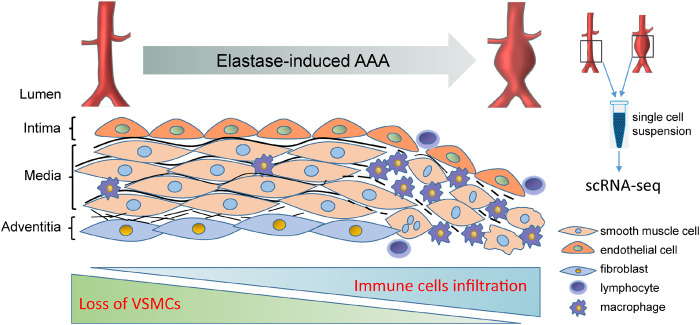
Abstract
Aims
The artery contains numerous cell types which contribute to multiple vascular diseases. However, the heterogeneity and cellular responses of these vascular cells during abdominal aortic aneurysm (AAA) progression have not been well characterized.
Methods and results
Single-cell RNA sequencing was performed on the infrarenal abdominal aortas (IAAs) from C57BL/6J mice at Days 7 and 14 post-sham or peri-adventitial elastase-induced AAA. Unbiased clustering analysis of the transcriptional profiles from >4500 aortic cells identified 17 clusters representing nine-cell lineages, encompassing vascular smooth muscle cells (VSMCs), fibroblasts, endothelial cells, immune cells (macrophages, T cells, B cells, and dendritic cells), and two types of rare cells, including neural cells and erythrocyte cells. Seurat clustering analysis identified four smooth muscle cell (SMC) subpopulations and five monocyte/macrophage subpopulations, with distinct transcriptional profiles. During AAA progression, three major SMC subpopulations were proportionally decreased, whereas the small subpopulation was increased, accompanied with down-regulation of SMC contractile markers and up-regulation of pro-inflammatory genes. Another AAA-associated cellular response is immune cell expansion, particularly monocytes/macrophages. Elastase exposure induced significant expansion and activation of aortic resident macrophages, blood-derived monocytes and inflammatory macrophages. We also identified increased blood-derived reparative macrophages expressing anti-inflammatory cytokines suggesting that resolution of inflammation and vascular repair also persist during AAA progression.
Conclusion
Our data identify AAA disease-relevant transcriptional signatures of vascular cells in the IAA. Furthermore, we characterize the heterogeneity and cellular responses of VSMCs and monocytes/macrophages during AAA progression, which provide insights into their function and the regulation of AAA onset and progression.
Keywords: Single-cell RNA sequencing, Abdominal aortic aneurysm, Lineage heterogeneity, Vascular smooth muscle cell, Macrophage
Graphical Abstract
1. Introduction
Abdominal aortic aneurysm (AAA) is a life-threatening cardiovascular disease that is characterized by permanent dilatation of the arterial wall of 50% or more compared to the normal diameter.1,2 More than 80% of AAA occurs in the infrarenal aorta proximal to the aortic bifurcation.1,2 The main pathological features of AAA include extracellular matrix (ECM) degradation,3 loss of medial contractile vascular smooth muscle cells (VSMCs),4 and immune cells infiltration and activation in the adventitia and tunica media,5,6 all contributing to vascular remodelling and weakening of the aortic wall. During the past decade, the development of in vivo lineage tracing systems has enabled the unbiased fate mapping and identification of vascular cells during the progression of cardiovascular diseases.7–9 However, current approaches to characterize the cell types using a small number of preselected markers do not reflect their in vivo functional states and transcriptional profiles.
Although the major aortic cell types in the whole aorta are well known,10,11 the heterogeneity and relative contribution of different vascular cells in healthy and aneurysmal aortas are poorly understood. Recently, the development and use of single-cell RNA sequencing (scRNA-seq) provides an opportunity for a comprehensive and unbiased characterization of the molecular profile and heterogeneity of large numbers of individual cells in healthy or diseased aortas. Recent studies have used scRNA-seq to depict the landscapes of endothelial cells (ECs),11 VSMCs,10 adventitial cells,12 perivascular adipose tissue stem cells,13 immune cells,14,15 and macrophages8 within the aorta. Most of these studies isolated the single cells from the whole aorta or thoracic aorta, which are susceptible to atherosclerosis, and demonstrated the transcriptional and functional heterogeneity of vascular cells during the progression of atherosclerosis.
Understanding the heterogeneous identities, diverse functional states and subpopulation fractions within healthy and aneurysmal aortas will be fundamental to understanding aetiology and progression of AAA as to better define potential targets for early intervention. Here, we present the first comprehensive analysis of the lineage heterogeneity, altered transcriptomic profiles and functional states of vascular cells from healthy and aneurysmal mouse infrarenal abdominal aortas (IAAs) using scRNA-seq. By clustering analysis of the IAA cells, we identified 17 clusters and nine distinct cell types. We also report the changes of cellular subpopulations, fractions and transcriptomic profiles in response to elastase-induced AAA model.
2. Methods
Expanded methods are provided in Supplementary material online.
2.1 Mouse model of elastase-induced AAA
Elastase peri-adventitial exposure was used to induce AAA in 10-week-old C57BL/6J male mice.16,17 In brief, the mice were anaesthetized by intraperitoneal injection of a mixture of ketamine (100 mg/kg) and xylazine (5 mg/kg), and IAAs were exposed, isolated, and wrapped with sterile cotton soaked with 30 μL of elastase (E1250, lot No. SLBV9311, MilliporeSigma) or heat-inactivated elastase (to serve as Sham group) for 30 min. Then the elastase-soaked cotton was removed, and the abdominal cavity was washed with saline twice before closing the surgical incision. IAAs were harvested 7, 14, and 28 days after elastase exposure or 14 days after heat-inactivated elastase exposure (Sham). All animal care and experimental procedures were approved by the Institutional Animal Care & Use committee (IACUC) at the University of Michigan and complied with the National Institutes of Health (NIH) Guidelines for the care and use of laboratory animals.
2.2 Aortic dissociation and single cell preparation
At the end of experiments, mice were anaesthetized by using carbon dioxide (CO2) in accordance with the NIH Guidelines for the euthanasia of animals, and then perfused with 10 mL of PBS through left ventricular puncture. Preparation of the single-cell suspension of the aortic cells was performed following a previously described enzymatic digestion protocol.11,18 In brief, the IAAs pooled from five mice in each group were cut and digested with an enzyme solution [450 U/mL collagenase type I (Gibco, cat# 17100-017), 125 U/mL collagenase type XI (Millipore Sigma, cat# C7657), 60 U/mL hyaluronidase type I-s (Millipore Sigma, cat# H3506), and 60 U/mL DNase I (Millipore Sigma, cat# DN25)] for 1.5 h at 37°C. The cell suspension was strained through a 70 μm filter and washed thrice with PBS. The cells were resuspended in opti-MEM with 10% FBS, and the cell viability was greater than 70%, as confirmed by trypan blue staining. The resuspended cells were subjected to scRNA-seq.
2.3 Single-cell RNA sequencing
Standard 10× Chromium Single Cell 3′ Solution v2 (10× Genomics Gemcode Technology) protocols were followed for scRNA-seq.
2.4 Processing of scRNA-seq data
The raw scRNA-seq data was processed using the 10× Genomics Cell Ranger software. The CellRanger mkfastq command was used to generate Fastq files. Subsequently, data were mapped to the pre-build mouse reference genome (mm10, version 1.2.0).
2.5 Scrna-seq data analysis
After aggregation of the samples from IAA cells, R package Seurat v3.0 was used for cell filtration, normalization, principal component analysis, variable genes finding, clustering analysis, and Uniform Manifold Approximation and Projection (UMAP) dimensional reduction. The Seurat functions Vlnplot, FeaturePlot, DotPlot, and DoHeatmap were used to visualize the gene expression with violin plot, feature plot, dot plot, and heatmap, respectively. Markers for a specific cluster against all remaining cells were found by using the Seurat function FindAllMarkers.
2.6 Statistical analysis
The data corresponding to aortic diameters, elastin degradation grade, and inflammatory cells (including leucocytes and macrophages) infiltration in the aortic wall are presented as mean ± standard error of the mean. Statistical analysis was performed using GraphPad Prism 7.0 Software (GraphPad Software, San Diego, CA, USA). All data were tested for normality and similar variance. The Kruskal–Wallis (non-parametric) test was used to compare the elastin degradation grade. One-way analysis of variance followed by Tukey’s post hoc analysis was used to compare the aortic diameters and inflammatory cells infiltration among Sham, Elastase7d, Elastase14, and Elastase28d.
3. Results
3.1 Single-cell RNA sequencing revealed 17 cell clusters representing nine-cell lineages in the infrarenal abdominal aorta
The elastase-induced AAA model was performed on C57BL/6J mice (Supplementary material online, Figure S1A–F), as previously reported.16,17 During the 4 weeks after elastase exposure, no rupture or mortality occurred in this model. At Days 7, 14, and 28 after elastase exposure (Elastase7d, Elastase14d, and Elastase28d), the IAAs were time-dependently expanded in the elastase-treated groups compared with the heat-inactivate elastase (Sham) group (Supplementary material online, Figure S1A and B). Verhoff-Van Gieson staining performed on serial sections of the IAAs demonstrated that elastin degradation was slightly increased in Elastase7d and significantly increased in Elastase14d and Elastase28d, albeit without significant changes between Elastase14d and Elastase28d (Supplementary material online, Figure S1C and D). Moreover, leucocyte (Cd45+) and macrophage (Mac2+) infiltration to the aortic wall was markedly increased in the elastase-treated groups compared with the sham group (Supplementary material online, Figure S1E). Meanwhile, immunofluorescence staining of the smooth muscle cell (SMC) marker SM22α showed greater SMC loss after elastase exposure (Supplementary material online, Figure S1F). Based on the similarity of the pathological features related to aneurysm formation, including aortic dilation, aortic wall degradation, inflammatory cells infiltration in the aortic walls, and loss of medial SMCs, between Elastase14d and Elastase28d (Supplementary material online, Figure S1A–F) which are consistent with stabilization of the lesions as previously reported,19 only the IAAs from Sham, Elastase7d, and Elastase14d were subjected to scRNA-seq.
