Abstract
Background
Seasonal human coronaviruses (hCoVs) broadly circulate in humans. Their epidemiology and effect on the spread of emerging coronaviruses has been neglected thus far. We aimed to elucidate the epidemiology and burden of disease of seasonal hCoVs OC43, NL63, and 229E in patients in primary care and hospitals in Belgium between 2015 and 2020.
Methods
We retrospectively analysed data from the national influenza surveillance networks in Belgium during the winter seasons of 2015–20. Respiratory specimens were collected through the severe acute respiratory infection (SARI) and the influenza-like illness networks from patients with acute respiratory illness with onset within the previous 10 days, with measured or reported fever of 38°C or greater, cough, or dyspnoea; and for patients admitted to hospital for at least one night. Potential risk factors were recorded and patients who were admitted to hospital were followed up for the occurrence of complications or death for the length of their hospital stay. All samples were analysed by multiplex quantitative RT-PCRs for respiratory viruses, including seasonal hCoVs OC43, NL63, and 229E. We estimated the prevalence and incidence of seasonal hCoV infection, with or without co-infection with other respiratory viruses. We evaluated the association between co-infections and potential risk factors with complications or death in patients admitted to hospital with seasonal hCoV infections by age group. Samples received from week 8, 2020, were tested for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Findings
2573 primary care and 6494 hospital samples were included in the study. 161 (6·3%) of 2573 patients in primary care and 371 (5·7%) of 6494 patients admitted to hospital were infected with a seasonal hCoV. OC43 was the seasonal hCoV with the highest prevalence across age groups and highest incidence in children admitted to hospital who were younger than 5 years (incidence 9·0 [95% CI 7·2–11·2] per 100 000 person-months) and adults older than 65 years (2·6 [2·1–3·2] per 100 000 person-months). Among 262 patients admitted to hospital with seasonal hCoV infection and with complete information on potential risk factors, 66 (73·3%) of 90 patients who had complications or died also had at least one potential risk factor (p=0·0064). Complications in children younger than 5 years were associated with co-infection (24 [36·4%] of 66; p=0·017), and in teenagers and adults (≥15 years), more complications arose in patients with a single hCoV infection (49 [45·0%] of 109; p=0·0097). In early 2020, the Belgian SARI surveillance detected the first SARS-CoV-2-positive sample concomitantly with the first confirmed COVID-19 case with no travel history to China.
Interpretation
The main burden of severe seasonal hCoV infection lies with children younger than 5 years with co-infections and adults aged 65 years and older with pre-existing comorbidities. These age and patient groups should be targeted for enhanced observation when in medical care and in possible future vaccination strategies, and co-infections in children younger than 5 years should be considered during diagnosis and treatment. Our findings support the use of national influenza surveillance systems for seasonal hCoV monitoring and early detection, and monitoring of emerging coronaviruses such as SARS-CoV-2.
Funding
Belgian Federal Public Service Health, Food Chain Safety, and Environment; Belgian National Insurance Health Care (Institut national d'assurance maladie-invalidité/Rijksinstituut voor ziekte-en invaliditeitsverzekering); and Regional Health Authorities (Flanders Agentschap zorg en gezondheid, Brussels Commission communautaire commune, Wallonia Agence pour une vie de qualité).
Introduction
Since its emergence in China in late-2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in more than 88 million cases of COVID-19 and more than 1·8 million deaths as of Jan 11, 2021.1 Its devastating worldwide effect on public health and the economy underscores the threat that members of the Coronaviridae viral family represent for global health. In addition to SARS-CoV-2, this family includes two other highly pathogenic viruses of zoonotic origin that have previously caused severe global epidemics but have not achieved continuous transmission in the human population (SARS-CoV and Middle East respiratory syndrome coronavirus).1 Four other human coronaviruses (hCoVs) are endemic in humans worldwide (ie, hCoV-OC43, hCoV-229E, hCoV-NL63, and hCoV-HKU1), typically follow a seasonal pattern, and are together estimated to be the second most frequent cause of common cold worldwide.2 Because these seasonal hCoVs are historically associated with mild disease, their epidemiology and burden of disease has been poorly characterised.3 Seasonal hCoVs can, however, also cause severe lower respiratory tract infections, such as pneumonia or bronchiolitis, which can lead to further complications requiring hospital admission and even death, especially in those who are immunocompromised, infants, and older adults.2
Research in context.
Evidence before the study
Overspills of zoonotic coronaviruses, such as severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus, and in 2019, SARS-CoV-2, have caused severe epidemics or pandemics in the past two decades. Few studies have investigated the burden of less pathogenic seasonal human coronaviruses, members of the same viral family. We searched PubMed from database inception up until June 19, 2020, using the search terms “seasonal human coronaviruses 229E OC43 NL63”, which yielded 59 articles in English. An update of the search from Nov 7, 2020, identified five additional relevant articles. Most studies focused on the prevalence of seasonal coronavirus in children using data mainly collected in primary care settings during one season. We also searched PubMed on June 19, 2020, using the search terms “(Severe acute respiratory syndrome coronavirus-2 OR SARS-CoV-2 OR COVID-19 OR 2019-nCoV) AND (influenza surveillance)” and obtained 143 articles in English. Two of them described the use of primary care influenza surveillance for detection of SARS-CoV-2, one of them retrospectively, while the other did not report virological confirmation. A new literature search on PubMed with the same terms was done on Nov 7, 2020, and yielded more than 250 additional articles in English, but only 14 were based on data collected through influenza surveillance systems or emphasising the potential role of these networks for SARS-CoV-2 monitoring.
Added value of this study
We provide important insights into the epidemiology of seasonal human coronaviruses 229E, OC43, and NL63, and their burden of disease in children and adults in Belgium across five winter seasons. Our data showed that the main burden of disease is caused by OC43 in children younger than 5 years, where co-infections with other respiratory viruses contribute significantly to morbidity. Moreover, we analysed the effect of potential risk factors and showed that pre-existing comorbidities and, in particular, immunodeficiency are associated with complications or death from infection with seasonal human coronaviruses. Our study highlights the usefulness of national influenza surveillance networks to monitor seasonal human coronaviruses, as well as to detect the emergence of zoonotic coronaviruses, such as SARS-CoV-2.
