Abstract
Corynebacterium glutamicum is a prominent production host for various value-added compounds in white biotechnology. Gene repression by dCas9/clustered regularly interspaced short palindromic repeats (CRISPR) interference (CRISPRi) allows for the identification of target genes for metabolic engineering. In this study, a CRISPRi-based library for the repression of 74 genes of C. glutamicum was constructed. The chosen genes included genes encoding enzymes of glycolysis, the pentose phosphate pathway, and the tricarboxylic acid cycle, regulatory genes, as well as genes of the methylerythritol phosphate and carotenoid biosynthesis pathways. As expected, CRISPRi-mediated repression of the carotenogenesis repressor gene crtR resulted in increased pigmentation and cellular content of the native carotenoid pigment decaprenoxanthin. CRISPRi screening identified 14 genes that affected decaprenoxanthin biosynthesis when repressed. Carotenoid biosynthesis was significantly decreased upon CRISPRi-mediated repression of 11 of these genes, while repression of 3 genes was beneficial for decaprenoxanthin production. Largely, but not in all cases, deletion of selected genes identified in the CRISPRi screen confirmed the pigmentation phenotypes obtained by CRISPRi. Notably, deletion of pgi as well as of gapA improved decaprenoxanthin levels 43-fold and 9-fold, respectively. The scope of the designed library to identify metabolic engineering targets, transfer of gene repression to stable gene deletion, and limitations of the approach were discussed.
Keywords: CRISPR interference, carotenoids, CRISPRi, library, metabolic engineering, terpenoids, Corynebacterium glutamicum
1. Introduction
Metabolic engineering offers the possibility to construct production strains that overproduce a valuable compound of interest. Besides the overproduction of native products, also non-native products can be accessed by introducing heterologous pathways. The ability of rational strain engineering holds tremendous value for molecular biology and biotechnology and requires methods to precisely and predictably target genes for expression or repression [1,2].
The clustered regularly interspaced short palindromic repeats (CRISPR) system provides a method for targeted gene editing and gene regulation [3,4]. The CRISPR system from Streptococcus pyogenes is applied most often. One approach, called CRISPR interference (CRISPRi) [5], allows for controlled gene repression. It relies on modified protein dCas9 that is catalytically dead due to two amino acid changes in its RuvC and HNH endonuclease domains (D10A and H841A) [5]. The dCas9–sgRNA complex binds to the 20 bp complementary DNA target sequence of the sgRNA, which results in the inhibition of transcription by either blocking transcription elongation or inhibiting transcription initiation [6]. In contrast to a stable gene knockout (deletion), a gene knockdown temporarily suppresses the RNA level of the target gene, and it is a powerful technique to repress genes where a knockout would be lethal [7]. CRISPRi enables fast and robust, but reversible repression of genes. CRISPRi is well suited for functional characterization of essential genes as the knockdown results in reduced, but not abolished activity [7,8,9].
Metabolic engineering targets comprise enzymes that are beneficial for a chosen metabolic trait when their activities are either decreased or increased. The latter can be identified by screening gene overexpression libraries, and the former by screening a collection of deletion mutants or a CRISPRi library for gene repression. Identification of new bottlenecks often requires detailed genetic and biochemical information about the metabolism [10,11]. In this context, the construction of genetic libraries is highly efficient to screen for favorable/unfavorable strain characteristics in a systematic approach. There are genome-wide CRISPRi screenings with 92,000 different sgRNAs in the Escherichia coli genome [12,13]. The limiting step of library-based screening is often the readout for the desired phenotype [10], which typically relies on laborious quantifications.
Carotenoids are yellow-to-red-colored natural pigments, and their easy visual readout is suited for phenotypic screening approaches [14,15,16]. Due to their beneficial effects on health and their possible pharmaceutical and nutraceutical applications, carotenoids are also important products for different industries, such as the feed and health industries [17,18]. The global carotenoid market value is expected to reach US$2.0 billion by 2022, with naturally derived carotenoids on the rise.
Corynebacterium glutamicum is an excellent platform organism of the bioindustry because of its advantageous traits, such as rapid growth, genetic stability, well-studied genetic background, and the genetic tools for recombinant engineering [19,20]. C. glutamicum has already been engineered for the production of a variety of natural and non-natural products from renewable biomass resources, e.g., amino acids [21,22,23]. For the past decades, the industry has relied on the ability of the soil organism C. glutamicum to synthesize and secrete amino acids [24]. The central carbon metabolism of C. glutamicum has been characterized biochemically and by carbon flux analysis, genetic analysis, and genome-wide studies [25,26]. The metabolic engineering of genes in the central metabolism has been shown to increase fermentative production by C. glutamicum, e.g., of l-lysine [27,28,29]. C. glutamicum naturally produces the yellow C50 carotenoid decaprenoxanthin and its glucosides [30]. Over the past years, its terpenoid metabolism has been engineered for the production of various isoprenoids, like astaxanthin [31], β-carotene [31], decaprenoxanthin [32], lycopene [33], α-pinene [34], sesquarterpenes [35], patchoulol [36], α-farnesene [37], and (+)-valencene [38]. C. glutamicum synthesizes isopentenyl pyrophosphates (isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP)) via the 2-methylerythritol 4-phosphate (MEP) pathway [39], and its genome contains a carotenogenic operon encoding the enzymes responsible for terminal decaprenoxanthin biosynthesis starting from the isoprenoid pyrophosphates [40,41]. Metabolic engineering of the central metabolism increases carotenoid production in other host organisms [42,43,44,45], but this has not yet been attempted in C. glutamicum.
CRISPR genome editing and CRISPR base editors have been adopted for use in this bacterium [46,47,48,49]. The first CRISPR application to C. glutamicum was CRISPRi [50]. CRISPRi was used in several metabolic engineering approaches [7,50,51,52,53,54,55,56], e.g., to improve the production of the amino acids lysine and glutamate [50], as well as butyrate [54] and PHB [52]. The aim of this study was to construct a CRISPRi library for C. glutamicum targeting the central metabolism, which should make it useful for the fast identification of targets for rational metabolic engineering to improve the production of a desired compound. The CRISPRi-based library constructed in this study comprised 74 genes, including genes encoding enzymes of glycolysis, the pentose phosphate pathway, and the tricarboxylic acid cycle, regulatory genes, as well as genes of the MEP and carotenoid biosynthesis pathways. As a first application, the CRISPRi library was successfully used to identify targets affecting decaprenoxanthin biosynthesis.
2. Materials and Methods
Strains and plasmids used in this study are listed in Table 1. Chemicals were delivered by Carl Roth (Karlsruhe, Germany) if not stated differently. E. coli DH5α cells were used for cloning and were cultivated at 37 °C in LB medium. CRISPRi library experiments were carried out in the prophage-cured MB001 strain [57]. Experiments adapting the results of the library were carried out in the wild-type ATCC 13032. Precultures of C. glutamicum strains were grown in the brain heart infusion (BHI) complex medium (37 g L−1) supplemented with 10 g L−1 glucose overnight. Main cultures for CRISPRi library screening were grown in CGXII minimal medium [58] supplemented with 40 g L−1 of glucose supplemented with 1 mM IPTG and 0.25 µg mL−1 of anhydrotetracycline (aTc) for induction after washing in minimal medium. Cultures were inoculated to an initial OD600nm of 1 using a Shimadzu UV-1202 spectrophotometer (Duisburg, Germany). Cultivations were performed in 1 mL in the Biolector®flowerplate microcultivation system (m2p-labs GmbH, Baesweiler, Germany) at 1100 rpm and 30 °C. C. glutamicum WT and C. glutamicum WT ΔsdhCAB were cultivated in 50 mL of CGXII plus 40 g L−1 of glucose in baffled shake flasks. C. glutamicum WT and C. glutamicum WT ∆aceE were cultivated in 50 mL of CGXII plus 40 g L−1 of glucose and 20 g L−1 of potassium acetate in baffled shake flasks. C. glutamicum WT and strains WT Δpgi, WT ΔgapA, and WT ΔsugR were cultivated in 1 mL of CGXII plus 40 g L−1 of glucose in Duetz plates at 30 °C and 220 rpm. As an antibiotic, chloramphenicol (VWR, Darmstadt, Germany) was added to the CRISPRi plasmids in concentrations of 7.5 μg mL−1 for C. glutamicum cultures and 30 μg mL−1 for E. coli cultures.
Table 1.
Strains and plasmids used in this study.
| Strain | Characteristics | Reference |
|---|---|---|
| Corynebacterium glutamicum strains | ||
| Wild type (WT) | Wild-type ATCC 13032 | [59] |
| WT ΔaceE | aceE (cg2466) deletion mutant of WT | [60] |
| WT ΔgapA | gapA (cg1791) deletion mutant of WT | [61] |
| WT Δpgi | pgi (cg0973) deletion mutant of WT | [62] |
| WT ΔsdhCAB | sdhCAB (cg0445/0447/0448) deletion mutant of WT | This work |
| WT ΔsugR | sugR (cg2115) deletion mutant of WT | [25] |
| MB001 | Prophage-cured, genome-reduced WT | [57] |
| MB001 ΔcrtR | crtR (cg0725) deletion mutant of MB001 | [63] |
| E. coli strains | ||
| E. coli DH5α | F-thi−1 endA1 hsdr17(r-, m-) supE44 ΔlacU169 (Φ80lacZΔM15) recA1 gyrA96 | [64] |
| Plasmids | ||
| pK19mobsacB | Mobilizable E. coli vector used for the construction of insertion and deletion mutants of C. glutamicum (oriV, sacB, lacZα); KanR | [65] |
| pK19mobsacB-ΔsdhCAB | pK19mobsacB for the deletion of sdhCAB (cg0445/0447/0448); KanR | This work |
| pRG_dCas9 | E. coli/C. glutamicum shuttle clustered regularly interspaced short palindromic repeats interference (CRISPRi) vector, anhydrotetracycline (aTc)- and IPTG-inducible; CmR | [56] |
| piCas | E. coli/B. methanolicus shuttle CRISPRi vector, mannitol-inducible; CmR | [66] |
| pS_dCas9 | pRG_dCas9 carrying the dCas9 handle followed by the terminator from S. pyogenes; CmR | This work |
| pS_dCas9_aceA | pS_dCas9 plasmid carrying the aceA (cg2560) sgRNA | This work |
| pS_dCas9_aceB | pS_dCas9 plasmid carrying the aceB (cg2559) sgRNA | This work |
| pS_dCas9_aceE | pS_dCas9 plasmid carrying the aceE (cg2466) sgRNA | This work |
| pS_dCas9_ackA | pS_dCas9 plasmid carrying the ackA (cg3047) sgRNA | This work |
| pS_dCas9_acn | pS_dCas9 plasmid carrying the acn (cg1737) sgRNA | This work |
