Abstract
This cross-sectional study was conducted between January and June 2020, in five large poultry slaughter slabs in Dar es Salaam, Tanzania. Purposive sampling was used to select broilers and spent layers, from which meat and cloaca swabs were collected to determine the occurrence of multidrug resistant (MDR) Escherichia coli. Identification of isolates was done using API 20E, and antimicrobial susceptibility testing was performed as per CLSI (2018) guidelines. EBSL (CTX-M, TEM, SHV) and plasmid mediated quinolone (qnrA, qnrB, qnrS and aac(6′)-Ib-cr) were screened using PCR. Out of 384 samples, 212 (55.2%) were positive for E. coli, of which 147 (69.3%) were resistant to multiple drugs (MDR). Highest resistance was detected to tetracycline (91.9%), followed by sulfamethoxazole-trimethoprim (80.5%), ampicillin (70.9%), ciprofloxacin (40.2%) and 25% cefotaxime, gentamycin (10.8%) and imipenem (8.6%) (95% CI, p < 0.01). Out of the E. coli-positive samples, ten (10/212) (4.7%) were ESBL producing E. coli, of which CTX-M was detected in two isolates and quinolones resistant gene (qnrS) in eight, while TEM, SHV, qnrA, qnrB and aac(6′)-lb-cr were not detected. The high level of resistance and multidrug resistance imply these antibiotics are ineffective, add unnecessary cost to poultry farmers and certainly facilitate emergence and spread of resistance.
Keywords: multidrug resistant, Escherichia coli, chicken meat, cloaca, Dar es Salaam, Tanzania
1. Introduction
In Tanzania, the demand for chicken meat was projected to increase from 130,000 tons in 2017 to 465,600 tons in 2020 [1], largely due to an increase in urbanization and trade liberation of live animals and products [2]. Dar es Salaam, which is the commercial city of the country, with an estimated population of 4,364,541 people, is by far the largest consumer of poultry meat in Tanzania [3].
Poultry farming in Dar es Salaam is done both in urban and peri-urban areas, often in overcrowded and unhygienic conditions [4]. Such conditions are frequently associated with occurrence of diseases and use of excessive antimicrobials. Several studies conducted in Tanzania have shown both overuse of antibiotics and high levels of resistant organisms in poultry production systems [2,5,6]. Antibiotics are commonly used for disease prevention and therapeutic purposes in poultry production systems, are obtained over the counter and are administered without advice of veterinary officers [7]. The knowledge of most poultry keepers on prudent use of antibiotics and their effect is low, and antimicrobial prescribers and unregistered veterinary drug dealers also have little prescription knowledge, which all together create an environment for emergence and spread of antimicrobial resistance [5]. Metaphylaxis is also very common among poultry keepers [8], exposing even healthy chickens to unnecessary antimicrobials.
We conducted this study in Dar es Salaam, where the demand for poultry meat and products is the highest in the country and the use of antimicrobials among poultry keepers is very high [5,9]. We determined the occurrence of multidrug-resistant E. coli in raw chicken meat and in cloaca as well as the occurrence of extended spectrum beta lactamase, specifically CTX-M, TEM and SHV, and plasmid mediated quinolone-resistant genes (qnrA, qnrB, qnrS and aac(6′)-Ib-cr).
2. Results
2.1. Prevalence of E. coli in Raw Chicken Meat and Cloaca in Broiler and Spent Layers
A total of 384 chicken meat and cloaca swabs samples were collected in the five selected poultry slabs in Dar es Salaam. Out of these samples, 212 (55.2%) were positive for E. coli. Of the isolated strains, 147 (69.3%) were resistant to more than three tested antibiotics of different classes. The slab with the highest proportion of MDR isolates was at Stereo in the Temeke District (18/19, 94.7%), followed by Shekilango in the Ubungo district (37/43, 86%), Manzese in the Ubungo district (28/40, 70%), Mtambani in the Kinondoni district (14/20, 70%) and Kisutu in the Ilala District (50/90, 55.6%) (Table 1).
Table 1.
Frequency of MDR and non-MDR Escherichia coli isolated from the selected poultry slabs in Dar es Salaam (n = 384).
| Poultry Slabs | MDR | Not MDR | ||
|---|---|---|---|---|
| n | % | n | % | |
| Stereo | 18 | 94.7 | 1 | 5.3 |
| Manzese | 28 | 70.0 | 12 | 30.0 |
| Mtambani | 14 | 70.0 | 6 | 30.0 |
| Shekilango | 37 | 86.0 | 6 | 14.0 |
| Kisutu | 50 | 55.6 | 40 | 44.4 |
| Total | 147 | 69.3 | 65 | 30.7 |
MDR—multidrug resistant.
