Summary
Sphingolipid biosynthesis occurs in both the endoplasmic reticulum (ER) and the Golgi apparatus. Ceramide synthesized in the ER is transported to the Golgi and incorporated into complex sphingolipids. Here, we present a step-by-step protocol to analyze sphingolipid metabolism in budding yeast. Ceramide and inositolphosphorylceramide (IPC) are classes of sphingolipids present in yeast and are metabolically labeled with radioactive precursors. This protocol for metabolic labeling can be used to investigate ceramide transport in an in vivo environment.
For complete details on the use and execution of this protocol, please refer to Ikeda et al. (2020).
Subject areas: Cell Biology, Metabolism, Model Organisms, Molecular Biology
Graphical abstract

Highlights
-
•
A step-by-step procedure for analyzing sphingolipid metabolism in budding yeast
-
•
Quantification and statistical analysis of sphingolipid metabolism assay data
-
•
The protocol can be applied to other cells synthesizing inositol-containing sphingolipids
Sphingolipid biosynthesis occurs in both the endoplasmic reticulum (ER) and the Golgi apparatus. Ceramide synthesized in the ER is transported to the Golgi and incorporated into complex sphingolipids. Here, we present a step-by-step protocol to analyze sphingolipid metabolism in budding yeast. Ceramide and inositolphosphorylceramide (IPC) are classes of sphingolipids present in yeast and are metabolically labeled with radioactive precursors. This protocol for metabolic labeling can be used to investigate ceramide transport in an in vivo environment.
Before you begin
This protocol was used in a recent publication (Ikeda et al., 2020) to study the roles of tricalbins in the transport of ceramide in living Saccharomyces cerevisiae cells. Ceramide is delivered to the luminal side of the Golgi apparatus and converted into IPC by IPC synthase catalyzing the transfer of phosphoinositol from phosphatidylinositol to ceramide. Therefore, synthesis of IPC depends on the enzymatic activity of IPC synthase and the production of phosphatidylinositol and ceramide precursors, otherwise it depends on ceramide transport. Here, we provide a detailed protocol for measuring IPC synthesis, IPC synthase activity and ceramide synthesis.
Preparation of medium
Timing: 4 h
Prior to the experiment, prepare the medium and buffers to be used for cell culture and metabolic labeling.
-
1.
Prepare the Trace Elements (1000×), Sol. A (50×), Sol. B (50×) and Vitamins with inositol (Vitamins + inositol) (100×) or without (Vitamins – inositol) (100×) to be used for preparation of semi-synthetic defined (semi-SD) medium and SD medium without inositol (SD – inositol medium).
-
2.
Prepare the semi-SD medium to be used for cell culture.
-
3.
Prepare the SD – inositol medium to be used for metabolic labeling with Myo-[2-3H(N)]-inositol ([3H]myo-inositol).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Glucose | Nacalai Tesque | Cat#16806-54 |
| Uracil | Nacalai Tesque | Cat#35824-82 |
| Adenine | Nacalai Tesque | Cat#06398-82 |
| L-Histidine | Nacalai Tesque | Cat#18116-92 |
| L-Leucine | Nacalai Tesque | Cat#20327-62 |
| L-Lysine | MilliporeSigma | Cat#L5626 |
| L-Methionine | Nacalai Tesque | Cat#21719-02 |
| L-Tryptophan | Nacalai Tesque | Cat#35607-32 |
| Difco™ Yeast Extract | Thermo Fisher Scientific | Cat#210934 |
| Boric acid | Nacalai Tesque | Cat#05215-05 |
| Copper (II) sulfate pentahydrate | Nacalai Tesque | Cat#09605-04 |
| Potassium iodide | MilliporeSigma | Cat#P8256 |
| Iron (III) chloride hexahydrate | MilliporeSigma | Cat#F2877 |
| Zinc chloride | Nacalai Tesque | Cat#36920-24 |
| Potassium dihydrogen phosphate | Kanto Chemical | Cat#32379-00 |
| Dipotassium hydrogen phosphate | Nacalai Tesque | Cat#23727-95 |
| Sodium chloride | Nacalai Tesque | Cat#31320-05 |
| Ammonium sulfate | Kanto Chemical | Cat#01322-00 |
| Magnesium chloride hexahydrate | MilliporeSigma | Cat#M2670 |
| Calcium chloride dihydrate | Wako Chemical | Cat#031-00435 |
| d-Biotin | MilliporeSigma | Cat#B4501 |
| Calcium pantothenate | MilliporeSigma | Cat#C8731 |
| Nicotinic acid | MilliporeSigma | Cat#N4126 |
| 4-Aminobenzoic acid | MilliporeSigma | Cat#A9878 |
| Pyridoxin | MilliporeSigma | Cat#P9755 |
| Thiamine | MilliporeSigma | Cat#T4625 |
| Riboflavin | MilliporeSigma | Cat#R4500 |
| Folic acid | MilliporeSigma | Cat#F7876 |
| Myo-inositol | MilliporeSigma | Cat#I5125 |
| C2-Ceramide (N-acetoyl-D-erythro-sphingosine) | Merck KGaA | Cat#860502P |
| Myo-[2-3H(N)]-inositol (250 μCi) | PerkinElmer | Cat#NET114250UC |
| D-Erythro-[4,5-3H]-dihydrosphingosine | American Radiolabeled Chemicals | Cat#ART0460 |
| Sodium fluoride | MilliporeSigma | Cat#S1504 |
| Sodium azide | MilliporeSigma | Cat#S2002 |
| Chloroform | Wako Chemicals | Cat#038-02601 |
| Methanol | Nacalai Tesque | Cat#21915-93 |
| 1-Butanol | Nacalai Tesque | Cat#060-16 |
| Distilled water | N/A | N/A |
| Deionized water | N/A | N/A |
| Potassium chloride | Nacalai Tesque | Cat#28514-75 |
| Ammonium hydroxide | MilliporeSigma | Cat#221228 |
| Acetic acid | MilliporeSigma | Cat#01-0280-5 |
| Sodium hydroxide (Optional) | Wako Chemicals | Cat#197-02125 |
| Acetic acid (Optional) | MilliporeSigma | Cat#01-0280-5 |
| Clear-sol I | Nacalai Tesque | Cat#091-35 |
| Experimental models: Organisms/strains | ||
| S. cerevisiae: strain FKY2928 Mat α; ura3-52 leu2-3,112 his3-11 trp1-1 lys2-810 bar1-1 | Ikeda et al., 2020 | N/A |
| S. cerevisiae: strain FKY2927 Mat a; ura3-52 leu2-3,112 his3-11 trp1-1 lys2-810 bar1-1 tcb1Δ::TRP1 tcb2Δ::HIS3 tcb3Δ::LEU2 | Ikeda et al., 2020 | N/A |
| S. cerevisiae: strain FKY2960 Mat a; ura3-52 leu2-3,112 his3-11 lys2-810 bar1-1 sec12-4(ts) | Ikeda et al., 2020 | N/A |
| S. cerevisiae: strain FKY4892 Mat α; ura3-52 leu2-3,112 his3-11 trp1-1 lys2-810 ade2-101 dga1Δ::KanMX lro1Δ::KanMX | Ikeda et al., 2020 | N/A |
| Software and algorithms | ||
| Image reader for FLA-7000 | Fujifilm | N/A |
| Multi Gauge | Fujifilm | N/A |
| Other | ||
| Incubator (SLI-1200) | EYELA | Cat#197960 |
| Shaker (MMS-3020) | EYELA | Cat#267870 |
| Cell density meter (Ultrospec 2100 pro) | GE Healthcare | Cat#80-2112-21 |
| Water bath shaker (NTS-4000B) | EYELA | Cat#211890 |
| Vortex | N/A | N/A |
| 50 mL Conical sterile polypropylene centrifuge tubes | Thermo Fisher Scientific | Cat#339652 |
