Abstract
Objective
Cisplatin resistance is one of the main reasons for treatment failure in ovarian cancer (OC). Here, the effects of LINC00184 on cisplatin-resistant OC were studied.
Patients and Methods
LINC00184, miR-1305 and CNTN1 expression in tissues from 70 OC patients was determined by qRT-PCR, in situ hybridization and Western blot. OC cell lines and OC cisplatin-resistant cell lines were cultured. Cells were transfected using Lipofectamine 2000 and treated with 100 nM cisplatin. Cell proliferation and apoptosis were researched by the CCK-8 assay and flow cytometry. A dual-luciferase reporter gene assay and RNA pull-down were performed to explore the relationship between two genes. LINC00184, miR-1305 and CNTN1 expression in cells was detected by qRT-PCR and Western blot. An in vivo experiment was conducted using nude mice. Ki67 and CNTN1 expression and apoptosis of xenograft tumors were investigated using immunohistochemistry and a TUNEL assay.
Results
LINC00184 was up-regulated in OC clinical tissues and OC cells, especially in cisplatin-resistant OC patients and cells (p<0.01 or p<0.0001). LINC00184 overexpression significantly enhanced OC cell proliferation and cisplatin resistance, and inhibited OC cell apoptosis (p<0.05 or p<0.01). LINC00184 elevated CNTN1 expression via sponging miR-1305. LINC00184 overexpression markedly exacerbated the malignant phenotype of OC cells and cisplatin-resistant OC cells via the miR-1305/CNTN1 axis (p<0.01). Silencing of LINC00184 significantly suppressed OC cell growth and cisplatin resistance in vivo (p<0.01). LINC00184 silencing inhibited Ki67 and CNTN1 expression and promoted apoptosis of xenograft tumors. CNTN1 overexpression promoted proliferation and cisplatin resistance, and reduced apoptosis of OC cells (p<0.05 or p<0.01).
Conclusion
LINC00184 promoted OC cell proliferation and cisplatin resistance by elevating CNTN1 expression via sponging miR-1305.
Keywords: OC, cisplatin resistance, LINC00184, miR-1305, CNTN1
Introduction
Ovarian cancer (OC) poses a serious threat to women’s health and lives. It is one of the most common malignancies in gynecology.1 According to previous epidemiological studies, there are about 200,000 new OC cases all over the world each year, with a mortality rate more than 60%.2 At present, surgical removal of lesion tissues and chemotherapy are the main treatment strategies for most human solid malignancies, including OC. But unfortunately, more than 70% of OC patients show resistance to chemotherapy drugs, which leads to a serious adverse prognosis, such as shortened survival or even loss of life.2,3 Cisplatin is a commonly used chemotherapy drug for clinical treatment of OC. However, resistance to cisplatin is still a major issue in chemotherapy for OC.4 Currently, one of the reasons for treatment failure in OC is clinical cisplatin resistance.5 Hence, addressing cisplatin resistance is one of the keys to improving the prognosis of OC patients.
Long non-coding RNAs (lncRNAs) are emerging as new players in tumor drug resistance.6 LncRNAs are a type of non-coding RNA with a length of more than 200 nucleotides. LncRNAs have been found to be involved in cisplatin resistance in OC. It has been reported that activated autophagy could result in cisplatin resistance in human cancers,7 and lncRNA HOTAIR decreased the cisplatin sensitivity in OC by suppressing cisplatin-induced autophagy.8 Miao et al9 demonstrated that the silencing of lncRNA ANRIL enhanced OC cell apoptosis and cisplatin sensitivity by targeting the let-7a/HMGA2 axis. MALAT-1 knockdown promoted cisplatin sensitivity in OC cells by suppressing the Notch1 signaling pathway.10 Moreover, lncRNA CCAT1 has been discovered to enhance OC cisplatin resistance by sponging miR-454/survivin.11 These findings suggest that lncRNAs could be used as targets for intervention in cisplatin-resistant OC.
LINC00184 is a novel lncRNA that has only recently been reported in 2019. It has been identified as a prognostic marker for breast cancer. High expression of LINC00184 is associated with a worse 10-year survival for patients with breast cancer.12 Furthermore, Li et al13 observed that LINC00184 expression was up-regulated in esophageal cancer. After down-regulation of LINC00184 expression, esophageal cancer cell proliferation and tumor growth in vivo were both suppressed. LINC00184 was therefore suggested as an intervention target for esophageal cancer. However, the effects of and mechanism by which LINC00184 regulates OC development and cisplatin resistance remain uninvestigated. The discovery of more lncRNAs regulating OC cisplatin resistance will provide more options for OC treatment. Thus, LINC00184 was selected as the research objective of this article. The function of LINC00184 in OC progression and cisplatin resistance was then explored in this study. Importantly, online bioinformatics predicted that LINC00184 possessed binding sites for miR-1305, and miR-1305 had binding sites for CNTN1. Therefore, this study researched the mechanism of LINC00184 in regulating OC development and cisplatin resistance with miR-1305/CNTN1 as the axis. In our preliminary study, we detected LINC00184 expression in four OC cell lines, namely, A2780, SKOV3, OVCAR-3 and CoC1. We noticed that A2780 and SKOV3 cell lines had much higher LINC00184 expression than OVCAR-3 and CoC1 cell lines. Therefore, in this study, we selected A2780 and SKOV3 cell lines for investigation. The findings from this article will provide a novel molecular target and accurate molecular mechanism for the treatment of OC.
Methods
Clinical Tissues
Tumor tissues and adjacent normal tissues from 70 OC patients were collected during surgery. All patients were admitted to The Second Affiliated Hospital of Nantong University from July 2014 to November 2015. All specimens were stored in liquid nitrogen. Among these patients, 38 cases were resistant to cisplatin while 32 cases were sensitive to cisplatin. Sensitive and resistant patients were defined as follows: patients who developed progressive disease within 6 months after initial treatment were defined as cisplatin resistant; the other patients were defined as cisplatin sensitive.14 Adjacent normal tissues served as the N group (n=70). Tumor tissues obtained from cisplatin-resistant patients were used as the R group (n=38). Tumor tissues collected from cisplatin-sensitive patients were named the S group (n=32). The expression of LINC00184 in these tissues was determined by quantitative real-time polymerase chain reaction (qRT-PCR).
This study was approved by The Second Affiliated Hospital of Nantong University ethics committee in accordance with the guidelines of the Declaration of Helsinki. Written informed consent was obtained from all participants.
In Situ Hybridization (ISH)
The expression of LINC00184 in clinical specimens was detected by an ISH Kit (Boster, Wuhan, Hubei, China), in line with the manual. In brief, clinical tissues were dehydrated with ethanol, followed by being embedded in paraffin and cut into sections (5 μm). After being dewaxed and rehydrated, sections were treated with Triton X-100 to enhance probe penetration. Paraformaldehyde (4%) was used to fix the sections. Phosphate-buffered saline (PBS) was used to wash the sections. The sections were prehybridized for 4 h at 40°C. Thereafter, the sections were hybridized with hybridization solution containing digoxin-labeled-LINC00184-specific probe (300 ng/mL) for 12 h at 42°C. PBS was used to wash the sections three times. Sections were treated with sheep serum blocking solution for 1 h at 37°C. Then, the sections were treated with anti-digoxigenin–alkaline phosphatase antibody (Roche, Berlin, Germany) for 12 h at 4°C. NBT/BCIP solution (Roche, Berlin, Germany) was used to stain the sections. The expression of LINC00184 was observed under a microscope (Olympus, Tokyo, Japan).13,15
Cell Lines
A human normal ovarian epithelial cell line (IOSE80), human OC cell lines (A2780, SKOV3) and OC cisplatin-resistant cell lines (A2780-DDP, SKOV3-DDP) were provided by the Shanghai Institute of Cell Biology (Shanghai, China). Cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) in a humidified incubator at 37°C, 5% CO2.
