Summary
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer death worldwide. However, the pathogenesis of HCC is complicated, and the drugs used for HCC treatment are limited. The following protocol combines a genetically engineered mouse model (GEMM) with a sleeping beauty system to establish an in vivo liver tumorigenesis model. By using this model, the impact of genes of interest in liver tumorigenesis and progression can be studied. This model can also be applied to develop new therapeutic drugs for HCC.
For complete details on the use and execution of this protocol, please refer to He et al. (2020).
Subject areas: Cancer, Genetics, Model Organisms, Molecular Biology, Gene Expression
Graphical Abstract
Highlights
-
•
This protocol describes a mouse model for liver tumorigenesis
-
•
This method combines sleeping beauty strategy and a genetically engineered mouse model
-
•
An effective approach to investigate genetic interplay in liver tumorigenesis
-
•
This method provides a platform for drug screen and efficacy evaluation for HCC
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer death worldwide. However, the pathogenesis of HCC is complicated, and the drugs used for HCC treatment are limited. The following protocol combines a genetically engineered mouse model (GEMM) with a sleeping beauty system to establish an in vivo liver tumorigenesis model. By using this model, the impact of genes of interest in liver tumorigenesis and progression can be studied. This model can also be applied to develop new therapeutic drugs for HCC.
Before you begin
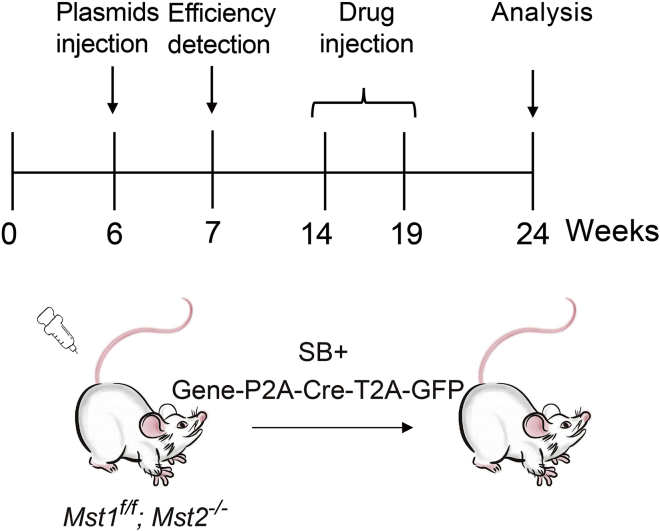
The disorder of the Hippo pathway has been linked to liver tumorigenesis (Hong et al., 2015). Liver-specific deletion of Mst1/2 leads to the continuous liver enlargement and ultimately tumor formation predominantly through sustained YAP activation (Lu et al., 2010; Song et al., 2010; Zhou et al., 2009). However, traditional Mst1/2 knockout in liver induced by Alb-Cre spreads almost the whole liver, and the liver quickly enlarges and shows tumor formation. Here, we combine the transposase system with traditional Mst1/2 knockout transgenic mice. Thus, the sporadic loss of the interested gene can better resembles the process in human liver cancer. With this model, we can study the factors involved in liver cancer process and screen therapeutic drugs for HCC.
Breeding cage setup and genotyping
Timing: 9–10 weeks
-
1.
Set up three breeding cages (Mating Mst1f/f, Mst2-/- with Mst1f/f, Mst2-/-).
CRITICAL: All mice are kept in a specific pathogen-free (SPF) environment at the Shanghai Institute of Biochemistry and Cell Biology and treated in strict accordance with protocols approved by the Institutional Animal Use Committee of the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
-
2.
Once the female is pregnant, separate it from male.
-
3.
Number the pups by cutting toes, and cut a little bit tail for genotyping when they are ten days old.
-
4.
Place identified pups into new cages when they are aged at four weeks and keep female mice till six-week old prior to the following experiments.
Plasmids preparation
Timing: 4–7 days
-
5.