Implications of all the available evidence
The current SARS-CoV-2 pandemic calls attention to the threat that emerging zoonotic coronaviruses place on global health. More research is needed to clarify the role of circulating seasonal coronaviruses in population susceptibility and its spread. The high burden of seasonal coronaviruses in children younger than 5 years raises questions about why this age group is only mildly affected by SARS-CoV-2. Our work also underlines the importance of considering co-infections and potential risk factors in the clinical management of patients with seasonal coronavirus. We show that national influenza surveillance networks could be leveraged to rapidly incorporate continuous monitoring of SARS-CoV-2 in the post-pandemic phase and could be integrated in early warning systems for future emerging coronaviruses.
Of the studies assessing seasonal hCoV burden, most have focused on children and few on healthy adults or adults at high risk.4, 5, 6, 7 One large study reporting results from multiple cohorts found that annual infection rates with 229E and OC43 ranged from 2·8% to 26·0% in healthy adults and that prevalence ranged from 3·3% to 11·1% in patients who were admitted to hospital.7 The same study also reported that asymptomatic infection and viral co-infection was significantly more common with 229E than with OC43.
Pre-existing humoral immunity towards seasonal hCoVs could affect the response to and spread of emerging coronaviruses.8 Pre-existing data for the seasonality and cross-reactive immunity of these hCoVs has been used to model the spread of SARS-CoV-2 in the post-pandemic period.9 High prevalence of serum IgG (91–100%) and mucosal IgA (8–31%) against the four seasonal hCoVs was observed in a cohort of American patients aged 50 years and older with chronic pulmonary disease, and overall antibody titres were higher in older (≥60 years) than in younger (21–40 years) adults.10, 11 Besides overall high seroprevalence in the population, a 2020 retrospective analysis of serum samples from ten adult men across more than three decades found that re-infection with all four seasonal hCoVs occurred frequently 12 months after an infection and was linked to an intermediate reduction of antibody concentrations.12
The objective of the present study was to improve our understanding of the epidemiology and burden of disease of seasonal hCoVs in the general population in Belgium. We used the data obtained from samples collected through the national surveillance systems of influenza-like illness and severe acute respiratory infection (SARI) during the past five influenza seasons. The National Influenza Centre established multiplex quantitative RT-PCR (RT-qPCR) testing for respiratory viruses alongside influenza virus in 2015—which includes testing for seasonal hCoVs OC43, NL63, and 229E—and was further extended to routinely incorporate SARS-CoV-2 upon its recorded emergence in early 2020. Here, we estimate the prevalence and burden of disease from seasonal hCoVs OC43, NL63, and 229E during the winters of 2015–20 in primary care and hospitals in Belgium. We further aimed to assess the capacity of existing surveillance networks for the early detection and continuous monitoring of emerging pathogens. We also showed the timely detection of SARS-CoV-2 during its emergence in Belgium in samples collected through the influenza surveillance system.
Methods
Study design and data collection
We did a retrospective analysis focusing on seasonal hCoVs from the surveillance data routinely collected through the national influenza surveillance networks in Belgium during the winter seasons (December to April) of 2015–16 to 2019–20 (appendix pp 10–12). Respiratory specimens collected through these networks were sent to the National Influenza Centre (Brussels, Belgium), along with detailed clinicoepidemiological data, such as information on age, sex, symptoms, and potential risk factors such as pregnancy or comorbidities (ie, chronic respiratory diseases, asthma, chronic cardiovascular diseases, obesity, diabetes, hepatic or renal insufficiency, immunodeficiency, and neuromuscular disease). Patients admitted to hospital were followed up for the occurrence of complications (ie, detection of pneumonia based on chest radiography, development of acute respiratory distress syndrome, requirement for respiratory assistance or for extracorporeal membrane oxygenation, admission to an intensive care unit), or death from any cause during hospital admission.
Patients were included in the study population if they met the following study case definition: acute respiratory illness with onset within the past 10 days, with measured or reported fever of 38°C or greater, cough, or dyspnoea, or a combination of these symptoms, and hospital admission for at least one night for the patients admitted to hospital. Samples were excluded if the patient's date of birth or age was missing. Furthermore, samples from children (aged <15 years) received through the general practice (GP) network were excluded, as they were heavily under-represented in the sampling strategy.
All authors had full access to the surveillance network databases, which were used to select the study population. Official statistics on the Belgian national population counts were obtained from Statbel, the Belgian statistical office.
Informed consent was obtained from all participants or their parents or guardians. The study surveillance protocol was approved by central ethical committees in Brussels, Belgium (reference AK/12-02-11/4111; Centre Hospitalier Universitaire St-Pierre in 2011 and Universitair Ziekenhuis Vrije Universiteit Brussel since 2014) and the local ethical committees of each participating hospital.
Procedures
Respiratory samples were analysed at the Belgian National Influenza Centre. Viral nucleic acids were extracted and in-house multiplex RT-qPCRs were done to detect type A and B influenza viruses; type 1, 2, 3, or 4 parainfluenzaviruses; type A and B respiratory syncytial viruses; human metapneumoviruses; adenoviruses; picornaviruses (Rhinovirus and Enterovirus genera); specific enterovirus D68; parechovirus; bocavirus; and seasonal hCoVs OC43, 229E, and NL63. Detection of SARS-CoV-2 was done with use of the protocol by Corman and colleagues.13 Material used, primer and probe sequences, and PCR conditions are detailed in the appendix (pp 8–10).
All samples received through the influenza surveillance networks in the study period were tested for the presence of respiratory viruses by the Belgian National Influenza Centre.