Plasmid and Transfection
A2780-DDP, SKOV3-DDP, A2780 and SKOV3 cells were plated in six-well plates with 1×105 cells per well. Serum-free DMEM (1 mL) was added to each well. The siRNA targeting LINC00184 (siLINC00184-1 and siLINC00184-2) and scrambled negative control siRNA (siNC) (GenePharma, Shanghai, China) were transfected into A2780-DDP and SKOV3-DDP cells using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA). The full length of LINC00184 cDNA was synthesized and ligated into the pcDNA3.1 vector by GenePharma (Shanghai, China). Plasmid vectors, including pcDNA3.1-LINC00184 vector (oeLINC00184) and pcDNA3.1-empty vector (oeNC), were each transfected into A2780-DDP, SKOV3-DDP, A2780 and SKOV3 cells. miR-1305 mimic and negative control (miR-NC) (GenePharma, Shanghai, China) were applied to transfect A2780 and SKOV3 cells. In addition, miR-1305 inhibitor and its negative control (NC-inh) were purchased from GenePharma (Shanghai, China). A2780-DDP cells underwent cotransfection with siNC and NC-inh, or siLINC00184 and NC-inh, or siLINC00184 and miR-1305 inhibitor. Meanwhile, A2780 cells were cotransfected with oeNC and miR-NC, or oeLINC00184 and miR-NC, or oeLINC00184 and miR-1305 mimic. Moreover, pcDNA3.1-CNTN1 vector (oeCNTN1) and pcDNA3.1-empty vector (oeCtrl) (GenePharma, Shanghai, China) were respectively transfected into A2780 and SKOV3 cells. A2780-DDP and SKOV3-DDP cells were respectively transfected with CNTN1 siRNA (siCNTN1) and the corresponding negative control (siCtrl) (GenePharma, Shanghai, China). All transfections were performed using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA). After 6 h incubation at 37°C, 5% CO2, cells were cultured for 48 h with DMEM containing 10% FBS.
Cell Treatment With Cisplatin
A2780-DDP and SKOV3-DDP cells that were transfected with siLINC00184, siNC, siCNTN1 and siCtrl were collected. These cells were plated in six-well plates with 1×105 cells per well. Meanwhile, A2780 and SKOV3 cells that were transfected with oeLINC00184, oeNC, oeCNTN1 and oeCtrl were also plated in six-well plates (1×105 cells/well). DMEM containing 10% FBS and 100 nM cisplatin (final concentration) was added to each well. Cells were incubated at 37°C, 5% CO2, for subsequent experiments.16 Previous research showed that 100 nM cisplatin could obviously reduce tumor growth in neuroblastoma.17 Based on this, we performed a preliminary study and found that 100 nM cisplatin has an obvious inhibitory effect on OC cells. Thus, in this study, 100 nM cisplatin was used to treat OC cells.
Cell Counting Kit-8 (CCK-8) Assay
Cells were collected after transfection or cisplatin treatment, followed by being seeded in 96-well plates (1×104 cells/well). DMEM (10% FBS) with or without 100 nM cisplatin was added to each well. Cells were cultured for 24, 48 and 72 h in the incubator at 37°C, 5% CO2. Thereafter, a total of 20 μL CCK-8 solution was added to each well. The plates were placed at 37°C for 4 h. A microplate reader (Biotek, Winooski, VT, USA) was then used to measure the optical density (OD) value at 450 nm wavelength. Higher OD450 values meant stronger cell proliferation ability.
In addition, the cisplatin dose that inhibited cell growth by 50% (IC50) of OC cells was researched using the CCK-8 assay, as described in the previous paragraph. Notably, OC cells were cultured with DMEM containing 10% FBS and a series of cisplatin concentration (0.09765625, 0.1953125, 0.390625, 0.78125, 1.5625, 3.125, 6.25, 12.5, 25, 50, 100, 200, 400, 800 and 1600 μM) for 48 h. The IC50 was calculated using the LOGIT method.
Flow Cytometry
Cells were harvested after 48 h of transfection or cisplatin treatment. PBS was used to wash cells three times. After centrifugation, cells were added to a tube and dispersed in 1× binding buffer. FITC-annexin V (5 μL) and propidium iodide (PI) (10 μL) were then added to the tube. Cells in the tube were incubated for 5 min in darkness. Thereafter, PBS (2 mL) was added to the flow tube. Apoptosis was assessed by a flow cytometer (FACScan; BD Biosciences, Mountain View, CA, USA) equipped with Cell Quest software (BD Biosciences, Mountain View, CA, USA).
Dual-Luciferase Reporter Gene Assay
In this study, 293T cells (Shanghai Institute of Cell Biology, Shanghai, China) were used for the dual-luciferase reporter gene assay. In brief, 293T cells underwent transfection with miR-1305 mimic or its negative control using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Successfully transfected 293T cells were collected and seeded in six-well plates (1×104 cells/well). DMEM without FBS was added to each well (1 mL/well). The wild-type (WT)-LINC00184-3ʹ-UTR segments, mutant-type (Mut)-LINC00184-3ʹ-UTR segments, WT-CNTN1-3ʹ-UTR segments and Mut-CNTN1-3ʹ-UTR segments were designed and synthesized by GenePharma (Shanghai, China). The four segments were cloned into the luciferase reporters (Promega, Madison, WI, USA) according to the instructions. Subsequently, 293T cells, which had been transfected with miR-1305 mimic or its negative control, were further cotransfected with the four kinds of luciferase reporters. After 6 h incubation at 37°C, 5% CO2, DMEM with 10% FBS was added to each well. Cells were cultured for 48 h. The relative luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
RNA Pull-Down
The binding relationship between LINC00184 and miR-1305 was explored by an RNA pull-down assay. In detail, LINC00184 RNA was synthesized using T7 RNA polymerase (Ambion Company, Austin, TX, USA). Then, LINC00184 RNA was purified with the RNeasy Mini kit (Qiagen Company, Hilden, Germany) and DNase I (Qiagen Company, Hilden, Germany). The 3ʹ-terminal of the purified RNA was labeled with biotin RNA-labeled mixture (Ambion, Austin, TX, USA). SKOV3 cells were collected at a confluence of 80% and were incubated with cell lysis buffer (Solarbio, Beijing, China) for 30 min at 4°C. After centrifugation at 4°C (12,000×g, 10 min), the supernatant was collected and added to an RNase-free centrifuge tube. RIP buffer with a volume of 500 μL was added to the biotinylated RNA. Then, the mixture was added to the supernatant. Streptavidin beads were added for 4 h incubation at 4°C. The beads were washed and incubated with Proteinase K for 30 min. RNA was extracted using phenol–chloroform extraction, and the eluted miR-1305 was quantified by qRT-PCR.
qRT-PCR
Total RNA in clinical specimens and OC cells were extracted using TRIzol reagent (Solarbio, Beijing, China). The concentration of RNA samples was assessed using an ultraviolet spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) at 260 nm and 280 nm. Samples with A260/A280 ratio from 1.8–2.0 were available for subsequent analysis. Then, 5 μg of each RNA sample was collected for reverse transcription using the PrimeScript RT Reagent Kit (Takara, Dalian, Liaoning, China). The PCR experiment was conducted using the iQ5 quantitative PCR system (Bio-Rad, Hercules, CA, USA) according to the following conditions: pre-denaturation (95°C, 10 min), and 40 cycles of denaturation (95°C, 10 s), annealing (60°C, 20 s) and extension (72°C, 34 s). Primers were designed and synthesized by GenePharma (Shanghai, China), as follows: LINC00184, sense: 5ʹ-GGACCACCTATGGGGAAAGG-3ʹ, antisense: 5ʹ-GATGCCTTGCTTGACCCAAC-3ʹ; miR-1305, sense: 5ʹ-ACAGGCCGGGACAAGTGCAATA-3ʹ, antisense: 5ʹ-GCTGTCAACGATACGCTACGTAACG-3ʹ; U6, sense: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ, antisense: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ; CNTN1, sense: 5ʹ-CAACAAAACCATATCCTGCTGA-3ʹ, antisense: 5ʹ-AGATCACTGCCTATGTCCACCT-3ʹ; β-actin, sense: 5ʹ-TCACCCACACTGTGCCCATCTACGA-3ʹ, antisense: 5ʹ-CAGCGGAACCGCTCATTGCCAATGG-3ʹ. U6 was used as an internal control for LINC00184 and miR-1305.18 β-Actin was set as an internal control for CNTN1. The relative expression of LINC00184, miR-1305 and CNTN1 was determined by the 2-ΔΔCt method.
In Vivo Experiment
All animal experiments in this study were approved by the animal ethics committee of The Second Affiliated Hospital of Nantong University. Animal welfare met the five standards of National Institutes of Health Guide for the Care and Use of Laboratory Animals: freedom from hunger and thirst; freedom from physical discomfort; freedom from pain, injury or disease; freedom from fear and uneasiness; and freedom to express all natural behavior. Animals were provided by Shanghai Experimental Animal Center of the Chinese Academy of Sciences (Shanghai, China).
