Single-atom zero-valent copper was biosynthesized by a copper-resistant bacterium isolated from a copper mine in Brazil.
Abstract
The chemical synthesis of monoatomic metallic copper is unfavorable and requires inert or reductive conditions and the use of toxic reagents. Here, we report the environmental extraction and conversion of CuSO4 ions into single-atom zero-valent copper (Cu0) by a copper-resistant bacterium isolated from a copper mine in Brazil. Furthermore, the biosynthetic mechanism of Cu0 production is proposed via proteomics analysis. This microbial conversion is carried out naturally under aerobic conditions eliminating toxic solvents. One of the most advanced commercially available transmission electron microscopy systems on the market (NeoArm) was used to demonstrate the abundant intracellular synthesis of single-atom zero-valent copper by this bacterium. This finding shows that microbes in acid mine drainages can naturally extract metal ions, such as copper, and transform them into a valuable commodity.
INTRODUCTION
The dielectric, magnetic, optical, antimicrobial, imaging, and catalytic properties of copper make this element a candidate for applications in photoelectrochemical cells, sensors, solar cells, inks, and antimicrobial coatings (1, 2). Recent studies have also found that many microorganisms, such as bacteria (3, 4) and fungi (5), can produce inorganic nanoparticles (NPs), such as Ag (6), Au (7), Cu (8, 9), CuO (10, 11), and magnetite (12), among others (13). A few studies described the synthesis of Cu NPs (8, 9) ranging from 10 to 40 nm intra- and extracellularly by a sustainable method using bacteria. These studies described the involvement of reductase enzymes, such as NADPH (reduced form of nicotinamide adenine dinucleotide phosphate)–dependent enzymes with redox potential for metal ion reduction (14–16). Yet, we are not aware of any report of a natural synthesis of single-atom Cu0 (size in the range of 170 to 179 pm) using microorganisms or their mechanisms of biosynthesis and stabilization.
The importance of producing monoatomic metallic copper is the potential of single atoms in catalysis, doping, and energy applications to maximize the efficiency of metal-atom utilization (17, 18). However, the single-atom synthesis is still a challenge, mainly because atom isolation is rather complicated and requires complex synthesis involving toxic chemicals (19, 20). Alternative methods, such as chemical vapor deposition, sputtering, and femtosecond laser ablation, among others, are complex, and the efficiency is still limited (2, 17, 21, 22). Here, we discovered an environmental bacterium found in a copper mine that can synthesize single atoms of Cu0 in a sustainable manner. Its biosynthetic mechanism was proposed using the proteomics approach. This sustainable synthesis of Cu0 is carried out in microbial growth media, eliminating the use of toxic solvents. The isolation of the monoatomic copper directly from naturally occurring copper mines (Pará/Brazil) (latitude 6°27′ 15.848″S and longitude 50°4′37.507″W) suggests that microorganisms can potentially extract ions and metals from the environment and transform in a highly valuable commodity. This finding, by itself, implies that microbes can extract copper from contaminated sites and, thus, improve sustainability.
RESULTS AND DISCUSSION
The discovery of an isolate from the genus Bacillus that is able to produce monoatomic copper inside the cells was recorded initially based on the visual observation of color changes in the bacterium growth media with CuSO4 after 48 hours (Fig. 1A). The change in the growth media from green (CuSO4 + bacteria) to “orange color” represented the transformation from CuSO4 to Cu0 (Fig. 1A). The confirmation of the presence of monoatomic copper was demonstrated using aberration-corrected atomic resolution transmission electron microscopy (TEM). The cross-sectional TEM image (Fig. 1B) shows the bacteria after 48-hour incubation with CuSO4 [copper sulfate (100 mg/liter)]. Lower- and higher-magnification TEM images, respectively, inside the bacterial cross section are shown in Fig. 1 (C and D). Furthermore, it is possible to observe the individual copper atoms, particularly in the inset in Fig. 1D.
Fig. 1. Cu atom synthesis and TEM characterization.
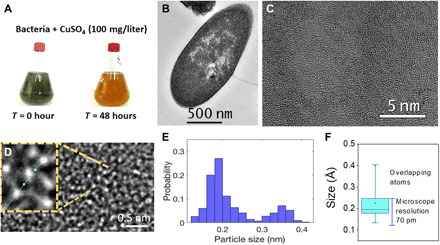
(A) Flasks of Bacillus sp. strain 105 containing CuSO4 (100 mg/liter) at time 0 and after 48-hour growth showing bioproduction of the copper atoms. (B) Electron microscopy of cross-sectional view of Bacillus sp. strain 105. (C) Low-magnification image inside the bacterial cross section. (D) Inset zoom out of (C) showing the actual copper atoms under TEM with sizes in the range of 170 to 179 pm and (E) the respective measurement conducted in the inset (D), which shows that the atoms have sizes between 1.6 and 1.85 Å. (F) Box plot summary of all the atoms measured in several samples with over 75% of them falling within the copper’s theoretical atomic radii or within this size and the microscope’s resolution (70 pm); the other 25% corresponds to overlapping atoms.
