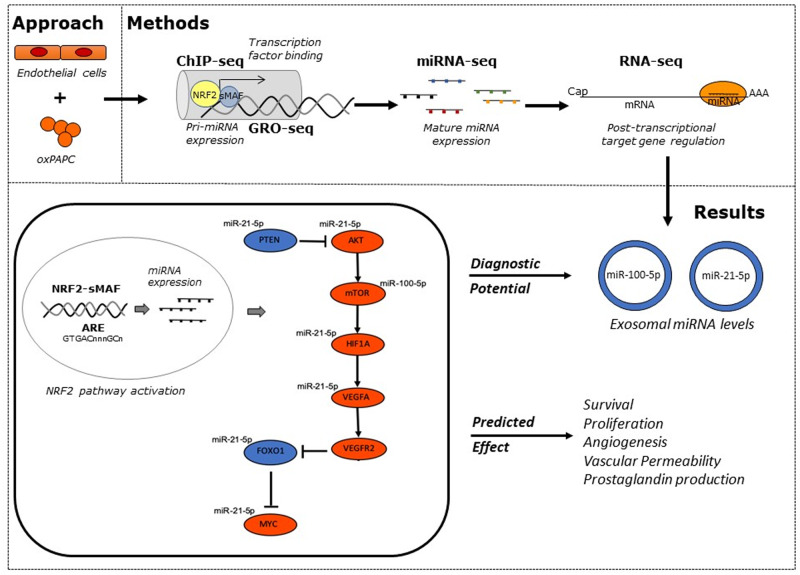
Abstract
Aims
Oxidized phospholipids and microRNAs (miRNAs) are increasingly recognized to play a role in endothelial dysfunction driving atherosclerosis. NRF2 transcription factor is one of the key mediators of the effects of oxidized phospholipids, but the gene regulatory mechanisms underlying the process remain obscure. Here, we investigated the genome-wide effects of oxidized phospholipids on transcriptional gene regulation in human umbilical vein endothelial cells and aortic endothelial cells with a special focus on miRNAs.
Methods and results
We integrated data from HiC, ChIP-seq, ATAC-seq, GRO-seq, miRNA-seq, and RNA-seq to provide deeper understanding of the transcriptional mechanisms driven by NRF2 in response to oxidized phospholipids. We demonstrate that presence of NRF2 motif and its binding is more prominent in the vicinity of up-regulated transcripts and transcriptional initiation represents the most likely mechanism of action. We further identified NRF2 as a novel regulator of over 100 endothelial pri-miRNAs. Among these, we characterize two hub miRNAs miR-21-5p and miR-100-5p and demonstrate their opposing roles on mTOR, VEGFA, HIF1A, and MYC expressions. Finally, we provide evidence that the levels of miR-21-5p and miR-100-5p in exosomes are increased upon senescence and exhibit a trend to correlate with the severity of coronary artery disease.
Conclusion
Altogether, our analysis provides an integrative view into the regulation of transcription and miRNA function that could mediate the proatherogenic effects of oxidized phospholipids in endothelial cells.
Keywords: miR-21-5p • miR-100-5p • NRF2 • Atherosclerosis
Graphical Abstract
1. Introduction
Atherosclerosis is an inflammatory disease, which causes lipid accumulation and plaque formation in the endothelium and intimal layers along the inner walls of the arteries, narrowing the lumen and reducing blood flow to the target tissue. It is an underlying condition to many cardiovascular disorders, such as coronary artery disease (CAD), stroke, and peripheral vascular disease, and thus, responsible for an immense burden of morbidity and mortality worldwide. The pathogenesis of atherosclerosis is a bundle of several different cell types and environmental risk factors, but the most critical cells are the endothelial cells that form the inner lining of the vascular wall, as the activation of these cells from their quiescent state is a prerequisite for the onset of atherosclerosis.1 Proinflammatory microenvironment, turbulent blood flow, and high amounts of oxidized lipid products, such as oxidized 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (oxPAPC),2,3 result in endothelial dysfunction increasing vascular permeability and promoting lipid accumulation into the vessel wall. Furthermore, accumulation and subsequent oxidization of lipids stimulate the expression of chemokines and adhesion molecules, which attract leukocytes on the site and facilitate their migration under the endothelial layer, thereby initiating the formation of an atherosclerotic plaque.1
MicroRNAs (miRNAs) contribute to both prevention and progression of atherosclerosis by regulating various aspects of atherogenesis, from early atherosclerotic lesion formation to plaque erosion and rupture.4,5 They elicit their function at the post-transcriptional level by fine-tuning the expression of protein-coding genes through suppression of protein synthesis or initiation of mRNA degradation. miRNAs can become selectively enriched in secretory nanovesicles, exosomes, and mediate communication with remote cells affecting thus disease pathogenesis.6 However, these circulating miRNAs are also considered as novel biomarkers for disease manifestation and progression.7 miRNAs act co-operatively and can target full pathways and gene networks instead of just one gene, which can be beneficial when treating multifaceted diseases like atherosclerosis.8 Although the recent years have added to knowledge on miRNA function substantially, many aspects remain understudied, including regulation of miRNA expression. In order to ensure the safety of the therapeutic miRNA applications, some of which have already entered clinical trials,9,10 detailed understanding of miRNA biology is crucial.
Nuclear factor E2-related factor 2 (NRF2) is a known mediator of cellular response to oxidized phospholipids that represent a hallmark of lipid load and inflammatory microenvironment of the atherosclerotic plaques. Using miRNA array upon 4 h oxPAPC treatment, we have previously identified 19 significantly (P < 0.01) regulated mature miRNAs of which half were confirmed to be oxPAPC-responsive also on transcriptional level based on GRO-seq data, and five of the loci were predicted to be NRF2-regulated by previously published NRF2-binding model.11 From those, we chose to characterize miR-93 and demonstrated that NRF2-regulated miR-93 controls glycolysis and proliferation of endothelial cells.12 Here, we sought to expand on this previous research to obtain a more complete picture of the NRF2-regulated miRNome by integration of transcriptional profiles from GRO-Seq (pri-miRNA transcription), small RNA-Seq (mature miRNA levels), and RNA-Seq (target mRNAs), epigenomic information from H3K27ac ChIP-Seq and ATAC-Seq [regulatory activity in predicted NRF2 antioxidant response element (ARE)-binding sites] and direct or indirect (detected by Hi-C) NRF2 binding at the pri-miRNA promoter. Using our approach, we were able to identify NRF2 as an important regulator of over 100 endothelial miRNAs and further characterized the role of miR-21-5p and miR-100-5p on target mRNA expression and evaluated their usability as diagnostic biomarkers in atherosclerosis.
2. Methods
2.1 Cell culture
Human umbilical vein endothelial cells (HUVECs) were extracted with collagenase (0.3 mg/mL) digestion from umbilical cords obtained from the maternity ward of the Kuopio University Hospital with the approval of the institution and the Research Ethics Committee of the Northern Savo Hospital District, Kuopio, Finland. Prior written consent was obtained from the participants and the experiments were performed according to the relevant regulations and Helsinki declaration. HUVECs of 11 different donors were used in the study, and the essential results were repeated on at least three donor batches. Unless indicated otherwise, passages 4–6 were used for experiments. Human aortic endothelial cells (HAECs), human aortic smooth muscle cells (HASMC), and peripheral blood CD14+ monocytes were purchased from Lonza (CC-2535, CC-2571, and 2 W-400A, respectively). HUVECs and HAECs were cultivated in Endothelial Cell Basal Medium (Lonza, Basel, SUI) with recommended supplements (EGM SingleQuot Kit Supplements & Growth Factors, Lonza, Basel, SUI), HASMC were grown in Medium 231 (Cascade Biologics®, Waltham, MA, USA) supplemented with smooth muscle growth supplement, and CD14+ monocytes were cultivated in Medium RPMI 1640, supplemented with 10% FBS, 100 μg/mL streptomycin, 100 U/mL penicillin, and 2 mM glutamine, 1% Na-pyruvate, 1% NEAA, and further supplemented with rHu M-CSF (50 ng/mL; Thermo Fisher Scientific, Waltham, MA, USA) to initiate the differentiation into macrophages.
2.2 Patient samples and sample preparation
Pericardial fluid samples (2–27 mL) were collected during open-heart surgery and processed immediately after sample collection as described in Ref.13 A written informed consent was obtained from each participant and the Research Ethics Committee of the Hospital District of Northern Savo, Kuopio, Finland approved the study protocol in accordance with Helsinki declaration. Briefly, pericardial fluid samples were collected from 41 subjects with CAD, mitral valve insufficiency, aortic stenosis, or aortic valve insufficiency. See Supplementary material online for the summary of the clinical characteristics of the patients. New York Heart Association functional classification (NYHA class) was used to estimate the extent of heart failure (HF): patients with cardiac disease but no HF symptoms and no limitations in ordinary physical activity were assigned to group I (n = 2), patients with mild symptoms in group II (n = 18), patients with marked limitation in activity due to symptoms in group III (n = 10), and patients with severe limitations and symptoms even while resting, in group IV (n = 11). Samples were processed directly after collection using three-step centrifugation. Samples were first centrifuged at 300 g for 10 min at RT to remove cells. Then, the supernatants were collected and centrifuged again at 16 500 g for 20 min at 4°C to remove cell debris. The collected liquid fractions were centrifuged a final time at 20 000 g for 15 min at 4°C to remove other microparticles, leaving exosomes and protein-bound miRNAs to the supernatants. The supernatants were transferred to RNase-free tubes, snap-frozen with liquid nitrogen, and stored at −80°C.
2.3 Stimulus
1-Palmitoyl-2-archidonoyl-sn-glycero-3-phophocholine (PAPC, 10 mg/mL) was purchased from Avanti Polar Lipids Inc (Alabaster, AL, USA) oxidized to oxPAPC by exposure to air for 40 h, dissolved in chloroform and stored at −70°C. Upon use, the chloroform was evaporated with nitrogen (g) and the lipids were re-suspended in growth medium. For experiments, concentration of 30 μg/mL in medium with 1% FBS was used.
OA-NO2 was prepared as previously described.14 The synthetic nitration product used was an equimolar mixture of 9- and 10-nitro-octadec-9-enoic acid. For experiments, cells were treated with 3 µM OA-NO2 or vehicle (0.06% meOH) for 2 or 12 h in basal medium.
2.4 Global run-on sequencing
HUVECs, HAECs, HASMCs, and CD14+ macrophages were grown to 80% confluency on 15 cm plates and treated with oxPAPC (30 μg/mL) for 6 h in 1% FBS EGM or with fresh 1% FBS EGM (control). To collect nuclei for GRO-Seq, cells were washed once with PBS, trypsinized for 30 s, neutralized with ice-cold swelling buffer (10 mM Tris–HCl, 2 mM MgCl2, 3 mM CaCl2, and 2 U/mL SUPERase Inhibitor, Thermo Fisher Scientific, Waltham, MA, USA), and collected by scraping. Nuclei were prepared as described in Ref.15 and approximately five million nuclei were recovered and subjected to run-on reaction. GRO-seq libraries were prepared as previously described with minor modifications.16 Briefly, the run-on products were base hydrolysed (RNA fragmentation reagent, Thermo Fisher Scientific, Waltham, MA, USA), end-repaired using polynucleotide kinase and immunoprecipitated with anti-BrdU agarose beads (Santa Cruz, CA, USA) in binding buffer (0.5XSSPE, 1 mM EDTA, 0.05% Tween-20) for 1 h at RT. Beads were washed twice with ice-cold binding buffer and low salt buffer (0.2 × SSPE, 1 mM EDTA, 0.05% Tween-20), once with high salt buffer (0.5% SSPE, 1 mM EDTA, 0.05% Tween-20, 150 mM NaCl) and twice with TET buffer (TE pH 7.4, 0.05% Tween-20). The captured RNA was eluted with 3 × 100 μl elution buffer (20 mM DTT, 300 mM NaCl, 5 mM Tris pH 7.5, 1 mM EDTA, and 0.1% SDS), purified using Trizol-LS protocol (Thermo Fisher Scientific, Waltham, MA, USA), ethanol precipitated, and subjected to a second round of immunoprecipitation. The immunopurified RNA was eluted as above and ethanol precipitated. The following day, a poly-A tailing reaction (PolyA polymerase, New England Biolabs, Ipswich, MA, USA), cDNA synthesis (Superscript III, Thermo Fisher Scientific, Waltham, MA, USA), Exonuclease I (New England Biolabs, Ipswich, MA, USA) treatment, purification using ChIP DNA Clean & Concentrator Kit (Zymo Research Corporation, Irvine, CA, USA), RNaseH treatment, and circularization (CircLigase, Illumina, San Diego, CA, USA), were performed. The libraries were amplified for 13 cycles and library band was excised (∼200–350 bp) from 10% NOVEX TBE gel (Thermo Fisher Scientific, Waltham, MA, USA). The libraries were quantified (Qubit dsDNA HS Assay Kit on a Qubit fluorometer, Thermo Fisher Scientific, Waltham, MA, USA) and pooled for 50-bp single-end sequencing with Illumina Hi-Seq2000 (GenCore, EMBL Heidelberg, Germany).