Statistical analysis
The prevalence of seasonal hCoVs OC43, NL63, and 229E infection was estimated by sex and by defined age group (0–4, 5–14, 15–64, and ≥65 years), across all five winter seasons from 2015 to 2020 in each patient group. Differences in sex and age group distribution between patients with seasonal hCoV infection as compared with infection with other respiratory viruses was determined using Pearson's χ2 test (appendix p 12). A p value of less than 0·05 was considered statistically significant.
To estimate the burden of disease of seasonal hCoV infection, the study group was narrowed down to the patients who had a positive seasonal-hCoV RT-qPCR test result. Incidence estimates were calculated per 100 000 person-months, per seasonal hCoV and season in each patient group. They were calculated for single seasonal hCoV infections as well as for seasonal hCoV infection with co-infection with other respiratory viruses by age group across all five winter seasons.
To obtain the denominator, the catchment populations of the hospital sentinel system and GP sentinel system were calculated by age group across the five analysed seasons (2015–20), based on WHO recommendations for seasonal influenza (appendix pp 10–11).14 To obtain the person-month, the number of weeks during each surveillance season was multiplied by 7 days and divided by the mean number of days in a month during the winter period (divided by a mean of 30 days per month during the winter period). The 95% CIs of all incidence estimates were calculated using the Rothman–Greenland method.15
The study group was further narrowed down to include only the patients admitted to hospital with complete information on follow-up. The proportion of complications was determined by age group, taking into account the presence or absence of potential risk factors or co-infections. Pearson's χ2 test and Fisher's exact test were used to compare differences in potential risk factors or co-infection between patients with and without complications (appendix p 12). Pearson's χ2 test for trend was used to assess an association between the number of potential risk factors and the occurrence of complications. A p value of less than 0·05 was considered statistically significant. All data analyses and visualisations were done in R Studio, version 3.6.3 (appendix p 12).
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
3168 samples from primary care and 8931 samples from patients admitted to hospital were submitted to the National Influenza Centre in Belgium during the winter seasons (December to April) of 2015–16 up until 2019–20. Of those, 2573 (81·2%) of 3168 primary care and 6494 (72·7%) of 8931 hospital samples had complete information on patient age, met the study case definition, and were included in the seasonal hCoV study population (figure 1 ).
Figure 1.
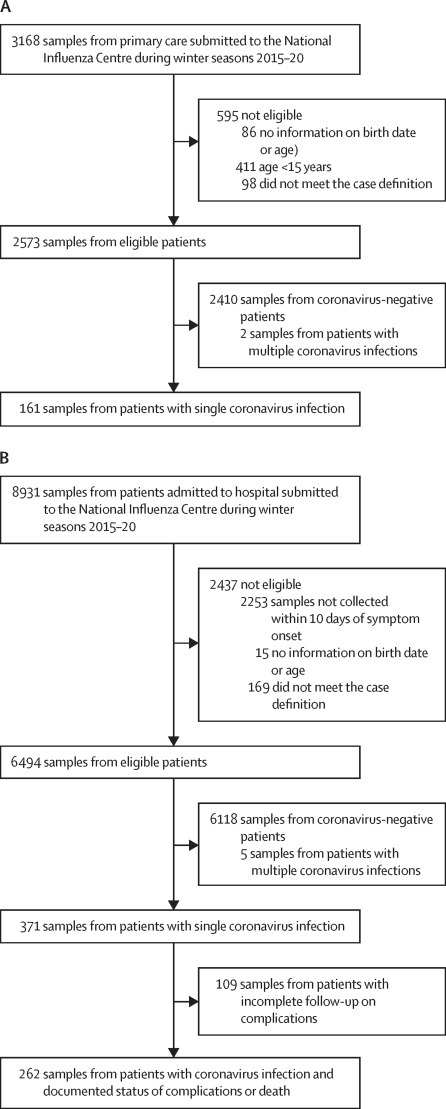
Study profile
During the winter seasons from 2015–16 to 2019–20, respiratory specimens from primary care (A) and patients admitted to one of the six sentinel hospitals (B) with acute respiratory infections were routinely collected through the corresponding sentinel surveillance networks in Belgium and submitted to the National Influenza Centre to be tested for the presence of respiratory viruses by multiplex PCR assay.
Among the primary care patients, 87 (3·4%) of 2573 tested positive for OC43, 39 (1·5%) for 229E, and 35 (1·4%) for NL63, with 161 (6·3%) positive for a single seasonal hCoV (figure 1, appendix p 1). Of the primary care patients with a single seasonal hCoV infection, 143 (88·8%) of 161 were 15–64 years old and 18 (11·2%) were 65 years or older (table 1 ). Of the patients admitted to hospital, 196 (3·0%) of 6494 tested positive for OC43, 91 (1·4%) for 229E, and 84 (1·3%) of 6494 for NL63, with 371 (5·7%) positive for a single seasonal hCoV (appendix p 1). Of patients admitted to hospital with a single seasonal hCoV infection, 147 (39·6%) of 371 were younger than 15 years, 74 (19·9%) were 15–64 years, and 150 (40·4%) were aged 65 years and older (table 1).
Table 1.
