Abstract
Canthin-6-one, one of the main alkaloid compounds extracted from Ailanthus altissima, has recently attracted increasing interest for its antifungal activity. To evaluate the potential of canthin-6-one in controlling plant fungal diseases, we investigated the antifungal activity of canthin-6-one isolated from A. altissima against Fusarium oxysporum f. sp. cucumerinum (Foc) in vitro. The mycelial growth rate and micro-broth dilution were used to test antifungal activity. Furthermore, label-free quantitative proteomics and parallel reaction monitoring (PRM) techniques were applied to analyze the antifungal mechanism. It was found that canthin-6-one significantly inhibited the growth of Foc, and had higher inhibitory action than chlorothalonil at the same concentration. Proteomic analysis showed that the expression of 203 proteins altered significantly after canthin-6-one treatment. These differentially expressed proteins were mainly involved in amino acid biosynthesis and nitrogen metabolism pathways. These results suggest that canthin-6-one significantly interferes with the metabolism of amino acids. Therefore, it affects nitrogen nutrients and disturbs the normal physiological processes of fungi, and ultimately leads to the death of pathogens. This study provides a natural plant antifungal agent and a new perspective for the study of antifungal mechanisms.
Introduction
Ailanthus altissima (Mill.) Swingle, belonging to the family Simaroubaceae, is a fast-growing deciduous tree that is native to Asia [1]. Phytochemical investigations have revealed that alkaloids, flavonoids, terpenoids, steroids, and many other biological components exist in A. altissima [2]. Among these compounds, alkaloids are the main and most common bioactive substances found in A. altissima [3]. Canthin-6-one, a subclass of β-carboline alkaloids with an additional D-ring, is one of the main alkaloids in A. altissima [4]. Many studies have shown that canthin-6-one possesses antiviral, anticancer, antimicrobial, and enzyme inhibitory properties [5]. In particular, the high antimicrobial activity of canthine-6-one has attracted extensive attention [6–8]. However, most of these studies have been associated with the medical field, and only a few studies have reported the effects of this compound on plant pathogens.
Plant diseases, particularly fungal diseases, can cause crop losses and exert serious biological pressure on food security [9, 10]. Fusarium is one of the most serious pathogenic fungi and can cause wilt, rot, and canker diseases in many plants [11, 12]. At present, chemical sterilization is the main control method for Fusarium disease. However, this method causes a series of problems, including chemical resistance, chemical residues, and environmental and health hazards [13]. Secondary metabolites isolated from plants have recently attracted increasing interest for their antifungal activity. The compounds derived from plants generally have the characteristics of biodegradation and low toxicity residues, which are safer to human health than synthetic chemicals [14]. In a preliminary study, we found that canthin-6-one isolated from A. altissima exhibited antifungal activity against some plant pathogenic fungi, such as F. graminearum, F. oxysporum f. sp. benincasae, F. oxysporum f. sp. cucumerinum (Foc), and Gloeosporium sp. Nevertheless, the antifungal activity and mechanism of canthin-6-one as a fungicide remain unclear. In this study, Foc was used as the fungal strain to investigate the inhibitory effect of canthin-6-one on mycelial growth. In addition, proteomics techniques have been used to determine the pathway of action of canthin-6-one.
Materials and methods
Preparation of canthin-6-one
Canthin-6-one was extracted and isolated from the stem bark of A. altissima, which was purchased from Jiaxu Pharmaceutical Co., Ltd., Bozhou City, Anhui Province, China, in June 2017. The specimen (No. 2017-AA02) was deposited in the Laboratory of Plant Resources, College of Chemistry and Life Science, Chifeng University, Inner Mongolia, China. The extraction and isolation of canthin-6-one was carried out using previously published methods [3]. The structure of canthin-6-one was determined using physical (m. p.) and spectral analyses (13C-NMR and 1H-NMR). The purity of canthin-6-one was evaluated using high-performance liquid chromatography (HPLC), and found to have a purity greater than 98%. Canthin-6-one was then dissolved in dimethyl sulfoxide (DMSO, 1% v/v).
Culture of fungal mycelia and preparation of spores
The fungal strain of Foc (No. 2532) was obtained from the China Center of Industrial Culture Collection (CICC), Beijing, China. The strain was inoculated in potato dextrose agar (PDA) medium (6 g L-1 potato extract, 20 g L-1 glucose, and 20 g L-1 agar) and cultured at 25°C for 6 days. Mycelia were used to prepare spore suspensions and to set up the antifungal activity assays. Mycelium disks (6 mm in diameter) were cut from 6-day-old cultures and suspended in potato dextrose broth (PDB) medium (6 g L-1 potato extract, 20 g L-1 glucose). After oscillation, filtration, and microscopic examination, the concentration of the suspension was maintained at approximately 1 × 106 spores mL-1.
Evaluation of the antifungal activity
MIC assay
The minimal inhibitory concentration (MIC) was determined using the microdilution method [15–17]. The canthin-6-one samples were diluted with PDB to prepare stock solutions of different concentrations. The stock solution (50 μL) and spore suspension (50 μL) were dispensed into each well of 96-well plates, and the canthin-6-one concentrations were adjusted to 1.0, 2.0, 4.0, 8.0, 16.0, 32.0, 64.0, and 128.0 μg mL-1. Wells without canthin-6-one were used as the blank controls. The plates were covered and incubated at 25°C. The MIC was defined as the lowest concentration at which no visual growth of fungal mycelium was observed when the fungal mycelium appeared in the control medium [18–20].
Inhibitory activity assay
Inhibitory activity against the fungal mycelium was tested using a previously described protocol with a few modifications [14]. PDA medium plates (90 mm in diameter) containing canthin-6-one at different concentrations (1.0, 2.0, 4.0, 8.0, and 16.0 μg mL-1) were prepared. Equal amounts of chlorothalonil and DMSO were added to PDA medium as a positive control and blank control, respectively. One mycelium disk (6 mm in diameter) was cut from the edge of 6-day-old mycelia cultures, placed in the center of each PDA plate, and incubated upside down at 25°C for 72 h. Each test was performed in triplicate. The crisscross method was used to measure the diameters of the fungal colonies. The mycelium growth inhibitory rate (%) was determined as follows: I (%) = (C—T)/(C—K) × 100, where C and T are the average diameters of the fungal colonies in the control and treatment, respectively, and K is the average diameter of the inoculum disk. Then, the inhibitory rate-dose response curve and linear regression equation were obtained. The effective concentration for 50% inhibition (EC50) was calculated using the regression equation [21].
