Abstract
During embryonic development, neural stem/progenitor cells generate hundreds of different cell types through the combination of intrinsic and extrinsic cues. Recent data obtained in mouse and human cortical neurogenesis provide novel views about this interplay and how it evolves with time, whether during irreversible cell fate transitions that neural stem cells undergo to become neurons, or through gradual temporal changes of competence that lead to increased neuronal diversity from a common stem cell pool. In each case the temporal changes result from a dynamic balance between intracellular states and extracellular signalling factors. The underlying mechanisms are mostly conserved across species, but some display unique features in human corticogenesis, thereby linking temporal features of neurogenesis and human brain evolution.
Current Opinion in Neurobiology 2021, 66:195–204
This review comes from a themed issue on Developmental neuroscience
Edited by Denis Jabaudon and Alain Chédotal
For a complete overview see the Issue and the Editorial
Available online 5th January 2021
https://doi.org/10.1016/j.conb.2020.12.006
0959-4388/© 2020 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
The cerebral neocortex is parcellated into numerous layers and areas, each comprising dozens of distinct types of neurons defined by specific patterns of gene expression, morphology, and most importantly, hodological properties [1, 2, 3, 4, 5]. It is no surprise then that the developing cortex has long been a fascinating model to study the mechanisms that control how to generate neurons and specify their diverse identities.
Cortical neurogenesis starts as the neuroepithelial cells, the primordial stem cells at the origin of all cortical neurons, transform into radial glial cells (RGC) - the major type of stem/progenitor cells that also serve as scaffolds for neuronal migration [6, 7, 8, 9]. Cortical pyramidal neurons are then produced, either directly from the RGC, or indirecty via additional progenitor populations, the intermediate progenitor cells (IPC) present abundantly in the cortex of all mammals, or the outer radial glia cells (oRGC), particularly amplified in those with expanded cortex such as the anthropoid primates [7,10].
A fundamental feature of cortical neurogenesis is temporal patterning [11]. This means that RGC change of competence to produce sequentially different types of neurons over the course of corticogenesis. As a result, the first neurons generated from RGC will populate the deepest layers of the cortex and express specific gene expression profiles and hodological properties, while those that are born at later stages of corticogenesis will acquire different identities, corresponding to progressively more superficial layers, together with distinct gene expression and connectivity patterns [12,13].
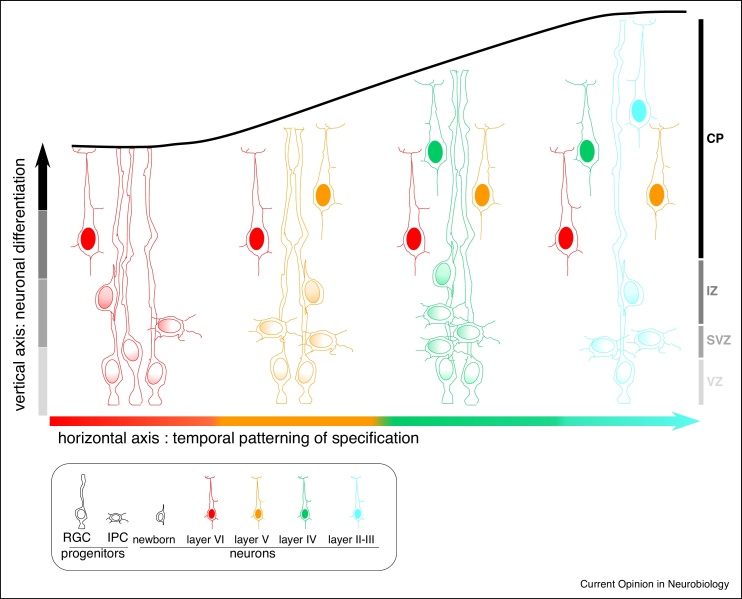
In this frame it can be useful to conceptualize corticogenesis as a timely progression along two orthogonal temporal axes (Figure 1), that RGC follow to undergo two main fate transitions: one the one hand, neuronal generation (‘vertical’ axis of neurogenesis), and on the other hand neuronal subtype specification (‘horizontal’ axis of temporal patterning of specification). Along the vertical axis of differentiation, RGC undergo major fate switches to become IPC and/or neurons, followed by further differentiation/maturation of postmitotic neurons. Along the ‘horizontal’ axis of temporal patterning, RGC and their progeny change their competence as embryonic development proceeds, thereby generating distinct types of neurons that will populate specific cortical layers.
Figure 1.
Two orthogonal temporal axes to define the corticogenesis process.
Radial glial cells (RGC) differentiate into neurons directly or indirectly through the generation of progenitor subpopulations such as intermediate progenitors (IPC), defining a ‘vertical’ temporal axis of neurogenesis. Another ‘horizontal’ temporal axis is defined by temporal changes taking place in RGC and their progeny over embryonic development, leading to the sequential generation of distinct neuronal subtypes forming the different cortical layers.
The vertical temporal axis of neurogenesis: from mitosis towards irreversible neuronal fate commitment
The first axis of neurogenesis starts as a dramatic change in cell identity, by which RGC stop self-renewing and dividing, to convert eventually to postmitotic neurons. This process was captured in real-time at the cellular level almost twenty years ago using organotypic slices of the mouse developing cortex [9], and involves a complex cellular metamorphosis, including delamination of the cell, a transient loss of its epithelial polarity, followed by initiation of radial migration and growth of neuritic processes. Recently, single-cell RNA sequencing technologies led to reconstruct this massive fate transition process at the transcriptional level [14,15]. However, the logic linking the molecular and cellular events still remains to be fully uncovered. In other words how does the structure of the underlying developmental gene regulatory network actually controls the neurogenic transition?