Baseline characteristics and co-infection status
|
Primary care |
Hospital |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All total (N=2573) | Single seasonal hCoV infection |
Seasonal hCoV-negative total (N=2410) | All total (N=6494) | Single seasonal hCoV infection |
Seasonal hCoV-negative total (N=6118) | |||||||||
| Total (N=161) | hCoV-229E (n=39) | hCoV-NL63 (n=35) | hCoV-OC43 (n=87) | Total (N=371) | hCoV-229E (n=91) | hCoV-NL63 (n=84) | hCoV-OC43 (n=196) | |||||||
| Sex | .. | .. | .. | .. | .. | 0·60* | .. | .. | .. | .. | .. | 0·28* | ||
| Female | 1261 (49·0%) | 85 (52·8%) | 25 (64·1%) | 22 (62·9%) | 38 (43·7%) | 1175 (48·8%) | 2996 (46·1%) | 159 (42·9%) | 42 (46·2%) | 37 (44·0%) | 80 (40·8%) | 2835 (46·3%) | ||
| Male | 1206 (46·9%) | 74 (46·0%) | 14 (35·9%) | 13 (37·1%) | 47 (54·0%) | 1131 (46·9%) | 3317 (51·1%) | 198 (53·4%) | 46 (50·5%) | 42 (50·0%) | 110 (56·1%) | 3116 (51·0%) | ||
| Missing | 106 (4·1%) | 2 (1·2%) | 0 | 0 | 2 (2·3%) | 104 (4·3%) | 181 (2·8%) | 14 (3·8%) | 3 (3·3%) | 5 (6·0%) | 6 (3·0%) | 167 (2·7%) | ||
| Age group, years | .. | .. | .. | .. | .. | 0·17* | .. | .. | .. | .. | .. | 0·019* | ||
| 0-4 | .. | .. | .. | .. | .. | .. | 2026 (31·2%) | 137 (36·9%) | 12 (13·2%) | 46 (54·8%) | 79 (40·3%) | 1888 (30·9%) | ||
| 5-14 | .. | .. | .. | .. | .. | .. | 336 (5·2%) | 10 (2·7%) | 3 (3·3%) | 2 (2·4%) | 5 (2·6%) | 326 (5·3%) | ||
| 15-64 | 2367 (92·0%) | 143 (88·8%) | 36 (92·3%) | 31 (88·6%) | 76 (87·4%) | 2222 (92·2%) | 1270 (19·6%) | 74 (19·9%) | 25 (27·5%) | 15 (17·9%) | 34 (17·3%) | 1196 (19·5%) | ||
| ≥65 | 206 (8·0%) | 18 (11·2%) | 3 (7·7%) | 4 (11·4%) | 11 (12·6%) | 188 (7·8%) | 2862 (44·1%) | 150 (40·4%) | 51 (56·0%) | 21 (25·0%) | 78 (39·8%) | 2708 (44·3%) | ||
| Co-infection with other respiratory viruses | ||||||||||||||
| All age groups, years | .. | 43 (26·7%) | 17 (43·6%) | 7 (20·0%) | 19 (21·8%) | .. | .. | 161 (43·4%) | 44 (48·4%) | 34 (40·5%) | 83 (42·3%) | .. | ||
| 0-4 | .. | .. | .. | .. | .. | .. | .. | 89 (24·0%) | 10 (11·0%) | 25 (29·8%) | 54 (27·6%) | .. | ||
| 5-14 | .. | .. | .. | .. | .. | .. | .. | 6 (1·6%) | 1 (1·1%) | 1 (1·2%) | 4 (2·0%) | .. | ||
| 15-64 | .. | 38 (23·6%) | 15 (38·5%) | 6 (17·1%) | 17 (19·5%) | .. | .. | 24 (6·5%) | 13 (14·3%) | 3 (3·6%) | 8 (4·1%) | .. | ||
| ≥65 | .. | 5 (27·8%) | 2 (5·1%) | 1 (2·9%) | 2 (2·3%) | .. | .. | 42 (11·3%) | 20 (22·0%) | 5 (6·0%) | 17 (8·7%) | .. | ||
| Influenza | .. | 34 (21·1%) | 14 (35·9%) | 5 (14·3%) | 15 (17·2%) | .. | .. | 67 (18·1%) | 26 (28·6%) | 8 (9·5%) | 33 (16·8%) | .. | ||
| Respiratory syncytial virus | .. | 3 (1·9%) | 0 | 0 | 3 (3·4%) | .. | .. | 37 (10·0%) | 7 (7·7%) | 6 (7·1%) | 24 (12·2%) | .. | ||
| Picornaviruses | .. | 2 (1·2%) | 1 (2·6%) | 1 (2·9%) | 0 | .. | .. | 38 (10·2%) | 9 (9·9%) | 9 (10·7%) | 20 (10·2%) | .. | ||
| Adenovirus | .. | 0 | 0 | 0 | 0 | .. | .. | 26 (7·0%) | 4 (4·4%) | 7 (8·3%) | 15 (7·7%) | .. | ||
| hMPV | .. | 4 (2·5%) | 3 (7·7%) | 0 | 1 (1·1%) | .. | .. | 19 (5·1%) | 0 | 8 (9·5%) | 11 (5·6%) | .. | ||
| Parainfluenza viruses | .. | 1 (0·6%) | 0 | 0 | 1 (1·1%) | .. | .. | 10 (2·7%) | 7 (7·7%) | 2 (2·4%) | 1 (0·5%) | .. | ||
| Bocavirus | .. | 1 (0·6%) | 0 | 1 (2·9%) | 0 | .. | .. | 9 (2·5%) | 0 | 6 (7·2%) | 3 (1·6%) | .. | ||
Data are n (%) unless specified. hCoV=human coronavirus. hMPV=human metapneumovirus.
χ2 p value.
The distribution of age groups between patients admitted to hospital with a single seasonal hCoV infection and patients who were seasonal hCoV-negative was significantly different (χ2 p=0·019; table 1). In particular, the proportion of children younger than 5 years who were admitted to hospital and were seasonal hCoV-positive (137 [36·9%] of 371) was higher than among those who were seasonal hCoV-negative (1888 [30·9%] of 6118; χ2 p=0·017).
43 (26·7%) of 161 primary care patients and 161 (43·4%) of 371 patients admitted to hospital with seasonal hCoV infection were co-infected with other respiratory viruses (table 1). Co-infection was more common in patients infected with 229E than in those infected with NL63 or OC43 (table 1). In primary care and hospital patients aged 15 years or older, influenza virus was the most common co-infection for all three seasonal hCoVs (appendix p 2). Among patients admitted to hospital, children younger than 5 years carried the highest proportion of co-infections, which most frequently included respiratory syncytial viruses, picornaviruses, and adenovirus (appendix p 2).