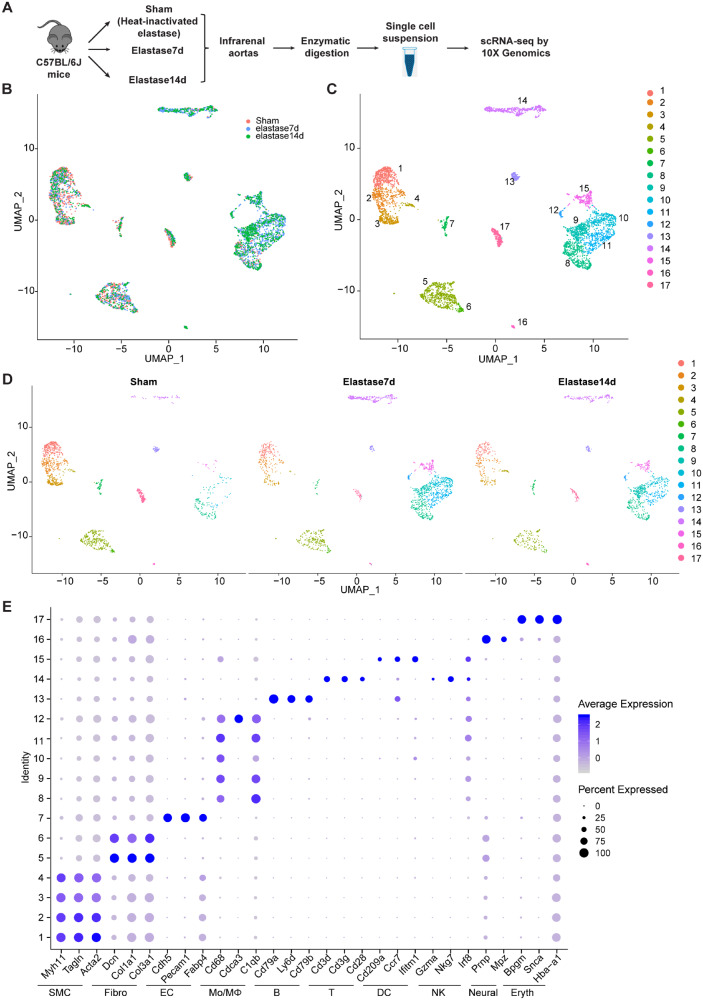
Single cells from three samples from each treatment group, isolated as described in the methods, were subsequently individually bar-coded and sequenced by using the 10× Genomics Chromium platform (Figure 1A). The individual samples yielded similar median number of genes (median genes) per cell after processing the datasets with Cell Ranger (Supplementary material online, Figure S1G). The transcriptional profiles of 1509, 1737, and 1396 cells for Sham, Elastase7d, and Elastase14d, respectively, were included in the subsequent analysis after implementing the quality control filtering described in the methods (Supplementary material online, Figures S1H–J and S2). Data integration and unbiased clustering identified a total of 17 clusters were singled out in IAA cells of the integrated datasets (Figure 1B–D).
Figure 1.
Identification of cell clusters present in the mouse infrarenal abdominal aortas (IAA) by single-cell RNA sequencing (scRNA-seq). (A) Schematic diagram indicating the procedure for scRNA-seq. Sham, 14 days after heat-inactivated elastase-exposure; Elastase7d, 7 days after elastase-exposure; Elastase14d, 14 days after elastase exposure. For each experimental condition, the IAA cells were pooled from 5 mice in each group. (B) Uniform Manifold Approximation and Projection (UMAP) plot of aggregate IAA cells from Sham, Elastase7d and Elastase14d. Colours denote different conditions. After quality control, 1509, 1737, and 1396 cells from Sham, Elastase7d, and Elastase14d, respectively, were captured for clustering analysis. (C) UMAP plot of aggregate IAA cells with colours denoting different clusters. (D) UMAP plot of cell clusters in IAA cells across the indicated conditions. Colours denoting different clusters. (E) Dot plot of selected marker genes for each cluster and lineage in aggregate IAA cell clusters. Dot size indicates the percentage of cells expressing each gene, and dot colour indicates expression level. B, B cells; DC, dendritic cells; EC, endothelial cells; Eryth, erythrocytes; Fibro, fibroblasts; Mono/MΦ, monocytes/macrophages; Neural, neural cells; NK, natural killer cells; SMC, smooth muscle cells; T, T cells.
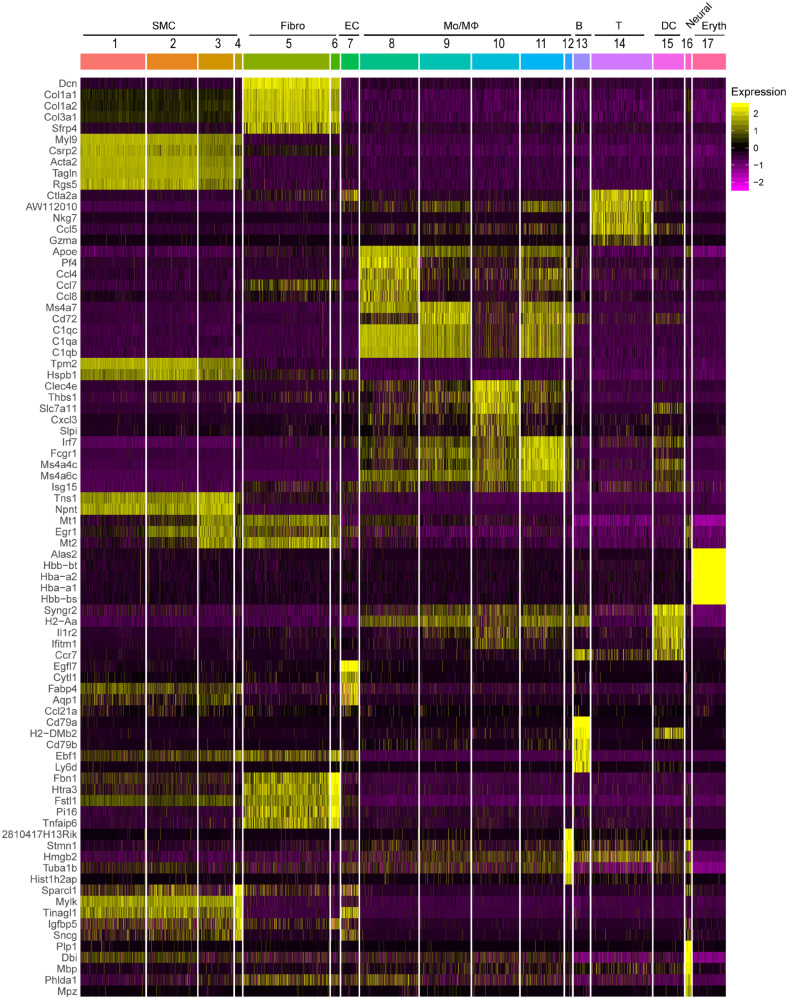
The top 10 marker genes for a cluster were defined [by average log(fold change)] relative to all other clusters (Supplementary material online, Table S_I). The cell type-specific canonical markers for each cell lineage were used to distinguish cell lineages (Figure 1E). Similar to the separate clustering data, nine distinct cell lineages were identified in the IAA cells from the integrated data (Supplementary material online, Figures S2 and S3 and Figure 2). Major cell types comprised: (i) SMCs (Clusters 1, 2, 3, 4), which highly expressed the SMC canonical markers, Myh11, Tagln, and Acta2; (ii) fibroblasts (Fibro, Clusters 5, 6), which featured the expression of collagens/collagen-binding proteins, Dcn, Col1a1, and Col3a1; (iii) ECs (Cluster 7), which was marked by the expression of Cdh5, Pecam1, and Fabp4; (iv) monocytes and macrophages (Mo/MΦ, Clusters 8, 9, 10, 11, and 12), which demonstrated high expression of Cd68, Cdca3, and C1qb; (v) B cells (B, Cluster 13), which highly expressed Cd79a, Ly6d, and Cd79b; (vi) T cells (T, Cluster 14), which showed high level of Cd3d, Cd3g and Cd28; (vii) dendritic cells (DCs, Cluster 15), which highly expressed the marker genes, Cd209a, Ccr7, and Ifitm1 (Figure 1E and Supplementary material online, Figure S3). Of note, although the expression of natural killer cell marker genes Gzma, Nkg7, and Irf8 were also detected in Cluster 14, no natural killer cells were singled out, and this cluster showed highest percentage of expression of the T-cell markers Cd3d and Cd3g; additionally, (viii) a small cluster of neural cells (Neural, Cluster 16) with marker genes Prnp, and (ix) a small population of erythrocyte cells (Eryth, Cluster 17) with high expression of Bpgm and Snca (Figure 1E and Supplementary material online, Figure S2) were also singled out in IAA cells. We also identified the top 5 marker genes (sorted by P-value) for each cluster relative to all other clusters, and these genes were plotted using heatmap (Figure 2 and Supplementary material online, Table S_II). Collectively, distinct gene expression patterns across all clusters were observed with unbiasedly identified marker genes for each cluster listed in Supplementary material online, Table S_III.
Figure 2.
Heat map of the top 5 [by average Log(fold change)] genes with specific expression for each cell cluster in aggregate IAA cells. The top5 cluster-specific markers for each cluster were selected from the all markers of each cluster based on the average Log(fold change). The FindAllmarkers function in Seurat v3.0 was used with the parameters test. use = wilcox, min.pct = 0.25, thresh.use = 0.25, only.positive = TRUE and return.thresh = 0.01, to find all markers for each cluster.
