Lipoprotein(a) is a plasma lipoprotein composed of a low-density lipoprotein (LDL) particle linked to apolipoprotein(a). The risk of coronary artery disease (CAD) associated with lipoprotein(a) has been associated with both molar concentration and apolipoprotein(a) isoform size, which are inversely correlated properties and highly heritable.1 Similar to LDL, lipoprotein(a) harbours apolipoprotein B and is associated with increased risk of CAD. Apolipoprotein B is believed to be a key unifying feature of atherogenic lipoproteins.2 However, it is unclear to what extent non-apolipoprotein B-mediated mechanisms of lipoprotein(a) contribute to the pathogenicity of CAD in humans.1 Here, we investigated whether like LDL cholesterol (LDL-C),2 apolipoprotein B is sufficient to explain the risk of CAD associated with lipoprotein(a) particles.
We used individual level data from the UK Biobank and Jackson Heart Study to investigate the observational relationship between lipoprotein(a) mass and lipoprotein(a) molar concentration with apolipoprotein B. Secondary use of UK Biobank and Jackson Heart Study data was approved by the Massachusetts General Hospital Institutional Review Board. The UK Biobank is a prospective observational study of adults aged 40–69 years recruited from across the UK between 2006 and 2010. Alternatively, the Jackson Heart Study is a community-based cohort of African Americans in the Jackson, Mississippi area consisting of a baseline examination (2000–04) and two follow-up examinations (2005–08 and 2009–13).3 Jackson Heart Study data were obtained from dbGaP (dataset accessions: pht008792.v1.p2 and pht001945.v2.p2). All analyses were performed using R version 4.0.2 [R Core Team (2020)].
Firstly, we investigated the association between baseline apolipoprotein B with LDL-C or lipoprotein(a) molar concentration in UK Biobank participants without prevalent CAD and not using cholesterol-lowering medication at enrolment (n=327 929).4 The mean age of individuals included in this study was 56.5 years (SD=8.1 years) and 58.4% of individuals were women. In contrast to the strong correlation between measured apolipoprotein B and LDL-C (Spearman correlation Rho: 0.96, P<0.001), the correlation between apolipoprotein B and lipoprotein(a) molar concentration was modest (Rho: 0.13, P<0.001). We also observed a modest correlation between measured apolipoprotein B and lipoprotein(a) mass (Rho: 0.16, P<0.001, n=721) relative to measured apolipoprotein B and LDL-C among African Americans in the Jackson Heart Study (Rho: 0.93, P<0.001). The correlation between apolipoprotein B and LDL-C was modestly attenuated when LDL-C was adjusted for lipoprotein in both the UK Biobank (Rho: 0.90) and Jackson Heart Study [Rho: 0.87; assuming lipoprotein(a) mass contributes to ∼30% of measured LDL-C)].
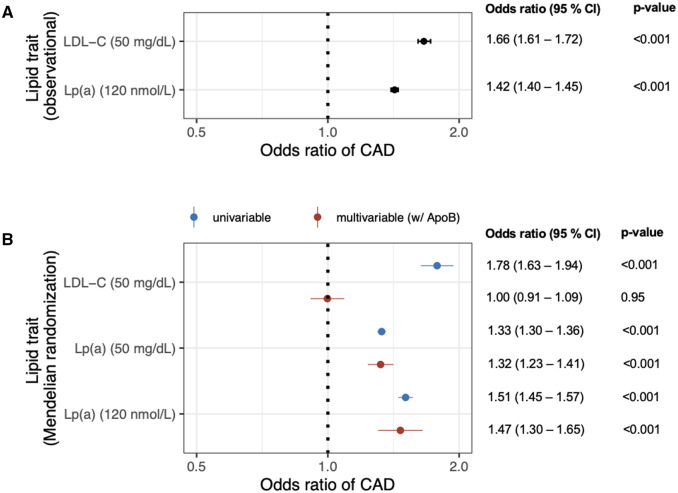
Secondly, we assessed the association of measured LDL-C and lipoprotein(a) with risk of incident CAD among UK Biobank participants. CAD was defined as myocardial infarction or coronary revascularization occurring between enrolment and 31 March 2020 [median follow-up (interquartile range): 11.1 years (1.4 years)]. The age- and sex-adjusted odd ratios for CAD were 1.66 for a 50 mg/dL increase in LDL-C levels (95% CI: 1.61–1.72, P<0.001) and 1.42 for a 120 nmol/L increase in lipoprotein(a) (120 nmol/L ∼ 50 mg/dL; 95% CI: 1.40–1.45, P<0.001) (Figure 1A). When LDL-C was adjusted for lipoprotein(a), the adjusted odds ratio for CAD per a 50 mg/dL increase in LDL-C was modestly attenuated to 1.47 (95% CI: 1.42–1.52, P<0.001).