| pS_dCas9_crtB | pS_dCas9 plasmid carrying the crtB (cg0721) sgRNA | This work |
| pS_dCas9_crtE | pS_dCas9 plasmid carrying the crtE (cg0723) sgRNA | This work |
| pS_dCas9_crtEb | pS_dCas9 plasmid carrying the crtEb (cg0717) sgRNA | This work |
| pS_dCas9_crtI | pS_dCas9 plasmid carrying the crtI (cg0720) sgRNA | This work |
| pS_dCas9_crtR | pS_dCas9 plasmid carrying the crtR (cg0725) sgRNA | This work |
| pS_dCas9_crtX | pS_dCas9 plasmid carrying the crtX (cg0730) sgRNA | This work |
| pS_dCas9_crtYe | pS_dCas9 plasmid carrying the crtYe (cg0719) sgRNA | This work |
| pS_dCas9_deoC | pS_dCas9 plasmid carrying the deoC (cg0458) sgRNA | This work |
| pS_dCas9_dxr | pS_dCas9 plasmid carrying the dxr (cg2208) sgRNA | This work |
| pS_dCas9_dxs | pS_dCas9 plasmid carrying the dxs (cg2083) sgRNA | This work |
| pS_dCas9_eno | pS_dCas9 plasmid carrying the eno (cg1111) sgRNA | This work |
| pS_dCas9_fba | pS_dCas9 plasmid carrying the fba (cg3068) sgRNA | This work |
| pS_dCas9_fbp | pS_dCas9 plasmid carrying the fbp (cg1157) sgRNA | This work |
| pS_dCas9_fixB | pS_dCas9 plasmid carrying the fixB (cg1386) sgRNA | This work |
| pS_dCas9_fum | pS_dCas9 plasmid carrying the fum (cg1145) sgRNA | This work |
| pS_dCas9_gapA | pS_dCas9 plasmid carrying the gapA (cg1791) sgRNA | This work |
| pS_dCas9_gltA | pS_dCas9 plasmid carrying the gltA (cg0949) sgRNA | This work |
| pS_dCas9_glxR | pS_dCas9 plasmid carrying the glxR (cg0350) sgRNA | This work |
| pS_dCas9_gnd | pS_dCas9 plasmid carrying the gnd (cg1643) sgRNA | This work |
| pS_dCas9_icd | pS_dCas9 plasmid carrying the icd (cg0766) sgRNA | This work |
| pS_dCas9_idsA | pS_dCas9 plasmid carrying the idsA (cg2384) sgRNA | This work |
| pS_dCas9_IIdD | pS_dCas9 plasmid carrying the IIdD (cg3227) sgRNA | This work |
| pS_dCas9_ispE | pS_dCas9 plasmid carrying the ispE (cg1039) sgRNA | This work |
| pS_dCas9_ispF | pS_dCas9 plasmid carrying the ispF (cg2944) sgRNA | This work |
| pS_dCas9_ispG | pS_dCas9 plasmid carrying the ispG (cg2206) sgRNA | This work |
| pS_dCas9_ispH | pS_dCas9 plasmid carrying the ispH (cg1164) sgRNA | This work |
| pS_dCas9_ldh | pS_dCas9 plasmid carrying the ldh (cg3219) sgRNA | This work |
| pS_dCas9_malE | pS_dCas9 plasmid carrying the malE (cg3335) sgRNA | This work |
| pS_dCas9_mdh | pS_dCas9 plasmid carrying the mdh (cg2613) sgRNA | This work |
| pS_dCas9_mmpl | pS_dCas9 plasmid carrying the mmpl (cg0722) sgRNA | This work |
| pS_dCas9_odhA | pS_dCas9 plasmid carrying the odhA (cg1280) sgRNA | This work |
| pS_dCas9_odx | pS_dCas9 plasmid carrying the odx (cg1458) sgRNA | This work |
| pS_dCas9_opcA | pS_dCas9 plasmid carrying the opcA (cg1779) sgRNA | This work |
| pS_dCas9_pck | pS_dCas9 plasmid carrying the pck (cg3169) sgRNA | This work |
| pS_dCas9_pfkA | pS_dCas9 plasmid carrying the pfkA (cg1409) sgRNA | This work |
| pS_dCas9_pgi | pS_dCas9 plasmid carrying the pgi (cg0973) sgRNA | This work |
| pS_dCas9_pgk | pS_dCas9 plasmid carrying the pgk (cg1790) sgRNA | This work |
| pS_dCas9_pgl | pS_dCas9 plasmid carrying the pgl (cg1780) sgRNA | This work |
| pS_dCas9_pgm | pS_dCas9 plasmid carrying the pgm (cg2800) sgRNA | This work |
| pS_dCas9_ppc | pS_dCas9 plasmid carrying the ppc (cg1787) sgRNA | This work |
| pS_dCas9_ppsA | pS_dCas9 plasmid carrying the ppsA (cg0644) sgRNA | This work |
| pS_dCas9_pqo | pS_dCas9 plasmid carrying the pqo (cg2891) sgRNA | This work |
| pS_dCas9_pta | pS_dCas9 plasmid carrying the pta (cg3048) sgRNA | This work |
| pS_dCas9_ptsG | pS_dCas9 plasmid carrying the ptsG (cg1537) sgRNA | This work |
| pS_dCas9_pyc | pS_dCas9 plasmid carrying the pyc (cg0791) sgRNA | This work |
| pS_dCas9_pyk | pS_dCas9 plasmid carrying the pyk (cg2291) sgRNA | This work |
| pS_dCas9_ramB | pS_dCas9 plasmid carrying the ramB (cg0444) sgRNA | This work |
| pS_dCas9_rpe | pS_dCas9 plasmid carrying the rpe (cg1801) sgRNA | This work |
| pS_dCas9_rpi | pS_dCas9 plasmid carrying the rpi (cg2658) sgRNA | This work |
| pS_dCas9_rsdA | pS_dCas9 plasmid carrying the rsdA (cg0697) sgRNA | This work |
| pS_dCas9_rshA | pS_dCas9 plasmid carrying the rshA (cg0877) sgRNA | This work |
| pS_dCas9_sdhA | pS_dCas9 plasmid carrying the sdhA (cg0446) sgRNA | This work |
| pS_dCas9_sdhB | pS_dCas9 plasmid carrying the sdhB (cg0447) sgRNA | This work |
| pS_dCas9_sdhCD | pS_dCas9 plasmid carrying the sdhCD (cg0445) sgRNA | This work |
| pS_dCas9_sigA | pS_dCas9 plasmid carrying the sigA (cg2092) sgRNA | This work |
| pS_dCas9_sigB | pS_dCas9 plasmid carrying the sigB (cg2102) sgRNA | This work |
| pS_dCas9_sigC | pS_dCas9 plasmid carrying the sigC (cg0309) sgRNA | This work |
| pS_dCas9_sigD | pS_dCas9 plasmid carrying the sigD (cg0696) sgRNA | This work |
| pS_dCas9_sigE | pS_dCas9 plasmid carrying the sigE (cg1271) sgRNA | This work |
| pS_dCas9_sigH | pS_dCas9 plasmid carrying the sigH (cg0876) sgRNA | This work |
| pS_dCas9_sigM | pS_dCas9 plasmid carrying the sigM (cg3420) sgRNA | This work |
| pS_dCas9_sucC | pS_dCas9 plasmid carrying the sucC (cg2837) sgRNA | This work |
| pS_dCas9_sucD | pS_dCas9 plasmid carrying the sucD (cg2836) sgRNA | This work |
| pS_dCas9_sugR | pS_dCas9 plasmid carrying the sugR (cg2115) sgRNA | This work |
| pS_dCas9_tal | pS_dCas9 plasmid carrying the tal (cg1776) sgRNA | This work |
| pS_dCas9_thiE | pS_dCas9 plasmid carrying the thiE (cg2236) sgRNA | This work |
| pS_dCas9_tkt | pS_dCas9 plasmid carrying the tkt (cg1774) sgRNA | This work |
| pS_dCas9_tpi | pS_dCas9 plasmid carrying the tpi (cg1789) sgRNA | This work |
| pS_dCas9_zwf | pS_dCas9 plasmid carrying the zwf (cg1778) sgRNA | This work |
2.1. Construction of the CRISPRi Vector System
For construction of the CRISPRi library plasmid pS_dCas9, the vector pRG_dCas9 [46] was restricted with PstI and SalI (NEB, Frankfurt, Germany) and dephosphorylated (Antarctic phosphatase, New England Biolabs, Frankfurt, Germany). The sequences of the dCas9 handle and terminator from S. pyogenes were amplified by high-fidelity PCR (All-in HiFi, Kraichtal, Germany) from the plasmid piCas [66] with the oligonucleotides vgag and vgam (Table S1), and the PCR amplicon was purified with a PCR and gel extraction kit (Macherey-Nagel, Düren, Germany). The PCR product was cloned into the PstI- and SalI-restricted, dephosphorylated vector pRG_dCas9 by Gibson Assembly [67], resulting in plasmid pS_dCas9. Standard genetic procedures were performed as described previously [68].
2.2. Construction of the CRISPRi Library
The sgRNAs were designed to contain a 20 bp region homologous to the non-template strand of the chosen DNA targets. The genome sequence of C. glutamicum ATCC 13,032 [39] was used as a basis for the selection of the 20 bp targeting sequences using the CRISPy-web tool [69]. CRISPRi library plasmids were constructed in E. coli DH5α through the annealing oligo method, with single-stranded oligonucleotides covering sgRNA (20 bp) and 20 bp overlaps with plasmid pS_dCas9. Equal volumes (5 µL) of equimolar oligonucleotides (100 µM) were mixed with the annealing buffer (990 µL), and the mixture was incubated at 95 °C for 5 min and then cooled down to room temperature. The CRISPRi library plasmid pS_dCas9 was restricted with PstI (NEB, Frankfurt, Germany) and dephosphorylated (Antarctic phosphatase, New England Biolabs, Frankfurt, Germany) before the double-stranded oligonucleotides were annealed by the Gibson Assembly method [67]. The concentration of DNA was measured with an ND-1000 spectrophotometer (Thermo Fisher Scientific, Schwerte, Germany). The oligonucleotides (Table S1) used in this study were obtained from Metabion (Planegg/Steinkirchen, Germany). E. coli DH5α cells were transformed by heat shock after preparation of CaCl2-competent cells [70]. Transformants were screened by colony PCR, and plasmids were isolated by a plasmid miniprep kit (GeneJET, Thermo Fisher Scientific, Schwerte, Germany). Library vectors were confirmed by sequencing with oligonucleotides vgai and vgaj (Table S1). C. glutamicum cells were transformed by electroporation [71].
2.3. Construction of C. glutamicum Deletion Mutants
For deletion of the sdhCAB operon, the suicide vector pK19mobsacB was used [65]. The genomic flanking regions of sdhCAB were amplified from the genomic DNA of C. glutamicum WT using the oligonucleotide pairs del-sdhCAB1/del-sdhCAB2 and del-sdhCAB3/del-sdhCAB4 (Table S1). The PCR amplicons were purified, linked by crossover PCR, and subsequently cloned into a SmaI-restricted pK19mobsacB. The resulting deletion vector pK19mobsacB-sdhCAB was introduced into C. glutamicum via trans conjugation with E. coli S17-1 [65]. Deletion of sdhCAB was achieved by two-step homologous recombination using the respective deletion vector, as previously described [70]. Integration of the vector into one of the gene-flanking regions represents the first recombination event and was selected via kanamycin resistance. Integration of the vector into the genome results in sucrose sensitivity due to levansucrase, encoded by sacB. Selection for the second recombination event, loss of the vector, was carried out via sucrose resistance. Deletion of sdhCAB was verified via sequencing with primers del-sdhCAB-5 and del-sdhCAB-6 (Table S1).
2.4. Quantification of the mRNA Levels of Targeted Cells by CRISPRi
RNA levels were determined by quantitative reverse transcription PCR (qRT-PCR). Total RNA was isolated from C. glutamicum strains growing exponentially in CGXII medium. Biological triplicates were analyzed. Aliquots of 500 µL were centrifuged at 14,000 rpm for 15 s (Eppendorf centrifuge 5810 R), and the pellets were immediately frozen in liquid nitrogen and stored at −80 °C until further use. For RNA isolation, the samples were homogenized by resuspending the cells in 100 μL of TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8) containing 5 mg mL−1 of lysozyme. After incubation at 37 °C for 30 min, total RNA was extracted using a NucleoSpin® RNA kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions. After extraction, RNA samples were treated with DNase restriction using RNase-free DNase Set and RNeasy MinElute kits (Qiagen, Hilden, Germany) to eliminate possible genomic DNA contamination. The total RNA concentration was measured using a spectrophotometer (NanoDrop®, ND-1000; ThermoFisher Scientific, Schwerte, Germany). Quality control was performed to determine the purity and integrity of isolated RNA.
Equal amounts of 50 ng of each sample were used to perform cDNA synthesis. qRT-PCR was performed using the SensiFASTTM SYBR® No-ROX One-Step Kit (Bioline, London, UK) and the CFX96 cycler system (Bio-Rad, Hercules, CA, USA). The temperature profile was (1) 45 °C for 10 min; (2) 95 °C for 2 min; (3) 40 cycles of 95 °C for 5 s, 56 °C for 15 s, and 72 °C for 15 s; and (4) melt curve analysis with measures between 65 °C and 95 °C. The used primers for qRT-PCR are listed in Table S1. The ΔCq method was used in calculations [72,73].