3.2 Decreased SMCs and expansion of immune cells in the aneurysmal infrarenal abdominal aorta
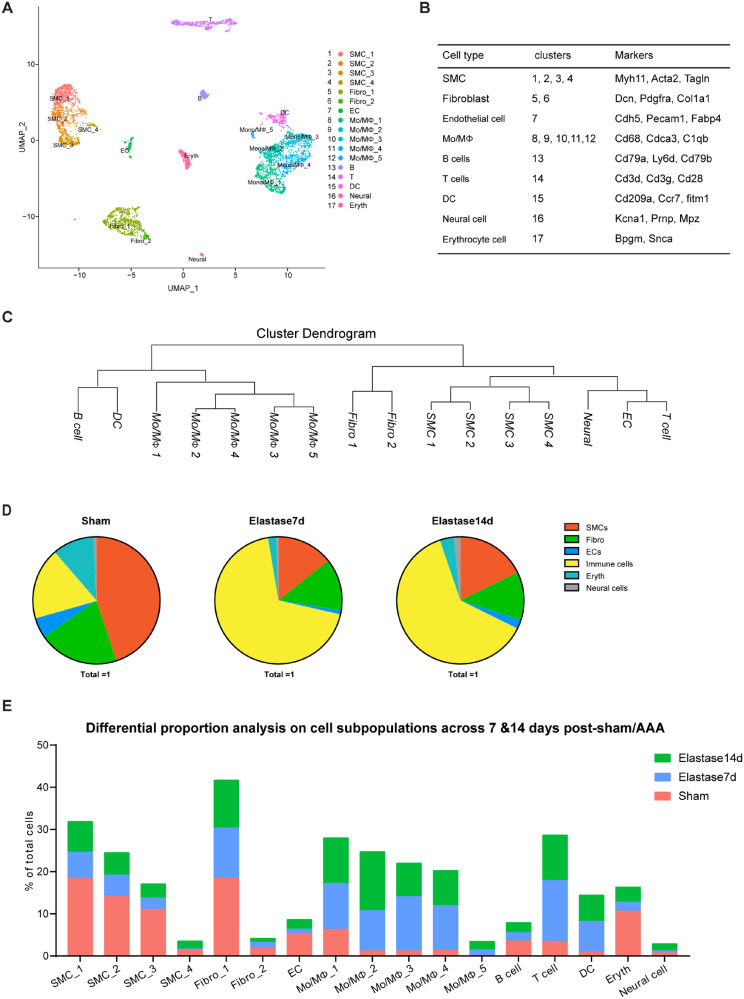
After characterizing the cell clusters identified by the cell type-specific markers, we then examined the cell subpopulations within the IAA (Figure 3A and B). Among the identified cell types, SMCs, fibroblasts, and monocyte/macrophages were clustered into multiple subpopulations: four SMC clusters (1, SMC_1; 2, SMC_2; 3, SMC_3; 4, SMC_4), two fibroblast clusters (5, Fibro_1; 6, Fibro_2), and five Mo/MΦ clusters (8, Mo/MΦ_1; 9, Mo/MΦ_2; 10, Mo/MΦ_3; 11, Mo/MΦ_4; 12, Mo/MΦ_5) (Figure 3A–C).
Figure 3.
Categorization of mouse IAA cell populations. (A) UMAP plot of aggregate IAA cells with identified cell lineages and subpopulations. Sham, n = 1509 cells. Elastase7d, n = 1737 cells. Elastase14d, n = 1396 cells. (B) Clusters and major cell types correspondence. (C) Dendrogram of the major cell types and subpopulations in IAA cells according to average RNA expression. (D) Fraction of each cell type in IAA cells across conditions (Control, elastase7d, and elastase14d). Sham, n = 1509 cells. Elastase7d, n = 1737 cells. Elastase14d, n = 1396 cells. (E) Cell population percentages across conditions.
We also observed aortic aneurysm-associated cellular responses of the IAA cells at the single-cell level, including decrease of SMC population and expansion of immune cell populations (Figure 3D), which is consistent with the pathological changes in SMCs and immune cells determined by immunofluorescence staining (Supplementary material online, Figure S1E and F). Within the IAA cells from Sham, SMCs are the largest population, accounting for 44.9% of all cells (Supplementary material online, Figure S2B and Figure 3D and E). Fibroblasts and total immune cells have similar proportions, accounting for 20.3% and 18.1%, respectively (Figure 3D and E). Among the immune cells, Mo/MΦ are the largest population, which accounts for 55.7% of the immune cells (Figure 3D and E).
During AAA progression, vascular SMCs undergo phenotypic modulation or apoptosis.4,20–22 Consistent with the pathologic changes seen in human AAAs,21,22 the SMC populations significantly decreased in the aneurysmal aortas relative to the sham aortas (Sham, 44.9% vs. Elastase7d, 14.3% vs. Elastase14d, 17.8%, Figure 3B, D, E). Additionally, at Day 7, before aneurysms were evident, the elastase-treated aorta had a 68.2% reduction of the total SMCs, compared with the sham aorta, and the reduction of SMC population (60.5%) remained at 14 days after AAA formation (Supplementary material online, Figure S1A and B and Figure 3D and E). We also observed immune cells expansion (Sham, 18.1% vs. Elastase7d, 68.8%, vs. Elastase14d, 62.5%) and decrease of fibroblasts (Sham, 20.3% vs. Elastase7d, 13.2%, vs. Elastase14d, 12.2%) during AAA progression (Figure 3D and E). Among the immune cells, Mo/MΦ, T cells and DCs expanded during AAA progression (Mo/MΦ, Sham, 10.1% vs. Elastase7d, 45.2%, vs. Elastase14d, 43.1%; T cells, Sham, 3.5% vs. Elastase7d, 14.5%, vs. Elastase14d, 10.8%; DCs, Sham, 1% vs. Elastase7d, 7.2%, vs. Elastase14d, 6.2%, of total cells sequenced in each treatment) (Figure 3E). Additionally, we analysed pooled T cells by UMAP and identified three T-cell subpopulations (Supplementary material online, Figure S4A). T_1 was the largest subpopulation, accounting for more than 50% of the total T cells and highly expressing Ly6c2, Cd8a, and Cd8b1 (Supplementary material online, Figure S4B–E). T_2 expressed Gzma, which encodes a cytotoxic T-cell and natural killer cell-specific serine protease, granzyme A (Supplementary material online, Figure S4C–E). T_3 expressed Cd4 and Il2ra, encoding CD25, which preferentially marks regulatory T cells (Supplementary material online, Figure S4D and E). During AAA progression, the proportions of T_1 and T_3 were decreased at Day 7 and were restored at Day 14 (Supplementary material online, Figure S4B). On the contrary, the proportion of T_2 was increased at Day 7 and was restored at Day14 (Supplementary material online, Figure S4B).
3.3 VSMC transcriptomes reflect phenotypic and functional heterogeneity within aneurysmal IAA
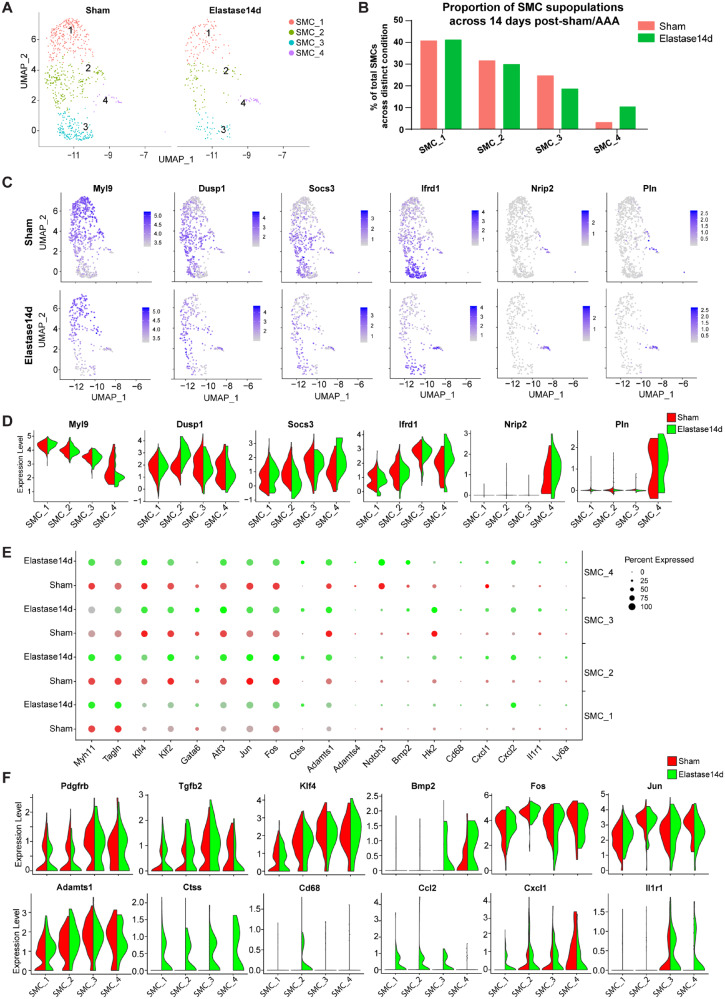
We identified four distinct SMC clusters both in the sham and aneurysmal IAA (Figure 4A, and Supplementary material online, Figure S5A). SMC_1, SMC_2, and SMC_3 account for more than 98% of the SMC population in the sham group, whereas SMC_4 only represented less than 2% of the total SMCs (Figure 4B). Of note, even the total SMCs significantly decreased after AAA formation, SMC_1, SMC_2, and SMC_3 were proportionally decreased and demonstrated similar ratios during AAA development (Figure 4B and Supplementary material online, Figure S5B). In contrast, SMC_4 increased from less than 2% of total SMCs in Sham and Elastase7d, to 10% in the Elastase14d (Figure 4B and Supplementary material online, Figure S5B).
Figure 4.