Figure 1.
Apolipoprotein B is sufficient to explain the risk of CAD associated with LDL-C but not lipoprotein(a). (A) The epidemiological estimates for the association of a 50 mg/dL increase in measured LDL-C or 120 nmol/L increase in lipoprotein(a) (∼50 mg/dL) with risk incident CAD are shown for UK Biobank participants (n=327 929). Data are displayed as odds ratios with 95% confidence intervals (calculated by logistic regression adjusted for age and sex). (B) Causal estimates for the relationship between a 50 mg/dL increase in genetically associated LDL-C or lipoprotein(a) mass with risk of CAD are shown as odds ratios with 95% confidence intervals (calculated using an inverse weighted variance Mendelian randomization analysis). Causal estimates are also depicted for the relationship between a 120 nmol/L increase in genetically associated lipoprotein(a) molar concentration and risk of CAD. Mendelian randomization results are depicted as univariable analyses and multivariable analyses that included ApoB. ApoB, apolipoprotein B; CAD, coronary artery disease; LDL-C, low-density lipoprotein cholesterol; Lp(a), lipoprotein(a).
Thirdly, we used Mendelian randomization to provide causal inference for the genetic association between LDL-C or lipoprotein(a) and risk of CAD when adjusted for apolipoprotein B. Genetic associations with apolipoprotein B, LDL-C, lipoprotein(a) mass, and CAD were obtained from the UK Biobank (n=308 897 Europeans, excluding individuals using cholesterol-lowering medication),4 Million Veterans Program (n=227 817 Europeans),4 Coronary Heart Disease Exome+ Consortium (n=34 276 Europeans),5 and CARDIoGRAMplusC4D Consortium (60 801 CAD cases and 123 504 controls; predominately European, South Asian, and East Asian), which does not include UK Biobank.6 To identify valid genetic instruments, we applied the Lasso method to genetic associations that displayed significant, conditional associations with LDL-C7 or lipoprotein(a)5 at a genome-wide threshold of statistical significance. We identified 161 valid instruments for LDL-C and 30 valid instruments for lipoprotein(a). Univariable and multivariable Mendelian randomization analyses were performed using the inverse weighted variance method. Pleiotropic effects were assessed, and if present, corrected for using MR-PRESSO.8 In univariable Mendelian randomization analyses, the odds ratios for CAD were 1.78 for a 50 mg/dL increase in LDL-C levels (95% CI: 1.63–1.94, P<0.001) and 1.33 for a 50 mg/dL increase in lipoprotein(a) mass (95% CI: 1.30–1.36, P<0.001) (Figure 1B). The odds ratio for CAD for a 120 nmol/L increase in genetically associated lipoprotein(a) was 1.51 [95% CI: 1.45–1.57; using UK Biobank summary statistics for lipoprotein(a) molar concentration], which suggests that assays for both lipoprotein(a) mass and molar concentration provide similar results. Alternatively, in multivariable analyses with apolipoprotein B, the odds ratio of CAD for a 50 mg/dL increase in LDL-C was markedly attenuated [odds ratio (95% CI): 1.00 (0.91–1.09), P=0.95], while the odds ratio for a 50 mg/dL increase in lipoprotein(a) mass was unchanged (Figure 1B).
