Abstract
Background
We identified a global chemical pattern of volatile organic compounds in exhaled breath capable of discriminating between COVID-19 patients and controls (without infection) using an electronic nose.
Methods
The study focused on 42 SARS-CoV-2 RT-qPCR positive subjects as well as 42 negative subjects. Principal component analysis indicated a separation of the study groups and provides a cumulative percentage of explanation of the variation of 98.3%.
Results
The canonical analysis of principal coordinates model shows a separation by the first canonical axis CAP1 (r2 = 0.939 and 95.23% of correct classification rate), the cut-off point of 0.0089; 100% sensitivity (CI 95%:91.5–100%) and 97.6% specificity (CI 95%:87.4–99.9%). The predictive model usefulness was tested on 30 open population subjects without prior knowledge of SARS-CoV-2 RT-qPCR status. Of these 3 subjects exhibited COVID-19 suggestive breath profiles, all asymptomatic at the time, two of which were later shown to be SARS-CoV-2 RT-qPCR positive. An additional subject had a borderline breath profile and SARS-CoV-2 RT-qPCR positive. The remaining 27 subjects exhibited healthy breath profiles as well as SARS-CoV-2 RT-qPCR test results.
Conclusions
In all, the use of olfactory technologies in communities with high transmission rates as well as in resource-limited settings where targeted sampling is not viable represents a practical COVID-19 screening approach capable of promptly identifying COVID-19 suspect patients and providing useful epidemiological information to guide community health strategies in the context of COVID-19.
Keywords: COVID-19 screening, Electronic nose, Volatile organic compounds, Exhaled breath, Asymptomatic
1. Introduction
The World Health Organization declared COVID-19 disease caused by SARS-CoV-2 a pandemic on March 11th, 2020; this has spread globally and caused more than 137 million infections and an excess of 2.95 million deaths (as of April 13th, 2021). Disease control efforts have included social restrictions, increased respiratory cautions, nucleic acid-based molecular detection of infected cases, transmission control through non-pharmacological strategies, the evaluation of novel drug treatments, as well as the search for clinical severity markers.
Timely and accurate detection of COVID-19 is paramount to the prevention and effective control of the pandemic [1]. In this context, the golden standard for diagnosis is based on reverse transcriptase-polymerase chain reaction (RT-qPCR), which aims to detect SARS-CoV-2 RNA in nasopharyngeal and oropharyngeal swab samples. However, the collection of these specimens relies on an invasive procedure, is expensive, a cold chain of transportation of samples is needed and, exposes health personnel to biohazardous sampling [2]. In addition, nucleic acid-based molecular testing for SARS-CoV-2 relies on proper clinical suspicion and may incur in false negative results which depend on both technical variables relating to specimen collection, transportation and processing as well as to individual variations in viral behavior [3].
An interesting test concept proposed by Lamote et al. is breath analysis, which can be an alternative strategy capable of identifying characteristic chemical patterns in patients with COVID-19 and could be a useful screening tool [4].
Human exhaled breath is a complex composition of gases in which a large variety of chemical compounds have been identified, including small inorganic compounds (such as NO, O2, CO2), volatile organic compounds (VOCs, such as hydrocarbons, alcohols, ketones, aldehydes, esters) and non-volatile organic compounds (isoprostanes, cytosines, leukotrienes and hydrogen peroxide) [5]. This mix of organic compounds result from cellular metabolism, and as they have low solubility in the blood, they are easily exhaled and are thus amenable to a breath analysis.
Several studies have shown that chemical patterns exist in the exhaled breath which are specific to certain diseases. Our research group has extensive experience in the identification of exhaled breath biomarkers in lung diseases, breast cancer, diabetes, among others [6], [7], [8], [9].
These chemical patterns, referred to as volatilome, are the result of normal physiological health conditions and specific physiopathological conditions, which allows this technique to be considered as low-cost, timely, non-invasive test and can be used to evaluate rapidly spreading diseases, such as COVID-19 [10]. This concept is based on previous studies of differential diagnosis of viruses such as influenza, respiratory syncytial virus and rhinovirus [11], [12]. Therefore, we undertook an exploratory study with the appropriate biosafety conditions to identify a global chemical pattern of VOCs in exhaled breath samples capable of being used to discriminate between COVID-19 patients and individuals without COVID-19 (control group) through the implementation of olfactory technology.