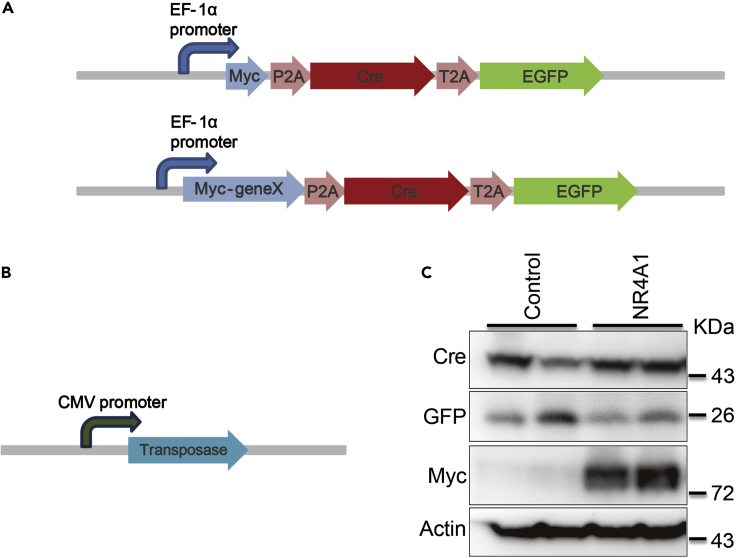
If you aim to study the function of a specific gene (termed as “X”) in tumorigenesis, clone it into the pT3-Myc-P2A-Cre-T2A-eGFP vector shown in Figure1A.
-
6.
Extract pT3-Myc-P2A-Cre-T2A-EGFP, pT3-Myc-geneX-P2A-Cre-T2A-EGFP and SB plasmids (Figure 1B).
Pause point: The extracted plasmids can be stored at −20°C.
Note: To extract high quality-concentrated plasmid, we recommend using a NucleoBond Xtra Midi Plus Kit [MN, cat.740412.50]. Please check the website (https://www.mn-net.com/nucleobond-xtra-midi-plus-kit-for-transfection-grade-plasmid-dna-740412.50) for the detailed extraction protocol.
CRITICAL: Plasmid DNA solutions must be sterile, endotoxin free and highly concentrated (1–3 μg/μL).
Figure 1.
Constructs used in sleeping beauty transposase method to induce HCC in mice
(A) Generic plasmid maps of pT3-Myc-P2A-Cre-T2A-EGFP and pT3-Myc-geneX-P2A-Cre-T2A-EGFP used to generate HCC in mice.
(B) Generic plasmid maps of SB transposase.
(C) Western blotting analysis of the expression level of Cre, GFP and Myc-tagged NR4A1 expression in HEK293 T cells transfected with pT3-P2A-Cre-T2A-EGFP control vector or pT3-Myc-NR4A1-P2A-Cre-T2A-EGFP plasmid.
Test the expression of plasmid
Timing: 4–5 days
-
7.
Transfect the pT3-Myc-P2A-Cre-T2A-EGFP and pT3-Myc-geneX-P2A-Cre-T2A-EGFP plasmids in 293 T (100,000 cells per well in 6 well plate) respectively.
-
8.
Run SDS-PAGE to test the expression of Cre, GFP and Myc-tagged-geneX with their primary antibodies (Figure1C). See Troubleshooting 3. The work dilution of antibodies: Cre, 1:1000; GFP, 1:1000; Myc, 1:5000;
Set up equipment
Timing: 30 min
-
9.Check the equipment to be used.
-
a.Heating lamp
-
b.Restrainer tube
-
c.Scale
-
d.Goggles
-
e.High-speed tissue homogenizer
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-MYC | Proteintech | Cat# 60003-2-Ig, RRID:AB_2734122 |
| Mouse anti-GFP | Santa Cruz Biotechnology | Cat# sc-9996; RRID:AB_627695 |
| Rabbit anti-Cre | Abcam | Cat# ab188568 |
| Mouse anti-GAPDH | Proteintech | Cat# 60004-1-Ig, RRID:AB_2107436 |
| Chemicals, peptides, and recombinant proteins | ||
| Normal saline | Beyotime Biotechnology | Cat#ST341-500ml |
| Protease Inhibitors Cocktail | MCE | Cat#HY-K0010 |
| Chloral hydrate | Sigma-Aldrich | Cat#15307; CAS: 302-17-0 |
| Isoflurane | RWD | Cat#902-0000-522; CAS: 26675-46-7 |
| Trichostatin A | MedChemExpress | Cat#HY-15144; CAS: 58880-19-6 |
| Tris base | Sigma | Cat# 648311; CAS: 77-86-1 |
| Hydrochloric acid (HCl) | Sinopharm Chemical Reagent | Cat# 10011018; CAS: 2647-01-0 |