To determine the size of the radii of the generated copper atoms, we analyzed approximately 13,000 atoms from 13 TEM microstructures (similar to those in Fig. 1E). The size of each atoms was within the predicted atomic radii of copper in the literature (1.7 and 1.85 Å) (Fig. 1E). The box plot summarizes the actual population distribution and sizes of all the measured atoms in the different TEM images considered (Fig. 1F). Results showed that over 75% of the copper atoms fall within 1.89 ± 0.19 Å, which corresponds to metallic copper’s theoretical atomic radii ± the microscope’s resolution (70 pm) (23, 24). Therefore, our result is clearly within the theoretical/experimental size of the metallic copper atom with an standard deviation way below that of the microscope resolution. Larger sizes observed were considered overlapping atoms that are further demonstrated in the double Gaussian histogram for the single and double atoms (Fig. 1E). The identified atoms fall within the expected range for a neutral atom (zero valence, Cu “0”). The ionic radii for copper Cu1+, Cu2+, and Cu3+ are in the range of 0.54 to 0.63 Å (23–25), which are notably smaller than the atomic copper. Therefore, the presence of copper in the samples must be only from the actual metallic copper.
The distribution of the elements in the cells was determined by energy dispersive spectroscopy (EDS) (Fig. 2). We selected a molybdenum grid to ensure that all copper signals detected by EDS belong solely to those in the bacteria (Fig. 2). A similar logic was followed for the electron energy-loss spectroscopy (EELS) analysis (Fig. 3). The backscattered image (Fig. 2) shows a clear contrast between the heavier and lighter atoms, in this case, copper versus carbon, nitrogen, and oxygen. The EDS map for copper shows a highly homogeneous distribution, and there is no significant agglomeration, as demonstrated in the statistical analysis (Fig. 1, D and E). Similarly, the EDS spectra (Fig. 2) reveal the actual chemical makeup of the sample. These results demonstrate that copper is one of the most abundant species in the sample, and the type of copper is zero-valent (Cu0) as was further demonstrated by x-ray photoelectron spectroscopy (XPS) analyses (Fig. 3).
Fig. 2. EDS results showing the chemical composition of the sample.
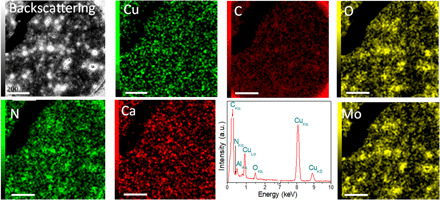
Backscattering image of Bacillus sp. strain 105 cross section (Fig. 1) and its respective EDS map for the different elements (Cu, C, O, N, and Ca) in the sample. The EDS map of Cu shows the presence of copper atoms within the sample. The Mo map corresponds to the grid used for the analysis. a.u., arbitrary units. Scale bar for all images, 200 nm.
Fig. 3. Spectroscopic characterizations of the Cu atoms.
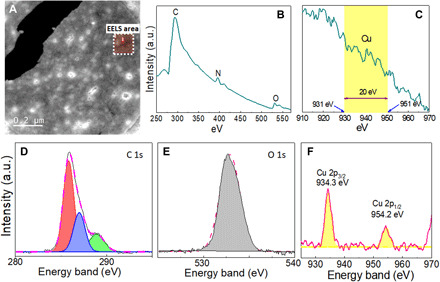
(A) Backscattering image of the region analyzed with EELS (red square). EELS of (B) carbon, nitrogen, and oxygen and (C) the copper region. XPS analysis of (D) C 1s, (E) O 1s, and (F) Cu 2p regions.