Ongoing randomized clinical trials are investigating whether lowering plasma lipoprotein(a) levels can reduce the risk of recurrent CAD. Importantly, this study highlights the need to identify the key features of lipoprotein(a) particles that causally contribute to CAD. Here, we demonstrate that unlike LDL-C,2 apolipoprotein B alone was insufficient to explain the risk of CAD associated with lipoprotein(a). On an equimolar basis, it has been hypothesized that lipoprotein(a) is more atherogenic than conventional apolipoprotein B particles due to its atherogenic (similar to LDL), pro-inflammatory (i.e. propensity to carry oxidized phospholipids), and prothrombotic properties (apolipoprotein(a)-mediated). However, it has also been suggested that apolipoprotein(a) isoform size does not associate with increased risk of CAD independent of the molar concentration of lipoprotein(a).1 Future studies are needed to address if other atherogenic and prothrombotic characteristics of lipoprotein(a), such as oxidated phospholipids and apolipoprotein(a) (which has no traditional lipid binding sites and may impart atherothrombotic effects), are causally related to increased risk of CAD. Additionally, this work suggests that current strategies of extrapolating the dose–dependent relationship between LDL-C and risk of CAD, which is dependent on apolipoprotein B, to estimate the potential benefit of lipoprotein(a)-lowering strategies in randomized clinical trials or primary prevention may have limitations.5
Limitations of this study include (i) inter-study differences in assays used to measure lipoprotein(a), (ii) the generalizability of study results to non-European populations, (iii) ascertainment bias associated with volunteer recruitment for the observational associations of lipid traits with CAD, and (iv) epidemiologic associations with CAD were assessed using diagnosis and operational billing codes. In summary, the plasma concentration of apolipoprotein B can fully explain the association of LDL-C with CAD, but not lipoprotein(a). This work does not suggest that lipoprotein(a) is a more superior biomarker for cardiovascular risk prediction than apolipoprotein B.
Acknowledgements
This work was supported by UK Biobank application 7089, and the authors would like to thank the UK Biobank study staff and participants.
Conflict of interest: P.N. reports grant support from Amgen, Apple, and Boston Scientific, consulting income from Apple, Blackstone Life Sciences, Genentech, and Novartis, and spousal employment at Vertex, all unrelated to the present work. The other authors do not report any disclosures.
Funding
P.N. was supported by grants from the National Heart, Lung, and Blood Institute (R01HL142711, R01HL148565, R01HL148050) and Fondation Leducq (TNE-18CVD04), and a Hassenfeld Scholar Award from the Massachusetts General Hospital. S.M.Z. was supported by the NIH National Heart, Lung, and Blood Institute (1F30HL149180-01) and the NIH Medical Scientist Training Program Training Grant (T32GM136651).
Data Availability
Summary statistics used in this study can be obtained from the CARDIoGRAMplusC4D Consortium for CAD (http://www.cardiogramplus4d.org), Million Veterans Program for LDL-C (Supplementary Table 9 of PMID: 30275531 or dbGaP Study Accession: phs001672.v4.p1), Coronary Heart Disease Exome+ Consortium lipoprotein (a) for lipoprotein (a) mass (eTable 3 of PMID: 29926099), UK Biobank for lipoprotein(a) molar concentration (Dr. Benjamin Neale's laboratory, http://www.nelab.ls/uk-biobank/). Individual level data that support the findings of this study are available from the Jackson Heart Study through dbGaP (dbGaP Study Accession: phs000286.v3.p1) or from the UK Biobank (http://www.ukbiobank.ac.uk/about-biobank-uk). Restictions apply to the availability of these data. Data are available for bona fide researchers upon application to dbGaP for Jackson Heart Study data or the UK Biobank.