| Sodium hydroxide (NaOH) | China Reagent | Cat# 1310-73-2; CAS: 1310-73-2 |
| EDTA | Sigma-Aldrich | Cat#U3620; CAS: 60-00-4 |
| Sodium dodecyl sulfate (SDS) | Sigma-Aldrich | Cat#L3771;CAS: 151-21-3 |
| 1,4-Dithiothreitol (DTT) | Sigma-Aldrich | Cat#3488-12-3 |
| Critical commercial assays | ||
| NucleoBond Xtra Midi Plus | MN | Cat#740412.50 |
| Experimental models: Cell lines | ||
| HEK293 T cell ATCC | ATCC | ATCC ACS-4500 |
| Experimental models: Organisms/strains | ||
| Mouse: 129/Sv, Mst1f/f, Mst2-/- | (Du et al., 2014) | (Du et al., 2014) |
| Recombinant DNA | ||
| pT3-Myc-P2A-Cre-T2A-EGFP | This paper |
http://www.brics.ac.cn/plasmid/template/plasmid/plasmid_list.html, catalog number SP-2939 |
| pT3-Myc-NR4A1-P2A-Cre-T2A-EGFP | This paper | N/A |
| pCMV-Transposase (SB) | This paper |
http://www.brics.ac.cn/plasmid/template/plasmid/plasmid_list.html, catalog number SP-2940 |
| Other | ||
| 1 mL Syringe | Solarbio | Cat#YA0550 |
| 2 mL Syringe | Solarbio | Cat#YA0556 |
| Sonication tubes | SARSTEDT | Cat#72.730 |
| Protein extraction beads (0.9–1.1 mm) | Titan | Cat#02048529 |
| Protein extraction beads (2.8–3.2 mm) | Titan | Cat#02048534 |
| High-speed tissue homogenizer | OMNI | OptimateTM Bead Mill Operating System Model:NE486LL/A |
| Heating lamp | AUPU | Cat#E27 |
| Restrainer tube | Bruderer experimental equipment | Cat#GGQ-5 |
| Anaesthesia machine | Renyi instrument | Cat#MSS-3S |
Materials and equipment
Protein extraction buffer (add fresh protease inhibitor before use)
| Reagent | Final concentration | Stock concentration | Amount |
|---|---|---|---|
| Tris-HCl (pH7.5) | 50 mM | 1M | 50 mL |
| EDTA (pH8) | 0.5 mM | 0.5M | 1 mL |
| SDS | 1% | 10% (m/v) | 100 mL |
| DTT | 1 mM | 1M | 1 mL |
| H2O | 848 mL |
The buffer can be stored at 20°C–25°C for six months.
Step-by-step method details
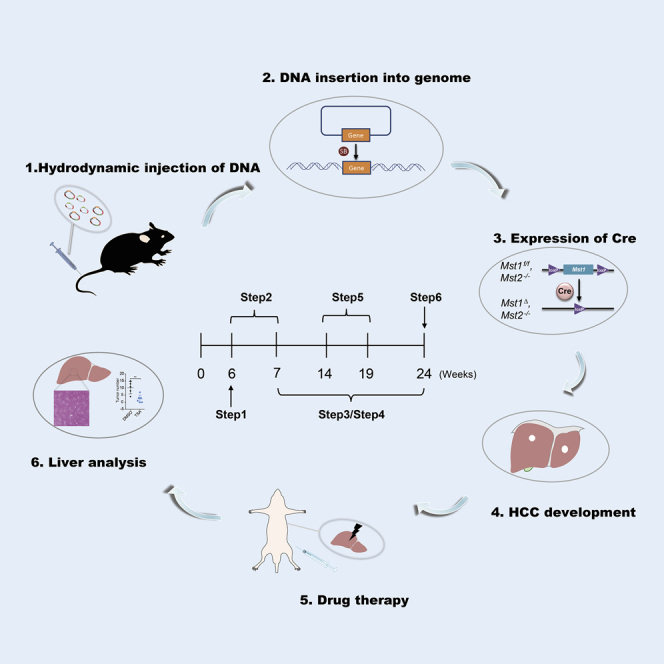
Hydrodynamic injection
Timing: 1 day
Please see the Methods video S1 for the detailed operation.
-
1.
Mix the SB and pT3-Myc-P2A-Cre-T2A-EGFP plasmids together with 0.9% saline (SB: 1 μg/mL, pT3:10 μg/mL).
Note: Prepare about 2 mL DNA solution (containing 20 μg pT3 plasmids and 2 μg SB plasmids) for each mouse, and each group should contain more than 5 mice. So we suggest preparing DNA solution between 10 mL to 15 mL for each group.
Note: If the goal is to test the effectiveness of the drug in treating liver cancer, inject the mice with pT3-Myc-P2A-Cre-T2A-EGFP and SB plasmid. If it is to test the function of the genes of interest in liver tumorigenesis, inject pT3-Myc-P2A-Cre-T2A-EGFP or pT3-Myc-geneX-P2A-Cre-T2A-EGFP along with SB plasmids.
