Abstract
The aim of the present study was to assess the technological and safety potential of 207 lactic acid bacteria (LAB) and 195 yeast strains isolated from spontaneously fermented Greek wheat sourdoughs. More accurately, the amylolytic, proteolytic, lipolytic, phytase and amino acid decarboxylase activities, along with the production of exopolysaccharides and antimicrobial compounds by the LAB and yeast isolates, were assessed. A well diffusion assay revealed seven proteolytic LAB and eight yeast strains; hydrolysis of tributyrin was evident only in 11 LAB strains. A further Sodium Dodecyl Sulphate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) indicated partial hydrolysis of gluten. Lipolysis kinetics over 21 days was applied, exhibiting that lipolytic activity ranged from 6.25 to 65.50 AU/mL. Thirteen LAB inhibited Penicillium olsonii and Aspergillus niger growth and 12 yeast strains inhibited Pe. chrysogenum growth. Twenty-one Lactiplantibacillus plantarum strains exhibited inhibitory activity against Listeria monocytogenes, as well as several sourdough-associated isolates. The structural gene encoding plantaricin 423 was detected in 19 Lcb. plantarum strains, while the structural genes encoding plantaricins NC8, PlnE/F, PlnJ/K, and S were detected in two Lcb. plantarum strains. None of the microbial strains tested exhibited exopolysaccharide (EPS) production, amino acid decarboxylase, amylolytic or phytase activity. The technological and safety potential of the Lcb. plantarum and Wickerhamomyces anomalus strains was highlighted, since some of them exhibited proteolytic, lipolytic, antibacterial and antimould activities.
Keywords: proteolysis, lipolysis, antimicrobial compounds, plantaricins, Lactiplantibacillus plantarum, Wickerhamomyces anomalus
1. Introduction
Within the past few years, the ever-increasing consumer demand for “clean label” products has shifted the technological interest of baking industries towards the development of more ecologically friendly methods of preserving foods, such as sourdough fermentation [1]. The incorporation of sourdough into bread making imparts positive effects on all aspects of bread quality, namely technological, sensorial, safety and nutritional attributes. The production of microbial metabolites, which further affect the quality of the end product, is highly dependent on the contribution of lactic acid bacteria (LAB) and yeast strains, which form the sourdough microecosystem [2]. Thus, suitable starter cultures with defined metabolic properties should be carefully selected to assure the reproducibility of the process at industrial level and, at the same time, develop a bakery product with the desired sensorial traits.
As far as the technological properties are concerned, amylase activity in wheat-based sourdoughs is involved in starch hydrolysis, with concomitant liberation of fermentable sugars, namely maltose, glucose and maltodextrines [3]. Despite the fact that α-amylase is regularly absent in flours—meaning its supplementation is necessary in order to increase the presence of fermentable sugars—excessive levels of amylase have been reported as undesirable [4]. However, for the majority of the LAB species derived from wheat-based fermentations, amylase activity has not been considered a common enzymatic property. Apart from amylase activity, amino acids and peptides liberated during proteolysis affect leavened baked goods as both flavor and bioactive compounds [5]. Initial protein degradation to oligopeptides is carried out by cereal proteases, namely aspartic proteases, which are activated under acidic conditions [3]. Further proteolysis is dependent on strain-specific intracellular peptidases of LAB, with the subsequent release of free amino acids [6,7]. The combination of sourdough LAB with mould enzymes has been reported to lead to complete protein hydrolysis, with increased levels of free amino acids [8]. Regarding lipolysis in sourdoughs, only scarce literature is available [2,8]. Although wheat and rye lipids constitute a small part of the corresponding flours, their degradation into flavor precursors, such as aldehydes and alcohols, strongly affects the rheological properties of the final product. As far as homofermentative lactobacilli are concerned, an increase in products from lipid oxidation has been reported [3]. Another enzymatic activity with a nutritional impact, namely phytase activity, results in the degradation of phytic acid present in the cereal grain structure [9,10]. Phytic acid is considered an antinutritional factor due to its ability to create insoluble complexes with essential minerals, namely Ca2+, Zn2+, Fe2+, and Mg2+, thus preventing mineral bioavailability [11]. Apart from flour endogenous phytases, several bacterial and yeast isolates have been reported to possess phytase activity, thus contributing to decreased levels of phytate content as well [12,13]. The capacity of sourdough LAB to produce exopolysaccharides (EPS) is another trait with technological significance; this is due to their application as a replacement for commercial hydrocolloids in the bread making process, with subsequent improvements in water absorption of the dough, bread rheology, texture, and shelf life [14]. Apart from technological contributions, EPS from sourdough LAB have been associated with biofilm formation, pathogen exclusion, and prebiotic activity, too. Generally, sourdough LAB have been reported to produce homopolysaccharides (HoPS), while only few a isolates have been associated with heteropolysaccharide (HePS) production [15].
The screening of sourdough microorganisms for antimould and antibacterial activity has been an area of increasing focus, aiming to select appropriate starters that can further improve the shelf life and safety of end products, with respect to consumer demands for less chemical preservatives [16,17]. The capacity of LAB and non-conventional yeast strains to control mould spoilage caused by species of the genera Aspergillus, Penicillium and Fusarium has been thoroughly studied, and is mainly attributed to the synergistic effect of different organic acids, peptides, hydroxyl fatty acids, and phenolic compounds [18,19]. As far as antibacterial capacity is concerned, bacteriocin production has gained much attention the past few years as an alternative biopreservation technique [20]. Although LAB strains producing antibacterial compounds are not intended for extending the shelf life of sourdough fermented cereal foods, metabolic products synthesized during the fermentation process have been reported to enhance the stability of sourdough, thus leading to the production of microbiologically safer products [8]. An additional safety attribute to consider is the inability of starters to form biogenic amines (BAs) during sourdough fermentations [21,22]. In food fermentations, LAB are the main BAs producers through their decarboxylase activity, thus, an increase in BAs accumulation has been reported [23]. In the case of bakery products, although relatively low levels of BAs have been reported [24], the capacity of sourdough microorganisms to form them has not been adequately assessed.
Several authors have studied the properties of yeast and LAB sourdough isolates. However, in the majority of these studies, only a limited set of properties are included, thus offering a restricted interpretation of the capacity of the isolates and the effect they may have on the quality of the final product. The aim of the present study was to evaluate the technological and safety potential of 207 LAB and 195 yeast isolates. More accurately, the amylolytic, proteolytic, lipolytic, phytase and amino acid decarboxylase activities, along with the production of exopolysaccharides and antimicrobial compounds by the LAB and yeast isolates, were assessed.
2. Materials and Methods
2.1. Microbial Strains and Culture Conditions
A total of 207 lactic acid bacteria and 195 yeast isolates were obtained from thirteen Greek spontaneously fermented wheat sourdoughs [25]. The lactic acid bacteria isolates were identified as follows: Lactiplantibacillus plantarum (70 isolates); Levilactobacillus brevis (71 isolates); Companilactobacillus paralimentarius (30 isolates); Lvb. zymae (1 isolate); Latilactobacillus curvatus (6 isolates); Ltb. sakei (12 isolates); Leuconostoc citreum (1 isolate); Ln. mesenteroides (1 isolate); Lactococcus lactis (3 isolates); and Fructilactobacillus sanfranciscensis (12 isolates). The yeast isolates were identified as follows: Saccharomyces cerevisiae (161 isolates); Kazachstania humilis (2 isolates); Pichia fermentans (8 isolates); Pi. membranifaciens (18 isolates); and Wickerhamomyces anomalus (6 isolates). All isolates were stored at −20 °C in a Nutrient broth supplemented with 50% glycerol (Applichem, Darmstadt, Germany). Before experimental use, lactic acid bacteria and yeast isolates were grown twice in de Mann, Rogosa, and Sharpe (MRS) broth, and in Brain Heart Infusion (BHI) broth, and their purity was examined through streaking in MRS agar and BHI agar, respectively. All substrates were from LAB M (Lancashire, UK).
2.2. Technological Properties
2.2.1. Production of Exopolysaccharides (EPS)
Screening for EPS production was performed according to Smitinont et al. [26]. More accurately, overnight bacteria cultures were used to inoculate MRS agar containing 2% glucose, fructose, maltose, or sucrose (Applichem). Incubation took place at 30 °C for 3 days. Production of the slimy phenotype was indicative of EPS production.