2.2. Antibiotic Resistance Rates in E. coli Isolates
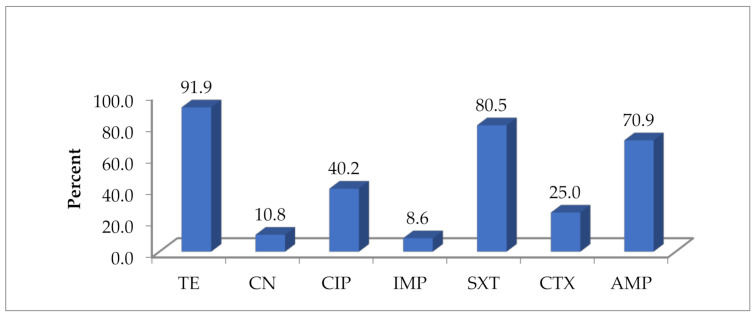
Overall, the highest resistance was detected in tetracycline (91.9%), followed by trimethoprim-sulfamethoxazole (80.5%), ampicillin (70.9%), ciprofloxacin (40.2%), cefotaxime (22.5%), 10.8% gentamycin (10.8%) and imipenem (3.3%) (Figure 1).
Figure 1.
Overall antibiotic resistance pattern of E. coli isolates in five poultry slabs (n = 212). TE = tetracycline, CN = gentamycin, CIP = ciprofloxacin, IMP = imipenem, SXT = trimethoprim-sulfamethoxazole, CTX = cefotaxime, AMP = ampicillin.
Of the 147 MDR E. coli isolates, 49% showed resistance to at least three classes of antibiotics, 33.3% to at least four classes, 14.3% resistant to five classes, 2.7% resistant to six classes, and one isolate (0.7%) was resistant to all seven tested antibiotics (Table 2 and Table 3).
Table 2.
Classes of antimicrobial patterns resisted n (%).
| MDR E. coli Isolates | Classes of Antibiotics | ||||
|---|---|---|---|---|---|
| 3 | 4 | 5 | 6 | 7 | |
| 147 | 72(49) | 49(33.3) | 21(14.3) | 4(2.7) | 1(0.7) |
Table 3.
Antimicrobial resistance pattern of multidrug-resistant E. coli.
| Antibiotic Combination | Number of Isolates | % | Number of Antibiotics Classes |
|---|---|---|---|
| TE, CN, CIP | 1 | 0.7 | 3 |
| TE, CN, SXT | 1 | 0.7 | 3 |
| TE, CIP, SXT | 17 | 11.6 | 3 |
| TE, CN, AMP | 1 | 0.7 | 3 |
| TE, IMP, SXT | 3 | 2.0 | 3 |
| TE, CIP, AMP | 8 | 5.4 | 3 |
| TE, CN, CIP, SXT | 3 | 2.0 | 4 |
| TE, IMP, AMP | 1 | 0.7 | 3 |
| TE, SXT, AMP | 39 | 26.5 | 3 |
| TE, CIP, IMP, SXT | 1 | 0.7 | 4 |
| CIP, SXT, AMP | 1 | 0.7 | 3 |
| TE, CN, SXT, AMP | 4 | 2.7 | 4 |
| TE, CIP, IMP, AMP | 1 | 0.7 | 4 |
| TE, CIP, SXT, AMP | 29 | 19.7 | 4 |
| TE, IMP, SXT, AMP | 1 | 0.7 | 4 |
| TE, CN, CIP, IMP, AMP | 1 | 0.7 | 5 |
| TE, IMP, CTX, AMP | 1 | 0.7 | 4 |
| TE, CN, CIP, SXT, AMP | 4 | 2.7 | 5 |
| TE, SXT, CTX, AMP | 9 | 6.1 | 4 |
| TE, CIP, IMP, SXT, AMP | 3 | 2.0 | 5 |
| TE, CN, SXT, CTX, AMP | 3 | 2.0 | 5 |
| TE, CIP, SXT, CTX, AMP | 9 | 6.1 | 5 |
| TE, CN, CIP, IMP, SXT, AMP | 2 | 1.4 | 6 |
| TE, IMP, SXT, CTX, AMP | 1 | 0.7 | 5 |
| TE, CN, CIP, SXT, CTX, AMP | 2 | 1.4 | 6 |
| TE, CN, CIP, IMP, SXT, CTX, AMP | 1 | 0.7 | 7 |
TE = tetracycline, AMP = ampicillin, SXT = trimethoprim-sulfamethoxazole, CIP = ciprofloxacin, CTX = cefotaxime, CN = gentamycin, IMP = imipenem.
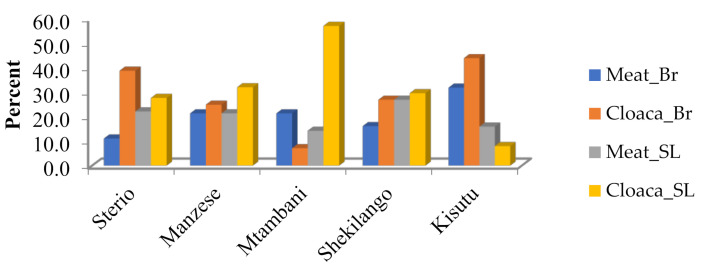
As shown in Figure 2, the isolation of MDR E. coli was higher in cloaca than chicken meat in both types of chicken.
Figure 2.
Multiple drug resistance of E. coli by location of poultry slab.