Neurogenesis is regulated by a complex interplay between extrinsic and intrinsic cues acting on neural progenitors, which control the balance between their differentiation and self-renewal. The proneural transcription factors, most strikingly Neurogenins in the cerebral cortex, constitute the key intrinsic factors promoting neurogenesis, through reciprocal antagonism with the Notch pathway [16,17]. On the other hand, most classical morphogens, including Wnts [18, 19, 20, 21], Sonic Hedgehog [22,23], and Fibroblast Growth factors [24, 25, 26], act as extrinsic cues that globally tend to favour self-renewal over neurogenesis (reviewed in Ref. [27]) (Figure 2). Adding another layer of complexity is the fact that each round of RGC division can be symmetrical, whether it is proliferative or differentiating, or asymmetrical [28,29]. Altogether this interplay aims at generating neurons in the right number and type, through the timed balance of differentiation versus self-renewal.
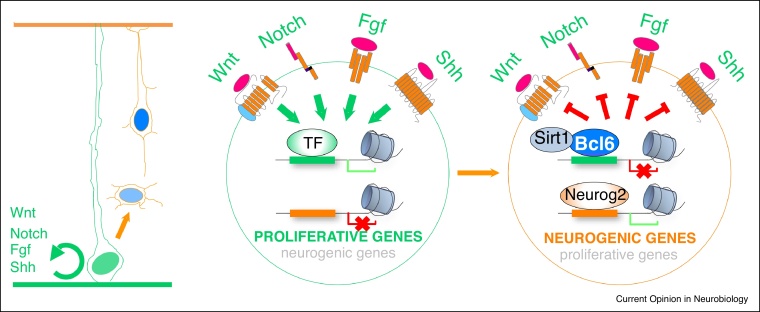
Figure 2.
Neurogenesis and the balance of extrinsic and intrinsic cues.
During differentiation, progenitors become insulated from extracellular proliferative cues (such as Notch, Wnts, FGFs, SHH), to ensure an irreversible commitment towards neuronal identity. Mechanistically, the switch from proliferation to differentiation of cortical progenitors is promoted by proneural factors like Neurogenin 2, together with Bcl6 repressor activity that ensures the insulation from the extrinsic proliferative cues.
One fundamental question is therefore: what drives the switch from a proliferative behaviour of cortical progenitor cells to cell-cycle exit and the terminal differentiation of neuronal progeny? Intriguingly, it has been long proposed that postmitotic cells ongoing neuronal differentiation become insulated from extrinsic signaling [30], which could ensure irreversible conversion to neuronal fate despite the presence of morphogen cues. But this has remained largely hypothetical and the molecular mechanisms have remained unknown.
The extrinsic cues that act on cortical progenitors to promote expansion and self-renewal come from many different sources (Figure 2): these include in particular the embryonic cerebrospinal fluid through secretion from the choroid plexus [31], but also cortical progenitors and neurons themselves in the cortical plate [27]. Thus, while the delamination of the progenitors outside their niche in the ventricular zone might partly enable them to escape from the influence of these cues, one would predict additional mechanisms by which differentiating cortical cells become insulated from these proliferative pathways. The Bcl6 transcriptional repressor was recently shown to fulfill this role during mouse cortical neurogenesis [32•]. Bcl6 was previously identified as a proneurogenic factor, acting as potently as conventional proneural factors to convert RGC into neurons in vitro, through direct repression of Notch downstream effectors Hes genes [33]. Recently a more complete spectrum of Bcl6 targets was determined by combining transcriptome and ChIP-seq analyses downstream of Bcl6 in cortical cells [32•]. This led to the surprising finding that Bcl6 acts a potent and direct transcriptional repressor of a selective repertoire of genes belonging to signaling components of all extrinsic cues known to promote self-renewal: Notch, Shh, Fgf/Igf, and most strikingly the Wnt pathway. Bcl6 appears to target genes corresponding to all levels of signalling, from morphogen ligands and receptors to downstream effectors, including some, such as Cyclind1/2 genes, that act as critical effectors at the convergence of all these pathways (Figure 2). These data suggest a model whereby Bcl6 ensures irreversible differentiation initiated by proneural factors, by making the differentiating cells unable to respond to the cues that would maintain them in a proliferative state (Figure 2). At the molecular level, Bcl6 repression involves the recruitment of the Sirtuin-1 (Sirt1) deacetylase, followed by specific Histone deacetylation, thereby providing a stable and selective epigenetic repression of Bcl6 targets, leading to differentiation.
Could such a model be generalized to other systems? Bcl6 was also found as a proneurogenic factor in the cerebellum, where it promotes granule neuron fate through the repression of Shh effectors Gli1/2 [34], but on the other hand it is absent from most other brain regions, suggesting that either only some classes of progenitors need such an intrinsic system to undergo irreversible neurogenesis, or that other molecular actors could be at play. These may include the transcription factor Myt1l, recently found to act in a similar way as a pan-repressor of non-neuronal fate during neuronal differentiation and reprogramming [35], but also additional chromatin remodelling factors, such as the BAF chromatin remodeling complexes that repress the Wnt pathway in cortical progenitors undergoing neurogenesis [36].
One puzzling feature of Bcl6 is that it is expressed in progenitors (RGC and IPC) but at even higher levels in developing newborn neurons [33]. One could speculate that this sustained expression in neurons prevents de-differentiation, even though at least some of the mechanisms ensuring terminal differentiation can be imprinted in stem/progenitor cells [37]. Alternatively this could suggest that part of the neurogenic fate commitment process is still occurring after mitosis. Earlier studies suggest that the decision to become a neuron is largely taken before mitosis, in particular during the G1 phase of the mother cell, the length of which can influence the outcome of the next division [38,39]. Could it be that part of the decision to become a neuron is also made after mitosis? This would be coherent with the mechanisms of asymmetric cell fate acquisition that by definition occur postmitotically [28,29].