Comparison of SMC subpopulations in IAA from Sham and AAA. (A) UAMP plot of the SMC subpopulations (1, SMC_1; 2, SMC_2; 3, SMC_3; 4, SMC_4) from Sham and Elastase14d. Sham, n = 678 SMCs. Elastase14d, n = 248 SMCs. (B) The percentages of the SMC subpopulations from Sham and Elastase14d. For Sham, SMC_1, n = 276 cells, SMC_2, n = 214 cells, SMC_3, n = 167 cells, SMC_4, n = 21 cells. For Elastase14d, SMC_1, n = 102 cells, SMC_2, n = 74 cells, SMC_3, n = 46 cell, SMC_4, n = 26 cell. (C) Expression of selected marker genes for the SMC subpopulations from Sham or Elastase14d as visualized by feature plots. (D) Expression of selected marker genes for SMC subpopulations from Sham and Elastase14d as visualized by Violin plots. (E) Dot plot of selected marker genes for each SMC subpopulation. Dot size indicates the percentage of cells within the group expressing each gene. Dot colour intensity indicates the gene expression level. With the limitation of Seurat V3.0, the colour scale bar for the average expression cannot be shown in on dotplot that have been split by different experimental conditions. (F) Violin plot of selected marker genes for each SMC subpopulation with colour denoting experimental conditions.
Although all of the four clusters were identified as SMCs, they still had different expression profiles (Figures 1E, 4C andD and Supplementary material online, Figure S5C and D). Next, we determined the differentially expressed genes among the four clusters and found 34 marker genes showing significant differential expression using the Wilcoxon rank-sum test with Bonferroni correction (P < 0.001, avg_logFC > 1) (Supplementary material online, Table S_IV). Both SMC_1 and SMC_2 showed consistently high expression of the SMC-specific contractile markers (Myh11, Acta2, Tagln, and Myl9) and low expression of collagen and cytokine genes (Col1a1, Col1a2, Col3a1, Ccl4, Cxcl1, Bmp2, and Tgfb2), signalling receptor gene (Pdgfrb), as well as stem cell marker gene Ly6a (encoding Sca1) (Supplementary material online, Table S_III, Figures 2 and 4E and Supplementary material online, Figure S5E). Additionally, SMC_1 and SMC_2 displayed similar gene expression patterns after elastase-treatment, including slight down-regulation of Myh11 and Tagln, and significant up-regulation of Ctss, Adamts1, Cxcl2, and Ccl2 (Figure 4E and F and Supplementary material online, Figure S5D and E). In agreement with previous reports,4,23,24 the quiescent, contractile VSMCs in the media can undergo phenotypic switching to a pro-inflammatory phenotype during AAA progression. Importantly, the marker genes for SMC_2 distinguishing it from other three SMC clusters included Fos, Jun, Klf2, and Atf3, all important for regulation of cell growth and proliferation (Supplementary material online, Table S_IV). These growth-regulated genes were over-represented in SMC_2, and elastase exposure further up-regulated their expression (Figure 4E and F, and Supplementary material online, Figure S5D and E), strongly suggesting a growth function and activated state of SMC_2 even in the absence of aortic aneurysm. Of note, Dusp1, which encodes mitogen-activated protein kinase phosphatase-1 and suppresses VSMC proliferation by inactivating MAPK,25 was also highly represented in SMC_2 (Figure 4C and D and Supplementary material online, Figure S5C), which may contribute to maintain the phenotypic homeostasis of SMC_2. Here, we termed SMC_1 as quiescent, contractile SMCs and SMC_2 as proliferative, contractile SMCs, respectively, and referred to them as distinct cell entities, without prejudice about their origin, fate or contractile nature.
Although they expressed the SMC contractile markers Myh11, Tagln and Acta2, SMC_3 and SMC_4 displayed different expression patterns compared with SMC_1 and SMC_2 (Figures 2, 4C and E and Supplementary material online, Figure S5C and D). Ifrd1, which encodes interferon related developmental regulator 1 (IFRD1) and negatively regulates the thermogenic and mitochondrial gene expression in brown adipocytes,26 was enriched in both SMC_3 and SMC_4 (Figure 4C and D and Supplementary material online, Figure S5C). Nrip2, encoding nuclear receptor interacting protein 2, and Pln, encoding phospholamban which acts as a negative regulator of the sarcoplasmic reticulum Ca(2+)-pumping ATPase,27 were selectively expressed in SMC_4 (Figure 4C and D). Further analysis of SMC_3 and SMC_4 revealed some novel transcription factors that have been directly linked to VSMC phenotype or have documented roles in cardiovascular disease, inflammation and VSMC biology, specifically on proliferation and migration (Supplementary material online, Table S_IV). For example, one of the transcriptional repressors of SMC differentiation, Klf4 is highly expressed in SMC_3 and SMC_4 both in basal and elastase-treated conditions (Figure 4F and Supplementary material online, Figure S5E). Other genes showing similar expression profiles in SMC_3 and SMC_4, include factors involved in SMC proliferation (such as Atf3) and migration (such as Klf2, Ctss, Adamts1), as well as inflammation (Cxcl2, Ccl2) (Figure 4E and F and Supplementary material online, Figure S5D and E).
To further characterize the cellular functions that differ between SMC_3 and SMC_4, we investigated the expression profiles enriched for each subpopulation (Supplementary material online, Table S_IV). Among the marker genes of SMC_3, Mt1 and Mt2, which encode the antioxidant metallothionein1 and 2, respectively, displayed selective expression (Figure 2 and Supplementary material online, Table S_II). On the other hand, SMC_3 showed significantly greater expression of Hk2 (Figure 4E and Supplementary material online, Figure S5D), which encodes the glycolytic enzyme hexokinase 2. Hk2 elevated expression potentially drives energetic switch from oxidative phosphorylation to glycolysis in response to energetic failure.28 Therefore, SMC_3 may have an alternative bioenergetics profile compared to the other three SMC clusters. Additionally, Gata6, which is the only member of the GATA family expressed in VSMC and regulates their growth and adaptation,29 is also specifically and highly expressed in SMC_3 (Figure 4E and Supplementary material online, Figure S5D). Based on the lowest expression of contractile markers and alterative metabolic profile in SMC_3, this cluster may be termed as dedifferentiated SMCs. For SMC_4, multiple maker genes were selectively and highly expressed, including Sparcl1, Igfbp5, Sncg, and Thbs1 (Figure 2). Among the differentially expressed genes, Thbs1 is also highly expressed in macrophages (Figure 2). Additionally, Notch3, a cell-autonomous regulator of arterial differentiation and maturation of VSMCs,30 is also specifically and highly expressed in SMC_4 (Figure 4E and Supplementary material online, Figure S5D). In response to elastase-exposure, SMC_4 showed higher expression of pro-inflammatory factors, including Cd68, Cxcl1, Cxcl2, and Il1r1 (Figure 4E and F and Supplementary material online, Figure S5D and E). Notably, immunofluorescence staining showed that a small number of medial SMCs expressed the macrophage marker (Mac2) (Supplementary material online, Figure S1F). In addition, the SMC-specific contractile markers were highly expressed in SMC_4 in the sham group, then decreased at 7 days and restored at 14 days after elastase exposure (Figure 4E and Supplementary material online, Figure S5D). Thus, SMC_4 displayed a pro-inflammatory phenotype and were increased at the late stage of AAA development, hence termed as inflammatory-like SMCs.
3.4 Both aortic resident and blood-derived macrophages are involved in AAA progression
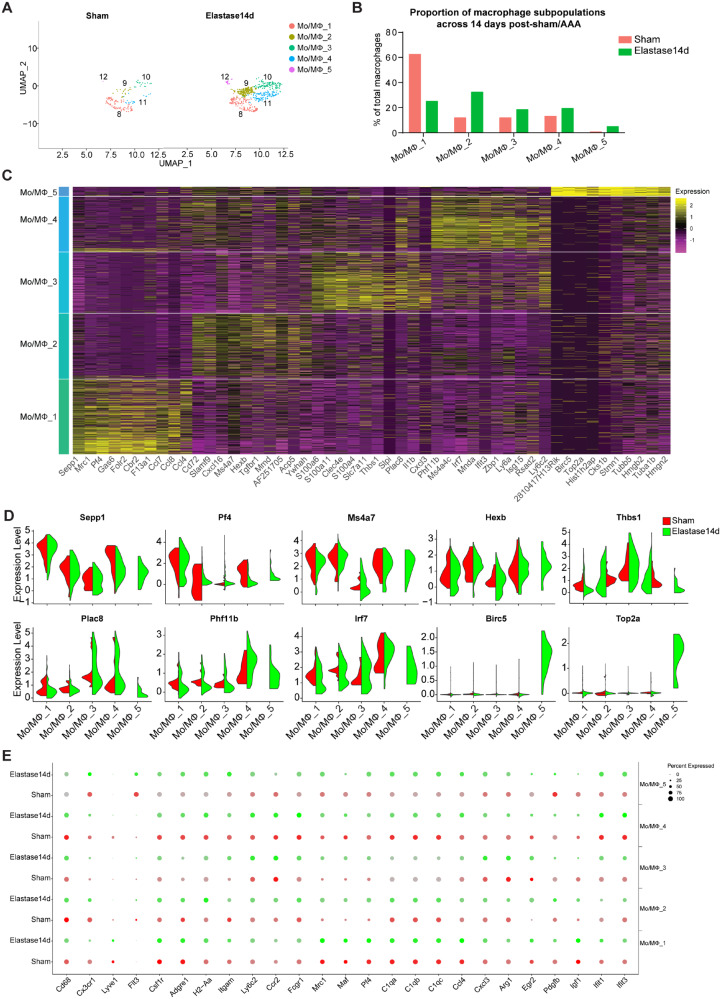
In the previous clustering analysis, five clusters of macrophages (Mo/MΦ_1, Mo/MΦ_2, Mo/MΦ_3, Mo/MΦ_4, and Mo/MΦ_5) were singled out in the sham and elastase-treated IAA cells (Figures 3A and B, 5A and Supplementary material online, Figure S6A). In the sham group, 62.5% of Mo/MΦ cells corresponds to Mo/MΦ_1, with the other four clusters only accounting for 37.5% (11.8%, 11.8%, 13.2%, 0.7%, respectively) (Figure 5B). Of note, although the canonical macrophage markers, including Cd14, Cd68, Adgre1 (encoding F4/80), H2-Aa (encoding MHC-II), and Fcgr1 (encoding CD11b), were expressed in the five Mo/MΦ clusters, albeit at varying proportions and levels, multiple genes were differentially expressed among them (Figure 2, Supplementary material online, Table S_V and Figure 5C and D). For example, Sepp1, encoding selenoprotein P, and Pf4, encoding a small cytokine platelet factor 4 and also known as CXCL4, were selectively expressed by Mo/MΦ_1. Both Mo/MΦ_2 and Mo/MΦ_4 expressed Ms4a7 (encoding membrane-spanning 4-domains subfamily A member 7) and Hexb (encoding hexosaminidase subunit beta), whereas Phf11b (encoding PHD finger protein 11) and Irf7 (encoding interferon regulatory factor 7) were selectively enriched in Mo/MΦ_4. In Mo/MΦ_3, Thbs1 (encoding thrombospondin 1) and Plac8 (encoding placenta specific 8) were highly expressed. For the smallest cluster, Mo/MΦ_5 exhibited highest expression of Birc5 (encoding baculoviral inhibitor of apoptosis repeat-containing 5) and Top2a (encoding DNA topoisomerase 2-alpha). In addition, there is an overlap in gene expression in the macrophage clusters and DC cluster (marked by Cd209a, H2-Aa and H2-DMb2), showing high expression of H2-Aa (Figure 2). Moreover, elastase-exposure induced significant expansion of the total macrophage population compared to the sham group, and the expansion continued during AAA progression (Figure 3E, Sham, 10.1% vs. Elastase7d, 45.2%, and Elastase14d, 43.1%). Interestingly, among the identified macrophage clusters, there was an increased proportion of Mo/MΦ_2, Mo/MΦ_3, Mo/MΦ_4 and Mo/MΦ_5 and a decreased proportion of Mo/MΦ_1 in elastase-treated IAA (Figure 5B and Supplementary material online, Figure S6B), suggesting a unique origin or function of Mo/MΦ_1.