2.3. Extended Spectrum Beta Lactamase (ESBL) Producing E. coli
Out of 212 identified E. coli, 10 (4.7%) isolates were screened and confirmed to be ESBL-producing E. coli (Table 4) All 10 isolates were found to be MDR. All (100%) ESBL producers were resistant to cefotaxime and ampicillin, 90% were resistant to tetracycline and trimethoprim-sulfamethoxazole, 40% were resistant to ciprofloxacin and 10% were resistant to imipenem. However, all 10 (100%) E. coli isolates were susceptible to gentamycin (Table 4). All confirmed ESBL-producing E. coli were isolated from one poultry slab at Stereo in Temeke district, and were mostly from spent layers.
Table 4.
Antimicrobial resistance of ESBL-producing E. coli isolates (n = 10).
| Chicken Category | |||||||
|---|---|---|---|---|---|---|---|
| Antibiotic | Isolates (n) | % | Meat SL | Cloaca SL | Meat Br | Cloaca Br | Total |
| TE | 9/10 | 90 | 3 | 4 | 1 | 1 | 9 |
| CN | 0/10 | 0 | 0 | 0 | 0 | 0 | 0 |
| CIP | 4/10 | 40 | 3 | 1 | 0 | 0 | 4 |
| IMP | 1/10 | 10 | 1 | 0 | 0 | 0 | 1 |
| SXT | 9/10 | 90 | 3 | 4 | 1 | 1 | 9 |
| CTX | 10/10 | 100 | 4 | 4 | 1 | 1 | 10 |
| AMP | 10/10 | 100 | 4 | 4 | 1 | 1 | 10 |
Meat SL = spent layers’ meat, cloaca SL = spent layers’ cloaca, cloaca meat Br = broiler meat, Br = broiler cloaca, TE = tetracycline, CN = gentamycin, CIP = ciprofloxacin, IPM = imipenem, SXT = trimethoprim-sulfamethoxazole, CTX = cefotaxime, AMP = ampicillin.
2.4. Detection of CTX-M, TEM, SHV and PMQR Genes (qnrA, qnrB, qnrS and aac(6′)-lb-cr)
Plasmid mediated quinolones-resistant genes were detected in 8/10 ESBL-producing E. coli either as single genes or in combination with CTX-M, while TEM and SHV were not detected. The qnrS were present in eight (80%) of the isolates isolated from four spent layers’ meat, two spent layers’ cloaca, one broiler meat and one cloaca of broilers. All eight isolates with detected qnrS were resistant to ampicillin, sulfamethoxazole-trimethoprim and cefotaxime, seven of the eight were resistant to tetracycline, four of the eight were resistant to ciprofloxacin and three of the eight were resistant to imipenem. PMQR determinants qnrA, qnrB and aac(6′)-lb-cr were not detected in any of the E. coli isolates tested (Table 5, Figure 3 and Figure 4).
Table 5.
Distribution of ESBL- and PMQR-encoding genes by PCR (n = 10).
| Detected Genes |
E. coli No (%) |
Spent Layers Meat | Spent Layers Cloaca | Broiler Meat | Broiler Cloaca |
|---|---|---|---|---|---|
| CTX-M | 2/10 (20) | 1 | 1 | 0 | 0 |
| TEM | 0/10 (0.0) | 0 | 0 | 0 | 0 |
| SHV | 0/10 (0.0) | 0 | 0 | 0 | 0 |
| QnrA | 0/10 (0.0) | 0 | 0 | 0 | 0 |
| QnrB | 0/10 (0.0) | 0 | 0 | 0 | 0 |
| QnrS | 8/10 (80) | 4 | 2 | 1 | 1 |
| aac(6′)-Ib-cr | 0/10 (0.0) | 0 | 0 | 0 | 0 |
| Total | 10/10(100) | 5 | 3 | 1 | 1 |
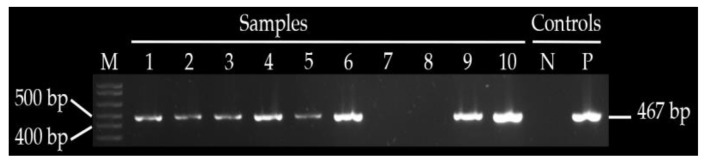
Figure 3.
Shows amplified qnrS gene in sample 1–6, 9 and 10, M—1 kb ladder, NC—negative control, PC—positive control.
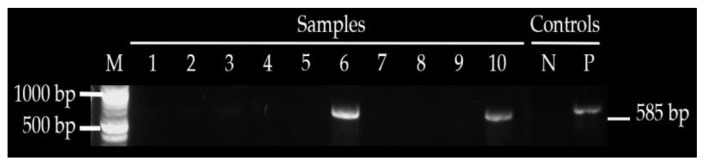
Figure 4.
Shows amplified CTX—M in sample 6 and 10, M—1 kb ladder, NC—negative control and PC—positive control.