References
- 1. Gudbjartsson DF, Thorgeirsson G, Sulem P, Helgadottir A, Gylfason A, Saemundsdottir J, Bjornsson E, Norddahl GL, Jonasdottir A, Jonasdottir A, Eggertsson HP, Gretarsdottir S, Thorleifsson G, Indridason OS, Palsson R, Jonasson F, Jonsdottir I, Eyjolfsson GI, Sigurdardottir O, Olafsson I, Danielsen R, Matthiasson SE, Kristmundsdottir S, Halldorsson BV, Hreidarsson AB, Valdimarsson EM, Gudnason T, Benediktsson R, Steinthorsdottir V, Thorsteinsdottir U, Holm H, Stefansson K.. Lipoprotein(a) concentration and risks of cardiovascular disease and diabetes. J Am Coll Cardiol 2019;74:2982–2994. [DOI] [PubMed] [Google Scholar]
- 2. Richardson TG, Sanderson E, Palmer TM, Ala-Korpela M, Ference BA, Smith GD, Holmes MV.. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: a multivariable Mendelian randomisation analysis. PLoS Med 2020;17:e1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zekavat SM, Ruotsalainen S, Handsaker RE, Alver M, Bloom J, Poterba T, Seed C, Ernst J, Chaffin M, Engreitz J, Peloso GM, Manichaikul A, Yang C, Ryan KA, Fu M, Johnson WC, Tsai M, Budoff M, Vasan RS, Cupples LA, Rotter JI, Rich SS, Post W, Mitchell BD, Correa A, Metspalu A, Wilson JG, Salomaa V, Kellis M, Daly MJ, Neale BM, McCarroll S, Surakka I, Esko T, Ganna A, Ripatti S, Kathiresan S, Natarajan P; NHLBI TOPMed Lipids Working Group. Deep coverage whole genome sequences and plasma lipoprotein(a) in individuals of European and African ancestries. Nat Commun 2018;9:2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, Cortes A, Welsh S, Young A, Effingham M, McVean G, Leslie S, Allen N, Donnelly P, Marchini J.. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burgess S, Ference BA, Staley JR, Freitag DF, Mason AM, Nielsen SF, Willeit P, Young R, Surendran P, Karthikeyan S, Bolton TR, Peters JE, Kamstrup PR, Tybjærg-Hansen A, Benn M, Langsted A, Schnohr P, Vedel-Krogh S, Kobylecki CJ, Ford I, Packard C, Trompet S, Jukema JW, Sattar N, Angelantonio ED, Saleheen D, Howson JMM, Nordestgaard BG, Butterworth AS, Danesh J; European Prospective Investigation Into Cancer and Nutrition–Cardiovascular Disease (EPIC-CVD) Consortium. Association of LPA variants with risk of coronary disease and the implications for lipoprotein(a)-lowering therapies: a Mendelian randomization analysis. JAMA Cardiol 2018;3:619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nikpay M, Goel A, Won H-H, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, Webb TR, Zeng L, Dehghan A, Alver M, Armasu SM, Auro K, Bjonnes A, Chasman DI, Chen S, Ford I, Franceschini N, Gieger C, Grace C, Gustafsson S, Huang J, Hwang S-J, Kim YK, Kleber ME, Lau KW, Lu X, et al. A comprehensive 1000 Genomes–based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, Gagnon DR, DuVall SL, Li J, Peloso GM, Chaffin M, Small AM, Huang J, Tang H, Lynch JA, Ho Y-L, Liu DJ, Emdin CA, Li AH, Huffman JE, Lee JS, Natarajan P, Chowdhury R, Saleheen D, Vujkovic M, Baras A, Pyarajan S, Di Angelantonio E, Neale BM, Naheed A, Khera AV, Danesh J, Chang K-M, Abecasis G, Willer C, Dewey FE, Carey DJ, Concato J, Gaziano JM, O’Donnell CJ, Tsao PS, Kathiresan S, Rader DJ, Wilson PWF, Assimes TL; Global Lipids Genetics Consortium. Genetics of blood lipids among ∼300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet 2018;50:1514–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Verbanck M, Chen C-Y, Neale B, Do R.. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018;50:693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Summary statistics used in this study can be obtained from the CARDIoGRAMplusC4D Consortium for CAD (http://www.cardiogramplus4d.org), Million Veterans Program for LDL-C (Supplementary Table 9 of PMID: 30275531 or dbGaP Study Accession: phs001672.v4.p1), Coronary Heart Disease Exome+ Consortium lipoprotein (a) for lipoprotein (a) mass (eTable 3 of PMID: 29926099), UK Biobank for lipoprotein(a) molar concentration (Dr. Benjamin Neale's laboratory, http://www.nelab.ls/uk-biobank/). Individual level data that support the findings of this study are available from the Jackson Heart Study through dbGaP (dbGaP Study Accession: phs000286.v3.p1) or from the UK Biobank (http://www.ukbiobank.ac.uk/about-biobank-uk). Restictions apply to the availability of these data. Data are available for bona fide researchers upon application to dbGaP for Jackson Heart Study data or the UK Biobank.