Figure 5.
Comparison of monocyte and macrophage (Mo/MΦ) subpopulations in IAA from Sham and AAA. (A) UAMP plot of Mo/MΦ subpopulations from Sham and Elastase14d. Sham, n = 152 Mo/MΦs. Elastase14d, n = 602 Mo/MΦs. (B) The percentages of macrophage subpopulations from Sham and Elastase14d. For Sham, Mo/MΦ_1, n = 95 cells, Mo/MΦ_2, n = 18 cells, Mo/MΦ_3, n = 18 cells, Mo/MΦ_4, n = 20 cells, Mo/MΦ_5, n = 1 cells. For Elastase14d, Mo/MΦ_1, n = 151 cells, Mo/MΦ_2, n = 195 cells, Mo/MΦ_3, n = 111 cells, Mo/MΦ_4, n = 116 cells, Mo/MΦ_5, n = 29 cells. (C) Heat map of the top 10 [by average Log(fold change)] genes for each Mo/MΦ cell subpopulation. (D) Expression of selected marker genes for Mo/MΦ subpopulations as visualized by Violin plot, with colour denoting experimental conditions. (E) Dot plot of selected marker genes for Mo/MΦ subpopulations from Control and Elastase14d, where the dot size corresponds to the percentage of cells within the group expressing each gene, and dot colour intensity corresponds to the gene expression level.
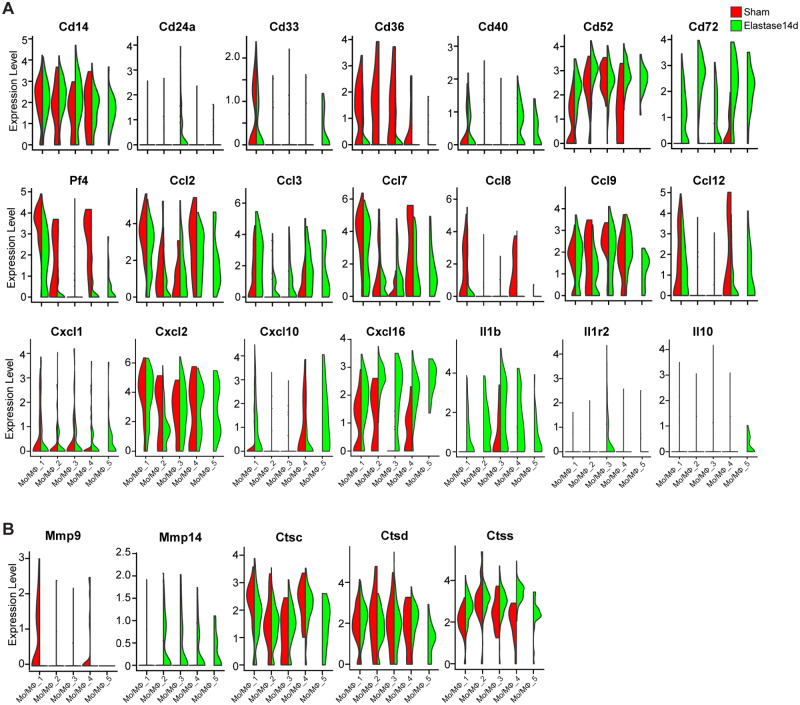
Based on the signature of Cx3cr1+ and Flt3+, Mo/MΦ_1 and Mo/MΦ_5 were singled out as aortic-resident macrophage (AR-MΦ), likely originated from Cx3cr1+ progenitors and Flt3 progenitors, respectively (Figure 5E). Since the tissue resident-macrophages are maintained mainly through local proliferation, with little input from the bone marrow, there is a resultant decrease of Mo/MΦ_1 fraction after elastase-induced increase of the bone marrow-derived Mo/MΦ fraction (Figure 5B). Notably, Mo/MΦ_5, the smallest subpopulation in the sham group (0.7% of total macrophages) was time-dependently increased after elastase-exposure (Elastase7d, 2.9% of total macrophages; Elastase14d, 4.8% of total macrophages) (Figure 5B and Supplementary material online, Figure S6B), which indicates a high proliferative activity of Mo/MΦ_5. Furthermore, Mo/MΦ_1 exhibited an expression signature characterized by Cx3cr1+Adgre1(F4/80)highItgam(Cd11b)lowLy6c2lowCcr2− (Figure 5E and Supplementary material online, Figure S6D). Both in basal and elastase-treated conditions, Mo/MΦ_1 showed high expression of the inflammatory macrophage markers Mrc1, Maf4 and Pf4, which are involved in phagocytosis, and C1 complement genes C1qa, C1qb and C1qc, as well as the pro-inflammatory cytokines Ccl2, Ccl4, Ccl7, Ccl8, Ccl9, Ccl12, Cxcl2, Cxcl16, all of which are involved in recruitment of new inflammatory cells (Figures 5C, E, 6A and Supplementary material online, Figure S6D and E). In addition, Mo/MΦ_1 also highly expressed proteolysis genes, including Mmp9 and cathepsins (Ctsc, Ctsd, and Ctss) (Figure 6B and Supplementary material online, Figure S6F), which play a crucial role in vascular remodelling and aortic aneurysm formation.31,32 In addition, we discerned another AR-MΦ subpopulation (Mo/MΦ_5) arising from Flt3+ precursor cells with the signature Flt3highItgam(Cd11b)lowLy6c2lowCcr2− (Figure 5E and Supplementary material online, Figure S6D). Additionally, Mo/MΦ_5 showed a proliferative signature with high expression of genes involved in cell proliferation, such as topoisomerase II α (Top2a) and Stmn1, and genes associated with microtubule activity, such as Tubb5 and Tuba1b33 (Figure 5C and D). Intriguingly, Mo/MΦ_5 exhibited co-expression of inflammatory genes, such as Ccl2, Ccl3, Ccl7, Ccl12, Cxcl2, and Il1b, and the anti-inflammatory gene, Il10 after elastase exposure (Figure 6A), which indicates that the aortic resident macrophage Mo/MΦ_5 may contribute to the resolution of inflammation during AAA progression.
Figure 6.
Violin plot of the differential expression of inflammation- and extracellular matrix (ECM) degradation-associated genes by Mo/MΦ subpopulations. (A) The inflammation-associated genes, including cluster of differentiation (Cd), C-C motif chemokine ligand (Ccl), C-X-C motif chemokine ligand (Cxcl), and interleukin (Il), were expressed by each Mo/MΦ subpopulations with colour denoting experimental conditions. (B) The ECM degradation-associated genes, including Mmp-9, Mmp-14, Cathepsin C (Ctsc), Ctsd, and Ctss, were expressed by each Mo/MΦ subpopulations with colour denoting experimental conditions.
The blood-derived monocytes and monocytes (Mo)-derived macrophages also contribute to the Mo/MΦ pool in the tissue, and they are distinguished by the MΦ markers including Adgre1 (F4/80), Itgam (Cd11b) and H2-Aa (MHC-II). Accordingly, the Mo/MΦ_2 was identified as blood-derived monocytes with the signature Ccr2+Ly6c2lowAdgre1(F4/80) lowItgam(Cd11b) lowH2-Aalow, while Mo/MΦ_3 and Mo/MΦ_4 were identified as Mo-derived macrophages with the signature Ccr2highLy6c2highAdgre1(F4/80) highItgam (Cd11b)+H2-Aahigh (Figure 5C, E and Supplementary material online, Figure S6D). Additionally, Mo/MΦ_2 showed high expression of the complement genes (C1qa, C1qb, and C1qc) and pro-inflammatory cytokines (Ccl2, Ccl3, Ccl9, Ccl12, Cxcl2, Cxcl10, and Il1b), which are involved in recruitment of new inflammatory cells (Figures 5E, 6A and Supplementary material online, Figure S6D and E). Recent evidences suggest that the traditional markers of M1 and M2 activation states are not mutually exclusive.34,35 Therefore, we examined the expression of M1 markers, such as Ccl2, Ccl3, Cxcl10, and Il1b, M2 markers, such as Arg1, Egr2, Il1r2, and Il10. Upon comparison, Mo/MΦ_3 exhibited high expression of Arg1, Egr2, and Il1r2, while Mo/MΦ_4 highly expressed inflammatory genes, such as Ccl2, Ccl3, Ccl7, Ccl8, Ccl12, Cxcl10, Cxcl16, and Il1b, and proteolysis genes, such as Mmp9, Ctsc, Ctsd, and Ctss (Figures 5C, E, 6A and Supplementary material online, Figure S6E). Accordingly, the two subpopulations were defined as reparative macrophages and inflammatory macrophages, respectively. We observed that Mo/MΦ_3 also contained cells that expressed pro-inflammatory genes, such as Ccl9, Cxcl2, Cxcl3, and Il1b and Mo/MΦ_4 contained cells that expressed the M2 markers (Figures 5C, 6A and Supplementary material online, Figure S6E). These data indicate that the expression of M1 and M2 markers is not mutually exclusive and that a panel of traditional markers are not sufficiently sensitive to characterize the programing states of macrophages in vivo, consistent with the new concepts regarding M1 and M2 states.34,35 In addition, the proportion of Mo/MΦ_3 was increased after elastase-induced vascular injury (Figure 5B and Supplementary material online, Figure 6SB), indicating that during AAA progression, the reparative macrophage-mediated resolution of inflammation and vascular repair still persist.