Nude mice (n=24, 4–5 weeks old) were randomly divided into the shNC group, shLINC00184 group, shNC + cisplatin group and shLINC00184 + cisplatin group. There were six mice in each group. The shRNA targeting LINC00184 (sh-LINC00184) and negative control (shNC) were purchased from GenePharma (Shanghai, China). A2780-DDP cells were transfected with sh-LINC00184 or shNC using Lipofectamine 2000. Cells were cultured to a confluence of 80% at 37°C, 5% CO2. Then, cells were collected and dispersed in PBS (1×107 cells/mL). Cells (20 μL cell suspension, 2×105 cells) transfected with shNC were subcutaneously inoculated into the center of the back of mice in the shNC group. At the same time, cells (20 μL cell suspension, 2×105 cells) transfected with sh-LINC00184 were subcutaneously injected into mice in the shLINC00184 group at the same site. Mice in the shNC + cisplatin group and shLINC00184 + cisplatin group were subcutaneously inoculated with A2780-DDP cells (20 μL cell suspension, 2×105 cells) transfected with shNC and sh-LINC00184 respectively. Notably, cisplatin (5 mg/kg) was intraperitoneally injected into mice in the two groups every day. The maximum diameter (a) and minimum diameter (b) of xenograft tumors were measured every 7 days. Tumor volume was calculated by the formula (a × b2/2). After 4 weeks, mice were euthanized, and xenograft tumors were stripped and weighed.
Immunohistochemistry
Xenograft tumors embedded in paraffin were prepared into sections (5 μm thickness). Xylene and gradient alcohol were used, respectively, for dewaxing and dehydrating sections. Citrate buffer (0.01 M) was added to sections for antigen retrieval in a repair kit. Sections were treated with H2O2 (3%) for 15 min, followed by being blocked with 5% goat serum for 1 h. Mouse anti-human Ki67 monoclonal antibody (1:100; Santa Cruz, CA, USA) and rabbit anti-human CNTN1 antibody (1:100; Shybio, Shanghai, China) was added to sections for 12 h incubation at 4°C. Thereafter, secondary antibody (1:200; Boster, Wuhan, Hubei, China) was used to incubate sections for 30 min at room temperature. The sections were washed three times with PBS. Then, sections were stained with diaminobenzidine (DAB) and counterstained with hematoxylin. Neutral resin was used to seal the sections. The Ki67-positive cells were observed under a microscope. Brownish yellow particles were considered as positive signals.
TUNEL Assay
Xenograft tumor sections were treated with xylene for dewaxing. Gradient alcohol was applied to rehydrate them. Paraformaldehyde (4%) was used to fix the sections for 30 min. Sections were treated with proteinase K solution (20 μg/mL) for 30 min at room temperature. TUNEL reaction buffer was used to treat the sections for 1 h at 37°C. After washing twice with PBS, sections were stained with DAB and hematoxylin for 2 min at room temperature. Dehydration of the sections was performed using gradient ethanol. After treatment with xylene, sections were sealed in neutral resin. TUNEL-positive cells (brownish yellow particles) were observed under a microscope. More TUNEL-positive cells meant more apoptotic cells.
Western Blot
Cells were collected at 80% confluence and washed twice with PBS. Cell lysate (Boster, Wuhan, Hubei, China) was added to cells for 30 min incubation on ice. In addition, xenograft tumors were ground into powder in liquid nitrogen, followed by incubation with cell lysate for 30 min on ice. The mixture of each sample was centrifuged for 10 min at 4°C, 3000 rev/min. Total proteins in the supernatant were then collected. The concentration of total proteins was determined by a BCA kit (Beyotime, Jiangsu, China). Each total protein sample and 2× loading buffer were mixed in a ratio of 1:1. Then, the mixture was boiled in boiling water for 10 min. Total protein samples (30 μg) underwent 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred onto a polyvinylidene fluoride (PVDF) membrane. The membrane was placed in 5% skimmed milk powder for 1 h blocking at room temperature. Thereafter, the membrane was maintained in rabbit anti-human CNTN1 (1:1000; Abcam, Cambridge, UK) and rabbit anti-mouse β-actin (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) primary antibody for 12 h at 4°C. TBST was used to wash the membrane three times, for 5 min each time. The membrane was then kept in horseradish peroxidase-labeled rabbit anti-goat IgG secondary antibody (1:5000; Boster, Wuhan, Hubei, China) for 1 h at room temperature. Enhanced chemiluminescence reagent was added to the membrane. After being exposed in the gel imager, the blots were captured using a Bio-Rad image analysis system (Bio-Rad, Hercules, CA, USA). The relative expression of CNTN1 protein was evaluated using Quantity One software (Bio-Rad, Hercules, CA, USA). β-Actin was used as the internal control.
Statistical Analysis
All experiments in this article were repeated independently three times. SPSS 19.0 was used for statistical analysis. Data were presented in the form of mean ± standard deviation. The expression level relationship between LINC00184 and miR-1305 or between miR-1305 and CNTN1 was researched with Pearson’s correlation analysis. Student’s t-test and one-way analysis of variance were used for comparisons between two groups and among at least three groups separately. p<0.05 was set as the threshold for statistical significance.
Results
LINC00184 Was Up-Regulated in OC, Especially in Cisplatin-Resistant OC Patients and Cell Lines
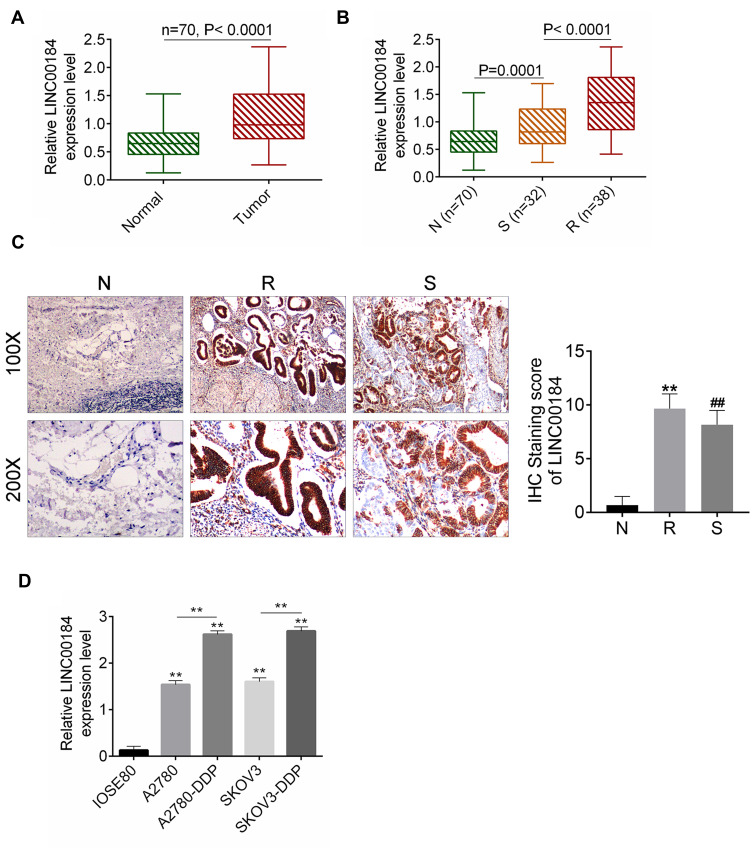
qRT-PCR was performed on 70 cases of OC tissues and adjacent normal tissues. The results showed that LINC00184 expression was significantly up-regulated in comparison with paired normal tissues (p<0.0001) (Figure 1A). Meanwhile, tumor tissues of the S group had significantly greater up-regulated LINC00184 expression than those of the N group (p=0.0001). Tumor tissues of the R group exhibited remarkably more up-regulated LINC00184 expression than those of the S group (p<0.0001) (Figure 1B). Similar results were also found in LINC00184 expression among the three groups according to ISH (Figure 1C). Patients’ characteristics are shown in Table 1. High LINC00184 expression was related to advanced FIGO stage, high histological grade, positive lymph-node invasion and cisplatin resistance. In addition, LINC00184 expression in A2780, SKOV3, A2780-DDP and SKOV3-DDP cell lines was remarkably more up-regulated than that in the IOSE80 cell line (p<0.01). Notably, A2780-DDP and SKOV3-DDP cell lines exhibited markedly more up-regulated LINC00184 expression than that in A2780 and SKOV3 cell lines (p<0.01) (Figure 1D). These results verified the high expression of LINC00184 in OC, especially in cisplatin-resistant OC patients and cell lines.
Figure 1.
LINC00184 was up-regulated in OC, especially in cisplatin-resistant OC patients and cell lines. (A) qRT-PCR for LINC00184 expression in OC tissues and paired adjacent normal tissues of 70 cases. (B) qRT-PCR for LINC00184 expression in normal tissues (N group), and tumor tissues from cisplatin-resistant patients (R group) and cisplatin-sensitive patients (S group). (C) ISH for LINC00184 expression in the N group, R group and S group (magnification: 100× and 200×). **p<0.01 relative to N group. ##p<0.01 relative to R group. (D) qRT-PCR for LINC00184 expression in human normal ovarian epithelial cell line (IOSE80), OC cell lines (A2780, SKOV3) and OC cisplatin-resistant cell lines (A2780-DDP, SKOV3-DDP). **p<0.01 relative to IOSE80 cell line. All experiments were performed in triplicate.