Observation of hyphal morphology
The spore suspension was cultured in PDB medium at 25°C. After 48 h, canthin-6-one was added to the suspension. The final concentrations of canthin-6-one were 0, 8.0, 16.0, 32.0, 64.0, and 128.0 μg mL-1. The mixed solution was then incubated at 25°C for 48 h. The resulting mycelia were collected, cleaned with PBS (0.1 M, pH 7.0), and observed under a light microscope (Olympus BX53F, Olympus, Tokyo, Japan) at 400× magnification.
Stability test of antifungal activity
The canthin-6-one solutions (4.2 μg ml-1, EC50) were treated with ultraviolet radiation, acid-base treatment, and varying temperatures and storage time. The inhibition rate of mycelial growth was used to investigate the antimicrobial stability of canthin-6-one after different treatments. Ultraviolet irradiation treatment was carried out with a 30 W ultraviolet lamp for 0, 3, 6, 12, and 24 h. Temperature treatment was performed in a refrigerator (4°C), water bath (25°C, 50°C, 100°C), and a high-temperature sterilizing oven (121°C) for 1 h. The pH of the PDA medium containing canthin-6-one was adjusted to pH 3, 4, 5, 6, 7, 8, 9, and 10 with sodium hydroxide or hydrochloric acid, and the PDA medium containing DMSO with the same acid base was used as a control. The storage conditions were as follows: canthin-6-one solutions were stored in the dark at 25°C for 30, 60, 90, 120, and 180 d. The mycelium growth inhibitory rate was determined according to the method of inhibition activity assay.
Quantitative proteomics experiments
Protein extraction and digestion
The spore suspension was cultured in PDB medium at 25°C. After 24 h, a certain amount of canthin-6-one was added to the suspension, and the final concentration of canthin-6-one was 4.2 μg mL-1 (EC50). An equal amount of DMSO was added as a blank control. The mixed solution was then incubated at 25°C for 24 h. The mycelia were collected and washed three times with 1× PBS for protein extraction. Three biological replicates were used for each sample. Protein extraction and digestion were performed according to a previously described method [22]. Proteins were extracted using SDT buffer (4% sodium dodecyl sulfate, 0.1 M dithiothreitol, 100 mM Tris/HCl, pH 7.6), and then quantified using the BCA protein assay reagent (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The filter-aided proteome preparation (FASP) method [23] was used to trypsinize (Promega, Madison, WI, USA), the appropriate amount of protein from each sample, and then the C18 cartridge (Empore™ SPE, bed I.D. 7 mm, volume 3 mL; Sigma, Kawasaki, Japan) was used to desalinize the hydrolyzed peptide. Dissolution buffer (40 μL, 0.1% trifluoroacetic acid) was added to the lyophilized peptides for reconstitution, and the peptide content was estimated using UV spectroscopy at 280 nm.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis
LC-MS/MS experiments were performed on a Q Exactive mass spectrometer coupled to an Easy nLC system (Thermo Fisher Scientific, Waltham, MA, USA), and the procedure was performed according to Li et al. [22]. First, the peptides of each sample were injected into a C18 column (Easy, 10 cm long, 75 μm I.D., 3 μm resin; Thermo Scientific) in buffer A (0.1% formic acid), and then separated with buffer B (0.1% formic acid and 84% acetonitrile) at a flow rate of 250 nL min-1. The solvent gradient was as follows: 0–35% buffer B for 50 min, 35–100% buffer B for 8 min, and then 100% buffer B for 3 min. MS analysis was performed using Q Exactive (Thermo Fisher Scientific). The survey scan range of precursor ions was 300–1800 m/z, and the 10 most abundant polypeptide fragments were dynamically selected. The duration of dynamic exclusion was 40.0 s. The target value was determined using an automatic gain control (AGC). Survey scans were obtained at a resolution of 70,000 at m/z 200, and the resolution for the HCD spectra was 17,500 at m/z 200. The normalized collision energy was 30 eV and the underfill ratio was 0.1%. The MS data were compared with the UniProt Fusarium oxysporum database (342953 sequences, downloaded May 2019) using MaxQuant software (v.1.5.5.1). Trypsin was used as a cleavage enzyme, allowing a maximum of two missing cleavages. The peptide mass tolerance was set to 20 ppm, and the main search used a mass window of 6 ppm. Carbamidomethylation of cysteine and oxidation of methionine were defined as fixed and variable modifications, respectively. The false discovery rate (FDR) for peptide and protein identification was set at ≤0.01.
Bioinformatics methods
Gene ontology (GO) annotations and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed using the software Omicsbean (http://www.omicsbean.cn/) to identify differentially expressed proteins [24]. Protein-protein interaction (PPI) networks were analyzed using the STRING database (https://string-db.org/). Cytoscape open-source software (https://cytoscape.org/) was used for visualization, and the orphan protein was deleted.
Parallel Reaction Monitoring (PRM) analysis
Additional quantification was performed by liquid chromatography parallel reaction monitoring mass spectrometry (LC-PRM/MS) to further check the changes in protein expression. Protein extraction and digestion were performed according to the procedure described in “Protein extraction and digestion”. The PRM analysis was performed using a Q Exactive mass spectrometer and an Easy nLc system (Thermo Fisher Scientific, Bremen, Germany). The digested peptide and iRT-Kit peptide (Biognosys AG, Zurich, Switzerland) were mixed in equal amounts as an internal standard. Each sample was detected by LC-PRM/MS, and PRM data analysis was performed using the Skyline software (3.5.0) [22]. Significant differences were determined by Student’s t-test at a significance level of p < 0.05.