The incidence of seasonal hCoV infections in primary care or in patients admitted to hospital varied between seasons and seasonal hCoV species (figure 2A, B ). Among patients admitted to hospital, the incidence of NL63 infections remained similar from year to year for the five seasons assessed, whereas 229E and OC43 showed increases in incidence for the winter seasons of 2016–17 and 2018–19 (appendix p 3).
Figure 2.
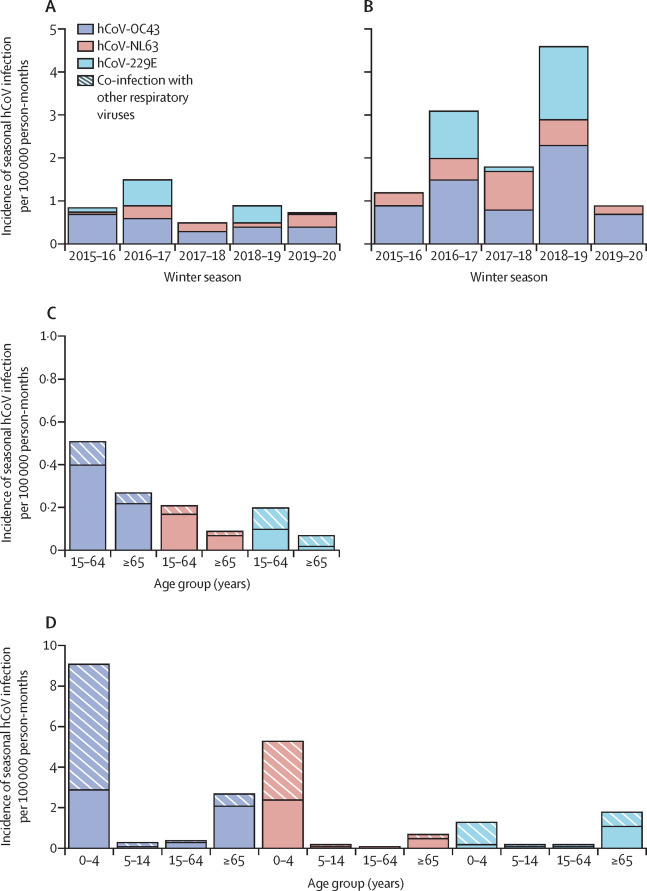
Incidence of seasonal hCoV infections in Belgium, 2015–20, per season and age group
The incidence of three circulating seasonal hCoV species (OC43, NL63, and 229E) in patients in primary care (A) and hospital (B) per 100 000 person-months was calculated by winter season across all ages (A and B), and by age group across five winter seasons, taking into account infection with single seasonal hCoV species only or co-infection with other respiratory viruses (C and D). For incidence point estimates and 95% CIs, see appendix (pp 3–5). hCoV=human coronavirus.
In patients in primary care, when looking at the overall incidence from 2015 to 2020, OC43 infections had the highest incidence. The incidence of infection for all three seasonal hCoVs was less than 0·5 in patients aged 15–64 years and less than 0·3 for those aged 65 years or older (figure 2C, appendix pp 4–5). The incidence for infections with seasonal hCoVs only was equal to or higher than that for co-infections with other respiratory viruses, for all age groups and all seasonal hCoVs, except for 229E in adults aged 65 years or older.
Among children admitted to hospital aged younger than 5 years, the highest burden was associated with OC43, followed by NL63, with incidence greater than 5·0 (figure 2D, appendix pp 4–5). Co-infections contributed notably to the burden of disease in children younger than 5 years, constituting more than half of the incidence estimates for all seasonal hCoVs investigated. Among adults admitted to hospital aged 65 years and older, the highest burden was associated with OC43, followed by 229E, with incidence of infection greater than 1·7. The incidence of infection for patients aged 5–14 years admitted to hospital and those aged 15–64 years were less than 0·3 for all three seasonal hCoVs (figure 2D, appendix pp 4–5).
Complete follow-up of complications or death during hospital admission was available for 262 (70·6%) of 371 patients who were infected with a seasonal hCoV (figure 1B). Overall, 90 (34·4%) of 262 adults developed complications or died (table 2 ). The most common complications were pneumonia, requirement for respiratory assistance, and intensive care unit transfer. Acute respiratory distress syndrome and death were less common, and extracorporeal membrane oxygenation was not required for any patient. Seven (4·7%) of 149 teenagers and adults (aged ≥15 years) and one (1·0%) of 105 children aged younger than 5 years died (table 2). The only complication recorded among children aged 5–14 years was pneumonia (in four [50%] of eight children).
Table 2.
Complications or death in patients admitted to hospital with seasonal hCoV infection in Belgium, 2015–20
| Any complication or death | Pneumonia | Respiratory assistance | Intensive care unit | Acute respiratory distress syndrome | Death | ||
|---|---|---|---|---|---|---|---|
| Patients admitted with complete follow-up data (n=262) | 90 (34·4%) | 64 (24·4%) | 37 (14·1%) | 19 (7·3%) | 9 (3·4%) | 8 (3·1%) | |
| Per age group, years | |||||||
| 0–4 (n=105) | 29 (27·6%) | 17 (16·2%) | 13 (12·4%) | 2 (1·9%) | 3 (2·9%) | 1 (1·0%) | |
| 5–14 (n=8) | 4 (50·0%) | 4 (50·0%) | 1 (12·5%) | 0 | 0 | 0 | |
| 15–64 (n=48) | 24 (50·0%) | 18 (37·5%) | 9 (18·8%) | 7 (14·6%) | 4 (8·3%) | 2 (4·2%) | |
| ≥65 (n=101) | 33 (32·7%) | 25 (24·8%) | 14 (13·9%) | 10 (9·9%) | 2 (2·0%) | 5 (5·0%) | |
| Potential risk factor | |||||||
| Yes (n=161) | 66 (41·0%) | 46 (28·6%) | 27 (16·8%) | 17 (10·6%) | 8 (5·0%) | 6 (3·7%) | |
| No (n=101) | 24 (23·8%) | 18 (17·8%) | 10 (9·9%) | 2 (2·0%) | 1 (1·0%) | 2 (2·0%) | |
| Co-infection | |||||||
| Single seasonal hCoV plus other (n=110) | 33 (30·0%) | 21 (19·1%) | 14 (12·7%) | 4 (3·6%) | 1 (0·9%) | 1 (0·9%) | |
| Single seasonal hCoV (n=152) | 57 (37·5%) | 43 (28·3%) | 23 (15·1%) | 15 (9·9%) | 8 (5·3%) | 7 (4·6%) | |
Data are n (%). Complications or death are not mutually exclusive. hCoV=human coronavirus.