The characterization of copper using EELS from the backscattered image in Fig. 3A is shown in Fig. 3 (B and C). In the first EELS spectrum (Fig. 3B), one can see the presence of carbon, nitrogen, and oxygen that are typical components of organic matter, which are part of the bacteria. The respective energy level for each is 290, 400, and 530 eV. Those values are in direct agreement with those reported by the EELS manufacturer database (Gatan). Copper is usually detected in the 931- to 953-eV range. In the present work, copper is untraceable by EELS (Fig. 3C) and is attributed to the monoatomic copper thickness, which is a known limitation of this technique. The identification of copper with EDS, XPS, and even visually (48 hours; Fig. 1A) but not with EELS is not a trivial finding. This finding is the key to demonstrate that the frameworks of copper present in our sample are in sizes below the traceable limits for EELS. The EELS result further demonstrates that the single atoms of copper are likely homogeneously distributed throughout the bacterium, and the presence of copper agglomerates is improbable, unlike the other elements (e.g., C, N, and O) that were clearly identified by EELS. This finding confirms the TEM results (Fig. 1, C to F).
The XPS data confirmed the EDS observation. The XPS results for C 1s, O 1s, and Cu 2p core-level XPS spectra obtained from the Cu synthesized by Bacillus sp. strain 105 after 48 hours are shown in Fig. 3 (D to F). The C 1s spectrum (Fig. 3D) presents three components at 285.8, 287, and 288.9 eV, corresponding to the bonds between the protein/peptides and the atomic copper (8). The region of the O 1s core-level XPS spectra (Fig. 3E) shows a binding-energy (BE) maximum at 532.8 eV corresponding to the carboxylic groups, which belong to proteins on the surface of the atomic copper (26). The Cu 2p presents two peaks at 932.3 and 952.0 eV, which correspond to the binding energies of the 2p3/2 and 2p½ electrons of Cu0 (Fig. 3F). Furthermore, the spectrum did not show the corresponding shake-up satellites, corroborating the absence of Cu2+ or CuO (8, 19). These results confirm that the type of copper synthesized by the bacterium is monoatomic Cu0.
To understand the mechanism of single-atom synthesis by Bacillus sp. strain 105, a proteomics approach was used. The bacterium proteins were identified in two different growth conditions: cultivation without copper sulfate (control) and with copper sulfate (100 mg/liter). In the control experiment, 652 proteins were expressed, however, when the strain was grown in the presence of copper sulfate, and 458 proteins were expressed, as shown in tables S1 and S2. From these 458 total proteins, 313 proteins were equally expressed in both growth conditions, and 145 proteins were only expressed in the presence of copper sulfate (fig. S1). To understand the biosynthesis mechanism and the stabilization of the copper atoms by the bacterium, we mainly focused on the uniquely expressed proteins in the presence of copper sulfate. Most of the proteins (102 proteins) were involved in the primary metabolism (carbon and energy), suggesting that the exposure to Cu can affect negatively the cell by triggering the production of more energy to survive metal stress (27, 28). Fifteen proteins were involved in resistance and stress functions, and three of them had functions in copper transport and uptake by the cell. The identified proteins involved in the copper pathway were the iron-regulated ABC transporter membrane component SufB, the copper-translocating P-type adenosine triphosphatase (ATPase), and Copz (protein 110 in table S2). Copz is a chaperone protein with a role of intracellular sequestration and transport of Cu+ from the cytoplasm to the periplasm (29). The p-type ATPase proteins are commonly described to be involved in bacterial resistance and toxicity mechanisms related to heavy metals (30). The remaining 11 proteins identified under growth conditions in the presence of copper sulfate were proteins that may be involved in biosynthesis and stabilization of the monoatomic copper. These proteins were mainly reductases. Six of the 11 proteins, i.e., thiol-disulfide isomerase/thioredoxin, thioredoxin reductase (TRXR), 4Fe-4S dicluster, 4Fe-4S ferredoxin, TlpA family protein disulfide reductase, and sulfate adenylyltransferase, have been described to reduce either sulfate or metals. In this case, on the basis of their function, they were probably reducing the sulfate from CuSO4, leaving free toxic copper (Cu2+) inside the cells.
The production of Se NPs by Stenotrophomonas maltophilia showed a possible association with an alcohol dehydrogenase. In this present study, two homolog proteins were identified in Bacillus sp.: NADH (reduced form of nicotinamide adenine dinucleotide)–dependent butanol dehydrogenase A and a putative NADH-dependent butanol dehydrogenase, indicating that these proteins could be participating in the biogenic synthesis of monoatomic Cu. Furthermore, the Ferritin Dps family protein and starvation-inducible DNA binding protein (Dps) were also expressed by Bacillus sp. strain 105. The proteins ferritin and starvation-inducible protein (Dps protein family) are reported to have a ferroxidase core (Fig. 4). The NP synthetic routes with these proteins were described to involve autoxidation, hydroxylation, or reduction processes; however, detailed mechanism is still unknown (31). Similar studies have reported biosynthesis of NPs using these proteins (purified and synthetic ones) with different metals such as Co (32), Ni and Cr (33), Ag (34), Au (35), and Fe (36), among others. Ensign et al. (37) further demonstrated the successful NP production of Cu0 from Cu2+ using commercial ferritin and photocatalysis. Here, we suggest the potential use of in vivo ferritin protein in the conversion of Cu2+ to Cu0. The combination of this protein with other proteins expressed in Bacillus sp. strain 105 in the media supplemented with copper sulfate seems to be directly connected to the biosynthetic pathway of monoatomic copper (Fig. 4).