Results
Inhibition of canthin-6-one against Fusarium oxysporium f. sp. cucumerinum
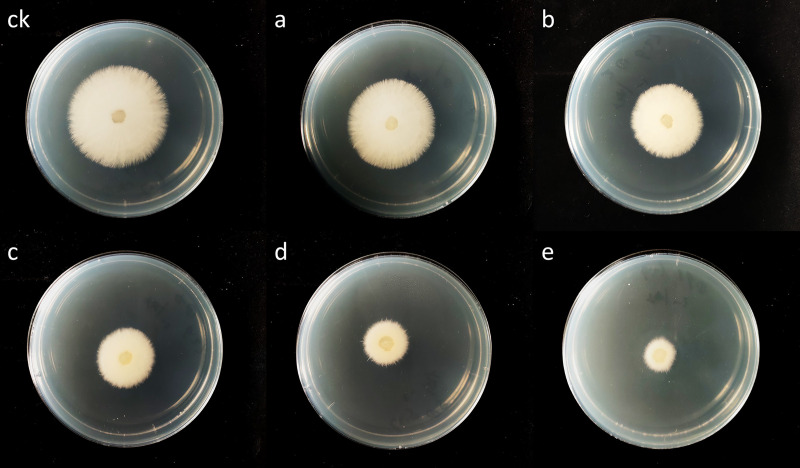
The MIC was used to quantitatively evaluate the antifungal activity. In this study, the MIC value of canthin-6-one against Foc was detected through fungal mycelium growth or no growth, with a value of 32.0 μg mL-1. The results showed that canthin-6-one exhibited strong antifungal activity against the tested fungal strains. The inhibitory effect of canthin-6-one on mycelial growth in vitro was concentration-dependent. As shown in Fig 1, the higher the concentration of canthin-6-one, the more obvious the inhibition of mycelial growth.
Fig 1. Inhibition effect of canthin-6-one on mycelium growth (72 h).
ck, blank control; a–e, treatment with 1.0, 2.0, 4.0, 8.0, 16.0 μg mL-1 canthin-6-one, respectively.
The inhibition rate reached over 81% at the highest concentration of canthin-6-one (16.0 μg mL-1), which was clearly superior to that of chlorothalonil (40.1%) (Table 1). The EC50 value calculated by the regression equation was approximately 4.2 μg mL-1.
Table 1. Effect of canthin-6-one on mycelium growth.
| Treatment | Concentration (μg mL-1) | Colony diameter (mm) | Inhibition rate (%) |
|---|---|---|---|
| Canthin-6-one | 0.0 | 39.2 ± 0.2a | |
| 1.0 | 33.2 ± 0.3b | 15.5 ± 0.94f | |
| 2.0 | 26.4 ± 0.6c | 32.8 ± 1.42e | |
| 4.0 | 20.5 ± 0.5e | 47.9 ± 0.98c | |
| 8.0 | 13.0 ± 0.2f | 66.8 ± 0.65b | |
| 16.0 | 7.3 ± 0.2g | 81.5 ± 0.61a | |
| Chlorothalonil | 16.0 | 22.9 ± 0.6d | 40.1 ± 1.55d |
Note: The values are the mean of three replicates ± SD (n = 3). Different letters after means indicate significant differences at p < 0.05.
Hyphal morphology was observed using a light microscope at 400× magnification. Compared with the control, the fungal hyphal grown in PDB containing alkaloids showed alterations in morphology (Fig 2). After treatment with canthin-6-one, the hyphal became thinner, branching decreased, and spore formation was reduced. At the highest concentration (128.0 μg mL-1), the morphology of hyphal changed significantly. These results suggested that canthin-6-one significantly reduced the hyphal branching, and destroyed the morphology of the hyphal.
Fig 2. The effect of canthin-6-one on hyphal morphology.
ck, blank control; a–e, treatment with 8.0, 16.0, 32.0, 64.0, 128.0 μg mL-1 canthin-6-one, respectively.
Analysis of the antifungal stability of canthin-6-one
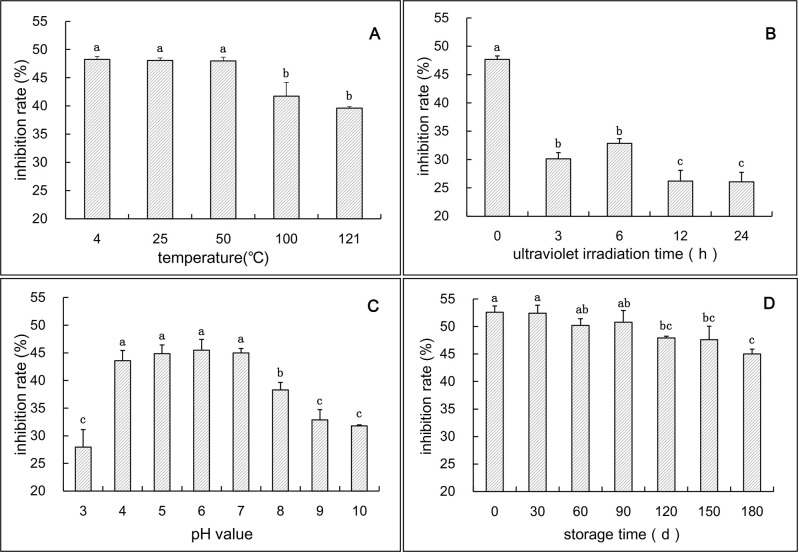
The antimicrobial ability of active compounds is usually affected by certain conditions, such as temperature, ultraviolet radiation, pH, and storage time. In this study, the mycelium growth inhibitory rate of canthin-6-one treated with different external factors was determined. The results showed that the antifungal activity of canthin-6-one was not significantly altered at lower temperatures (4°C, 25°C, and 50°C), and the inhibition rates were 48.27%, 48.07%, and 47.97%, respectively (Fig 3A). After high-temperature treatment (100°C and 121°C), the antifungal activity decreased to 41.75% and 39.61%, respectively, indicating that canthin-6-one was thermally unstable and high-temperature treatment should be avoided. As shown in Fig 3B, the antifungal activity of canthin-6-one decreased after UV irradiation treatment (p < 0.05). Compared with the control, the mycelial growth inhibition rate decreased by 37–45% after 3–24 h of UV irradiation. Under different pH conditions, canthin-6-one showed different antifungal activity. In this study, the effect of acid or alkali treatment on the antifungal activity of this compound was analyzed. The results showed that the antifungal ability of canthin-6-one was significantly obvious under the condition of pH 4–7 compared with pH 3 or pH 8–10 (p < 0.05), and the mycelial growth inhibition rate reached 43.57–45.5% (Fig 3C). The validity period is an important indicator of antifungal agents, and the effect of storage time on the antifungal activity of canthin-6-one was analyzed. The results indicated that the mycelium growth inhibition rate did not change significantly between 0–3 months. In the 3–6 month storage period, the inhibition rate decreased slightly (Fig 3D).