Most patients admitted to hospital with seasonal hCoV infection and who had complications or died had at least one potential risk factor (66 [73·3%] of 90), such as chronic cardiac disease, chronic respiratory disease, or immunodeficiency, among others (table 3 ). However, 24 (23·8%) of 101 patients without any potential risk factor had complications or died (table 2). We found that having a potential risk factor was associated with developing complications or the patient dying (p=0·0064; table 3). Specifically, immunodeficiency was associated with complications or death (p=0·0028; table 3). We also observed an increased risk of complications or death with an increase in the number of potential risk factors (χ2 test for trend p=0·041).
Table 3.
Potential risk factors among patients admitted to hospital with seasonal coronavirus and association with complications or death in Belgium, 2015–20
| Patients with documented status of complications or death (n=262) | Patients who had complications or died (n=90) | χ2 p value | ||
|---|---|---|---|---|
| Any potential risk factor | 161 (61·5%) | 66 (73·3%) | 0·0064 | |
| Potential risk factor by age group, years | ||||
| 0–4 | 23 (8·8%) | 11 (12·2%) | .. | |
| 5–14 | 4 (1·5%) | 2 (2·2%) | .. | |
| 15–64 | 42 (16·0%) | 21 (23·3%) | .. | |
| ≥65 | 92 (35·1%) | 32 (35·6%) | .. | |
| Number of potential risk factors per patient | .. | .. | 0·041* | |
| One comorbidity | 61 (23·3%) | 25 (27·8%) | .. | |
| Two comorbidities | 41 (15·6%) | 18 (20·0%) | .. | |
| Three or more comorbidities | 40 (15·3%) | 15 (16·7%) | .. | |
| Type of potential risk factor | ||||
| Chronic cardiac disease | 66 (25·2%) | 23 (25·6%) | 0·17 | |
| Chronic respiratory disease | 45 (17·2%) | 18 (20·0%) | 0·071 | |
| Immunodeficiency | 41 (15·6%) | 21 (23·3%) | 0·0028 | |
| Renal insufficiency | 36 (13·7%) | 13 (14·4%) | 0·22 | |
| Diabetes | 28 (10·7%) | 10 (11·1%) | 0·30 | |
| Neuromuscular disease | 22 (8·4%) | 10 (11·1%) | 0·072 | |
| Obesity | 12 (4·6%) | 3 (3·3%) | 1·0† | |
| Asthma | 13 (5·0%) | 6 (6·7%) | 0·10† | |
| Hepatic insufficiency | 9 (3·4%) | 5 (5·6%) | 0·052† | |
| Pregnancy | 3 (1·1%) | 1 (1·1%) | 0·57† | |
Data are n (%) unless otherwise specified.
Test for trend.
Fisher's exact test.
33 (36·7%) of 90 patients admitted to hospital who had complications or died were co-infected with a seasonal hCoV and other respiratory viruses (table 2). In children younger than 5 years, five (12·8%) of 39 with single seasonal hCoV infection versus 24 (36·4%) of 66 co-infected with a seasonal hCoV and other respiratory viruses developed complications (χ2 p=0·017). By contrast, in patients aged 15 years or older, 49 (45·0%) of 109 with a single seasonal hCoV infection versus eight (20%) of 40 co-infected with a seasonal hCoV and other respiratory viruses developed complications (χ2 p=0·0097; appendix p 6).
Upon the emergence of SARS-CoV-2 in Europe, the Belgian National Influenza Centre implemented the RT-qPCR protocol to test samples collected through the routine primary care and hospital surveillance systems for the presence of the virus. The first positive samples were detected in week 10 for hospital and week 11 for primary care surveillance. Samples were assessed for SARS-CoV-2 from week 8 for hospitals and week 9 for primary care. Prevalence of SARS-CoV-2-positive samples rose quickly, from 1·3% (two of 156) to 52·9% (83 of 157) in weeks 10–13 in hospital samples and from 18·2% (eight of 44) to 27·3% (six of 22) in weeks 11–12 in primary care samples, before surveillance was interrupted by the measures taken by the Belgian authorities to contain the spread of the virus. The number of GP visits by epidemiological week revealed a double peak, with a first peak at week 5 (influenza) and a second peak at week 12 (SARS-CoV-2; appendix p 7).
Discussion
In this study, we made use of the national influenza GP (influenza-like illness) and hospital (SARI) surveillance networks to assess the epidemiology and the burden of seasonal hCoVs in Belgium in the past five winter seasons (2015–20). We also provided evidence of the usefulness of influenza surveillance networks for the early detection and monitoring of emerging coronaviruses such as SARS-CoV-2.
The most common seasonal hCoV detected over the past five winters in Belgium was OC43, responsible for the highest burden of disease in children younger than 5 years and adults aged 65 years and older. OC43 and 229E showed a biennial peak in cases over the study period. For all five winter seasons, the incidence of seasonal hCoV infections was low among adult patients admitted to hospital, and higher among children admitted to hospital who were aged younger than 5 years. Similar patterns of seasonality and age distribution have been reported in other European and North American populations.16, 17
We found that almost half of patients older than 15 years with a single seasonal hCoV infection had a complication or died. This finding is not in line with the common belief that seasonal hCoVs cause mild infections and suggests that their burden of disease is worth investigating. We observed that complications in children admitted to hospital aged younger than 5 years with seasonal hCoV infection were significantly associated with co-infections with other respiratory viruses, highlighting the interest of manifold diagnostic tools, such as multiplex RT-qPCR. In patients older than 15 years, we also detected a high rate of co-infection with influenza viruses. A crucial role for co-infections in the severity of COVID-19 has been proposed, but is not well described thus far.18 Co-infection with SARS-CoV-2 and influenza viruses during the 2020–21 winter season has been discussed as a serious threat to public health.19
Nevertheless, surveillance reports from the USA and countries in the southern hemisphere (Australia, Chile, and South Africa) indicated relatively little influenza activity during the COVID-19 epidemic, probably due to changes in health-care-seeking behaviours, surveillance capacity, and widespread implementation of mitigation measures.20 The possibility of interference of SARS-CoV-2 with influenza virus circulation needs to be further investigated.