Fig. 4. Proposed mechanism of monoatomic copper synthesis assisted by the ferritin protein.
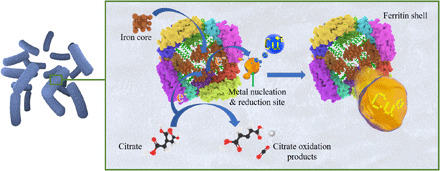
The protein contains an iron core that transfers the electrons to Cu2+ ions bound to the nucleation sites on the exterior of the ferritin protein shell to produce Cu0. To restore the hole, citrate is oxidized, and the electron is able to continue to flow to reduce additional Cu2+ ions present inside the bacterium.
In conclusion, a bacterium isolated from a copper mine demonstrated the ability to produce monoatomic copper inside the cell during the stationary phase (fig. S2). The employment of atomic resolution techniques allowed us to characterize the formation of monoatomic copper in the sample, which was further confirmed by EELS. The XPS helped demonstrate that copper is thoroughly zero-valent. This approach would open up exciting strategies to produce in large-scale atomic copper via sustainable manufacturing and at low cost for existing and future applications, such as in electronics, catalysis, functionalization, and antimicrobial processes. This finding also implies that microorganisms found in copper mines can not only play a role in environmental remediation but also facilitate sustainable synthesis of copper. Furthermore, this study opens a new field of research of environmental microorganisms that potentially are able to synthesize other monoatomic metals for applications in science, technology, engineering, and medicine.
MATERIALS AND METHODS
Materials
Reagents for the media used in this work were acquired from Sigma-Aldrich. Copper sulfate (CuSO4·5H2O), ethanol, glutaraldehyde, uranyl acetate, propylene oxide, and osmic acid were also purchased from Sigma-Aldrich. All reagents were used as received.
Methods
Microorganism isolation and identification
The bacterial strain used in this work, Bacillus sp. strain 105, was isolated from a copper mine in Brazil (Sossego-PA). The isolation was performed using 1.0 g of a sediment sample, containing about 600 parts per million (ppm) of copper sulfate, in Luria-Bertani (LB) with different copper sulfate concentrations (60 to 700 mg/liter) at 28°C and 160 rpm for 1 week. After that, according to Avanzi et al. (38), plates with fresh solid medium and different copper sulfate concentrations were used for the isolation of single colonies.
Genomic DNA was obtained from the isolate with the Illustra bacteria genomicPrep (GE Healthcare) as per the manufacturer’s instruction. The isolate was cultivated in LB medium for 8 hours before extraction. The 16S ribosomal RNA universal primers used to amplify the microorganism were 341F (5′-CCTACGGGNGGCNGCA-3′) and 826R (5′-CCTACGGGNGGCNGCA-3′). The polymerase chain reaction protocol was performed as described by Rodrigues et al. (39). The sequencing of the amplicons was obtained with an ABI 3730 DNA Analyzer (Applied Biosystems) and BigDye Terminator v3.1 (Applied Biosystems) as per the manufacturer’s instructions. The sequence was deposited in the National Center for Biotechnology Information (NCBI) database with the accession number (SUB3797756 105_copper MH080573). The bacterium presented a sequence similarity of 98.32% to Bacillus niacini strain CP-2MN.
Monoatomic copper biosynthesis
The synthesis occurred during the cultivation of the bacterium in 1 liter of the medium composed of 900 ml of LB and 100 ml of copper sulfate (CuSO4 solution) in a shaker at 180 rpm and 28°C (in triplicate). The final concentration of copper sulfate in the sample was 100 mg/liter. After 48 hours, the cells were harvested by centrifugation (Hitachi Koki Himac CR22N High-Speed Refrigerated Centrifuge) at 21,000g for 10 min. The growth medium was removed, and the cells were washed once with phosphate buffer solution (pH 7.4) adjusted with NaOH or HCl [NaCl (8.0 g/liter), KCl (200 mg/liter), Na2HPO4 (1.44 g/liter), and KH2PO4 (245 mg/liter)]. Then, the pellet was frozen at −80°C and lyophilized in a FreeZone 6 Liter Benchtop Freeze Dryer (−84°C and 0.22 mbar). The sample was kept in the dark to characterize the atoms.