2. Material and methods
2.1. Study design
The study was approved by the state health research ethics committee of San Luis Potosí, Mexico, in compliance with national regulations for the execution of health research projects in humans. The study design was analytical cross-sectional, with a targeted sampling of positive and negative subjects with RT-qPCR test from the Research Center for Health Sciences and Biomedicine (CICSAB) and the School of Medicine of the Autonomous University of San Luis Potosi (UASLP). To establish a minimum expected correlation of 0.5 and a power of 0.8, in the response of the electronic nose as a function of the RT-qPCR result for COVID-19 we use a moderate Cohen's effect 0.5, the minimum sample size corresponded to an expected correlation of 0.5, a level of 0.05 and a power of 0.8, resulting in a minimum of 29 people per group, plus the expected 20% of losses established that the minimum number for the study was 35 [13].
Inclusion criteria for the group of COVID-19 positive patients were: i) 18 to 70 years of age, both sexes; ii) symptomatic (patients presenting, headache, sore throat, body aches, general discomfort, loss of taste and smell, among other typical symptoms), and asymptomatic, and asymptomatic; iii) SARS-CoV-2 specific gene RT-qPCR Ct below 38 to be considered as positive. The non-inclusion criteria were: i) pregnant patients; ii) patients with confirmed pulmonary infection other than COVID-19 (influenza, tuberculosis or other infectious diseases). Criteria for elimination included: i) subjects who withdrew informed consent and, ii) subjects who in the course of sampling acquired an infectious pathology.
For the control group (SARS-CoV-2 negative subjects), the inclusion criteria were: i) 18 to 70 years of age; ii) both sexes; iii) negative test for SARS-CoV-2 by RT-qPCR and iv) subjects having at least 7 days without apparent COVID-19 symptoms.
2.2. Nasopharyngeal swab and exhaled breath sample collection
Nasopharyngeal swab collection was carried out on all participating subjects based on CDC’s Interim Guidelines for Collecting and Handling of Clinical Specimens for COVID-19 Testing (Feb. 26, 2021). Exhaled breath sample collection was based on previous studies by our research group [6]. Participants were instructed to rest prior to sampling after which they would take three deep inhalations to finally exhale into a valve connected to a hermetically sealed metallized breath collector bag previously purged with ultrapure nitrogen. After that, the breath collector bag was hermetically sealed and placed on a protector cover for its transport and further analysis (Fig. 1 . Supplementary material). Subjects were provided with the following instructions before participation: i) minimum fasting of 4 h, ii) minimum smoking withdrawal before the study of 2 h, iii) avoid dental and oral hygiene in the previous hours and, iv) avoid taking any type of medication before the test. In both study groups, a clinical questionnaire was applied. In addition, an environmental control sample was collected to eliminate possible interferences during sample collection. Health workers involved in sample collection used personal protective equipment exceeding CDC recommendations including surgical scrubs, liquid-impermeable hooded coverall, neoprene boots, nitrile gloves and either Powered Air Purifying Respirators (PAPR) for those directly involved in patient contact for nasopharyngeal swabs and breath collection, or goggles and N95 respirators for sampling station helpers. The samples (both nasopharyngeal swabs and breath bags) were stored and transported in IATA compliant triple packaging and referred to the Viral and Human Genomics Laboratory, UASLP Faculty of Medicine for processing under biosafety level 3 (BSL-3) precautions.
Fig. 1.