This issue was examined recently in a different context, involving mitochondria dynamics, the process by which mitochondria undergo fission and fusion, that has recently emerged as an instructive modulator during cell fate decisions in many systems [40, 41, 42]. Using live cell imaging in vitro and a novel system of high-throughput tracking of mitotic RGC, it was found that right after RGC have divided, their daughter cells can display a dual pattern of mitochondria dynamics, which is tightly correlated with cell fate: cells that will become neurons display high levels of mitochondria fission, while those that will undergo self-renewal as cortical stem cells will undergo rapid mitochondria fusion [43•]. Most importantly, the forced induction of mitochondria fusion shortly after mitosis, whether in vitro or in the mouse embryonic cortex in vivo, alters the balance of cell fate, leading most RGC to undergo self-renewal instead of neuronal differentiation. These data identify a postmitotic critical period of plasticity during which fate switch can still occur, thus revising the classical view that the decision for neural stem cells to commit to neuronal fate precedes mitosis [30,38]. Interestingly the effect of mitochondria on neurogenesis appears to be mediated in part through the NAD+ sensor Sirt1, also involved in Bcl6-mediated neurogenesis as above mentioned. It will be interesting to test further the implied model that the final, postmitotic, commitment to neuronal fate occurs as a result of the coincident detection of metabolic signals from the mitochondria and nuclear gene expression.
The horizontal axis of neurogenesis: temporal patterning of competence, commitment, and plasticity
These data overall point to the importance of the interplay of intrinsic and extrinsic components along the ‘vertical’ temporal axis of neurogenesis. But how about the second ‘horizontal’ axis, that is, the temporal patterning of cortical neuron subtype specification? It has been debated whether the specification of neurons was either predefined early in progenitors (premitotic model) or defined at the progenitor-neuron transition (postmitotic model) [44]. Although many genes were identified allowing to distinguish neurons of distinct layers [1], it had remained difficult to decipher a temporal transcriptional code in cortical progenitors [45,46]. Single-cell RNA-seq analyses of mouse and human cortical progenitors and neurons at high temporal resolution along the two axes of neurogenesis [14,47•], have now provided substantial evidence for the premitotic model. Indeed single-cell RNA-seq of timed RGC reveal specific temporal patterns of gene expression across different embryonic stages, which then appear to be transmitted to the daughter neurons for further postmitotic refinement [47•]. Importantly, the detected changes are not just bystanders of the temporal patterning, as they can actually influence fate acquisition: for instance the Ezh2 epigenetic factor, found to be expressed at higher levels in early than late RGC, is required for the proper temporal specification of early cortical neurons [47•]. Finally, one core difference between early and late RGC resides in the expression of genes related to extrinsic signalling, which tend to be upregulated in late progenitors, suggesting that late progenitors tend to be more ‘extraverted’, that is, more responsive and exposed to extrinsic cues as cortical development proceeds [47•] (Figure 3).
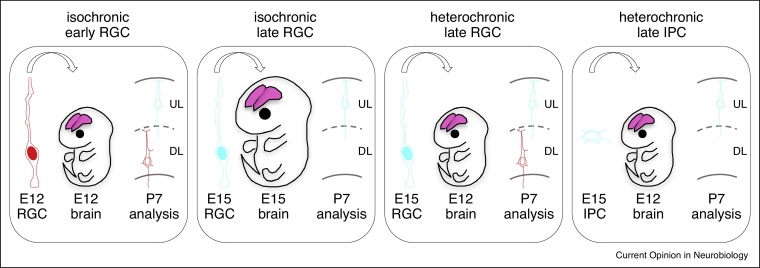
Figure 3.
Timing of fate specification revealed by heterochronic transplantations.
Isochronic and heterochronic transplantation experiments reveal that late radial glial cells can regain their ability to generate early deep-layer neurons while intermediate progenitors have a fixed identity that cannot be respecified by transplantation.
As the differentiation progresses, RGC temporal patterns are then transferred to neurons: these likely include post-transcriptional mechanisms through which genes already transcribed in RGC become only active in neurons by increased stability or translation [48, 49, 50].
In addition and importantly, this nascent developmental program of neuronal specification will be then further refined to drive the neurons towards their final identity [47•], indicating influence of specification cues also on postmitotic neurons. For instance, the final laminar fate acquisition of deep layer neurons may be influenced by Wnt signals emanating from pioneer subplate neurons [51,52], while the identity of neurons of the upper layers can be influenced by their final position within the cortical plate [53]. Thus the final identity of pyramidal neurons of the developing cerebral cortex can still be influenced significantly at postmitotic stages. Interestingly such a mixed premitotic and postmitotic model is also proposed as the source of diversity of the other population of cortical neurons, the GABAergic inhibitory interneurons, despite their distinct progenitor origins in the ganglionic eminences [54,55].
The core program of cortical neuron specification is initiated in the RGC, followed by further refinement in postmitotic neurons. But is the temporal patterning intrinsic to the RGC, or is it driven by extrinsic cues in the ever-changing embryonic brain environment? And secondly, are these changes irreversible, or can for instance the competence of late RGC be rejuvenated to generate normally early born neurons? These questions have long been puzzling the field. On the one hand, in vitro experiments using cortical progenitors derived from the mouse brain [56] or pluripotent stem cells (PSC) [57] demonstrated that cortical progenitors, even cultured as single cells [57], can generate sequentially pyramidal neurons corresponding to all six layers identity (although with a different proportion, biased towards deep layer fates), indicating that the whole process of cortical neuron specification is encoded within the cell lineage. At the other edge of the experimental spectrum, heterochronic transplantations experiments in the ferret had concluded that cortical progenitors could be respecified by the embryonic environment, but in a time-specific way: early progenitors could be respecified following transplantation at later stages, but not the other way around, suggesting that temporal patterning was under extrinsic influence, and this was in part the result of a restriction in the fate potential of late RGC [58,59].
These aspects were recently re-examined using heterochronic transplantation experiments in the mouse, combined with exhaustive characterization of the transplanted cells [60•] (Figure 3). This revealed that the RGC can actually display a higher degree of plasticity than previously thought, as late RGC transplanted in the early mouse cortex could regain competence to generate neurons displaying all cellular and molecular properties of early generated deep layer neurons. But importantly, this plastic behaviour could not be detected among transplanted IPC, consistent with the notion that the commitment to a specific neuronal fate is acquired irreversibly in these cells [61]. As IPC are much more abundant in higher mammals like the ferret cortex, this may explain the discrepancy with earlier experiments, where most of the transplanted cells at late stages might have been committed IPC [58,59]. It would be interesting to test further this hypothesis by performing heterochronic transplantation in the ferret like previously, but followed by a modern analysis of the molecular and hodological properties of the transplanted neurons.