| 1.5 mL Snap cap low retention microcentrifuge tubes | Thermo Fisher Scientific | Cat#3448 |
| Pipette tip (1000 μL) | AS ONE | Cat#2-3976-05 |
| Pipette tip (2–200 μL) | AS ONE | Cat# 2-3976-03 |
| Pipette tip 1–200uL for gel loading | AS ONE | Cat# 62-7022-46 |
| Low-speed centrifuge | TOMY DIGITAL BIOLOGY | LC-200 |
| Desktop micro-cooling centrifuge | KUBOTA Corporation | 5320 |
| Glass beads (ϕ0.5 mm, treated with low alkali, and sold as in a dried state) | Yasui Kikai Corporation | YGBLA05 |
| Ultrasonic cleaner | AS ONE | US-2R |
| Dry block bath | EYELA | MG-2200 |
| Pressured gas blowing concentrator | EYELA | MGS-2200 |
| Super polyethylene vial 20 mL | PerkinElmer | Cat#6008117 |
| Liquid scintillation counter | Aloka | LSC-5100 |
| TLC silica gel 60 | Merck Millipore | Cat#1.05553.0001 |
| TLC development tank | N/A | N/A |
| Fujifilm BAS IP TR 2040 (tritium-sensitive imaging plate) | Cytiva | Cat#28956481 |
| Fujifilm BAS Cassette 2040 | N/A | N/A |
| Typhoon FLA 7000 | GE Healthcare | N/A |
Materials and equipment
Semi-SD medium, SD – inositol medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Glucose | 2% w/v | 20 g |
| Trace Elements (1000×) - See Preparations Below | 0.1% v/v | 1 mL |
| Sol. A (50×) - See Preparations Below | 2% v/v | 20 mL |
| Sol. B (50×) - See Preparations Below | 2% v/v | 20 mL |
| ∗1 Difco Yeast Extract | 2 g/L | 2 g |
| Uracil | 80 mg/L | 80 mg |
| Adenine | 80 mg/L | 80 mg |
| L-Histidine | 80 mg/L | 80 mg |
| L-Leucine | 80 mg/L | 80 mg |
| L-Lysine | 80 mg/L | 80 mg |
| L-Methionine | 80 mg/L | 80 mg |
| ∗2 Tryptophan (8 mg/mL) | 80 mg/L | 10 mL |
| ∗3 Vitamins + inositol or Vitamins – inositol | 1% v/v | 10 mL |
| Deionized Water | N/A | Up to 1 L |
| Total | N/A | 1 L |
∗1Add Yeast Extract to make semi-SD medium but not SD – inositol medium.
∗2After autoclaving, add 10 mL of Tryptophan solution (8 mg/mL) sterilized by filtration.
∗3After autoclaving, add 10 mL of Vitamins + inositol solution (100×) sterilized by filtration to make semi-SD medium. After autoclaving, add 10 mL of Vitamins – inositol solution (100×) sterilized by filtration to make SD – inositol medium.
Note: Solubilize the mixture completely by a stir plate and a magnetic bar and sterilize by autoclaving. After added filtrated solutions, store at 15–25°C for up to 1 month.
Trace Elements (1000×)
| Reagent | Final concentration | Amount |
|---|---|---|
| Boric Acid | 500 mg/L | 500 mg |
| Copper (II) Sulfate Pentahydrate | 40 mg/L | 40 mg |
| Potassium Iodide | 100 mg/L | 100 mg |
| Iron (III) Chloride Hexahydrate | 200 mg/L | 200 mg |
| Zinc Chloride | 340 mg/L | 340 mg |
| Deionized Water | N/A | Up to 1 L |
| Total | N/A | 1 L |
Note: Solubilize the mixture completely by a stir plate and a magnetic bar and sterilize by filtration. Store at –20°C indefinitely.
Sol. A (50×)
| Reagent | Final concentration | Amount |
|---|---|---|
| Potassium Dihydrogen Phosphate | 43.756 g/L | 43.756 g |
| Dipotassium Hydrogen Phosphate | 6.25 g/L | 6.25 g |
| Sodium Chloride | 5 g/L | 5 g |
| Ammonium Sulfate | 250 g/L | 250 g |
| Deionized Water | N/A | Up to 1 L |
| Total | N/A | 1 L |
Note: Solubilize the mixture completely by a stir plate and a magnetic bar and sterilize by autoclaving. Store at 15–25°C indefinitely.
Sol. B (50×)
| Reagent | Final concentration | Amount |
|---|---|---|
| Magnesium Chloride Hexahydrate | 25 g/L | 25 g |
| Calcium Chloride Dihydrate | 5 g/L | 5 g |
| Deionized Water | N/A | Up to 1 L |
| Total | N/A | 1 L |
Note: Solubilize the mixture completely by a stir plate and a magnetic bar and sterilize by autoclaving. Store at 15–25°C indefinitely.
Vitamins + inositol (100×), vitamins – inositol (100×)
| Reagent | Final concentration | Amount |
|---|---|---|
| d-Biotin | 2 mg/L | 2 mg |
| Calcium Pantothenate | 200 mg/L | 200 mg |
| Nicotinic Acid | 40 mg/L | 40 mg |
| 4-Aminobenzoic Acid | 20 mg/L | 20 mg |
| Pyridoxin | 40 mg/L | 40 mg |
| Thiamine | 40 mg/L | 40 mg |
| Riboflavin | 20 mg/L | 20 mg |
| Folic Acid | 0.2 mg/L | 0.2 mg |
| ∗4 Myo-inositol | 1000 mg/L | 1000 mg |
| Deionized Water | N/A | Up to 1 L |
| Total | N/A | 1 L |
∗4 Do not add this to make SD – inositol medium.
Note: Solubilize the mixture completely by a stir plate and a magnetic bar and sterilize by filtration. Store at –20°C indefinitely.
10 mM C2-ceramide
| Reagent | Final concentration | Amount |
|---|---|---|
| C2-ceramide | 10 mM | 5 mg |
| Ethanol (Not specified) | N/A | 1.5 mL |
| Total | N/A | 1.5 mL |
500 mM NaF
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium Fluoride | 500 mM | 21 mg |
| Distilled Water | N/A | 1 mL |
| Total | N/A | 1 mL |
CRITICAL: Sodium fluoride is an acutely toxic (by oral exposure) and corrosive substance, and a skin, eye, and respiratory tract irritant, and is an environmental toxin. When handling this material, we recommend to use the appropriate personal protective equipment (PPE) such as goggles, appropriate gloves (low breakthrough), a mask, a lab coat, and a draft chamber.
500 mM NaN3
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium Azide | 500 mM | 32.51 mg |
| Distilled Water | N/A | 1 mL |
| Total | N/A | 1 mL |
CRITICAL: Sodium azide is an acutely toxic substance (by ingestion, oral, or dermal exposure). When handling this material, we recommend to use the appropriate PPE such as goggles, appropriate gloves (low breakthrough), a mask, a lab coat, and a draft chamber.
Chloroform-methanol (CM, 1/1, v/v)
| Reagent | Final concentration | Amount |
|---|---|---|
| Chloroform | 50% v/v | 10 mL |
| Methanol | 50% v/v | 10 mL |
| Total | N/A | 20 mL |
CRITICAL: The use of chloroform solvent is a health hazard because the solvent is considered to be toxic for both chronic and acute exposures, is considered to be a corrosive substance, a reproductive toxin, a carcinogenic substance, and a skin, eye, and respiratory irritant. When handling this solvent, we recommend to use the appropriate PPE such as goggles, appropriate gloves (low breakthrough), a mask, a lab coat, and a draft chamber.
Chloroform-methanol-water (CMW, 10/10/3, v/v/v)
| Reagent | Final concentration | Amount |
|---|---|---|
| Chloroform | 43.5% v/v | 10 mL |
| Methanol | 43.5% v/v | 10 mL |
| Distilled Water | 13.0% v/v | 3 mL |
| Total | N/A | 23 mL |
CRITICAL: The use of chloroform solvent is a health hazard. When handling this solvent, we recommend using the appropriate PPE.