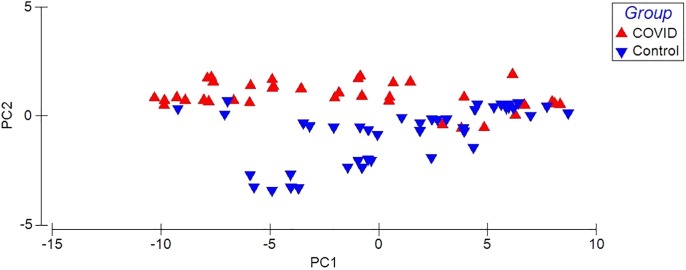
Principal Components Analysis of VOCs in the exhaled breath of patients with COVID-19 (red triangle) and control group (blue triangle). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Biosafety considerations
Upon arrival at the Viral & Human Genomics Laboratory, tertiary containers were disinfected using 70% ethanol and secondary containers introduced to the BSL-3 area. BSL-3 area is physically isolated from the main laboratory and kept at 0.4 in. of negative water pressure differential and directional inward airflow thanks to a HEPA exhausted air handling unit. All swab samples, as well as breath collector bags, were opened and processed inside a class II type A2 biological safety cabinet. All work carried out in the BSL3 area made use of surgical scrubs, liquid and gas impermeable coveralls, plastic apron, neoprene boots and PAPR or filter cartridge full-facepiece. After processing, all materials coming into contact with patient samples as well as sample specimen swabs, tubes and breath bags were decontaminated with 0.5% NaOCl allowing a contact time of 15 min before removing from the biological safety cabinet. Final decontamination of biological waste followed institutional guidelines on biohazardous waste disposal.
2.4. Molecular detection of SARS-CoV-2
Viral RNA was extracted from collected swabs using the QIAamp Viral RNA Mini kit (Qiagen) and subsequently tested for SARS-CoV-2 through a qualitative one-step RT-qPCR strategy using the COVID-19 PLUS RealAmp Kit (GeneFinder, OSANG Healthcare Co. Ltd.) on an Applied Biosystems 7500 real-time PCR instrument (Life Technologies, Thermo Fisher Scientific Corp.).
2.5. Exhaled breath analysis by electronic nose
The Cyranose 320 (Sensigent) was employed to determine the breath-print of the study groups. This technology is equipped with 32 polymer-based sensors (chemiresistors), all with different sensitivities to several VOCs increasing the electrical resistance of each sensor. Each chemiresistor has different VOCs adsorption properties that produce varying degrees of response owing to their polymeric composition (polyvinyl butyral, polyvinyl acetate, polystyrene and polyethylene oxide) and the conductive nanoparticles (black carbon and carbon nanotubes) of which they are composed.
The electronic nose setup consisted of a constant flow rate of 120 ml/min for 40 s of baseline recording with ultra-pure nitrogen with a sample analysis period of 46 s, then a flow rate of 180 ml/min of ultra-pure nitrogen was increased for the sample line purge and air inlet, with a substrate temperature of 40 °C. During the analysis, the instrument recorded the increase in electrical resistance of each chemiresistor as a result of the adsorption of VOCs on the sensors. As a biosafety measure at the end of the sample reading, ethanol vapors were passed through the instrument to disinfect the equipment and a container with sodium hypochlorite solution was placed at the outlet in the exhaust of the electronic nose.
2.6. Statistical analysis
A multivariate statistical analysis was performed employing the increase in resistance of the 32 sensors obtained from the fractional difference: ΔR/Ro = (Rmax-Ro)/Ro where R is the maximum system response of each sensor, and Ro is the reference reading of each sensor (ultrapure nitrogen). Principal component analysis (PCA) and canonical analysis of principal coordinates (CAP) were performed to evaluate the breath-prints of the study groups. The CAP was obtained through the multivariate data cloud which was the best to discriminate between the predefined groups.
Cross-validation with exclusion to predict group associations to obtain overall classification success rates was included for the CAP procedure. Also, this procedure included a leave-one-out cross-validation (using a K-fold of n = 8) to predict group associations to obtain overall classification success rates, with a value of m = 28. The statistical model was based on Euclidean distance matrices calculated from normalized (X- mean/SD) and log (X + 1) transformed data pre-processed from the 32-sensor R delta data; the difference between groups was calculated using 9999 permutations, this is performed for each observation in the data set, then the proportion of observations that were misclassified is calculated [14]. Also, the CAP model was used to predict the grouping of the new samples. The associations of the 32 sensors analyzed with the CAP 1 axis were evaluated by Pearson correlation. External validation was performed by selecting 70% of the population for cluster definition and the other 30% was randomly selected to validate the model.