2.2.2. Amylase Activity
Overnight yeast and bacterial cultures were spot-inoculated on the surface of modified BHI and MRS agar, in which glucose was replaced by 2% soluble starch (Applichem) and incubated at 30 °C for 5 days. Then, the substrate was flooded with Gram’s iodine solution (Sigma-Aldrich, St. Louis, MO, USA). The presence of a clarification halo around each colony was indicative of amylase activity.
2.2.3. Proteolytic Activity
Proteolytic activity was assessed through an agar well diffusion assay. More accurately, in freshly prepared lawns of a medium consisting of 0.5% tryptone (LAB M), 0.25% yeast extract (LAB M), 0.1% glucose (Applichem), 1% gluten (Sigma-Aldrich), and 1.5% agar (LAB M), wells were aseptically punched. Overnight yeast and bacterial cultures were centrifuged (12,000× g; 15 min; 4 °C) to obtain cell-free supernatants (CFS). Then, 25 μL of each CFS was added in each well. Incubation took place at 30 °C for 5 days. Then, the substrate was stained with 0.05% (w/v) Coomassie Brilliant Blue G-250 (Applichem). The presence of a clarification halo around each well was indicative of proteolytic activity.
The strains that exhibited proteolytic activity were subjected to further study. More accurately, the overnight culture was centrifuged (12,000× g; 15 min; 4 °C), washed twice with sterile saline, and mixed with a dough made of 10 g wheat flour (T70) and 20 mL tap water. Incubation took place at 30 °C for 24 h. Uninoculated doughs were used as controls. After incubation, albumins, globulins, gliadins, and glutenins fractions were obtained according to Di Cagno et al. [27]. Decomposition of the gluten fractions was assessed through Sodium Dodecyl Sulphate-Polyacrylamide Gel Electrophoresis (SDS-PAGE), in 12% polyacrylamide gel, according to Paramithiotis et al. [28].
2.2.4. Lipolytic Activity
Lipolytic activity was assessed through an agar well diffusion assay. More accurately, the medium consisted of 0.5% peptone (LAB M), 0.3% meat extract (LAB M), 0.5% lecithin (Serva, Heidelberg, Germany), 1% tributyrin (Merck, Darmstadt, Germany), and 1.5% agar, with the addition of 2.5 mM CaCl2 (Applichem) and 5 mM MgSO4 (Applichem), according to Carrazco-Palafox et al. [29]. Then, wells were aseptically punched. Overnight yeast and bacterial cultures were centrifuged (12,000× g; 15 min; 4 °C) to obtain CFS. Twenty-five (25) μL of each CFS were added in each well. Incubation took place at 30 °C for 10 days. The presence of a clarification halo around each well was indicative of lipolytic activity.
Strains that exhibited lipolytic activity were subjected to further study. More accurately, overnight culture was centrifuged (12,000× g; 15 min; 4 °C), washed twice with sterile saline, and used to inoculate flasks containing the above medium without agar addition. Incubation took place under shaking (200 rpm) at 30 °C for 21 days. The pH value and total titratable acidity of the samples were determined at days 3, 6, 9, 12, 15, 18 and 21. Uninoculated flasks served as controls. Lipolytic activity was expressed in AU/mL; one arbitrary unit was defined as the amount of enzyme that catalyzed the release of 1 μmol of fatty acids.
2.2.5. Phytase Activity
The protocol described by Anastasio et al. [30] was used to detect phytase activity. In brief, overnight yeasts and bacteria cultures were inoculated into Chalmers broth that was made without neutral red, and with the addition of 1% sodium phytate (Sigma-Aldrich), and incubated at 30 °C for 48 h. Then, a loopful was spotted on the surface of the Chalmers agar that was made without calcium carbonate, and with the addition of 1% hexacalcium phytate (Sigma-Aldrich), and was incubated at 30 °C for 48 h. After incubation, the plates were flooded with 2% (w/v) cobalt chloride (Sigma-Aldrich) for 20 min. The presence of a clarification halo around each colony was indicative of phytase activity.
2.3. Safety Properties
2.3.1. Production of Antimicrobial Compounds
The well diffusion assay was employed to assess the production of antimould compounds by lactic acid bacteria and yeasts. LAB isolates were further tested for antibacterial activity. Overnight cultures were centrifuged to obtain a CFS that was consequently neutralized and treated with catalase (Sigma-Aldrich). Antibacterial activity was assessed against a mixture of five strains of the foodborne pathogens Listeria monocytogenes, Staphylococcus aureus, Escherichia coli O157:H7, and Salmonella serovars. Incubation took place at 37 °C for 24 h. Growth inhibition of the indicator strains around the wells, exceeding 5 mm, was indicative of the presence of antibacterial substances in the cell free supernatant. The strains that exhibited antibacterial activity against the pathogens were further assessed against a mixture of sourdough isolates, namely: Lcb. plantarum (LQC 2328, 2330, 2343, 2385, 2462, five isolates); Lvb. brevis (LQC 2368, 2429, 2484, 2509, 2518, five isolates); Cb. paralimentarius (LQC 2323, 2381, 2399, 2517, 2537, five isolates); Ltb. sakei (LQC 2448, 2452, 2456, 2470, 2473, five isolates); Ltb. curvatus (LQC 2472, 2475, 2476, 2497, 2498, five isolates); Fb. sanfranciscensis (LQC 2402, 2408, 2419, 2425, 2428, five isolates); Lvb. zymae (2394, one isolate); Lc. lactis (LQC 2375, 2499, 2510, three isolates); Ln. citreum (LQC 2508, one isolate); and Ln. mesenteroides (LQC 2512, one isolate). Antimould activity was examined against Penicillium chrysogenum, Pe. olsonii, and Aspergillus niger (mouldy bread isolates), as described above, with the exception that incubation took place at 30 °C for 5 days.
The protein nature of the antimicrobial substances was examined by assessing the effect of proteolytic enzymes on antimicrobial activity. More accurately, aliquots of 160 μL CFS were mixed with 40 μL 50 mM phosphate buffer pH 7.5 containing 2 mg mL−1 proteinase (Sigma-Aldrich) and 40 μL 50 mM phosphate buffer pH 7.0 containing 2 mg mL−1 trypsin (Sigma-Aldrich), and incubated at 37 °C for 1 h. Then, the antimicrobial activity of the CFS was examined as described above.
To determine the effect of pH on the stability of antimicrobial compounds, the pH value of CFS was adjusted to 2, 4, 6, 8 and 10 with 3 M HCl and 3 M NaOH, and incubated at 37 °C for 1 h. To evaluate the thermal stability of the antimicrobial compounds, CFS were treated at 60, 80 and 100 °C for 10 and 30 min, respectively. Untreated CFS were used as controls.
Specific PCR was employed for the detection of the plantaricin structural genes. DNA of the Lcb. plantarum strains under study was extracted according to Paramithiotis et al. [2]. The protocol described by Omar et al. [31] was used for the detection of the structural genes encoding plantaricins PlnA, PlnE/F, PlnJ/K, PlnN, NC8, S, and W. Detection of the structural gene of plantaricin 423 was performed in a 20 μL final reaction volume containing 0.2 mM dNTPs, 1.5 mM MgCl2, 0.5 μM each primer (413F: TGT GGT AAA CAT TCC TGC TCT G; 413R: CAC TTT CCA TGA CCG AAG TTA GC), and 1 U Taq polymerase (Kappa Biosystems, Boston, MA, USA); the resulting 86 bp amplicon was detected by electrophoresis in a 2% agarose gel.