0.6 N NaOH in methanol
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium Hydroxide | 0.6 N | 240 mg |
| Methanol | N/A | 10 mL |
| Total | N/A | 10 mL |
0.6 N AcOH in methanol
| Reagent | Final concentration | Amount |
|---|---|---|
| Acetic Acid | 0.6 N | 360.3 mg |
| Methanol | N/A | 10 mL |
| Total | N/A | 10 mL |
Solvent systems (I, II, III)
CRITICAL: Preparation of solvent systems should be done in a draft chamber.
Note: Blending of a small volume of a high concentration salt solutions into another solution that contains a large proportion of organic solvent often leads to salt precipitation. To avoid the salt precipitation, the aqueous potassium chloride solution or ammonium hydroxide solution is measured into a glass bottle at first, and then methanol and chloroform are added in order. Use a glass graduated measuring cylinder to measure the volume of aqueous solution, methanol and chloroform, transfer them into a glass bottle and mix completely by a stir plate and a magnetic bar for 16–18 hours.
Note: You can use the prepared solvent within a few days. It should not be used when separated into two layers due to composition changes by the volatilization of the organic solvent.
Solvent system I
Chloroform-methanol-0.25% KCl (55/45/10, v / v / v) solvent mixture
| Reagent | Final concentration | Volume |
|---|---|---|
| Chloroform | 50% v/v | 110 mL |
| Methanol | 41% v/v | 90 mL |
| 0.25% Aqueous Potassium Chloride Solution | 9% v/v | 20 mL |
| Total | N/A | 220 mL |
CRITICAL: The use of chloroform solvent is a health hazard. When handling this solvent, we recommend using the appropriate PPE.
Solvent system II
Chloroform-methanol-4.2N ammonium hydroxide (9/7/2, v/v/v) solvent mixture
| Reagent | Final concentration | Volume |
|---|---|---|
| Chloroform | 50% v/v | 135 mL |
| Methanol | 39% v/v | 105 mL |
| Aqueous 4.2N Ammonium Hydroxide Solution | 11% v/v | 30 mL |
| Total | N/A | 270 mL |
CRITICAL: The use of chloroform solvent and ammonium hydroxide solution is a health hazard. When handling these solvents, we recommend using the appropriate PPE.
Solvent system III
Chloroform-methanol-4.2N ammonium hydroxide (40/10/1, v/v/v) solvent mixture
| Reagent | Final concentration | Volume |
|---|---|---|
| Chloroform | 78% v/v | 200 mL |
| Methanol | 20% v/v | 50 mL |
| Aqueous 4.2N Ammonium Hydroxide Solution | 2% v/v | 5 mL |
| Total | N/A | 255 mL |
CRITICAL: The use of chloroform solvent and ammonium hydroxide solution is a health hazard. When handling these solvents, we recommend using the appropriate PPE.
Step-by-step method details
Protocol for measuring IPC synthesis
Timing: 1 week
Timing: 2 daysand 1 h for cell culture and preparation
Timing: 1.5–5 h for metabolic labeling for IPC synthesis
Timing: 3 h for lipid extraction
Timing: 3 h for NaOH treatment
Timing: 2 h for butanol purification
Timing: 4 h for lipid separation
Timing: 1 day or more for analysis
Metabolic labeling with [3H]myo-inositol specifically labels phosphatidylinositol, which is the precursor of IPC (Smith and Lester, 1974). Thus [3H]myo-inositol is used to analyze the biosynthesis of IPC and other inositol-containing lipids. This section describes how to label yeast cells with [3H]myo-inositol and analyze radiolabeled IPC by thin-layer chromatography (TLC).
[3H]myo-inositol is a radiolabeled compound that contains the radioisotope tritium (3H). Thus, be sure to comply with the laws and regulations regarding the use, storage, disposal, and other handlings of radioactive isotopes established by the national, state, and agency. The work described in this protocol must be performed as follow the CRITICAL Notes below.
CRITICAL: Experimental operation must be performed in a laboratory space that is appropriately designated for radiolabeled work.
CRITICAL: Experimental operation must be performed by authorized staffs that have formal institutional training in the handling, use, disposal, cleanup, and decontamination of radiolabeled substances.
CRITICAL: Depending on federal, state, local, and institutional policies, handling with the level of radiation (250 μCi) present in the commercial [3H]myo-inositol stock solution used for this work may require special environmental testing requirements (i.e., wipe testing of the benchtop); consultation with the institutional radiation safety officer before beginning work is highly recommended.
CRITICAL: All vials, tubes, pipette tips, discarded medium, discarded water washes, cell debris, TLC plates, solvent systems that contain [3H]myo-inositol is considered radioactive and should be discarded appropriately. Refer to the researcher's federal, state, local, and institutional policies and procedures.
-
1.Cell culture and preparation (Figure 1)
 CRITICAL: To avoid contamination, all steps in this section will be carried out in a clean bench.
CRITICAL: To avoid contamination, all steps in this section will be carried out in a clean bench. CRITICAL: The cells remaining on the plate after inoculation and the cells remaining after dispensing for labeling should be sterilized by autoclave and then discarded in the prescribed manners.
CRITICAL: The cells remaining on the plate after inoculation and the cells remaining after dispensing for labeling should be sterilized by autoclave and then discarded in the prescribed manners. CRITICAL: Before and after using, wipe the clean bench with 70% ethanol and sterilize the inside with a UV lamp.
CRITICAL: Before and after using, wipe the clean bench with 70% ethanol and sterilize the inside with a UV lamp.-
a.Inoculate yeast cells from a single colony grown on solid medium (Not specified) in 10 mL of semi-SD liquid medium in a 50 mL glass Erlenmeyer flask and grow at 25°C with gyratory shaking at 160 rpm for 16–18 hours.
-
b.Dilute the culture with a 20 mL volume of semi-SD liquid medium to an optical density at 600 nm (OD600) of 0.01–0.02 and grow cells in a 100 mL glass Erlenmeyer flask at 25°C with gyratory shaking at 160 rpm for 16–18 hours.
-
c.When the OD600 of culture is 0.2–0.6, transfer the culture to a 50 mL conical tube.
-
d.Spin down yeast cells by swinging bucket centrifuge at 1,580 g for 3 min at 15–25°C and remove the supernatant.
-
e.Wash the cells with 20 mL SD – inositol medium, spin down at 1,580 g for 3 min at RT and remove the supernatant. Repeat this step at least three times.
-
f.Spin down at 1,580 g for 3 min at RT, and remove the supernatant completely.
-
g.Resuspend the cells in SD – inositol medium to get an OD600 of 10 (3.0 × 108 cells).
-
h.Transfer a 0.5 mL volume of the cell suspension to a new 50 mL conical tube.
-
a.
-
2.Metabolic labeling for IPC synthesis (Figure 1)
-
a.Preincubate the 50 mL conical tube containing 0.5 mL of the cell suspension between 20 and 90 min within a given temperature range (basically between 24°C and 38°C) in a shaking water bath at 130 rpm.
-
b.Add 10 μCi of [3H]myo-inositol to the cell suspension and incubate between 60 and 180 min within a given temperature range (basically between 24°C and 38°C) in a shaking water bath at 130 rpm.Optional: For a pulse-chase analysis, add 1.5 mL SD + inositol medium to the cell suspension after pulse labeling and incubate for further 120 min.
-
c.To stop metabolic labeling, put the 50 mL conical tube on ice and add a 10 μL volume of a 500 mM NaF solution, and a 10 μL volume of a 500 mM NaN3 solution.
 CRITICAL: Sodium fluoride and sodium azide are toxic substances. When handling these materials, we recommend using the appropriate PPE.
CRITICAL: Sodium fluoride and sodium azide are toxic substances. When handling these materials, we recommend using the appropriate PPE.
-
a.
-
3.Lipid extraction (Figure 1)
-
a.Spin down cells in the 50 mL conical tube at 1,580 g for 3 min at RT.