2.5. Carotenoid Quantification
Carotenoid production was analyzed by high-performance liquid chromatography (HPLC) analysis. Carotenoids were extracted from the cell fraction using a methanol:acetone (7:3) mixture. Extraction was performed at 60 °C and 600 rpm for 30 min. After centrifugation at 14,000 rpm and 10 min, the supernatant was used for HPLC analysis. The Agilent 1200 series system (Agilent Technologies, Waldbronn, Germany) was used with a reversed-phase precolumn (LiChrospher 100 RP18 EC-5, 40 × 4 mm) (CS-Chromatographie, Langerwehe, Germany) and a reversed-phase main column (LiChrospher 100 RP18 EC-5, 125 × 4 mm) (CS-Chromatographie, Langerwehe, Germany), and methanol (A) and methanol:water (9:1) (B) were used as mobile phases. Carotenoids were detected with a diode array detector (DAD) by recording of the UV–visible (Vis) spectrum. A gradient at a flow rate of 1.5 mL min−1 was used as follows: 0 min B: 0%, 10 min B: 100%, and 32.5 min B: 100%. The decaprenoxanthin measured and presented in the Results section (Section 3) is di-glycosylated decaprenoxanthin. For quantification, the extracted wavelength chromatogram at λmax of 470 nm was used and standardized with β-carotene (Sigma-Aldrich, Steinheim, Germany).
3. Results
3.1. Design and Initial Testing of a CRISPRi Library for Gene Repression in C. glutamicum
This work aimed to design, construct, and test a CRISPRi library suitable for gene repression screening in C. glutamicum. In principle, the approach is generalizable, but here, we focused on scoring the effect of repressing genes of the central carbon metabolism and carotenogenesis as well as regulatory genes on the biosynthesis of decaprenoxanthin, the natural pigment of C. glutamicum.
3.1.1. Construction of a Vector CRISPRi Library for C. glutamicum
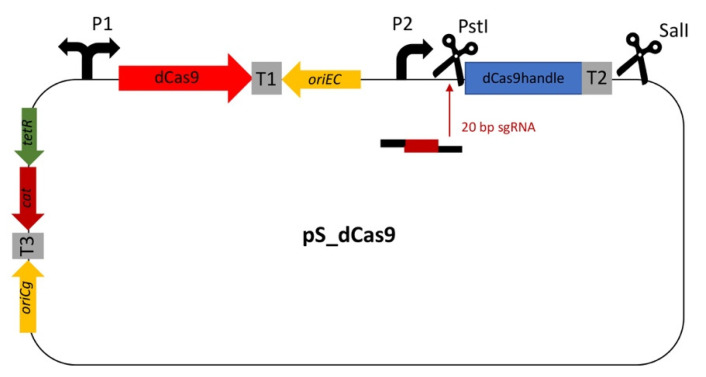
The CRISPRi library was based on pRG_dCas9 [56]. To ease library preparation, the dCas9 handle followed by the terminator from S. pyogenes was amplified from plasmid piCas [66] and inserted between the PstI and SalI restriction sites, resulting in vector pS_dCas9 (Figure 1). The various gene-specific 20 bp sgRNA sequences of the target library (Table S2) were generated from oligonucleotides by the annealing oligo method before being inserted into the PstI cloning site.
Figure 1.
Construction of the dual-inducible CRISPRi expression plasmid pS_dCas9. Adapted pRG_dCas9 plasmid carrying the dCas9 handle followed by the terminator from S. pyogenes amplified from plasmid piCas between the PstI and SalI restriction sites. A 20 bp sgRNA sequence can be inserted in the PstI cloning site. For multiplexing of more than one specific sgRNA, the restriction site SalI after the first sgRNA-cs can be used. It has chloramphenicol resistance. P1: tetR/tetA promotor; P2: tac promotor; T1: rrnB T1 terminator; T2: terminator from S. pyogenes; T3: lambda terminator; oriEc: p15A; oriCg: pCG1; cat: chloramphenicol resistance.
3.1.2. Testing of the CRISPRi Library Vector for the Repression of crtR
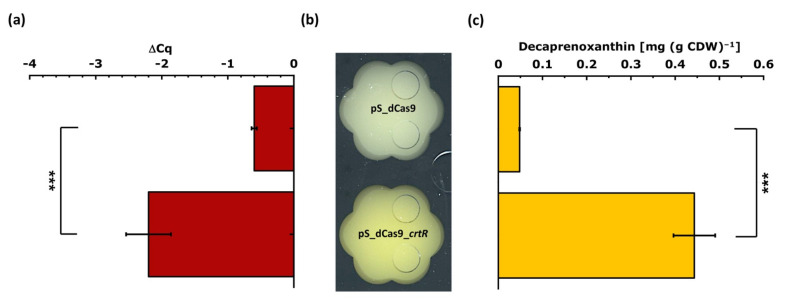
To test the sensitivity and function of the CRISPRi library vector, the repression of crtR coding for the transcriptional repressor CrtR of the carotenogenic crt operon [63] was chosen. We have shown previously that deletion of crtR in C. glutamicum MB001 increased the accumulation of the native carotenoid pigment decaprenoxanthin about 30-fold [63]. Therefore, we assumed that targeting crtR by CRISPRi will increase decaprenoxanthin biosynthesis. The sgRNA sequence for crtR (and later for all tested genes) was chosen according to the following strategy: the sgRNA (i) was identified using CRISPy-web [69] targeting the non-template strand, (ii) is located in the coding sequence of the target gene for inhibition of transcription elongation, (iii) preferably is in the 5′ proximal region of the coding sequence, and (iv) is unique in the C. glutamicum genome. The plasmid pS_dCas9_crtR and the empty vector pS_dCas9 were used to transform C. glutamicum MB001, and the resulting recombinant strains were cultivated in glucose minimal medium with 1 mM IPTG for induction of the sgRNA and with 0.25 µg mL−1 of aTc for induction of the dCas9 gene. Exponentially growing cells were harvested for RNA extraction, while the decaprenoxanthin content was quantified after cultivation for 28 h. After qRT-PCR, the ΔCq value was calculated using the vegetative RNA polymerase sigma factor gene sigA as a reference. The qRT-PCR analysis revealed a statistically significant and about fourfold lower crtR RNA level upon targeting crtR using pS_dCas9_crtR (Figure 2a).
Figure 2.
Testing the CRISPRi system for identification of metabolic engineering targets relevant for carotenoid production. Strain C. glutamicum MB001(pS_dCas9_crtR) for CRISPRi-mediated repression of crtR was compared to the empty vector carrying control strain MB001(pS_dCas9) with respect to (a) crtR RNA levels, (b) color phenotypes, and (c) cellular decaprenoxanthin content. For qRT-PCR analysis, exponentially growing cells were harvested and sigA was used as a reference (a). The color phenotype (b) was judged by visual inspection after growth in the Biolector®flowerplate microcultivation system. Cells were grown in 40 g L−1 of glucose CGXII minimal medium for 28 h and induced at 0 h with 1 mM IPTG and 0.25 µg mL−1 of aTc. The cellular decaprenoxanthin content (c) is given as ß-carotene equivalents, as determined by HPLC analysis. Mean values and standard deviations of three biological replicates are given. The p-value of <0.001 (***) was calculated using Student’s t-test (two sided, unpaired).
Phenotypically, C. glutamicum MB001(pS_dCas9_crtR) indeed showed a more intense yellow pigmentation than strain MB001(pS_dCas9) (Figure 2b). Accordingly, HPLC analysis revealed an about ninefold, significantly (p-value < 0.001) higher decaprenoxanthin content (0.44 ± 0.05 mg (g CDW)−1) for C. glutamicum MB001(pS_dCas9_crtR) as compared to the empty vector carrying the control strain MB001(pS_dCas9) (0.05 ± 0.00 mg (g CDW)−1; Figure 2c). Thus, based on visual observation of pigmentation, qRT-PCR analysis of crtR RNA levels, and decaprenoxanthin quantification, the CRISPRi system proved suitable to score the effect that repression of a gene of interest has on carotenogenesis. To test the duration of the inhibitory effect of the CRISPRi targeting crtR, serial transfers from a culture grown for 6 generations in the presence of inducers to a medium without inducers were performed. CRISPRi targeting of crtR visibly increased decaprenoxanthin levels to above those of the empty vector control strain in both serial cultures. However, the effect faded gradually from serial transfer one (5 generations without inducers) to serial transfer two (8 generations without inducers) (Figure S1).
3.2. Characterization of a CRISPRi Library to Interrogate 74 Target Genes with Potential Relevance for Carotenogenesis in C. glutamicum
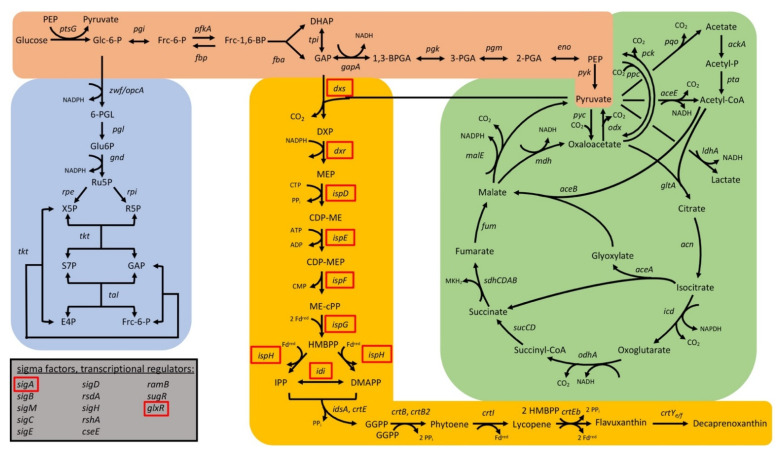
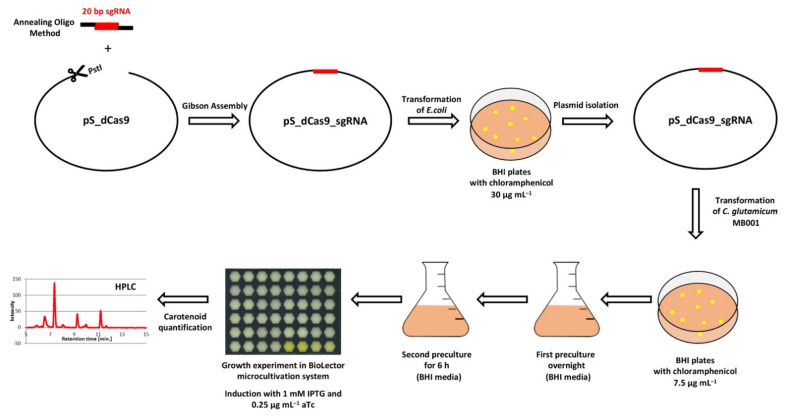
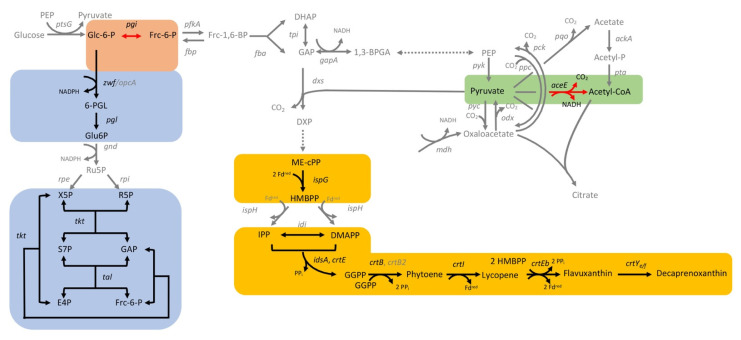
In total, 74 target genes from C. glutamicum were repressed by CRISPRi; the growth parameters of the respective strains are listed in Table S3. The CRISPRi library comprising a subset of genes from central carbon metabolism as well as regulatory genes was selected to cover glycolysis (19 gene targets), the pentose phosphate pathway (8 gene targets), and the tricarboxylic acid (TCA) cycle (17 gene targets) for general use in metabolic engineering (Figure 3). With the goal to improve isoprenoid and carotenoid production as the chosen application example in this study, genes were selected to cover the MEP pathway of isoprenoid pyrophosphate biosynthesis (6 genes targets) as well as terminal carotenogenesis (8 gene targets) (Figure 3). The CRISPRi library approach is commensurate with multivariate modular metabolic engineering [74]. The workflow of the approach is illustrated in Figure 4.