2.3.2. Biogenic Amine Production
The protocol described by Bover-Cid and Holzapfel [32] was used to assess the amino acid decarboxylase activity of the yeast and bacteria isolates. More accurately, overnight yeast and bacteria cultures were spot-inoculated on the surface of a medium consisting, per liter, of 5 g yeast extract, 5 g tryptone, 5 g meat extract, 5 g glucose, 2.5 g sodium chloride, 2 g ammonium citrate, 1 g Tween 80, 0.2 g MgSO4, 0.05 g MnSO4, 0.04 g FeSO4, 0.01 g thiamine, 2 g K2PO4, 0.1 g CaCO3, 0.05 g pyridoxal-5-phosphate, 0.06 g bromocresol purple, and 20 g agar, and supplemented with 10 g lysine, tyrosine, ornithine, or histidine, pH 5.3. Incubation took place at 30 °C for 48 h. Occurrence of color change around the colonies, from yellow to purple, was indicative of decarboxylation of the respective amino acid. All chemicals were from Sigma-Aldrich.
2.4. Statistical Analysis
One-way ANOVA was employed to assess the statistical significance of the differences observed in the lipolysis kinetics between the microbial strains.
3. Results
3.1. Assessment of Technological Properties
3.1.1. Proteolytic Activity
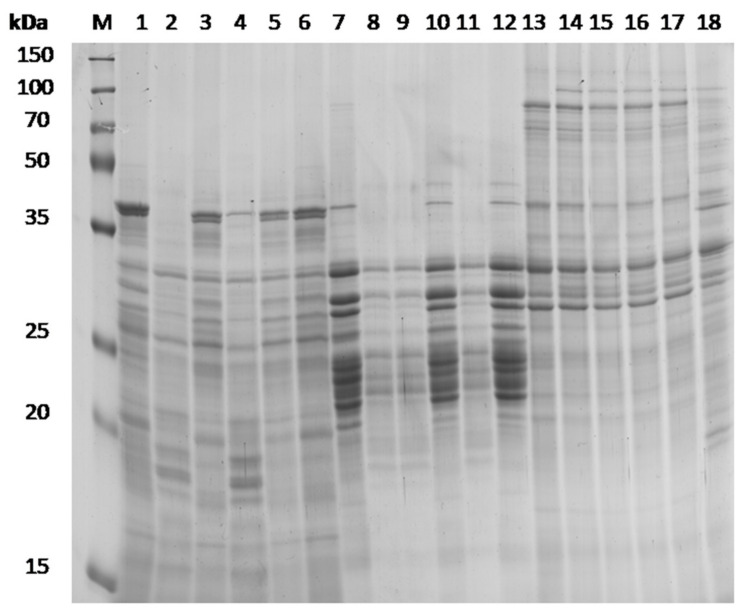
The agar well diffusion assay revealed that, of the 207 LAB and 195 yeast strains initially screened for proteolytic activity, seven and eight strains, respectively, were able to hydrolyze gluten. A clear halo around the wells was present for these 15 strains, indicating the presence of proteolytic capacity. Regarding LAB isolates, four belonged to Lcb. plantarum (LQC 2320, 2372, 2464, 2520) and three to Lvb. brevis (LQC 2469, 2474, 2493). In the case of yeasts, four isolates were assigned to S. cerevisiae (LQC 10343, 10378, 10398, 10402), three to W. anomalus (LQC 10346, 10353, 10360), and one to Pi. fermentans (LQC 10349). The proteolytic capacity of these 15 microbial strains was further evaluated with SDS-PAGE analysis. The SDS-PAGE confirmed the gluten degrading potential of three LAB strains, namely LQC 2320, 2469, and 2520. More accurately, sourdough fermentation with the specific monocultures of LAB strains resulted in significant hydrolysis of albumins and gliadins (Figure 1). Hydrolysis was partial or absent towards glutenins. Compared to the albumin control, sourdough fermentation with LAB strains LQC 2520 and 2320 resulted in complete and partial hydrolysis, respectively, of the albumin band with molecular weight (Mw) of ca. 40 kDa. In addition, the band intensity of albumins with Mw ranging between 25–35 kDa decreased, while the disappearance of the albumin bands in the range of 20–25 kDa, due to extensive hydrolysis, was also evident. The appearance of additional albumin bands of lower Mw, ranging between 15–20 kDa and belonging to doughs fermented by LAB strains LQC 2520 and 2320, respectively, was also shown in the electrophoretic analysis.
Figure 1.
Representative SDS-PAGE analysis of gluten fractions extracted from a mixture of wheat flour and water (1:2), incubated for 24 h at 30 °C, after inoculation with different LAB and yeast strains, which previously displayed positive proteolytic activity. M, protein marker; Lane 1, albumin fraction extracted from dough without inoculum (control); Lanes 2–6, albumin fractions extracted from doughs inoculated with microbial strains LQC 2320, 2474, 2520, 2469, and 10343, respectively; Lane 7, gliadin fraction extracted from dough without inoculum (control); Lanes 8–12, gliadin fractions extracted from doughs inoculated with microbial strains LQC 2320, 2520, 10343, 2469, and 2474, respectively; Lane 13, glutenin fraction extracted from dough without inoculum (control); Lanes 14–18, glutenin fractions extracted from doughs inoculated with microbial strains LQC 2320, 2469, 2474, 10343, and 2520, respectively.
In the case of gliadins, LAB strains LQC 2320, 2520, and 2469 revealed similar profiles of proteolysis. Compared to the control, complete hydrolysis of gliadin bands corresponding to ca. 40 and 20 kDa was detected in SDS-PAGE. After sourdough fermentation with each of the aforementioned LAB strains, the intensity of gliadin bands with Mw between 20–35 was significantly decreased. As far as the third gluten fraction was concerned, glutenins extracted from dough previously inoculated with LAB strain LQC 2520 revealed additional bands between 35–70, and below 25 kDa. The rest of the glutenins extracted were not affected by LAB fermentation.
3.1.2. Lipolytic Activity
The agar well diffusion assay revealed that, of the 207 LAB strains tested, 11 were found to be lipase positive, among which six were identified as Lcb. plantarum (LQC 2320, 2321, 2385, 2397, 2516, and 2520), four as Lvb. brevis (LQC 2374, 2411, 2416, and 2432), and one as Cb. paralimentarius (LQC 2410). None of the 195 yeast strains tested were able to hydrolyze tributyrin. A further lipolysis kinetics over 21 days was performed, with lipase activity expressed in AU/mL, as shown in Table 1. Data obtained revealed that Lcb. plantarum LQC 2321 and 2397 and Lvb. brevis LQC 2416 presented the highest enzyme activities of 62.75, 65.40, and 55.75, respectively, at the day 18 of incubation, with lipase activities remaining almost constant until day 21 of incubation.
Table 1.
Lipolysis kinetics of 11 bacterial strains during 21 days of incubation, at 30 °C.