-
b.Resuspend the cell pellet in the supernatant and transfer the labeled cells to a 1.5 mL Eppendorf microcentrifuge tube (tube #1).
-
c.Add a 0.5 mL volume of cold water to the 50 mL conical tube after removing most of the cells, mix by pipetting, and transfer the remaining cells to the tube #1.
-
d.Collect yeast cells by a microcentrifuge at 20,600 g for 3 min at 4°C, and remove the supernatant.
-
e.Add a 1 mL volume of cold water to the cell pellet, vortex well, spin down at 20,600 g for 3 min at 4°C and remove the supernatant. Repeat this step at least three times.
-
f.Adjust volume of cell suspension to 66 μL with cold water and resuspend the cells by vortex mixing.
-
g.Add 66 μL of glass beads and vortex vigorously 3–5 times for 30 sec to lyse cells, each time keeping the cells on ice for 1–2 min between vortexings.Note: Glass beads are low-alkali treated and are sold dry. Scoop with something like a small spoon and add to the tube #1.Note: Do not allow glass beads to get into the gap between the tube and the cap, because they may cause leakage of organic solvent during vortex mixing.
-
h.Add 400 μL of chloroform-methanol (CM; 1/1, v/v) to the cell lysate and vortex well at RT.
 CRITICAL: The use of chloroform solvent is a health hazard. When handling this solvent, we recommend using the appropriate PPE.Note: To avoid the loss of the sample attached to the pipette tip, use the same tip repeatedly for each sample.
CRITICAL: The use of chloroform solvent is a health hazard. When handling this solvent, we recommend using the appropriate PPE.Note: To avoid the loss of the sample attached to the pipette tip, use the same tip repeatedly for each sample. -
j.In order to extract completely the remaining radiolabeled lipids in the protein pellet in the tube #1, add a 200 μL volume of chloroform-methanol-water (CMW, 10/10/3, v/v/v) to the tube #1 and sonicate for 10–15 min in bath-type ultrasonic cleaner until the pellet is completely resuspended.
-
k.Centrifuge the tube #1 at 20,600 g for 3 min at RT and transfer the supernatant to the tube #2.
-
l.Add a 100 μL volume of CMW to the tube #1, vortex vigorously, centrifuge at 20,600 g for 3 min at RT, and transfer the supernatant to the tube #2.
-
m.Dry the combined supernatants in the tube #2 completely with N2 gas using pressure gas blowing concentrator at 30°C.
 CRITICAL: Drying the CMW in the samples with N2 gas must be done in a draft chamber, to prevent the radioactive-labeled lipid contained in the organic solvents from being aerosolized and contaminating the laboratory space.
CRITICAL: Drying the CMW in the samples with N2 gas must be done in a draft chamber, to prevent the radioactive-labeled lipid contained in the organic solvents from being aerosolized and contaminating the laboratory space. CRITICAL: Confirm with facility staff that maintains the building infrastructure that the draft chamber is equipped with an appropriate engineering control (i.e., trap) that limits radioactivity exposure to ensure that radioactive contamination of N2 or potential exhaust lines for the building does not occur.
CRITICAL: Confirm with facility staff that maintains the building infrastructure that the draft chamber is equipped with an appropriate engineering control (i.e., trap) that limits radioactivity exposure to ensure that radioactive contamination of N2 or potential exhaust lines for the building does not occur. Pause point: Samples can be stored at –20°C (for about 1 month).
Pause point: Samples can be stored at –20°C (for about 1 month).
-
a.
-
4.
NaOH treatment (Optional)
To differentiate complex sphingolipids from glycerophospholipids, lipid extracts can be subjected to mild base hydrolysis with NaOH, which selectively deacylates glycerophospholipids (Puoti et al., 1991).-
a.Add 100 μL of CMW to the tube #2, vortex well and centrifuge at 20,600 g for 1 min at RT.
-
b.Add 16 μL of 0.6 N NaOH (in methanol), vortex well and incubate for 2 h at 30°C.
-
c.Centrifuge at 20,600 g for 1 min at RT.
-
d.Add 16 μL of 0.6 N AcOH (in methanol), vortex well and centrifuge at 20,600 g for 1 min at RT.
-
e.Dry the samples in the tube #2 completely with N2 gas using pressure gas blowing concentrator at 30°C.
 CRITICAL: Drying the CMW in the samples with N2 gas must be done in a draft chamber.
CRITICAL: Drying the CMW in the samples with N2 gas must be done in a draft chamber. Pause point: Samples can be stored at –20°C (for about 1 month).
Pause point: Samples can be stored at –20°C (for about 1 month).
-
a.
-
5.Butanol purification (Figure 1)
-
a.To desalt, add 100 μL of water to the tube #2, vortex well and spin down at 20,600 g for 1 min at RT.
-
b.Add 100 μL of water-saturated 1-butanol to the tube #2, vortex well, centrifuge at 20,600 g for 3 min at RT and transfer the butanol (upper) phase containing complex sphingolipids to a new 1.5 mL Eppendorf microcentrifuge tube (tube #3).
 CRITICAL: Butanol is a highly flammable and corrosive solvent that causes skin, eye, and respiratory tract irritation. When handling this solvent, we recommend using the appropriate PPE such as goggles, gloves (low breakthrough), a mask, a lab coat, and a draft chamber.
CRITICAL: Butanol is a highly flammable and corrosive solvent that causes skin, eye, and respiratory tract irritation. When handling this solvent, we recommend using the appropriate PPE such as goggles, gloves (low breakthrough), a mask, a lab coat, and a draft chamber. -
c.In order to collect completely the remaining radiolabeled lipids in the water phase in the tube #2, add 100 μL of water-saturated 1-butanol to the tube #2, vortex well, centrifuge at 20,600 g for 3 min at RT and transfer the butanol phase to the tube #3. Repeat this step two more times.Note: To avoid the loss of the sample attached to the pipette tip, use the same tip repeatedly for each sample.
-
d.Add 100 μL of water to the tube #3, vortex well, centrifuge at 20,600 g for 3 min at RT and transfer the butanol phase to a new 1.5 mL tube (tube #4).
-
e.In order to collect the remaining radiolabeled lipids in the tube #3, add 200 μL of butanol to the tube #3, vortex well, centrifuge at 20,600 g for 3 min at RT and transfer the butanol phase to tube #4.
-
f.Dry the combined supernatants in the tube #4 completely with N2 gas using pressure gas blowing concentrator at 30°C.
 CRITICAL: Drying the butanol in the samples with N2 gas must be done in a draft chamber, to prevent the radioactive-labeled lipid contained in the organic solvents from being aerosolized and contaminating the laboratory space.
CRITICAL: Drying the butanol in the samples with N2 gas must be done in a draft chamber, to prevent the radioactive-labeled lipid contained in the organic solvents from being aerosolized and contaminating the laboratory space. Pause point: Samples can be stored at –20°C (for about 1 month).
Pause point: Samples can be stored at –20°C (for about 1 month).
-
a.
-
6.Lipid separation (Figure 2)Note: Prepare a developing solvent I (chloroform-methanol-0.25% KCl (55/45/10, v / v / v) solvent mixture) (Puoti et al., 1991) in a medium bottle by the day before, and stir for 16–18 hours with a stirrer and a stirrer chip.Note: Add the solvent to the tank (lined with filter papers) till about 1 cm from the bottom 2 h before TLC development, and fill the tank with the volatilized solvent.
 CRITICAL: The use of chloroform solvent is a health hazard. When handling this solvent, we recommend using the appropriate PPE.
CRITICAL: The use of chloroform solvent is a health hazard. When handling this solvent, we recommend using the appropriate PPE.-
a.Add 25 μL of CMW to the tube #4 and vortex well.Note: Rotate the tube #4 to dissolve the lipids on the upper wall of the tube.
-
b.Centrifuge at 20,600 g for 1 min at 4°C
-
c.To calculate cpm (count per minute)/μL of samples, add 1μL of sample to the polyethylene vial containing 5 mL of Clear-sol I.