Only the CAP1 axis was evaluated using the ROC curve (receiver operating characteristic curve) because it represented 100% of the data. With a 95% confidence interval (CI) and the threshold value or cut-off point was selected with the highest specificity/sensitivity ratio [15].
Statistical significance was determined using 5 and 1%. The statistical analysis was carried through the use of GraphPad Prism 5.0 and the Primer 7 + Permanova add-on software package (v7.0.12 and v1.0.6; PRIMER-E Lt.), for ROC and multivariate analysis, respectively.
2.7. Predictive model value in an open population: Pilot study
A pilot study was conducted in three populations to verify the usefulness of the model for the global identification of the chemical breath-print of COVID-19 in the open population with high incidence of COVID-19 but limited access to SARS-CoV-2 tests, the study participants were evaluated by RT-PCR test based on the previously described inclusion criteria.
The pilot study was conducted in three different scenarios: i) Stonemasons. Located in the community of Escalerillas, this area is the main quarrying region in the state of San Luis Potosi, Mexico (22°06′40″N 101°04′36″O); with a total population of 6226 inhabitants, the locality presents a high degree of marginalization. ii) Brickmakers. Located in the brick zone “Las terceras” in San Luis Potosi, Mexico (22°12′04″N, 100°51′26″W); the municipality has a population of 824,229 inhabitants and iii) Milpillas. Located in the suburbs of the state of San Luis Potosi, Mexico, (22°18′36′’N, 101°13′00′’W).
3. Results
The study included 84 subjects, 42 being SARS-CoV-2 positive and 42 negative individuals as confirmed by RT-qPCR. Patient gender were 33% female and 77% male, with an average age of participants being 38 ± 14 years, the average weight of 74 ± 15 kg, the average height of 1.6 ± 0.1 m and an average body mass index of 26.7 ± 4.6 kg/m2. For SARS-CoV-2 RT-qPCR positive subjectsCOVID-19 symptom onset ranged from 4 to 8 days.
The data obtained from the breath samples were analyzed through a PCA (Fig. 1), a natural separation of the study groups is observed, indicating different chemical breath-prints of VOCs, PC1 presents a percentage of explanation of 93.1% and PC2 of 5.2% reaching a cumulative percentage of explanation of the variation of 98.3%.
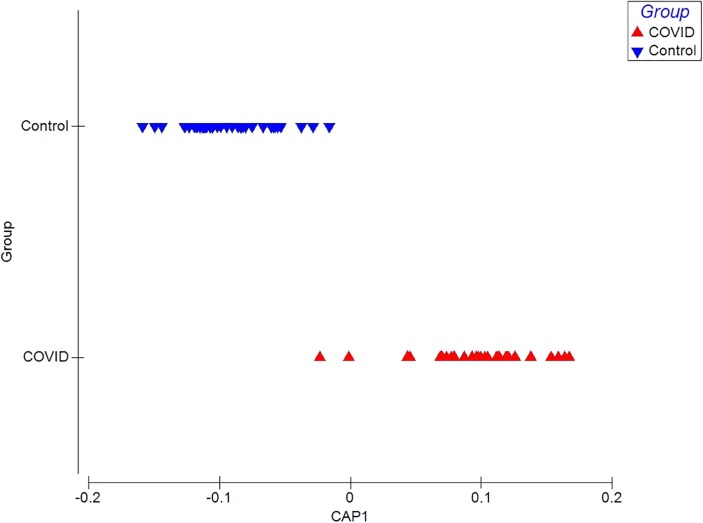
The CAP model shows a separation between the chemical breath-prints of patients with COVID-19 and control group by the first canonical axis CAP1 with r2 of 0.9392 and 95.23% of correct classification rate (Fig. 2 ). Also, the values of the external validation of the CAP model obtained a percentage of correct prediction of 100%.
Fig. 2.
Canonical Analysis of Principal Coordinates (CAP) of VOCs in patients with COVID-19 and control group. The chemical print of each patient with COVID-19 is shown in a red triangle and each control is shown as blue triangles. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To evaluate the contribution of the sensors to the discrimination between groups, a correlation of the 32 sensors was carried out with the values of the discrimination model (CAP), it is observed that for the control group the 28 sensors are positively associated with CAP 1, except for sensor 5, 11, 23 and, 31, with respect of the COVID-19 group, 26 sensors are negatively associated with CAP 1 except for sensors 6, 8, 14, 20, 23 and, 24 (Table 1 ).