| Sourdough LAB Strains | Days | ||||||
|---|---|---|---|---|---|---|---|
| 3 | 6 | 9 | 12 | 15 | 18 | 21 | |
|
Lcb. plantarum LQC 2320 |
8.75 ± 1.06 e,E | 23.75 ± 1.06 g,G | 34.00 ± 0.00 g,C | 35.50 ± 0.71 d,D | 36.00 ± 0.00 e,D,E | 37.50 ± 0.71 e,E | 37.25 ± 0.35 e,E |
|
Lcb. plantarum LQC 2321 |
2.50 ± 0.71 ab,AB | 11.50 ± 0.71 e,E | 42.00 ± 0.00 h,C | 52.50 ± 0.71 g,G | 59.50 ± 0.71 i,E | 62.75 ± 1.06 i,F | 63.00 ± 0.00 i,F |
|
Lvb. brevis LQC 2374 |
3.00 ± 0.00 a,ABC | 3.25 ± 0.35 ab,A | 3.50 ± 0.00 abc,AB | 4.00 ± 0.00 bc,A | 4.25 ± 0.35 c,B | 6.50 ± 0.71 d,C | 6.25 ± 0.35 d,C |
|
Lcb. plantarum LQC 2385 |
1.50 ± 0.71 a,A | 5.20 ± 0.42 b,B | 9.55 ± 0.78 bc,C | 14.50 ± 0.71 b,B | 19.50 ± 0.71 d,E | 22.50 ± 0.71 d,F | 22.50 ± 0.71 d,F |
|
Lcb. plantarum LQC 2397 |
5.50 ± 0.71 d,D | 11.25 ± 1.06 e,E | 49.00 ± 1.41 i,C | 57.50 ± 0.71 h,H | 61.50 ± 0.71 k,E | 65.40 ± 0.85 j,F | 65.50 ± 0.71 j,F |
|
Cb. paralimentarius LQC 2410 |
3.00 ± 0.00 abc,ABC | 7.50 ± 0.71 cd,CD | 8.45 ± 0.78 b,B | 14.25 ± 1.06 b,B | 16.00 ± 0.00 c,D | 17.25 ± 0.35 c,D | 17.00 ± 0.00c,D |
|
Lvb. brevis LQC 2411 |
9.00 ± 0.71 e,E | 13.25 ± 1.06 f,F | 19.75 ± 0.35 e,C | 34.50 ± 0.71 d,D | 37.50 ± 0.71 f,E | 48.60 ± 0.85 g,F | 47.75 ± 1.06 g,F |
|
Lvb. brevis LQC 2416 |
5.50 ± 0.71 d,D | 10.50 ± 0.71 e,E | 42.75 ± 0.35 h,C | 49.75 ± 0.35 f,F | 55.25 ± 0.35 h,E | 55.75 ± 0.35 h,E | 55.75 ± 0.35 h,E |
|
Lvb. brevis LQC 2432 |
1.75 ± 0.06 a,A | 13.50 ± 0.71 f,F | 23.75 ± 1.06 f, C | 41.50 ± 0.71 e,E | 41.75 ± 0.35 g,D | 45.50 ± 0.71 f,E | 46.00 ± 0.71 f,E |
|
Lcb. plantarum LQC 2516 |
3.50 ± 0.71 bc,BC | 6.25 ± 0.35 bc,BC | 10.50 ± 0.71 c,C | 14.50 ± 0.71 b,B | 14.50 ± 0.71 b,D | 14.75 ± 0.35 b,D | 14.50 ± 0.71 b,D |
|
Lcb. plantarum LQC 2520 |
4.50 ± 0.71 cd,CD | 8.00 ± 0.71 d,D | 15.50 ± 0.71 d,C | 17.25 ± 0.35 c,C | 18.50 ± 0.71 d,D | 22.50 ± 0.71 d,E | 22.75 ± 0.35 d,E |
The lipase activity is expressed in AU/mL. Cb.: Companilactobacillus; Lcb.: Lactiplantibacillus; Lvb.: Levilactobacillus. Statistically significant inter-species and intra-species differences are expressed with different letters—a–j and A–F, respectively.
Similarly, Lvb. brevis LQC 2374, Cb. paralimentarius LQC 2410, and Lcb. plantarum 2516 exhibited the lowest lipase activities until day 21 of incubation (6.25, 17.00 and 14.50 AU/mL, respectively). The other LAB strains exhibited moderate lipase activities, ranging from 22.50 to 47.75, until day 21 of incubation.
None of the microbial strains tested exhibited EPS production, amylolytic, or phytase activity.
3.2. Assessment of Safety Properties
3.2.1. Antimicrobial Activity
Among the 207 bacterial and 195 yeast strains initially evaluated against Pe. chrysogenum, Pe. olsonii, and A. niger by the agar well diffusion assay, 13 and 12, respectively, were able to inhibit mould growth. In the case of LAB, the following showed extensive inhibition against Pe. olsonii and A. niger: two Lcb. plantarum (LQC 2321 and 2327), two Lvb. brevis (LQC 2523 and 2529), two Ltb. sakei (LQC 2448 and 2454), one strain identified as Fb. sanfranciscensis (LQC 2419), Cb. paralimentarius (LQC 2392), Ltb. curvatus (LQC 2476), Lvb. zymae (LQC 2394), Lc. lactis (LQC 2499), Ln. mesenteroides (LQC 2512), and Ln. citreum (LQC 2508). As far as yeasts were concerned, seven S. cerevisiae (LQC 10286, 10298, 10307, 10393, 10404, 10421, and 10469), three W. anomalus (LQC 10346, 10353, and 10360), and two Pi. fermentans (LQC 10355 and 10356) presented inhibition against Pe. chrysogenum. LAB strains exhibited no inhibitory activity against Pe. chrysogenum, while Pe. olsonii and A. niger showed great resistance against all yeast strains.
The antibacterial capacity of LAB strains was also tested against foodborne pathogens L. monocytogenes, S. aureus, E. coli O157:H7, and Salmonella serovars. Twenty-one Lcb. plantarum strains (LQC 2320, 2384, 2422, 2441, 2442, 2443, 2444, 2445, 2446, 2447, 2485, 2486, 2487, 2488, 2489, 2490, 2491, 2492, 2506, 2516, and 2520) exhibited inhibitory properties against a mixture of L. monocytogenes 4b strains. No inhibitory potential of LAB strains evaluated against S. aureus, E. coli O157:H7, and Salmonella serovars was observed. These 21 Lcb. plantarum strains were further shown to inhibit sourdough isolates of Lcb. plantarum, Lvb. brevis, Cb. paralimentarius, Ltb. sakei, Ltb. curvatus, Fb. sanfranciscensis, and Lc. lactis. No inhibitory activity against Lvb. zymae, Ln. citreum, and Ln. mesenteroides was observed.
The structural gene encoding plantaricin 423 was detected in nineteen Lcb. plantarum strains. On the contrary, the structural genes encoding plantaricins NC8, PlnE/F, PlnJ/K, and S were detected in strains LQC 2320 and 2520.
None of the LAB and yeast isolates presented production of biogenic amines.
3.2.2. Effect of Proteolytic Enzymes, pH, and Temperature on the Stability of Antimould and Antibacterial Compounds Produced by Sourdough LAB and Yeasts
The CFS obtained from LAB and yeast strains, which previously demonstrated antimicrobial activity, were further assessed for their stability upon enzymatic, pH, and thermal treatment. The antibacterial compounds towards L. monocytogenes produced by 21 Lcb. plantarum strains were of proteinaceous nature, as the inhibitory action was abolished after treatment with at least one of the proteolytic enzymes employed, namely proteinase and trypsin. As far as the effect of thermal and pH treatment was concerned, these antibacterial substances demonstrated a highly thermostable and pH tolerant profile, since activity was retained after any temperature or pH treatment was applied. The antimould compounds produced by both LAB and yeast strains were non proteinaceous, as their activity remained even after enzymatic treatment. Regarding thermal stability, the CFS from all microbial strains tested completely lost their antimould capacity after thermal treatment. On the contrary, antimould compounds from both LAB and yeast strains exhibited a pH tolerant profile at all different pH values, ranging from 2 to 10.
4. Discussion
In the past few years, increasing demand for large-scale and controllable bread production has driven both scientific and industrial attention towards a more careful selection of sourdough starters. Desired technological and safety properties of candidate starters are those determining the potential use of sourdough microbial strains in the production of baked goods.
Proteolysis during sourdough fermentation has been considered a key process for determining the dough rheology and overall quality of the final product. Thus, the presence of proteolytic activity is a selection criterium of paramount importance among candidate sourdough starters [33,34]. Primary gluten hydrolysis is dependent on cereal proteases, while secondary proteolysis is carried out by strain-specific peptidases of sourdough LAB [35]. Except for LAB acidification, increased levels of thiol groups in the gluten proteins, produced via the glutathione reductase activity of heterofermentative lactobacilli, contribute to the depolymerization of glutenin macropolymer, thus increasing their susceptibility to enzymatic degradation [8]. In the present study, the initial test revealed seven proteolytic LAB strains and eight proteolytic yeast strains. However, the SDS-PAGE confirmed the proteolytic capacity for only three LAB strains. Similar results are often reported and may be attributed to insufficient incubation time [36,37,38]. Lancetti et al. [34] have already reported the proteolytic capacity of Lcb. plantarum ES137 and Pd. acidilactici ES22, previously isolated from Argentinian grains, by applying SDS-PAGE analysis. More accurately, sourdoughs inoculated with Lcb. plantarum ES137 and Pd. acidilactici ES22 presented a different protein profile in electrophoretic gel, compared to the controls, which is characterized by a decreased band number and intensity. Another study reported that the application of SDS-PAGE for characterization of the protein content of sourdoughs, inoculated with a mixed culture of Lcb. plantarum and yeast, revealed extensive hydrolysis of higher molecular weight protein bands, with the concomitant appearance of additional bands of lower molecular weight [39]. In the case of yeasts, decreased proteolytic activity has been reported by the majority of researchers [33]. Generally, yeasted doughs are characterized by decreased levels of amino acids compared to doughs fermented with LAB [40].