Note: From this step, keep aseptic operations unless otherwise indicated.
CRITICAL: The saline must be sterile, otherwise it will decrease the survival rate of mice. See Troubleshooting 2.
CRITICAL: Thaw the plasmids at 65°C before use, otherwise the plasmid with high concentration will be too sticky to pipet.
-
2.
Take 10 μL solution and coat it to bacterial colony culture dish which is without bacteria and antibiotics for sterility test. We can go to next step as long as there are not colony growths on the dish.
-
3.
Pipet and vortex the mixture. Maintain mixture at 20°C–25°C for half an hour prior to injection.
-
4.
Sterilize the devices with 75% alcohol in accordance with SPF procedure.
-
5.
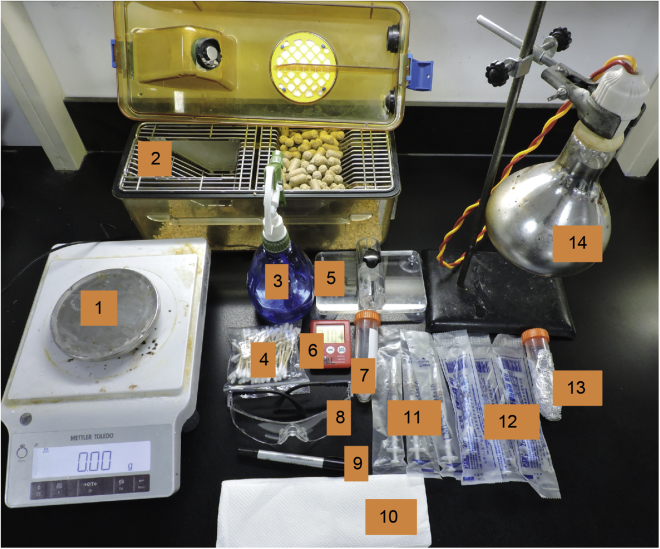
Enter the mouse room with prepared reagents and devices (Figure 2).
-
6.
Take the mice, devices and reagents to the operation room.
-
7.
Weigh the mouse aged at 6 weeks and record their weight.
Note: The weight of 6-week-old female mouse is about 20 g.
CRITICAL: Eliminate mouse weighting over 25 g. See Troubleshooting 5.
-
8.
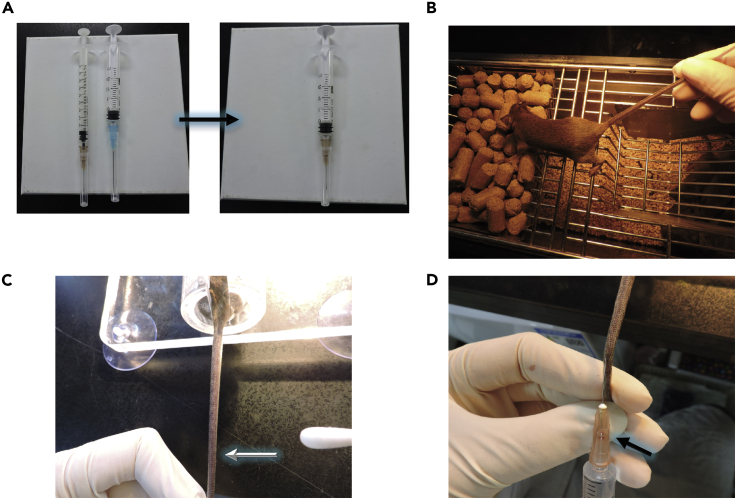
Place a needle from 1 mL syringe on 2 mL syringe (Figure 3A).
-
9.
Load the syringe with the DNA solution [10% (volume/body weight)]. For example: a 20 g mouse will be injected with 2 mL of DNA solution.
Note: The operator should put on the goggles before using the syringe to protect eyes.
-
10.
Remove the air from the syringe by tapping it and pressing the plunger slowly, then put syringe aside until injection.
-
11.
Anesthetize mice with isoflurane-loaded anaesthesia machine.
Optional: The mice also can be anesthetized by intraperitoneal injection with anesthetic. If you choose this way, the mouse should be anesthetized between steps 7 and 8.
-
12.
Place the tail of the mouse under a heat lamp for 1 min until the lateral vein of the mouse tail is visualized (Figure 3B).
Note: This step will increase the vascular volume of the tail and make the injection easier. If the mouse’s tail vein is easy to distinguish, skip to next step.
CRITICAL: Do not overheat the mice. When noticing the mice hide in the corners of the cage, adjust the height of the lamp to the cage or close the lamp for a while.