The molecular mechanisms underlying the plasticity of RGC remain to be fully understood, but appear to implicate Wnt signalling, which displays a higher tonus in early RGC, and is required for the respecification of late RGC by the early cortical environment [60•]. Interestingly, effects of the Wnt pathway on cortical neuron laminar specification were also observed in human in vitro systems (in monolayer cultures and organoids), though at later stages, suggesting multiple levels of regulation by this pleiotropic morphogen family [51,62].
Links between the timing of neurogenesis and human cortical evolution
A major biological question related to cortical neurogenesis is human brain evolution. While many of the above-discussed mechanisms are likely to be largely conserved in all mammals, neurogenesis is expected to display species-specific features as well. Indeed the human brain is characterized by an increase in size and complexity of the cerebral cortex, which is thought to be linked in part to changes in the developmental program of neurogenesis [10,63,64]. Species differences in cortical neurogenesis have been linked in particular to increased diversity of cortical progenitors, including IPC that are thought to constitute an important drive of cortical expansion in mammals compared with non-mammalian amniotes devoid of neocortex and that also lack IPC [65], and most strikingly the oRGC that display highly increased expansion potential in the human cortex [7,10]. On the other hand, the timing of cortical neurogenesis is considerably prolonged in human corticogenesis - from the initial proliferation of the neuroepithelial cells, to subsequent neurogenesis from RGC, which typically takes about a week in the mouse but almost four months in the human embryonic cortex for instance (Figure 4). This has been proposed as another important mechanism of cortical size increase, by enabling more rounds of self-renewing divisions to occur, and thereby increase neuronal production during corticogenesis. What could be at the origin of these different temporal patterns of neurogenesis across species? Interestingly, the prolonged timescale of human corticogenesis compared with other species appears to be largely conserved in in vitro systems from pluripotent stem cells (PSC), whether using two dimensional monocultures or mixed cultures of cortical progenitors [66,67]. Even more strikingly, early human cortical progenitors transplanted in the mouse neonatal cortex will only start to generate upper-layer neurons several weeks following transplantation, thus following their own scale of temporal patterning, suggesting that intrinsic cues may be central to control the timescale of cortical neurogenesis [66]. Several comparative genomic, epigenomic and transcriptomic analyses have revealed divergent gene expression patterns in the developing human cortex compared with non-human primate species, using fetal tissue and PSC-derived organoids [68•,69•,70, 71, 72]. These studies nicely documented a prolonged developmental time line in human cortical cells, thus confirming at the gene expression and chromatin levels what was known at the cellular level. They have also started to reveal species-specific gene regulatory mechanisms that could underlie some of the characteristics of human corticogenesis, such as the specific upregulation of the mTOR pathway in human cortical progenitors (in particular oRGC) compared with non-human primate counterparts [68•], which may underlie some of their unique expansion capacities.
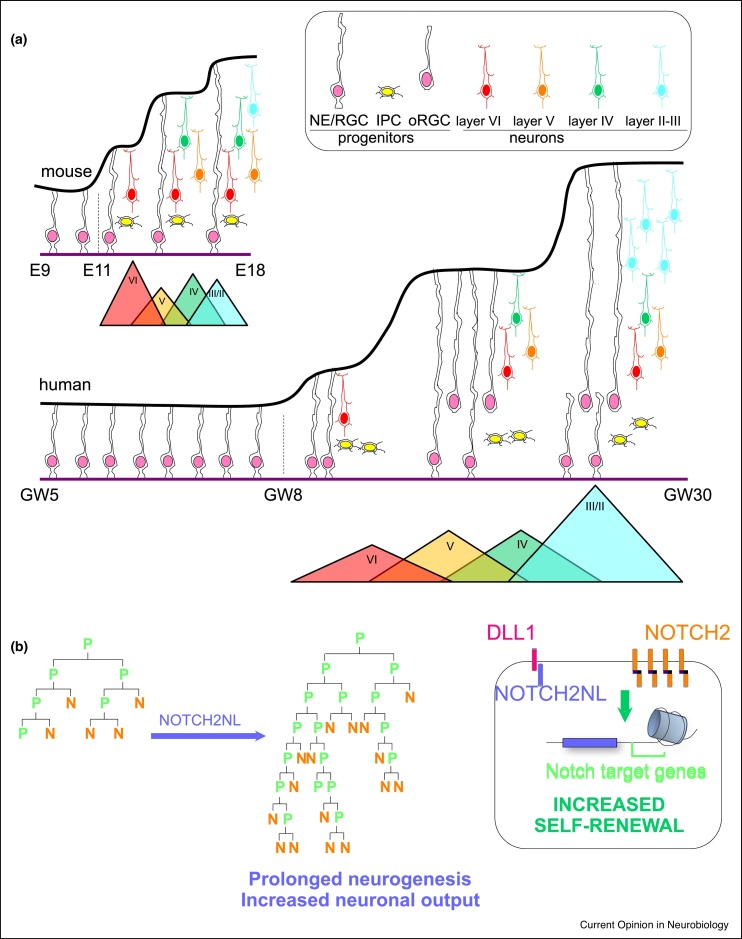
Figure 4.
Species differences in the developmental timing of cortical neurogenesis.
(a) Cortex expansion may rely on increased number and diversity of progenitor subpopulations (IPC: intermediate progenitor cells and oRGC: outer radial glial cells), as well as protacted steps of initial proliferation of stem/progenitor cells (NE: neuroepithelial cells, RGC: radial glial cells) and neurogenesis. (b) The latter is partly controlled by the human-specific NOTCH2NL genes that increase the self-renewal of cortical progenitors and their subsequent neuron output, by stimulating Notch signaling through a cell-autonomous mechanism.