-
d.Count the cpm of samples by using Liquid scintillation counter.
 CRITICAL: To prevent health hazards, the steps from here will be carried out in a draft chamber.Note: For the reason that CMWs are prone to evaporation, the required amount should be loaded onto the TLC plate immediately after counting the cpm. The remaining sample can be stored at –20°C.
CRITICAL: To prevent health hazards, the steps from here will be carried out in a draft chamber.Note: For the reason that CMWs are prone to evaporation, the required amount should be loaded onto the TLC plate immediately after counting the cpm. The remaining sample can be stored at –20°C. -
e.Load 1,000,000 cpm of each sample on TLC plate with a pipette tip 2 - 200 μL for gel loading.Note: The spots are lined up at intervals of about 1 cm. Hairdryer may be useful to dry solvent after loading the sample.
-
f.Place the plate in a glass TLC developing tank and develop with chloroform-methanol-0.25% KCl (55/45/10, v/v/v) solvent mixture.
-
g.When wetting front reaches within 1 cm of top of TLC plate (it takes about 90 to 120 min), remove the plate from the tank and dry it completely at RT or with cold air of hairdryer.
-
a.
-
7.Analysis (Figure 2)
-
a.Set the TLC plate in an exposure cassette and expose it to a tritium-sensitive imaging plate for a day or a few days.
-
b.Capture images with a FLA-7000 image analyzer.
-
c.Analyze the captured images and quantify the signals of bands with Multi Gauge software by FUJIFILM.
-
a.
Figure 1.
Illustration of several steps of lipid labeling, extraction, and butanol purification
Figure 2.
Illustration of several steps of lipid separation and analysis
Protocol for measuring IPC synthase activity
Timing: 1 week
Timing: 2 days and 1 h for cell culture and preparation
Timing: 1.5–3.5 h for metabolic labeling for C2-IPC
Timing: 3 h for lipid extraction
Timing: 2 h for butanol purification
Timing: 4 h for lipid separation
Timing: 1 day or more for analysis
This section describes how to measure IPC synthase activity in vivo. For this purpose, a short-chain ceramide, C2-ceramide (N-acetoyl-D-erythro-sphingosine) that reaches the Golgi through a diffusion-mediated or an endocytic route from the plasma membranes when added exogenously to cells can be used (Kajiwara et al., 2008). As exogenous C2-ceramide is incorporated into C2-IPC by IPC synthase, C2-IPC synthesis depends on IPC synthase activity and synthesis of phosphatidylinositol precursor. Thus, IPC synthase activity can be assayed using non-radiolabeled C2-ceramide and [3H] myo-inositol.
[3H]myo-inositol is a radiolabeled compound that contains the radioisotope tritium (3H). Thus, be sure to comply with the laws and regulations regarding the use, storage, disposal, and other handlings of radioactive isotopes established by the national, state, and agency. The work described in this protocol must be performed as follow the CRITICAL Notes below.
CRITICAL: Experimental operation must be performed in a laboratory space that is appropriately designated for radiolabeled work.
CRITICAL: Experimental operation must be performed by authorized staffs that have formal institutional training in the handling, use, disposal, cleanup, and decontamination of radiolabeled substances.
CRITICAL: Depending on federal, state, local, and institutional policies, handling with the level of radiation (250 μCi) present in the commercial [3H]myo-inositol stock solution used for this work may require special environmental testing requirements (i.e., wipe testing of the benchtop); consultation with the institutional radiation safety officer before beginning work is highly recommended.
CRITICAL: All vials, tubes, pipette tips, discarded medium, discarded water washes, cell debris, TLC plates, solvent systems that contain [3H]myo-inositol is considered radioactive and should be discarded appropriately. Refer to the researcher's federal, state, local, and institutional policies and procedures.
-
8.Cell culture and preparation
-
a.Same as described in step 1. for Cell culture and preparation.
-
a.
-
9.Metabolic labeling for C2-IPC (Figure 1)
-
a.Add a 10 μL volume of 10 mM C2-ceramide to the 50 mL conical tube containing a 0.5 mL volume of the cell suspension (OD600 of 10 (3.0 × 108 cells)) and preincubate for 20 min within a given temperature range (basically between 24°C and 38°C) in a shaking water bath at 130 rpm.
-
b.Add 10 μCi of [3H]myo-inositol to the cell suspension and incubate between 60 and 180 min within a given temperature range (basically between 24°C and 38°C) in a shaking water bath at 130 rpm.Optional: For a pulse-chase analysis, add 1.5 mL SD + inositol medium to the cell suspension after pulse labeling and incubate for further 120 min.
-
c.To stop metabolic labeling, put the 50 mL conical tube on ice and add a 10 μL volume of a 500 mM NaF solution, and a 10 μL volume of a 500 mM NaN3 solution.
 CRITICAL: Sodium fluoride and sodium azide are toxic substances. When handling these materials, we recommend using the appropriate PPE.
CRITICAL: Sodium fluoride and sodium azide are toxic substances. When handling these materials, we recommend using the appropriate PPE.
-
a.
-
10.Lipid extraction
-
a.Same as described in step 3. for Lipid extraction.
-
a.
-
11.Butanol purification
-
a.Same as described in step 5. for Butanol purification.
-
a.
-
12.Lipid separation
-
a.Same as described in step 6. for Lipid separation.
-
a.
-
13.Analysis
-
a.Same as described in step 7. for Analysis.
-
a.
Protocol for measuring ceramide synthesis
Timing: 2 weeks
Timing: 2 days and 1 h for cell culture and preparation
Timing: 2.5h for metabolic labeling for ceramide synthesis
Timing: 5 h for lipid extraction and butanol purification
Timing: 4 h for lipid separation (1st)
Timing: 1 day or more for analysis (1st)
Timing: 5 h for ceramide extraction
Timing: 4 h for lipid separation (2nd)
Timing: 1 day or more for analysis (2nd)
This section describes how to measure ceramide synthesis by metabolic labeling with a radioactive ceramide precursor, D-erythro-[4,5-3H]-dihydrosphingosine ([3H]DHS) (Oh et al., 1997; Kajiwara et al., 2014). This protocol also allows to analyze acylceramide synthesis (Voynova et al., 2012).
[3H]DHS is a radiolabeled compound that contains the radioisotope tritium (3H). Thus, be sure to comply with the laws and regulations regarding the use, storage, disposal, and other handlings of radioactive isotopes established by the national, state, and agency. The work described in this protocol must be performed as follow the CRITICAL Notes below.
CRITICAL: Experimental operation must be performed in a laboratory space that is appropriately designated for radiolabeled work.
CRITICAL: Experimental operation must be performed by authorized staffs that have formal institutional training in the handling, use, disposal, cleanup, and decontamination of radiolabeled substances.
CRITICAL: Depending on federal, state, local, and institutional policies, handling with the level of radiation (250 μCi) present in the commercial [3H]DHS stock solution used for this work may require special environmental testing requirements (i.e., wipe testing of the benchtop); consultation with the institutional radiation safety officer before beginning work is highly recommended.
CRITICAL: All vials, tubes, pipette tips, discarded medium, discarded water washes, cell debris, TLC plates, solvent systems that contain [3H]DHS are considered radioactive and should be discarded appropriately. Refer to the researcher's federal, state, local, and institutional policies and procedures.
-
14.Cell culture and preparation (Figure 1)
 CRITICAL: To avoid contamination, all steps in this section will be carried out in a clean bench.
CRITICAL: To avoid contamination, all steps in this section will be carried out in a clean bench. CRITICAL: The cells remaining on the plate after inoculation and the cells remaining after dispensing for labeling should be sterilized by autoclave and then discarded in the prescribed manners.
CRITICAL: The cells remaining on the plate after inoculation and the cells remaining after dispensing for labeling should be sterilized by autoclave and then discarded in the prescribed manners. CRITICAL: Before and after using, wipe the clean bench with 70% ethanol and sterilize the inside with a UV lamp.