Control experiments without copper sulfate were performed in parallel under the same conditions. The procedure was repeated three times in triplicates in two different laboratories in two different countries for reproducibility assurance. In total, the synthesis procedure was tested at least 18 times.
Proteomics analysis
Protein extraction and trypsin digestion
To identify the proteins involved in the monoatomic copper synthesis, the bacterium was grown as described above with or without copper sulfate (100 mg/liter) for 48 hours using a close flask. The proteins were extracted according to Gracioso et al. (27). After the extraction, proteins were digested in peptides using the Trypsin Singles kit (Sigma-Aldrich, USA) according to the manufacturer’s instruction. After digestion, the peptide mixture was dried with a SpeedVac (Hetodrywinner-MaxydryIyo).
Nanoflow liquid chromatography–electrospray ionization–tandem mass spectrometry analysis
The dried peptide mixture was resuspended in 2% acetonitrile (ACN) with 0.1% formic acid and analyzed on a Dionex UltiMate 3000 RSLCnano HPLC System (Thermo Fisher Scientific, Bremen, Germany) interfaced to an Impact II quadrupole time-of-flight (Q-TOF) mass spectrometer equipped with a Captive Spray nanoelectrospray source with an ACN-enriched nanoBooster dopant system for increased sensitivity (Bruker Daltonics, Bremen, Germany). A volume of 5 μl was injected in an Acclaim PepMap 100 nano-trap column (Dionex-C18, 100 Å, 100 μm by 2 cm by 5 μm; Thermo Fisher Scientific) to preconcentrate the peptides. Sample loading was done for 10 min at a flow rate of 5 μl/min with 0.1% trifluoroacetic acid (TFA). Separation was performed on an Acclaim PepMap RSLC analytical column (Dionex-C18, 100 Å, 75 μm by 15 cm) with an increasing gradient from 2 to 35% of solvent B (ACN in 0.1% TFA) within 100 min and a flow rate of 300 nl/min. The temperature of the column was maintained at 40°C.
The eluted peptides were analyzed with an Impact II Q-TOF mass spectrometer operated in positive mode. Data were acquired using data-dependent automatic tandem mass spectrometry (auto-MS/MS). A high-resolution TOF-MS scan over a mass range of 50 to 3000 m/z (mass/charge ratio) at an acquisition rate of five spectra/s was followed by auto-MS/MS scans of the top 20 most intense precursor ions per cycle of fragmentation. Dynamic exclusion of precursor ions was set to 30-s duration after acquisition of one spectrum, with a window of 0.05 Th. Acquisition rate for auto-MS/MS was set to 2 Hz (MS/MS, adjusted by precursor ion intensity). The collision energy was dynamically adjusted between 7 and 70 eV (Bruker Compass 1.9 Otof Control Version 4.0, Build 60.11, Top20 Auto MS/MS method).
Homology database searches
At the end of nanoflow liquid chromatography–electrospray ionization–MS/MS (nanoLC-ESI-MS/MS) analysis, the raw files (.d) were loaded into the Peaks Studio 8.5 software (Bioinformatics Solution Inc., Waterloo, Canada) and processed with de novo peptide sequencing, PeaksDB, PeaksPTM, and SPIDER tools for identification of the proteins through a multiround database search (40). First, the MS/MS spectra were searched against the peptide sequences of the Bacillus (genus) database from UniProtKB/Swiss-prot (Uniref90—sequence clusters with identity of 90%) with 8154 reviewed protein entries. Then, nonmatched MS/MS spectra were searched using the homology database against the Bacillus (genus) from UniProtKB/TrEMBL (Uniref90) with 1,085,071 unreviewed protein entries, as a second round database search. The following parameters were used: precursor mass tolerance = 25 ppm; fragment mass tolerance = 0.025 Da; trypsin was set as the specific digestion enzyme, and two missed cleavage sites and one nonspecific were allowed. Carbamidomethylation (Cys) was set as a fixed modification, and oxidation (Met) was set as a variable modification. A false discovery rate threshold of 1% on the peptide-spectrum match was applied to filter out the peptide sequences. Protein identification required at least one unique peptide. Proteins were considered to be identified in a sample when they matched at least two peptides in the database.