Table 1.
Association Between LINC00184 Expression and Clinical Characteristics of Ovarian Cancer Patients
| Variables | N | LINC00184 Expression | p Value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | ||||
| <50 | 27 | 15 | 12 | >0.05 |
| ≥50 | 43 | 24 | 19 | |
| FIGO stage | ||||
| I–II | 48 | 28 | 20 | <0.05 |
| III–IV | 22 | 5 | 17 | |
| Histological grade | ||||
| Low | 39 | 27 | 12 | >0.05 |
| High | 31 | 13 | 18 | |
| Lymph-node invasion | ||||
| Negative | 42 | 29 | 13 | <0.05 |
| Positive | 28 | 12 | 16 | |
| Chemotherapeutic response | ||||
| Sensitive | 32 | 22 | 10 | <0.01 |
| Resistant | 38 | 17 | 21 | |
LINC00184 Facilitated Proliferation and Cisplatin Resistance, and Inhibited Apoptosis of OC Cells and Cisplatin-Resistant OC Cells
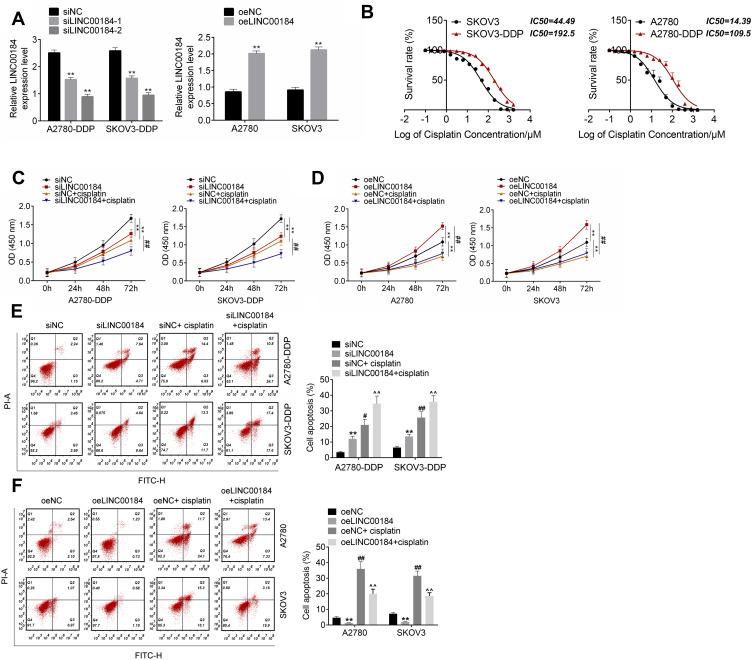
qRT-PCR was used to detect the transfection efficiency of OC cells. A2780-DDP and SKOV3-DDP cells of the siLINC00184-1 group and siLINC00184-2 group showed much lower LINC00184 expression than those of the siNC group (p<0.01). Meanwhile, A2780 and SKOV3 cells of the oeLINC00184 group exhibited much higher LINC00184 expression than those of the oeNC group (p<0.01) (Figure 2A). Thus, the expression of LINC00184 in OC cells was successfully regulated by transfection. It should be noted that A2780-DDP and SKOV3-DDP cells of the siLINC00184-2 group had lower LINC00184 expression than those of the siLINC00184-1 group. Therefore, A2780-DDP and SKOV3-DDP cells of the siLINC00184-2 group were used in subsequent experiments and renamed the siLINC00184 group. The IC50 of OC cells was investigated. As shown in Figure 2B, SKOV3, SKOV3-DDP, A2780 and A2780-DDP cells were resistant to cisplatin in a dose-dependent manner. Notably, the IC50 values for SKOV3, SKOV3-DDP, A2780 and A2780-DDP cells were 44.49, 192.5, 14.39 and 109.5 μM, respectively.
Figure 2.
LINC00184 facilitated proliferation and cisplatin resistance, and inhibited apoptosis of OC cells and cisplatin-resistant OC cells. (A) After transfection, qRT-PCR was used to detect the transfection efficiency of OC cells. **p<0.01 compared with the siNC group or oeNC group. (B) CCK-8 assay was used to detect cisplatin resistance in OC cells. The IC50 was calculated using the LOGIT method. (C) CCK-8 assay for A2780-DDP and SKOV3-DDP cell proliferation. **p<0.01 compared with the siNC group. ##p<0.01 compared with the siNC + cisplatin group. (D) CCK-8 assay for A2780 and SKOV3 cell proliferation. **p<0.01 compared with the oeNC group. ##p<0.01 compared with the oeNC + cisplatin group. (E) Flow cytometry for A2780-DDP and SKOV3-DDP cell apoptosis. **p<0.01, #p<0.05 and ##p<0.01 compared with the siNC group. ^^p<0.01 compared with the siNC + cisplatin group. (F) Flow cytometry for A2780 and SKOV3 cell apoptosis. **p<0.01 and ##p<0.01 compared with the oeNC group. ^^p<0.01 compared with the oeNC + cisplatin group. All experiments were performed in triplicate.
Cell proliferation was explored by the CCK-8 assay. For A2780-DDP and SKOV3-DDP cells, obviously decreased proliferation was found in the siLINC00184 group and siNC + cisplatin group compared with the siNC group (p<0.01). The proliferation in the siLINC00184 + cisplatin group was much lower than that in the siNC + cisplatin group (p<0.01) (Figure 2C). For A2780 and SKOV3 cells, relative to the oeNC group, the proliferation was greatly increased in the oeLINC00184 group and significantly deceased in the oeNC + cisplatin group (p<0.01). In comparison with the oeNC + cisplatin group, the proliferation of the oeLINC00184 + cisplatin group was significantly increased (p<0.01) (Figure 2D).
Thereafter, apoptosis of cells in each group was assessed by flow cytometry. For A2780-DDP and SKOV3-DDP cells, the siLINC00184 group and siNC + cisplatin group exhibited markedly higher apoptotic ratios than the siNC group (p<0.05 or p<0.01). Compared with the siNC + cisplatin group, an obviously higher apoptotic ratio was observed in the siLINC00184 + cisplatin group (p<0.01) (Figure 2E). For A2780 and SKOV3 cells, compared to the oeNC group, the apoptotic ratio was remarkably decreased in the oeLINC00184 group and markedly increased in the oeNC + cisplatin group (p<0.01). Relative to the oeNC + cisplatin group, a much lower apoptotic ratio was found in the oeLINC00184 + cisplatin group (p<0.01) (Figure 2F).
miR-1305 Was Sponged by LINC00184
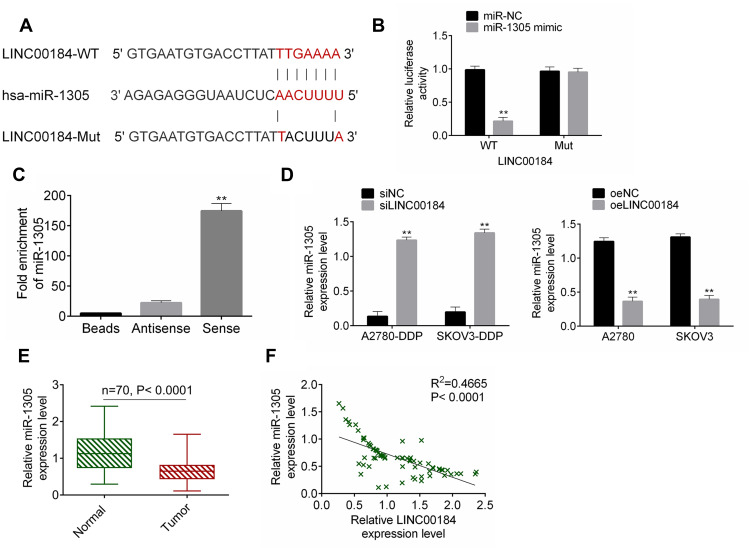
The binding site of LINC00184 and miR-1305 was predicted by miRDB (http://mirdb.org/cgi-bin/custom_predict/customDetail.cgi). The LINC00184-WT and LINC00184-Mut fragments containing the binding site for miR-1305 were designed and synthesized (Figure 3A). The accurate regulatory relationship between LINC00184 and miR-1305 was verified by the dual-luciferase reporter assay. The luciferase activity of LINC00184-WT reporter was seriously reduced in the miR-1305 mimic group in comparison with the miR-NC group (p<0.01). No statistically significant differences were observed in the luciferase activity of LINC00184-Mut reporter between the miR-NC group and miR-1305 mimic group (Figure 3B). An RNA pull-down experiment was performed and the enrichment of miR-1305 was then measured by qRT-PCR. The results indicated that miR-1305 was significantly enriched by LINC00184 (p<0.01) (Figure 3C). A2780-DDP and SKOV3-DDP cells of the siLINC00184 group showed significantly increased miR-1305 expression compared with those of the siNC group (p <0.01). The expression of miR-1305 in A2780 and SKOV3 cells of the oeLINC00184 group was greatly decreased compared with those of the oeNC group (p<0.01) (Figure 3D). Detection in clinical specimens revealed significantly down-regulated miR-1305 expression in tumor tissues compared with that in normal tissues (p<0.01) (Figure 3E). A significant negative correlation was found between miR-1305 and LINC00184 expression in tumor tissues (p<0.0001) (Figure 3F).