2.5 RNA sequencing and miRNA sequencing
To reflect the lag between the nascent transcription and the transcription of mature RNAs, the collection of RNA samples started 1 h after the GRO-Seq collection, at 7 h. For the RNA-seq, cells were treated with cycloheximide at a concentration of 0.1 mg/mL for 10 min before being washed with PBS and scraped into lysis buffer [1× Mammalian Polysome Buffer (Epicentre, Madison, WI, USA), 1% Triton X-100, 1 mM DTT, 250 U/mL SUPERase Inhibitor, 7.1 U/mL Turbo DNase (Thermo Fisher Scientific, Waltham, MA, USA), and 0.1 mg/mL cycloheximide] on ice. A 22- to 25-gauge needle was used to ensure the complete lysis of the cells. The cleared lysate was treated with 10% SDS, snap-frozen in liquid nitrogen, and stored at −80°C. Larger mRNAs (>200nt) were extracted using Zymo RNA Clean and Conc kit (Zymo Research, Irvine, CA, USA) and rRNAs were excluded using the Ribo-Zero Gold rRNA Removal Kit (Illumina, San Diego, CA, USA). Subsequently, RNA was fragmentated (RNA fragmentation reagent, Thermo Fisher, Waltham, MA, USA), dephosphorylated, and prepared to library as GRO-Seq but omitting the immunoprecipitation steps. The libraries were amplified by 11–16 cycles of PCR, size selected (190–350 bp), quantified (Qubit dsDNA HS Assay Kit on a Qubit fluorometer, Thermo Fisher, Waltham, MA, USA), and sequenced using Illumina Hi-Seq2000 (single end 50 bases) at EMBL GeneCore (Heidelberg, Germany). Smaller RNA-fraction (17–200 bases) was obtained at the same time as the larger mRNAs by adding ethanol to the flow-through from Zymo RNA Clean and Conc kit and passing the RNA through a new column. Libraries were prepared following TruSeq small RNAseq protocol at Finnish Microarray and Sequencing Centre (FMSC) in Turku Centre for Biotechnology (Finland) or Exiqon (Vedbæk, Denmark) using small RNA protocol.
2.6 Transductions
For transduction, HUVECs were seeded onto 6-well plates at the density of 180 000 cells/well and allowed to adhere for 24 h. The cells were transduced in serum-free conditions with AdCMV,17 AdNRF2,17 or AdKEAP1.12 The multiplicity of infection (MOI) was 100 in all experiments. After an hour, cell culture supplements were added, and after additional 16 h the transduced cells were washed with PBS and fresh medium with full supplements was added. Experiments were performed 48 h after transductions.
2.7 miRNA overexpression and silencing
HUVECs were seeded onto 6-well plates and transfected at 70% confluency with MISSION miRNA Mimics Negative Control No. 1 (HMC0002, Sigma–Aldrich, St. Louis, MO, USA), hsa-miR-21-5p mimic (HMI0371, Sigma–Aldrich, St. Louis, MO, USA), MISSION Synthetic microRNA Inhibitors Negative control I (NCSTUD001, Sigma–Aldrich, St. Louis, MO, USA), and hsa-miR-100-5p inhibitor (HSTUD0023, Sigma–Aldrich, St. Louis, MO, USA) at a final concentration of 25 nM for mimic/mimic control and 1 nm for inhibitor/inhibitor control using Oligofectamine (Invitrogen, Carlsbad, CA, USA). Supplements were added to transfected cells 4 h after transfections. On the next day, cells were washed with PBS and fresh EBM medium with full supplements was added. After 48 h, cells were treated with oxPAPC for 8 h and collected for qRT–PCR analysis.
2.8 RNA extraction, cDNA synthesis, and qRT–PCR
Exosomes were extracted using miRCURY Exosome Isolation Kit (Exiqon, Vedbæk, Denmark). Exosomal and cellular RNA were isolated using miRCURY RNA isolation kit for cells and plants (Exiqon, Vedbæk, Denmark). Total fluid RNA and non-exosomal RNA, which was extracted from the medium after exosome isolation, were extracted using miRCURY RNA Isolation Kit for Biofluids (Exiqon, Vedbæk, Denmark). After transfection, HUVEC miRNAs and mRNAs were purified and separated using RNEasy Mini Kit (QIAGEN, Hilden, Germany). Cells were disrupted using 350 μL of buffer RLT followed by the same volume of 70% ethanol. The samples were placed into an RNeasy Mini spin column and centrifuged for 15 s at 8000 g. The flow-through was kept to extract the miRNA while the column purification was continued for the larger mRNA (>200nt). To purify the miRNA fraction, the flow-through was diluted by adding 0.65 volume of 100% ethanol and loaded into a new RNEasy Mini Kit (QIAGEN, Hilden, Germany) column.
miRNAs were reverse transcribed using miRCURY LNA Universal RT miRNA PCR, polyadenylation and cDNA synthesis kit (Exiqon, Vedbæk, Denmark), and mRNA with Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, SUI) or using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s protocol for first-strand cDNA Synthesis, followed by RNAseH treatment at 37°C for 20 min (Ribonuclease H, Ambion®, Thermo Fisher Scientific, Waltham, MA, USA).
cDNA templates were assayed in 10/17 µL PCR reactions with 10 pmol/µL of each primer and 2 µL of template cDNA with either LightCycler 96 System or LightCycler 480 Real-Time PCR System (Roche, Basel, SUI) according to the protocol of miRCURY LNA Universal RT miRNA PCR for miRNA samples and the protocol of Fast Start Universal Probe Master (Rox) (Roche, Basel, SUI) for mRNA samples. miRNA levels were determined using hsa-miR-21-5p (No. 204230, Exiqon, Vedbæk, Denmark), and hsa-miR-100-5p (No. 205689, Exiqon, Vedbæk, Denmark) LNA™ PCR primer sets.
mRNA levels were determined using Universal Probe Library System (Roche, Basel, SUI), Taqman Assays (Applied Biosystems, Foster City, CA, USA), or SYBR green [FastStart Universal SYBR Green Master (Roche, Basel, SUI)]. Specific primer information is listed in Supplementary material online. For analysis, Roche LC Software was used for Cp determination (by the second-derivative method) and for melting curve analysis. SNORD48 (No. 203903, Exiqon, Basel, SUI,), GAPDH, RPLP0, ATP5F1, and PPIA were used for normalization.
miRNA levels in human pericardial fluid (total fluid) and exosomes were assayed by Exiqon Services (Exiqon, Vedbæk, Denmark). Shortly, miRNAs were reverse transcribed into cDNA and quantified with miRCURY LNA Universal RT miRNA PCR Human panels I and II (v2) with SYBR Green mastermix.13 Reactions with several melting points or with melting points that were not within the specifications, as well as reactions with amplification efficiency <1.6 were removed from the dataset. Moreover, reactions giving Cp values that were within 5 Cp values of the negative controls reaction or their detection was >37 Cp, were removed from the dataset (this explains any discrepancy between the n stated in the figure legends compared to shown data points). To normalize the data, the average Cp of the assays that had signal in all the samples was used. This is described as global mean, and has been shown to be the best normalization method.18 For each sample, the normalized Cp corresponds to the average Cp minus the sample Cp.
2.9 Data analysis
2.9.1 Processing, mapping, and quantification
Raw-sequencing reads were processed (poly(A) trimming and quality filter—for 50-bp read requiring a minimum 97% of all bases to have a minimum phred quality score of 10) with the FastQC tool.19 miRNA reads were trimmed to 21 bp. GRO-seq reads were aligned to GRCh37/hg19 reference genome using bowtie,20 one alignment for each read was reported, allowing up to two mismatches. Alignment of RNA- and miRNA-Seq reads to GRCh37/hg19 genome were performed using STAR v2.5.4b,21 according to ENCODE standard options for long RNA-Seq pipeline and small RNA-Seq pipeline for RNA-Seq and miRNA-Seq reads, respectively.22 Subsequently, GRO-seq and RNA-seq tag directories were created with HOMER v4.923 makeTagDirectory.pl algorithm, using default parameters and fragment length of 75 bp. In RNA-Seq tag directories, the maximum number of tags per base pair was set to 3. Raw counts were quantified using the HOMER analyzeRepeats.pl algorithm. Coordinate files were set as following: hg19 for RNA-seq, custom annotation file including de novo identified pri-miRNA coordinates for GRO-seq (Supplementary material online, Table S3)15 and miRbase v22 for miRNA-seq. Data were visualized using UCSC Genome Browser24 and WashU Epigenome Browser (Hi-C data).25
2.9.2 Primary miRNA detection
Pri-miRNA coordinates were identified as previously reported.15 Shortly, publicly available and in-house GRO-seq data for 27 human cell types were pooled per cell type and augmented with matching ChIP-seq (histone markers H3K4ME1, H3K4ME3 and/or H3K27AC) and CAGE-seq data. De novo identification of primary transcripts from GRO-seq nascent transcripts was performed with the HOMER findPeaks.pl algorithm using parameters -groseq, -uniqmap, as well as three separate parameter settings for varying detection sensitivity and specificity. Transcription start site (TSS) coordinates were set with single nucleotide-specificity based on CAGE-seq peaks within ±500 bp of detected transcripts and collapsed for unique across cell types. In addition to a CAGE-seq peak, promoter histone methylation levels >10 cpm H3K4me3 and five-fold H3K4me3>H3K4me1 were required from all putative TSSs. Transcript end coordinates were set based on de novo transcripts clustering by adjacency and change point analysis. The final primary transcript coordinates represent non-overlapping transcripts between TSSs across individual loci and thus allow for multiple TSSs per locus. Pre-miRNA locations were set based on GENCODE (v19) and miRBase (v20) existing annotations and pri-miRNA transcript coordinates were identified by overlapping de novo transcripts with the pre-miRNA transcripts. Before fold change and statistics calculations, samples were filtered by discarding low read counts by filtering for RPKM > 0.5 in at least 3 samples, in order to improve the sensitivity and precision of the differential expression analysis.26
Non-mappable coordinates, exons of coding genes, and ribosomal RNA regions were removed from pri-miRNA transcripts prior to quantification using BEDTools (subtractBed) to exclude regions causing problems specifically in quantification of GRO-seq data. The non-overlapping regions of each pri-miRNA transcript were quantified with HOMER (analyzeRepeats.pl with parameters -strand + -noadj -noCondensing -pc 3), in order to obtain TV-specific expression values, and the lengths of the quantified region and total read counts per sample were used to report normalized expression values (RPKM). To determine the contribution of each TSS (TSSi) to the overall transcriptional activity in a given locus, the signal level at the upstream element (TSSi + 1) was subtracted, based on the RPKM values: RPKMi = RPKMi−RPKM(i + 1).