Table 1.
Pearson's correlation between sensor responses and CAP 1 values.
| Sensors | Healthy controls | COVID-19 |
|---|---|---|
| S1 | 0.4019 | −0.3073 |
| S2 | 0.3938 | −0.3213 |
| S3 | 0.404 | −0.3391 |
| S4 | 0.3911 | −0.3232 |
| S5 | – | −0.3366 |
| S6 | 0.3624 | – |
| S7 | 0.4056 | −0.3293 |
| S8 | 0.4171 | |
| S9 | 0.3828 | −0.3488 |
| S10 | 0.407 | −0.3212 |
| S11 | – | −0.3517 |
| S12 | 0.395 | −0.3338 |
| S13 | 0.3973 | −0.3139 |
| S14 | 0.3918 | – |
| S15 | 0.3908 | −0.3641 |
| S16 | 0.409 | −0.3343 |
| S17 | 0.3922 | −0.3521 |
| S18 | 0.3875 | −0.3207 |
| S19 | 0.407 | −0.3321 |
| S20 | 0.4249 | – |
| S21 | 0.3863 | −0.3447 |
| S22 | 0.4087 | −0.3297 |
| S23 | – | – |
| S24 | 0.422 | – |
| S25 | 0.4011 | −0.3399 |
| S26 | 0.3236 | −0.3551 |
| S27 | 0.405 | −0.3221 |
| S28 | 0.3777 | −0.3092 |
| S29 | 0.3857 | −0.3353 |
| S30 | 0.3634 | −0.3661 |
| S31 | – | −0.3385 |
| S32 | 0.3895 | −0.3462 |
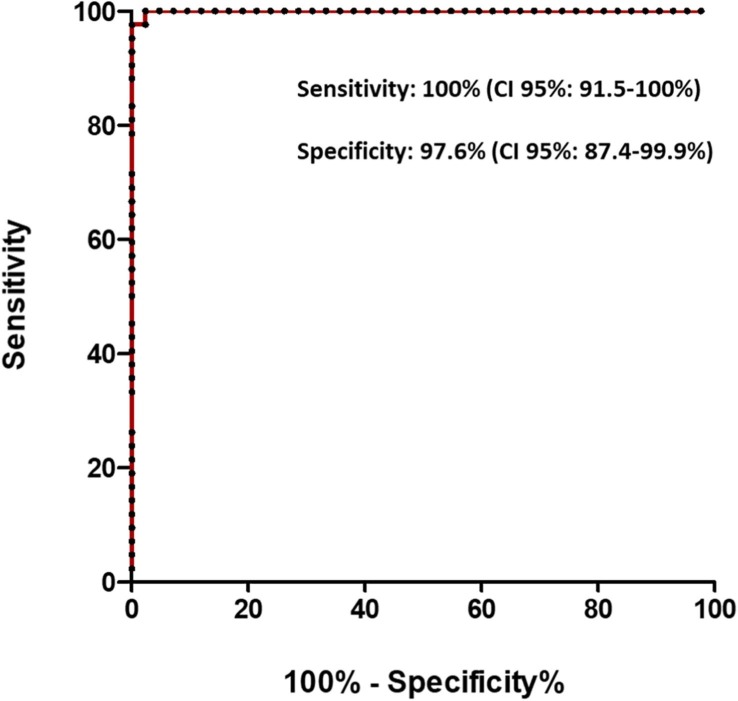
Furthermore, with the values created in the CAP 1 score, the cut-off point of 0.0089 was established, which provided 100% sensitivity (95% CI: 91.5–100%) and 97.6% specificity (95% CI: 95%: 87.4–99.9%) (Fig. 3 ).
Fig. 3.
ROC curve for the diagnosis of COVID-19 when using the CAP1 axis. An AUC of 0.9994 (CI 95%: 0.997–1.0) was obtained when using a cut-off point of 0.0089.