-
13.
Place the mouse into the restrainer tube and wipe the tail with an alcohol pad.
-
14.
Adjust the position of mice until the lateral side of tail is up (Figure 3C).
-
15.
Inject the needle into the vein at the site of 2 cm from the tip of the tail.
CRITICAL: If you are not sure that you can succeed at the first time, it is strongly recommended to inject from the tip of the tail. Once the needle is in the wrong position, adjust it to the upper position and continue to inject.
CRITICAL: Turn the needle to keep the bevel points up and make the scales on the syringe barrel visible.
-
16.
Once you see a blood backflow into the needle, press on plunger of the syringe evenly (Figure 3D).
CRITICAL: The plunger usually moves smoothly. But if the plunger does not move easily, remove the needle and reinsert it up to the previous injection site and continue. If all of the attempt is made, try the opposite side of the tail.
CRITICAL: If the plunger stops moving before the injection has been finished, this mouse should be removed from your experiment. Because this delivery will not be efficient as others.
-
17.
Control the whole injection time within 4–8 s. See Troubleshooting 3.
-
18.
When finished, remove the needle and press on the injection site until the bleeding stops.
-
19.
Repeat steps 7–18 with the next mouse and record the accurate injection volume and date. See Troubleshooting 1.
Note: Several mice without injection should be kept as negative control.
-
20.
Put the mouse to the recovery cage at 37°C until they are recovered to normal activity.
Note: The mice will be recovered within about 30 min after hydrodynamic injection.
-
21.
Monitor the health status of the mice after injection.
Figure 2.
Reagents and devices for hydrodynamic injection
1. Scale; 2. mouse cage and mice; 3. 75% Alcohol; 4. Alcohol pads; 5. Mouse restrainer tube; 6. Timer; 7. DNA solution; 8. Goggles; 9. Mark pen. 10. Sterile paper; 11. 1 mL syringes; 12. 2 mL syringes; 13. anesthetic drug; 14. Heat lamp.
Figure 3.
Detailed procedure of hydrodynamic injection
(A) Picture of 2 mL syringe with a needle from 1 mL syringe.
(B) Picture of mouse under a heat lamp to make its tail vein visualized.
(C) Picture of mouse with its lateral side of tail vein up.
(D) Picture showing blood backflow into the needle at the injection time.
Detection of the efficiency of DNA delivery and insertion
Timing: 2–3 days
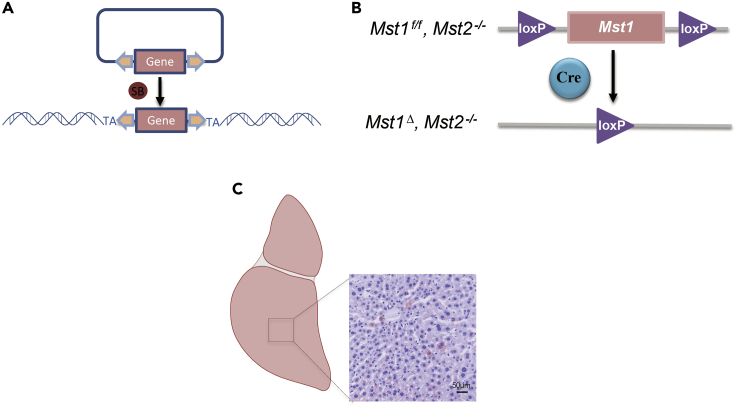
One week after injection, the injected DNA should have inserted into chromosome and express Cre protein to induce knockout of Mst1 (Figures 4A and 4B).
Figure 4.
Schematic diagram of sleeping beauty model inducing tumorigenesis
(A) Transposase SB helps target sequences to randomly integrate into the host genomic DNA.
(B) Expressed Cre in hepatocytes induces knockout of Mst1.
(C) Immunohistochemical analysis of GFP expression in liver tissues of Mst1f/f;Mst2-/-. Scale bar, 50 μm.
Day 1
-
22.
Fill the 1.5 mL sonication tubes with a mix of two kinds of protein extraction beads (five beads each type; see Key Resources Table).
-
23.
Add protease inhibitor cocktail (100× stock) to the cold protein extraction buffer to get a 1× dilution.
-
24.
Add 500 μL cold protein extraction buffer to the tubes with beads.
-
25.
Take two mice from each group (including blank control group) to detect their expression of target genes in liver.
-
26.
Weigh mice and calculate the dosage of anesthetic [10 fold (volume/body weight)]. For example, inject 200 μL anesthetic for a 20 g mouse.