Figure 3.
Scheme of the central carbon metabolism and carotenogenesis in C. glutamicum with glycolysis (orange shading), the pentose phosphate pathway (blue shading), the TCA cycle (green shading), as well as the MEP pathway and the carotenogenesis (yellow shading). Gene names are given next to the reactions catalyzed by their gene products. The corresponding gene identifiers can be found in Table 1. Essential genes are depicted with a red box. aceA: isocitrate lyase; aceB: malate synthase; aceE: pyruvate dehydrogenase E1 component; ackA: acetate kinase; acn: aconitase; crtB: phytoene synthase; crtB2: phytoene synthase 2; crtE: geranylgeranyl-diphosphate synthase; crtEb: lycopene elongase; crtI: phytoene desaturase; crtI2: phytoene desaturase; crtYe/f: C50 carotenoid epsilon cyclase; cseE: anti-sigma factor E; dxr: 1-deoxy-D-xylulose 5-phosphate reductoisomerase; dxs: 1-deoxyxylulose-5-phosphate synthase; eno: enolase; fba: fructose-1,6-bisphosphate aldolase; fbp: fructose 1,6-bisphosphatase; fum: fumarase; gapA: glyceraldehyde-3-phosphate dehydrogenase A; gltA: citrate synthase; glxR: global transcriptional regulator; gnd: 6-phosphogluconate dehydrogenase; icd: isocitrate dehydrogenase; idi: isopentenyldiphosphate isomerase; idsA: geranylgeranyl diphosphate synthase; ispD: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; ispE: 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; ispF: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; ispG: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase; ispH: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; ldh: NAD-dependent L-lactate dehydrogenase; malE: malic enzyme; mdh: malate dehydrogenase; odhA: oxoglutarate dehydrogenase subunit A; odx: oxaloacetate decarboxylase; opcA: glucose-6-phosphate dehydrogenase; pck: phosphoenolpyruvate carboxykinase; pfkA: 6-phosphofructokinase; pgi: glucose-6-phosphate isomerase; pgk: phosphoglycerate kinase; pgl: 6-phosphogluconolactonase; pgm: phosphoglucomutase; ppc: phosphoenolpyruvate carboxylase; pqo: pyruvate quinone oxidoreductase; pta: phosphotransacetylase; ptsG: glucose-specific enzyme II BC component of PTS; pyc: pyruvate carboxylase; pyk: pyruvate kinase; ramB: transcriptional regulator of acetate metabolism A; rpe: ribulose-5-phosphate epimerase; rpi: phosphopentose isomerase; rsdA: anti-sigma factor D; rshA: anti-sigma factor H; sdhABDC: succinate dehydrogenase subunits A, B, C, and D; sigA: sigma factor A; sigB: sigma factor B; sigC: sigma factor C; sigD: sigma factor D; sigE: sigma factor E; sigH: sigma factor H; sigM: sigma factor M; sucCD: succinyl-CoA synthetase beta and alpha subunits; sugR: transcriptional regulators of sugar metabolism; tal: transaldolase; tkt: transketolase; tpi: triosephosphate isomerase; zwf: glucose-6-phosphate 1-dehydrogenase. 1,3-BPGA: 1,3-bisphosphate glycerate; 2-PGA:2-phosphate glycerate; 3-PGA: 3-phosphate glycerate; 6-PGI: 6-phosphogluconolactone; acetyl-P: acetyl-phosphate; CDP-ME: 4-diphosphocytidyl-2-methylerythritol; CDP-MEP: 4-diphosphocytidyl-2-methylerythritol 2-phosphate; DHAP: dihydroxyacetone phosphate; DMAPP: dimethylallyl diphosphate; DXP: 1-deoxy-D-xylulose 5-phosphate synthase; E4P: erythrose-4-phosphate; Frc-1,6-BP: fructose-1,6-bisphosphate; Frc-6-P: fructose-6-phosphate; GAP: glyceraldehyde 3-phosphate; GGPP: geranylgeranyl diphosphate; Glc-6-P: glucose-6-phosphate; Glu6P: 6-phosphogluconate; HMBPP: (E)-4-hydroxy-3-methyl-but-2-enyl diphosphate; IPP: isopentenyl diphosphate; ME-cPP: 2-methylerythritol 2,4-cyclodiphosphate; MEP: 2-methylerythritol 4-phosphate; PEP: phosphoenolpyruvate; R5P: ribose-5-phosphate; Ru5P: ribulose-5-phosphate; S7P: sedoheptulose 7-phosphate;X5P: xylulose-5-phosphate.
Figure 4.
Workflow of CRISPRi library construction and screening in C. glutamicum MB001.
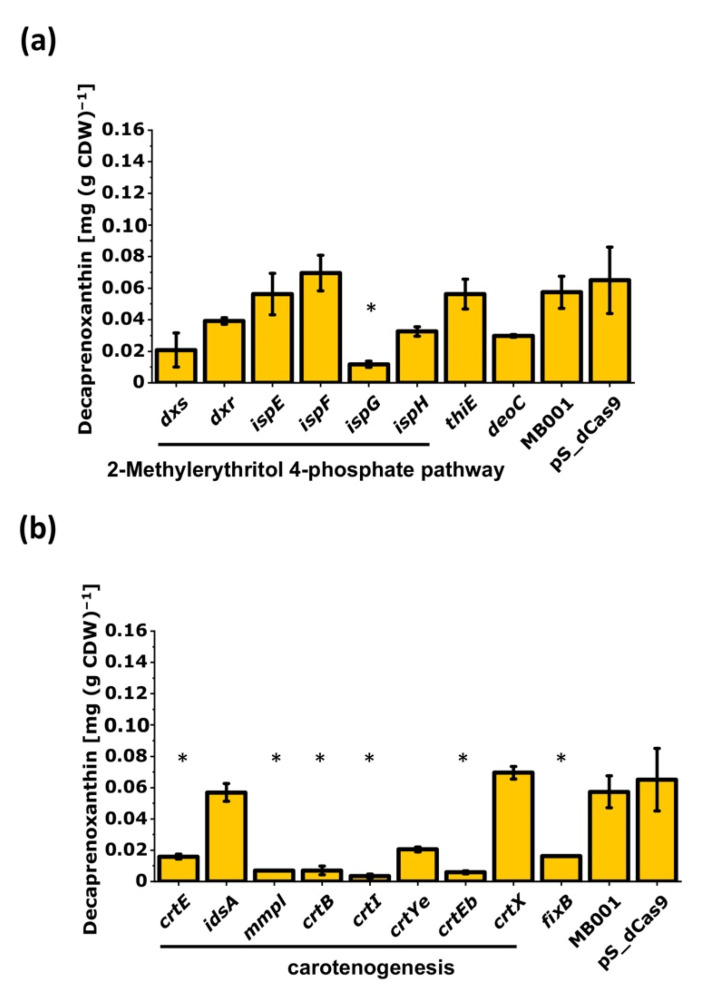
3.2.1. CRISPRi-Based Repression of Genes of the MEP Pathway and of Carotenogenesis-Reduced Decaprenoxanthin Pigmentation
Repression of genes of the MEP pathway and of carotenogenesis was expected to reduce pigmentation. To test this hypothesis, the respective CRISPRi library transformants were analyzed for decaprenoxanthin production. Indeed, repression of ispG lowered the cellular decaprenoxanthin content significantly, whereas repression of the other MEP pathway genes did not affect decaprenoxanthin content in a statistically significant manner, albeit some reduction was observed (Figure 5a). CRISPRi targeting of the crt operon genes crtE, mmpL, crtB, crtI, and crtEb reduced decaprenoxanthin biosynthesis significantly (Figure 5b). This was observed neither upon targeting idsA nor upon targeting crtX. These genes are not part of the crt operon. While CRISPRi repression of crtX did not reduce the decaprenoxanthin level (Figure 5b), unglucosylated instead of diglucosylated decaprenoxanthin accumulated, which was not observed for all other strains. This finding was commensurate with CrtX functioning as decaprenoxanthin glucosyltransferase [33].
Figure 5.
Influence of CRISPRi-mediated repression of genes of the MEP pathway (a) or of carotenogenesis (b) on decaprenoxanthin production by C. glutamicum. In addition, thiE, deoC, and fixB were analyzed. Mean values of biological duplicates are given. Statistical analysis was calculated with ANOVA against all measured decaprenoxanthin production of C. glutamicum MB001 from all Biolector®flowerplates and is marked by a star (*). As a reference, the decaprenoxanthin production of the empty vector strain C. glutamicum MB001 (pS_dCas9) in biological duplicates of the corresponding experiment is shown. For abbreviations, see Figure 3.
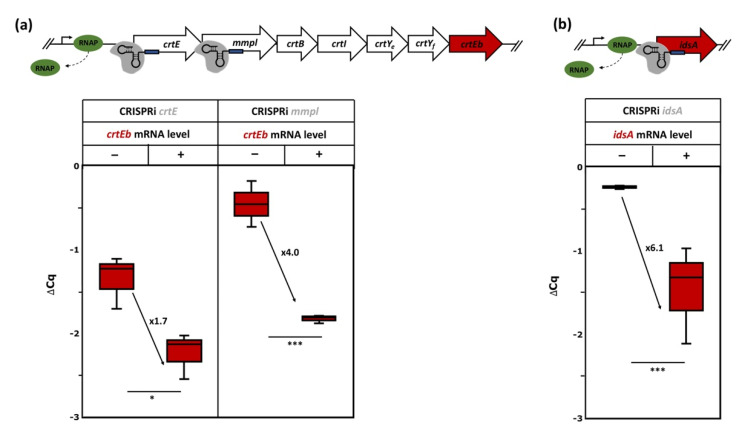
In the case of idsA, qRT-PCR analysis revealed that repression reduced the mRNA level by about sixfold (Figure 6), while decaprenoxanthin biosynthesis was hardly affected. Possibly, CrtE compensates for the loss of IdsA, since crtE and idsA both encode geranylgeranyl diphosphate (GGPP) synthases [75]. The reduced decaprenoxanthin level observed upon CRISPRi repression of crtE may either be due to IdsA not being able to compensate for reduced CrtE levels or be due to polar effects on downstream crt operon genes, since crtE is the first gene in the crt operon. To directly test for downstream effects, qRT-PCR analysis of the RNA level of crtEb, the last gene of the crtE-mmpL-crtB-crtI-crtYe-crtYf-crtEb operon, was performed in strains where CRISPRi repressed either the first or the second crt operon gene (crtE or mmpL). Indeed, the crtEb mRNA level was significantly reduced upon CRISPRi targeting of crtE as well as mmpL (Figure 6); thus, the observed reduced decaprenoxanthin biosynthesis upon CRISPRi repression of crtE and mmpL is, at least in part, not due to lowered CrtE or MmpL levels but due to repression of the whole crt operon.
Figure 6.
Schematic representation of the crt operon and idsA and qRT-PCR analysis of crtEb RNA levels upon CRISPRi targeting of crtE or mmpL (a) or of idsA RNA levels upon CRISPRi targeting of idsA (b). Cells exponentially growing in 40 g L−1 of glucose CGXII minimal medium with (+) or without (−) induction using 1 mM IPTG and 0.25 µg mL−1 of aTc were analyzed. Mean values and standard deviations of three biological replicates are given. The p-values of <0.001 (***), and <0.05 (*) were calculated using Student’s t-test (two sided, unpaired). For abbreviations, see Figure 3.
Three further genes were analyzed. Since the MEP pathway provides precursors also for thiamin biosynthesis, repression of the first gene of the thiamin biosynthesis operon thiEOSGF coding for thiamine-phosphate pyrophosphorylase ThiE was studied. The gene for deoxyribose-phosphate aldolase (deoC) that catalyzes a condensation reaction similar to that of the MEP pathway enzyme Dxs was also chosen. Finally, fixB coding for a protein of the electron transfer flavoprotein (ETF) family with some resemblance to a novel β-cyclase enzyme CruA from Chlorobium tepidum [76] was repressed. Upon CRISPRi targeting of thiE, deoC, and fixB, decaprenoxanthin levels were unaffected, somewhat reduced, and significantly decreased, respectively (Figure 5).