2.9.3 Differential gene expression analysis
Differential gene expression analysis was performed on replicates treated with oxPAPC using DESeq227 (GSE136813 and GSE103530 in Ref.12). Correlation between OXPAPC and NO2-OA stimuli was performed on the differentially expressed genes (FDR < 0.05) in HUVECs. Ingenuity Pathway Analysis (IPA, Qiagen, Redwood City, www.qiagen.com/ingenuity) was used to perform the upstream regulator analysis on the differentially expressed genes.
2.9.4 NRF2-binding analysis at regulated genes
OxPAPC-regulated genes detected in GRO-seq were compared to NRF2 ChIP-seq peaks detected in HAEC after OxPAPC stimulation.28 HOMER’s command ‘mergePeaks’ was used to identify the NRF2 peak that was bound at the promoter (±2 kb) or to nearby regions (±50 kb from TSS) and ‘annotatePeaks’ using the option ‘-hist’ was ran to display the distribution of the reads. The pausing was calculated as the average of the pausing ratio obtained for each replicate after using HOMER’s command ‘analyzeRepeats’ with the ‘-count pausing’ option.
2.9.5 Motif analysis
In order to increase the sensitivity of the transcript detection, the tag directories of each individual cell type were pooled together using ‘makeTagDirectory’ command from HOMER v4.9. All transcribed regions were detected for each cell type individually using HOMER algorithm ‘findPeaks.pl’ with ‘–style groseq’ option. Transcripts detected were then quantified using HOMER ‘annotatePeaks.pl’ for each tag directory individually. After a filtering where only reads showing a RPKM value >3 in at least 4 samples were kept, differential expression was performed with HOMER v4.9 using ‘getDiffExpression.pl’ algorithm. For each peak up-regulated at least two-fold in any condition compared to control, a region of 400 bp around the TSS was analysed for motif enrichment using ‘findMotifsGenome.pl’ against GRCh37/hg19 reference genome and using all the TSSs detected for the analysed cell type as background.
2.10 Statistical analyses
All statistical analyses were performed using the GraphPad Prism 8 software or R. All experiments were performed at least three times with at least three biological replicates per experiment. Statistical significance was evaluated with unpaired, two-tailed Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). Results are expressed as mean ± SD.
3. Results
3.1 Global characterization of NRF2-regulated transcriptional mechanisms in human vascular endothelial cells
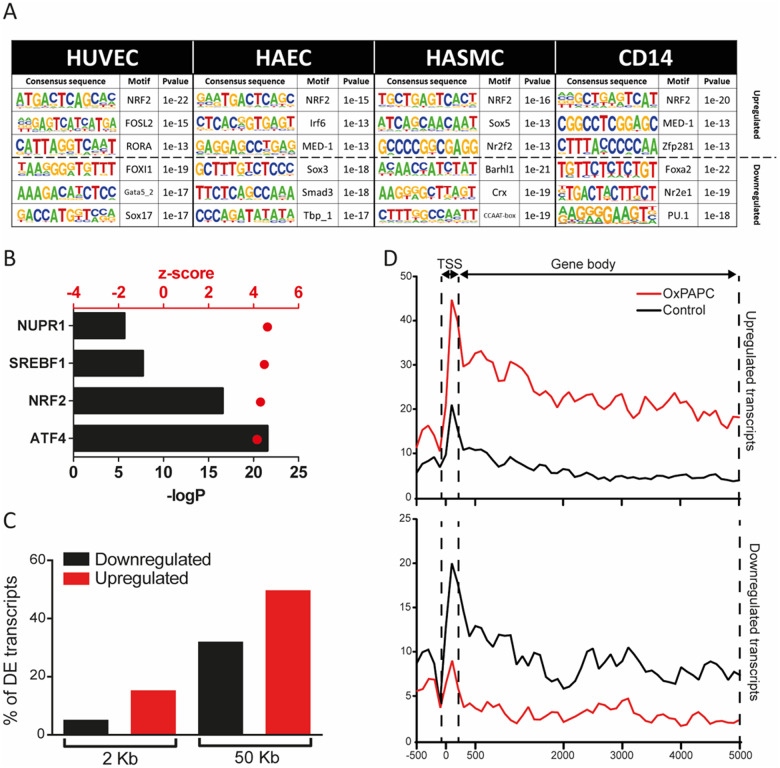
To confirm the important role of NRF2 in oxPAPC-mediated transcriptional response, we first sought to identify the transcription factor motifs that were enriched within the TSSs of up-regulated (>2-fold) transcripts upon oxPAPC stimulus. To this end, we performed GRO-Seq in HUVECs and HAECs upon 6 h exposure to oxPAPC. Expectedly, the results showed NRF2-binding sequence, ARE, as one of the top motifs enriched (Figure 1A and Supplementary material online, Table S1). In contrast, the down-regulated transcripts (Figure 1A and Supplementary material online, Table S1), showed cell type-specific transcription factor motifs, such as GATA and SOX for endothelial cells and PU.1 for macrophages.
Figure 1.
Global characterization of NRF2-regulated transcriptional mechanisms in human vascular endothelial cells. (A) Motif enrichment in up-regulated and down-regulated transcripts from the GRO-seq data in HUVECs, HAECs, HASMCs, and CD14+ macrophages under oxPAPC stimuli. (B) Ingenuity pathway upstream regulator analysis of differentially expressed genes (FDR < 0.05) in HUVECs after OXPAPC stimulation, generated with IPA (QIAGEN). (C) Percentage of down-regulated (black) and up-regulated (red) genes found in the vicinity (2 or 50 kb) of NRF2 peaks detected by ChIP-Seq in HAECs (GSM2394418) under OxPAPC condition. (D) Reads distribution among TSS and gene body of up- and down-regulated genes proximal to NRF2 ChIP-Seq peaks (GSM2394418) under oxPAPC and normoxia.
The central role of NRF2 was further confirmed by the Ingenuity Pathway Upstream regulator analysis that predicted NRF2 as one of the top transcriptional regulators responsible for the gene expression changes in HUVECs (Figure 1B and Supplementary material online, Table S2). Among the top transcriptional regulators exhibiting an activation z-score >4, we also identified ATF4, SREBF1, and NUPR1, of which ATF4 has been shown to be activated by oxidized phospholipids and cooperate with NRF2 to orchestrate downstream gene activation.29 Moreover, we correlated the gene expression changes resulting from oxPAPC to our in-house generated data from a more specific NRF2-activator, electrophilic fatty acid-derivative nitro-oleic acid (NO2-OA) (2 and 12 h) and found a strong correlation between the two treatments, suggesting that short exposure to oxPAPC does largely reflect NRF2-mediated response (Supplementary material online, Figure S1A).
Next, we analysed the relationship of NRF2-binding and the direction of transcriptional gene regulation. To achieve this, we analysed GRO-Seq and NRF2 ChIP-Seq28 data from oxPAPC-treated HAECs. Interestingly, we found that 15% and 50% of the up-regulated genes after oxPAPC treatment had a detected NRF2 peak ∼2 and 50 kb of their TSS, respectively, while only 5% and 32% of the down-regulated genes were found to have NRF2 peaks ∼2 and 50 kb of their TSS, respectively (Figure 1C). This suggested that up-regulation involves likely more direct NRF2-binding compared to down-regulation.
Transcriptional activation and repression could be mediated by changes in initiation or elongation of transcription.30 There is ample evidence suggesting that spatio-temporal control of gene activity often involves the controlled release of paused polymerase into productive RNA synthesis.31 In order to gain more insights of the NRF2-mediated regulation, we studied the distribution of GRO-Seq reads at the TSSs and at the gene bodies of the differentially expressed genes that bound NRF2 at their vicinity, allowing us to estimate the pausing ratio. Our results demonstrated that only 11.5% (26/226) of the induced and 16.9% (36/213) of the repressed genes exhibited over two-fold change in the pausing ratio and majority of the regulated genes exhibited and equal change at the TSS and at the gene body (Figure 1D and Supplementary material online, Figure S1B). Altogether, this suggests that majority of the NRF2-bound genes could be controlled at the level of transcriptional initiation.
3.2 Identification of NRF2-regulated miRNome in vascular cells
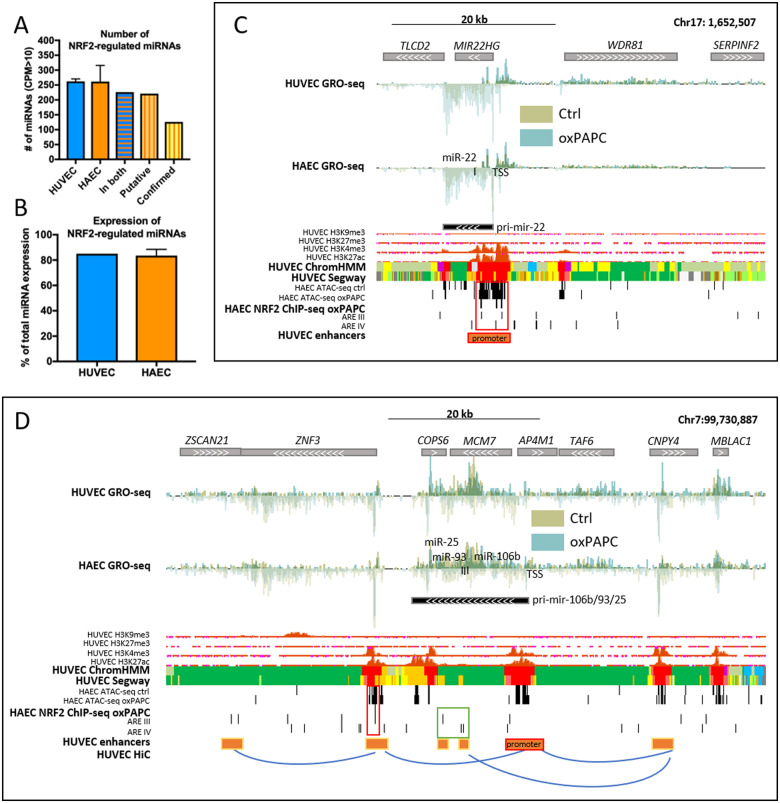
We next sought to determine the extent of NRF2-regulated miRNome in endothelial cells using a combination of next generation-sequencing data and NRF2-binding site annotations. First, we determined the global expression of HUVEC and HAEC pri-miRNAs and mature miRNAs with GRO-Seq and miRNA-seq, respectively. The total number of mature miRNAs expressed (CPM > 10) in oxPAPC-treated HUVECs and HAECs were 261 and 279 miRNAs, respectively, 226 of these miRNAs being expressed in both cell types (Figure 2A). Secondly, we utilized our previously validated approach11,12 and combined genome-wide NRF2-binding site annotations11 with cell-type-specific chromatin accessibility maps and regulatory landscapes gained from histone marker ChIP-Seq and ATAC-Seq datasets. Then, we intersected the overlapping areas with GRO-Seq defined pri-miRNA loci.15 Using this approach, we were able to confirm that the vast majority (221/226, 98%) of active regulatory sites close to miRNA loci contained NRF2-binding sites (Figure 2A and Supplementary material online, Table S3). After intersecting endothelial NRF2 ChIP-Seq peaks with these loci, 126 miRNAs (126/226, 56%) remained (Figure 2A and Supplementary material online, Table S3). All these 126 miRNAs had NRF2 binding on miRNA promoter or enhancer(s) looping to the promoter. Among these miRNAs, were both novel targets, such as miR-22 (Figure 2C), as well as previously validated targets, such as mir-106b/25/93 cluster (Figure 2D).12 Taken together, we identified 116 novel and confirmed 10 previously published NRF2 target miRNAs11,12,33–35 in human endothelial cells (Table 1). Altogether these miRNAs make up over 80% of the total miRNA expression in endothelial cells (Figure 2B).