Moreover, highly conserved pathways may be modified in a species-specific way to allow more prolonged patterns of neurogenesis. This is exemplified by recent studies exploring the impact of the NOTCH2NL gene family [73•,74•,75] (Figure 4). These comprise four hominid-specific genes (NOTCH2NLA/B/C/D), three of which are human-specific, which emerged very recently as segmental, partial duplications of the ancestral NOTCH2 gene [73•,74•]. Two of the NOTCH2NL paralogs (A/B) are highly expressed in human cortical progenitors (both RGC and oRGC). Furthermore they are found within a specific chromosomal interval of 1q21.1, close to an intriguing locus highly enriched in human-specific genes, and where copy number variants (CNV) are associated with pathological changes in brain size, with microduplications associated with macrocephaly, and microdeletions associated with microcephaly [76].
Functional studies with PSC-derived human cortical cells, taking advantage of clonal analyses, revealed that NOTCH2NL paralogs significantly expand human cortical progenitors by extending their self-renewal, which ultimately results in an increased neuronal production at the clonal level [74•]. The impact of NOTCH2NL genes thus strikingly parallels the expected features of human corticogenesis compared with non-human primates, that is, prolonged self-renewal and increased neuronal output. Conversely, loss of function of two of the NOTCH2NL paralogs resulted in accelerated development of neural organoids [73•]. Mechanistically, NOTCH2NL gene products interact directly with the Notch pathway and increase Notch signaling in a cell-autonomous fashion. Finally, fine mapping of 1q21.1 microdeletions and microduplications found in patients with microcephaly and macrocephaly further revealed that the recombination breakpoints were located at the level of the same two NOTCH2NLA/B genes [73•]. Overall these findings point to the role of human-specific, cell-intrinsic modifiers of the highly conserved Notch pathway that may be in part responsible for the expansion of human cortical neurogenesis, and thereby increased cortical size.
Additional timing mechanisms beyond human-specific genes must distinguish human neurogenesis form other species. For instance, when examining the impact of mitochondria on RGC fate choice as discussed above [43•], the critical period of postmitotic fate plasticity was found to be doubled in the human compared with the mouse (6 hours instead of 3 hours). While the significance of these findings remains to be explored further, it is tempting to speculate that the observed differences in mitochondria-dependent fate plasticity may be linked to species-specific developmental timing of cortical neurogenesis. This would echo with known species differences in global metabolic rates, that are so far linked to differences in life span and aging [77], and with species differences in protein turnover that have been recently linked to mouse versus human differences in developmental timing of motor neuron differentiation and somite segmentation clock [78, 79, 80].
As a conclusion, cortical neurogenesis articulates along two major temporal axes of neuronal differentiation and specification, which result from the close interplay of intrinsic and extrinsic cues in both neural progenitors and their differentiated progeny. A major challenge will be to extract new mechanistic hypotheses from the massive amount of molecular data now available from deep-sequencing studies in different species and systems [81], and to test these functionally in vivo and using cellular models of corticogenesis, in order to elucidate the molecular logic linking gene expression, cell dynamics, and fate acquisition.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
Acknowledgements
We wish to apologise to the authors whose work could not be discussed due to space constraints. Work from the authors described here was funded by the European Research Council (ERC Adv Grant GENDEVOCORTEX), the EOS Programme, the Fondation ROGER DE SPOELBERCH, the Belgian FWO, the Belgian FRS/FNRS, the WELBIO Programme, the AXA Research Fund, the Belgian Queen Elizabeth Foundation, and the Fondation ULB.
Contributor Information
Jérôme Bonnefont, Email: jerome.bonnefont@ulb.ac.be.
Pierre Vanderhaeghen, Email: pierre.vanderhaeghen@kuleuven.vib.be.
References
- 1.Greig L.C., Woodworth M.B., Galazo M.J., Padmanabhan H., Macklis J.D. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Economo M.N., Viswanathan S., Tasic B., Bas E., Winnubst J., Menon V., Graybuck L.T., Nguyen T.N., Smith K.A., Yao Z. Distinct descending motor cortex pathways and their roles in movement. Nature. 2018;563:79–84. doi: 10.1038/s41586-018-0642-9. [DOI] [PubMed] [Google Scholar]
- 3.Tasic B., Yao Z., Graybuck L.T., Smith K.A., Nguyen T.N., Bertagnolli D., Goldy J., Garren E., Economo M.N., Viswanathan S. Shared and distinct transcriptomic cell types across neocortical areas. Nature. 2018;563:72–78. doi: 10.1038/s41586-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saunders A., Macosko E.Z., Wysoker A., Goldman M., Krienen F.M., de Rivera H., Bien E., Baum M., Bortolin L., Wang S. Molecular diversity and specializations among the cells of the adult mouse brain. Cell. 2018;174:1015–1030. doi: 10.1016/j.cell.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeisel A., Hochgerner H., Ernfors P., Marklund U., Linnarsson S., Lö P., Johnsson A., Memic F., Van Der Zwan J., Hä Ring M. Molecular architecture of the mouse nervous system resource molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kriegstein A., Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrell V., Götz M. Role of radial glial cells in cerebral cortex folding. Curr Opin Neurobiol. 2014;27:39–46. doi: 10.1016/j.conb.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Malatesta P., Hartfuss E., Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 9.Noctor S.C., Flint A.C., Weissman T.A., Dammerman R.S., Kriegstein A.R. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 10.Lui J.H., Hansen D.V., Kriegstein A.R. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohwi M., Doe C.Q. Temporal fate specification and neural progenitor competence during development. Nat Rev Neurosci. 2013;14:823–838. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberst P., Agirman G., Jabaudon D. Principles of progenitor temporal patterning in the developing invertebrate and vertebrate nervous system. Curr Opin Neurobiol. 2019;56:185–193. doi: 10.1016/j.conb.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Rossi A.M., Fernandes V.M., Desplan C. Timing temporal transitions during brain development. Curr Opin Neurobiol. 2017;42:84–92. doi: 10.1016/j.conb.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowakowski T.J., Bhaduri A., Pollen A.A., Alvarado B., Mostajo-Radji M.A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S.J., Velmeshev D. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telley L., Govindan S., Prados J., Stevant I., Nef S., Dermitzakis E., Dayer A., Jabaudon D. Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science. 2016;351:1443–1446. doi: 10.1126/science.aad8361. [DOI] [PubMed] [Google Scholar]