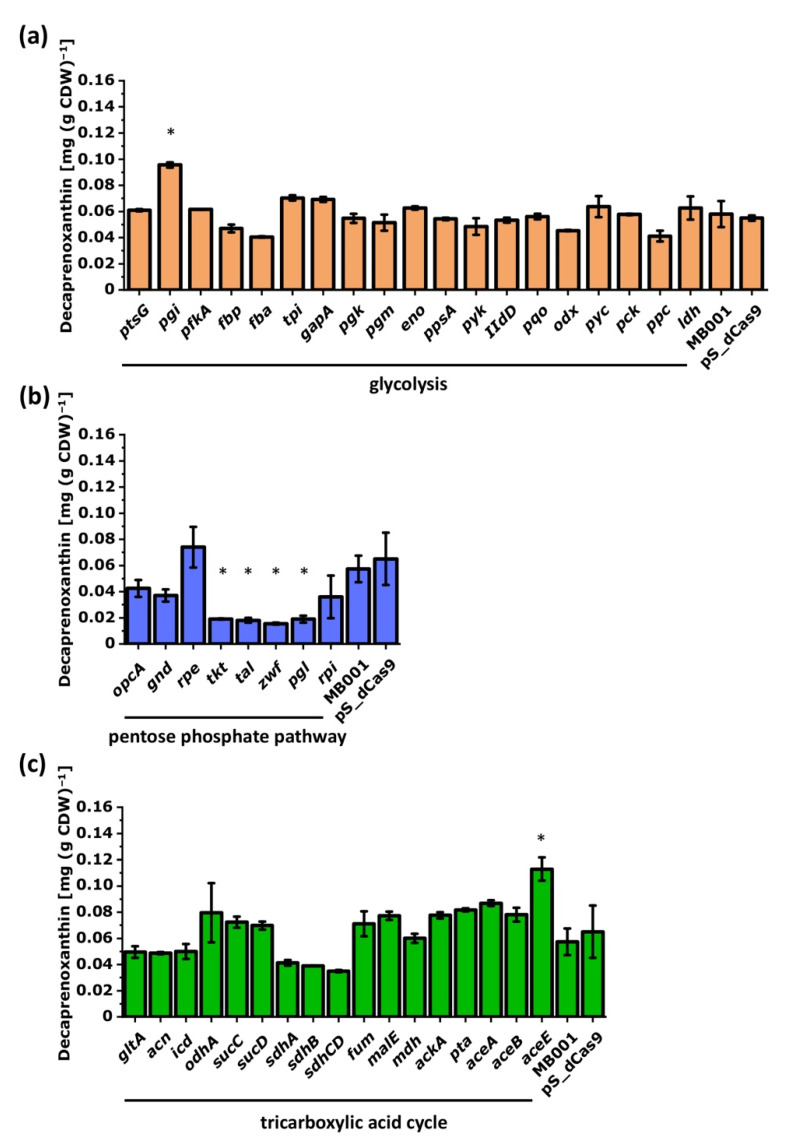
3.2.2. CRISPRi-Based Repression of Genes of the Central-Carbon-Metabolism-Identified Supply of GAP and Entry into the Pentose Phosphate Pathway as Potential Bottlenecks in Decaprenoxanthin Biosynthesis
To identify the influence of the central metabolism on the decaprenoxanthin production, selected genes were repressed by CRISPRi and decaprenoxanthin was quantified (Figure 7). While the repression of the glycolytic gene pgi improved decaprenoxanthin production significantly, CRISPRi targeting of the pentose phosphate pathway genes tkt, tal, zwf, and pgl negatively affected decaprenoxanthin pigmentation (Figure 7b). Repression of aceE encoding a subunit of the pyruvate dehydrogenase complex that is relevant for carbon entry into the TCA cycle improved decaprenoxanthin biosynthesis (Figure 7c). CRISPRi targeting of the TCA cycle genes sdhA, sdhB, and sdhCD lowered decaprenoxanthin accumulation (Figure 7c) in each case; however, these changes were not statistically significant.
Figure 7.
Influence of CRISPRi-mediated repression of genes of glycolysis (a), the pentose phosphate pathway (b), or the TCA cycle and glyoxylate shunt (c) on decaprenoxanthin biosynthesis. Mean values of biological duplicates are given. Statistical analysis was calculated with ANOVA against all measured decaprenoxanthin production of C. glutamicum MB001 from all Biolector®flowerplates and is marked by a star (*). As a reference, the decaprenoxanthin production of the empty vector strain C. glutamicum MB001 (pS_dCas9) in biological duplicates of the corresponding experiment is shown. For abbreviations, see Figure 3.
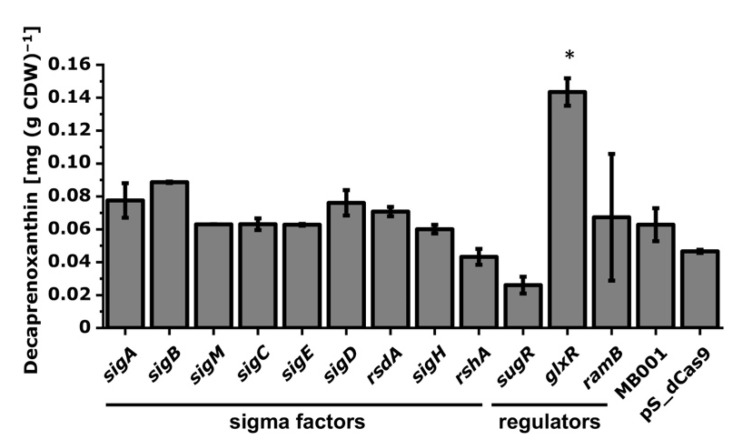
3.2.3. Interrogation of Regulatory Genes by CRISPRi with Respect to Carotenoid Biosynthesis in C. glutamicum
C. glutamicum possesses seven RNA polymerase sigma factors, which in part are regulated by their cognate anti-sigma factors [39]. The deletion of the sigma factor gene sigB is known to increase carotenoid production [77]. To identify the influence of regulatory genes on carotenoid production in C. glutamicum, RNA polymerase sigma factor and anti-sigma factor genes and the genes for the transcriptional regulators of carbon metabolism SugR, GlxR, and RamB were chosen. CRISPRi targeting of sigB and sugR increased and decreased, respectively, the cellular decaprenoxanthin content; however, the effects were not statistically relevant (Figure 8). Notably, CRISPRi-mediated repression of glxR significantly increased the cellular decaprenoxanthin by about twofold (Figure 8).
Figure 8.
Influence of CRISPRi-mediated repression of RNA polymerase sigma factor and transcriptional regulator genes on decaprenoxanthin biosynthesis. Mean values of biological duplicates are given. Statistical analysis was calculated with ANOVA against all measured decaprenoxanthin production of C. glutamicum MB001 from all Biolector®flowerplates and is marked by a star (*). As a reference, the decaprenoxanthin production of the empty vector strain C. glutamicum MB001 (pS_dCas9) in biological duplicates of the corresponding experiment is shown. For abbreviations, see Figure 3.
3.3. Deletion of Selected Target Genes Identified by CRISPRi Repression
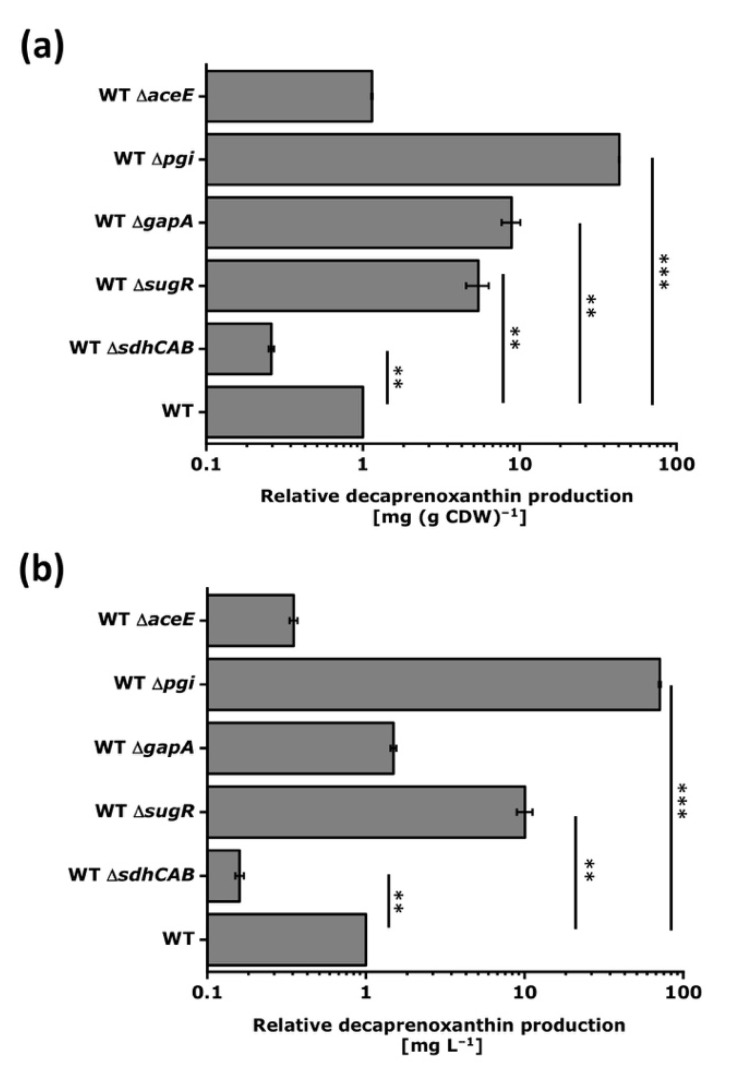
Gene deletion is a favored metabolic engineering strategy since the constructed strains are genetically stable (except when suppressor mutations occur in trans). Gene deletion results in the complete loss of function (knockout), while CRISPRi-mediated repression reduces gene function (knockdown). A gene can be repressed by CRISPRi, but not deleted, if its function is essential. The MEP pathway genes, the RNA polymerase sigma factor A gene sigA, and, according to some reports, the regulatory gene glxR cannot be deleted in C. glutamicum. Deletion of the crt operon is known to abolish decaprenoxanthin biosynthesis [33,40], thus supporting the evidence obtained by CRISPRi (Figure 5). Deletion of crtX is known to result in accumulation of unglucosylated decaprenoxanthin [32]. In addition, we chose other genes to study the effect of gene deletion on decaprenoxanthin production, i.e., pgi, gapA, and aceE, as their deletion was expected to increase decaprenoxanthin levels, as well as sdhABCD and sugR, since their deletion was expected to reduce carotenoid biosynthesis (Figure 9).
Figure 9.
Relative decaprenoxanthin production by C. glutamicum strains carrying various deletions. Cells were grown in 40 g L−1 of glucose CGXII minimal medium. C. glutamicum WT ΔaceE was grown with 20 g L−1 of potassium acetate and 40 g L−1 of glucose in CGXII minimal medium. Decaprenoxanthin production was determined by HPLC analysis. Due to the medium differences, decaprenoxanthin contents in mg g−1 CDW (a) and concentrations in mg L−1 (b) were normalized to the values obtained with the parental WT strain. Mean values and standard deviations of triplicates are given. The p-values of <0.001 (***), and <0.01 (**) were calculated using Student’s t-test (two sided, unpaired). For abbreviations, see Figure 3.
In line with the CRISPRi results, deletion of the sdhABCD operon significantly reduced decaprenoxanthin levels in comparison to C. glutamicum WT, while deletion of pgi and gapA improved the cellular decaprenoxanthin content significantly (Figure 9a). Since the gapA deletion mutant showed impaired growth, the decaprenoxanthin concentration in the culture was not increased (Figure 9b). Therefore, the pleiotropic effects of gapA deletion nullified the increased decaprenoxanthin content per cell biomass as the biomass concentration was reduced. SugR represses many target genes and binds their promoter DNA regions with different affinities in vitro [78]; thus, besides pleiotropic effects, gradual regulatory differences may be expected as well. In contrast to the CRISPRi results, deletion of sugR increased the decaprenoxanthin content in comparison to the control strain fivefold. Although not understood mechanistically, these results indicate that the complete absence of SugR upon gene deletion is beneficial, while SugR levels below those of the WT strain upon CRISPRi targeting may limit decaprenoxanthin biosynthesis.