CRITICAL: Before and after using, wipe the clean bench with 70% ethanol and sterilize the inside with a UV lamp.-
a.Inoculate yeast cells from a single colony on solid media in 10 mL semi-SD liquid medium in a 50 mL glass Erlenmeyer flask and grow at 25°C with gyratory shaking at 160 rpm for 16–18 hours.
-
b.Dilute the culture with 20 mL semi-SD liquid medium to an OD600 of 0.01–0.02 and grow cells in a 100 mL glass Erlenmeyer flask at 25°C with gyratory shaking at 160 rpm for 16–18 hours.
-
c.When the OD600 of culture is 0.2–0.6, transfer the culture to a 50 mL conical tube.
-
d.Spin down yeast cells by swinging bucket centrifuge at 1,580 g for 3 min at RT and remove the supernatant.
-
e.Wash the cell with 20 mL semi-SD medium, spin down at 1,580 g for 3 min at RT and remove the supernatant.
-
f.Spin down at 1,580 g for 3 min at RT, and remove the supernatant completely.
-
g.Resuspend the cells in semi-SD medium to get an OD600 of 10 (3.0 × 108 cells).
-
h.Transfer 0.5 mL of the cell suspension to a new 50 mL conical tube.
-
a.
-
15.Metabolic labeling for ceramide synthesis (Figure 1)
-
a.Preincubate the 50 mL conical tube containing 0.5 mL of the cell suspension between 20 and 90 min within a given temperature range (basically between 24°C and 38°C) in a shaking water bath at 130 rpm.
-
b.Add 10 μCi of [3H]DHS to the cell suspension and incubate between 60 and 180 min within a given temperature range (basically between 24°C and 38°C) in a shaking water bath at 130 rpm.
-
c.To stop metabolic labeling, put the 50 mL conical tube on ice and add 10 μL of 500 mM NaF and 10 μL of 500 mM NaN3.
-
a.
CRITICAL: Sodium fluoride and sodium azide are toxic substances. When handling these materials, we recommend using the appropriate PPE.
-
16.Lipid extraction and butanol purification
-
a.Same as described in step 3. for Lipid extraction.
-
b.Same as described in step 5. for Butanol purification.
-
a.
-
17.Lipid separation (1st) (Figure 2)Note: Prepare a developing solvent II (chloroform-methanol-4.2N ammonium hydroxide (9/7/2, v/v/v) solvent mixture) (Mandala et al., 1995) in a medium bottle by the day before, and stir for 16–18 hours with a stirrer and a stirrer chip.Note: Add the solvent to the tank (lined with filter papers) till about 1 cm from the bottom 2 hours before TLC development, and fill the tank with the volatilized solvent.
 CRITICAL: The use of chloroform solvent and ammonium hydroxide solution is a health hazard. When handling this solvent, we recommend using the appropriate PPE.
CRITICAL: The use of chloroform solvent and ammonium hydroxide solution is a health hazard. When handling this solvent, we recommend using the appropriate PPE.-
a.Add 20μL of CMW to the 1.5 mL Eppendorf microcentrifuge tube containing radiolabeled lipids dried completely with N2 gas using pressure gas blowing concentrator after butanol purification and vortex well.Note: Rotate the tube to dissolve the lipids on the upper wall of the tube.
-
b.Centrifuge at 20,600 g for 1 min at 4°C.
-
c.Load all of each sample on TLC plate with a pipette tip 2 - 200 μL for gel loading.Note: The spots are lined up at intervals of about 1 cm. Hairdryer may be useful to dry solvent after loading the sample.
-
d.Place the plate in glass TLC developing tank and develop with chloroform-methanol-4.2N ammonium hydroxide (9/7/2, v/v/v) solvent mixture.
-
e.When wetting front reaches within 1 cm of top of TLC plate, remove the plate from the tank and dry it completely at RT or with cold air of hairdryer.
-
a.
-
18.Analysis (1st)
-
a.Same as described in step 7. for Analysis.
-
a.
-
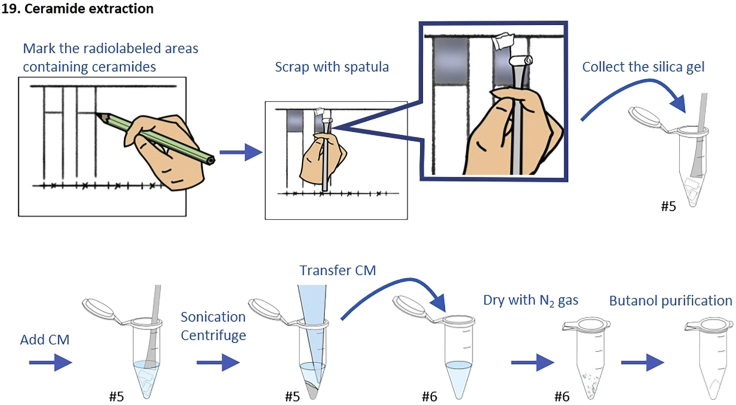
19.Ceramide extraction (Figure 3)
-
a.After visualized by a FLA-7000 image analyzer, mark the radiolabeled areas containing ceramide on TLC plate with a pencil.
-
b.Wet the marked area with a few drops of water.
-
c.Collect the silica gel of the area by scrapping with a spatula and transfer to a new 1.5 mL Eppendorf microcentrifuge tube (tube #5).
-
d.Add 400 μL of CM to the tube #5.
-
e.Sonicate for 10–15 min in a bath-type ultrasonic cleaner until the silica is completely suspended.
-
f.Centrifuge the tube #5 at 20,600 g for 5 min at RT and transfer the supernatant to a new 1.5 mL Eppendorf microcentrifuge tube (tube #6).
-
g.Add 200 μL of CM to the pellet in the tube #5, vortex well, centrifuge at 20,600 g for 5 min at RT, and transfer the supernatant to the tube #6.Note: To avoid the loss of the sample attached to the pipette tip, use the same tip repeatedly for each sample.
-
h.Dry the combined supernatants in the tube #6 completely with N2 gas using pressure gas blowing concentrator at 30°C.
 CRITICAL: Drying the CMW in the samples with N2 gas must be done in a draft chamber.
CRITICAL: Drying the CMW in the samples with N2 gas must be done in a draft chamber. Pause point: Samples can be stored at –20°C (for about 1 month).
Pause point: Samples can be stored at –20°C (for about 1 month). Pause point: Samples can be stored at –20°C (for about 1 month).
Pause point: Samples can be stored at –20°C (for about 1 month).
-
a.
-
20.Lipid separation (2nd)Note: Prepare a developing solvent III (chloroform-methanol-4.2N ammonium hydroxide (40/10/1, v/v/v) solvent mixture) (Haak et al., 1997) in a medium bottle by the day before, and stir for 16–18 hours with a stirrer and a stirrer chip.Note: Add the solvent to the tank (lined with filter papers) till about 1 cm from the bottom 2 hours before TLC development, and fill the tank with the volatilized solvent.
 CRITICAL: The use of chloroform solvent and ammonium hydroxide solution is a health hazard. When handling these solvents, we recommend using the appropriate PPE.
CRITICAL: The use of chloroform solvent and ammonium hydroxide solution is a health hazard. When handling these solvents, we recommend using the appropriate PPE.-
a.Same as described in step 17. for Lipid separation (1st) except developing solvent.
-
a.
-
21.Analysis (2nd)
-
a.Same as described in step 7. for Analysis.
-
a.
Figure 3.