TEM characterization
To analyze the samples with high-resolution TEM (HRTEM), the samples were fixed using the following protocol. A solution of 2% glutaraldehyde was reacted with the sample for 2 hours. Then, the samples were washed with buffer and reacted with 2% osmic acid for 2 hours. After that, the samples were rewashed with buffer. For staining, 1% uranyl acetate was added and kept overnight. For dehydration, the sample was exposed for 10 min in each of the following steps: 70% ethanol twice, 95% ethanol twice, 100% ethanol for four times, and propylene oxide twice. Propylene oxide with resin was added (1:1) for 6 hours and then replaced by pure resin for 6 more hours for inclusion.
The identification of the single-atom copper produced inside the bacterium was determined via aberration-corrected atomic resolution TEM using a JEOL NEOARM microscope operated at 80 kV in a molybdenum grid. The copper signal was collected with EDS and EELS coupled to the electron microscope. The HRTEM images were analyzed using ImageJ and MATLAB. For the investigation, 13 TEM images were obtained. Approximately, 1000 atoms in each TEM image were counted with ImageJ. The shape factor of the analyzed features in the microstructures was set to 1 to elucidate the sphericity of the atoms, and no limits were set for the size. The number of particles was expressed in terms of probability in a histogram by using the MATLAB software. OriginLab was used for data analysis and image representation.
XPS analysis
For the XPS analysis, the samples were drop-casted on gold substrates. A PHI Quantera SXM Scanning X-ray Microprobe with Al Kα (1486.6 eV), as the excitation source, was used for the measurements. High-resolution spectra of C 1s, O 1s, and Cu 2p core-level spectra with a pass energy of 23.5 eV were obtained for the samples, an energy step size of 0.2 eV, and a time step of 50 ms. The background for the XPS spectra was corrected using the Shirley algorithm, and their respective binding energies were aligned to Au s at 84 eV. The calibration of the binding energy was done by setting the adventitious carbon (corresponding to C─C bonds) to 284.8 eV. The MultiPak software was used for the XPS data analysis.
Acknowledgments
We want to thank H. Nguyen for drawing Fig. 4. Funding: We gratefully acknowledge Vale, BNDES, CNPq-INCT (Brazilian National Council of Scientific and Technological Development), and FAPESP 2013/50218-2 (São Paulo Research Foundation) for the financial and technical support. L.H.G. thank FAPESP for fellowships granted under processes 2013/11020-2 and 2015/18089-3. M.A.J. thanks the Sao Paulo State Research Support Foundation for the financial support (FAPESP—projects 18/13588-0). We acknowledge W. Conde and D. Assis from Bruker for the support on operating Q-TOF mass spectrometer and protein identification. We acknowledge the JEOL Ltd. Japan and USA, particularly to Z. Marek and P. Phillips, for outstanding support with the NeoARM analysis. We acknowledge FAPESP for a research grant (FAPESP proc. 2018/22790-7).This work was supported by the NSF BEINM grant number 1705511 and the Robert A. Welch Foundation award number (E-2011-20190330). We wish also to thank the College of Technology at the University of Houston for the financial support for electron microscopy, particularly F. Merchant (Chair) and A. Ambler (Dean). The findings achieved herein are solely the responsibility of the authors. Author contributions: L.H.G. contributed with the isolation of the bacterium, the biosynthesis of the Cu atoms, proteomics analysis to elucidate the biosynthetic pathway, and manuscript preparation. J.P.-B. contributed with the XPS measurements, HRTEM analysis with ImageJ and MATLAB software, manuscript preparation, and data analysis. B.B.B. contributed with biosynthesis and protein identification. B.K. collected the samples in the mine for the bacterial isolation. M.A.J. contributed with the nanoLC-ESI-MS/MS analysis. E.A.P. and C.A.O.N. were responsible for overseeing the sample collections and isolation of the bacterium in Brazil. They were responsible for the sequencing of the bacterium for identification. H.H. collected the HRTEM and EELS images and data for the project. F.C.R.H. contributed with the HRTEM and EELS analysis, discussion of the results, and preparation of the manuscript. D.F.R. was responsible for the original idea, the conceptualization of the study, designing the experiments and analyses, managing all the experimental activities, and preparing the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The isolated microorganism can be obtained by request, pending scientific review of the request and a completed material transfer agreement. Requests for the isolated microorganism should be submitted to D. F. Rodrigues. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/17/eabd9210/DC1