3. Discussion
In this study, isolation frequency of E. coli was more than half (55.2%), and most of the isolates (69.3%) were resistant to multiple drugs (MDR), some up to seven classes of antibiotics. The most frequent resistant combination was from tetracycline, ampicillin, sulfamethoxazole-trimethoprim and ciprofloxacin, which unfortunately are the most commonly used antibiotics in both humans and animals [10,11]. We found no significant differences in MDR E. coli between broilers and spent layers, even though broilers are raised in a relatively short period (four to six weeks) compared with spent layers (up to two years). This could be explained by the fact that antibiotics are used more intensely for growth promotion and prophylaxis in raising broilers than in spent layers [12]. For both types of chicken, cloaca had higher isolation frequency of MDR E. coli (25.2% to 32%) than in meat samples (20.4% to 22.4%), a trend that has also been observed in duck fecal samples [13], which indicates the epidemiological significance of chicken droppings in contaminating the environment, and acting as a potential driver of AMR spread [11,14]. We found significant difference in antimicrobial resistance rates between poultry slabs, indicating possible contribution of the slaughtering environment in contaminating poultry meat with MDR bacteria, which has been cited to be a factor [15,16,17]. However, in this study we did not investigate the sources of contamination.
For individual antibiotics, highest resistance was to tetracycline (91.9%), followed by sulfamethoxazole-trimethoprim (80.5%), ampicillin (70.9%) and ciprofloxacin (40.2%), a pattern that has also been reported previously [7,13,18]. These antibiotics are relatively cheap and are easily obtained over the counter [19,20,21,22], and often farmers do not follow withdrawal period [4,5].
On the other hand, cefotaxime, and especially imipenem, which are not commonly used [23], were less resistant. ESBL-producing E. coli were detected in 10/212 (4.7%) and quinolone resistance genes in 80% of them, supporting observations of several studies that have found a strong association between qnr-positive and ESBL-positive isolates [24,25,26,27]. Among the ESBL-producing, we only found CTX-M and not TEM or SHV, and unlike a study done in Niger that showed several qnr genes (qnrA, qnrB and qnrS) [28], we only found qnrS, and did not find qnrA, qnrB or aac(6′)-lb-cr in any of the ESBL isolates. Collectively, these findings suggest significant geographical differences in the occurrence of ESBL and quinolone resistance genes, supporting the need for their continuous surveillance in different settings.
The finding of quinolone resistance, which can rapidly spread along the food chain and in other ecosystems through plasmids [29,30,31], is significant, requiring very strict regulation in their use and, if possible, banning their use in animal food production. Tanzania has a number of acts and policies that are intended to control the quality of livestock production. Unfortunately, the Meat Industry Act of 2006 that gives a legal backing to support meat inspection to ensure quality does not explicitly address issues of drug residues in meat and meat products. Equally, the National Livestock Policy of 2006 and the National Agriculture Policy of 2013 do not address issues of antimicrobial use (AMU) and antimicrobial resistance (AMR) in livestock and agriculture sectors, respectively.
In all five slabs, we found improper handling of chicken carcasses and unregulated waste disposal from slaughter poultry slabs including blood, feces and wastewater disposed into municipal drains without either monitoring or treatment. Unfortunately, the National Environmental Policy of 1997 that is supposed to ensure food security through the promotion of production systems that are environmentally sound does not address the issues of environmental contamination with antimicrobials. Likewise, the Animal Diseases Act of 2003, which makes provisions for monitoring of production of animal products for disposal of animal carcasses, is silent on issues related to antimicrobials.
We strongly suggest the existing acts and policies, some of which are more than ten years old, be critically reviewed by stakeholders from human health, veterinary and environment sectors in order to curb AMU and AMR in livestock production and for protection of humans and the environment. The revised acts and policies should be reinforced through legislation. We also advocate for judicious use of antimicrobials in poultry, through improved hygiene, vaccinations and provision of extensive farmers’ education.
4. Materials and Methods
4.1. Study Area
The study was conducted in Dar es Salaam, the commercial city of Tanzania, which has a population of 4,364,541 people [3], with the highest production and consumption of chicken meat and eggs in Tanzania. The study involved five large poultry slabs in four Districts (Ilala, Ubungo, Temeke and Kinondoni). Approximately 20,000 chicken are slaughtered daily in these five poultry slaughter slabs, which provides about 80% of the chicken consumed in Dar es Salaam.
4.2. Study Design
This was a cross-sectional study conducted between January and June 2020 in four districts, which have the largest poultry slabs in Dar es Salaam. The slabs were Manzese, and Shekilango in the Ubungo district, Kisutu in the Ilala district, Mtambani in the Kinondoni district and Stereo in the Temeke district. In this study we targeted broilers and spent layers because they are raised intensively in overcrowded environments, and use of antimicrobials for prophylaxis, growth promotion and in management of infections is very high. Other types of poultry such as indigenous chickens were excluded from the study.
4.3. Sampling Technique
Using the purposive sampling technique, we selected 96 broilers and 96 spent layers, making a total of 192 chickens in all the five poultry slabs. Two samples (i.e., cloaca and meat swab) were collected from each chicken, making a total of 384 samples. Cloaca swabs were collected before chickens were slaughtered (at the entry point), while chicken meat swabs were collected after chickens were slaughtered (at the poultry slabs).