Lipolysis has been extensively studied in fermented dairy, non-dairy and meat products [38,41,42]. Despite the fact that a certain level of lipolysis is a desirable attribute of microbial strains, in terms of synthesis of flavor precursors, only a few pieces of scientific literature have reported the microbial screening for lipolytic activity of sourdough microorganisms [2]. At first, lipases hydrolyze triacylglycerols into free fatty acids [43]. Then, unsatturated fatty acids, namely linoleic acid, the major component of cereal lipids, are further degraded into peroxides through either autoxidation during flour storage or cereal lipoxygenase activity during dough mixing [44]. Finally, degradation of peroxides into aldehydes takes place, with the latter being further reduced to alcohols via heterofermentative lactobacilli during sourdough fermentation. The agar well diffusion assay applied in the present study revealed 11 lipase positive LAB strains of the 207 tested; the lipolysis kinetics further applied over 21 days revealed that Lcb. plantarum LQC 2321 and LQC 2397 and Lvb. brevis LQC 2416 presented the highest lipolytic activity on day 21 of incubation. None of the yeasts tested exhibited lipolytic activity. To our knowledge, no previous study has reported the presence of lipolytic microbial strains isolated from spontaneously fermented sourdoughs; except for Paramithiotis et al. [28], who reported the presence of Yarrowia lipolytica in Greek sourdough microbiota. Y. lipolytica strains are known for their strong lipolytic and proteolytic activity [45]. Regarding LAB, they are considered as weak lipolytic compared to other bacterial species, such as Pseudomonas spp., Acinetobacter spp., and Flavonobacterium spp. Indeed, the absence of lipolytic activity among strains of Lcb. plantarum and Lvb. brevis was reported by Kamiloğlu et al. [46] and Ebadi Nehzad et al. [47]. However, another study revealed that, of the 137 bacterial strains initially screened for proteolytic and lipolytic activity, seven exhibited both [41]. Among them, two belonged to Lcb. plantarum. Zymography was further applied to isolate the proteins responsible for exhibiting lipolytic activity; however, no lipolytic proteins were detected in the case of Lcb. plantarum. Regarding Cb. paralimentarius, this is the first study to report its lipolytic activity.
Sourdough fermented with antimould LAB and yeast strains has been the epicenter of intensive study over the past few years; it is an alternative biopreservation approach in line with consumer demand for clean label products [48]. The most common moulds spoiling bread products belong to the genera Aspergillus, Penicillium, Fusarium, and Cladosporium, and their presence constitutes a major concern for baking industries due to the potential of mycotoxin production, with concomitant long term health risks [1]. Antimould metabolites synthesized by sourdough LAB and yeast strains are responsible for biopreservation effects. In the present study, 13 LAB inhibited Pe. olsonii and A. niger growth and 12 yeast strains inhibited Pe. chrysogenum growth. The antimould activity of the majority of LAB species tested in the present study has been previously reported against numerous moulds which commonly spoil bread. More accurately, Fraberger et al. [1] reported the strain-dependent antimould potential of wheat and rye sourdough-derived lactobacilli; the majority of the isolates belonging to Lcb. plantarum presented wide antimould activity against all mould strains tested. In the case of Lvb. brevis., isolate S4.5 strongly inhibited Pe. roqueforti, while the mould remained resistant against Lvb. brevis S13.18, S14.3, and S6.13. Isolates belonging to Fb. sanfranciscensis, Ltb. sakei S4.19, and Ltb. curvatus S4.14, S5.22, S6.15 exhibited very strong inhibition against F. graminearum. In addition, Cb. paralimentarius S7.5 exhibited very strong inhibitory activity against A. fumigatus and F. graminearum. An additional study by Manini et al. [49] reported the antimould activity of wheat bran sourdough-originating Lcb. plantarum, Lvb. brevis, Ltb. curvatus, and Ltb. sakei against A. oryzae and A. niger. Ln. mesenteroides and Ln. citreum strains were also able to inhibit both Aspergillus species. As far as Lc. lactis was concerned, strain CH179 derived from chia flour fermentation presented inhibitory activity against A. niger and Pe. roqueforti [50]. In the case of Lvb. zymae, the antimould activity of a sourdough-derived strain has not been reported so far, while few studies have documented Lvb. zymae as inactive against mould growth [51]. Regarding yeasts, the literature is focused on their leavening capacity, flavor development, and mutualistic interaction with LAB, while only few scientific data have reported their role as antimould agents [18,52]. More accurately, Jin et al. [53] reported the absence of antimould activity of sourdough-derived S. cerevisiae against A. flavus. In addition, its combination with antimould Pd. pentosaceus, and their application as starters in sourdough fermentations, presented the greatest inhibitory effect against A. flavus. Regarding W. anomalus, a strong inhibitory activity against a range of moulds has been reported when used as starter in dough fermentation. Sourdough fermented with a combination of W. anomalus with Lcb. plantarum exhibited slightly decreased antimould activity; however, sourdough bread delayed mould growth until 28 days of storage. The antimould peptides and ethyl acetate synthesized by Lcb. plantarum and W. anomalus, respectively, were responsible for the extension of the mould-free shelf life of bread. With respect to Pi. fermentans, no previous study has reported the antimould activity of this sourdough originating yeast. However, the antimould potential of coffee fruit-derived strain LPBYB13 was reported against an ochratoxigenic strain of A. westerdijkiae on agar tests, and a further inhibition of ochratoxin A production in coffee beans was performed [54].
The antibacterial potential of LAB is a significant criterium for the selection of more competitive starters in sourdough fermentations, and could determine the microbiological stability and safety of the baked products. Despite the fact that the antibacterial compounds produced by sourdough LAB, namely bacteriocins, bacteriocin-like inhibitory substances (BLIS), and antibiotics, do not extend the mould-free shelf life of the end products, they positively affect their microbiological safety by counteracting food contamination during processing [55]. L. monocytogenes, S. aureus, E. coli, and Salmonella serovars are the most common foodborne pathogens. In the present study, of 207 LAB isolates evaluated against food borne pathogens, 21 Lcb. plantarum strains exhibited inhibitory activity against a mixture of L. monocytogenes 4b strains. These 21 Lb. plantarum strains further inhibited several sourdough-associated LAB. In agreement with the present data, hull-less barley sourdough-originating Lcb. plantarum SAB15 exhibited a very strong inhibitory potential against B. subtillis, B. cereus, and E. coli, based on agar diffusion assay [9]. In addition, Demirbas et al. [56] reported the antibacterial activity of Turkish wheat sourdough-derived Lcb. plantarum ED10 against B. cereus, S. aureus, Y. enterocolitica, and E. coli. The DNA sequence of the structural genes encoding eight plantaricins, namely NC8, PlnA, PlnE/F, PlnJ/K, PlnN, W, S, and 423, has been verified so far. The latter is plasmid-encoded, while the rest are chromosomally-encoded [57,58]. Their characteristics and applications have been also reviewed for the majority of them [20,59]. To our knowledge, this is the first study reporting the presence of plantaricins NC8, PlnE/F, PlnJ/K, S, and 423 from sourdough-derived Lcb. plantarum isolates. Previous studies have reported only the production of plantaricin ST31 [60] and plantaricin A [61] from sourdough-derived strains of Lcb. plantarum.