Figure 2.
Identification of NRF2-regulated miRNome in vascular cells. (A) Global expression of HUVEC and HAEC miRNAs, those found in both, and putative and confirmed NRF2-regulated miRNAs. (B) Percentages of NRF2-regulated miRNAs of the total miRNA expression in HUVECs and HAECs. For all: n = 2, mean ± SD. Genomic loci of mir-22 (C) and mir-106b/25/93 cluster (D). Histone and chromatin segmentation data are ENCODE data32 from UCSC Genome Browser.24 Chromatin segmentation track shows promoters in red, enhancers in orange, and active chromatin regions in green. AREs are determined using previously published tool.11 HiC interactions were visualized with WashU Epigenome Browser.25
Table 1.
NRF2-regulated miRNAs
| Genes | Primary miRNA | Expressed mature miRNAs |
|---|---|---|
| MIRLET7A2, MIR100, MIR125B1 | pri-mir-let-7a-2/mir-100/125b-1 | miR-125b-1-3p, miR-125b-5p, miR-100-5p, let-7a-2-3p |
| MIRLET7A3, MIRLET7B, MIR3619 | pri-mir-let-7a-3/7b/mir-3619 | let-7b-5p, let-7b-3p |
| MIRLET7C, MIR125B2, MIR99A | pri-mir-let-7c/mir-125b-2/99a | let-7c, miR-99a-5p, miR-125b-2-3p |
| MIR106B, MIR25, MIR93 | pri-mir-106b/25/93 | miR-25-3p, miR-93-5p, miR-106b-3p, miR-106b-5p, miR-93-3p |
| MIR126 | pri-mir-126 | miR-126-3p, miR-126-5p |
| MIR128-1 | pri-mir-128-1 | miR-128 |
| MIR1301 | pri-mir-1301 | miR-1301 |
| MIR130A | pri-mir-130a | miR-130a-3p |
| MIR130B, MIR301B | pri-mir-130b/301b | miR-130b-3p, miR-130b-5p, miR-301b |
| MIR132 | pri-mir-132 | miR-132-3p, miR-132-5p |
| MIR139 | pri-mir-139 | miR-139-3p, miR-139-5p |
| MIR143, MIR145 | pri-mir-143/145 | miR-143-3p |
| MIR146A, MIR3142 | pri-mir-146a/3142 | miR-146a-5p |
| MIR146B | pri-mir-146b | miR-146b-5p |
| MIR148A | pri-mir-148a | miR-148a-3p |
| MIR148B | pri-mir-148b | miR-148b-3p |
| MIR149 | pri-mir-149 | miR-149-5p |
| MIR151A | pri-mir-151a | miR-151a-3p |
| MIR152 | pri-mir-152 | miR-152 |
| MIR181C, MIR181D | pri-mir-181c/181d | miR-181c-3p, miR-181c-5p, miR-181d |
| MIR185 | pri-mir-185 | miR-185-5p |
| MIR192, MIR194-2 | pri-mir-192/194-2 | miR-192-5p |
| MIR193A, MIR365B, MIR4725 | pri-mir-193a/365b/mir-4725 | miR-193a-3p, miR-193a-5p, miR-365b-5p |
| MIR193B, MIR365A | pri-mir-193b/365a | miR-193b-3p, miR-365a-5p |
| MIR195, MIR497 | pri-mir-195/497 | miR-195-3p, miR-195-5p |
| MIR197 | pri-mir-197 | miR-197-3p |
| MIR21 | pri-mir-21 | miR-21-3p, miR-21-5p |
| MIR210 | pri-mir-210 | miR-210 |
| MIR216A, MIR217 | pri-mir-216a/217 | miR-216a-3p, miR-216a-5p, miR-217 |
| MIR22 | pri-mir-22 | miR-22-3p, miR-22-5p |
| MIR221, MIR222 | pri-mir-221/222 | miR-221-3p, miR-221-5p, miR-222-3p, miR-222-5p |
| MIR23A, MIR24-2, MIR27A | pri-mir-23a/24-2/27a | miR-23a-3p, miR-27a-3p, miR-24-2-5p, miR-27a-5p |
| MIR23B, MIR27B, MIR24-1 | pri-mir-23b/27b/24-1 | miR-27b-3p, miR-23b-3p, miR-27b-5p |
| MIR26B | pri-mir-26b | miR-26b-5p, miR-26b-3p |
| MIR28 | pri-mir-28 | miR-28-3p, miR-28-5p |
| MIR29A, MIR29B1 | pri-mir-29a/29b-1 | miR-29a-3p, miR-29b-1-5p |
| MIR29B2, MIR29C | pri-mir-29b-2/29c | miR-29c-3p, miR-29c-5p |
| MIR301A, MIR454 | pri-mir-301a/454 | miR-454-3p |
| MIR30A, MIR30C2 | pri-mir-30a/30c-2 | miR-30a-3p, miR-30a-5p, miR-30c-2-3p |
| MIR31 | pri-mir-31 | miR-31-5p, miR-31-3p |
| MIR32 | pri-mir-32 | miR-32-5p |
| MIR320A | pri-mir-320a | miR-320a |
| MIR324 | pri-mir-324 | miR-324-3p, miR-324-5p |
| MIR326 | pri-mir-326 | miR-326 |
| MIR328 | pri-mir-328 | miR-328 |
| MIR330 | pri-mir-330 | miR-330-3p, miR-330-5p |
| MIR331, MIR3685 | pri-mir-331/3685 | miR-331-3p, miR-331-5p |
| MIR339 | pri-mir-339 | miR-339-3p, miR-339-5p |
| MIR33A | pri-mir-33a | miR-33a-5p |
| MIR340 | pri-mir-340 | miR-340-3p, miR-340-5p |
| MIR34A | pri-mir-34a | miR-34a-5p |
| MIR3615 | pri-mir-3615 | miR-3615 |
| MIR3676, MIR4521 | pri-mir-3676/4521 | miR-3676-5p, miR-4521 |
| MIR374A, MIR374B, MIR421, MIR545 | pri-mir-374a/374b/421/545 | miR-374a-3p, miR-374a-5p, miR-374b-5p, miR-421 |
| MIR423 | pri-mir-423 | miR-423-3p, miR-423-5p |
| MIR424, MIR450A1, MIR450A2, MIR450B, MIR503, MIR542 | pri-mir-424/450a-1/450a-2/450b/503/542 | miR-424-3p, miR-424-5p, miR-450b-5p, miR-503-5p, miR-542-3p |
| MIR425 | pri-mir-425 | miR-191-5p, miR-425-3p, miR-425-5p |
| MIR548K | pri-mir-548k | miR-548k |
| MIR574 | pri-mir-574 | miR-574-3p |
| MIR582 | pri-mir-582 | miR-582-3p |
| MIR584 | pri-mir-584 | miR-584-5p |
| MIR589 | pri-mir-589 | miR-589-5p |
| MIR641 | pri-mir-641 | miR-641 |
| MIR671 | pri-mir-671 | miR-671-3p, miR-671-5p |
| MIR769 | pri-mir-769 | miR-769-5p |
| MIR874 | pri-mir-874 | miR-874 |
| MIR887 | pri-mir-887 | miR-887 |
| MIR92B | pri-mir-92b | miR-92b-3p |
Previously identified NRF2-regulated miRNAs are highlighted in blue.
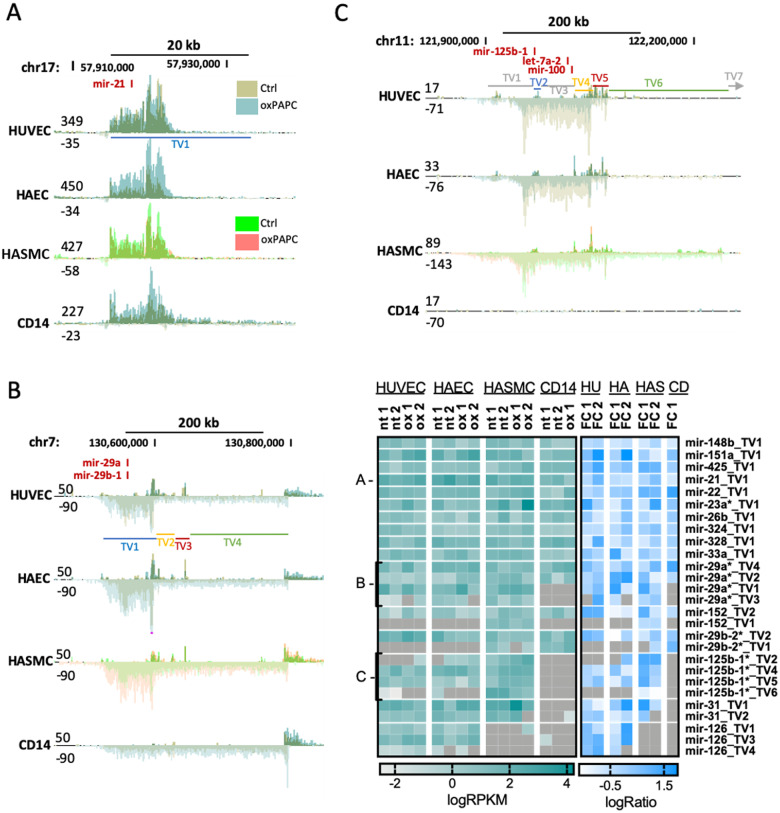
NRF2-binding sequence was also found top enriched in the active regulatory elements of oxPAPC-treated HASMC and CD14+ macrophages (Figure 1A) which together with vascular endothelial cells are implicated in atherosclerosis. Therefore, we set out to decipher the expression patterns of the NRF2-regulated pri-miRNAs in these two additional cell types using GRO-Seq (Figure 3 and Supplementary material online, Figures S2 and S3). Out of the 126 NRF2-regulated miRNAs in endothelial cells, 73% showed similar TSS-usage and transcript variant expression also in smooth muscle cells and macrophages (Figure 3A), 21% showed similar patterns in either smooth muscle cells or macrophages (Figure 3B), and 6% were either unique to endothelial cells (Supplementary material online, Figure S3) or had differential TSS-usage or expression patterns from endothelial cells (Figure 3C). Thus, most of the NRF2-regulated endothelial miRNAs were similarly transcribed also in smooth muscle cells and macrophages.
Figure 3.
Most NRF2-regulated miRNAs have similar expression patterns in other vascular cell types. GRO-seq signals for mir-21 (A), mir-29a/29b-1 (B), and mir-125b-1/let-7a-2/miR-100 (C) loci are shown. TVs mark the different transcript variants as determined in Ref.15 Heatmap shows transcript variant expression based on GRO-seq data for the listed miRNAs.
3.3 NRF2-regulated miRNAs are predicted to promote atherosclerosis and include several hub miRNAs
To study the biological effects of the NRF2-regulated miRNAs under oxPAPC stimuli, we measured the target gene expression with RNA-seq and utilized MicroRNA Target Filter of Ingenuity Pathway Analysis (IPA, Qiagen Redwood City, www.qiagen.com/ingenuity) to link the miRNAs with their targets in both HUVECs and HAECs separately. We filtered the data to include only those miRNA target genes that showed opposite expression trends with their regulator miRNAs and thus could be under canonical post-transcriptional regulation of the miRNAs. Expectedly, all miRNAs had more than one target gene, and most target genes were regulated by multiple miRNAs (Supplementary material online, Figure S4A). The Pearson correlation coefficient for the target gene transcription (GRO-seq) with expression (RNA-seq) was 0.4 and 0.3 (P < 0.0001) in HUVECs and HAECs, respectively, indicating a substantial role for post-transcriptional gene regulation in the expression of the genes (Supplementary material online, Figure S4B).