4.4. Specimen Collection
Chicken meat and cloaca swabs were collected aseptically using sterile cotton swabs and placed into a sterile tube containing 5 mL of Cary Blair transport medium (Oxoid, Basingstoke, UK). The collected samples were transported in a cool box at 2 to 8 °C containing a thermometer and were processed within 2 h of collection in the Microbiology Teaching Laboratory of the Muhimbili University and Allied Sciences (MUHAS).
4.5. Isolation and Identification of Enterobacteria
In the laboratory, swabs were inoculated onto the MacConkey agar (Oxoid, Basingstoke, UK) without antibiotics and incubated aerobically at 37 °C for 24 h. Identification of E. coli was done using colonial morphology, lactose fermentation and Gram stain. Lactose fermenters were subjected to conventional phenotypical identification using a set of biochemical tests including triple sugar iron agar (TSI), sulphur indole motility (SIM) agar and citrate utilization test. Confirmation was done using API 20E identification system for Enterobacteriaceae according to the instructions of the manufacturer (BioMérieux, Marcy-Etoile, France).
4.6. Screening and Confirmation of ESBL Production
Confirmed E. coli isolates were inoculated onto MacConkey agar containing 2 mg/L cefotaxime for preliminary screening of ESBL production. ESBL producers were confirmed using a combination disk method of cefotaxime 30 µg alone, combination with clavulanic acid (10 µg) and ceftazidime 30 µg alone and combination with clavulanic acid 10 µg. The difference of inhibition zone of more than or equal to 5 mm was confirmed as ESBL-positive [32]. Klebsiella pneumoniae ATCC 700603 was used as a positive control and E. coli ATCC 25922 as a negative strain.
4.7. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was done using the Kirby-Bauer disc diffusion method on Mueller Hinton Agar (Oxoid, Basingstoke, UK) based on CLSI 2018 guidelines [32]. Seven antibiotics from different classes were used, which included ampicillin (10 µg), tetracycline (30 µg), gentamycin (10 µg), ciprofloxacin (5 µg), imipenem (10 µg), sulfamethoxazole-trimethoprim (1.25/23.5 µg) and cefotaxime 30 µg [32].
Colonies of lactose fermenters identified as E. coli were emulsified into sterile saline to achieve turbidity equivalent to 0.5 McFarland standard, which is equivalent to 108 cfu/mL [32]. Suspensions were spread onto Muller Hinton agar (MHA) using sterile cotton swabs and incubated aerobically at 37 °C for 16 to 18 h. The inhibition zone of each antibiotic was measured after 16 to 18 h of incubation, and results were interpreted according to the 2018 CLSI guidelines [32]. E. coli strain ATCC 29522 was used as a control strain. A strain was referred to be multidrug resistant (MDR) if it exhibited resistance to at least three different antibiotic classes [32].
4.8. Polymerase Chain Reaction (PCR)
4.8.1. DNA Extraction
ESBL-producing E. coli isolates were inoculated on nutrient agar and incubated aerobically at 37 °C for 24 h. DNA was extracted by boiling in a water bath at 100 °C for 10 min, followed by centrifugation at 1500 rpm for 3 min. The supernatant containing DNA was transferred into sterile Eppendorf PCR tube (Eppendorf AG, Hamburg, Germany), and centrifugation and separation of supernatant were repeated three times. The concentration of DNA was determined by Nano drop spectrophotometer (Biochrom LTD, Cambridge, England) at 260/280 and 260/230 wavelength. DNA was stored at −20 °C, before being used for detection of ESBL genes (CTX—M, TEM and SHV) and PMQR genes (qnrA, qnrB, qnrS and aac(6′)-Ib-cr).
The Dream Tag DNA polymerase kit (Sigma-Aldrich Chemie GmbH, Taufkirchen, German) was used in detection of resistance genes. Total PCR reaction volumes were 25 µL, consisting of 10× dream Tag Buffer 5 µL, dNTP 2 mM 5 µL, forward and reverse primers were 1 µL each, DNA extract was 2 µL, Dream Tag DNA Polymerase (1.25 U) 1 µL and nuclease free water 10 µL. The primers used in amplification of respective E. coli resistance genes are listed in Table 6.
Table 6.
List of primers used.
| Gene | Primer Set | Amplicon Size | Reference |
|---|---|---|---|
| CTX-M | F: SCSATGTGCAGYACCAGTAA R: ACCAGAAYVAGCGGBGC |
585 bp | [33,34] |
| QnrA | F: TCAGCAAGAGGATTTCTCA R: GGCAGCACTATTACTCCCA |
627 bp | [35] |
| QnrB | F: GGMATHGAAATTCGCCACTG R: TTTGCYGYYCGCCAGTCGAA |
264 bp | [35] |
| QnrS | F: ATGGAAACCTACAATCATAC R: AAAAACACCTCGACTTAAGT |
467 bp | [35] |
| aac(6′)-Ib-cr | F: TTGCGATGCTCTATGAGTGGCTA R: CTCGAATGCCTGGCGTGTTT |
482 bp | [34,36] |
| TEM | F: ATGAGTATTCAACATTTCCG R: CTGACAGTTACCAATGCTTA |
867 bp | [37] |
| SHV | F: GGTTATGCGTTATATTCGCC R: TTAGCGTTGCCAGTGCTC |
867 bp | [37] |
4.8.2. Molecular Detection of CTX-M Genes
All ESBL-producing E. coli isolates were screened for the CTX-M gene using Uniplex PCR-based technique [33]. The process involved initial denaturation at 94 °C for 10 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 60 s and final extension at 72 °C for 5 min [34].