Note: We use 5% chloral hydrate dissolved in ddH2O (m/v) as anesthetic.
-
27.
Inject anesthetic intraperitoneally and wait for 5 min until the mouse is fully anesthetized.
-
28.
Separate the liver from the mouse quickly.
Note: In order to obtain a complete liver tissue, firstly, fully expose the liver by cutting along the mid-abdomen of the mouse, and then cut the connective tissue between the liver and the thoracic diaphragm, stomach and intestines. Dissected tissues can be frozen in liquid nitrogen.
CRITICAL: Minimize the time of tissue collection to prevent protein degradation.
-
29.
Put a piece of the liver tissue into cold 500 μL protein extraction buffer (with fresh proteinase inhibitor cocktail).
-
30.
Extract the proteins with a high-speed tissue homogenizer.
Note: We recommend the speed of 4 m/s with 2 cycles of 15 s on and 15 s off for liver.
CRITICAL: In order to prevent the sample from overheating and protein degradation, put the sample on ice as soon as possible after homogenization.
-
31.
Take 100 μL protein sample and boil it at 100°C for 10 min.
-
32.
Transfer the supernatant to a new 1.5 mL tube and centrifuge all of the samples at 16,800 × g for 10 min at 4°C.
-
33.
Transfer the supernatant containing soluble proteins to a new tube and take an aliquot for protein quantification.
Days 2–3
-
34.
Analyze the expression of Cre, eGFP and Myc-tagged geneX by Western blotting. GAPDH is used as a control (work dilution: 1:10000).
Optional: The expression efficiency also can be detected by immunohistochemistry staining of the livers with GFP antibody (work dilution: 1:200) (Figure 4C).
Intraperitoneal drug injection (optional)
Timing: 6 weeks
-
35.
Prepare sterile drugs and vehicle (control) to be injected.
Note: The drugs are recommended to be stored in sterile multi-dose vials.
Note: Optimize the concentration of drugs before use.
Optional: Prepare drugs that can inhibit liver tumorigenesis as positive control.
-
36.
Load the 1 mL syringe with drugs.
-
37.
Take the mouse (injected with pT3-Myc-P2A-Cre-T2A-EGFP and SB plasmids) from the cage and restrain it gently.
-
38.
Insert needle with bevel point and the numbers on the syringe barrel facing up, and inject the mouse with a volume about 100 μL.
CRITICAL: Control the insertion depth of needle to avoid damage to the abdominal organs.
-
39.
Press the injection site until the bleeding stops.
-
40.
Inject the mouse with drug one by one.
-
41.
Change the syringe with different drugs.
-
42.
Repeat steps 35–41 once a week for 6 weeks (Figure 5).
Note: The frequency and duration of drug injection can be optimized.
Figure 5.
Schematic diagram of sleeping beauty model
Analysis of mouse liver tumors
Timing: 1 day
-
43.
Inject anesthetic intraperitoneally and wait for 5 min until the mouse is fully anesthetized.
-
44.
Weigh and record the whole body of mice.
-
45.
Dissect the liver from the mouse quickly.
CRITICAL: Put the mouse liver in the cold PBS from this step.
-
46.
Weigh the dissected liver and record.
-
47.
Repeat steps 43–46.
-
48.
Count the liver tumors and measure the tumor diameter with calipers.
Note: According to tumor size, tumors are classified into two grades: grade I represents the tumor with a diameter of less than 5 mm, and grade II represents the tumor with a diameter of more than 5mm.
-
49.
Take a picture of the liver with a ruler (Figure 6A).
-
50.
Take a piece of liver for protein sample extraction.
-
51.
Perform statistical analysis of liver-over-body weight ratio and tumor size of each group (Figure 6B).
-
52.
Test the expression of delivered genes by Western blotting.
-
53.
Make paraffin-embedded 5 μm sections using the rest of the liver.
-
54.
Stain the slides with hematoxylin-eosin (H&E) for histologic examination (Figure 6C).
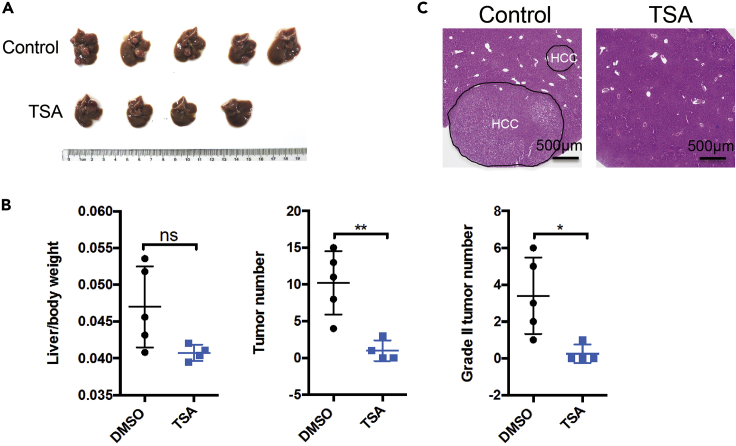
Figure 6.