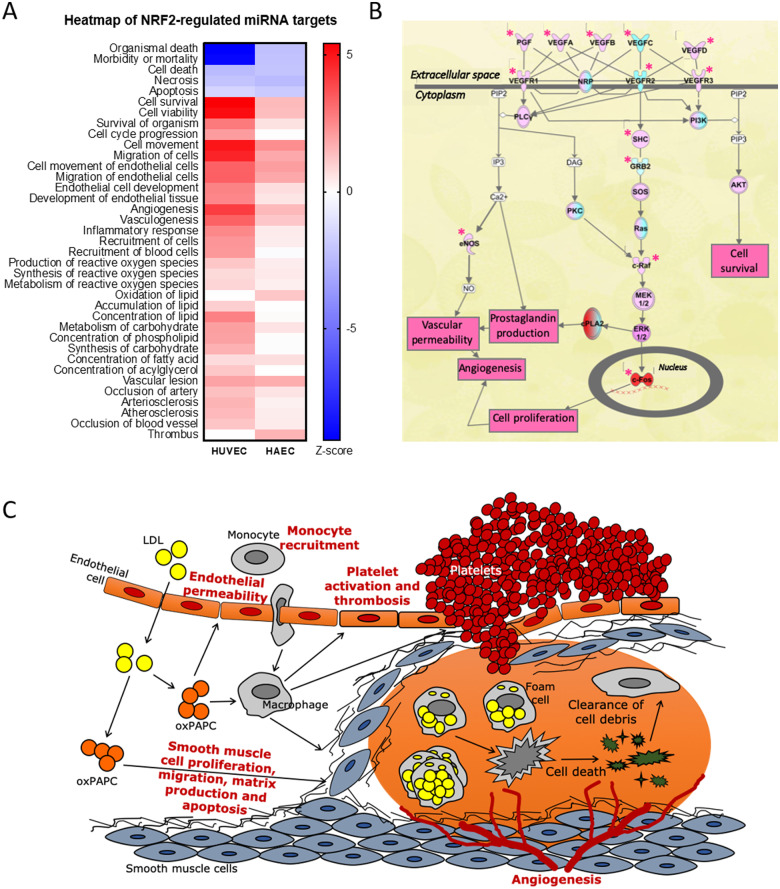
In general, the predicted functional effects of miRNAs on target mRNAs were similar in HUVECs and HAECs (Figure 4A). The perceived changes suggested proatherogenic changes in the cell function, such as increased activation and movement, reduced barrier function and increased endothelial permeability, increased inflammatory response and monocyte recruitment, increased production of reactive oxygen products and lipid oxidation, increased lesion formation and platelet activation and thrombosis. One major pathway contributing to these changes was the VEGF pathway, which was activated in response to oxPAPC stimuli and miRNA function (Figure 4B). The overall changes are summarized in Figure 4C.
Figure 4.
NRF2-regulated miRNAs involved in atherosclerosis-related functions. (A) IPA’s comparison analysis for HUVECs and HAECs showing the changes in atherosclerosis-related functions. (B) IPA’s VEGF pathway from HUVEC data. Stars mark the molecules directly affected by miRNAs. Red marks up-regulation and cyan down-regulation. (C) Overview of the observed changes in atherosclerosis context.
To find miRNAs with the most pronounced effects in vascular context, we utilized miRNet36,37 and determined the hub miRNAs by creating miRNA-target interaction networks for the NRF2-regulated miRNAs based on experimentally validated miRNA-target data from human umbilical vein tissue (Supplementary material online, Figure S5). Among the 126 NRF2-regulated miRNAs, 20 were designated as hub miRNAs and three of these, namely miR-21-5p, miR-126-3p, miR-100-5p were among the three most abundant endothelial hub miRNAs accounting for 71% and 45% of total miRNA expression in HUVECs and HAECs, respectively. All of these three miRNAs have been shown to be relevant for atherosclerosis in mouse knockout models.38–43 Since miR-126 has been previously validated as NRF2-regulated miRNA,11 we decided to select the other two hub miRNAs, miR-21-5p, and miR-100-5p for further validations.
3.4 Detailed analysis of NRF2-regulated miR-21-5p and miR-100-5p expressions
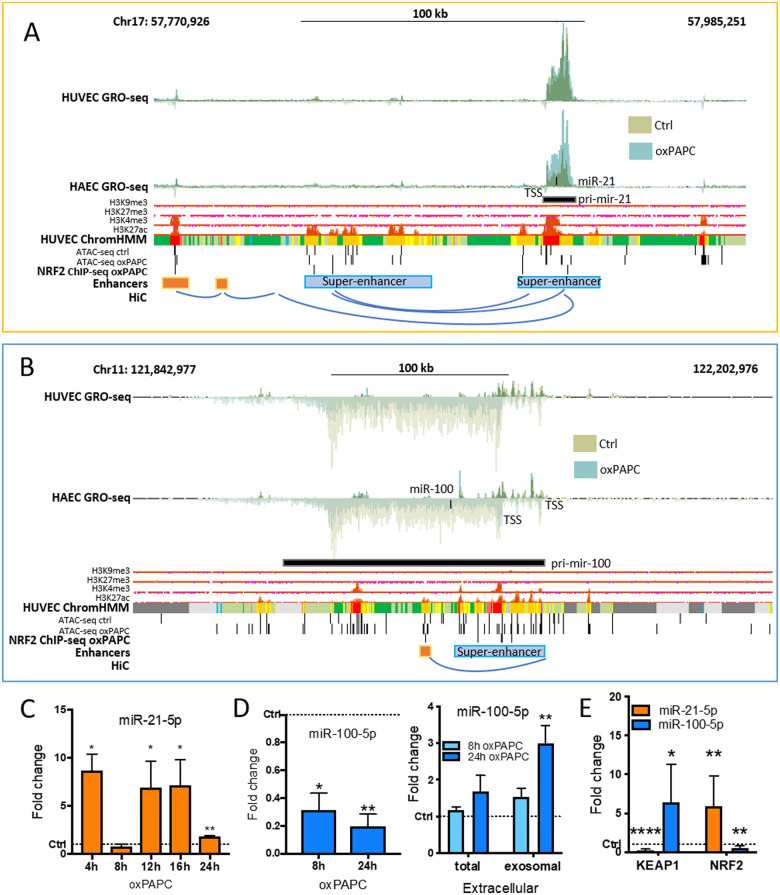
First, we confirmed that mature miR-21-5p and miR-100-5p expression was in line with pri-miRNA transcription (Figure 5). In qPCR measurements, miR-21-5p expression was unchanged in oxPAPC-treated cells at 8 h compared to control cells consistently with HUVEC GRO-seq signal at 6 h (Figure5A andC). However, in other time points miR-21-5p was up-regulated in oxPAPC-treated cells compared to control cells. miR-100-5p, on the other hand, was down-regulated in all timepoints as in GRO-seq data at 6 h (Figure5B andD). Analysis of the mature and pri-miRNA expression of all the miRNAs in the mir-125b-1/let-7a-2/mir-100 cluster revealed that oxPAPC-mediated repression of pri-miRNA transcription led to lower expression of all mature miRNAs, both in HAECs and HUVECs (Supplementary material online, Figure S6). Importantly, the binding of NRF2 to the promoter of miR-100 and nearby enhancer regions of miR-21 and miR-100 was confirmed with ChIP-Seq (Figure5A andB).
Figure 5.
Detailed analysis of NRF2-regulated miR-21-5p and miR-100-5p expressions. Genomic loci of mir-21 (A) and mir-125b-1/let-7a-2/mir-100 cluster (B). Histone and chromatin segmentation data are ENCODE data32 from UCSC Genome Browser.24 Chromatin segmentation track shows promoters in red, enhancers in orange, and active chromatin regions in green. AREs are determined using previously published tool.11 HiC interactions were visualized with WashU Epigenome Browser.25 (C) miR-21-5p expression in HUVECs under oxPAPC stimuli for indicated times compared to control samples. (D) miR-100-5p expression in HUVECs under oxPAPC stimuli for indicated times compared to control samples, and extracellular miRNA expression measured from the growth media of oxPAPC-treated cells and from the exosomes extracted from the growth media compared to control samples. (E) miRNA expression in KEAP1 and NRF2 overexpressing cells. (n = 3, mean ± SD, unpaired two-tailed t-test, ****P < 0.0001, **P < 0.01, *P < 0.05).
In addition to transcriptional down-regulation, miR-100-5p was also shown to be enriched, to some extent, in the growth media of the oxPAPC-treated cells most likely due to its active release from the cells upon oxPAPC stimuli (Figure 5D). The levels were most significantly up-regulated in the exosomal fraction of the media. Finally, we confirmed that miR-21-5p and miR-100-5p expression responds to NRF2 pathway activation and inhibition by overexpressing NRF2 or inhibiting its function through KEAP1 overexpression, which blocks the NRF2 pathway activation.44 Overexpression of NRF2 up-regulated miR-21-5p and down-regulated miR-100-5p consistently with the oxPAPC results. KEAP1 overexpression had the opposite effect, decreasing miR-21-5p and increasing miR-100-5p expression (Figure 5E and Supplementary material online, Figure S7). Thus, the results suggest that NRF2 mediates the oxPAPC-response of these miRNAs by up-regulating miR-21-5p and down-regulating miR-100-5p expression.
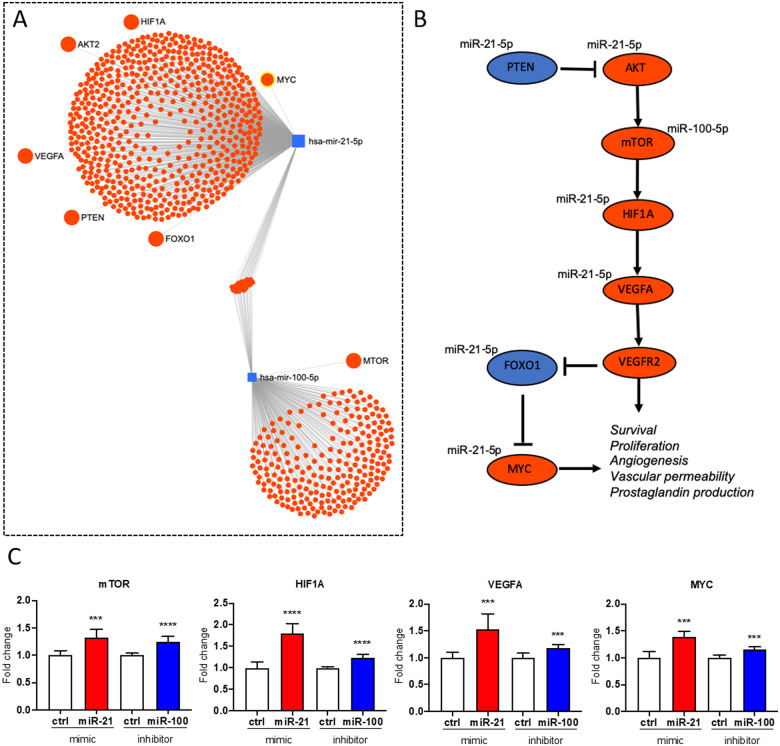
3.5 miR-21-5p and miR-100-5p mediate oxPAPC effects on VEGFA/MYC pathway
NRF2 has been previously shown to mediate the effects of oxPAPC on endothelial proliferation and energy metabolism.12 Here, NRF2-regulated miRNAs were shown to have similar net effect on several functions controlled by the VEGFA signalling (Figure 4). Therefore, we sought to investigate the individual effects of miR-21-5p up-regulation and miR-100-5p down-regulation on the VEGFA pathway. miRNet36,37 analysis in human umbilical vein tissue revealed several direct interactions between miR-21-5p and the upstream and downstream components of the VEGFA pathway (Figure6A andB). None of the targets was shared with miR-100-5p, which was only shown to interact with mTOR. Based on these interaction results, miR-100-5p would likely down-regulate mTOR and the downstream cascade of the VEGF pathway, but miR-21-5p might have more complex effects (Figure 6B). To decipher the effects of altered miR-21-5p and miR-100-5p expression on the VEGFA pathway in cellular context, we overexpressed miR-21-5p and silenced miR-100-5p in oxPAPC-treated endothelial cells (Supplementary material online, Figure S8). In line with the pathway predictions (Figure 6B), up-regulation of miR-21-5p and down-regulation of miR-100-5p both activated the VEGFA pathway leading to up-regulation of mTOR, HIF1A, VEGFA, and MYC (Figure 6C), which could stimulate endothelial survival, proliferation, and angiogenesis and thereby could promote proatherogenic changes.