- 16.Guillemot F., Hassan B.A. Beyond proneural: emerging functions and regulations of proneural proteins. Curr Opin Neurobiol. 2017;42:93–101. doi: 10.1016/j.conb.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Kageyama R., Ohtsuka T., Shimojo H., Imayoshi I. Dynamic regulation of Notch signaling in neural progenitor cells. Curr Opin Cell Biol. 2009;21:733–740. doi: 10.1016/j.ceb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Munji R.N., Choe Y., Li G., Siegenthaler J.A., Pleasure S.J. Wnt signaling regulates neuronal differentiation of cortical intermediate progenitors. J Neurosci. 2011;31:1676–1687. doi: 10.1523/JNEUROSCI.5404-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou C.J., Borello U., Rubenstein J.L., Pleasure S.J. Neuronal production and precursor proliferation defects in the neocortex of mice with loss of function in the canonical Wnt signaling pathway. Neuroscience. 2006;142:1119–1131. doi: 10.1016/j.neuroscience.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Chenn A., Walsh C.A. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 21.Mutch C.A., Funatsu N., Monuki E.S., Chenn A. Beta-catenin signaling levels in progenitors influence the laminar cell fates of projection neurons. J Neurosci. 2009;29:13710–13719. doi: 10.1523/JNEUROSCI.3022-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rash B.G., Grove E.A. Patterning the dorsal telencephalon: a role for sonic hedgehog? J Neurosci. 2007;27:11595–11603. doi: 10.1523/JNEUROSCI.3204-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L., Hou S., Han Y.G. Hedgehog signaling promotes basal progenitor expansion and the growth and folding of the neocortex. Nat Neurosci. 2016;19:888–896. doi: 10.1038/nn.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raballo R., Rhee J., Lyn-Cook R., Leckman J.F., Schwartz M.L., Vaccarino F.M. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J Neurosci. 2000;20:5012–5023. doi: 10.1523/JNEUROSCI.20-13-05012.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rash B.G., Lim H.D., Breunig J.J., Vaccarino F.M. FGF signaling expands embryonic cortical surface area by regulating notch-dependent neurogenesis. J Neurosci. 2011;31:15604–15617. doi: 10.1523/JNEUROSCI.4439-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahara S., O’Leary D.D. Fgf10 regulates transition period of cortical stem cell differentiation to radial glia controlling generation of neurons and basal progenitors. Neuron. 2009;63:48–62. doi: 10.1016/j.neuron.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tiberi L., Vanderhaeghen P., van den Ameele J. Cortical neurogenesis and morphogens: diversity of cues, sources and functions. Curr Opin Cell Biol. 2012;24:269–276. doi: 10.1016/j.ceb.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Delaunay D., Kawaguchi A., Dehay C., Matsuzaki F. Division modes and physical asymmetry in cerebral cortex progenitors. Curr Opin Neurobiol. 2017;42:75–83. doi: 10.1016/j.conb.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Lancaster M.A., Knoblich J.A. Spindle orientation in mammalian cerebral cortical development. Curr Opin Neurobiol. 2012;22:737–746. doi: 10.1016/j.conb.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edlund T., Jessell T.M. Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell. 1999;96:211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- 31.Fame R.M., Lehtinen M.K. Emergence and developmental roles of the cerebrospinal fluid system. Dev Cell. 2020;52:261–275. doi: 10.1016/j.devcel.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Bonnefont J., Tiberi L., van den Ameele J., Potier D., Gaber Z.B., Lin X., Bilheu A., Herpoel A., Velez Bravo F.D., Guillemot F. Cortical neurogenesis requires Bcl6-mediated transcriptional repression of multiple self-renewal-promoting extrinsic pathways. Neuron. 2019;103:1096–1108. doi: 10.1016/j.neuron.2019.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that the insulation of cortical progenitors from extracellular proliferative cues by transcriptional repression ensures irreversible commitment towards neuronal fate.
- 33.Tiberi L., Van Den Ameele J., Dimidschstein J., Piccirilli J., Gall D., Herpoel A., Bilheu A., Bonnefont J., Iacovino M., Kyba M. BCL6 controls neurogenesis through Sirt1-dependent epigenetic repression of selective Notch targets. Nat Neurosci. 2012;15:1627–1635. doi: 10.1038/nn.3264. [DOI] [PubMed] [Google Scholar]
- 34.Tiberi L., Bonnefont J., van den Ameele J., Le Bon S.D., Herpoel A., Bilheu A., Baron B.W.B.W., Vanderhaeghen P. A BCL6/BCOR/SIRT1 complex triggers neurogenesis and suppresses medulloblastoma by repressing sonic hedgehog signaling. Cancer Cell. 2014;26:797–812. doi: 10.1016/j.ccell.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 35.Mall M., Kareta M.S., Chanda S., Ahlenius H., Perotti N., Zhou B., Grieder S.D., Ge X., Drake S., Euong Ang C. Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature. 2017;544:245–249. doi: 10.1038/nature21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen H., Kerimoglu C., Pirouz M., Pham L., Kiszka K.A., Sokpor G., Sakib M.S., Rosenbusch J., Teichmann U., Seong R.H. Epigenetic regulation by BAF complexes limits neural stem cell proliferation by suppressing Wnt signaling in late embryonic development. Stem Cell Rep. 2018;10:1734–1750. doi: 10.1016/j.stemcr.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mora N., Oliva C., Fiers M., Ejsmont R., Soldano A., Zhang T.T., Yan J., Claeys A., De Geest N., Hassan B.A. A temporal transcriptional switch governs stem cell division, neuronal numbers, and maintenance of differentiation. Dev Cell. 2018;45:53–66. doi: 10.1016/j.devcel.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 38.Dehay C., Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 39.Salomoni P., Calegari F. Cell cycle control of mammalian neural stem cells: Putting a speed limit on G1. Trends Cell Biol. 2010;20:233–243. doi: 10.1016/j.tcb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Pernas L., Scorrano L. Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu Rev Physiol. 2016;78:505–531. doi: 10.1146/annurev-physiol-021115-105011. [DOI] [PubMed] [Google Scholar]
- 41.Khacho M., Clark A., Svoboda D.S., Azzi J., MacLaurin J.G., Meghaizel C., Sesaki H., Lagace D.C., Germain M., Harper M.E. Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Cell Stem Cell. 2016;19:232–247. doi: 10.1016/j.stem.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 42.Beckervordersandforth R., Ebert B., Schäffner I., Moss J., Fiebig C., Shin J., Moore D.L., Ghosh L., Trinchero M.F., Stockburger C. Role of mitochondrial metabolism in the control of early lineage progression and aging phenotypes in adult hippocampal neurogenesis. Neuron. 2017;93:560–573. doi: 10.1016/j.neuron.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Iwata R., Casimir P., Vanderhaeghen P. Mitochondrial dynamics in postmitotic cells regulate neurogenesis. Science. 2020;862:858–862. doi: 10.1126/science.aba9760. [DOI] [PubMed] [Google Scholar]; This paper demonstrates that cortical neurogenesis is controlled by mitochondria dynamics shortly after the last neural stem cell division, thus identifying an unexpected postmitotic critical period of fate plasticity.