Analysis of tumorigenesis in mice liver after Trichostatin A treatment
(A) Macroscopic picture of livers gross from DMSO group (n = 5) and Trichostatin A (TSA) group (n = 4).
(B) Statistical analysis of liver-over-body weight, total tumor number and grade II tumor number.
(C) H&E staining analysis of liver tissue. The black dotted line points out the area of liver with hepatocellular carcinoma (HCC). Scale bars, 500 μm.
Expected outcomes
The plasmids delivery should be smooth. One-week post plasmids injection, the expression of GFP will be detected by Western blotting or immunofluorescence staining in each group (Figure 4C). Compared with the blank control group, there should be positive signal (1%–5%) in the injected group of mice. The group of mice without GFP expression can be discarded, because this indicates that the hydrodynamic injection failed. Eighteen-week post plasmids injection, the mice injected with pT3-Myc-P2A-Cre-T2A-eGFP and SB plasmids will show tumor burden in liver, while the mice treated by positive drug show less tumor foci (Figure 6A). H&E analysis will also reveal tumor presence in the liver (Figure 6C). In the meantime, the tumors should also continue to express the genes injected.
Quantification and statistical analysis
Comparisons between groups were analyzed using an unpaired Student’s t test by GraphPad Prism. Differences are considered statistically significant at ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. ns means no significance. All data were presented as mean ± SEM.
Limitations
This procedure requires skilled operators to conduct hydrodynamic injection, or it will result in uneven tumor formation of mouse from the same group. See Troubleshooting 5.
Hydrodynamic transfection delivers target DNA in a low percentage of hepatocytes (approximately 1%–5%). Thus, it will take a long time to induce tumor formation.
Although liver is the main organ for plasmid DNA uptake after hydrodynamic tail vein injections, about 0.1% DNA will be also delivered in other organs, such as heart, lung, kidney.
Troubleshooting
Problem 1
Tail vein injection is not smooth (hydrodynamic injection, steps 15 and 16)
Potential solution
We strongly recommend that the users practice the operation of hydrodynamic injection for several times before start. Following the step 12 by placing the tail of the mouse under a heat lamp until the tail vein is visualized will also be helpful.
Problem 2
The mouse is dead quickly after injection (hydrodynamic injection, step 21)
Potential solution
There are several reasons that cause the mouse dead after injection.
Reason 1, push the syringe too fast: The large volume of DNA solution will enter directly into the inferior vena cava, which will lead to cardiac congestion and drive the solution into the liver in retrograde. Injection time less than 4 s or uneven force will lead to serious heart damage and death of mouse. So injecting the DNA sample into the tail vein within 4–8 s with even force is recommended.
Reason 2, the plasmids are not “sterile and endotoxin free”: Use the kit recommended to extract plasmids, and keep aseptic operations during injection.
Reason 3, incomplete removal of the air from the syringe. Air entering the vein may cause air embolism, so it is important to follow step 10.
Problem 3
There is no tumorigenesis after 18 weeks from injection (analysis of mouse liver tumors, step 48)
Potential solution
Normally, there will be visible tumor foci in mouse liver 14 weeks after injection. At 18 weeks, the tumor grows to a diameter of 5 mm–8 mm. If no obvious tumor is observed in mouse liver at 18 weeks after injection, it may be due to no or low expression of Cre, so it is necessary to detect the gene expression level prior to injection. In addition, it may be due to the low efficiency of plasmid delivery and insertion in genome. Follow the “4–8 s” principle to enhance the delivery efficiency when give an injection. Besides, the users could also extent the time of tumorigenesis, for example, analyze the liver tumor 22 weeks after injection.
Problem 4
The drug used as positive control does not inhibit tumorigenesis (intraperitoneal drug injection, steps 35–42)
Potential solution
Even though the results are stable by using this system to screen drugs, it is still a bit risky assessing the therapeutic effect by sacrificing mice after drug treatment. To overcome this, we encourage the users to visualize the tumors during the stage of drug treatment to confirm the volume of tumors. To achieve this, we recommend ultrasound imaging with a Vevo 3100 scanner (FUJIFILM Visualsonics, Toronto, ON, Canada) (Fagerland et al., 2020). Besides, the users also can choose the fluorescent imaging by using an IVIS imaging system (PerkinElmer). Thus, the tumor expressed with GFP will be detective. Please note that the users need to optimize the protocols to get a reliable result. Based on this, increasing the frequency and duration of drug treatment will reduce failure rate.