Fig 3. Antifungal activity of canthin-6-one alkaloid treated by different factors.
A, temperature treatment; B, ultraviolet irradiation treatment; C, acid or alkali treatment; D, storage time. Different letters indicate significant differences (p < 0.05).
Quantitative proteomics analysis
Changes in Foc protein expression after canthin-6-one treatment
Quantitative proteomics analysis was carried out using a label-free approach to elucidate the possible antifungal mechanism of canthin-6-one against Foc. Based on the database of Fusarium oxysporum, 1389 proteins with quantitative information were identified. The expression of 1186 proteins (85.4%) of mycelia did not show obvious differences between the treatment (canthin-6-one) and control (ck) groups. Compared with the control groups, the expression of 203 proteins in treatment groups changed, 123 of which were down-regulated, and 80 of them were up-regulated (fold changes > 1.5, p < 0.05) (S1 Fig, S1 Table). The screening of these differentially expressed proteins was reasonable and could effectively distinguish the comparison groups according to the hierarchical clustering heatmap (S2 Fig). These results indicated that canthin-6-one stimulated physiological responses and induced different expression of some proteins in the tested fungal strain.
Bioinformatics analysis of differentially expressed proteins
The properties and functions of these differentially expressed proteins were elucidated using bioinformatics analyses (S2 Table). GO annotations were performed in biological process (BP), cellular component (CC), and molecular function (MF) [25, 26]. According to the BP, these proteins were mainly involved in the organic acid metabolic process, oxoacid metabolic process, small molecule metabolic process, carboxylic acid metabolic process, and organonitrogen compound metabolic process. CC analysis showed that masses of proteins were localized intracellularly, mainly in the cytoplasm. Meanwhile, a large number of these proteins participated in pyridoxal phosphate binding, transferase activity, transferring glycosyl groups, cofactor binding, and antioxidant activity, according to the analysis of MF (S3 Fig).
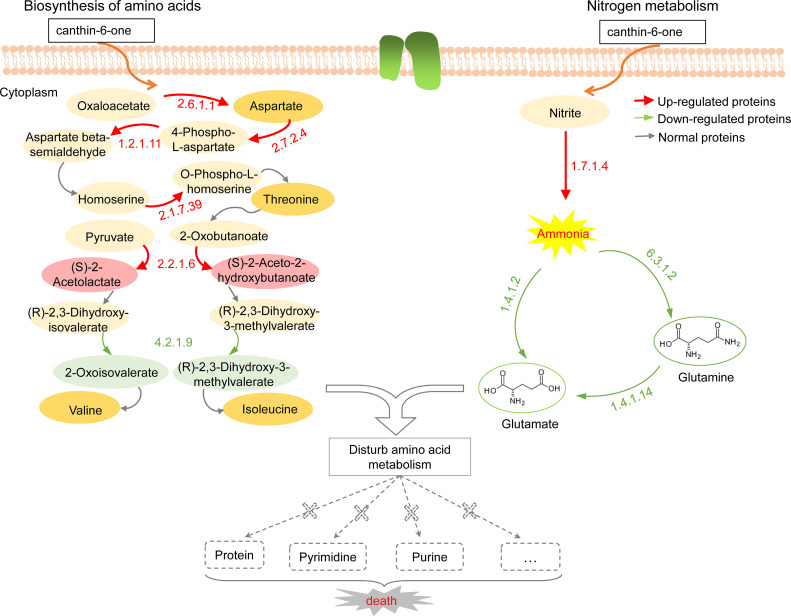
The KEGG database was used to identify the pathways involved in differentially expressed proteins [27]. Through KEGG pathway analysis, two main metabolic pathways, amino acid biosynthesis and nitrogen metabolism pathways, were locked (S4 Fig). These pathways are known to play a significant role in amino acid metabolism. The detailed pathway is shown in Fig 4. In the pathway of amino acid biosynthesis, the expression of many enzymes was changed, upregulated, or downregulated, which could affect normal amino acid metabolism. For example, the expression of acetolactate synthase (EC 2.2.1.6) was upregulated, which greatly increased the biosynthesis of (S)-2-Acetolactate and (S)-2-Aceto-2-hydroxybutanoate. However, in the subsequent step, the downregulated expression of dihydroxyacid dehydratase (EC 4.2.1.9) significantly inhibited the normal synthesis of the intermediates 2-Oxoisovalerate and (S)-3-Methyl-2-oxopentanoate, which resulted in the blockage of this synthetic route and the shortage of valine and isoleucine. Similarly, overexpressed proteins were also found in threonine synthesis, potentially causing imbalances in these intermediate metabolites and further disrupting normal metabolic pathways. In another pathway, nitrogen metabolism is disturbed by the expression of some enzymes. The expression of nitrite reductase (EC 1.7.1.4) was upregulated, but glutamate synthetase (EC 1.4.1.14), glutamate dehydrogenase (EC 1.4.1.2), and glutamine synthetase (EC 6.3.1.2) were downregulated in subsequent steps. The change in the expression of these enzymes may lead to an excessive accumulation of ammonia and a reduction of glutamate and glutamine. Amino acids are key materials for the synthesis of proteins, nucleic acids, phospholipids, and vitamins, among others. Ammonia is an important substance in nitrogen nutrition, but excessive ammonia is harmful to mycelial growth. The reduction of glutamate and glutamine directly disrupts the synthesis of purine and pyrimidine, which further affects nucleic acid metabolism. Thus, canthin-6-one could significantly regulate the expression of some critical enzymes, which disturbed the normal amino acid metabolism process. These consequences might further damage the metabolism of proteins, nucleic acids, or other substances, resulting in the death of pathogens.