- 44.Telley L., Jabaudon D. A mixed model of neuronal diversity. Nature. 2018;555:452–454. doi: 10.1038/d41586-018-02539-4. [DOI] [PubMed] [Google Scholar]
- 45.Kawaguchi A., Ikawa T., Kasukawa T., Ueda H.R., Kurimoto K., Saitou M., Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- 46.Okamoto M., Miyata T., Konno D., Ueda H.R., Kasukawa T., Hashimoto M., Matsuzaki F., Kawaguchi A. Cell-cycle-independent transitions in temporal identity of mammalian neural progenitor cells. Nat Commun. 2016;7:11349. doi: 10.1038/ncomms11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Telley L., Agirman G., Prados J., Amberg N., Fièvre S., Oberst P., Bartolini G., Vitali I., Cadilhac C., Hippenmeyer S. Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science. 2019;364:eaav2522. doi: 10.1126/science.aav2522. [DOI] [PubMed] [Google Scholar]; This paper identifies temporal modules of gene expression in radial glial cells that instruct the specification of cortical neurons, thereby unveiling part of the long-sought transcriptional network for cortical temporal patterning.
- 48.Yoon K.J., Ringeling F.R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., Su Y., Kim N.S., Zhu Y., Zheng L. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell. 2017;171:877–889. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zahr S.K., Yang G., Kazan H., Borrett M.J., Yuzwa S.A., Voronova A., Kaplan D.R., Miller F.D. A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron. 2018;97:520–537. doi: 10.1016/j.neuron.2017.12.045. [DOI] [PubMed] [Google Scholar]
- 50.Nowakowski T.J., Rani N., Golkaram M., Zhou H.R., Alvarado B., Huch K., West J.A., Leyrat A., Pollen A.A., Kriegstein A.R. Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development. Nat Neurosci. 2018;21:1784–1792. doi: 10.1038/s41593-018-0265-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozair M.Z., Kirst C., van den Berg B.L., Ruzo A., Rito T., Brivanlou A.H. hPSC modeling reveals that fate selection of cortical deep projection neurons occurs in the subplate. Cell Stem Cell. 2018;23:60–73. doi: 10.1016/j.stem.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 52.Ohtaka-Maruyama C., Okamoto M., Endo K., Oshima M., Kaneko N., Yura K., Okado H., Miyata T., Maeda N. Synaptic transmission from subplate neurons controls radial migration of neocortical neurons. Science. 2018;360:313–317. doi: 10.1126/science.aar2866. [DOI] [PubMed] [Google Scholar]
- 53.Oishi K., Nakagawa N., Tachikawa K., Sasaki S., Aramaki M., Hirano S., Yamamoto N., Yoshimura Y., Nakajima K. Identity of neocortical layer 4 neurons is specified through correct positioning into the cortex. eLife. 2016;5:1–26. doi: 10.7554/eLife.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayer C., Hafemeister C., Bandler R.C., Machold R., Batista Brito R., Jaglin X., Allaway K., Butler A., Fishell G., Satija R. Developmental diversification of cortical inhibitory interneurons. Nature. 2018;555:457–462. doi: 10.1038/nature25999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mi D., Li Z., Lim L., Li M., Moissidis M., Yang Y., Gao T., Hu T.X., Pratt T., Price D.J. Early emergence of cortical interneuron diversity in the mouse embryo. Science. 2018;360:81–85. doi: 10.1126/science.aar6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen Q., Wang Y., Dimos J.T., Fasano C.A., Phoenix T.N., Lemischka I.R., Ivanova N.B., Stifani S., Morrisey E.E., Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 57.Gaspard N., Bouschet T., Hourez R., Dimidschstein J., Naeije G., Van Den Ameele J., Espuny-Camacho I., Herpoel A., Passante L., Schiffmann S.N.S.N. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- 58.McConnell S.K., Kaznowski C.E. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.1925583. [DOI] [PubMed] [Google Scholar]
- 59.Frantz G.D., McConnell S.K. Restriction of late cerebral cortical progenitors to an upper-layer fate. Neuron. 1996;17:55–61. doi: 10.1016/S0896-6273(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 60•.Oberst P., Fièvre S., Baumann N., Concetti C., Bartolini G., Jabaudon D. Temporal plasticity of apical progenitors in the developing mouse neocortex. Nature. 2019;573:370–374. doi: 10.1038/s41586-019-1515-6. [DOI] [PubMed] [Google Scholar]; This paper uses heterochronic transplantation to reveal that late radial glial cells transplanted in earlier embryonic cortex can regain their ability to differentiate into early generated neurons, while intermediate progenitor cells are committed and lack this retrograde plasticity.