4. Discussion
The mammalian artery is composed of a multitude of multifunctional cell populations, with each of them distinctly involved in cardiovascular diseases, such as atherosclerosis and aortic aneurysm. Various studies have used the scRNA-seq technique to delineate the heterogeneity of vascular cells, including VSMCs,10 ECs,11 macrophages,8 and aortic adventitial cells12 in the healthy and atherosclerotic arteries. However, the cellular heterogeneity and aneurysm-relevant transcriptional signatures remain largely unknown. Here, we present the scRNA-seq data from >4600 individual cells from IAA after exposure to inactivated or active elastase. We identified 17 clusters and 9 distinct cell lineages within IAA, and also delineated the transcriptional profiles, functional states and fractional changes of VSMC and Mo/MΦ in the sham and aneurysmal IAA. Thus, our study identified the aneurysm-relevant transcriptional signatures in the major vascular cell types of IAA in the elastase model for the first time, with implications for understanding the functional significance of these heterogeneous cells in AAA.
Diverse AAA models have been developed to address the main pathophysiological determinants of aneurysm, including genetics, alterations in ECM, VSMC apoptosis, endothelial damage, and the contributions of innate and adaptive immune responses.36 Currently, no animal model faithfully recapitulates the chronic pathology of human AAA, but rather diverse animal models provide complementary insights on specific mechanistic aspects that need to be systematically addressed and contextually analysed. Therefore, systematic studies at the single-cell level in different animal models for AAA are necessary to aid in our understanding of the diversity of factors contributing to AAA. Our findings here provide strong evidence that the elastase-induced AAA model is better suited to study the role of vascular inflammation, ECM degradation and SMCs loss in the development of human AAA formation, and are in agreement with the recognized advantages of this model.17,19,36
The need for complementary studies in the field is further highlighted when comparing the results here to those in a previous study from Hadi et al.37 that addressed the cellular heterogeneity in the suprarenal abdominal aorta from the angiotensin II-induced AAA model in the ApoE−/− mouse using scRNA-seq. Consistently, both vascular resident cells, including SMCs, fibroblasts and ECs, and infiltrated immune cells, including macrophages, B cells, T cells, and DCs, were singled out in that report as well. Among the immune cells, macrophages are the largest population in the two AAA models. Compared with our study, the major difference is the proportion of immune cells and fibroblasts. In the Hadi’s study, the largest population is fibroblasts, while the immune cells account for no more than 10% of the total cells in the AngII-induced AAA model. However, in our study, macrophages were the largest population and immune cells accounted for more than 60% of the total single cells both at Days 7 and 14 in the elastase-induced AAA model. However, the neutrophil population identified in the angiotensin II-induced suprarenal AAA was not evident in the elastase-induced infrarenal AAA. Besides, the proportion of each cell population was different between the two AAA models. In our study, immune cells accounted for more than 60% of total cells isolated from elastase-treated IAAs, while in the AngII-induced AAA model, the proportion of immune cells is no more than 10% of total aortic cells with fibroblasts as the largest population in that model. These variations are likely the result of the intrinsic differences in the pathophysiological aspects of the two models discussed above and, to some extent, may be associated with minor technical aspects, including the enzymatic cocktails used for single-cell release and the approaches for data analysis and visualization.
In agreement with previous studies on VSMC heterogeneity within and between different vascular regions using lineage tracing and scRNA-seq approaches,10,23 we singled out four distinct VSMC subpopulations within the murine IAA and identified their distinct gene expression patterns suggesting corresponding functional properties. We identified two clusters of contractile VSMCs (SMC_1 and SMC_2) within IAA, characterized by the expression of the canonical VSMC markers such as Myh11, Tagln, Acta2, and Cnn1. However, there are still pronounced differences in the expression profiles between them for genes involved in cell proliferation, migration and inflammation. Unlike the other SMC subpopulations, SMC_1 demonstrated highest expression of contractile marker genes and lowest expression of genes involved in cell proliferation, migration and inflammation, even upon elastase exposure. This difference suggests the functional specialization of the maturely differentiated SMC_1 in maintaining the stable vascular structure. Based on the differential transcriptional profiles, we observed that SMC_2 highly expressed transcription factors which contribute to SMC dedifferentiation, inflammation (e.g. Klf2, Klf4, Gata6, Atf3, Jun and Fos) and provide evidence for the functional heterogeneity of the two contractile SMC subpopulations. These differentially expressed genes are involved in VSMC biology, including phenotypic switching,20,29 apoptosis,38 proliferation,39 migration,40,41 and inflammation, consistent with previous reports. We also describe a novel dedifferentiated VSMC subpopulation (SMC_3) with the lowest expression of contractile markers and an alternative metabolic profile. The lowest proportion subpopulation of VSMCs, SMC_4, showed increased expression of genes related to inflammation and ECM remodelling, including Ccl2 and Il1r1. Interestingly, SMC_4 is the only subpopulation that showed an increase in their relative proportion at the late stage of AAA development. These results indicate the likely phenotypic transition of SMC_4 to inflammatory SMCs and a potential underlying role in regulating vascular inflammation and remodelling, which could be targeted for therapy. Notably, previous studies demonstrated that abrogation of transforming growth factor-β signalling in SMCs results in thoracic aortic aneurysm and induces reprograming of normal SMCs into an inflammatory phenotype through activation of the NF-κB/interleukin (IL)-1β pathway.42,43 Thus, such heterogeneity of VSMCs in IAA is noteworthy as it indicates the existence of specific subsets of VSMCs with particular disease-relevant properties. Sca1 (mesenchymal-stem cell marker, encoded by Ly6a)-expressing cells in the vessel wall have been reported previously.44 Accordingly, we also demonstrated Sca1+ VSMCs in the normal IAA and found that Sca1 expression was up-regulated in three subpopulations (SMC_1, SMC_2, and SMC_3) upon exposure to elastase for 7 days and subsequently decreased at 14 days after elastase treatment. These data suggest that Sca1 expression during AAA development could mark an intermediate VSMC state, which might give rise to phenotypically distinct SMCs within the aneurysmal tissue. In line with previous studies,3,6elastase-treatment induced increased expression of pro-inflammatory cytokines and proteinase, including Cd68, Ccl2, Cxcl1, Il1r1, Adamts1, and Ctss. In addition, Bmp2, whose function in vascular calcification has been well characterized,45 was only highly expressed in SMC_3 and SMC_4, especially in the elastase-treated SMCs. So our scRNA-seq data performed on the sham and aneurysmal IAAs showed phenotypic diversity of VSMCs and provided an unprecedented depth in the characterization of the VSMC heterogeneity in normal and diseased blood vessels.
During AAA progression, one dramatic change in the adventitia is Mo/MΦ infiltration in the aneurysmal tissue.5 The Mo/MΦs within the vascular wall have diverse functions, including amplification of the local inflammatory response by secreting pro-inflammatory cytokines, chemokines and producing proteases and reactive oxygen.5,6,46 A number of lineage tracing experiments and scRNA-seq data have demonstrated the heterogeneity of Mo/MΦs within the atherosclerotic lesion and arteries.8,14,15 Based on the scRNA-seq data herein, we now characterized the transcriptional profiles of the Mo/MΦ lineages within the murine IAA and identified their original and functional heterogeneities. Like other organs, the adult arteries contain resident MΦs, which have embryonic and postnatal origins (e.g. Cx3cr1+ progenitors in the yolk sac, Flt3+ foetal liver precursor cells and post-natal Flt3+ bone marrow cells) before the onset of definitive haematopoiesis.47,48 Regardless of their origins, these tissue resident MΦs can maintain themselves through self-renewal, with little input from the bone marrow during homeostasis and injury.48,49 Using the scRNA-seq analysis, we found that the IAA contains two distinct aortic-resident macrophage (AR-Mac) subpopulations (Mo/MΦ_1 and Mo/MΦ_5), likely originating from Cx3cr1+ yolk sac progenitors and Flt3 foetal liver monocytes, respectively, consistent with the previous fate-mapping studies in mice.49 Although both AR-Macs exhibited high expression of pro-inflammatory cytokines, the observed unique transcriptional profiles for the two subpopulations further demonstrate their functional specialization. Upon comparison, Mo/MΦ_1, the more abundant AR-Mac subpopulation, likely has roles in antigen presentation, stimulation of other immune cells responses and phagocytosis, whereas Mo/MΦ_5 was identified as a proliferative AR-Mac subpopulation, which consistent with their known self-renewing properties. During AAA progression, Mo/MΦ_5 continue to proliferate and show co-expression of the inflammatory and anti-inflammatory genes, raising the possibility that cells in this subpopulation are programmed for the resolution of inflammation. Thus, our data provide the first transcriptional signatures of the proliferative AR-Mac subpopulation during both basal and aortic aneurysm conditions.