Fig 4. Schematic of amino acid biosynthesis and nitrogen metabolism pathways in response to canthin-6-one.
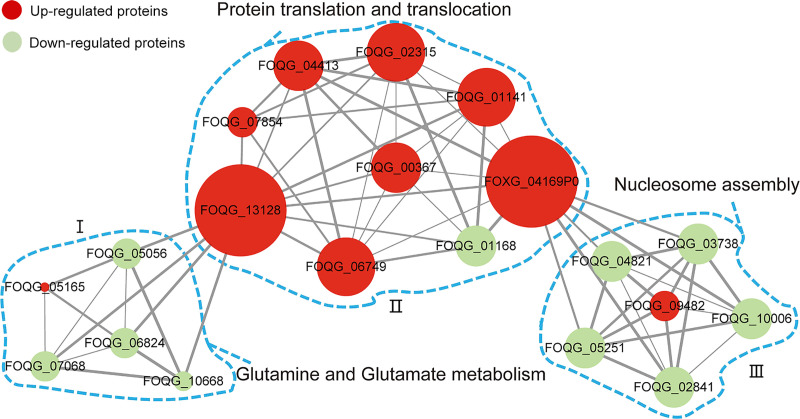
To further understand the functions and associations of these differentially expressed proteins, protein-protein interaction networks were established using the STRING database [28]. In this section, the 20 highest-scoring differentially expressed proteins with strong interactions were screened for interaction network analysis. As shown in Fig 5, the proteins were divided into three different clusters. The connections of Part I indicated that the proteins involved in amino acid biosynthesis were significantly disturbed. The process of protein translation and post-translational translocation was also seriously affected in Part II. In addition, the connections of Part III indicated that proteins involved in nucleosome assembly might be triggered by protein folding and degradation systems, which affect the nucleic acid metabolism pathway. These interactions suggest that canthin-6-one may significantly affect various physiological processes and cause pathogen death.
Fig 5. Analysis of protein-protein interaction network for differentially expressed proteins.
The nodes represent proteins (down-regulated and up-regulated), and edges represent interaction between proteins. Node size and edge width indicate the number of connected nodes and the strength of interaction, respectively.
Quantitative analysis of protein by PRM
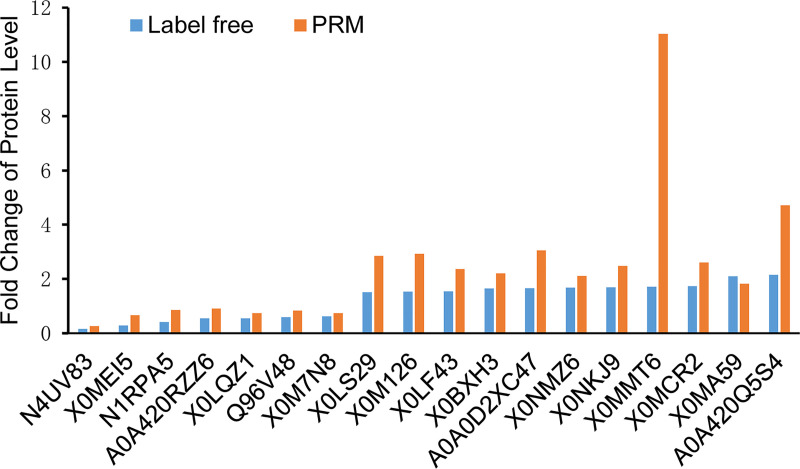
The detection of related gene transcription and PRM are considered the two main methods to further evaluate the expression levels of target proteins [29]. Compared with gene transcription detection, the PRM technique is more accurate and comprehensive in quantitative protein analysis [30–33]. Therefore, PRM was used to further verify the results of label-free quantitative proteomics in this study. According to the results of the bioinformatics analysis, 18 differentially expressed proteins were selected for PRM analysis (S3 Table). The fold change of protein levels analyzed using PRM and label-free quantitative proteomics is shown in Fig 6. The results showed that the tendency of 18 proteins identified using PRM was similar to the label-free quantitative proteomics data, demonstrating that the label-free results were reliable.
Fig 6. PRM analysis for 18 selected differentially expressed proteins.
Discussion
Plants are an important biological resource and contain many compounds that play an important role in metabolic disorders [34]. Owing to their biodegradability and low toxicity, plant-derived compounds are widely used in the development of biological pesticides. A. altissima is a resourceful tree that contains many bioactive substances. Canthin-6-one is an important bioactive substance in A. altissima, and its antifungal ability is worthy of attention. In this study, the antifungal activity and mechanism of canthin-6-one isolated from A. altissima against Foc were evaluated.
Canthin-6-one from A. altissima inhibits the growth of Foc
Antifungal activity analysis showed that canthin-6-one is able to inhibit the growth of Foc pathogen, with an MIC value of 32 μg mL-1. The level of antifungal activity observed here is similar to that reported for canthin-6-one isolated from Zanthoxylum chiloperone var. angustifolium against Candida albicans, Aspergillus fumigatus, and Trichophyton mentagrophytes var. interdigitale [35]. Mycelium growth is an important indicator of fungi. In this study, we analyzed the effect of canthin-6-one on the mycelial growth and morphology of Foc. The results indicated that canthin-6-one could significantly inhibit the growth and damage the structure of Foc (Table 1, Fig 2). It was also found that the inhibition rate of canthin-6-one was superior to chlorothalonil at the same concentration. Based on these preliminary results, canthin-6-one may be a natural antifungal candidate for further exploration and development in future studies.
The antifungal activity of canthin-6-one from A. altissima is relatively stable
Antifungal stability analysis (Fig 3) showed that canthin-6-one was relatively stable. The stability of antifungal activity at 4–50°C (Fig 3A) suggested that canthin-6-one has a wide range of temperature applications and is not affected by temperature in agricultural production. In neutral and acidic environments, canthin-6-one maintained a high antifungal activity (Fig 3C). In recent years, soil acidification has become an increasingly serious issue owing to the influence of acid rain and chemical fertilizers. Under acidic soil conditions, canthin-6-one may be advantageous as an antifungal agent. In addition, the antifungal activity of canthin-6-one decreased slightly in the dark at 25°C within 6 months (Fig 3D). In contrast, canthin-6-one was sensitive to ultraviolet irradiation (Fig 3B). Therefore, canthin-6-one needs to be stored away from light. The degradability of canthin-6-one under ultraviolet light is beneficial for its application in eco-friendly agriculture.