The presence of phytase and amylase positive sourdough microorganisms is of paramount importance from a technological perspective. During sourdough fermentation, phytases dephosphorylate phytic acid into myo-inositol and phosphoric acid, leading to increased mineral bioavailability and further improvement of the nutritional characteristics of the final product. Regarding amylases, they are responsible for starch hydrolysis, with concomitant formation of fermentable sugars. In the present study, no phytase and amylase positive LAB and yeast strains were detected. In line with the present data, Paramithiotis et al. [2] detected no amylolytic activities of sourdough LAB and yeast strains tested. On the other hand, several studies have reported the presence of sourdough-derived microbial strains with phytase activity [62,63]. Another property with technological impact, however, not detected in the present study, was EPS production by LAB strains. On the contrary, previous studies by Milanović et al. [64] and Manini et al. [49] reported the presence of EPS producing LAB strains, derived from cereal based substrates and wheat bran sourdoughs, respectively. A last safety attribute assessed in the present study, but what was not detected in either LAB or yeast strains tested was the ability to form BAs. Within permissible limits, BAs exert no adverse health effects on consumers; however, when accumulated in food products, they can possibly present a health risk. Compared to other fermented foods, BA production in sourdough fermentations is of less concern, with BA levels ranging far below those posing health risks [24]. In agreement with the previous statement, Bartkiene et al. [65] reported on BA production in the solid-state fermentation of flaxseed; however, the levels were relatively low to pose health issues.
5. Conclusions
The results obtained in the present study clearly reveal the technological and safety potential of several bacterial and yeast strains. Two Lcb. plantarum strains, LQC 2320 and 2520, exhibited proteolytic, lipolytic and antibacterial capacity. The presence of structural genes encoding plantaricins NC8, PlnE/F, PlnJ/K, and S was also detected in both LAB strains. In the case of yeasts, three W. anomalus strains, LQC 10343, 10353, and 10360, exhibited both proteolytic and antimould potential. These properties deserve further research due to the impact they may have on the quality of sourdough bread.
Author Contributions
Conceptualization, E.H.D., P.N.S., M.M. and S.P.; application of analytical methods, M.K.S., S.T., E.-E.P., S.-D.M. and S.P.; data curation, M.K.S., M.M. and S.P.; writing—original draft preparation, M.K.S., S.P., M.M. and L.B.; writing—review and editing, M.K.S., S.P., M.M., L.B., E.H.D. and P.N.S.; project administration, M.M.; funding acquisition, M.M., S.P., E.H.D. and P.N.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research has been co-financed by the European Union and Greek national funds. through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH—CREATE—INNOVATE (T1EDK-05339).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fraberger V., Ammer C., Domig K.J. Functional Properties and Sustainability Improvement of Sourdough Bread by Lactic Acid Bacteria. Microorganisms. 2020;8:1895. doi: 10.3390/microorganisms8121895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paramithiotis S., Tsiasiotou S., Drosinos E.H. Comparative Study of Spontaneously Fermented Sourdoughs Originating from Two Regions of Greece: Peloponnesus and Thessaly. Eur. Food Res. Technol. 2010;231:883–890. doi: 10.1007/s00217-010-1345-0. [DOI] [Google Scholar]
- 3.Gänzle M.G. Enzymatic and Bacterial Conversions during Sourdough Fermentation. Food Microbiol. 2014;37:2–10. doi: 10.1016/j.fm.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Struyf N., Verspreet J., Courtin C. The Effect of Amylolytic Activity and Substrate Availability on Sugar Release in Non-yeasted Dough. J. Cereal Sci. 2016;69:111–118. doi: 10.1016/j.jcs.2016.02.016. [DOI] [Google Scholar]
- 5.Pétel C., Onno B., Prost C. Sourdough Volatile Compounds and Their Contribution to Bread: A Review. Trends Food Sci. Technol. 2017;59:105–123. doi: 10.1016/j.tifs.2016.10.015. [DOI] [Google Scholar]
- 6.Gänzle M.G., Loponen J., Gobbetti M. Proteolysis in Sourdough Fermentations: Mechanisms and Potential for Improved Bread Quality. Trends Food Sci. Technol. 2008;19:513–521. doi: 10.1016/j.tifs.2008.04.002. [DOI] [Google Scholar]
- 7.Gobbetti M., Rizzello C.G., Di Cagno R., De Angelis M. How the Sourdough May Affect the Functional Features of Leavened Baked Goods. Food Microbiol. 2014;37:30–40. doi: 10.1016/j.fm.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Gänzle M., Gobbetti M., editors. Handbook on Sourdough Biotechnology. Springer; New York, NY, USA: 2013. Physiology and Biochemistry of Lactic Acid Bacteria; pp. 183–216. [Google Scholar]
- 9.Çakır E., Arici M., Durak M.Z. Biodiversity and Techno Functional Properties of Lactic Acid Bacteria in Fermented Hull-less Barley Sourdough. J. Biosci. Bioeng. 2020;130:450–456. doi: 10.1016/j.jbiosc.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Fekri A., Torbati M., Yari Khosrowshahi A., Bagherpour Shamloo H., Azadmard-Damirchi S. Functional Effects of Phytate-degrading, Probiotic Lactic Acid Bacteria and Yeast Strains Isolated from Iranian Traditional Sourdough on the Technological and Nutritional Properties of Whole Wheat Bread. Food Chem. 2020;306:125620. doi: 10.1016/j.foodchem.2019.125620. [DOI] [PubMed] [Google Scholar]
- 11.De Angelis M., Minervini F., Siragusa S., Rizzello C.G., Gobbetti M. Wholemeal Wheat Flours Drive the Microbiome and Functional Features of Wheat Sourdoughs. Int. J. Food Microbiol. 2019;302:35–46. doi: 10.1016/j.ijfoodmicro.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Ispirli H., Demirbaş F., Yüzer M.O., Dertli E. Identification of Lactic Acid Bacteria from Spontaneous Rye Sourdough and Determination of Their Functional Characteristics. Food Biotechnol. 2018;32:222–235. doi: 10.1080/08905436.2018.1507913. [DOI] [Google Scholar]
- 13.Karaman K., Sagdic O., Durak M.Z. Use of Phytase Active Yeasts and Lactic Acid Bacteria Isolated from Sourdough in the Production of Whole Wheat Bread. LWT-Food Sci. Technol. 2018;91:557–567. doi: 10.1016/j.lwt.2018.01.055. [DOI] [Google Scholar]
- 14.Zhang G., Zhang W., Sun L., Sadiq F.A., Yang Y., Gao J., Sang Y. Preparation Screening, Production Optimization and Characterization of Exopolysaccharides Produced by Lactobacillus sanfranciscensis Ls-1001 Isolated from Chinese Traditional Sourdough. Int. J. Biol. Macromol. 2019;139:1295–1303. doi: 10.1016/j.ijbiomac.2019.08.077. [DOI] [PubMed] [Google Scholar]
- 15.Ispirli H., Özmen D., Yılmaz M.T., Sağdıç O., Dertli E. Impact of Glucan Type Exopolysaccharide (EPS) Production on Technological Characteristics of Sourdough Bread. Food Control. 2020;107:106812. doi: 10.1016/j.foodcont.2019.106812. [DOI] [Google Scholar]
- 16.Axel C., Zannini E., Arendt E.K. Mold Spoilage of Bread and Its Biopreservation: A Review of Current Strategies for Bread Shelf Life Extension. Crit. Rev. Food Sci. Nutr. 2017;57:3528–3542. doi: 10.1080/10408398.2016.1147417. [DOI] [PubMed] [Google Scholar]