In the course of many pathological conditions, including atherosclerosis and aortic aneurysm, an early response to local or systemic inflammatory stimuli is the extravasation of leucocytes, including monocyte.18,50 After the monocytes enter the arterial walls, they differentiate further and subsequently acquire an activated macrophage phenotype.51 Beyond the AR-Macs, our scRNA-seq data also identified blood-derived monocytes (Mo/MΦ_2) and monocyte-derived inflammatory macrophages (Mo/MΦ_4). The inflammatory macrophages, Mo/MΦ_4, highly expressed pro-inflammatory factors, such as Ccl2, Cxcl2, and Il1b, as well as secreted multiple proteases, such as, Mmp9, Ctsc, Ctsd, and Ctss, which contribute to ECM degradation and tissue destruction.3,32 During injury repair and inflammation resolution, the blood-derived monocytes recruited to the blood vessels and accumulating at the inflammatory sites, can also give rise to the reparative macrophages.51 Accordingly, we identified a subpopulation of monocyte-derived reparative macrophage (Mo/MΦ_3), with specific and high expression of the genes Arg1 and Il-10. It has been generally accepted that the alternatively activated reparative macrophages are responsible for tissue repair. However, the function of reparative macrophages in regression or delay of AAA progression remains unclear. Notably, we determined the expression of M1 and M2 macrophage markers in all of the five monocyte and macrophage subpopulations. In fact, the five subpopulations exhibited dual expression of pro-inflammatory genes and anti-inflammatory genes, albeit at varying levels. Specifically, although Arg1 has been used to assign the M2 phenotype to macrophages, we show that the subpopulation that expressed Arg1 also exhibited high expression of pro-inflammatory genes. Thus, a panel of traditional markers for M1 or M2 macrophages are not sufficiently specific to functionally characterize the cells that express them.34,35 Together, these analyses suggest that the traditional M1-vs.-M2 dichotomy derived from in vitro studies is inadequate in elaborating the molecular basis for the dynamic programmes of activated macrophages in vivo.
Amongst the inflammatory cells infiltrated in aneurysmal tissues, macrophages are the main population with their role in the pathogenesis of AAA well-described in both mouse and human.5,37 Furthermore, the presence of T cells, B cells, DCs, and natural killer cells in aneurysmal tissues was identified at the single-cell level, in agreement with previous immunohistochemical studies of AAA.6,52 Previously, it was demonstrated that both CD4+ T cells and CD8+ T cells promote AAA formation.52,53 Consistently, the identification of CD4+ T cells and CD8+ T cells in our study indicates that antigen-dependent activation of naïve T cells and adaptive immune response should be involved in the elastase-induced AAA progression. By demonstrating the increase of these immune cells, our study supports that the elastase-induced AAA model is suited to study the role of vascular inflammation in the development of human AAA formation.
Previous studies indicated a pathogenic role for neutrophils in AAA.16,54 However, the neutrophil recruitment to the aortic wall was mainly to the perivascular adipose tissue and to a lower extent to periphery of the adventitia, as shown by Ly6G immunofluorescence staining.16 The perivascular adipose tissue was removed from the IAAs in this study; thus, the neutrophils are not represented. Of note, neutrophils infiltration to the media is more relevant to the intraluminal elastase-induced AAA model, where intraluminal thrombosis (ILT) is also present.54
In summary, our study revealed the hallmarks of infrarenal aortic cell heterogeneity in an unbiased manner. Using scRNA-seq, we identified the response signatures of VSMCs and Mo/MΦ during elastase-induced AAA progression. These findings may provide insight into the function and regulation of AAA onset and progression and pave the way for selective targeting of causative cell populations in vascular diseases.
Data availability
The scRNA-seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under the accession code GSE152583. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
G.Z., H.L., Z.C., and Y.Z. performed experiments. G.Z. and H.L. performed data analysis. G.Z. and J.Z. wrote the article. T.Z., L.C., Y.G. and M.T.G.-B. provided technical support and contributed to the discussion of the project and article. M.T.G.-B. did critical editing. Y.E.C. and J.Z. designed research and discussed results.
Supplementary Material
Acknowledgements
We thank Judith S. Opp and other staff members at the University of Michigan Advanced Genomics Core for their support on single-cell RNA sequencing.
Conflict of interest: None.
Funding
This work was supported by the National Institutes of Health (HL138139 to J.Z., HL068878 and HL134569 to Y.E.C.) and American Heart Association Grant (20P0ST35110064 to G.Z.).
Translational perspective
The pathophysiology of human AAA is not yet completely understood. In this study, we identified 17 clusters representing 9 cell lineages in the elastase-induced murine AAA samples by using the single-cell RNA sequencing technology. It revealed the AAA disease-relevant transcriptional signatures in these cells during AAA development. This study provides strong evidence that the elastase-induced AAA is a better model to study the role of vascular inflammation and smooth muscle cell loss in human AAA progression. Therefore, our findings will be fundamental at the cellular level to understand the aetiology of AAA and explore the potential targets for early intervention.
References
- 1. Davis FM, Daugherty A, Lu HS.. Updates of recent aortic aneurysm research. Arterioscler Thromb Vasc Biol 2019;39:e83–e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nordon IM, Hinchliffe RJ, Loftus IM, Thompson MM.. Pathophysiology and epidemiology of abdominal aortic aneurysms. Nat Rev Cardiol 2011;8:92–102. [DOI] [PubMed] [Google Scholar]
- 3. Rabkin SW. The role matrix metalloproteinases in the production of aortic aneurysm. Prog Mol Biol Transl Sci 2017;147:239–265. [DOI] [PubMed] [Google Scholar]
- 4. Zhao G, Fu Y, Cai Z, Yu F, Gong Z, Dai R, Hu Y, Zeng L, Xu Q, Kong W.. Unspliced XBP1 confers VSMC homeostasis and prevents aortic aneurysm formation via FoxO4 interaction. Circ Res 2017;121:1331–1345. [DOI] [PubMed] [Google Scholar]
- 5. Raffort J, Lareyre F, Clement M, Hassen-Khodja R, Chinetti G, Mallat Z.. Monocytes and macrophages in abdominal aortic aneurysm. Nat Rev Cardiol 2017;14:457–471. [DOI] [PubMed] [Google Scholar]
- 6. Rizas KD, Ippagunta N, Tilson MD 3rd. Immune cells and molecular mediators in the pathogenesis of the abdominal aortic aneurysm. Cardiol Rev 2009;17:201–210. [DOI] [PubMed] [Google Scholar]
- 7. Biddy BA, Kong W, Kamimoto K, Guo C, Waye SE, Sun T, Morris SA.. Single-cell mapping of lineage and identity in direct reprogramming. Nature 2018;564:219–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cochain C, Vafadarnejad E, Arampatzi P, Pelisek J, Winkels H, Ley K, Wolf D, Saliba AE, Zernecke A.. Single-cell RNA-Seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res 2018;122:1661–1674. [DOI] [PubMed] [Google Scholar]
- 9. Feil S, Fehrenbacher B, Lukowski R, Essmann F, Schulze-Osthoff K, Schaller M, Feil R.. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ Res 2014;115:662–667. [DOI] [PubMed] [Google Scholar]
- 10. Dobnikar L, Taylor AL, Chappell J, Oldach P, Harman JL, Oerton E, Dzierzak E, Bennett MR, Spivakov M, Jørgensen HF.. Disease-relevant transcriptional signatures identified in individual smooth muscle cells from healthy mouse vessels. Nat Commun 2018;9:4567–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kalluri AS, Vellarikkal SK, Edelman ER, Nguyen L, Subramanian A, Ellinor PT, Regev A, Kathiresan S, Gupta RM.. Single-cell analysis of the normal mouse aorta reveals functionally distinct endothelial cell populations. Circulation 2019;140:147–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gu W, Ni Z, Tan YQ, Deng J, Zhang SJ, Lv ZC, Wang XJ, Chen T, Zhang Z, Hu Y, Jing ZC, Xu Q.. Adventitial cell atlas of wt (wild type) and ApoE (Apolipoprotein E)-deficient mice defined by single-cell RNA sequencing. Arterioscler Thromb Vasc Biol 2019;39:1055–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gu W, Nowak WN, Xie Y, Le Bras A, Hu Y, Deng J, Issa Bhaloo S, Lu Y, Yuan H, Fidanis E, Saxena A, Kanno T, Mason AJ, Dulak J, Cai J, Xu Q.. Single-cell RNA-sequencing and metabolomics analyses reveal the contribution of perivascular adipose tissue stem cells to vascular remodeling. Arterioscler Thromb Vasc Biol 2019;39:2049–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Winkels H, Ehinger E, Vassallo M, Buscher K, Dinh HQ, Kobiyama K, Hamers AAJ, Cochain C, Vafadarnejad E, Saliba AE, Zernecke A, Pramod AB, Ghosh AK, Anto Michel N, Hoppe N, Hilgendorf I, Zirlik A, Hedrick CC, Ley K, Wolf D.. Atlas of the immune cell repertoire in mouse atherosclerosis defined by single-cell RNA-sequencing and mass cytometry. Circ Res 2018;122:1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Papalexi E, Satija R.. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 2018;18:35–45. [DOI] [PubMed] [Google Scholar]