4.8.3. Detection of TEM and SHV Genes
Extended spectrum beta lactamase TEM and SHV genes were screened by uniplex PCR-based assay [37] with the following amplification conditions: initial denaturation at 96 °C for 5 min, followed by 35 cycles of denaturation at 96 °C for 1 min, annealing at 58 °C (TEM) and at 60 °C (SHV) for 1 min, extension at 72 °C for 1 min and final extension at 72 °C for 10 min [37].
4.8.4. Detection of PMQR Genes (qnrA, qnrB and qnrS)
PMQR-resistant genes (qnrA, qnrB and qnrS) were amplified and detected using multiplex PCR assay [14]. The process involved initial denaturation at 94 °C for 5 min, followed by 32 cycles of denaturation at 94 °C for 45 s, annealing at 53 °C for 1 min, extension at 72 °C for 1 min and final extension at 72 °C for 10 min [14].
4.8.5. Detection of aac(6′)-lb-cr Gene
aac(6′)–lb-cr genes were screened by uniplex PCR-based assay [35] using the following amplification conditions: initial denaturation at 94 °C for 5 min, followed by 34 cycles of denaturation at 94 °C for 45 s, annealing at 55 °C for 45 s, extension at 72 °C for 45 s and final extension at 72 °C for 10 min [36].
4.9. Data Analysis
The data were entered into Microsoft Excel; proportions were analyzed by Chi-square test. A paired t-test assuming unequal variance was used for comparing overall prevalence and comparing resistance rate among tested antibiotics in SPSS version 16 software. A p-value (<0.05) was considered to be statistically significant.
5. Conclusions
The high levels of resistance to antibiotics seen in this study has several implications: (i) there is over use of antibiotics in poultry production; (ii) these agents are ineffective in either prophylaxis or treatement of infections in poultry farming; (iii) there is a serious public health threat (through antimicrobial residues in meat); and (iv) increased possibility of development and spread of antimicrobial resistance in the environment. Therefore, urgent measures are required to reduce the use of antibiotics in poultry production at the farm level and to improve hygiene practices at poultry slaughter. In addition, the present acts and policies governing animal food production should be revised to provide legislation to enforce judicious use of antimicrobial agents.
Acknowledgments
We would like to thank the Dar es Salaam regional and district authorities including the Regional Administrative Secretary, District Executive Directors, and District Veterinary Officers for their permission to conduct the study. We acknowledge technical support provided by staff of the Departments Microbiology at Muhimbili University of Health and Allied Sciences and Sokoine University of Agriculture.
Author Contributions
Designed the study, data collection, data analysis, interpretation of data, generated the first draft and wrote the Manuscript, F.X.M.; Reviewed the first draft and final draft of manuscript, A.P.M.; Designed the study, outlines of manuscript, reviewed the first and final draft of manuscript and provided the relevant editing, M.I.M.; Designed outline for data analysis, reviewed the first draft to final work and provided the relevant editing, A.S.H. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by World Bank supported Eastern and Southern Africa Higher Education Centers of Excellence Project Grant P151847.
Institutional Review Board Statement
The permit to conduct this study was given by the Sokoine University of Agriculture Ethical Review Committee with reference number: SUA/DPRTC/R/186 VOL.III, dated 2/12/2019.
Conflicts of Interest
The authors declare to have no conflicts of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.United Republic of Tanzania Ministry of Livestock and Fisheries and International Livestock Research Institute . Tanzania Livestock Master Plan. Ministry of Livestock and Fish Development; Dar es Salaam, Tanzania: 2018. pp. 1–104. [Google Scholar]
- 2.Hounmanou Y.M.G., Mdegela R.H. Current situation for antimicrobial use, antimicrobial resistance and antimicrobial residues in the food and agriculture sectors in Tanzania: A review. Tanz. Vet. J. 2017;35:58–62. [Google Scholar]
- 3.Tanzania National Bureau of Statistics (NBS) Population and Housing Census, Population Distribution by Administrative Areas. National Bureau of Statistics, Ministry of Finance; Dar es Salaam, Tanzania: 2012. p. 56. [Google Scholar]
- 4.Kimera Z.I., Mshana S.E., Rweyemamu M.M., Mboera L.E.G., Matee M.I.N. Antimicrobial use and resistance in food-producing animals and the environment: An African perspective. Antimicrob. Resist. Infect. Control. 2020;9:37. doi: 10.1186/s13756-020-0697-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ezekiel P.M., Francis S., Martin E.K., Joram J.B. Antimicrobial use in the poultry industry in Dar-es-Salaam, Tanzania and public health implications. Am. J. Resear. Commun. 2014;2:51–63. [Google Scholar]