REFERENCES AND NOTES
- 1.Dagher S., Haik Y., Ayesh A. I., Tit N., Synthesis and optical properties of colloidal CuO nanoparticles. JOL 151, 149–154 (2014). [Google Scholar]
- 2.Haas I., Shanmugam S., Gedanken A., Pulsed sonoelectrochemical synthesis of size-controlled copper nanoparticles stabilized by poly(N-vinylpyrrolidone). J. Phys. Chem. B 110, 16947–16952 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Tiwari M., Jain P., Hariharapura R. C., Narayanan K., Bhat K U., Udupa N., Rao J. V., Biosynthesis of copper nanoparticles using copper-resistant Bacillus cereus, a soil isolate. Process Biochem. 51, 1348–1356 (2016). [Google Scholar]
- 4.Fayaz A. M., Girilal M., Rahman M., Venkatesan R., Kalaichelvan P. T., Biosynthesis of silver and gold nanoparticles using thermophilic bacterium Geobacillus stearothermophilus. Process Biochem. 46, 1958–1962 (2011). [Google Scholar]
- 5.Khan A. U., Malik N., Khan M., Cho M. H., Khan M. M., Fungi-assisted silver nanoparticle synthesis and their applications. Bioprocess Biosyst. Eng. 41, 1–20 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Li G., He D., Qian Y., Guan B., Gao S., Cui Y., Yokoyama K., Wang L., Fungus-mediated green synthesis of silver nanoparticles using Aspergillus terreus. Int. J. Mol. Sci. 13, 466–476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawle B. D., Salimath B., Deshpande R., Bedre M. D., Prabhakar B. K., Venkataraman A., Biosynthesis and stabilization of Au and Au–Ag alloy nanoparticles by fungus, Fusarium semitectum. Sci. Technol. Adv. Mater. 9, 035012 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramanathan R., Field M. R., O’Mullane A. P., Smooker P. M., Bhargava S. K., Bansal V., Aqueous phase synthesis of copper nanoparticles: A link between heavy metal resistance and nanoparticle synthesis ability in bacterial systems. Nanoscale 5, 2300–2306 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Kimber R. L., Lewis E. A., Parmeggiani F., Smith K., Bagshaw H., Gianolio D., Haigh S. J., Pattrick R. A. D., Turner N. J., Lloyd J. R., Biosynthesis and characterization of copper nanoparticles using Shewanella oneidensis: Application for click chemistry. Small 14, 10.1002/smll.201703145, (2018). [DOI] [PubMed] [Google Scholar]
- 10.Gunalan S., Sivaraj R., Venckatesh R., Aloe barbadensis Miller mediated green synthesis of mono-disperse copper oxide nanoparticles: Optical properties. Spectrochim. Acta A Mol. Biomol. Spectrosc. 97, 1140–1144 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Singh A. V., Patil R., Anand A., Milani P., Gade W. N., Biological synthesis of copper oxide nano particles using Escherichia coli. Curr. Nanosci. 6, 365–369 (2010). [Google Scholar]
- 12.Bharde A., Rautaray D., Bansal V., Ahmad A., Sarkar I., Yusuf S. M., Sanyal M., Sastry M., Extracellular biosynthesis of magnetite using fungi. Small 2, 135–141 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Li X., Xu H., Chen Z. S., Chen G., Biosynthesis of nanoparticles by microorganisms and their applications. J. Nanomater. 2011, 1–16 (2011).21808638 [Google Scholar]
- 14.Nangia Y., Wangoo N., Goyal N., Shekhawat G., Suri C. R., A novel bacterial isolate Stenotrophomonas maltophilia as living factory for synthesis of gold nanoparticles. Microb Cell Fact. 8, 39 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Ahmed S., Annu, Ikram S., Yudha S S., Biosynthesis of gold nanoparticles: A green approach. J. Photochem. Photobiol. B Biol. 161, 141–153 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Mittal A. K., Bhaumik J., Kumar S., Banerjee U. C., Biosynthesis of silver nanoparticles: Elucidation of prospective mechanism and therapeutic potential. J. Colloid Interface Sci. 415, 39–47 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Qu Y., Li Z., Chen W., Lin Y., Yuan T., Yang Z., Zhao C., Wang J., Zhao C., Wang X., Zhou F., Zhuang Z., Wu Y., Li Y., Direct transformation of bulk copper into copper single sites via emitting and trapping of atoms. Nat. Catal. 1, 781–786 (2018). [Google Scholar]