Figure 3.
miR-1305 was sponged by LINC00184. (A) The LINC00184-WT and LINC00184-Mut fragments containing the binding site for miR-1305. (B) Dual-luciferase reporter assay. **p<0.01. (C) An RNA pull-down experiment was performed and the enrichment of miR-1305 was measured by qRT-PCR. **p<0.01. (D) miR-1305 expression detection in cells by qRT-PCR. (E) qRT-PCR for miR-1305 expression in clinical specimens. **p<0.01. (F) Pearson’s correlation analysis for LINC00184 and miR-1305 expression in tumor tissues. All experiments were performed in triplicate.
CNTN1 Was a Target Gene of miR-1305
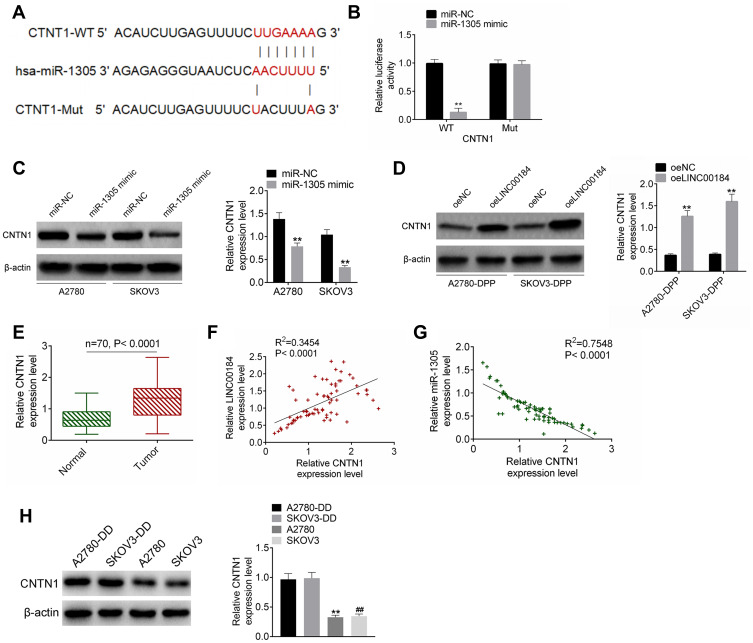
The TargetScan online prediction (http://www.targetscan.org/vert_71/) results showed the binding sites for miR-1305 and CNTN1. Fragments of CNTN1-WT and CNTN1-Mut containing the binding site for miR-1305 were designed accordingly (Figure 4A). The dual-luciferase reporter assay revealed that miR-1305 mimic significantly reduced the luciferase activity of CNTN1-WT reporter (p<0.01). However, there was no obvious difference in CNTN1-Mut reporter luciferase activity between the miR-1305 mimic group and miR-NC group (Figure 4B). For A2780 and SKOV3 cells of the miR-1305 mimic group, CNTN1 protein expression was aberrantly down-regulated compared with that of the miR-NC group (p<0.01) (Figure 4C). In contrast, seriously up-regulated CNTN1 protein expression was observed in A2780-DDP and SKOV3-DDP cells of the oeLINC00184 group relative to the oeNC group (p<0.01) (Figure 4D). qRT-PCR for clinical tissues indicated significantly up-regulated CNTN1 mRNA expression in tumor tissues compared with that in normal tissues (p<0.0001) (Figure 4E). In clinical tumor specimens, CNTN1 mRNA expression was positively correlated with LINC00184 expression (p<0.0001). However, a negative correlation was found between CNTN1 mRNA and miR-1305 expression (p<0.0001) (Figure 4F and G). CNTN1 protein expression in vitro was explored by Western blot. A2780-DDP cells had dramatically higher CNTN1 protein expression than A2780 cells (p<0.01). Simultaneously, SKOV3-DDP cells showed much higher CNTN1 protein expression than SKOV3 cells (p<0.01) (Figure 4).
Figure 4.
CNTN1 was a target gene of miR-1305. (A) Fragments of CNTN1-WT and CNTN1-Mut containing the binding site for miR-1305. (B) Dual-luciferase reporter assay. **p<0.01. (C) Western blot for CNTN1 protein expression in A2780 and SKOV3 cells after transfection. **p<0.01. (D) Western blot for CNTN1 protein expression in A2780-DDP and SKOV3-DDP cells after transfection. **p<0.01. (E) qRT-PCR for CNTN1 mRNA expression in clinical tissues. (F) Pearson’s correlation analysis for LINC00184 and CNTN1 mRNA expression in clinical tumor specimens. (G) Pearson’s correlation analysis for miR-1305 and CNTN1 mRNA expression in clinical tumor specimens. (H) Western blot was used to research CNTN1 protein expression in A2780, SKOV3, A2780-DDP and SKOV3-DDP cells. **p<0.01 relative to A2780-DDP cells. ##p<0.01 relative to SKOV3-DDP cells. All experiments were performed in triplicate.
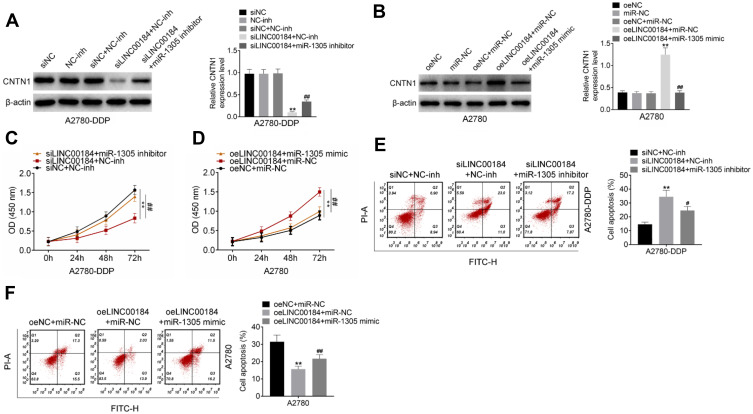
LINC00184 Exacerbated the Malignant Phenotype of OC Cells and Cisplatin-Resistant OC Cells by Regulating miR-1305/CNTN1 Axis
Rescue experiments were performed to verify the underlying molecular mechanism of LINC00184 affecting the malignant phenotype of OC cells. For A2780-DDP cells of the siLINC00184 + NC-inh group, the CNTN1 protein expression was markedly decreased compared with that of the siNC group, NC-inh group and siNC + NC-inh group (p<0.01). Relative to the siLINC00184 + NC-inh group, significantly increased CNTN1 protein expression was found in A2780-DDP cells of the siLINC00184 + miR-1305 inhibitor group (p<0.01) (Figure 5A). In contrast, A2780 cells of the oeLINC00184 + miR-NC group exhibited greatly increased CNTN1 protein expression compared with those of the oeNC group, miR-NC group and oeNC + miR-NC group (p<0.01). The CNTN1 protein expression was seriously reduced in the oeLINC00184 + miR-1305 mimic group relative to the oeLINC00184 + miR-NC group (p<0.01) (Figure 5B). Therefore, LINC00184 could elevate the expression of CNTN1 via sponging miR-1305.
Figure 5.