- 61.Mihalas A.B., Hevner R.F. Clonal analysis reveals laminar fate multipotency and daughter cell apoptosis of mouse cortical intermediate progenitors. Development. 2018;145:dev164335. doi: 10.1242/dev.164335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qian X., Su Y., Adam C.D., Deutschmann A.U., Pather S.R., Goldberg E.M., Su K., Li S., Lu L., Jacob F. Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell. 2020;26:766–781. doi: 10.1016/j.stem.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sousa A.M.M., Meyer K.A., Santpere G., Gulden F.O., Sestan N. Evolution of the human nervous system function, structure, and development. Cell. 2017;170:226–247. doi: 10.1016/j.cell.2017.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Astick M., Vanderhaeghen P. From human pluripotent stem cells to cortical circuits. Curr Top Dev Biol. 2018;129:67–98. doi: 10.1016/bs.ctdb.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 65.Cárdenas A., Villalba A., de Juan Romero C., Picó E., Kyrousi C., Tzika A.C., Tessier-Lavigne M., Ma L., Drukker M., Cappello S. Evolution of cortical neurogenesis in amniotes controlled by Robo signaling levels. Cell. 2018;174:590–606. doi: 10.1016/j.cell.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Espuny-camacho I., Michelsen K.A., Gall D., Linaro D., Hasche A., Bonnefont J., Bali C., Orduz D., Bilheu A., Herpoel A. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 67.Otani T., Marchetto M.C., Gage F.H., Simons B.D., Livesey F.J. 2D and 3D stem cell models of primate cortical development identify species-specific differences in progenitor behavior contributing to brain size. Cell Stem Cell. 2016;18:467–480. doi: 10.1016/j.stem.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68•.Pollen A.A., Bhaduri A., Andrews M.G., Nowakowski T.J., Meyerson O.S., Mostajo-Radji M.A., Di Lullo E., Alvarado B., Bedolli M., Dougherty M.L. Establishing cerebral organoids as models of human-specific brain evolution. Cell. 2019;176:743–756. doi: 10.1016/j.cell.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes human-specific transcriptional features of corticogenesis, including differentially expressed genes enriched for recent gene duplications and increased mTOR signalling in outer radial glial cells.
- 69•.Kanton S., Boyle M.J., He Z., Santel M., Weigert A., Calleja F.S., Sidow L., Fleck J., Guijarro P., Han D. Single-cell genomic atlas of great ape cerebral organoids uncovers human-specific features of brain development. Nature. 2019;574:418–422. doi: 10.1101/685057. [DOI] [PubMed] [Google Scholar]; Using pluripotent stem cell-derived cerebral organoids from human and non-human primates, this paper describes human-specific gene expression patterns associated with different chromatin accessibility and protracted neurogenesis.
- 70.Amiri A., Coppola G., Scuderi S., Wu F., Roychowdhury T., Liu F., Pochareddy S., Shin Y., Safi A., Song L. Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science. 2018;362:eaat6720. doi: 10.1126/science.aat6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de la Torre-Ubieta L., Stein J.L., Won H., Opland C.K., Liang D., Lu D., Geschwind D.H. The dynamic landscape of open chromatin during human cortical neurogenesis. Cell. 2018;172:289–304. doi: 10.1016/j.cell.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trevino A.E., Sinnott-Armstrong N., Andersen J., Yoon S.J., Huber N., Pritchard J.K., Chang H.Y., Greenleaf W.J., Pașca S.P. Chromatin accessibility dynamics in a model of human forebrain development. Science. 2020;367:eaay1645. doi: 10.1126/science.aay1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Fiddes I.T., Lodewijk G.A., Mooring M., Bosworth C.M., Ewing A.D., Mantalas G.L., Novak A.M., van den Bout A., Bishara A., Rosenkrantz J.L. Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell. 2018;173:1356–1369. doi: 10.1016/j.cell.2018.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]; These papers identify the crucial role of the NOTCH2NL human-specific genes, which act as human-specific modifiers of the Notch pathway to control human cortical expansion.
- 74•.Suzuki I.K., Gacquer D., Van Heurck R., Kumar D., Wojno M., Bilheu A., Herpoel A., Lambert N., Cheron J., Polleux F. Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell. 2018;173:1370–1384. doi: 10.1016/j.cell.2018.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]; These papers identify the crucial role of the NOTCH2NL human-specific genes, which act as human-specific modifiers of the Notch pathway to control human cortical expansion.
- 75.Florio M., Heide M., Pinson A., Brandl H., Albert M., Winkler S., Wimberger P., Huttner W.B., Hiller M. Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. eLife. 2018;7:e32332. doi: 10.7554/eLife.32332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brunetti-Pierri N., Berg J.S., Scaglia F., Belmont J., Bacino C.A., Sahoo T., Lalani S.R., Graham B., Lee B., Shinawi M. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fushan A.A., Turanov A.A., Lee S.G., Kim E.B., Lobanov A.V., Yim S.H., Buffenstein R., Lee S.R., Chang K.T., Rhee H. Gene expression defines natural changes in mammalian lifespan. Aging Cell. 2015;14:352–365. doi: 10.1111/acel.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsuda M., Hayashi H., Garcia-Ojalvo J., Kobayashi K.Y., Kageyama R., Yamanaka Y., Ikeya M., Toguchida J., Alev C., Ebisuya M. Species-specific oscillation periods of human and mouse segmentation clocks are due to cell autonomous differences in biochemical reaction parameters. Science. 2020;9:1450–1455. doi: 10.1126/science.aba7668. [DOI] [PubMed] [Google Scholar]
- 79.Rayon T., Stamataki D., Perez-Carrasco R., Garcia-Perez L., Barrington C., Melchionda M., Exelby K., Tybulewicz V., Fisher E., Briscoe J. Species-specific developmental timing is associated with global differences in protein stability in mouse and human. Science. 2020;369:eaba7667. doi: 10.1126/science.aba7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Iwata R., Vanderhaeghen P. Tempus fugit: How time flies during development. Science. 2020;369:1431–1432. doi: 10.1126/science.abe0953. [DOI] [PubMed] [Google Scholar]
- 81.Briscoe J., Marín O. Looking at neurodevelopment through a big data lens. Science. 2020;369:eaaz8627. doi: 10.1126/SCIENCE.AAZ8627. [DOI] [PubMed] [Google Scholar]