- 16. He L, Fu Y, Deng J, Shen Y, Wang Y, Yu F, Xie N, Chen Z, Hong T, Peng X, Li Q, Zhou J, Han J, Wang Y, Xi J, Kong W.. Deficiency of FAM3D (Family With Sequence Similarity 3, Member D), a novel chemokine, attenuates neutrophil recruitment and ameliorates abdominal aortic aneurysm development. Arterioscler Thromb Vasc Biol 2018;38:1616–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhamidipati CM, Mehta GS, Lu G, Moehle CW, Barbery C, DiMusto PD, Laser A, Kron IL, Upchurch GR Jr, Ailawadi G.. Development of a novel murine model of aortic aneurysms using peri-adventitial elastase. Surgery 2012;152:238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Butcher MJ, Herre M, Ley K, Galkina E.. Flow cytometry analysis of immune cells within murine aortas. J Vis Exp 2011;53:2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu G, Su G, Davis JP, Schaheen B, Downs E, Roy RJ, Ailawadi G, Upchurch GR Jr, A novel chronic advanced stage abdominal aortic aneurysm murine model. J Vasc Surg 2017;66:232–242.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ailawadi G, Moehle CW, Pei H, Walton SP, Yang Z, Kron IL, Lau CL, Owens GK.. Smooth muscle phenotypic modulation is an early event in aortic aneurysms. J Thorac Cardiovasc Surg 2009;138:1392–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rowe VL, Stevens SL, Reddick TT, Freeman MB, Donnell R, Carroll RC, Goldman MH.. Vascular smooth muscle cell apoptosis in aneurysmal, occlusive, and normal human aortas. J Vasc Surg 2000;31:567–576. [PubMed] [Google Scholar]
- 22. Zhang J, Schmidt J, Ryschich E, Schumacher H, Allenberg JR.. Increased apoptosis and decreased density of medial smooth muscle cells in human abdominal aortic aneurysms. Chin Med J (Engl) 2003;116:1549–1552. [PubMed] [Google Scholar]
- 23. Liu M, Gomez D.. Smooth muscle cell phenotypic diversity. Arterioscler Thromb Vasc Biol 2019;39:1715–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Petsophonsakul P, Furmanik M, Forsythe R, Dweck M, Schurink GW, Natour E, Reutelingsperger C, Jacobs M, Mees B, Schurgers L.. Role of vascular smooth muscle cell phenotypic switching and calcification in aortic aneurysm formation. Arterioscler Thromb Vasc Biol 2019;39:1351–1368. [DOI] [PubMed] [Google Scholar]
- 25. Shen J, Zhou S, Shi L, Liu X, Lin H, Yu H, Liang X, Tang J, Yu T, Cai X.. DUSP1 inhibits cell proliferation, metastasis and invasion and angiogenesis in gallbladder cancer. Oncotarget 2017;8:12133–12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park G, Horie T, Kanayama T, Fukasawa K, Iezaki T, Onishi Y, Ozaki K, Nakamura Y, Yoneda Y, Takarada T, Hinoi E.. The transcriptional modulator Ifrd1 controls PGC-1alpha expression under short-term adrenergic stimulation in brown adipocytes. FEBS J 2017;284:784–795. [DOI] [PubMed] [Google Scholar]
- 27. Sutliff RL, Hoying JB, Kadambi VJ, Kranias EG, Paul RJ.. Phospholamban is present in endothelial cells and modulates endothelium-dependent relaxation. Evidence from phospholamban gene-ablated mice. Circ Res 1999;84:360–364. [DOI] [PubMed] [Google Scholar]
- 28. Heiss EH, Schachner D, Donati M, Grojer CS, Dirsch VM.. Increased aerobic glycolysis is important for the motility of activated VSMC and inhibited by indirubin-3'-monoxime. Vascul Pharmacol 2016;83:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lepore JJ, Cappola TP, Mericko PA, Morrisey EE, Parmacek MS.. GATA-6 regulates genes promoting synthetic functions in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol 2005;25:309–314. [DOI] [PubMed] [Google Scholar]
- 30. Domenga V, Fardoux P, Lacombe P, Monet M, Maciazek J, Krebs LT, Klonjkowski B, Berrou E, Mericskay M, Li Z, Tournier-Lasserve E, Gridley T, Joutel A.. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev 2004;18:2730–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun J, Sukhova GK, Zhang J, Chen H, Sjoberg S, Libby P, Xia M, Xiong N, Gelb BD, Shi GP.. Cathepsin K deficiency reduces elastase perfusion-induced abdominal aortic aneurysms in mice. Arterioscler Thromb Vasc Biol 2012;32:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qin Y, Cao X, Yang Y, Shi GP.. Cysteine protease cathepsins and matrix metalloproteinases in the development of abdominal aortic aneurysms. Future Cardiol 2013;9:89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jung M, Shin MK, Jung YK, Yoo HS.. Modulation of macrophage activities in proliferation, lysosome, and phagosome by the nonspecific immunostimulator, mica. PLoS One 2015;10:e0117838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mould KJ, Jackson ND, Henson PM, Seibold M, Janssen WJ.. Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight 2019;4:e126556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nahrendorf M, Swirski FK.. Abandoning M1/M2 for a network model of macrophage function. Circ Res 2016;119:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lysgaard Poulsen J, Stubbe J, Lindholt JS.. Animal models used to explore abdominal aortic aneurysms: a systematic review. Eur J Vasc Endovasc Surg 2016;52:487–499. [DOI] [PubMed] [Google Scholar]
- 37. Hadi T, Boytard L, Silvestro M, Alebrahim D, Jacob S, Feinstein J, Barone K, Spiro W, Hutchison S, Simon R, Rateri D, Pinet F, Fenyo D, Adelman M, Moore KJ, Eltzschig HK, Daugherty A, Ramkhelawon B.. Macrophage-derived netrin-1 promotes abdominal aortic aneurysm formation by activating MMP3 in vascular smooth muscle cells. Nat Commun 2018;9:5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Salmon M, Spinosa M, Zehner ZE, Upchurch GR, Ailawadi G.. Klf4, Klf2, and Zfp148 activate autophagy-related genes in smooth muscle cells during aortic aneurysm formation. Physiol Rep 2019;7:e14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abe M, Hasegawa K, Wada H, Morimoto T, Yanazume T, Kawamura T, Hirai M, Furukawa Y, Kita T.. GATA-6 is involved in PPARgamma-mediated activation of differentiated phenotype in human vascular smooth muscle cells. Arterioscler Thromb Vasc Biol 2003;23:404–410. [DOI] [PubMed] [Google Scholar]
- 40. Wu J, Bohanan CS, Neumann JC, Lingrel JB.. KLF2 transcription factor modulates blood vessel maturation through smooth muscle cell migration. J Biol Chem 2008;283:3942–3950. [DOI] [PubMed] [Google Scholar]
- 41. Lv D, Meng D, Zou FF, Fan L, Zhang P, Yu Y, Fang J.. Activating transcription factor 3 regulates survivability and migration of vascular smooth muscle cells. IUBMB Life 2011;63:62–69. [DOI] [PubMed] [Google Scholar]
- 42. Chen PY, Qin L, Li G, Malagon-Lopez J, Wang Z, Bergaya S, Gujja S, Caulk AW, Murtada SI, Zhang X, Zhuang ZW, Rao DA, Wang G, Tobiasova Z, Jiang B, Montgomery RR, Sun L, Sun H, Fisher EA, Gulcher JR, Fernandez-Hernando C, Humphrey JD, Tellides G, Chittenden TW, Simons M.. Smooth muscle cell reprogramming in aortic aneurysms. Cell Stem Cell 2020;26:542–557.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Da Ros F, Carnevale R, Cifelli G, Bizzotto D, Casaburo M, Perrotta M, Carnevale L, Vinciguerra I, Fardella S, Iacobucci R, Bressan GM, Braghetta P, Lembo G, Carnevale D.. Targeting interleukin-1beta protects from aortic aneurysms induced by disrupted transforming growth factor beta signaling. Immunity 2017;47:959–973.e9. [DOI] [PubMed] [Google Scholar]
- 44. Psaltis PJ, Simari RD.. Vascular wall progenitor cells in health and disease. Circ Res 2015;116:1392–1412. [DOI] [PubMed] [Google Scholar]
- 45. Lagna G, Ku MM, Nguyen PH, Neuman NA, Davis BN, Hata A.. Control of phenotypic plasticity of smooth muscle cells by bone morphogenetic protein signaling through the myocardin-related transcription factors. J Biol Chem 2007;282:37244–37255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shimizu K, Mitchell RN, Libby P.. Inflammation and cellular immune responses in abdominal aortic aneurysms. Arterioscler Thromb Vasc Biol 2006;26:987–994. [DOI] [PubMed] [Google Scholar]
- 47. Honold L, Nahrendorf M.. Resident and monocyte-derived macrophages in cardiovascular disease. Circ Res 2018;122:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klapproth K, Lasitschka F, Rodewald HR.. Multilayered ancestry of arterial macrophages. Nat Immunol 2016;17:117–118. [DOI] [PubMed] [Google Scholar]
- 49. Ensan S, Li A, Besla R, Degousee N, Cosme J, Roufaiel M, Shikatani EA, El-Maklizi M, Williams JW, Robins L, Li C, Lewis B, Yun TJ, Lee JS, Wieghofer P, Khattar R, Farrokhi K, Byrne J, Ouzounian M, Zavitz CC, Levy GA, Bauer CM, Libby P, Husain M, Swirski FK, Cheong C, Prinz M, Hilgendorf I, Randolph GJ, Epelman S, Gramolini AO, Cybulsky MI, Rubin BB, Robbins CS.. Self-renewing resident arterial macrophages arise from embryonic CX3CR1(+) precursors and circulating monocytes immediately after birth. Nat Immunol 2016;17:159–168. [DOI] [PubMed] [Google Scholar]
- 50. Libby P, Hansson GK.. Inflammation and immunity in diseases of the arterial tree: players and layers. Circ Res 2015;116:307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bolego C, Cignarella A, Staels B, Chinetti-Gbaguidi G.. Macrophage function and polarization in cardiovascular disease: a role of estrogen signaling? Arterioscler Thromb Vasc Biol 2013;33:1127–1134. [DOI] [PubMed] [Google Scholar]
- 52. Galle C, Schandene L, Stordeur P, Peignois Y, Ferreira J, Wautrecht JC, Dereume JP, Goldman M.. Predominance of type 1 CD4+ T cells in human abdominal aortic aneurysm. Clin Exp Immunol 2005;142:519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sagan A, Mikolajczyk TP, Mrowiecki W, MacRitchie N, Daly K, Meldrum A, Migliarino S, Delles C, Urbanski K, Filip G, Kapelak B, Maffia P, Touyz R, Guzik TJ.. T cells are dominant population in human abdominal aortic aneurysms and their infiltration in the perivascular tissue correlates with disease severity. Front Immunol 2019;10:1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meher AK, Spinosa M, Davis JP, Pope N, Laubach VE, Su G, Serbulea V, Leitinger N, Ailawadi G, Upchurch GR Jr.. Novel role of IL (interleukin)-1β in neutrophil extracellular trap formation and abdominal aortic aneurysms. Arterioscler Thromb Vasc Biol 2018;38:843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under the accession code GSE152583. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.