LINC00184 exacerbated the malignant phenotype of OC cells and cisplatin-resistant OC cells by regulating the miR-1305/CNTN1 axis. (A) Western blot for CNTN1 protein expression in A2780-DDP cells. **p<0.01 relative to the siNC group, NC-inh group and siNC + NC-inh group. ##p<0.01 relative to the siLINC00184 + NC-inh group. (B) Western blot for CNTN1 protein expression in A2780 cells. **p<0.01 relative to the oeNC group, miR-NC group and oeLINC00184 + miR-NC group. (C) CCK-8 assay for proliferation of A2780-DDP cells. **p<0.01 compared with the siNC + NC-inh group or oeNC + miR-NC group. #p<0.05 and ##p<0.01 compared with the siLINC00184 + NC-inh group or oeLINC00184 + miR-NC group. (D) CCK-8 assay for proliferation of A2780 cells. **p<0.01 compared with the siNC + NC-inh group or oeNC + miR-NC group. #p<0.05 and ##p<0.01 compared with the siLINC00184 + NC-inh group or oeLINC00184 + miR-NC group. (E) Flow cytometry for apoptosis of A2780-DDP cells. **p<0.01 compared with the siNC + NC-inh group or oeNC + miR-NC group. #p<0.05 and ##p<0.01 compared with the siLINC00184 + NC-inh group or oeLINC00184 + miR-NC group. (F) Flow cytometry for apoptosis of A2780 cells. **p<0.01 compared with the siNC + NC-inh group or oeNC + miR-NC group. #p<0.05 and ##p<0.01 compared with the siLINC00184 + NC-inh group or oeLINC00184 + miR-NC group. All experiments were performed in triplicate.
The CCK-8 assay illustrated that, compared with the siNC + NC-inh group, the proliferation of A2780-DDP cells in the siLINC00184 + NC-inh group was significantly reduced (p<0.01). However, there was significantly elevated proliferation of A2780-DDP cells in the siLINC00184 + miR-1305 inhibitor group in comparison with the siLINC00184 + NC-inh group (p<0.01) (Figure 5C). On the other hand, for A2780 cells, the oeLINC00184 + miR-NC group showed obviously increased proliferation compared with the oeNC + miR-NC group (p<0.01). Meanwhile, the proliferation in the oeLINC00184 + miR-1305 mimic group was markedly reduced compared with that in the oeLINC00184 + miR-NC group (p<0.01) (Figure 5D).
According to results from flow cytometry, A2780-DDP cells of the siLINC00184 + NC-inh group had a much increased apoptotic ratio compared with the siNC + NC-inh group (p<0.01). An obviously reduced apoptotic ratio was presented in A2780-DDP cells of the siLINC00184 + miR-1305 inhibitor group relative to the siLINC00184 + NC-inh group (p<0.05) (Figure 5E). Conversely, for A2780 cells, a significantly decreased apoptotic ratio was found in the oeLINC00184 + miR-NC group compared with the oeNC + miR-NC group (p<0.01). Relative to the apoptotic ratio of A2780 cells in the oeLINC00184 + miR-NC group, it was remarkably increased in the oeLINC00184 + miR-1305 mimic group (p<0.01) (Figure 5F).
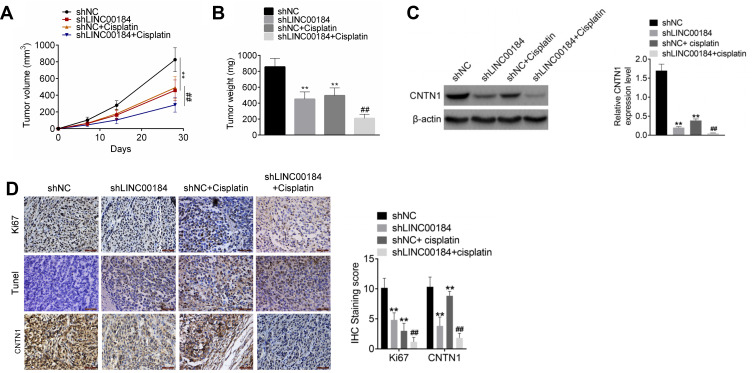
LINC00184 Silencing Suppressed OC Cell Growth and Cisplatin Resistance In Vivo
An in vivo experiment was carried out using nude mice. The volume and weight of xenograft tumors in the shLINC00184 group and shNC + cisplatin group were both decreased relative to the shNC group (p<0.01). Meanwhile, in comparison with the shNC + cisplatin group, dramatically decreased volume and weight of xenograft tumors were observed in the shLINC00184 + cisplatin group (p<0.01) (Figure 6A and B). CNTN1 protein expression in xenograft tumors was investigated by Western blot, and markedly lower CNTN1 protein expression was found in xenograft tumors of the shLINC00184 group and shNC + cisplatin group relative to the shNC group (p<0.01). CNTN1 protein expression in xenograft tumors of the shNC + cisplatin group was much higher than in the shLINC00184 + cisplatin group (p<0.01) (Figure 6C). Immunohistochemistry and TUNEL experiments illustrated that, compared with the shNC group, fewer Ki67 and CNTN1-positive cells (p<0.01) and more TUNEL-positive cells were observed in xenograft tumors of the shLINC00184 group and shNC + cisplatin group. Relative to the shNC + cisplatin group, xenograft tumors of the shLINC00184 + cisplatin group exhibited fewer Ki67 and CNTN1-positive cells (p<0.01) and more TUNEL-positive cells (Figure 6D).
Figure 6.
LINC00184 silencing suppressed OC cell growth and cisplatin resistance in vivo. (A) Volume ofxenograft tumors in nude mice. (B) Weight of xenograft tumors in nude mice. (C) Western blot was used to research CNTN1 protein expression in xenograft tumors. (D) Immunohistochemistry and TUNEL experiments for xenograft tumors in nude mice (magnification: 200×). **p<0.01 compared with the shNC group. ##p<0.01 compared with the shNC + cisplatin group. All experiments were performed in triplicate.
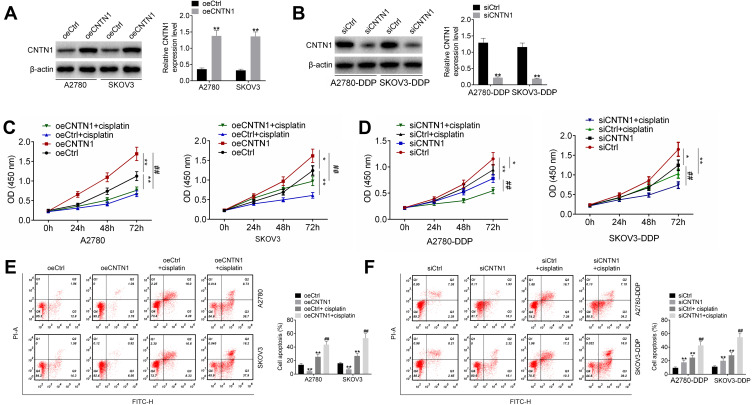
CNTN1 Overexpression Promoted Cisplatin Resistance in OC Cells
This study further researched the effect of CNTN1 on cisplatin resistance in OC cells. As shown in Figure 7A and B, A2780 and SKOV3 cells of the oeCNTN1 group exhibited much higher CNTN1 protein expression than those of the oeCtrl group (p<0.01). Conversely, much lower CNTN1 protein expression was found in A2780-DDP and SKOV3-DDP cells of the siCNTN1 group relative to the siCtrl group (p<0.01). Thus, CNTN1 protein expression in these cells was successfully regulated by transfection.
Figure 7.
CNTN1 overexpression promoted cisplatin resistance in OC cells. (A and B) Western blot was used to research the transfection efficiency of OC cells. (C and D) CCK-8 assay was performed to research OC cell proliferation. (E and F) Flow cytometry was used for the detection of OC cell apoptosis. *p<0.05 relative to the oeCtrl group or siCtrl group. **p<0.01 relative to the oeCtrl group or siCtrl group. ##p<0.01 relative to the oeCNTN1 group or siCNTN1 group. All experiments were performed in triplicate.
The CCK-8 assay showed that, relative to the oeCtrl group, A2780 and SKOV3 cells of the oeCNTN1 group had markedly higher proliferation (p<0.05 or p<0.01). However, A2780 and SKOV3 cells of the oeCtrl + cisplatin group displayed obviously lower proliferation than those of the oeCtrl group (p<0.01). Compared with the oeCNTN1 group, greatly decreased proliferation was observed in A2780 and SKOV3 cells of the oeCNTN1 + cisplatin group (p<0.01) (Figure 7C). In addition, the proliferation was remarkably decreased in A2780-DDP and SKOV3-DDP cells of the siCNTN1 group and siCtrl + cisplatin group relative to the siCtrl group (p<0.05 or p<0.01). In comparison with the siCNTN1 group, A2780-DDP and SKOV3-DDP cells of the siCNTN1 + cisplatin group presented much lower proliferation (p<0.01) (Figure 7D).
Apoptosis was explored by flow cytometry. For A2780 and SKOV3 cells, significantly lower apoptosis in the oeCNTN1 group and higher apoptosis in the oeCtrl + cisplatin group occurred compared with the oeCtrl group (p<0.01). Relative to the oeCNTN1 group, greatly increased apoptosis was observed in A2780 and SKOV3 cells of the oeCNTN1 + cisplatin group (p<0.01) (Figure 7E). For A2780-DDP and SKOV3-DDP cells, dramatically higher apoptosis was exhibited in the siCNTN1 group and siCtrl + cisplatin group compared with the siCtrl group (p<0.01). However, in comparison with the siCNTN1 group, apoptosis was distinctly increased in A2780-DDP and SKOV3-DDP cells of the siCNTN1 + cisplatin group (p<0.01) (Figure 7F).
