zhang J, Laird S, Wang CC, Chan TF, Li TC.. A comparison of transcriptomic profiles in endometrium during window of implantation between women with unexplained recurrent implantation failure and recurrent miscarriage. Reproduction 2017;153:749–758. [DOI] [PubMed] [Google Scholar]
- Jauniaux E, Farquharson RG, Christiansen OB, Exalto N.. Evidence-based guidelines for the investigation and medical treatment of recurrent miscarriage. Hum Reprod 2006;21:2216–2222. [DOI] [PubMed] [Google Scholar]
- Johnson W, Li C, Rabinovic A.. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–127. [DOI] [PubMed] [Google Scholar]
- Kelleher AM, Behura SK, Burns GW, Young SL, DeMayo F, Spencer TE. Determination of the Forkhead box A2 (FOXA2) Cistrome in the Human Endometrium, Gene Expression Omnibus, Accession number GSE119209. 2018.
- Kiyomizu M, Kitawaki J, Obayashi H, Ohta M, Koshiba H, Ishihara H, Honjo H.. Association of two polymorphisms in the peroxisome proliferator-activated receptor-gamma gene with adenomyosis, endometriosis, and leiomyomata in Japanese women. J Soc Gynecol Investig 2006;13:372–377. [DOI] [PubMed] [Google Scholar]
- Koot YEM, van Hooff SR, Boomsma CM, van Leenen D, Groot Koerkamp MJA, Goddijn M, Eijkemans MJC, Fauser BCJM, Holstege FCP, Macklon NS.. An endometrial gene expression signature accurately predicts recurrent implantation failure after IVF. Sci Rep 2016;6:19411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lédée N, Munaut C, Aubert J, Sérazin V, Rahmati M, Chaouat G, Sandra O, Foidart JM.. Specific and extensive endometrial deregulation is present before conception in IVF/ICSI repeated implantation failures (IF) or recurrent miscarriages. J Pathol 2011;225:554–564. [DOI] [PubMed] [Google Scholar]
- Lessey BA, Kim JJ.. Endometrial receptivity in the eutopic endometrium of women with endometriosis: it is affected, and let me show you why. Fertil Steril 2017;108:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Lv X, Yu H, Xu P, Ma R, Zou K.. In search of key genes associated with endometriosis using bioinformatics approach. Eur J Obstet Gynecol Reprod Biol 2015;194:119–124. [DOI] [PubMed] [Google Scholar]
- Long N, Liu N, Liu XL, Li J, Cai BY, Cai X.. Endometrial expression of telomerase, progesterone, and estrogen receptors during the implantation window in patients with recurrent implantation failure. Genet Mol Res 2016;15:gmr7849. [DOI] [PubMed] [Google Scholar]
- Lucas E S, Dyer N P, Murakami K, Hou Lee Y, Chan Y-W, Grimaldi G, Muter J, Brighton P J, Moore J D, Patel G. et al. Loss of Endometrial Plasticity in Recurrent Pregnancy Loss. Stem Cells 2016;34:346–356. 10.1002/stem.2222 [DOI] [PubMed] [Google Scholar]
- Marquardt RM, Kim TH, Shin JH, Jeong JW.. Progesterone and estrogen signaling in the endometrium: What goes wrong in endometriosis? Int J Mol Sci 2019;20: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miravet-Valenciano J, Ruiz-Alonso M, Gomez E, Garcia-Velasco JA.. Endometrial receptivity in eutopic endometrium in patients with endometriosis: it is not affected, and let me show you why. Fertil Steril 2017;108:28–31. [DOI] [PubMed] [Google Scholar]
- Murphy CR. Uterine receptivity and the plasma membrane transformation. Cell Res 2004;14:259–267. [DOI] [PubMed] [Google Scholar]
- Niederberger C, Pellicer A, Cohen J, Gardner DK, Palermo GD, O’Neill CL, Chow S, Rosenwaks Z, Cobo A, Swain JE. et al. Forty years of IVF. Fertil Steril 2018;110:185–324.e5. [DOI] [PubMed] [Google Scholar]
- Noyes RW, Hertig AT, Rock J.. Dating the endometrial biopsy. Am J Obstet Gynecol 1975;122:262–263. [DOI] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M.. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 1999;27:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BG, Lenk EE, Lebovic DI, Shu Y, Yu J, Taylor RN.. Pathogenesis of endometriosis: Interaction between Endocrine and inflammatory pathways. Best Pract Res Clin Obstet Gynaecol 2018;50:50–60. [DOI] [PubMed] [Google Scholar]
- Pathare ADS, Zaveri K, Hinduja I.. Downregulation of genes related to immune and inflammatory response in IVF implantation failure cases under controlled ovarian stimulation. Am J Reprod Immunol 2017;78:e12679. [DOI] [PubMed] [Google Scholar]
- Pathare ADS, Zaveri K, Hinduja I. Gene expression study of human endometrial receptivity in IVF implantation failure patients undergoing ovarian stimulation II, Gene Expression Omnibus, Accession number GSE92324. 2016.
- Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI.. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res 2020;48:D845–D855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanski LT, Baumgarten MN, Quenby S, Brosens J, Campbell BK, Raine-Fenning NJ. What exactly do we mean by “recurrent implantation failure”? A systematic review and opinion. Reprod Biomed Online. 2014;28:409–423. [DOI] [PubMed] [Google Scholar]
- Qiu W, Lee M-LT, Whitmore GA. sizepower: Sample Size and Power Calculation in Micorarray Studies. R-package2018;
- R Core Team. R: A language and environment for statistical computing. Vienna, Austria R Found Stat Comput.2016; Available from: https://www.R-project.org/.
- Rekker K, Altmäe S, Suhorutshenko M, Peters M, Martinez-Blanch J F, Codoñer F M, Vilella F, Simón C, Salumets A, Velthut-Meikas A.. A Two-Cohort RNA-seq Study Reveals Changes in Endometrial and Blood miRNome in Fertile and Infertile Women. Genes 2018;9:574. 10.3390/genes9120574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M, McCarthy D, Smyth G.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saare M, Laisk T, Teder H, Paluoja P, Palta P, Koel M, Kirss F, Karro H, Sõritsa D, Salumets A. et al. A molecular tool for menstrual cycle phase dating of endometrial samples in endometriosis transcriptome studies. Biol Reprod 2019;101:1–3. [DOI] [PubMed] [Google Scholar]
- Sebastian-Leon P, Garrido N, Remohí J, Pellicer A, Diaz-Gimeno P.. Asynchronous and pathological windows of implantation: Two causes of recurrent implantation failure. Hum Reprod 2018;33:626–635. [DOI] [PubMed] [Google Scholar]
- Sigurgeirsson B, Åmark H, Jemt A, Ujvari D, Westgren M, Lundeberg J, Gidlöf S. Comprehensive RNA sequencing of healthy human endometrium at two time points of the menstrual cycle, Gene Expression Omnibus, Accession number GSE86491. 2016. [DOI] [PubMed]
- Somigliana E, Vigano P, Busnelli A, Paffoni A, Vegetti W, Vercellini P. Repeated implantation failure at the crossroad between statistics, clinics and over-diagnosis. Reprod Biomed Online2018; [DOI] [PubMed]
- Stelzer G, Rosen R, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Iny ST, Nudel R, Lieder I, Mazor Y. et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analysis. Curr Protoc Bioinforma 2016;54:1.30.1. [DOI] [PubMed] [Google Scholar]
- Suhorutshenko M, Kukushkina V, Velthut-Meikas A, Altmäe S, Peters M, Mägi R, Krjutškov K, Koel M, Codoñer FM, Martinez-Blanch JF. et al. Endometrial receptivity revisited: Endometrial transcriptome adjusted for tissue cellular heterogeneity. Hum Reprod 2018;33:2074–2086. [DOI] [PubMed] [Google Scholar]
- Suhorutshenko M, Kukushkina V, Velthut-Meikas A, Mägi R, Peters M, Altmäe S, Krjutškov K, Martinez-Blanch JF, Codoñer FM, Vilella F. et al. Expression profile of early and mid-secretory healthy human endometrium revealed by RNA-seq, Gene Expression Omnibus, Accession number GSE98386. 2017.