- 18.Yang H., Wu Y., Li G., Lin Q., Hu Q., Zhang Q., Liu J., He C., Scalable production of efficient single-atom copper decorated carbon membranes for CO2 electroreduction to methanol. J. Am. Chem. Soc. 141, 12717–12723 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Wei W., Lu Y., Chen W., Chen S., One-pot synthesis, photoluminescence, and electrocatalytic properties of subnanometer-sized copper clusters. J. Am. Chem. Soc. 133, 2060–2063 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Park B. K., Jeong S., Kim D., Moon J., Lim S., Kim J. S., Synthesis and size control of monodisperse copper nanoparticles by polyol method. J. Colloid Interface Sci. 311, 417–424 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Ayyub P., Chandra R., Taneja P., Sharma A. K., Pinto R., Synthesis of nanocrystalline material by sputtering and laser ablation at low temperatures. App. Phys. A 73, 67–73 (2001). [Google Scholar]
- 22.Yang Z., Chen B., Chen W., Qu Y., Zhou F., Zhao C., Xu Q., Zhang Q., Duan X., Wu Y., Directly transforming copper (I) oxide bulk into isolated single-atom copper sites catalyst through gas-transport approach. Nat. Commun. 10, 3734 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh D. C., Biswas R., Theoretical calculation of absolute radii of atoms and ions. Part 1. The Atomic Radii. Int. J. Mol. Sci. 2002, 87–113 (2002). [Google Scholar]
- 24.Ghosh D. C., Biswas R., Theoretical calculation of absolute radii of atoms and ions. Part 2. The Ionic Radii. Int. J. Mol. Sci., 379–407 (2003). [Google Scholar]
- 25.Shannon R. D., Revised effective ionic radii and systematic studies of interatomic distances in halides and chalcogenides. Acta Crystallogr. Sect. A 32, 751–767 (1976). [Google Scholar]
- 26.Schiffrin A., Riemann A., Auwarter W., Pennec Y., Weber-Bargioni A., Cvetko D., Cossaro A., Morgante A., Barth J. V., Zwitterionic self-assembly of L-methionine nanogratings on the Ag(111) surface. Proc. Natl. Acad. Sci. U.S.A 104, 5279–5284 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gracioso L. H., Baltazar M. P. G., Avanzi I. R., Karolski B., Nascimento C. A. O., Perpetuo E. A., Analysis of copper response in Acinetobacter sp. by comparative proteomics. Metallomics 11, 949–958 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Poirier I., Jean N., Guary J. C., Bertrand M., Responses of the marine bacterium Pseudomonas fluorescens to an excess of heavy metals: Physiological and biochemical aspects. Sci. Total Environ. 406, 76–87 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Consortium T. U., UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Capeness M. J., Imrie L., Mühlbauer L. F., Le Bihan T., Horsfall L. E., Shotgun proteomic analysis of nanoparticle-synthesizing Desulfovibrio alaskensis in response to platinum and palladium. Microbiology 165, 1282–1294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimura H., Protein-assisted nanoparticle synthesis. Colloids Surfaces A Physicochem. Eng. Asp. 282, 464–470 (2006). [Google Scholar]
- 32.Douglas T., Stark V. T., Nanophase cobalt oxyhydroxide mineral synthesized within the protein cage of ferritin. Inorg. Chem. 39, 1828–1830 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Okuda M., Iwahori K., Yamashita I., Yoshimura H., Fabrication of nickel and chromium nanoparticles using the protein cage of apoferritin. Biotechnol. Bioeng. 84, 187–194 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Petrucci O. D., Hilton R. J., Farrer J. K., Watt R. K., A ferritin photochemical synthesis of monodispersed silver nanoparticles that possess antimicrobial properties. J. Nanomater. 2019, 9535708 (2019). [Google Scholar]
- 35.Keyes J. D., Hilton R. J., Farrer J., Watt R. K., Ferritin as a photocatalyst and scaffold for gold nanoparticle synthesis. J. Nanopart. Res. 13, 2563–2575 (2011). [Google Scholar]
- 36.Ilari A., Ceci P., Ferrari D., Rossi G. L., Chiancone E., Iron incorporation into Escherichia coli Dps gives rise to a ferritin-like microcrystalline core. J. Biol. Chem. 277, 37619–37623 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Ensign D., Young M., Douglas T., Photocatalytic synthesis of copper colloids from Cu (II) by the ferrihydrite core of ferritin. Inorg. Chem. 43, 3441–3446 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Avanzi I. R., Gracioso L. H., Baltazar M., Karolski B., Perpetuo E. A., do Nascimento C. A. O., Rapid bacteria identification from environmental mining samples using MALDI-TOF MS analysis. Environ. Sci. Pollut. Res. 24, 3717–3726 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Nguyen H. N., Castro-Wallace S. L., Rodrigues D. F., Acute toxicity of graphene nanoplatelets on biological wastewater treatment process. Environ. Sci. Nano 4, 160–169 (2017). [Google Scholar]
- 40.Zhang J., Xin L., Shan B., Chen W., Xie M., Yuen D., Zhang W., Zhang Z., Lajoie G. A., Ma B., PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics 11, M111.010587–M111.010587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/17/eabd9210/DC1