- 17.Sáez G., Saavedra L., Hebert E., Zárate G. Identification and Biotechnological Characterization of Lactic Acid Bacteria Isolated from Chickpea Sourdough in Northwestern Argentina. LWT Food Sci. Technol. 2018;93:249–256. doi: 10.1016/j.lwt.2018.03.040. [DOI] [Google Scholar]
- 18.De Vuyst L., Harth H., van Kerrebroeck S., Leroy F. Yeast Diversity of Sourdoughs and Associated Metabolic Properties and Functionalities. Int. J. Food Microbiol. 2016;239:26–34. doi: 10.1016/j.ijfoodmicro.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 19.Bartkiene E., Lele V., Ruzauskas M., Domig K.J., Starkute V., Zavistanaviciute P., Bartkevics V., Pugajeva I., Klupsaite D., Juodeikiene G., et al. Lactic Acid Bacteria Isolation from Spontaneous Sourdough and Their Characterization Including Antimicrobial and Antifungal Properties Evaluation. Microorganisms. 2020;8:64. doi: 10.3390/microorganisms8010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kareem R.A., Razavi S.H. Plantaricin Bacteriocins: As Safe Alternative Antimicrobial Peptides in Food Preservation—A Review. J. Food Saf. 2020;40:e12735. [Google Scholar]
- 21.Spano G., Russo P., Lonvaud-Funel A., Lucas P., Alexandre H., Grandvalet C., Coton E., Coton M., Barnavon L., Bach B., et al. Biogenic Amines in Fermented Foods. Eur. J. Clin. Nutr. 2010;64:S95–S100. doi: 10.1038/ejcn.2010.218. [DOI] [PubMed] [Google Scholar]
- 22.Li B., Lu S. The Importance of Amine-degrading Enzymes on the Biogenic Amine Degradation in Fermented Foods: A Review. Process Biochem. 2020;99:331–339. doi: 10.1016/j.procbio.2020.09.012. [DOI] [Google Scholar]
- 23.Ladero V., Calles-Enriquez M., Fernandez M., Alvarez M.A. Toxicological Effects of Dietary Biogenic Amines. Curr. Nutr. Food Sci. 2010;6:145–156. doi: 10.2174/157340110791233256. [DOI] [Google Scholar]
- 24.Brandt M.J. Industrial Production of Sourdoughs for the Baking Branch—An Overview. Int. J. Food Microbiol. 2019;302:3–7. doi: 10.1016/j.ijfoodmicro.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Syrokou M.K., Themeli C., Paramithiotis S., Mataragas M., Bosnea L., Argyri A., Chorianopoulos N.G., Skandamis P.N., Drosinos E.H. Microbial Ecology of Greek Wheat Sourdoughs Identified by Culture-dependent and Culture-independent Approach. Foods. 2020;9:1603. doi: 10.3390/foods9111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smitinont T., Tansakul C., Keeratipibul S., Nacarini L., Bosco M., Cescutti P. Exopolysaccharide-producing Lactic Acid Bacteria Strains from Traditional Fermented Food: Isolation, Identification and Exopolysaccharide Characterization. Int. J. Food Microbiol. 1999;15:105–111. doi: 10.1016/S0168-1605(99)00094-X. [DOI] [PubMed] [Google Scholar]
- 27.Di Cagno R., De Angelis M., Lavermicocca P., De Vincenzi M., Giovannini C., Faccia M., Gobbetti M. Proteolysis by Sourdough Lactic Acid Bacteria: Effects on Wheat Flour Protein Fractions and Gliadin Peptides Involved in Human Cereal Intolerance. Appl. Environ. Microbiol. 2002;68:623–633. doi: 10.1128/AEM.68.2.623-633.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paramithiotis S., Mueller M.R.A., Ehrmann M.A., Tsakalidou E., Seiler H., Vogel R., Kalantzopoulos G. 2000. Polyphasic Identification of Wild Yeast Strains Isolated from Greek Sourdoughs. Syst. Appl. Microbiol. 2000;23:156–164. doi: 10.1016/S0723-2020(00)80057-0. [DOI] [PubMed] [Google Scholar]
- 29.Carrazco-Palafox J., Rivera-Chavira B.E., Ramírez-Baca N., Manzanares-Papayanopoulos L.I., Nevárez-Moorillón G.V. Improved Method for Qualitative Screening of Lipolytic Bacterial Strains. MethodsX. 2018;5:68–74. doi: 10.1016/j.mex.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anastasio M., Pepe O., Cirillo T., Palomba S., Blaiotta G., Villani F. Selection and Use of Phytate-degrading LAB to Improve Cereal-based Products by Mineral Solubilization during Dough Fermentation. J. Food Sci. 2010;75:M28–M35. doi: 10.1111/j.1750-3841.2009.01402.x. [DOI] [PubMed] [Google Scholar]
- 31.Omar N.B., Abriouel H., Lucas R., Martínez-Cañamero M., Guyot J.-P., Gálvez A. Isolation of Bacteriocinogenic Lactobacillus plantarum Strains from Ben Saalga, a Traditional Fermented Gruel from Burkina Faso. Int. J. Food Microbiol. 2006;112:44–50. doi: 10.1016/j.ijfoodmicro.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 32.Bover-Cid S., Holzapfel W.H. Improved Screening Procedure for Biogenic Amine Production by Lactic Acid Bacteria. Int. J. Food Microbiol. 1999;53:33–41. doi: 10.1016/S0168-1605(99)00152-X. [DOI] [PubMed] [Google Scholar]
- 33.Fu W., Xue W., Liu C., Tian Y., Zhu Z. Screening of Lactic Acid Bacteria and Yeasts from Sourdough as Starter Cultures for Reduced Allergenicity Wheat Products. Foods. 2020;9:751. doi: 10.3390/foods9060751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lancetti R., Sciarini L., Pιrez G.T., Salvucci E. Technological Performance and Selection of Lactic Acid Bacteria Isolated from Argentinian Grains as Starters for Wheat Sourdough. Curr. Microbiol. 2020;78:255–264. doi: 10.1007/s00284-020-02250-6. [DOI] [PubMed] [Google Scholar]
- 35.Galli V., Mazzoli L., Luti S., Venturi M., Guerrini S., Paoli P., Vincezini M., Granchi L., Pazzagli L. Effect of Selected Strains of Lactobacilli on the Antioxidant and Anti-inflammatory Properties of Sourdough. Int. J. Food Microbiol. 2018;286:55–65. doi: 10.1016/j.ijfoodmicro.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 36.Kirilov N., Petkova T., Atanasova J., Danova S., Iliev I., Popov Y., Haertle T., Ivanova I.V. Proteolytic Activity in Lactic Acid Bacteria from Iraq, Armenia and Bulgaria. Biotechnol. Biotechnol. Equip. 2009;23:643–646. doi: 10.1080/13102818.2009.10818506. [DOI] [Google Scholar]
- 37.Dallas D.C., Citerne F., Tian T., Silva V.L.M., Kalanetra K.M., Frese S.A., Robinson R.C., Mills D.A., Barile D. Peptidomic Analysis Reveals Proteolytic Activity of Kefir Microorganisms on Bovine Milk Proteins. Food Chem. 2016;197:273–284. doi: 10.1016/j.foodchem.2015.10.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Syrokou M.K., Papadelli M., Ntaikou I., Paramithiotis S., Drosinos E.H. Sugary Kefir: Microbial Identification and Biotechnological Properties. Beverages. 2019;5:61. doi: 10.3390/beverages5040061. [DOI] [Google Scholar]
- 39.Yin Y., Wang J., Yang S., Feng J., Jia F., Zhang C. Protein Degradation in Wheat Sourdough Fermentation with Lactobacillus plantarum M616. Interdiscip. Sci. Comput. Life Sci. 2015;7:205–210. doi: 10.1007/s12539-015-0262-0. [DOI] [PubMed] [Google Scholar]
- 40.Thiele C., Gänzle M.G., Vogel R.F. Contribution of Sourdough Lactobacilli, Yeast, and Cereal Enzymes to the Generation of Amino Acids in Dough Relevant for Bread flavour. Cereal Chem. 2002;79:45–51. doi: 10.1094/CCHEM.2002.79.1.45. [DOI] [Google Scholar]
- 41.García-Cano I., Rocha-Mendoza D., Ortega-Anaya J., Wang K., Kosmerl E., Jiménez-Flores R. Lactic Acid Bacteria Isolated from Dairy Products as Potential Producers of Lipolytic, Proteolytic and Antibacterial Proteins. Appl. Microbiol. Biotechnol. 2019;103:5243–5257. doi: 10.1007/s00253-019-09844-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao Y., Liu Y., Chen C., Xie T., Li P. Effect of Lactobacillus plantarum and Staphylococcus xylosus on Flavour Development and Bacterial Communities in Chinese Dry Fermented Sausages. Food Res. Int. 2020;135:109247. doi: 10.1016/j.foodres.2020.109247. [DOI] [PubMed] [Google Scholar]