Canthin-6-one from A. altissima disturbs amino acid biosynthesis and nitrogen metabolism of Foc
Understanding the antifungal mechanism of canthin-6-one is key for the development and utilization of antifungal agents. The mechanism of action of canthin-6-one against Saccharomyces cerevisiae was previously reported to affect fatty acid metabolism by stimulating desaturase enzyme systems [7]. Further research has shown that canthin-6-one can permeate the cell membrane by binding to the sterol part of the membrane of S. cerevisiae, causing the death of the fungus [36]. These studies partially explained the antifungal mechanism of canthin-6-one. In recent years, quantitative proteomics has become a useful approach for revealing antimicrobial mechanisms [28, 37]. The combination of label-free quantitative proteomics and PRM-based targeted mass spectrometry is a sensitive and accurate method for protein quantification [22, 37].
In this study, label-free quantitative proteomics was applied to investigate the antifungal effect of canthin-6-one against Foc at the proteome level. The protein expression of two groups of samples, the control and samples treated with canthin-6-one, were detected by LC-MS/MS. The results showed that 203 proteins were differentially expressed between the two groups, indicating that canthin-6-one affected the expression of proteins. Through GO and KEGG bioinformatics analysis, these differentially expressed proteins were found to be mainly involved in amino acid biosynthesis and nitrogen metabolism pathways. Among these metabolic pathways, glutamine synthetase is considered the central and key enzyme for nitrogen assimilation and glutamine biosynthesis [38]. The expression of glutamine synthetase (EC 6.3.1.2) was significantly downregulated after canthin-6-one treatment. The downregulated expression of this enzyme disrupted the synthesis of glutamate and glutamine (Fig 4), and may further affect the metabolism of other amino acids, nucleic acids, or other substances. In addition, it was found that amino acid transporters, such as dicarboxylic acid amino acid permease, were strongly downregulated, which has the potential to interfere with the transport of amino acids. Protein-protein interaction analysis of these differentially expressed proteins also showed that canthin-6-one disrupted multiple physiological processes, amino acid metabolism, protein translation and post-translational translocation, and nucleosome assembly (Fig 5). Normal metabolism is essential for mycelial growth; however, the blockage or disturbance of the metabolic pathway can cause a series of problems, such as biochemical disorders, growth inhibition, and cell death. Some studies have shown that amino acids are the main source of nutrients for fungi, and their metabolic pathways and transported elements could be used as targets for antifungal drugs [39, 40]. Therefore, the metabolic process of amino acids is blocked, which can be fatal to the nutrition and growth of fungi. In this study, the synthesis of branched-chain amino acids was also found to be severely disturbed in the amino acid biosynthesis pathway, providing an insight into potential antifungal targets of canthin-6-one.
In summary, canthin-6-one effectively inhibited the growth of Foc, further confirming its antifungal potential against plant pathogens. Proteomic analysis revealed that canthin-6-one could significantly interfere with the metabolism of amino acids and affect the fungal nutrients, resulting in the disorder of normal physiological processes and the death of pathogens. Thus, amino acid metabolic pathways and transported elements may be targets for canthin-6-one action. This study provides an insight into the molecular mechanism of canthin-6-one against plant pathogens and provides a natural antifungal agent candidate derived from plants for the protection of plants during agricultural production. Nevertheless, more studies are still needed to evaluate the potential use of canthin-6-one as an antifungal agent and to elucidate the complete mechanism of its antifungal action.
Supporting information
Venn diagrams (a) and Volcano plot (b) of expressed proteins between control and treatment groups.
(TIF)
The relative abundance of proteins is represented by different colors, where red represents higher intensity and blue represents lower intensity.
(TIF)
GO analysis of biological process (a), cellular component (b), and molecular function (c).
(TIF)
The vertical axis represents the enriched KEGG classification (fold > 1.5, p < 0.05). The horizontal axis is the rich factor (rich factor ≤ 1), representing the ratio of the number of differentially expressed proteins to those identified in the KEGG pathway. The size of the circular area represents the number of differentially expressed proteins, and the circular color represents the enrichment p-value of the differentially expressed proteins under the KEGG classification.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The authors are grateful to the Chifeng University Microbiology Laboratory in Inner Mongolia Province, China for their assistance.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by Scientific and Technological Research project of Higher Learning Institutions in Inner Mongolia Autonomous Region of China (grant no.: NJZY19215). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Liao Y-Y, Guo Y-H, Chen J-M, Wang Q-F. Phylogeography of the widespread plant Ailanthus altissima (Simaroubaceae) in China indicated by three chloroplast DNA regions. J Syst Evol. 2014;52(2): 175–185. 10.1111/jse.12065 [DOI] [Google Scholar]