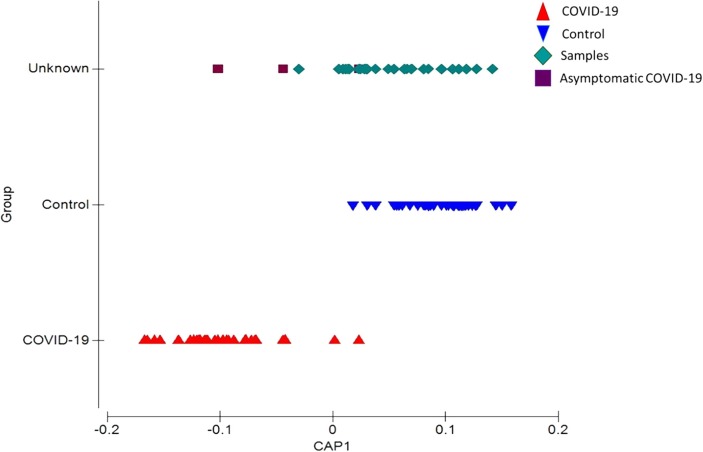
The predictive model usefulness pilot study evaluated 30 subjects from the open population, based on the RT-qPCR results, 3 people were identified with COVID-19 and 27 without the presence of the virus, they were also classified as asymptomatic, when applying the predictive model it was possible to identify 3 people with overall profiles similar to the COVID-19 group, two confirmed asymptomatic and 1 negative to RT-qPCR, 1 positive person was positioned in the control group (Fig. 4 ).
Fig. 4.
CAP prediction model of pilot sample positioning.
4. Discussion
The interaction of viruses with host cells has shown to trigger various biochemical processes that are reflected as changes in chemical signatures [16]. Studies with coronavirus 229E (HCoV-229E) in kidney and liver cell cultures have provided evidence of lipid metabolic remodelling [17]. These biochemical changes have been reported in patients with severe COVID-19, identifying altered metabolic pathways involving macrophage dysregulation, platelet degranulation and complement system pathways, and massive metabolic suppression [18]. SARS-CoV-2 binds to the angiotensin-converting enzyme receptor 2 (ACE2), leading to clearly distinct downstream pathways within infected cells [19], potentially followed by the formation of virus-specific VOCs even in the early stages of infection [4]. In this regard, Grassin-Delyle et al. were able to identify by gas chromatography-mass spectrometry four VOCs (methylpent-2-enal, 2,4-octadiene 1-chloroheptane, and nonanal) capable of distinguishing between COVID-19 and non-COVID-19 patients with acute respiratory distress syndrome (ARDS). The presence of these metabolites is indicative of products associated with the lipoperoxidation of cell membranes as a result of oxidative stress. The study involved 40 patients with ARDS (28 with COVID-19) and achieved a sensitivity of 90% and specificity of 94% [20].
Under our exploratory study, we were able to obtain a discrimination model between global chemical breath-prints of ambulatory subjects with COVID-19 and the control group. Shan et al., 2020, developed a discrimination model in exhaled breath in subjects from Wuhan China, by using a portable electronic nose analyzer equipped with 8 sensors of varied chemical nature coupled to spherical gold nanoparticles; in the breath sampling, is indicated that the study subjects breathed directly over an aperture of the instrument for 4 sec. The study included 49 patients with COVID-19 and 58 control subjects. With 100% sensitivity and 61% specificity, this system is promising for rapidly discriminating subjects with COVID-19 [21].
A study in the Netherlands conducted through the use of an electronic nose with 3 metal-oxide sensors, the study participants exhaled for 5 consecutive mins through a carbon filter and high-efficiency particulate matter (HEPA), their model included 219 participants of which 57 were COVID-19 positive, their model achieved a sensitivity of 86% and a negative predictive value of 92% [22]. De Vries et al., (2021), used an electronic nose equipped with 7 metal oxide semiconductor (MOS) sensors, its analysis processing consisted of a deep exhalation of the study subject to the equipment, the discrimination model was built with a group of 4,510 subjects of which 68 individuals were positive to COVID-19, their results indicate a sensitivity of 100% and specificity of 78% [23]. Our results are comparable with those reported in the literature, which demonstrates the potential of olfactory technology as a diagnostic tool. From our point of view, it should be considered that sampling represents an important risk of contagion, both for the analyst and the test subjects, since saliva droplets are the main sources of contagion [24], in this protocol, it is included the use of hermetic metallized breath collector bags, which reduces the risk of contagion for the analyst in addition to the biosafety conditions, this protocol should be considered in order to improve the analysis techniques. Likewise, when compared with other studies, differences were found such as avoiding cross-contamination by cleaning the equipment and the sensors with ethanol vapors (it was corroborated that the sensors did not present memory of the ethanol vapors previous to the reading of each sample).