Figure 6.
miR-21-5p and miR-100-5p mediate oxPAPC effects on VEGFA/MYC pathway. (A) miRNA-target network from miRNet.36,37 (B) Illustration of VEGF/MYC pathway. (C) Effects of miR-21-5p overexpression and miR-100-5p inhibition on MTOR, HIF1A, VEGFA, and MYC expressions. (n = 3, mean ± SD, unpaired two-tailed t-test, ****P < 0.0001, ***P < 0.001).
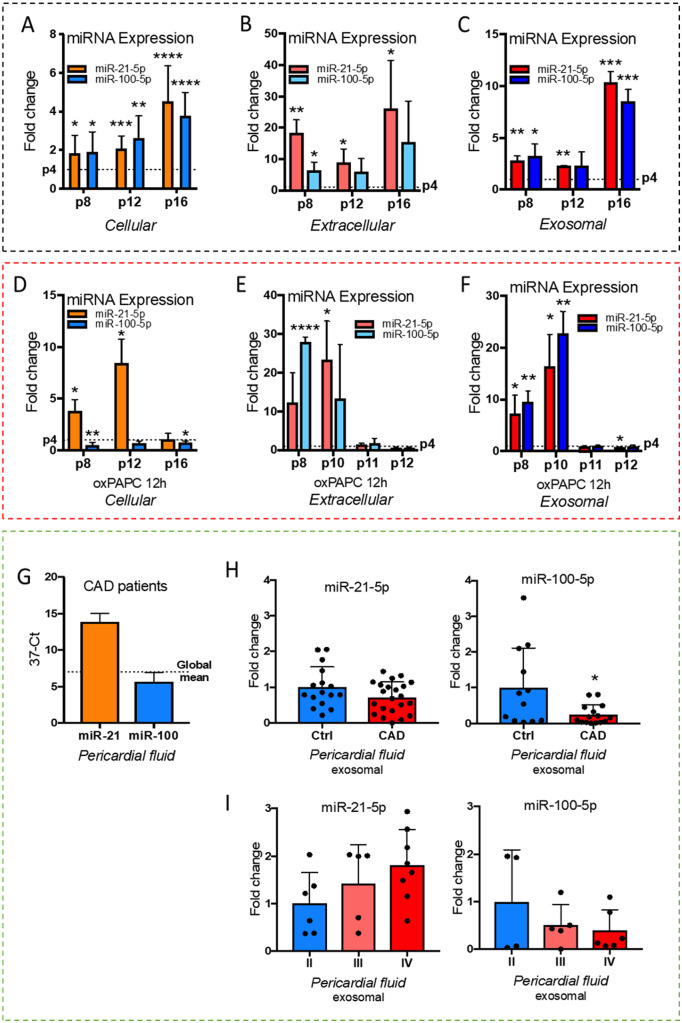
3.6 Senescence changes the cellular response to proatherogenic stimuli
miR-21-5p and miR-100-5p have both been shown to be abundant in senescent cells in previous studies.45,46 To decipher the impact of cellular senescence on oxPAPC-response of miR-21-5p and miR-100-5p, we measured their expression and secretion in young cells and all the way up to cellular senescence. Consistently with previous work, both miR-21-5p and miR-100-5p expression increased in older (p8–p16) cells compared to young (p4) (Figure 7A). In addition, extracellular levels of the miRNAs were also higher in p8–p16 cells compared to p4 suggesting increased release of miRNAs from the cells (Figure 7B). Most significant enrichment of the miRNAs was seen in the exosomal fraction of the growth media (Figure 7C). When treated with oxPAPC, the results for miR-100-5p response in the older cells remained consistent with the young cells (Figures5D and7D). However, for miR-21-5p, the response was consistent only until p12, after which the cells stopped responding to the stimuli (Figures5C and7D). Similarly, the release of both miR-21-5p and miR-100-5p from the cells dropped in oxPAPC-treated cells after p10 (Figure 7E). The same trend was seen both in the total media and in the exosomal fraction of the media (Figure7E andF). The drop was due to decreased abundance in the oxPAPC samples rather than increased abundance in the control samples. The results, hence, suggest that cellular senescence changes the cellular response to proatherogenic stimuli by affecting miR-21-5p response and the release of both miR-21-5p and miR-100-5p from the cells.
Figure 7.
miR-21-5p and miR-100-5p response during senescence and in coronary artery disease patients. (A) miRNA expression in passage (p) 8–16 cells compared to p4 cells. (B) miRNA levels in growth medium of p8–p16 cells compared to p4 cells. (C) miRNA levels in exosomes extracted from the growth medium of p8–p16 cells compared to p4 cells. (D) As in (A) but under oxPAPC stimuli. (E) As in (B) but under oxPAPC stimuli. (F) As in (C) but under oxPAPC stimuli. For (A–F) figures: n = 3, mean ± SD, unpaired two-tailed t-test, ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05. (G) miR-21-5p and miR-100-5p levels in human pericardial fluid of coronary artery disease (CAD) patients (n = 22, mean ± SD). (H) miRNA levels in pericardial fluid exosomes of control and CAD groups (n = 16 for control, n = 22 for CAD, mean ± SD, unpaired two-tailed t-test, *P < 0.05). (I) miRNA levels in pericardial fluid exosomes of CAD patients divided according to the severity of heart failure symptoms (NYHA classes I–IV), where I is no symptoms and IV severe symptoms (n = 4–6 for NYHA II, n = 5 for NYHA III and n = 6–8 for NYHA IV, mean ± SD).
3.7 Exosomal miR-21-5p and miR-100-5p levels in CAD patients
To estimate if the miRNAs could have a diagnostic value for atherosclerosis, we measured miRNA abundancies from the pericardial fluid of CAD patients. As vascular endothelial cells are a major contributor to the pericardial fluid miRNA composition,13 the results from the cell experiments would suggest miR-21 values to be higher than miR-100 values in the pericardial fluid of CAD patients, and according to our measurements, this is the case (Figure 7G). Based on our previous studies, miRNA levels in the exosomal fraction of the pericardial fluid may show better diagnostic value compared to total fluid miRNA values.12,13 Therefore, we measured the exosomal miRNA levels from the CAD patient and the control groups. Both miR-21-5p and miR-100-5p were lower in the CAD group compared to control group, although miR-21-5p was more abundant than miR-100-5p in the CAD samples (Figure 7H). However, when the measured values of the CAD group were further divided into subgroups based on the severity of the HF symptoms at the time of sample collection using New York Heart Association’s functional classification from mild (Class I) to severe (Class IV) symptoms, the miR-21-5p values showed a tendency to increase, while miR-100-5p values decreased with disease severity (Figure 7I).
4. Discussion
Atherosclerosis is a multifaceted disease, which affects millions of people worldwide. Despite the progress in its prevention and treatment, it continues to be a major financial burden to the health care system. Many risk factors for the disease have been recognized, but the molecular mechanisms driving the disease are still not sufficiently understood to enable prevention of plaque development and ultimately, reversal of the plaque formation. MicroRNAs are complex regulators of cellular functions, which provide means for manipulation of full pathways and gene networks instead of just one gene, and thus open interesting opportunities from the therapeutic point of view. In addition, extracellular miRNA levels could provide information on disease manifestation or progress and thus aid in disease diagnostics. Here, we set out to study miRNA regulation and expression under proatherogenic signalling in vascular endothelial cells, and to estimate the potential of proatherogenic miRNAs as diagnostic tools in CAD.
In order to identify the key transcription factors that mediate the effects of proatherogenic oxPAPC signalling in endothelial cells, we performed motif enrichment from active regulatory regions of human vascular smooth muscle cells, macrophages, and aortic and venous endothelial cells. The results showed a significant enrichment of NRF2-binding motif across all cell types, and further analysis predicted the binding of NRF2 to 56% of the active, oxPAPC-responsive miRNA regulatory regions with AREs. Altogether, we identified 126 NRF2-regulated miRNAs, out of which only 10 have been previously characterized. More detailed analysis of the miRNA expression patterns in endothelial cells, vascular smooth muscle cells and macrophages confirmed identical TSS-usage and transcript variant expression in all four cell types for 73% of the miRNA genes, and for additional 21% of the cases, the expression pattern was shared between three of the cell types thereby suggesting that NRF2 regulates the expression of most of the identified miRNAs also in vascular smooth muscle cells and macrophages. The findings are consistent with previous publications, as NRF2 is known to be a ubiquitous transcription factor, which has been recognized as the mediator of oxidized phospholipid signalling, and a key signalling molecule that is activated in response to hypoxia, in addition to oxidative and electrophilic stress.2,28,47 Furthermore, it is a putative global regulator of miRNA expression as it binds the AREs that have been shown to be enriched in miRNA promoters.48
Our data suggested that transcriptional activation by NRF2 likely involves direct binding to nearby regulatory elements whereas down-regulation is more indirect or involves tethered interactions with other transcription factors. Similar results have been described for other signal-dependent transcription factors like glucocorticoid receptor49 and peroxisome proliferator-activated receptor.50 Interestingly, the motif analysis and the IPA upstream regulator analysis further predicted ATF4, SREBF1, and NUPR1 as top regulators of the oxPAPC response. Previous studies have identified ATF4 cobinding with NRF2 to partake in the regulation of the NRF2-mediated gene activation in mammalian cells.51 Additionally, NUPR1 has been identified as an upstream regulator in mediating redox stress response and has been hypothesized to require NRF2 for the induction of the target gene expression.52 On the other hand, SREBF1, also known as SREBP1, has been shown to be down-regulated in the liver of Nrf2-deficient (Nrf2−/−) mice, and NRF2 binding to the ARE containing promoter of SREBP1 has been confirmed in mice liver, establishing a link of NRF2 in the regulation of hepatic lipid homeostasis.53 Future studies should address the roles of these potentially collaborative transcription factors in mediating the effects of NRF2 activation in different contexts.
Deeper analysis of the nascent RNA profiles further suggested that transcriptional initiation, leading to similar level of change at the TSS and at the gene body, represents the main mechanism by which NRF2-bound genes are regulated. This is in contrast to many other signal-dependent factors activated by inflammatory (LPS or TNFα) or angiogenic (VEGFA) stimulus, where transition into productive elongation represents the major mechanism of gene activation.16,54,55 This suggests that NRF2 could possibly recruit a different set of co-activators or chromatin remodelers to promote the recruitment of Pol II and the components of the general transcription machinery to form the pre-initiation complex at the promoters of target genes. Further experiments are required to evaluate such cooperation.