- 43.Pico J., Bernal J., Gómez M. Wheat Bread Aroma Compounds in Crumb and Crust: A Review. Food Res. Int. 2015;75:200–215. doi: 10.1016/j.foodres.2015.05.051. [DOI] [PubMed] [Google Scholar]
- 44.Gänzle M.G., Vermeulen N., Vogel R.F. Carbohydrate, Peptide and Lipid Metabolism of Lactic Acid Bacteria in Sourdough. Food Microbiol. 2007;24:128–138. doi: 10.1016/j.fm.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 45.Fröhlich-Wyder M.T., Arias-Roth E., Jakob E. Cheese Yeasts. Yeast. 2019;36:129–141. doi: 10.1002/yea.3368. [DOI] [PubMed] [Google Scholar]
- 46.Kamiloğlu A., Kaban G., Kaya M. Technological Properties of Autochthonous Lactobacillus plantarum Strains Isolated from Sucuk (Turkish Dry-fermented Sausage) Braz. J. Microbiol. 2020;51:1279–1287. doi: 10.1007/s42770-020-00262-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ebadi Nezhad S.J., Edalatian Dovom M.R., Habibi Najafi M.B., Yavarmanesh M., Mayo B. Technological Characteristics of Lactobacillus spp. Isolated from Iranian Raw Milk Motal Cheese. LWT. 2020;133:110070. doi: 10.1016/j.lwt.2020.110070. [DOI] [Google Scholar]
- 48.Debonne E., Van Schoors F., Maene P., Van Bockstaele F., Vermeir P., Verwaeren J., Eeckhout M., Devlieghere F. Comparison of the Antifungal Effect of Undissociated Lactic and Acetic Acid in Sourdough Bread and in Chemically Acidified Wheat Bread. Int. J. Food Microbiol. 2020;321:108551. doi: 10.1016/j.ijfoodmicro.2020.108551. [DOI] [PubMed] [Google Scholar]
- 49.Manini F., Casiraghi M.C., Poutanen K., Brasca M., Erba D., Plumed-Ferrer C. Characterization of Lactic Acid Bacteria Isolated from Wheat Bran Sourdough. LWT Food Sci. Technol. 2016;66:275–283. doi: 10.1016/j.lwt.2015.10.045. [DOI] [Google Scholar]
- 50.Maidana S.D., Ficoseco C.A., Bassi D., Cocconcelli P.S., Puglisi E., Savoy G., Vignolo G., Fontana C. Biodiversity and Technological-functional Potential of Lactic Acid Bacteria Isolated from Spontaneously Fermented Chia Sourdough. Int. J. Food Microbiol. 2020;316:108425. doi: 10.1016/j.ijfoodmicro.2019.108425. [DOI] [PubMed] [Google Scholar]
- 51.Rouxel M., Barthe M., Marchand P., Juin C., Mondamert L., Berges T., Blanc P., Verdon J., Berjeaud J.-M., Aucher W. Characterization of Antifungal Compounds Produced by Lactobacilli in Cheese-mimicking Matrix: Comparison between Active and Inactive Strains. Int. J. Food Microbiol. 2020;333:108798. doi: 10.1016/j.ijfoodmicro.2020.108798. [DOI] [PubMed] [Google Scholar]
- 52.Coda R., Cassone A., Rizzello C.G., Nionelli L., Cardinali G., Gobbetti M. Antifungal Activity of Wickerhamomyces anomalus and Lactobacillus plantarum during Sourdough Fermentation: Identification of Novel Compounds and Long-term Effect during Storage of Wheat Bread. Appl. Environ. Microbiol. 2011;77:3484–3492. doi: 10.1128/AEM.02669-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin J., Nguyen T.T.H., Humayun S., Park S., Oh H., Lim S., Mok I.-K., Li Y., Pal K., Kim D. Characteristics of Sourdough Bread Fermented with Pediococcus pentosaceus and Saccharomyces cerevisiae and Its Bio-preservative Effect against Aspergillus flavus. Food Chem. 2021;345:128787. doi: 10.1016/j.foodchem.2020.128787. [DOI] [PubMed] [Google Scholar]
- 54.Pereira G.V.M., Beux M., Pagnoncelli M.G.B., Soccol V.T., Rodrigues C., Soccol C.R. Isolation, Selection and Evaluation of Antagonistic Yeasts and Lactic Acid Bacteria against Ochratoxigenic Fungus Aspergillus westerdijkiae on Coffee Beans. Lett. Appl. Microbiol. 2016;62:96–101. doi: 10.1111/lam.12520. [DOI] [PubMed] [Google Scholar]
- 55.Minervini F., De Angelis M., Di Cagno R., Gobbetti M. Ecological Parameters Influencing Microbial Diversity and Stability of Traditional Sourdough. Int. J. Food Microbiol. 2014;171:136–146. doi: 10.1016/j.ijfoodmicro.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 56.Demirbas F., Ispirli H., Kurnaz A.A., Yilmaz M.T., Dertli E. Antimicrobial and Functional Properties of Lactic Acid Bacteria Isolated from Sourdoughs. LWT Food Sci. Technol. 2017;79:361–366. doi: 10.1016/j.lwt.2017.01.067. [DOI] [Google Scholar]
- 57.van Reenen C.A., Dicks L.M., Chikindas M.L. Isolation, Purification and Partial Characterization of Plantaricin 423, a Bacteriocin Produced by Lactobacillus plantarum. J. Appl. Microbiol. 1998;84:1131–1137. doi: 10.1046/j.1365-2672.1998.00451.x. [DOI] [PubMed] [Google Scholar]
- 58.Diep D.B., Straume D., Kjos M., Torres C., Nes I.F. An Overview of the Mosaic Bacteriocin Pln Loci from Lactobacillus plantarum. Peptides. 2009;30:1562–1574. doi: 10.1016/j.peptides.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Gonzalez N., Battista N., Prete R., Corsetti A. Health-promoting Role of Lactiplantibacillus plantarum Isolated from Fermented Foods. Microorganisms. 2021;9:349. doi: 10.3390/microorganisms9020349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Todorov S., Onno B., Sorokine O., Chobert J., Ivanova I., Dousset X. Detection and Characterisation of a Novel Antibacterial Substance Produced by Lactobacillus plantarum ST31 Isolated from Sourdough. Int. J. Food Microbiol. 1999;48:167–177. doi: 10.1016/S0168-1605(99)00048-3. [DOI] [PubMed] [Google Scholar]
- 61.Di Cagno R., De Angelis M., Calasso M., Vicentini O., Vernocchi P., Ndagijimana M., De Vincenzi M., Dessi M.R., Guerzoni M.E., Gobbetti M. Quorum Sensing in Sourdough Lactobacillus plantarum DC400: Induction of Plantaricin A (PlnA) under Cocultivation with Other Lactic Acid Bacteria and Effect of PlnA on Bacterial and Caco-2 Cells. Proteomics. 2010;10:2175–2190. doi: 10.1002/pmic.200900565. [DOI] [PubMed] [Google Scholar]
- 62.Palla M., Agnolucci M., Calzone A., Giovannetti M., Di Cagno R., Gobbetti M., Rizzello C.G., Pontonio E. Exploitation of Autochthonous Tuscan Sourdough Yeasts as Potential Starters. Int. J. Food Microbiol. 2019;302:59–68. doi: 10.1016/j.ijfoodmicro.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 63.Carrizo S.L., Montes de Oca C.E., Laiño J.E., Suarez N.E., Vignolo G., LeBlanc J.G., Rollán G. Ancestral Andean Grain Quinoa as Source of Lactic Acid Bacteria Capable to Degrade Phytate and Produce B-group Vitamins. Food Res. Int. 2016;89:488–494. doi: 10.1016/j.foodres.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 64.Milanović V., Osimani A., Garofalo C., Belleggia L., Maoloni A., Cardinali F., Mozzon M., Foligni R., Aquilanti L., Clementi F. Selection of Cereal-sourced Lactic Acid Bacteria as Candidate Starters for the Baking Industry. PLoS ONE. 2020;15:e0236190. doi: 10.1371/journal.pone.0236190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bartkiene E., Schleining G., Juodeikiene G., Vidmantiene D., Krungleviciute V., Rekstyte T., Basiskiene L., Stankevicius M., Akuneca I., Ragazinskiene O., et al. The Influence of Lactic Acid Fermentation on Biogenic Amines and Volatile Compounds Formation in Flaxseed and the Effect of Flaxseed Sourdough on the Quality of Wheat Bread. LWT Food Sci. Technol. 2014;56:445–450. doi: 10.1016/j.lwt.2013.11.033. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available in the manuscript.