- 2.Sladonja B, Susek M, Guillermic J. Review on Invasive Tree of Heaven (Ailanthus altissima (Mill.) Swingle) Conflicting Values: Assessment of Its Ecosystem Services and Potential Biological Threat. Environ Manage. 2015;56(4): 1009–1034. Epub 2015/06/15. 10.1007/s00267-015-0546-5 [DOI] [PubMed] [Google Scholar]
- 3.Kim HM, Lee JS, Sezirahiga J, Kwon J, Jeong M, Lee D, et al. A New Canthinone-Type Alkaloid Isolated from Ailanthus altissima Swingle. Molecules. 2016;21(5): 642. Epub 2016/05/20. 10.3390/molecules21050642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao F, Dai JK, Liu D, Wang SJ, Wang JR. Synthesis and Evaluation of Ester Derivatives of 10-Hydroxycanthin-6-one as Potential Antimicrobial Agents. Molecules. 2016;21(3): 390. Epub 2016/03/24. 10.3390/molecules21030390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai JK, Dan WJ, Li N, Du HT, Zhang JW, Wang JR. Synthesis, in vitro antibacterial activities of a series of 3-N-substituted canthin-6-ones. Bioorg Med Chem Lett. 2016;26(2): 580–583. Epub 2015/12/19. 10.1016/j.bmcl.2015.11.070 [DOI] [PubMed] [Google Scholar]
- 6.Omoto T, Sung YJSZ. Antimycotic Substances in the Crude Drugs Ⅱ. 1982;36: 307–314. [Google Scholar]
- 7.Lagoutte D, Nicolas V, Poupon E, Fournet A, Hocquemiller R, Libong D, et al. Antifungal canthin-6-one series accumulate in lipid droplets and affect fatty acid metabolism in Saccharomyces cerevisiae. Biomed Pharmacother. 2008;62(2): 99–103. Epub 2007/09/25. 10.1016/j.biopha.2007.07.014 [DOI] [PubMed] [Google Scholar]
- 8.Soriano-Agatón F, Lagoutte D, Poupon E, Roblot F, Fournet A, Gantier J-C, et al. Extraction, Hemisynthesis, and Synthesis of Canthin-6-one Analogues. Evaluation of Their Antifungal Activities. J Nat Prod. 2005;68(11): 1581–1587. 10.1021/np050250z [DOI] [PubMed] [Google Scholar]
- 9.Pennisi E. Armed and Dangerous. Science. 2010;327(5967): 804–805. 10.1126/science.327.5967.804 [DOI] [PubMed] [Google Scholar]
- 10.Osaki-Oka K, Suyama S, Sakuno E, Ushijima S, Nagasawa E, Maekawa N, et al. Antifungal activity of the volatile compound isovelleral produced by ectomycorrhizal Russula fungi against plant-pathogenic fungi. J Gen Plant Pathol. 2019;85(6): 428–435. 10.1007/s10327-019-00872-8 [DOI] [Google Scholar]
- 11.Ma L-J, Geiser DM, Proctor RH, Rooney AP, O’Donnell K, Trail F, et al. Fusarium Pathogenomics. Annu Rev Microbiol. 2013;67(1): 399–416. 10.1146/annurev-micro-092412-155650 [DOI] [PubMed] [Google Scholar]
- 12.Gao X, Li K, Ma Z, Zou H, Jin H, Wang J. Cucumber Fusarium wilt resistance induced by intercropping with celery differs from that induced by the cucumber genotype and is related to sulfur-containing allelochemicals. Sci Hortic. 2020; 271. 10.1016/j.scienta.2020.109475 [DOI] [Google Scholar]
- 13.Nguvo KJ, Gao X. Weapons hidden underneath: bio-control agents and their potentials to activate plant induced systemic resistance in controlling crop Fusarium diseases. J Plant Dis Prot. 2019;126(3): 177–190. 10.1007/s41348-019-00222-y [DOI] [Google Scholar]
- 14.Liu X, Ouyang C, Wang Q, Li Y, Yan D, Yang D, et al. Evaluation of antibacterial and antifungal properties of 9-oxo-10,11-dehydroageraphorone extracted from Eupatorium adenophorum. J Plant Dis Prot. 2016;123(2): 93–99. 10.1007/s41348-016-0006-3 [DOI] [Google Scholar]
- 15.Gulluce M, Adiguzel A, Ogutcu H, Sengul M, Karaman I, Sahin F. Antimicrobial effects of Quercus ilex L. extract. Phytother Res. 2004;18(3): 208–211. Epub 2004/04/23. 10.1002/ptr.1419 [DOI] [PubMed] [Google Scholar]
- 16.Gulluce M, Sahin F, Sokmen M, Ozer H, Daferera D, Sokmen A, et al. Antimicrobial and antioxidant properties of the essential oils and methanol extract from Mentha longifolia L. ssp. longifolia. Food Chem. 2007;103(4): 1449–1456. 10.1016/j.foodchem.2006.10.061 [DOI] [Google Scholar]
- 17.Li Y, Li J, Li Y, Wang XX, Cao AC. Antimicrobial Constituents of the Leaves of Mikania micrantha H. B. K. PLoS One. 2013;8(10): e76725. Epub 2013/10/08. 10.1371/journal.pone.0076725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helal GA, Sarhan MM, Abu Shahla AN, Abou El-Khair EK. Effects of Cymbopogon citratus L. essential oil on the growth, lipid content and morphogenesis of Aspergillus niger ML2-strain. J Basic Microbiol. 2006;46(6): 456–469. Epub 2006/12/02. 10.1002/jobm.200510106 [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Ouyang C, Li Y, Yang D, Fang W, Yan D, et al. Evaluation of the antimicrobial activity of 9-oxo-agerophorone against soil borne pathogens. J Plant Dis Prot. 2016;123(4): 163–170. 10.1007/s41348-016-0022-3 [DOI] [Google Scholar]
- 20.Zhang JH, Sun HL, Chen SY, Zeng L, Wang TT. Anti-fungal activity, mechanism studies on alpha-Phellandrene and Nonanal against Penicillium cyclopium. Bot Stud. 2017;58(1): 13. Epub 2017/05/17. 10.1186/s40529-017-0168-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiang L, Xue M, Yang L, Gong S, Yu D. Bionic fungicide physcion controls gray mold in tomato: possible modes of action. J Gen Plant Pathol. 2018;85(1): 57–65. 10.1007/s10327-018-0821-4 [DOI] [Google Scholar]
- 22.Li S, Yu Q, Han L, Zhang Y, Tian X, Zhao S. Effects of proteome changes on the tenderness of yak rumen smooth muscle during postmortem storage based on the label-free mass spectrometry. Food Res Int. 2019;116: 1336–1343. Epub 2019/02/06. 10.1016/j.foodres.2018.10.023 . [DOI] [PubMed] [Google Scholar]
- 23.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5): 359–362. Epub 2009/04/21. 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- 24.Zhao L, Zhang H, Wang J, Tian L, Li F, Liu S, et al. C60 Fullerols Enhance Copper Toxicity and Alter the Leaf Metabolite and Protein Profile in Cucumber. Environ Sci Technol. 2019;53(4): 2171–2180. Epub 2019/01/19. 10.1021/acs.est.8b06758 . [DOI] [PubMed] [Google Scholar]