Problem 5
The tumor development of mice within the group exhibits a large difference (analysis of mouse liver tumors, step 48)
Potential solution
Within the same group, differed age and weight of the mice will greatly affect the uniformity of tumor development. Besides, the plasmid was injected proportionally according to the weight of the mouse, but the efficiency of DNA delivery will not increase if the weight of mouse is over 25 g, so choose the mouse from similar age and weight less than 25 g. Secondly, the duration for the hydrodynamic injection of each mouse is unstable, resulting in inconsistent efficiency of gene delivery and insertion in genome. In order to reduce the difference of tumor burden between each mouse within the group, it is recommended that the users finish the hydrodynamic injection within a consistent time.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lei Zhang (rayzhang@sibcb.ac.cn).
Materials availability
Plasmids generated in this study have been deposited to http://www.brics.ac.cn/plasmid/template/plasmid/plasmid_list.html. Please see the catalog number SP-2939 for pT3-Myc-P2A-Cre-T2A-EGFP and catalog number SP-2940 for pCMV-Transposase (SB).
Data and code availability
This study did not generate any unique datasets or code.
Acknowledgments
We thank Wufan Tao for Mst1f/f; Mst2-/- mice. We also thank Lijian Hui for providing the pT3 and SB plasmids. This work is supported by the National Key Research and Development Program of China (grants 2019YFA0802001 and 2017YFA0103601 to L.Z.), National Natural Science Foundation of China (grants 32030025, 31530043, and 31625017 to L.Z.), ‘‘Strategic Priority Research Program’’ of Chinese Academy of Sciences (grant XDB19000000 to L.Z.), ‘‘Shanghai Leading Talents Program’’ (L.Z.), and China Postdoctoral Science Foundation (grant 2020M680059 to L.H.).
Author contributions
L.H. optimized the protocol and wrote the manuscript. W.Y. optimized the protocol. Y.L., J.X., and W.Z. edited the manuscript. L.H. and L.Z. supervised the project.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100445.
Contributor Information
Lingli He, Email: helingli2015@sibcb.ac.cn.
Lei Zhang, Email: rayzhang@sibcb.ac.cn.
References
- Du X., Shi H., Li J., Dong Y., Liang J., Ye J., Kong S., Zhang S., Zhong T., Yuan Z. Mst1/Mst2 regulate development and function of regulatory T cells through modulation of Foxo1/Foxo3 stability in autoimmune disease. J. Immunol. 2014;192:1525–1535. doi: 10.4049/jimmunol.1301060. [DOI] [PubMed] [Google Scholar]
- Fagerland S.-M.T., Berg S., Hill D.K., Snipstad S., Sulheim E., Hyldbakk A., Kim J., Davies C.d.L. Ultrasound-mediated delivery of chemotherapy into the transgenic adenocarcinoma of the mouse prostate model. Ultrasound Med. Biol. 2020;46:3032–3045. doi: 10.1016/j.ultrasmedbio.2020.07.004. [DOI] [PubMed] [Google Scholar]
- He L., Yuan L., Yu W., Sun Y., Jiang D., Wang X., Feng X., Wang Z., Xu J., Yang R. A regulation loop between YAP and NR4A1 balances cell proliferation and apoptosis. Cell Rep. 2020;33:108284. doi: 10.1016/j.celrep.2020.108284. [DOI] [PubMed] [Google Scholar]
- Hong L., Cai Y., Jiang M., Zhou D., Chen L. The Hippo signaling pathway in liver regeneration and tumorigenesis. Acta Biochim. Biophys. Sin. 2015;47:46–52. doi: 10.1093/abbs/gmu106. [DOI] [PubMed] [Google Scholar]
- Lu L., Li Y., Kim S.M., Bossuyt W., Liu P., Qiu Q., Wang Y., Halder G., Finegold M.J., Lee J.S. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. U S A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Mak K.K., Topol L., Yun K., Hu J., Garrett L., Chen Y., Park O., Chang J., Simpson R.M. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc. Natl. Acad. Sci. U S A. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Conrad C., Xia F., Park J.S., Payer B., Yin Y., Lauwers G.Y., Thasler W., Lee J.T., Avruch J. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.