Deletion of aceE is known to be possible, but the resulting mutant requires a source of acetyl-CoA, such as acetate, for growth [60]. Therefore, with glucose minimal medium, the positive effect observed of CRISPRi targeting aceE could not be tested by the deletion of aceE since aceE deletion is conditionally lethal under these conditions. To circumvent this problem, the aceE mutant was assayed for decaprenoxanthin biosynthesis in glucose medium supplemented with acetate. The decaprenoxanthin content per biomass of the aceE mutant grown with glucose and acetate was, however, not higher than that of the WT strain grown with glucose.
Taken together, these results highlight the advantage of CRISPRi screening over deletion analysis. On the one hand, it allows for fast target gene identification for metabolic engineering, and transfer to the construction of genetically stable strains by gene deletion often is straightforward (sdhCAB, pgi, gapA). On the other hand, CRISPRi is a well-suited method to analyze essential genes, conditionally lethal genes such as aceE, as well as pleiotropic genes or regulatory genes.
4. Discussion
In this study, the first C. glutamicum CRISPRi library for the repression of 74 genes (e.g., those for central metabolism, coding for global regulators and RNA polymerase sigma factors, besides the genes for carotenoid biosynthesis) was constructed and tested to identify targets affecting carotenogenesis. As expected, decaprenoxanthin levels were reduced when the MEP pathway or carotenogenesis genes were repressed. In addition, eight new targets to improve carotenoid production were identified. Using deletions instead of CRISPRi repression for five selected genes supported CRISPRi results for some genes (sdhABCD, pgi, gapA). This was not the case for the conditionally lethal aceE deletion and for the pleiotropic deletion of the global regulatory gene sugR.
CRISPRi screenings were useful to identify promising targets for increased carotenoid production in other organisms [79,80]. Clearly, CRISPRi repression is complementary to gene deletion, not only because CRISPRi allows functional analysis of essential genes by assigning phenotypes as a consequence of their repression, but also because regulatory genes can be titrated, resulting in phenotypes that are intermediate between the levels in the wild type and the null levels in a deletion mutant. In C. glutamicum systems biology, loss of function analysis by gene deletions is often coupled with transcriptome, proteome, metabolome, and fluxome studies to gain insight, for example, into regulons and modulons [81]. Similarly, CRISPRi libraries may contribute to a systems-level understanding of an organism when coupled with omics experiments, allowing one to assess responses to gradual rather than absence/presence perturbations imposed by gene deletions. Transcriptome analysis may unravel multiple levels of global gene expression patterns associated with down-regulated transcriptional regulator protein levels complementing insight from regulatory gene knockouts. The carbon metabolism regulator SugR binds to its target promoter DNA sequences with a wide range of affinities in vitro [25,82,83,84,85]. The differences observed in vivo between CRISPRi repression and deletion of the gene sugR with regard to decaprenoxanthin accumulation (Figure 8 and Figure 9) support this notion. This may call for an in-depth analysis of the effects of gradually reduced SugR protein levels between wild-type levels and zero in the deletion mutant using CRISPRi combined with genome-wide transcriptome analysis. These analogous vs. digital (CRISPRi vs. deletion) perturbation experiments may also provide insight into the gradual changes at the proteome, metabolome, or fluxome levels [86].
Systems metabolic engineering finely balances enzymes of the pathway of interest (including contributory pathways) using different promoters, ribosome-binding sites, or synthetic operon structures [2,87,88,89] in order to achieve high productivities. Recently, CRISPRi was used to adjust different levels of the arginine biosynthesis repressor ArgR in E. coli, which accelerated growth twofold as compared to the deletion of argR, while specific arginine production remained similar [90]. In a similar study, CRISPRi was applied to adjust cell growth by repression of key growth-related genes. Upon proper choice of sgRNAs, addition time, and induction level, carbon flux was precisely redistributed between biomass formation and synthesis of N-acetyl-glucosamine as the target product, which was produced up to about 90 g L−1 [91]. The approach described here may in the future be used for multiplexing since it is likely that CRISPRi multiplex screening will identify synergistic effects if two or more genes are repressed simultaneously by CRISPRi.
Decaprenoxanthin biosynthesis was significantly improved when pgi was repressed by CRISPRi or was deleted (Figure 7, Figure 9 and Figure 10). NADPH in C. glutamicum mainly derives from the pentose phosphate pathway [92] and is known to limit l-lysine production [93], especially during growth on carbon sources that support low PPP fluxes, such as acetate [20] or fructose [94,95]. Deletion of pgi is known to be beneficial for l-lysine production [96]. While production of 1 molecule of l-lysine requires 4 molecules of NADPH, 35 molecules of reduction equivalents (NADPH, reduced ferredoxin) are required per molecule of decaprenoxanthin. For the biosynthesis of the 10 C5 isoprenoid pyrophosphate precursors of decaprenoxanthin (2 DMAPP, 6 IPP, and 2 HMBPP) from GAP and pyruvate, 38 reduction equivalents are needed: 1 NADPH and 3 reduced ferredoxins per IPP or DMAPP (in the reactions of Dxr, IspG, and IspH) as well as 1 NADPH and 2 reduced ferredoxins per HMBPP (in the reactions of Dxr and IspG). Conversion of the 10 C5 precursors to the C50 decaprenoxanthin yields 3 reduced ferredoxins (1 in the reaction catalyzed by lycopene synthase CrtI and 2 in the elongation reactions catalyzed by lycopene elongase CrtEb; Figure 3). Thus, biosynthesis of decaprenoxanthin imposes a high demand for reduction equivalents to the cell. Improved NADPH provision may also be reached by overexpression of PPP genes, e.g., the genes coding for glucose-6-phosphate dehydrogenase (Zwf) [45], transketolase (Tkt), and transaldolase (Tal) [44]. Engineering of the PPP and the TCA cycle increased β-carotene production in E. coli by 64% [44] and in S. cerevisiae by 81.4% [45]. The importance of the PPP for decaprenoxanthin production was confirmed here when targeting the genes tkt, tal, zwf, and pgl by CRISPRi significantly decreased cellular decaprenoxanthin content (Figure 7b).
Figure 10.
Scheme of C. glutamicum metabolism with CRISPRi target genes that significantly improved (red arrows) or reduced (black arrows) decaprenoxanthin biosynthesis in C. glutamicum when repressed. Grey is used to depict all other reactions in glycolysis (orange shading), the pentose phosphate pathway (blue shading), the TCA cycle (green shading), as well as the MEP pathway and the carotenogenesis (yellow shading). Abbreviations are explained in Figure 3.
Supply of GAP and pyruvate as precursors for decaprenoxanthin biosynthesis was found to be crucial by the CRISPRi analysis (Figure 10). Repression of aceE (encoding subunit E1 of the pyruvate dehydrogenase complex that oxidatively decarboxylates pyruvate to acetyl-CoA) increased decaprenoxanthin. Although not statistically significant, CRISPRi of the fba gene coding for fructose-1,6-bisphosphate aldolase decreased decaprenoxanthin formation, while CRISPRi of the genes gapA and tpi encoding GAP converting glycolytic enzymes positively affected decaprenoxanthin formation (Figure 7). Notably, deletion of gapA increased the cellular decaprenoxanthin concentration significantly (Figure 9). However, CRISPRi repression and/or deletion of genes central to glycolysis do not come without side effects. For example, deletion of gapA cells showed an increased decaprenoxanthin content, but led to lower biomass concentrations; thus, the decaprenoxanthin titer was not higher (compare Figure 9a with Figure 9b). Moreover, deletion of aceE has been shown to abolish growth with glucose alone [60], and a positive effect on decaprenoxanthin production was not observable after growth on a glucose–acetate mixture (Figure 9). The results with aceE and gapA clearly showed that rerouting a major pathway of the central metabolism toward the production of decaprenoxanthin may critically interfere with growth. For example, the central carbon metabolism provides ATP, and indirectly CTP, which are required for isoprenoid pyrophosphate and carotenoid biosynthesis [15]. Thus, targets identified by CRISPRi library screening may require fine balancing of gene expression/repression to optimize decaprenoxanthin production without impairing growth.
Inherent problems in the interpretation of CRISPRi results when targeting an upstream gene of an operon exist due to polar effects on the downstream co-transcribed genes of that operon [97]. The crtE-mmpL-crtB-crtI-crtYe-crtYf-crtEb operon (Figure 6) consists of seven co-transcribed genes [30,33]. CRISPRi repression of the chosen genes of this operon reduced decaprenoxanthin biosynthesis (Figure 5). Besides gene-specific effects, polar effects have to be considered. CRISPRi targeting of the first and second crt operon genes (crtE and mmpl) significantly reduced mRNA levels of crtEb, the last gene in the operon (Figure 6). In particular, reduced decaprenoxanthin upon repression of crtE came as a surprise since deletion of crtE did not abolish carotenogenesis, while deletion of the crtE paralog idsA did [75]. Therefore, the finding that CRISPRi repression of idsA did not reduce decaprenoxanthin was not anticipated (Figure 5b). Since it has been shown that plasmid-borne overexpression of crtE compensates for the deletion of idsA [75], it is likely that partial repression of idsA by CRISPRi was compensated for by CrtE.
5. Conclusions
The C. glutamicum CRISPRi library designed in this study was shown to be suitable to score targets for improving decaprenoxanthin production, the application example chosen here. The inherent limitations of this screening approach with regard to the transfer of CRISPRi results to clean gene deletions became obvious as they reflect the change from a gradual analogous approach to a digital yes/no approach. With these limitations in mind, many applications of this CRISPRi library and its future extensions become evident, in particular when combined with systems approaches targeted at gaining a basic physiological understanding or at achieving superior biotechnological performance of C. glutamicum.
Acknowledgments
We thank Florian Meyer for the scientific discussion. We thank Jung-Won Youn for construction of the sdhCAB deletion strain. We also want to thank Michelle Celine Schwab for support in plasmid construction.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/4/670/s1: Figure S1: Duration of the inhibitory effect of the CRISPRi targeting crtR; Table S1: Oligonucleotides used in this study; Table S2: Annealing oligo primers for construction complementary regions of sgRNA templates; Table S3: Data generated with the CRISPRi library for all targets.