Illustration of several steps of ceramide extraction
Expected outcomes
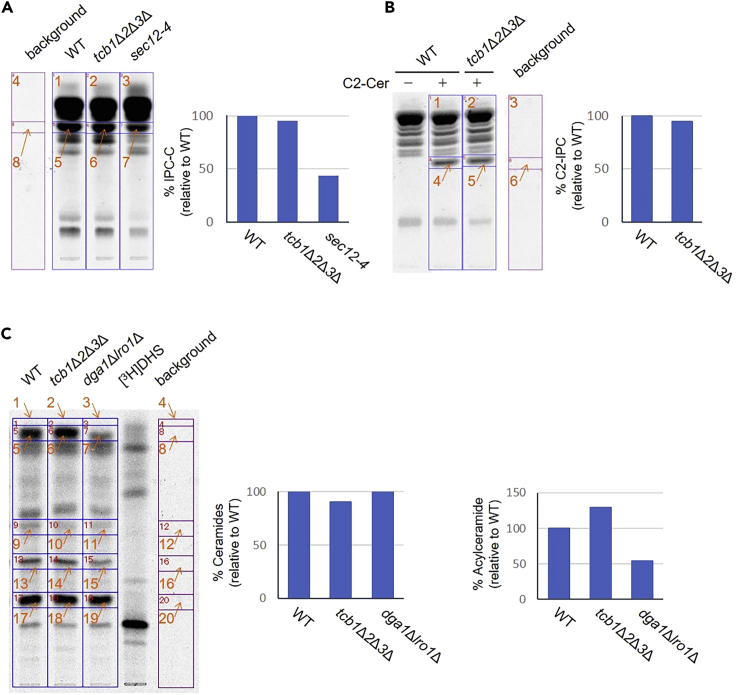
If all steps are followed correctly it is expected that the image analyzed by TLC should show clearly separated bands as shown in Figure 4. The actual value of the signal depends on the amount of sample loaded and the number of exposing days. Tables 1, 2, and 3 show examples of these values.
Figure 4.
Examples of TLC image data
(A) Examples of measuring complex sphingolipid synthesis. WT cells were grown at 25°C, sifted to 37°C for 20 min, and labeled with [3H]myo-inositol for 60 min. Labeled lipids were (+) or were not (−) mildly hydrolyzed with NaOH to deacylate glycophospholipids and detect base-resistant complex sphingolipids (IPC-C, MIPC and M(IP)2C). Samples were applied to a TLC plate and separated using solvent system I.
(B) Examples of measuring IPC synthase activity. WT cells were grown at 25°C, incubated without (−) or with (+) C2-ceramide for 20 min, and labeled with [3H]myo-inositol for 180 min. Labeled lipids were applied to a TLC plate and separated using solvent system I.
(C) Examples of measuring ceramide synthesis. WT cells were grown at 25°C, and labeled with D-erythro-[4,5-3H]-dihydrosphingosine ([3H]DHS) for 180 min. Labeled lipids were applied to a TLC plate and separated using solvent system II (left). Fractions containing ceramides and acylceramides on the TLC plate (in left, the area surrounded by a square) were collected by scrapping. After extraction from the silica, the lipids were analyzed by TLC using solvent system III (right).
(A–C) The figures are reprinted with permission from Ikeda et al., 2020.
Table 1.
Example of raw data measured on a multi gauge for Figure 5A
| No | Group | Name | PSL | Area(mm2) | PSL-BG |
|---|---|---|---|---|---|
| 1 | A | Total 1 | 216918.4 | 2202.13 | 214984.4 |
| 2 | A | Total 2 | 220726.8 | 2202.13 | 218792.8 |
| 3 | A | Total 3 | 229426.4 | 2202.13 | 227492.4 |
| 4 | A | Total BG | 1933.99 | 2202.13 | 0 |
| 5 | B | IPC-C 1 | 26307.55 | 129.31 | 26207.69 |
| 6 | B | IPC-C 2 | 25502.36 | 129.31 | 25402.5 |
| 7 | B | IPC-C 3 | 12228.84 | 129.31 | 12128.98 |
| 8 | B | IPC-C BG | 99.86 | 129.31 | 0 |
PSL: Photo Stimulated Luminescence; BG: Back Ground
Table 2.
Example of raw data measured on a multi gauge for Figure 5B
| No | Group | Name | PSL | Area(mm2) | PSL-BG |
|---|---|---|---|---|---|
| 1 | A | Total 1 | 87605.1 | 1898.33 | 81113.09 |
| 2 | A | Total 2 | 89810.78 | 1898.33 | 83318.77 |
| 3 | A | Total BG | 6492.01 | 1898.33 | 0 |
| 4 | B | C2-IPC 1 | 8420.19 | 126.68 | 7943.03 |
| 5 | B | C2-IPC 2 | 8217.62 | 126.68 | 7740.47 |
| 6 | B | C2-IPC BG | 477.15 | 126.68 | 0 |
PSL: Photo Stimulated Luminescence; BG: Back Ground
Table 3.
Example of raw data measured on a multi gauge for Figure 5C
| No | Group | Name | PSL | Area(mm2) | PSL-BG |
|---|---|---|---|---|---|
| 1 | A | Total 1 | 137119.74 | 3072.66 | 120626.72 |
| 2 | A | Total 2 | 143859.2 | 3072.66 | 127366.17 |
| 3 | A | Total 3 | 103697.34 | 3072.66 | 87204.32 |
| 4 | A | Total BG | 16493.02 | 3072.66 | 0 |
| 5 | B | Acyl-Cer 1 | 25266.83 | 177.44 | 24447.95 |
| 6 | B | Acyl-Cer 2 | 34158.01 | 177.44 | 33339.12 |
| 7 | B | Acyl-Cer 3 | 10376.7 | 177.44 | 9557.81 |
| 8 | B | Acyl-Cer BG | 818.89 | 177.44 | 0 |
| 9 | C | Cer-A,B 1 | 6747.93 | 177.44 | 5773.56 |
| 10 | C | Cer-A,B 2 | 5303.8 | 177.44 | 4329.43 |
| 11 | C | Cer-A,B 3 | 4458.77 | 177.44 | 3484.4 |
| 12 | C | Cer-A,B BG | 974.37 | 177.44 | 0 |
| 13 | D | Cer-B’ 1 | 10383.49 | 177.44 | 9479.86 |
| 14 | D | Cer-B’ 2 | 10701.07 | 177.44 | 9797.43 |
| 15 | D | Cer-B’ 3 | 6637.18 | 177.44 | 5733.54 |
| 16 | D | Cer-B’ BG | 903.63 | 177.44 | 0 |
| 17 | E | Cer-C 1 | 30991.38 | 177.44 | 29815.91 |
| 18 | E | Cer-C 2 | 30036.55 | 177.44 | 28861.08 |
| 19 | E | Cer-C 3 | 25431.34 | 177.44 | 24255.87 |
| 20 | E | Cer-C BG | 1175.47 | 177.44 | 0 |
PSL: Photo Stimulated Luminescence; BG: Back Ground
Using this protocol, we can study sphingolipid metabolism and transport in various mutant strains. If analysis in this protocol reveals that IPC synthesis activity and ceramide synthesis are normal, but IPC synthesis is reduced in a certain mutant strain, these findings suggest that the mutant cells exhibit a defect in ceramide transport from the ER to the Golgi apparatus.
Quantification and statistical analysis
Visualize the image exposed to a tritium-sensitive imaging plate with FLA-7000 Image Reader. Open the data with Multi Gauge software to quantify signal intensity. Figure 5 shows the process of quantification, and Tables 1, 2, and 3 show the examples of the numerical values obtained using the software. The numerical values of PSL-BG (Photo Stimulated Luminescence - Back Ground) after subtracting the background signals in Tables are used for quantification as net signal intensities. Incorporation (%) of [3H]myo-inositol into IPC-C or C2-IPC or of [3H]DHS into ceramide (Cer-A, -B, -B’ and C) or acylceramide is calculated using the following formula: % = signal intensities of each lipid/total signal intensities of all radiolabeled lipids × 100. Then, the relative amounts are expressed as percentages of the amounts in control cells. Usually, three or more independent experiments are performed, and data are expressed as mean ± standard deviation of multiple analyses. Statistical significance is also analyzed using Student's t-test.
Figure 5.
Illustration of how to quantify the image data
Image Data is quantified by using Multi Gauge software. Enclose the band in each lane with the Measure tool in Multi Gauge.