- 6.Rugumisa B.T., Call D.R., Mwanyika G.O., Mrutu R.I., Luanda C.M., Lyimo B.M., Subbiah M., Buza J.J. Prevalence of Antibiotic-Resistant Fecal Escherichia coli Isolates from Penned Broiler and Scavenging Local Chickens in Arusha, Tanzania. J. Food Prot. 2016;79:1424–1429. doi: 10.4315/0362-028X.JFP-15-584. [DOI] [PubMed] [Google Scholar]
- 7.Founou L.L., Amoako D.G., Founou R.C., Essack S.Y. Antibiotic Resistance in Food Animals in Africa: A Systematic Review and Meta-Analysis. Microb. Drug Resist. 2018;24:648–665. doi: 10.1089/mdr.2017.0383. [DOI] [PubMed] [Google Scholar]
- 8.Haritova A., Urumova V., Lutckanov M., Petrov V., Lashev L. Pharmacokinetic-pharmacodynamic indices of enrofloxacin in Escherichia coli O78/H12 infected chickens. Food Chem. Toxicol. 2011;49:1530–1536. doi: 10.1016/j.fct.2011.03.044. [DOI] [PubMed] [Google Scholar]
- 9.Shobrak M.Y., Abo-Amer A.E. Role of wild birds as carriers of multi-drug resistant Escherichia coli and Escherichia vulneris. Braz. J. Microbiol. 2014;45:1199–1209. doi: 10.1590/S1517-83822014000400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sindato C., Mboera L.E.G., Katale B.Z., Frumence G., Kimera S., Clark T.G., Legido-Quigley H., Mshana S.E., Rweyemamu M.M., Matee M. Knowledge, attitudes and practices regarding antimicrobial use and resistance among communities of Ilala, Kilosa and Kibaha districts of Tanzania. Antimicrob. Resist. Infect. Control. 2020;9:194. doi: 10.1186/s13756-020-00862-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimera Z.I., Frumence G., Mboera L.E.G., Rweyemamu M., Mshana S.E., Matee M.I.N. Assessment of Drivers of Antimicrobial Use and Resistance in Poultry and Domestic Pig Farming in the Msimbazi River Basin in Tanzania. Antibiotics. 2020;9:838. doi: 10.3390/antibiotics9120838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agunos A., Léger D.F., Carson C.A., Gow S.P., Bosman A., Irwin R.J., Reid-Smith R.J. Antimicrobial use surveillance in broiler chicken flocks in Canada, 2013–2015. PLoS ONE. 2017;12:e0179384. doi: 10.1371/journal.pone.0179384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kissinga D.H., Mwombeki F., Said K., Katakweba A.A.S., Nonga H.E., Muhairwa A.P. Antibiotic susceptibilities of indicator bacteria Escherichia coli and Enterococci spp. isolated from ducks in Morogoro Municipality, Tanzania. BMC Res. Notes. 2018;11:87. doi: 10.1186/s13104-018-3201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H., Duan G., Zhu J., Zhang W., Xi Y., Fan Q. Prevalence and characterization of plasmid-mediated quinolone resistance and mutations in the gyrase and topoisomerase IV genes among Shigella isolates from Henan, China, between 2001 and 2008. Int. J. Antimicrob. Agents. 2013;42:173–177. doi: 10.1016/j.ijantimicag.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 15.Chishimba K., Hang’ombe B.M., Muzandu K., Mshana S.E., Matee M.I., Nakajima C., Suzuki Y. Detection of Extended-Spectrum Beta-Lactamase-Producing Escherichia coli in Market-Ready Chickens in Zambia. Int. J. Microbiol. 2016;2016 doi: 10.1155/2016/5275724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis G.S., Waits K., Nordstrom L., Grande H., Weaver B., Papp K., Horwinski J., Koch B., Hungate B.A., Liu C.M., et al. Antibiotic-resistant Escherichia coli from retail poultry meat with different antibiotic use claims. BMC Microbiol. 2018;18:174–181. doi: 10.1186/s12866-018-1322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saud B., Paudel G., Khichaju S., Bajracharya D., Dhungana G., Awasthi M.S., Shrestha V. Multidrug-Resistant Bacteria from Raw Meat of Buffalo and Chicken, Nepal. Vet. Med. Inter. 2019;2019 doi: 10.1155/2019/7960268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamis Z., Tuntufye H., Shahada F. Antimicrobial Resistance Phenotypes of Escherichia coli Isolated from Tropical Free-Range Chickens. Int. J. Sci. Res. 2014;3:34–37. [Google Scholar]
- 19.Lupindu A.M., Dalsgaard A., Msoffe P.L., Ngowi H.A., Mtambo M.M., Olsen J.E. Transmission of antibiotic-resistant Escherichia coli between cattle, humans and the environment in peri-urban livestock keeping communities in Morogoro, Tanzania. Prev. Vet. Med. 2014;118:477–482. doi: 10.1016/j.prevetmed.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Caudell M.A., Quinlan M.B., Subbiah M., Call D.R., Roulette C.J., Roulette J.W., Roth A., Matthews L., Quinlan R.J. Antimicrobial Use and Veterinary Care among Agro-Pastoralists in Northern Tanzania. PLoS ONE. 2017;12:e0170328. doi: 10.1371/journal.pone.0170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katakweba A.A.S., Mtambo M.M.A., Olsen J.E., Muhairwa A.P. Awareness of human health risks associated with the use of antimicrobials among livestock keepers and factors that contribute to selection of antibiotic resistance bacteria within livestock in Tanzania. Livestock Rural Res. Dev. 2012;24:1–14. [Google Scholar]