Discussion
LncRNAs have emerged as novel biomarkers to distinguish between cisplatin-resistant and cisplatin-sensitive OC.19 Some lncRNAs have been identified as being involved in the regulation of OC cisplatin resistance. LncRNA UCA1 was found to be up-regulated in tumor tissues of OC cisplatin-resistant patients. In vitro research showed that lncRNA UCA1 enhanced OC cell proliferation and attenuated cisplatin-induced OC cell apoptosis.20 LncRNA EBIC was suggested as a potential target for OC treatment. The researchers discovered that lncRNA EBIC enhanced the cisplatin resistance of OC cells and predicted poor survival in OC patients.21 Conversely, Linc00312 expression was decreased in SKOV3/DDP cells. This may be a promising target for cisplatin-resistant OC, because overexpression of Linc00312 could improve the cisplatin sensitivity of SKOV3/DDP cells.22 Moreover, LINC01125 expression in cisplatin-resistant OC tissues and cells was observed to be reduced. Increased expression of LINC01125 could enhance the sensitivity of OC cells to cisplatin.23 This research indicated that LINC00184 expression was aberrantly up-regulated in OC patients. High LINC00184 expression was related to advanced FIGO stage, high histological grade, positive lymph-node invasion and cisplatin resistance. In vitro studies revealed that LINC00184 could promote OC cell proliferation and cisplatin resistance, and reduce apoptosis of OC cells. In terms of the mechanism, it may be that LINC00184 enhanced OC development by sponging the miR-1305/CNTN1 axis.
Previous studies have shown that lncRNAs exert their biological effects on human tumors mainly through mediating the specific miRNAs.24,25 In this paper, miR-1305 was sponged by LINC00184. LINC00184 silencing inhibited the malignant phenotype of A2780 and A2780-DDP cells, including reducing proliferation and enhancing apoptosis. However, the transfection with miR-1305 inhibitor reversed these effects on A2780 and A2780-DDP cells. miR-1305 was previously reported to regulate the progression of human tumors. For instance, decreased miR-1305 expression occurred in non-small cell lung cancer tissues, which was seriously associated with poor patient prognosis. Data from in vitro studies suggested that miR-1305 up-regulation could suppress proliferation and promote apoptosis of non-small cell lung cancer cells.26 Similar results have been found in other human malignancies, such as liver cancers and bladder cancer.27,28 In gynecological malignancies, miR-1305 expression was noticed to be reduced in cervical cancer. It acted as a tumor suppressor in cervical cancer by mediating the Wnt/β-catenin pathway.29 In addition, Andrade et al30 found that miR-1305 was an independent risk factor for the prognosis of triple-negative breast cancer. This study found that miR-1305 was down-regulated in OC. It is worth noting that miR-1305 inhibition reversed the inhibiting effect of LINC00184 knockdown on OC progression.
In this study, we noticed multiple targets of miR-1305 through TargetScan online prediction. Of these, the function of CNTN1 in OC has not been reported previously. Thus, CNTN1 was selected as the subject in this research. Our data illustrated that CNTN1 expression was directly inhibited by miR-1305. CNTN1 is a member of the CNTN family of neural cell-recognition molecules.31,32 Generally, CNTN1 is considered as a cancer-promoting gene in human tumors. CNTN1 was a promoting factor in prostate cancer, which promoted xenograft tumors growth in vivo.33 The aberrantly up-regulated CNTN1 mRNA and protein were discovered in hepatocellular carcinoma. High expression of CNTN1 was associated with poor overall survival and disease-free survival in hepatocellular carcinoma patients.31 CNTN1 was suggested as a potential biomarker and treatment target for thyroid cancer, because it could promote thyroid cancer cell proliferation.34 In breast cancer, CNTN1 facilitated the metastasis and growth of xenograft tumors.35 CNTN1 expression in OC has rarely been reported. This research indicated that CNTN1 expression was remarkably increased in OC tissues and cells. CNTN1 overexpression promoted OC cell proliferation and cisplatin resistance, and reduced apoptosis of OC cells. The opposite effect was observed in OC cells by knockdown of CNTN1. LINC00184 may promote OC cell proliferation and cisplatin resistance, and weaken apoptosis of OC cells by enhancing CNTN1 expression via sponging miR-1305. Previous studies found that CNTN1 could activate the PI3K/AKT signaling pathway, thereby enhancing cisplatin resistance and proliferation, and reducing apoptosis of tumor cells.36,37 The PI3K/AKT signaling pathway is one of the important tumor regulation signaling pathways. Studies have discovered that inhibition of the activity of the PI3K/AKT signaling pathway was conducive for the induction of apoptosis and inhibition of proliferation of OC cells.38,39 Moreover, it has been found that the activation of the PI3K/AKT signaling pathway can lead to the sustained growth and chemoresistance of OC cells.40 In this study, it may be that LINC00184 enhanced CNTN1 expression via sponging miR-1305. Thereafter, CNTN1 facilitated the activity of the PI3K/AKT signaling pathway, thereby enhancing proliferation and cisplatin resistance, and reducing apoptosis of OC cells.
There was a limitation to this study. To better clarify that LINC00184 promoted OC development by targeting the miR-1305/CNTN1 axis, in vivo rescue experiments should be performed. However, due to the limitations of the laboratory, such experiments could not be carried out in this study. In future research, we will conduct these in vivo rescue experiments.
Conclusions
This article reported the effects of LINC00184 on OC progression and cisplatin resistance. The results revealed that LINC00184 was up-regulated in OC, especially in cisplatin-resistant OC patients and cell lines. LINC00184 silencing could promote apoptosis and weaken proliferation and cisplatin resistance of OC cells both in vitro and in vivo. Regarding the mechanism, LINC00184 may enhance CNTN1 expression via sponging miR-1305. Thus, LINC00184 was suggested as a potential target for the cisplatin-resistant treatment of OC.