Here, we report that potential risk factors were significantly associated with adverse outcomes in seasonal hCoV infections. In particular, we saw a strong association in individuals who were immunocompromised. However, we did not observe clear evidence for an association with comorbidities described to increase the risk for severe COVID-19, such as respiratory system disease, cardiovascular disease, obesity, and diabetes.21, 22, 23 Regarding the association of immunodeficiency with COVID-19 severity, current data indicate that the nature of the immunodeficiency could be decisive and more studies are needed.24
Exposure to seasonal hCoVs has the capacity to elicit cross-reactive humoral and cellular immunity to SARS-CoV-2, as was shown especially for OC43, another member of the betacoronavirus genus.25, 26, 27 We found that OC43 is the most prevalent seasonal hCoV in Belgium and causes the highest burden of disease in populations such as young children and older adults who are vulnerable to severe disease. Overall, our data indicate that children can be highly susceptible to infection with seasonal hCoVs. It has been speculated that cross-reactive immunity acquired during childhood infections with seasonal hCoVs could underlie the low incidence of SARS-CoV-2 infections in children in the current pandemic.28 The precise role of children in SARS-CoV-2 transmission needs to be further clarified, but it could have a large effect on infection prevention and control measures, such as closure of schools and nurseries. Improved serological assays and population-wide seroprevalence studies of seasonal hCoV exposure are needed to assess the population immunity towards coronaviruses and the possible role in the spread of SARS-CoV-2. For example, the influenza surveillance networks could be extended to regularly collect blood samples, as is already implemented in Russia, to establish background seroprevalence of antibodies directed against seasonal hCoVs, as well as extended serosurveillance for SARS-CoV-2.29
The first known SARS-CoV-2 case in Belgium without travel history to China was confirmed on March 1, 2020, the same week that the first detected case within the SARI surveillance was collected, and 1 week before detection within the influenza-like illness surveillance. Our study confirms that influenza surveillance networks are valuable tools for timely detection of SARS-CoV-2, and can be used for its monitoring should there be persistent endemic, post-pandemic circulation, possibly even seasonal resurgence.
The main limitation of this study is that the full spectrum of seasonal hCoV infections was not captured. At the moment, no protocol has been established for routine testing for HKU-1 at the Belgian National Influenza Centre and we therefore cannot infer anything on its circulation in Belgium. Nickbakhsh and colleagues reported a very low overall prevalence of 0·3% for HKU-1 among patients with respiratory illness in Scotland, which later led to the discontinuation of testing for HKU-1.17 In addition, the current surveillance is only done during the winter months and therefore does not capture seasonal hCoV circulation during the rest of the year. The burden of mild disease caused by seasonal hCoVs, assessed through the influenza-like illness network, is likely to be underestimated in our study. Although the participating GPs were well distributed within the country to match the population density, only two samples per week per GP were collected through this network, with a sampling bias towards younger adults. In addition, the case definition for enrolment in the influenza-like illness surveillance is supposedly stringent for influenza (ie, fever and cough or dyspnoea), which is in agreement with our data for co-infection. Moreover, the network has insufficient reliable data for children seeking primary care, as there is currently no paediatric surveillance system for influenza in Belgium. By contrast, the burden of severe illness associated with seasonal hCoV is likely to be appropriately assessed in our study, as the Belgian SARI surveillance system covers the full age range of the entire population and captures a total of 10% of the Belgian population from all three regions of the country, with an exhaustive enrolment of the patients responding to the case definition.
To conclude, we showed that the national networks implemented for the surveillance of influenza viruses are valuable tools to estimate the burden of respiratory viruses, such as seasonal hCoVs. Co-infections in young children and potential risk factors are important elements in seasonal hCoV disease severity and should be considered for prevention, diagnosis, and treatment. More information on the relationship between influenza viruses and SARS-CoV-2 is needed to assess the possibility of successive epidemics as well as the clinical effect of co-circulation. Therefore, a common surveillance system could prove useful. We suggest that influenza surveillance systems, especially the SARI network, could be used as a tool for the surveillance of seasonal hCoVs and the continued surveillance of SARS-CoV-2 in the post-pandemic period. Moreover, their maintenance during future winter seasons is crucial, and even beyond, if SARS-CoV-2 circulation establishes itself outside of the winter season. Widening of the case definition could aid in the inclusion of other circulating respiratory viruses and could lead to an improved estimation of their true burden of disease. The potential role of pre-existing immunity towards seasonal hCoVs in the burden and severity of COVID-19 remains to be elucidated. To this aim, surveillance schemes including serology in addition to virology could be considered and broadly implemented.
Data sharing
All aggregated data are available either in the manuscript or as supplementary materials. Further requests might require ethical approval and should be discussed with the authors by contacting the corresponding author.
Acknowledgments
Acknowledgments
We thank the technical staff from the Belgian National Influenza Centre for their excellent technical assistance throughout the surveillance seasons and all the staff of the six hospitals who took part in the recruitment of the SARI cases, without whom the SARI surveillance would not be possible. We thank Aura Andreasen, Scientific Coordinator from the European Centre for Disease Prevention and Control, for her review of the manuscript. ND is a post-doctoral Clinical Master Specialist of the Fond National de la Recherche Scientifique. NF is a fellow in the European Public Health Microbiology Program funded through the European Programme for Intervention Epidemiology Training, European Centre for Disease Prevention and Control.