(A) Squares 1–3 are enclosed as Total, square 4 as a background signal of Total, squares 5–7 as IPC-C, and square 8 as a background signal of IPC-C. The signal intensities in the enclosed areas are shown in Table 1.
(B) Squares 1–2 are enclosed as Total, square 3 as a background signal of Total, squares 4–5 as C2-IPC, and square 6 as a background signal of C2-IPC. The signal intensities in the enclosed areas are shown in Table 2.
(C) Squares 1–3 are enclosed as Total, square 4 as a background signal of Total, squares 5–7 as acylceramide, square 8 as a background signal of aclyceramide, squares 9–11 are enclosed as ceramide-A and -B, square 12 as a background signal of ceramide-A and -B, squares 13–15 are enclosed as ceramide-B’, square 16 as a background signal of ceramide-B’, squares 17–19 are enclosed as ceramide-C, square 20 as a background signal of ceramide-C. The signal intensities in the enclosed areas are shown in Table 3. Radiolabel in ceramide-A, -B, -B’ and -C increases when cells are treated with aureobasidein A (2 μg/mL, for 1–4h), a specific inhibitor of IPC synthase (Kajiwara et al., 2012).
(A–C) The graphs show the quantified results as described in the Quantification and Statistical Analysis section. The figures are reprinted with permission from Ikeda et al., 2020.
Limitations
Although TLC following metabolic labeling is a widely used method to assess synthesis of lipids, it has some limitations. This protocol was optimized for the separation of subclasses of IPC (Ikeda et al., 2015) and ceramide (Haak et al., 1997). A developing solvent I (chloroform-methanol-0.25% KCl; 55/45/10, v/v/v) can separate IPC subclasses (IPC-A, -B, -B’, -C, -D) and C2-IPC. However, in this solvent system, most of the subclasses of MIPC and M(IP)2C could not be separated. An optional step of mild alkaline hydrolysis is recommended to detect IPC-A, -B or -B’ because their subclasses overlap with the bands corresponding to radiolabeled phosphatidylinositol. In addition, it is recommended to load 5–10 × 105 cpm per sample onto the TLC plate to easily detect individual radioactive signals. At a minimum, 1 × 105 cpm per sample may be required, but longer time exposures to a storage phosphor screen would be required to obtain enough signals. The amount of radiolabeled lipids is limited by the uptake and delivery of exogenous radioactive substrates to the sites of sphingolipid synthesis and the activity of the enzyme involved in the synthesis. To get a high amount of radiolabeled lipids, labeling times longer than 2h should be considered. Pulse-chase analysis might improve sphingolipid labeling. Another possible solution is the scale up of reaction mixtures, which can allow to get more radiolabeled lipids. Finally, mutant strains defective in substrate uptake and conditions that reduce metabolic activity such as low temperatures are inappropriate for lipid analysis by metabolic labeling.
Troubleshooting
Problem 1
Low radioactivity of extracted lipids.
Potential solution
Lipid extraction efficiency may be poor. For an efficient lipid extraction, lyse the cells completely using glass beads before adding CM, by increasing the number of beads beating cycles (step 3, g).
Problem 2
Very weak or undetectable signals on the images obtained using storage phosphor imaging systems.
Potential solution
Increase the amount of cpm for the sample to be applied to TLC plate (step 6, e) or the exposure time to a storage phosphor screen (step 6, h).
Problem 3
The TLC developed band is disturbed like wavy.
Potential solution
If room temperature or humidity is high, the sample separation may not be good and the shape of the band may bend. Keep the room temperature around 23°C and dry the room using an air conditioner. Also, incomplete purification of lipids with butanol (step 5) results in inefficient separation due to salt carry-in. Therefore, purify lipid sample further by butanol partitioning.
Problem 4
Lipid separation by TLC development does not work as expected.
Potential solution
The composition of developing solvents is liable to change due to the volatilization of organic solvent. Store the prepared solvent away from high temperatures with a parafilm around the lid of the bottle, and use it within a few days ("Materials and equipment" section).
Problem 5
Only the bands corresponding to phosphatidylinositol are strongly detected.
Potential solution
This might happen due to low incorporation of [3H]phosphatidylinositol into IPC. Increase labeling time. Additionally, this problem can be addressed with pulse-chase analysis (step 2, b, Optional).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kouichi Funato (kfunato@hiroshima-u.ac.jp).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate or analyze datasets or code.
Acknowledgments
This work was supported by the Grant-in-Aid for Scientific Research (KAKENHI) (19H02922 to K.F.).
Author contributions
A.I. performed experiments. A.I. and K.H. designed the figures. A.I. and K.F. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- Haak D., Gable K., Beeler T., Dunn T. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 1997;272:29704–29710. doi: 10.1074/jbc.272.47.29704. [DOI] [PubMed] [Google Scholar]
- Ikeda A., Muneoka T., Murakami S., Hirota A., Yabuki Y., Karashima T., Nakazono K., Tsuruno M., Pichler H., Shirahige K. Sphingolipids regulate telomere clustering by affecting the transcription of genes involved in telomere homeostasis. J. Cell Sci. 2015;128:2454–2467. doi: 10.1242/jcs.164160. [DOI] [PubMed] [Google Scholar]
- Ikeda A., Schlarmann P., Kurokawa K., Nakano A., Riezman H., Funato K. Tricalbins Are Required for Non-vesicular ceramide transport at ER-Golgi contacts and modulate lipid droplet biogenesis. iScience. 2020;23:101603. doi: 10.1016/j.isci.2020.101603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara K., Ikeda A., Aguilera-Romero A., Castillon G.A., Kagiwada S., Hanada K., Riezman H., Muñiz M., Funato K. Osh proteins regulate COPII-mediated vesicular transport of ceramide from the endoplasmic reticulum in budding yeast. J. Cell Sci. 2014;127:376–387. doi: 10.1242/jcs.132001. [DOI] [PubMed] [Google Scholar]
- Kajiwara K., Muneoka T., Watanabe Y., Karashima T., Kitagaki H., Funato K. Perturbation of sphingolipid metabolism induces endoplasmic reticulum stress-mediated mitochondrial apoptosis in budding yeast. Mol. Microbiol. 2012;86:1246–1261. doi: 10.1111/mmi.12056. [DOI] [PubMed] [Google Scholar]
- Kajiwara K., Watanabe R., Pichler H., Ihara K., Murakami S., Riezman H., Funato K. Yeast ARV1 is required for efficient delivery of an early GPI intermediate to the first mannosyltransferase during GPI assembly and controls lipid flow from the endoplasmic reticulum. Mol. Biol. Cell. 2008;19:2069–2082. doi: 10.1091/mbc.E07-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandala S.M., Thornton R.A., Frommer B.R., Curotto J.E., Rozdilsky W., Kurtz M.B., Giacobbe R.A., Bills G.F., Cabello M.A., Martín I. The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J. Antibiot. (Tokyo). 1995;48:349–356. doi: 10.7164/antibiotics.48.349. [DOI] [PubMed] [Google Scholar]
- Oh C.S., Toke D.A., Mandala S., Martin C.E. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- Puoti A., Desponds C., Conzelmann A. Biosynthesis of mannosylinositolphosphoceramide in Saccharomyces cerevisiae is dependent on genes controlling the flow of secretory vesicles from the endoplasmic reticulum to the Golgi. J. Cell. Biol. 1991;113:515–525. doi: 10.1083/jcb.113.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.W., Lester R.L. Inositol phosphorylceramide, a novel substance and the chief member of a major group of yeast sphingolipids containing a single inositol phosphate. J. Biol. Chem. 1974;249:3395–3405. [PubMed] [Google Scholar]
- Voynova N.S., Vionnet C., Ejsing C.S., Conzelmann A. A novel pathway of ceramide metabolism in Saccharomyces cerevisiae. Biochem. J. 2012;447:103–114. doi: 10.1042/BJ20120712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate or analyze datasets or code.