Author Contributions
V.L.G., P.P.-W., V.F.W. and N.A.H. designed the experiments. N.A.H. and V.F.W. acquired funding. V.F.W. and N.A.H. coordinated the study. V.L.G., I.S. and K.B. performed the experiments. All authors analyzed the data. V.L.G. drafted the manuscript. V.F.W. and N.A.H. finalized the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part by the European Regional Development Fund (ERDF) and the Ministry of Economic Affairs, Innovation, Digitalization and Energy of the State of North Rhine-Westphalia (grant number EFRE-0400184 (Bicomer) and grant number EFRE-0300095/1703FI04 (Cluster Industrial Biotechnology Kompetenzzentrum Biotechnologie CKB)). Support for the article processing charge by the Deutsche Forschungsgemeinschaft and the Open Access Publication Fund of Bielefeld University is acknowledged.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are present in the manuscript and its supplement.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Adli M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee J.-H., Wendisch V.F. Production of amino acids—Genetic and metabolic engineering approaches. Bioresour. Technol. 2017;245:1575–1587. doi: 10.1016/j.biortech.2017.05.065. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Horvath P., Barrangou R. CRISPR/Cas, the Immune System of Bacteria and Archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 5.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larson M.H., Gilbert L.A., Wang X., Lim W.A., Weissman J.S., Qi L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schultenkämper K., Brito L.F., Wendisch V.F. Impact of CRISPR interference on strain development in biotechnology. Biotechnol. Appl. Biochem. 2020;67:7–21. doi: 10.1002/bab.1901. [DOI] [PubMed] [Google Scholar]
- 8.Li S., Jendresen C.B., Landberg J., Pedersen L.E., Sonnenschein N., Jensen S.I., Nielsen A.T. Genome-wide CRISPRi-based identification of targets for decoupling growth from production. ACS Synth. Biol. 2020;9:1030–1040. doi: 10.1021/acssynbio.9b00143. [DOI] [PubMed] [Google Scholar]
- 9.Zhao H., Sun Y., Peters J.M., Gross C.A., Garner E.C., Helmann J.D. Depletion of undecaprenyl pyrophosphate phosphatases disrupts cell envelope biogenesis in Bacillus subtilis. J. Bacteriol. 2016;198:2925–2935. doi: 10.1128/JB.00507-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey J.E., Sburlati A., Hatzimanikatis V., Lee K., Renner W.A., Tsai P.S. Inverse Metabolic Engineering: A Strategy for Directed Genetic Engineering of Useful Phenotypes. Biotechnol. Bioeng. 1996;52:13. doi: 10.1002/(SICI)1097-0290(19961005)52:1<109::AID-BIT11>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Cheng H., Liu Y., Liu Y., Wen X., Zhang K., Ni X., Gao N., Fan L., Zhang Z., et al. In-situ generation of large numbers of genetic combinations for metabolic reprogramming via CRISPR-guided base editing. Nat. Commun. 2021;12:678. doi: 10.1038/s41467-021-21003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rousset F., Cui L., Siouve E., Becavin C., Depardieu F., Bikard D. Genome-wide CRISPR-dCas9 screens in E. coli identify essential genes and phage host factors. PLoS Gene. 2018;14:e1007749. doi: 10.1371/journal.pgen.1007749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui L., Vigouroux A., Rousset F., Varet H., Khanna V., Bikard D. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-04209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emmerstorfer-Augustin A., Moser S., Pichler H. Screening for improved isoprenoid biosynthesis in microorganisms. J. Biotechnol. 2016;235:112–120. doi: 10.1016/j.jbiotec.2016.03.051. [DOI] [PubMed] [Google Scholar]
- 15.Alper H., Jin Y.-S., Moxley J.F., Stephanopoulos G. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng. 2005;7:155–164. doi: 10.1016/j.ymben.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Park C.-S., Lee S.-W., Kim Y.-S., Kim E.-J., Sin H.-S., Oh D.-K., Kim S.-W., Um S.-J. Utilization of the recombinant human β-carotene-15,15′-monooxygenase gene in Escherichia coli and mammalian cells. Biotechnol. Lett. 2008;30:735–741. doi: 10.1007/s10529-007-9598-9. [DOI] [PubMed] [Google Scholar]
- 17.Armstrong G.A. Eubacteria Show Their True Colors: Genetics of Carotenoid Pigment Biosynthesis from Microbes to Plants. J. Bacteriol. 1994;176:4795–4802. doi: 10.1128/JB.176.16.4795-4802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belviranlı M., Okudan N. Well-Known Antioxidants and Newcomers in Sport Nutrition: Coenzyme Q10, Quercetin, Resveratrol, Pterostilbene, Pycnogenol and Astaxanthin. In: Lamprecht M., editor. Antioxidants in Sport Nutrition. CRC Press; Boca Raton, FL, USA: 2014. pp. 79–102. [PubMed] [Google Scholar]
- 19.Blombach B., Seibold G.M. Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of l-lysine production strains. Appl. Microbiol. Biotechnol. 2010;86:1313–1322. doi: 10.1007/s00253-010-2537-z. [DOI] [PubMed] [Google Scholar]
- 20.Wendisch V.F., de Graaf A.A., Sahm H., Eikmanns B.J. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: Comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 2000;182:3088–3096. doi: 10.1128/JB.182.11.3088-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker J., Wittmann C. Bio-based production of chemicals, materials and fuels—Corynebacterium glutamicum as versatile cell factory. Curr. Opin. Biotechnol. 2012;23:631–640. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Kogure T., Inui M. Recent advances in metabolic engineering of Corynebacterium glutamicum for bioproduction of value-added aromatic chemicals and natural products. Appl. Microbiol. Biotechnol. 2018;102:8685–8705. doi: 10.1007/s00253-018-9289-6. [DOI] [PubMed] [Google Scholar]
- 23.Kumagai H. Microbial Production of Amino Acids in Japan. In: Fiechter A., editor. History of Modern Biotechnology I. Volume 69. Springer; Berlin/Heidelberg, Germany: 2000. pp. 71–85. Advances in Biochemical Engineering/Biotechnology. [DOI] [PubMed] [Google Scholar]
- 24.Wendisch V.F. Metabolic engineering advances and prospects for amino acid production. Metab. Eng. 2020;58:17–34. doi: 10.1016/j.ymben.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Engels V., Wendisch V.F. The DeoR-Type Regulator SugR Represses Expression of ptsG in Corynebacterium glutamicum. J. Bacteriol. 2007;189:2955–2966. doi: 10.1128/JB.01596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wendisch V.F. Genome-wide expression analysis in Corynebacterium glutamicum using DNA microarrays. J. Biotechnol. 2003;104:273–285. doi: 10.1016/S0168-1656(03)00147-0. [DOI] [PubMed] [Google Scholar]
- 27.Becker J., Klopprogge C., Zelder O., Heinzle E., Wittmann C. Amplified expression of fructose 1,6-bisphosphatase in Corynebacterium glutamicum increases in vivo flux through the pentose phosphate pathway and lysine production on different carbon sources. AEM. 2005;71:8587–8596. doi: 10.1128/AEM.71.12.8587-8596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radmacher E., Eggeling L. The three tricarboxylate synthase activities of Corynebacterium glutamicum and increase of l-lysine synthesis. Appl. Microbiol. Biotechnol. 2007;76:587–595. doi: 10.1007/s00253-007-1105-7. [DOI] [PubMed] [Google Scholar]
- 29.van Ooyen J., Noack S., Bott M., Reth A., Eggeling L. Improved l-lysine production with Corynebacterium glutamicum and systemic insight into citrate synthase flux and activity. Biotechnol. Bioeng. 2012;109:2070–2081. doi: 10.1002/bit.24486. [DOI] [PubMed] [Google Scholar]
- 30.Krubasik P., Takaichi S., Maoka T., Kobayashi M., Masamoto K., Sandmann G. Detailed biosynthetic pathway to decaprenoxanthin diglucoside in Corynebacterium glutamicum and identification of novel intermediates. Arch. Microbiol. 2001;176:217–223. doi: 10.1007/s002030100315. [DOI] [PubMed] [Google Scholar]
- 31.Henke N., Heider S., Peters-Wendisch P., Wendisch V. Production of the Marine Carotenoid Astaxanthin by Metabolically Engineered Corynebacterium glutamicum. Mar. Drugs. 2016;14:124. doi: 10.3390/md14070124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heider S.A.E., Peters-Wendisch P., Netzer R., Stafnes M., Brautaset T., Wendisch V.F. Production and glucosylation of C50 and C40 carotenoids by metabolically engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2014;98:1223–1235. doi: 10.1007/s00253-013-5359-y. [DOI] [PubMed] [Google Scholar]
- 33.Heider S.A.E., Peters-Wendisch P., Wendisch V.F. Carotenoid biosynthesis and overproduction in Corynebacterium glutamicum. BMC Microbiol. 2012;12:198. doi: 10.1186/1471-2180-12-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang M.-K., Eom J.-H., Kim Y., Um Y., Woo H.M. Biosynthesis of pinene from glucose using metabolically-engineered Corynebacterium glutamicum. Biotechnol. Lett. 2014;36:2069–2077. doi: 10.1007/s10529-014-1578-2. [DOI] [PubMed] [Google Scholar]
- 35.Ravikumar S., Woo H.M., Choi J. Analysis of novel antioxidant sesquarterpenes (C35 terpenes) produced in recombinant Corynebacterium glutamicum. Appl. Biochem. Biotechnol. 2018;186:525–534. doi: 10.1007/s12010-018-2756-9. [DOI] [PubMed] [Google Scholar]
- 36.Henke N., Wichmann J., Baier T., Frohwitter J., Lauersen K., Risse J., Peters-Wendisch P., Kruse O., Wendisch V. Patchoulol Production with Metabolically Engineered Corynebacterium glutamicum. Genes. 2018;9:219. doi: 10.3390/genes9040219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim H., Park J., Woo H.M. Overexpression of the key enzymes in the methylerythritol 4-phosphate pathway in Corynebacterium glutamicum for improving farnesyl diphosphate-derived terpene production. J. Agric. Food Chem. 2020;68:10780–10786. doi: 10.1021/acs.jafc.0c04307. [DOI] [PubMed] [Google Scholar]
- 38.Binder D., Frohwitter J., Mahr R., Bier C., Grünberger A., Loeschcke A., Peters-Wendisch P., Kohlheyer D., Pietruszka J., Frunzke J., et al. Light-Controlled Cell Factories: Employing Photocaged Isopropyl-β-d-Thiogalactopyranoside for Light-Mediated Optimization of Lac Promoter-Based Gene Expression and (+)-Valencene Biosynthesis in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2016;82:6141–6149. doi: 10.1128/AEM.01457-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalinowski J., Bathe B., Bartels D., Bischoff N., Bott M., Burkovski A., Dusch N., Eggeling L., Eikmanns B.J., Gaigalat L., et al. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 2003;104:5–25. doi: 10.1016/S0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- 40.Krubasik P., Kobayashi M., Sandmann G. Expression and functional analysis of a gene cluster involved in the synthesis of decaprenoxanthin reveals the mechanisms for C50 carotenoid formation: Decaprenoxanthin formation. Eur. J. Biochem. 2001;268:3702–3708. doi: 10.1046/j.1432-1327.2001.02275.x. [DOI] [PubMed] [Google Scholar]
- 41.Krubasik P., Sandmann G. A carotenogenic gene cluster from Brevibacterium linens with novel lycopene cyclase genes involved in the synthesis of aromatic carotenoids. Mol. Gen. Genet. 2000;263:423–432. doi: 10.1007/s004380051186. [DOI] [PubMed] [Google Scholar]
- 42.Jung J., Lim J.H., Kim S.Y., Im D.-K., Seok J.Y., Lee S.-J.V., Oh M.-K., Jung G.Y. Precise precursor rebalancing for isoprenoids production by fine control of gapA expression in Escherichia coli. Metab. Eng. 2016;38:401–408. doi: 10.1016/j.ymben.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Farmer W.R., Liao J.C. Precursor balancing for metabolic engineering of lycopene production in Escherichia coli. Biotechnol. Prog. 2001;17:57–61. doi: 10.1021/bp000137t. [DOI] [PubMed] [Google Scholar]