Acknowledgments
The present study is supported by The Science and Technology Projects Fund of Nantong City (no. MS22016070, MS12018007 and MS12019004) and The Maternal and Child Health Research Projects Fund of Jiangsu Province (no. F201836).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Wang JY, Lu AQ, Chen LJ. LncRNAs in ovarian cancer. Clin Chim Acta. 2019;490:17–27. doi: 10.1016/j.cca.2018.12.013 [DOI] [PubMed] [Google Scholar]
- 2.Yan H, Xia JY, Feng FZ. Long non-coding RNA ENST00000457645 reverses cisplatin resistance in CP70 ovarian cancer cells. Genet Mol Res. 2017;16:gmr16019411. doi: 10.4238/gmr16019411 [DOI] [PubMed] [Google Scholar]
- 3.Huang KC, Yang J, Ng MC, et al. Cyclin A1 expression and paclitaxel resistance in human ovarian cancer cells. Eur J Cancer. 2016;67:152–163. doi: 10.1016/j.ejca.2016.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Risnayanti C, Jang YS, Lee J, Ahn HJ. PLGA nanoparticles co-delivering MDR1 and BCL2 siRNA for overcoming resistance of paclitaxel and cisplatin in recurrent or advanced ovarian cancer. Sci Rep. 2018;8:7498. doi: 10.1038/s41598-018-25930-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sajadpoor Z, Amini-Farsani Z, Teimori H, Shamsara M, Yadollahi F. Valproic acid promotes apoptosis and cisplatin sensitivity through downregulation of H19 noncoding RNA in ovarian A2780 cells. Appl Biochem Biotechnol. 2018;185:1132–1144. doi: 10.1007/s12010-017-2684-0 [DOI] [PubMed] [Google Scholar]
- 6.Chen QN, Wei CC, Wang ZX, Sun M. Long non-coding RNAs in anti-cancer drug resistance. Oncotarget. 2017;8:1925–1936. doi: 10.18632/oncotarget.12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sirichanchuen B, Pengsuparp T, Chanvorachote P. Long-term cisplatin exposure impairs autophagy and causes cisplatin resistance in human lung cancer cells. Mol Cell Biochem. 2012;364:11–18. doi: 10.1007/s11010-011-1199-1 [DOI] [PubMed] [Google Scholar]
- 8.Yu Y, Zhang X, Tian H, Zhang Z, Tian Y. Knockdown of long non-coding RNA HOTAIR increases cisplatin sensitivity in ovarian cancer by inhibiting cisplatin-induced autophagy. J BUON. 2018;23:1396–1401. [PubMed] [Google Scholar]
- 9.Miao JT, Gao JH, Chen YQ, Chen H, Meng HY, Lou LG. LncRNA ANRIL affects the sensitivity of ovarian cancer to cisplatin via regulation of let-7a/HMGA2 axis. Biosci Rep. 2019;39:BSR20182101. doi: 10.1042/BSR20182101 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Bai L, Wang A, Zhang Y, Xu X, Zhang X. Knockdown of MALAT1 enhances chemosensitivity of ovarian cancer cells to cisplatin through inhibiting the Notch1 signaling pathway. Exp Cell Res. 2018;366:161–171. doi: 10.1016/j.yexcr.2018.03.014 [DOI] [PubMed] [Google Scholar]
- 11.Wang DY, Li N, Cui YL. Long non-coding RNA CCAT1 sponges miR-454 to promote chemoresistance of ovarian cancer cells to cisplatin by regulation of surviving. Cancer Res Treat. 2020;52:798–814. doi: 10.4143/crt.2019.498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao Y, Zhang T, Qi L, et al. Integrated analysis of co-expression and ceRNA network identifies five lncRNAs as prognostic markers for breast cancer. J Cell Mol Med. 2019;23:8410–8419. doi: 10.1111/jcmm.14721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Huang K, Wen F, et al. LINC00184 silencing inhibits glycolysis and restores mitochondrial oxidative phosphorylation in esophageal cancer through demethylation of PTEN. EBioMedicine. 2019;44:298–310. doi: 10.1016/j.ebiom.2019.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Nie Y, Ding Y, Yang M. GRHL2 upregulation predicts a poor prognosis and promotes the resistance of serous ovarian cancer to cisplatin. Onco Targets Ther. 2020;13:6303–6314. doi: 10.2147/OTT.S250412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song X, Cao G, Jing L, et al. Analysing the relationship between lncRNA and protein-coding gene and the role of lncRNA as ceRNA in pulmonary fibrosis. J Cell Mol Med. 2014;18:991–1003. doi: 10.1111/jcmm.12243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R, Guo H, Lu S. MiR-335-5p restores cisplatin sensitivity in ovarian cancer cells through targeting BCL2L2. Cancer Med. 2018;7:4598–4609. doi: 10.1002/cam4.1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C, Tan J, Zhu J, Wang S, Wei G. YAP promotes tumorigenesis and cisplatin resistance in neuroblastoma. Oncotarget. 2017;8:37154–37163. doi: 10.18632/oncotarget.16209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee NK, Lee JH, Ivan C, et al. MALAT1 promoted invasiveness of gastric adenocarcinoma. BMC Cancer. 2017;17:46. doi: 10.1186/s12885-016-2988-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q, Zhang J, Zhou J, et al. lncRNAs are novel biomarkers for differentiating between cisplatin-resistant and cisplatin-sensitive ovarian cancer. Oncol Lett. 2018;15:8363–8370. doi: 10.3892/ol.2018.8433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Niu H, Qin Q, et al. lncRNA UCA1 mediates resistance to cisplatin by regulating the miR-143/FOSL2-signaling pathway in ovarian cancer. Mol Ther Nucleic Acids. 2019;17:92–101. doi: 10.1016/j.omtn.2019.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu QF, Tang YX, Wang X. LncRNA EBIC promoted proliferation, metastasis and cisplatin resistance of ovarian cancer cells and predicted poor survival in ovarian cancer patients. Eur Rev Med Pharmacol Sci. 2018;22:4440–4447. doi: 10.26355/eurrev_201807_15495 [DOI] [PubMed] [Google Scholar]
- 22.Zhang C, Wang M, Shi C, Shi F, Pei C. Long non-coding RNA Linc00312 modulates the sensitivity of ovarian cancer to cisplatin via the Bcl-2/Caspase-3 signaling pathway. Biosci Trends. 2018;12:309–316. doi: 10.5582/bst.2018.01052 [DOI] [PubMed] [Google Scholar]
- 23.Guo J, Pan H. Long noncoding RNA LINC01125 enhances cisplatin sensitivity of ovarian cancer via miR-1972. Med Sci Monit. 2019;25:9844–9854. doi: 10.12659/MSM.916820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Y, Dang W, Zhang S, et al. The role of exosomal noncoding RNAs in cancer. Mol Cancer. 2019;18:37. doi: 10.1186/s12943-019-0984-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdollahzadeh R, Daraei A. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J Cell Physiol. 2019;234:10080–10100. doi: 10.1002/jcp.27941 [DOI] [PubMed] [Google Scholar]
- 26.Cai Y, Hao Y, Ren H, et al. miR-1305 inhibits the progression of non-small cell lung cancer by regulating MDM2. Cancer Manag Res. 2019;11:9529–9540. doi: 10.2147/CMAR.S220568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei X, You X, Zhang J, Zhou C. MicroRNA-1305 inhibits the stemness of LCSCs and tumorigenesis by repressing the UBE2T-dependent Akt-signaling pathway. Mol Ther Nucleic Acids. 2019;16:721–732. doi: 10.1016/j.omtn.2019.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su Y, Feng W, Shi J, Chen L, Huang J, Lin T. circRIP2 accelerates bladder cancer progression via miR-1305/Tgf-β2/smad3 pathway. Mol Cancer. 2020;19:23. doi: 10.1186/s12943-019-1129-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W, Zhuang R, Feng S, et al. Long non-coding RNA ASB16-AS1 enhances cell proliferation, migration and invasion via functioning as a ceRNA through miR-1305/Wnt/β-catenin axis in cervical cancer. Biomed Pharmacother. 2020;125:109965. doi: 10.1016/j.biopha.2020.109965 [DOI] [PubMed] [Google Scholar]
- 30.Andrade F, Nakata A, Gotoh N, Fujita A. Large miRNA survival analysis reveals a prognostic four-biomarker signature for triple negative breast cancer. Genet Mol Biol. 2020;43:e20180269. doi: 10.1590/1678-4685-gmb-2018-0269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li GY, Huang M, Pan TT, Jia WD. Expression and prognostic significance of contactin 1 in human hepatocellular carcinoma. Onco Targets Ther. 2016;9:387–394. doi: 10.2147/OTT.S97367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohebiany AN, Harroch S, Bouyain S. New insights into the roles of the contactin cell adhesion molecules in neural development. Adv Neurobiol. 2014;8:165–194. [DOI] [PubMed] [Google Scholar]
- 33.Yan J, Ojo D, Kapoor A, et al. Neural cell adhesion protein CNTN1 promotes the metastatic progression of prostate cancer. Cancer Res. 2016;76:1603–1614. doi: 10.1158/0008-5472.CAN-15-1898 [DOI] [PubMed] [Google Scholar]
- 34.Shi K, Xu D, Yang C, et al. Contactin 1 as a potential biomarker promotes cell proliferation and invasion in thyroid cancer. Int J Clin Exp Pathol. 2015;8:12473–12481. [PMC free article] [PubMed] [Google Scholar]
- 35.Chen N, He S, Geng J, et al. Overexpression of Contactin 1 promotes growth, migration and invasion in Hs578T breast cancer cells. BMC Cell Biol. 2018;19:5. doi: 10.1186/s12860-018-0154-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang B, Yang X, Zhao T, et al. Upregulation of contactin-1 expression promotes prostate cancer progression. Oncol Lett. 2020;19:1611–1618. doi: 10.3892/ol.2019.11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang R, Sun S, Ji F, et al. CNTN-1 enhances chemoresistance in human lung adenocarcinoma through induction of epithelial-mesenchymal transition by targeting the PI3K/Akt pathway. Cell Physiol Biochem. 2017;43:465–480. doi: 10.1159/000480473 [DOI] [PubMed] [Google Scholar]
- 38.Zhou J, Jiang YY, Chen H, Wu YC, et al. Tanshinone I attenuates the malignant biological properties of ovarian cancer by inducing apoptosis and autophagy via the inactivation of PI3K/AKT/mTOR pathway. Cell Prolif. 2020;53:e12739. doi: 10.1111/cpr.12739 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Deng J, Bai X, Feng X, et al. Inhibition of PI3K/Akt/mTOR signaling pathway alleviates ovarian cancer chemoresistance through reversing epithelial-mesenchymal transition and decreasing cancer stem cell marker expression. BMC Cancer. 2019;19:618. doi: 10.1186/s12885-019-5824-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou L, Zhao Y. B7-H3 induces ovarian cancer drugs resistance through an PI3K/AKT/BCL-2 signaling pathway. Cancer Manag Res. 2019;11:10205–10214. doi: 10.2147/CMAR.S222224 [DOI] [PMC free article] [PubMed] [Google Scholar]