In animal models of atherosclerosis, the role of NRF2 in the disease development and progression has remained versatile and contradictory, and as a result, NRF2 has been deemed to be both pro- and anti-atherogenic.56 However, a recent study showed that unidirectional laminar flow (UF) in atherosclerosis activates NRF2 expression in endothelial cells, promoting adaptation to oxidative and nitrosative stress, as well as anti-inflammatory effects.57 The role that NRF2 plays in the regulation of angiogenesis has been consistently confirmed in many studies.58–60 Here, we selected two abundant hub miRNAs, one of which is up-regulated by NRF2 (miR-21-5p) and the other down-regulated (miR-100-5p) and validated their role in the regulation of the VEGFA/MYC pathway that promotes angiogenesis. Interestingly, both miR-21-5p up-regulation and miR-100-5p down-regulation activated the pathway, which leads to increased metabolic activity, proliferation, and angiogenesis of endothelial cells. This has been confirmed in functional studies, where down-regulation of miR-100 in HUVECs resulted in increased angiogenic tube formation as well as endothelial sprouting and proliferation.61 In another study, knockdown of miR-21 in HUVECs lead to decreased cell proliferation.62 Furthermore, glycolysis is a prerequisite for cell growth and proliferation and NRF2 has been shown to participate in this process in endothelial cells. In our previous studies, we have demonstrated that overexpression of miR-21 and miR-100, decreases the glycolysis rate in young endothelial cells, thus affecting NRF2-regulated functions, although the effect of oxPAPC in this process was not established.46 Altogether, our results provide the first evidence that NRF2 could mediate the global oxPAPC response of miRNAs and the subsequent effects on the regulation of genes controlling endothelial activation and angiogenesis that predisposes to thrombosis and plaque rupture.3
In this work, we provide characterization of two of the most abundant miRNAs, miR-100-5p, and miR-21-5p, in endothelial cells and demonstrate their regulation by NRF2. In the literature, miR-100-5p has been denoted as atheroprotective, as it inhibits the proliferation of endothelial cells and migration of vascular smooth muscle cells.63 These results suggest that therapeutic increase of miR-100-5p and lowering of miR-21-5p levels could improve endothelial function in atherosclerosis. For miR-100-5p recent evidence supports this, as miR-100-5p overexpression was shown to attenuate atherogenesis and decrease plaque area by 45% in a mouse model of atherosclerosis.38 However, the results for miR-21-5p are more complex: a global knockout of miR-21 in apoE−/− double knockout mice showed reduced atherosclerotic lesions, lower presence of macrophages and reduced smooth muscle cells and collagen content in the aorta,40 whereas local delivery of miR-21-5p has been shown to stabilize fibrous caps of vulnerable plaques in a double deficient (apoE−/− miR-21−/−) mouse model of advanced atherosclerosis.41 Moreover, endothelial-specific knockout of miR-21 in mice had vascular remodelling effects in the aorta, such as decreased diastolic pressure and impairment of endothelial-dependent relaxation of aorta, as well as decrease of elastin and collagen content and changes on Smad2/5/7 expression.42 Thus, these studies highlight the importance of gaining species, cell type, and cell state-specific knowledge on miRNA function.
Advanced age is a known risk factor for atherosclerosis and its adverse effects.64 One of the clinically most important underlying factors for the elevated risk is endothelial dysfunction, which happens early in vascular aging. To estimate the effect of cellular senescence on the proatherogenic miRNA expression, we measured the miR-21-5p and miR-100-5p levels from different endothelial passages. In accordance with the previously published data,46 both miRNAs had increased expression in higher cell passages. In addition, the secretion of these miRNAs was higher in older cells compared to young. However, when the cells were exposed to proatherogenic stimuli, both cellular expression and secretion of the miRNAs dropped to match the young cells indicating reduced responsiveness to oxPAPC stimuli. The subdued response to proatherogenic signalling might be partially due to NRF2 expression, as NRF2 levels have been shown to decrease in aging and cell senescence.65
Finally, to estimate the utility of the miRNAs in diagnostics, we used pericardial fluid samples of CAD patients and a matched control group. Consistent with the endothelial results, miR-21-5p levels were higher in the pericardial fluid of CAD patients than miR-100-5p. However, both miRNAs were lower in CAD group compared to control, which is consistent with the secretion data from senescent endothelial cells under proatherogenic stimuli. Conversely, when the CAD samples were further divided into subgroups based on severity of HF symptoms (NYHA I–IV), miR-21-5p levels were seen to increase and miR-100-5p levels to decrease with symptom severity, thus following more closely the oxPAPC-induced NRF2-mediated expression patterns. Our results are in line with measured miR-100-5p and miR-21-5p levels from human atherosclerotic plaque samples, where miR-21-5p has been shown to be up- and miR-100-5p down-regulated compared to control samples.66–68 Moreover, serum miR-21-5p levels have been shown to be elevated in acute coronary syndrome patients compared to stable CAD patients.69 However, future studies with a larger group of patients are needed to overcome the variability in the miRNA levels and confirm the prospective utility of exosomal miR-21-5p and miR-100-5p in cardiovascular disease diagnostics.
Taken together, we conclude that NRF2 is a key regulator of the endothelial miRNA expression under oxidized phospholipid stimulation as it is predicted to regulate the expression of majority of the expressed miRNAs and, especially, the expression of the key miRNAs miR-126-3p, miR-21-5p, and miR-100-5p.
Data availability
Sequencing data from this study have been submitted to NCBI Gene Expression Omnibus under accession number GSE136813. The public Hi-C and RNA-seq datasets used in this study have been deposited in the Gene Expression Omnibus (GEO)70 under accession numbers GSE89970 and GSE94872.
Translational Perspective
Our analysis provides deeper understanding of the transcriptional regulation of miRNAs and their target genes that could regulate the proatherogenic effects of oxidized phospholipids in endothelial cells. We further characterize the role of miR-21-5p and miR-100-5p in regulating the VEGF pathway upon proatherogenic stimuli. We demonstrate that exosomal levels of both miRNAs obtained from pericardial fluid of cardiovascular disease patients are correlated with disease severity. Thus, future studies should determine the utility of miR-21-5p and miR-100-5p for the diagnosis of cardiovascular disease.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
S.L.K. designed and performed majority of the experiments, analysed the data, and wrote the article; V.T.B. and PMO contributed to experimental validations, writing, and bioinformatic analysis. M.B.L., H.N., E.K., and A.K. performed the in vitro sequencing experiments; J.H., M.H., H.K., and P.T. collected and provided the human samples used in this study; A.L.L. supported writing of the article; M.U.K. contributed to the generation of sequencing libraries, writing of the article, and supervision of the project.
Supplementary Material
Acknowledgements
We thank the Sequencing Service GeneCore Sequencing Facility (EMBL,www.genecore.embl.de) for NGS library sequencing and UEF Bioinformatics Center for server infrastructure.
Conflict of interest: none declared.
Funding
This work was supported by the Academy of Finland (Grants Nos. 287478 and 294073 to M.U.K., 275147 to A.L.L. and 325510 to P.T.); Emil Aaltonen Foundation (to S.L.K.); the Finnish Foundation for Cardiovascular Research (to S.L.K., P.R.M., J.H. and M.U.K.); the Maud Kuistila Memorial Foundation (to S.L.K. and P.R.M.); the Orion Research Foundation (to S.L.K.); the Jane and Aatos Erkko Foundation (to M.U.K.); the Sigrid Jusélius Foundation (to S.L.K., P.T. and M.U.K.) and the Doctoral Program of Molecular Medicine of the University of Eastern Finland (to P.R.M.). This project was partly funded by the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (Grant No. 802825). Funding for open access charge: Academy of Finland.
References
- 1. Gimbrone MA, García-Cardeña G.. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res 2016;118:620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bochkov VN, Oskolkova OV, Birukov KG, Levonen A-L, Binder CJ, Stöckl J.. Generation and biological activities of oxidized phospholipids. Antioxid Redox Signal 2010;12:1009–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee S, Birukov KG, Romanoski CE, Springstead JR, Lusis AJ, Berliner JA.. Role of phospholipid oxidation products in atherosclerosis. Circ Res 2012;111:778–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andreou I, Sun X, Stone PH, Edelman ER, Feinberg MW.. miRNAs in atherosclerotic plaque initiation, progression, and rupture. Trends Mol Med 2015;21:307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nazari-Jahantigh M, Egea V, Schober A, Weber C.. MicroRNA-specific regulatory mechanisms in atherosclerosis. J Mol Cell Cardiol 2015;89:35–41. [DOI] [PubMed] [Google Scholar]
- 6. Yanan W, Yingyu X, Ao Z, Mingyang W, Zihan F, Junping Z.. Exosomes: an emerging factor in atherosclerosis. Biomed Pharmacother 2019;115:108951. [DOI] [PubMed] [Google Scholar]
- 7. Tomasetti M, Lee W, Santarelli L, Neuzil J.. Exosome-derived microRNAs in cancer metabolism: possible implications in cancer diagnostics and therapy. Exp Mol Med 2017;49:e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feinberg MW, Moore KJ.. MicroRNA regulation of atherosclerosis. Circ Res 2016;118:703–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee S-S.. Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol Ther Nucleic Acids 2017;8:132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mellis D, Caporali A.. MicroRNA-based therapeutics in cardiovascular disease: screening and delivery to the target. Biochem Soc Trans 2018;46:11–21. [DOI] [PubMed] [Google Scholar]
- 11. Kuosmanen SM, Viitala S, Laitinen T, Peräkylä M, Pölönen P, Kansanen E, Leinonen H, Raju S, Wienecke-Baldacchino A, Närvänen A, Poso A, Heinäniemi M, Heikkinen S, Levonen A-L.. The effects of sequence variation on genome-wide NRF2 binding-new target genes and regulatory SNPs. Nucleic Acids Res 2016;44:1760–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuosmanen S, Kansanen E, Kaikkonen M, Sihvola V, Hartikainen J, Pulkkinen K, Jyrkk H, Tuoresm P, Kokki H, Tavi P, Heikkinen S, Levonen AA-L, Jyrkkänen H-K, Tuoresmäki P, Hartikainen J, Hippeläinen M, Kokki H, Tavi P, Heikkinen S, Levonen AA-L.. NRF2 regulates endothelial glycolysis and proliferation with miR-93 and mediates the effects of oxidized phospholipids on endothelial activation. Nucleic Acids Res 2018;46:1124–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuosmanen SM, Hartikainen J, Hippeläinen M, Kokki H, Levonen A-L, Tavi P.. MicroRNA profiling of pericardial fluid samples from patients with heart failure. PLoS One 2015;10:e0119646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Woodcock SR, Bonacci G, Gelhaus SL, Schopfer FJ.. Nitrated fatty acids: synthesis and measurement. Free Radic Biol Med 2013;59:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bouvy-Liivrand M, Sande AD, Pölönen P, Mehtonen J, Vuorenmaa T, Niskanen H, Sinkkonen L, Kaikkonen MU, Heinäniemi M.. Analysis of primary microRNA loci from nascent transcriptomes reveals regulatory domains governed by chromatin architecture. Nucleic Acids Res 2017;45:9837–9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaikkonen MU, Niskanen H, Romanoski CE, Kansanen E, Kivelä AM, Laitalainen J, Heinz S, Benner C, Glass CK, Ylä-Herttuala S.. Control of VEGF-A transcriptional programs by pausing and genomic compartmentalization. Nucleic Acids Res 2014;42:12570–12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Levonen A-L, Inkala M, Heikura T, Jauhiainen S, Jyrkkänen H-K, Kansanen E, Määttä K, Romppanen E, Turunen P, Rutanen J, Ylä-Herttuala S.. Nrf2 gene transfer induces antioxidant enzymes and suppresses smooth muscle cell growth in vitro and reduces oxidative stress in rabbit aorta in vivo. Arterioscler Thromb Vasc Biol 2007;27:741–747. [DOI] [PubMed] [Google Scholar]