- 25.Song X, Li P, Li M, Yang A, Yu L, Luo L, et al. Synthesis and investigation of the antibacterial activity and action mechanism of 1,3,4-oxadiazole thioether derivatives. Pestic Biochem Physiol. 2018;147: 11–19. Epub 2018/06/24. 10.1016/j.pestbp.2017.10.011 [DOI] [PubMed] [Google Scholar]
- 26.Shi J, Yu L, Song B. Proteomics analysis of Xiangcaoliusuobingmi-treated Capsicum annuum L. infected with Cucumber mosaic virus. Pestic Biochem Physiol. 2018;149: 113–22. Epub 2018/07/24. 10.1016/j.pestbp.2018.06.008 [DOI] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42(Database issue): D199–205. Epub 2013/11/12. 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y-L, Huang X, Liu L-W, Wang P-Y, Long Q-S, Tao Q-Q, et al. Identification of Racemic and Chiral Carbazole Derivatives Containing an Isopropanolamine Linker as Prospective Surrogates against Plant Pathogenic Bacteria: In Vitro and In Vivo Assays and Quantitative Proteomics. J Agri Food Chem. 2019;67(26): 7512–7525. 10.1021/acs.jafc.9b02036 [DOI] [PubMed] [Google Scholar]
- 29.Long Q-S, Liu L-W, Zhao Y-L, Wang P-Y, Chen B, Li Z, et al. Fabrication of Furan-Functionalized Quinazoline Hybrids: Their Antibacterial Evaluation, Quantitative Proteomics, and Induced Phytopathogen Morphological Variation Studies. J Agri Food Chem. 2019;67(40): 11005–11017. 10.1021/acs.jafc.9b03419 [DOI] [PubMed] [Google Scholar]
- 30.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13(4): 227–232. Epub 2012/03/14. 10.1038/nrg3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Beyer A, Aebersold R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 2016;165(3): 535–550. Epub 2016/04/23. 10.1016/j.cell.2016.03.014 [DOI] [PubMed] [Google Scholar]
- 32.Rauniyar N. Parallel Reaction Monitoring: A Targeted Experiment Performed Using High Resolution and High Mass Accuracy Mass Spectrometry. Int J Mol Sci. 2015;16(12): 28566–28581. Epub 2015/12/04. 10.3390/ijms161226120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang BZ, Meng E, Zhang HJ, Zhang XM, Asgari S, Lin YP, et al. Combination of label-free quantitative proteomics and transcriptomics reveals intraspecific venom variation between the two strains of Tetrastichus brontispae, a parasitoid of two invasive beetles. J Proteomics. 2019;192: 37–53. Epub 2018/08/12. 10.1016/j.jprot.2018.08.003 [DOI] [PubMed] [Google Scholar]
- 34.Truba J, Stanislawska I, Walasek M, Wieczorkowska W, Wolinski K, Buchholz T, et al. Inhibition of Digestive Enzymes and Antioxidant Activity of Extracts from Fruits of Cornus alba, Cornus sanguinea subsp. hungarica and Cornus florida-A Comparative Study. Plants (Basel). 2020;9(1): 122. Epub 2020/01/23. 10.3390/plants9010122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thouvenel C, Gantier JC, Duret P, Fourneau C, Hocquemiller R, Ferreira ME, et al. Antifungal compounds from Zanthoxylum chiloperone var. angustifolium. Phytother Res. 2003;17(6): 678–680. Epub 2003/06/24. 10.1002/ptr.1137 [DOI] [PubMed] [Google Scholar]
- 36.Gazoni VF, Balogun SO, Arunachalam K, Oliveira DM, Filho VC, Lima SR, et al. Assessment of toxicity and differential antimicrobial activity of methanol extract of rhizome of Simaba ferruginea A. St.-Hil. and its isolate canthin-6-one. J Ethnopharmacol. 2018;223: 122–134. Epub 2018/05/18. 10.1016/j.jep.2018.05.014 [DOI] [PubMed] [Google Scholar]
- 37.Kang S, Kong F, Liang X, Li M, Yang N, Cao X, et al. Label-Free Quantitative Proteomics Reveals the Multitargeted Antibacterial Mechanisms of Lactobionic Acid against Methicillin-Resistant Staphylococcus aureus (MRSA) using SWATH-MS Technology. J Agric Food Chem. 2019;67(44): 12322–12332. Epub 2019/10/23. 10.1021/acs.jafc.9b06364 [DOI] [PubMed] [Google Scholar]
- 38.Margelis S, D’Souza C, Small AJ, Hynes MJ, Adams TH, Davis MA. Role of glutamine synthetase in nitrogen metabolite repression in Aspergillus nidulans. J Bacteriol. 2001;183(20): 5826–5833. Epub 2001/09/22. 10.1128/JB.183.20.5826-5833.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishida I, Watanabe D, Tsolmonbaatar A, Kaino T, Ohtsu I, Takagi H. Vacuolar amino acid transporters upregulated by exogenous proline and involved in cellular localization of proline in Saccharomyces cerevisiae. J Gen Appl Microbiol. 2016;62(3): 132–139. Epub 2016/06/02. 10.2323/jgam.2016.01.005 [DOI] [PubMed] [Google Scholar]
- 40.McCarthy MW, Walsh TJ. Amino Acid Metabolism and Transport Mechanisms as Potential Antifungal Targets. Int J Mol Sci. 2018;19(3): 909. Epub 2018/03/23. 10.3390/ijms19030909 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Venn diagrams (a) and Volcano plot (b) of expressed proteins between control and treatment groups.
(TIF)
The relative abundance of proteins is represented by different colors, where red represents higher intensity and blue represents lower intensity.
(TIF)
GO analysis of biological process (a), cellular component (b), and molecular function (c).
(TIF)
The vertical axis represents the enriched KEGG classification (fold > 1.5, p < 0.05). The horizontal axis is the rich factor (rich factor ≤ 1), representing the ratio of the number of differentially expressed proteins to those identified in the KEGG pathway. The size of the circular area represents the number of differentially expressed proteins, and the circular color represents the enrichment p-value of the differentially expressed proteins under the KEGG classification.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.