- Talbi S, Hamilton AE, Vo KC, Tulac S, Overgaard MT, Dosiou C, Le Shay N, Nezhat CN, Kempson R, Lessey BA. et al. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology 2006;147:1097–1121. [DOI] [PubMed] [Google Scholar]
- Talbi S, Hamilton AE, Vo KC, TulaC S, Overgaard MT, Dosiou C, Le Shay N, Kempson R, Lessey BA, Nayak NR. et al. Molecular phenotyping of human endometrium, Gene Expression Omnibus, Accession number GSE4888. 2006. [DOI] [PubMed]
- Tamaresis JS, Irwin JC, Goldfien GA, Rabban JT, Burney RO, Nezhat C, DePaolo LV, Giudice LC.. Molecular classification of endometriosis and disease stage using high-dimensional genomic data. Endocrinology 2014;155:4986–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaresis JS, Irwin JC, Goldfien GA, Rabban JT, Burney RO, Nezhat C, DePaolo LV, Giudice LC. Molecular Classification of Endometriosis and Disease Stage Using High-Dimensional Genomic Data, Gene Expression Omnibus, Accession number GSE51981. 2013. [DOI] [PMC free article] [PubMed]
- Tanbo T, Fedorcsak P.. Endometriosis-associated infertility: aspects of pathophysiological mechanisms and treatment options. Acta Obstet Gynecol Scand 2017;96:659–667. [DOI] [PubMed] [Google Scholar]
- Teder H, Koel M, Paluoja P, Jatsenko T, Rekker K, Laisk-Podar T, Kukuškina V, Velthut-Meikas A, Fjodorova O, Peters M. et al. TAC-seq: targeted DNA and RNA sequencing for precise biomarker molecule counting. NPJ Genom Me. 2018;3:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The American Fertility Society. Revised American Fertility Society classification of endometriosis: 1985. Fertil Steril 1985;43:351–352. [DOI] [PubMed] [Google Scholar]
- Tomassetti C, Meuleman C, Pexsters A, Mihalyi A, Kyama C, Simsa P, Tm D.. Endometriosis, recurrent miscarriage and implantation failure: Is there an immunological link? Reprod Biomed Online 2006;13:58–64. [DOI] [PubMed] [Google Scholar]
- Tran HTN, Ang KS, Chevrier M, Zhang X, Lee NYS, Goh M, Chen J.. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol 2020;21: [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hooff SR, Koot YE, Boomsma CM, van Leenen D, Groot Koerkamp MJ, Goddijn M, Eijkemans MJ, Fauser BC, Holstege FC, Macklon NS. The endometrial gene expression signature of recurrent implantation failure after IVF, Gene Expression Omnibus, Accession number GSE58144. 2014. [DOI] [PMC free article] [PubMed]
- Wang P-H, Fuh J-L, Chao H-T, Liu W-M, Cheng M-H, Chao K-C.. Is the surgical approach beneficial to subfertile women with symptomatic extensive adenomyosis? J Obs Gynaecol Res 2009;35:495–502. [DOI] [PubMed] [Google Scholar]
- Wang W, Vilella F, Alama P, Moreno I, Mignardi M, Isakova A, Pan W, Simon C, Quake S.. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat Med 2020;26:1644–1653. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant Graphics for Data Analysis. Springer, 2016. [Google Scholar]
- Wilcox AJ, Baird DD, Weinberg CR.. Time of Implantation of the Conceptus and Loss of Pregnancy. N Engl J Med 1999;340:1796–1799. [DOI] [PubMed] [Google Scholar]
- Yu W, Clyne M, Khoury MJ, Gwinn M.. Phenopedia and Genopedia: Disease-centered and Gene-centered Views of the Evolving Knowledge of Human Genetic Associations. Bioinformatics 2010;26:145–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepiridis L, Grimbizis G, Tarlatzis B.. Infertility and uterine fibroids. Best Pr Res Clin Obs Gynaecol 2016;34:66–73. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ruan L, Lu M, Yao X.. Analysis of key candidate genes and pathways of endometriosis pathophysiology by a genomics-bioinformatics approach. Gynecol Endocrinol 2019;35:576–581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw gene expression data and patient meta-data associated with the studies that were reprocessed and reanalysed in this work are freely available to download from GEO given their unique identifiers: GSE6364 (Giudice, 2007), GSE23339 (Creighton et al., 2011), GSE51981 (Tamaresis et al., 2014b), GSE58144 (van Hooff et al., 2015), GSE92324 (Pathare et al., 2019), GSE65099 (Brosens et al., 2015), GSE4888 (Talbi et al., 2006b), GSE29981 (Bradley et al., 2011), GSE98386 (Suhorutshenko et al., 2017), GSE86491 (Sigurgeirsson et al., 2016) and GSE119209 (Kelleher et al., 2018). Supplementary Tables SII and SIII of this article are available at the Figshare Repository, https://dx.doi.org/10.6084/m9.figshare.13643147