- 18. Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J.. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 2009;10:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krueger F, Andrews SR, Osborne CS.. Large scale loss of data in low-diversity illumina sequencing libraries can be recovered by deferred cluster calling. PLoS One 2011;6:e16607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Langmead B, Trapnell C, Pop M, Salzberg SL.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Ernst J.. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D.. The human genome browser at UCSC. Genome Res 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou X, Lowdon RF, Li D, Lawson HA, Madden PAF, Costello JF, Wang T.. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat Methods 2013;10:375–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sha Y, Phan JH, Wang MD. Effect of low-expression gene filtering on detection of differentially expressed genes in RNA-seq data. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS. 2015. pp. 6461–4. [DOI] [PMC free article] [PubMed]
- 27. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hogan NT, Whalen MB, Stolze LK, Hadeli NK, Lam MT, Springstead JR, Glass CK, Romanoski CE.. Transcriptional networks specifying homeostatic and inflammatory programs of gene expression in human aortic endothelial cells. Elife 2017;6:e22536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Afonyushkin T, Oskolkova OV, Philippova M, Resink TJ, Erne P, Binder BR, Bochkov VN.. Oxidized phospholipids regulate expression of ATF4 and VEGF in endothelial cells via NRF2-dependent mechanism: novel point of convergence between electrophilic and unfolded protein stress pathways. Arterioscler Thromb Vasc Biol 2010;30:1007–1013. [DOI] [PubMed] [Google Scholar]
- 30. Wade JT, Struhl K.. The transition from transcriptional initiation to elongation. Curr Opin Genet Dev 2008;18:130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Core L, Adelman K.. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation Leighton. Genes Dev 2019;33:960–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS, Hitz BC, Gabdank I, Narayanan AK, Ho M, Lee BT, Rowe LD, Dreszer TR, Roe G, Podduturi NR, Tanaka F, Hong EL, Cherry JM.. ENCODE data at the ENCODE portal. Nucleic Acids Res 2016;44:D726–D732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shah NM, Zaitseva L, Bowles KM, MacEwan DJ, Rushworth SA.. NRF2-driven miR-125B1 and miR-29B1 transcriptional regulation controls a novel anti-apoptotic miRNA regulatory network for AML survival. Cell Death Differ 2015;22:654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Joo MS, Lee CG, Koo JH, Kim SG.. miR-125b transcriptionally increased by Nrf2 inhibits AhR repressor, which protects kidney from cisplatin-induced injury. Cell Death Dis 2013;4:e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kurinna S, Schäfer M, Ostano P, Karouzakis E, Chiorino G, Bloch W, Bachmann A, Gay S, Garrod D, Lefort K, Dotto G-P, Beer H-D, Werner S.. A novel Nrf2-miR-29-desmocollin-2 axis regulates desmosome function in keratinocytes. Nat Commun 2014;5:5099. [DOI] [PubMed] [Google Scholar]
- 36. Fan Y, Xia J.. miRNet—functional analysis and visual exploration of miRNA–target interactions in a network context. Comput Cell Biol 2018;18:215–233. [DOI] [PubMed] [Google Scholar]
- 37. Fan Y, Siklenka K, Arora SK, Ribeiro P, Kimmins S, Xia J.. miRNet – dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res 2016;44:W135–W141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pankratz F, Hohnloser C, Bemtgen X, Jaenich C, Kreuzaler S, Hoefer I, Pasterkamp G, Mastroianni J, Zeiser R, Smolka C, Schneider L, Martin J, Juschkat M, Helbing T, Moser M, Bode C, Grundmann S.. MicroRNA-100 suppresses chronic vascular inflammation by stimulation of endothelial autophagy. Circ Res 2018;122:417–432. [DOI] [PubMed] [Google Scholar]
- 39. Canfrán‐Duque A, Rotllan N, Zhang X, Fernández‐Fuertes M, Ramírez‐Hidalgo C, Araldi E, Daimiel L, Busto R, Fernández‐Hernando C, Suárez Y.. Macrophage deficiency of miR‐21 promotes apoptosis, plaque necrosis, and vascular inflammation during atherogenesis. EMBO Mol Med 2017;9:1244–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gao L, Zeng H, Zhang T, Mao C, Wang Y, Han Z, Chen K, Zhang J, Fan Y, Gu J, Wang C.. MicroRNA-21 deficiency attenuated atherogenesis and decreased macrophage infiltration by targeting Dusp-8. Atherosclerosis 2019;291:78–86. [DOI] [PubMed] [Google Scholar]
- 41. Jin H, Li DY, Chernogubova E, Sun C, Busch A, Eken SM, Saliba-Gustafsson P, Winter H, Winski G, Raaz U, Schellinger IN, Simon N, Hegenloh R, Matic LP, Jagodic M, Ehrenborg E, Pelisek J, Eckstein H-H, Hedin U, Backlund A, Maegdefessel L.. Local delivery of miR-21 stabilizes fibrous caps in vulnerable atherosclerotic lesions. Mol Ther 2018;26:1040–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang XY, Shen BR, Zhang YC, Wan XJ, Yao QP, Wu GL, Wang JY, Chen SG, Yan ZQ, Jiang ZL.. Induction of thoracic aortic remodeling by endothelial-specific deletion of microRNA-21 in mice. PLoS One 2013;8:e59002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RTA, Heyll K, Noels H, Hristov M, Wang S, Kiessling F, Olson EN, Weber C.. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med 2014;20:368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kansanen E, Kuosmanen SM, Leinonen H, Levonen A-L.. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol 2013;1:45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuosmanen SM, Kansanen E, Sihvola V, Levonen A-L.. MicroRNA profiling reveals distinct profiles for tissue-derived and cultured endothelial cells. Sci Rep 2017;7:10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kuosmanen SM, Sihvola V, Kansanen E, Kaikkonen MU, Levonen A-L.. MicroRNAs mediate the senescence-associated decline of NRF2 in endothelial cells. Redox Biol 2018;18:77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Toth R, Warfel N.. Strange bedfellows: nuclear factor, erythroid 2-like 2 (Nrf2) and hypoxia-inducible factor 1 (HIF-1) in tumor hypoxia. Antioxidants 2017;6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jeong G, Lim Y-H, Kim Y-K.. Precise mapping of the transcription start sites of human microRNAs using DROSHA knockout cells. BMC Genomics 2016;17:908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang-Yen H-F, Chambard J-C, Sun Y-L, Smeal T, Schmidt TJ, Drouin J, Karin M.. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell 1990;62:1205–1215. [DOI] [PubMed] [Google Scholar]
- 50. Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK.. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature 2005;437:759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. He CH, Gong P, Hu B, Stewart D, Choi ME, Choi AMK, Alam J.. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J Biol Chem 2001;276:20858–20865. [DOI] [PubMed] [Google Scholar]
- 52. Narzt MS, Nagelreiter IM, Oskolkova O, Bochkov VN, Latreille J, Fedorova M, Ni Z, Sialana FJ, Lubec G, Filzwieser M, Laggner M, Bilban M, Mildner M, Tschachler E, Grillari J, Gruber F.. A novel role for NUPR1 in the keratinocyte stress response to UV oxidized phospholipids. Redox Biol 2019;20:467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang J, Tabbi-Anneni I, Gunda V, Wang L.. Transcription factor Nrf2 regulates SHP and lipogenic gene expression in hepatic lipid metabolism. Am J Physiol Gastrointest Liver Physiol 2010;299:1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hargreaves DC, Horng T, Medzhitov R.. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 2009;138:129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Escoubet-Lozach L, Benner C, Kaikkonen MU, Lozach J, Heinz S, Spann NJ, Crotti A, Stender J, Ghisletti S, Reichart D, Cheng CS, Luna R, Ludka C, Sasik R, Garcia-Bassets I, Hoffmann A, Subramaniam S, Hardiman G, Rosenfeld MG, Glass CK.. Mechanisms establishing TLR4-responsive activation states of inflammatory response genes. PLoS Genet 2011;7:e1002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mimura J, Itoh K.. Role of Nrf2 in the pathogenesis of atherosclerosis. Free Radic Biol Med 2015;88:221–232. [DOI] [PubMed] [Google Scholar]
- 57. Niu N, Xu S, Xu Y, Little PJ, Jin ZG.. Targeting mechanosensitive transcription factors in atherosclerosis. Trends Pharmacol Sci 2019;40:253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Valcarcel-Ares MN, Gautam T, Warrington JP, Bailey-Downs L, Sdosnowska D, Cabo R de, Losonczy G, Sonntag WE, Ungvari Z, Csiszar A.. Disruption of Nrf2 signaling impairs angiogenic capacity of endothelial cells: implications for microvascular aging. J Gerontol Ser A Biol Sci Med Sci 2012;67:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wei Y, Gong J, Thimmulappa RK, Kosmider B, Biswal S, Duh EJ.. Nrf2 acts cell-autonomously in endothelium to regulate tip cell formation and vascular branching. Proc Natl Acad Sci U S A 2013;110:E3910–E3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Florczyk U, Jazwa A, Maleszewska M, Mendel M, Szade K, Kozakowska M, Grochot-Przeczek A, Viscardi M, Czauderna S, Bukowska-Strakova K, Kotlinowski J, Jozkowicz A, Loboda A, Dulak J.. Nrf2 regulates angiogenesis: effect on endothelial cells, bone marrow-derived proangiogenic cells and hind limb ischemia. Antioxid Redox Signal 2014;20:1693–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Grundmann S, Hans FP, Kinniry S, Heinke J, Helbing T, Bluhm F, Sluijter JPG, Hoefer I, Pasterkamp G, Bode C, Moser M.. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation 2011;123:999–1009. [DOI] [PubMed] [Google Scholar]
- 62. Zhao D, Tu Y, Wan L, Bu L, Huang T, Sun X, Wang K, Shen B.. In vivo monitoring of angiogenesis inhibition via down-regulation of mir-21 in a VEGFR2-Luc murine breast cancer model using bioluminescent imaging. PLoS One 2013;8:e71472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shoeibi S. Diagnostic and theranostic microRNAs in the pathogenesis of atherosclerosis. Acta Physiol 2020;228:e13353. [DOI] [PubMed] [Google Scholar]
- 64. Tesauro M, Mauriello A, Rovella V, Annicchiarico-Petruzzelli M, Cardillo C, Melino G, Daniele N Di.. Arterial ageing: from endothelial dysfunction to vascular calcification. J Intern Med 2017;281:471–482. [DOI] [PubMed] [Google Scholar]
- 65. Zhang H, Davies KJA, Forman HJ.. Oxidative stress response and Nrf2 signaling in aging. Free Radic Biol Med 2015;88:314–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang R, Dong L-D, Meng X-B, Shi Q, Sun W-Y.. Unique MicroRNA signatures associated with early coronary atherosclerotic plaques. Biochem Biophys Res Commun 2015;464:574–579. [DOI] [PubMed] [Google Scholar]
- 67. Parahuleva MS, Lipps C, Parviz B, Hölschermann H, Schieffer B, Schulz R, Euler G.. MicroRNA expression profile of human advanced coronary atherosclerotic plaques. Sci Rep 2018;8:7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Raitoharju E, Lyytikäinen L-P, Levula M, Oksala N, Mennander A, Tarkka M, Klopp N, Illig T, Kähönen M, Karhunen PJ, Laaksonen R, Lehtimäki T.. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis 2011;219:211–217. [DOI] [PubMed] [Google Scholar]
- 69. Darabi F, Aghaei M, Movahedian A, Elahifar A, Pourmoghadas A, Sarrafzadegan N.. Association of serum microRNA-21 levels with Visfatin, inflammation, and acute coronary syndromes. Heart Vessels 2017;32:549–557. [DOI] [PubMed] [Google Scholar]
- 70. Edgar R, Domrachev M, Lash AE.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data from this study have been submitted to NCBI Gene Expression Omnibus under accession number GSE136813. The public Hi-C and RNA-seq datasets used in this study have been deposited in the Gene Expression Omnibus (GEO)70 under accession numbers GSE89970 and GSE94872.
Translational Perspective
Our analysis provides deeper understanding of the transcriptional regulation of miRNAs and their target genes that could regulate the proatherogenic effects of oxidized phospholipids in endothelial cells. We further characterize the role of miR-21-5p and miR-100-5p in regulating the VEGF pathway upon proatherogenic stimuli. We demonstrate that exosomal levels of both miRNAs obtained from pericardial fluid of cardiovascular disease patients are correlated with disease severity. Thus, future studies should determine the utility of miR-21-5p and miR-100-5p for the diagnosis of cardiovascular disease.