- 44.Zhao J., Li Q., Sun T., Zhu X., Xu H., Tang J., Zhang X., Ma Y. Engineering central metabolic modules of Escherichia coli for improving β-carotene production. Metab. Eng. 2013;17:42–50. doi: 10.1016/j.ymben.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Zhao X., Shi F., Zhan W. Overexpression of ZWF1 and POS5 improves carotenoid biosynthesis in recombinant Saccharomyces cerevisiae. Lett. Appl. Microbiol. 2015;61:354–360. doi: 10.1111/lam.12463. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J., Yang F., Yang Y., Jiang Y., Huo Y.-X. Optimizing a CRISPR-Cpf1-based genome engineering system for Corynebacterium glutamicum. Microb. Cell Fact. 2019;18:60. doi: 10.1186/s12934-019-1109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang Y., Qian F., Yang J., Liu Y., Dong F., Xu C., Sun B., Chen B., Xu X., Li Y., et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 2017;8:15179. doi: 10.1038/ncomms15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J., Wang Y., Lu Y., Zheng P., Sun J., Ma Y. Development of a CRISPR/Cas9 genome editing toolbox for Corynebacterium glutamicum. Microb. Cell Fact. 2017;16:205. doi: 10.1186/s12934-017-0815-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho J.S., Choi K.R., Prabowo C.P.S., Shin J.H., Yang D., Jang J., Lee S.Y. CRISPR/Cas9-coupled recombineering for metabolic engineering of Corynebacterium glutamicum. Metab. Eng. 2017;42:157–167. doi: 10.1016/j.ymben.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 50.Cleto S., Jensen J.V., Wendisch V.F., Lu T.K. Corynebacterium glutamicum Metabolic Engineering with CRISPR interference (CRISPRi) ACS Synth. Biol. 2016;5:375–385. doi: 10.1021/acssynbio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee S.S., Shin H., Jo S., Lee S.-M., Um Y., Woo H.M. Rapid identification of unknown carboxyl esterase activity in Corynebacterium glutamicum using RNA-guided CRISPR interference. Enzym. Microb. Technol. 2018;114:63–68. doi: 10.1016/j.enzmictec.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Li D., Lv L., Chen J.-C., Chen G.-Q. Controlling microbial PHB synthesis via CRISPRi. Appl. Microbiol. Biotechnol. 2017;101:5861–5867. doi: 10.1007/s00253-017-8374-6. [DOI] [PubMed] [Google Scholar]
- 53.Park J., Shin H., Lee S.-M., Um Y., Woo H.M. RNA-guided single/double gene repressions in Corynebacterium glutamicum using an efficient CRISPR interference and its application to industrial strain. Microb. Cell Fact. 2018;17:4. doi: 10.1186/s12934-017-0843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoon J., Woo H.M. CRISPR interference-mediated metabolic engineering of Corynebacterium glutamicum for homo-butyrate production. Biotechnol. Bioeng. 2018;115:2067–2074. doi: 10.1002/bit.26720. [DOI] [PubMed] [Google Scholar]
- 55.Zhang B., Liu Z.-Q., Liu C., Zheng Y.-G. Application of CRISPRi in Corynebacterium glutamicum for shikimic acid production. Biotechnol. Lett. 2016;38:2153–2161. doi: 10.1007/s10529-016-2207-z. [DOI] [PubMed] [Google Scholar]
- 56.Gauttam R., Seibold G.M., Mueller P., Weil T., Weiß T., Handrick R., Eikmanns B.J. A simple dual-inducible CRISPR interference system for multiple gene targeting in Corynebacterium glutamicum. Plasmid. 2019;103:25–35. doi: 10.1016/j.plasmid.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 57.Baumgart M., Unthan S., Rückert C., Sivalingam J., Grünberger A., Kalinowski J., Bott M., Noack S., Frunzke J. Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl. Environ. Microbiol. 2013;79:6006–6015. doi: 10.1128/AEM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eggeling L., Reyes O. Handbook of Corynebacterium glutamicum. CRC Press; Boca Raton, FL, USA: 2005. Experiments; pp. 3535–3566. [Google Scholar]
- 59.Abe S., Takayama K.-I., Kinoshita S. Taxonomical studies on glutamic acid-producing bacteria. J. Gen. Appl. Microbiol. 1967;13:279–301. doi: 10.2323/jgam.13.279. [DOI] [Google Scholar]
- 60.Schreiner M.E., Fiur D., Holátko J., Pátek M., Eikmanns B.J., Bacteriol J. E1 Enzyme of the Pyruvate Dehydrogenase Complex in Corynebacterium glutamicum: Molecular Analysis of the Gene and Phylogenetic Aspects. J. Bacteriol. 2005;187:6005–6018. doi: 10.1128/JB.187.17.6005-6018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reddy G.K., Lindner S.N., Wendisch V.F. Metabolic engineering of an ATP-neutral Embden-Meyerhof-Parnas pathway in Corynebacterium glutamicum: Growth restoration by an adaptive point mutation in NADH dehydrogenase. Appl. Environ. Microbiol. 2015;81:1996–2005. doi: 10.1128/AEM.03116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lindner S.N., Petrov D.P., Hagmann C.T., Henrich A., Krämer R., Eikmanns B.J., Wendisch V.F., Seibold G.M. Phosphotransferase system-mediated glucose uptake is repressed in phosphoglucoisomerase-deficient Corynebacterium glutamicum strains. Appl. Environ. Microbiol. 2013;79:2588–2595. doi: 10.1128/AEM.03231-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Henke N.A., Heider S.A.E., Hannibal S., Wendisch V.F., Peters-Wendisch P. Isoprenoid pyrophosphate-dependent transcriptional regulation of carotenogenesis in Corynebacterium glutamicum. Front. Microbiol. 2017;8:1–15. doi: 10.3389/fmicb.2017.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 65.Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbachb G., Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 66.Schultenkämper K., Brito L.F., López M.G., Brautaset T., Wendisch V.F. Establishment and application of CRISPR interference to affect sporulation, hydrogen peroxide detoxification, and mannitol catabolism in the methylotrophic thermophile Bacillus methanolicus. Appl. Microbiol. Biotechnol. 2019;103:5879–5889. doi: 10.1007/s00253-019-09907-8. [DOI] [PubMed] [Google Scholar]
- 67.Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 68.Sambrook J., Russell D. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; New York, NY, USA: 2001. [Google Scholar]
- 69.Blin K., Pedersen L.E., Weber T., Lee S.Y. CRISPy-Web: An online resource to design sgRNAs for CRISPR applications. Synth. Syst. Biotechnol. 2016;1:118–121. doi: 10.1016/j.synbio.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eggeling L., Bott M. Handbook of Corynebacterium glutamicum. 1st ed. CRC Press; Bota Raton, FL, USA: 2004. [Google Scholar]
- 71.van der Rest M.E., Lange C., Molenaar D. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 1999;52:541–545. doi: 10.1007/s002530051557. [DOI] [PubMed] [Google Scholar]
- 72.Higuchi R., Dollinger G., Walsh P.S., Griffith R. Simultaneous amplification and detection of specific DNA sequences. Nat. Biotechnol. 1992;10:413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 73.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 74.Ajikumar P.K., Xiao W.-H., Tyo K.E.J., Wang Y., Simeon F., Leonard E., Mucha O., Phon T.H., Pfeifer B., Stephanopoulos G. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heider S.A.E., Peters-Wendisch P., Beekwilder J., Wendisch V.F. IdsA is the major geranylgeranyl pyrophosphate synthase involved in carotenogenesis in Corynebacterium glutamicum. FEBS J. 2014;281:4906–4920. doi: 10.1111/febs.13033. [DOI] [PubMed] [Google Scholar]
- 76.Moise A.R., Al-Babili S., Wurtzel E.T. Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 2014;114:164–193. doi: 10.1021/cr400106y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taniguchi H., Henke N.A., Heider S.A.E., Wendisch V.F. Overexpression of the primary sigma factor gene SigA improved carotenoid production by Corynebacterium glutamicum: Application to production of β-carotene and the non-native linear C50 carotenoid bisanhydrobacterioruberin. Metab. Eng. Comm. 2017;4:1–11. doi: 10.1016/j.meteno.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Engels V., Lindner S.N., Wendisch V.F. The global repressor SugR controls expression of genes of glycolysis and of the l-lactate dehydrogenase LdhA in Corynebacterium glutamicum. J. Bacteriol. 2008;190:8033–8044. doi: 10.1128/JB.00705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mo X.-H., Zhang H., Wang T.-M., Zhang C., Zhang C., Xing X.-H., Yang S. Establishment of CRISPR interference in Methylorubrum extorquens and application of rapidly mining a new phytoene desaturase involved in carotenoid biosynthesis. Appl. Microbiol. Biotechnol. 2020;104:4515–4532. doi: 10.1007/s00253-020-10543-w. [DOI] [PubMed] [Google Scholar]
- 80.Lu Q., Liu J.-Z. Enhanced astaxanthin production in Escherichia coli via morphology and oxidative stress engineering. J. Agric. Food Chem. 2019;67:11703–11709. doi: 10.1021/acs.jafc.9b05404. [DOI] [PubMed] [Google Scholar]
- 81.Wendisch V.F., Bott M., Kalinowski J., Oldiges M., Wiechert W. Emerging Corynebacterium glutamicum Systems Biology. J. Biotechnol. 2006;124:74–92. doi: 10.1016/j.jbiotec.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 82.Toyoda K., Teramoto H., Inui M., Yukawa H. Molecular mechanism of SugR-mediated sugar-dependent expression of the ldhA gene encoding l-lactate dehydrogenase in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2009;83:315–327. doi: 10.1007/s00253-009-1887-x. [DOI] [PubMed] [Google Scholar]
- 83.Toyoda K., Teramoto H., Inui M., Yukawa H. Expression of the gapA gene encoding glyceraldehyde-3-phosphate dehydrogenase of Corynebacterium glutamicum is regulated by the global regulator SugR. Appl. Microbiol. Biotechnol. 2008;81:291–301. doi: 10.1007/s00253-008-1682-0. [DOI] [PubMed] [Google Scholar]
- 84.Tanaka Y., Teramoto H., Inui M., Yukawa H. Regulation of expression of general components of the phosphoenolpyruvate: Carbohydrate phosphotransferase system (PTS) by the global regulator SugR in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2008;78:309–318. doi: 10.1007/s00253-007-1313-1. [DOI] [PubMed] [Google Scholar]
- 85.Gaigalat L., Schlüter J.-P., Hartmann M., Mormann S., Tauch A., Pühler A., Kalinowski J. The DeoR-type transcriptional regulator SugR acts as a repressor for genes encoding the phosphoenolpyruvate:sugar phosphotransferase system (PTS) in Corynebacterium glutamicum. BMC Mol. Biol. 2007;8:104. doi: 10.1186/1471-2199-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Donati S., Kuntz M., Pahl V., Farke N., Beuter D., Glatter T., Gomes-Filho J.V., Randau L., Wang C.-Y., Link H. Multi-omics analysis of CRISPRi-knockdowns identifies mechanisms that buffer decreases of enzymes in E. coli metabolism. Cell Syst. 2021;12:56–67. doi: 10.1016/j.cels.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 87.Alper H.S., Avalos J.L. Metabolic Pathway Engineering. Synth. Syst. Biotechnol. 2018;3:1–2. doi: 10.1016/j.synbio.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blazeck J., Alper H.S. Promoter engineering: Recent advances in controlling transcription at the most fundamental level. Biotechnol. J. 2013;8:46–58. doi: 10.1002/biot.201200120. [DOI] [PubMed] [Google Scholar]
- 89.Park J.H., Lee S.Y., Kim T.Y., Kim H.U. Application of systems biology for bioprocess development. Trends Biotechnol. 2008;26:404–412. doi: 10.1016/j.tibtech.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 90.Sander T., Wang C.Y., Glatter T., Link H. CRISPRi-based downregulation of transcriptional feedback improves growth and metabolism of arginine overproducing E. coli. ACS Synth. Biol. 2019;8:1983–1990. doi: 10.1021/acssynbio.9b00183. [DOI] [PubMed] [Google Scholar]
- 91.Zhang Q., Hou Z., Ma Q., Mo X., Sun Q., Tan M., Xia L., Lin G., Yang M., Zhang Y., et al. CRISPRi-based dynamic control of carbon flow for efficient N -acetyl glucosamine production and its metabolomic effects in Escherichia coli. J. Agric. Food Chem. 2020;68:3203–3213. doi: 10.1021/acs.jafc.9b07896. [DOI] [PubMed] [Google Scholar]
- 92.Marx A., de Graaf A.A., Wiechert W., Eggeling L., Sahm H. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol. Bioeng. 1996;49:111–129. doi: 10.1002/(SICI)1097-0290(19960120)49:2<111::AID-BIT1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 93.Marx A., Striegel K., de Graaf A.A., Sahm H., Eggeling L. Response of the central metabolism of Corynebacterium glutamicum to different flux burdens. Biotechnol. Bioeng. 1997;56:168–180. doi: 10.1002/(SICI)1097-0290(19971020)56:2<168::AID-BIT6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 94.Kiefer P., Heinzle E., Zelder O., Wittmann C. Comparative metabolic flux analysis of lysine-producing Corynebacterium glutamicum cultured on glucose or fructose. AEM. 2004;70:229–239. doi: 10.1128/AEM.70.1.229-239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Georgi T., Rittmann D., Wendisch V.F. Lysine and glutamate production by Corynebacterium glutamicum on glucose, fructose and sucrose: Roles of malic enzyme and fructose-1,6-bisphosphatase. Metab. Eng. 2005;7:291–301. doi: 10.1016/j.ymben.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 96.Marx A., Hans S., Möckel B., Bathe B., de Graaf A.A. Metabolic phenotype of phosphoglucose isomerase mutants of Corynebacterium glutamicum. J. Biotechnol. 2003;104:185–197. doi: 10.1016/S0168-1656(03)00153-6. [DOI] [PubMed] [Google Scholar]
- 97.Goyal A., Myacheva K., Groß M., Klingenberg M., Duran Arqué B., Diederichs S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2016:1–12. doi: 10.1093/nar/gkw883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are present in the manuscript and its supplement.