Contributors
LS and CB were responsible for project conception. CB, NB, and IT were responsible for project administration. NF, LS, and CB were responsible for data curation and verification, and for formal analysis. NF, MR, MG, ND, PL, SD, BL, XH, KM, DJ, MB, BD, CB, and IT were responsible for investigations. MR, MG, ND, PL, SD, BL, XH, KM, DJ, MB, and BD were responsible for resources. NF was responsible for visualisation. NF, ND, LS, and CB were responsible for writing and preparing the original draft. All authors were responsible for review and editing of the manuscript. SQ and SVG were responsible for funding acquisition.
Declaration of interests
We declare no competing interests.
Supplementary Material
References
- 1.WHO Weekly operational update on COVID-19. 2021. https://www.who.int/publications/m/item/weekly-operational-update-on-covid-19---11-january-2021
- 2.Su S, Wong G, Shi W. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang JW, Lam TT, Zaraket H. Global epidemiology of non-influenza RNA respiratory viruses: data gaps and a growing need for surveillance. Lancet Infect Dis. 2017;17:e320–e326. doi: 10.1016/S1473-3099(17)30238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uddin SMI, Englund JA, Kuypers JY. Burden and risk factors for coronavirus infections in infants in rural Nepal. Clin Infect Dis. 2018;67:1507–1514. doi: 10.1093/cid/ciy317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talbot HKB, Crowe JE, Jr, Edwards KM. Coronavirus infection and hospitalizations for acute respiratory illness in young children. J Med Virol. 2009;81:853–856. doi: 10.1002/jmv.21443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heimdal I, Moe N, Krokstad S. Human coronavirus in hospitalized children with respiratory tract infections: a 9-year population-based study from Norway. J Infect Dis. 2019;219:1198–1206. doi: 10.1093/infdis/jiy646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh EE, Shin JH, Falsey AR. Clinical impact of human coronaviruses 229E and OC43 infection in diverse adult populations. J Infect Dis. 2013;208:1634–1642. doi: 10.1093/infdis/jit393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Che XY, Qiu LW, Liao ZY. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J Infect Dis. 2005;191:2033–2037. doi: 10.1086/430355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kissler SM, Tedijanto C, Goldstein E, Grad YH, Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020;368:860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorse GJ, Patel GB, Vitale JN, O'Connor TZ. Prevalence of antibodies to four human coronaviruses is lower in nasal secretions than in serum. Clin Vaccine Immunol. 2010;17:1875–1880. doi: 10.1128/CVI.00278-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorse GJ, Donovan MM, Patel GB. Antibodies to coronaviruses are higher in older compared with younger adults and binding antibodies are more sensitive than neutralizing antibodies in identifying coronavirus-associated illnesses. J Med Virol. 2020;92:512–517. doi: 10.1002/jmv.25715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edridge AWD, Kaczorowska J, Hoste ACR. Seasonal coronavirus protective immunity is short-lasting. Nat Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 13.Corman VM, Landt O, Kaiser M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO . World Health Organization; Geneva, Switzerland: 2015. A manual for estimating disease burden associated with seasonal influenza. [Google Scholar]
- 15.Rothman KJ, Greenland S, Lash TL. Lippincott Williams & Wilkins; Philadelphia, USA: 2008. Modern epidemiology. [Google Scholar]
- 16.Monto AS, DeJonge PM, Callear AP. Coronavirus occurrence and transmission over 8 years in the HIVE cohort of households in Michigan. J Infect Dis. 2020;222:9–16. doi: 10.1093/infdis/jiaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nickbakhsh S, Ho A, Marques DFP, McMenamin J, Gunson RN, Murcia PR. Epidemiology of seasonal coronaviruses: establishing the context for the emergence of coronavirus disease 2019. J Infect Dis. 2020;222:17–25. doi: 10.1093/infdis/jiaa185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox MJ, Loman N, Bogaert D, O'Grady J. Co-infections: potentially lethal and unexplored in COVID-19. Lancet Microbe. 2020;1:e11. doi: 10.1016/S2666-5247(20)30009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belongia EA, Osterholm MT. COVID-19 and flu, a perfect storm. Science. 2020;368 doi: 10.1126/science.abd2220. [DOI] [PubMed] [Google Scholar]
- 20.Olsen SJ, Azziz-Baumgartner E, Budd AP. Decreased influenza activity during the COVID-19 Pandemic—United States, Australia, Chile, and South Africa, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1305–1309. doi: 10.15585/mmwr.mm6937a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Zheng Y, Gou X. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int J Infect Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holman N, Knighton P, Kar P. Risk factors for COVID-19-related mortality in people with type 1 and type 2 diabetes in England: a population-based cohort study. Lancet Diabetes Endocrinol. 2020;8:823–833. doi: 10.1016/S2213-8587(20)30271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lighter J, Phillips M, Hochman S. Obesity in patients younger than 60 years is a risk factor for COVID-19 hospital admission. Clin Infect Dis. 2020;71:896–897. doi: 10.1093/cid/ciaa415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fung M, Babik JM. COVID-19 in immunocompromised hosts: what we know so far. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa863. published online June 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okba NMA, Müller MA, Li W. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wec AZ, Wrapp D, Herbert AS. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grifoni A, Weiskopf D, Ramirez SI. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489. doi: 10.1016/j.cell.2020.05.015. 501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludvigsson JF. Systematic review of COVID-19 in children shows milder cases and a better prognosis than adults. Acta Paediatr. 2020;109:1088–1095. doi: 10.1111/apa.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anna S, Burtseva E, Eropkin M. Influenza surveillance in Russia based on epidemiological and laboratory data from the period 2005 to 2012. Am J Infect Dis. 2013;9:77–93. doi: 10.3844/ajidsp.2013.77.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All aggregated data are available either in the manuscript or as supplementary materials. Further requests might require ethical approval and should be discussed with the authors by contacting the corresponding author.