- 22.Graham D.W., Bergeron G., Bourassa M.W., Dickson J., Gomes F., Howe A., Kahn L.H., Morley P.S., Scott H.M., Simjee S., et al. Complexities in understanding antimicrobial resistance across domesticated animal, human, and environmental systems. Ann. N. Y. Acad. Sci. 2019;1441:17–30. doi: 10.1111/nyas.14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdel Rahman M., Roshdy H., Samir A.H., Hamed E.A. Antibiotic resistance and extended-spectrum β-lactamase in Escherichia coli isolates from imported 1-day-old chicks, ducklings, and turkey poultry. Vet. World. 2020;13:1037–1044. doi: 10.14202/vetworld.2020.1037-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nordmann P., Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 2005;56:463–469. doi: 10.1093/jac/dki245. [DOI] [PubMed] [Google Scholar]
- 25.Jacoby G.A., Strahilevitz J., Hooper D.C. Plasmid-mediated quinolone resistance. Microbiol. Spectr. 2014:2. doi: 10.1128/microbiolspec.PLAS-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacoby G.A., Walsh K.E., Mills D.M., Walker V.J., Oh H., Robicsek A., Hooper D.C. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 2006;50:1178–1182. doi: 10.1128/AAC.50.4.1178-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robicsek A., Jacoby G.A., Hooper D.C. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 2006;6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 28.Moumouni A., Diagbouga S., Nadembèga C., Metuor Dabire A., Salah F., Obiri-Yeboah D., Soubéiga S.T., Ouattara A.K., Zohoncon T., Djigma F. Quinolone Resistance (qnr) genes in fecal carriage of extended Spectrum beta-lactamases producing Enterobacteria isolated from children in Niger. Curr. Res. Microbiol. Biotechnol. 2017;5:953–957. [Google Scholar]
- 29.Salah F.D., Soubeiga S.T., Ouattara A.K., Sadji A.Y., Metuor-Dabire A., Obiri-Yeboah D., Banla-Kere A., Karou S., Simpore J. Distribution of quinolone resistance gene (qnr) in ESBL-producing Escherichia coli and Klebsiella spp. in Lomé, Togo. Antimicrob. Resist. Infect. Control. 2019;8:104. doi: 10.1186/s13756-019-0552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sidjabat H.E., Townsend K.M., Lorentzen M., Gobius K.S., Fegan N., Chin J.J., Bettelheim K.A., Hanson N.D., Bensink J.C., Trott D.J. Emergence and spread of two distinct clonal groups of multidrug-resistant Escherichia coli in a veterinary teaching hospital in Australia. J. Med. Microbiol. 2006;55:1125–1134. doi: 10.1099/jmm.0.46598-0. [DOI] [PubMed] [Google Scholar]
- 31.Ben Sallem R., Ben Slama K., Rojo-Bezares B., Porres-Osante N., Jouini A., Klibi N., Boudabous A., Sáenz Y., Torres C. IncI1 plasmids carrying bla (CTX-M-1) or bla (CMY-2) genes in Escherichia coli from healthy humans and animals in Tunisia. Microb. Drug Resist. 2014;20:495–500. doi: 10.1089/mdr.2013.0224. [DOI] [PubMed] [Google Scholar]
- 32.CLSI . In: Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. Wayne P.A., editor. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. CLSI Supplement M100. [Google Scholar]
- 33.Valat C., Forest K., Billet M., Polizzi C., Saras E., Madec J.Y., Haenni M. Absence of co-localization between patho var-associated virulence factors and extended-spectrum β-lactamase (bla CTX-M) genes on a single plasmid. Veter. Microbiol. 2016;192:163–166. doi: 10.1016/j.vetmic.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Reich F., Atanassova V., Klein G. Extended-Spectrum ß-Lactamase and AmpC-Producing Enterobacteria in Healthy Broiler Chickens, Germany. Emerg. Infect. Dis. 2013;9:1253–1259. doi: 10.3201/eid1908.120879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moawad A.A., Hotzel H., Awad O., Tomaso H., Neubauer H., Hafez H.M., El-Adawy H. Occurrence of Salmonella enterica and Escherichia coli in raw chicken and beef meat in northern Egypt and dissemination of their antibiotic resistance markers. Gut Pathog. 2017;9:57–70. doi: 10.1186/s13099-017-0206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park C.H., Robicsek A., Jacoby G.A., Sahm D., Hooper D.C. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 2006;50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim K.T., Yasin R., Yeo C.C., Puthucheary S., Thong K.-L. Characterization of Multidrug Resistant ESBL-Producing Escherichia coli Isolates from Hospitals in Malaysia. BioMed Res. Int. 2009;2009 doi: 10.1155/2009/165637. [DOI] [PMC free article] [PubMed] [Google Scholar]