On the other hand, there are some limitations to our interpretation of the results, within the inclusion criteria for the selection of study subjects for the discrimination model, the use of medications that could alter VOCs was not entirely controlled, since the use of anti-inflammatory medications is generally recommended; although the results according to the sensitivity and specificity obtained from the ROC curve are promising, the size of the pilot sample was limited, and these observations will require confirmation with an external validation cohort due to the severity of the disease. As well as the absence of information on the specific metabolites that are produced in the exhaled breath by the interaction of SARS-CoV-2 and the host, and lead to the difference between the breath-prints of the two groups.
The motivating factor was the results obtained in the open population through the application of the exhaled breath model and confirmed with RT-qPCR, this gives us a direction for the massive application of olfactory technologies in sites with high transmission rates and where targeted sampling is not applied, these techniques for their simplicity, low-cost, short analysis time and non-invasive, could function as starting points for establishing actions to mitigate transmission. In all, the use of olfactory technologies in communities having high transmission rates as well as in resource-limited settings where targeted sampling is not viable represents a practical COVID-19 screening approach capable of promptly identifying COVID-19 suspect patients and providing useful epidemiological information to guide community health strategies in the context of COVID-19
Furthermore, our research group has demonstrated the use of this technology for lung diseases such as COPD, identifying molecules associated with inflammatory processes (aldehydes, ketones and carboxylic acids) [6], [7]; which opens a research line in which the use of olfactory technology is proposed in the evaluation of the so-called long COVID, which occurs mainly in manifestations of lung damage in symptomatic and asymptomatic people at least 2–6 months after COVID-19 onset [25], [26]. Currently, one of the recurrent ways to evaluate lung damage is computed tomography, nevertheless, this technology is costly, thus generating a fast, simple and low-cost screening test is now a priority [27].
CRediT authorship contribution statement
Maribel Rodríguez-Aguilar: Conceptualization, Analytical methods, writing. Lorena Díaz de León-Martínez: Conceptualization and Analytical methods, Writing and Editing. Blanca Nohemí Zamora-Mendoza: Conceptualization, Sampling. Andreu Comas-García: Supervision and validation. Sandra Elizabeth Guerra Palomares: Molecular testing of samples. Christian Alberto García-Sepúlveda: Sampling, biosafety supervision; Luz Eugenia Alcántara-Quintana: Sampling and Supervision. Fernando Díaz-Barriga: Conceptualization, Supervision, review and editing; Rogelio Flores-Ramírez: Conceptualization, Analytical methods, Writings, Editing and Funding.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Omar Ornelas Rebolledo who is director of the company LABINNOVA which financed the study and the Cyranose 320 equipment.
Acknowledgement
Addressing the cumulative risk from emerging biological, chemical and social threats that will exacerbate the second outbreak of COVID-19 of CEEPAC/UASLP 2020. LABINNOVA, Center of Investigation in Breath for early detection diseases.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cca.2021.04.015.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan J.-F.-W., Yuan S., Kok K.-H., To K.-K.-W., Chu H., Yang J., Xing F., Liu J., Yip C.-C.-Y., Poon R.-W.-S., Tsoi H.-W., Lo S.-K.-F., Chan K.-H., Poon V.-K.-M., Chan W.-M., Ip J.D., Cai J.-P., Cheng V.-C.-C., Chen H., Hui C.-K.-M., Yuen K.-Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. The Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng S., Yu F., Fan J., Zou Q., Xie G., Yang X., Chen W., Wang Q., Zhang D., Wang R. Saliva as a Diagnostic Specimen for SARS-CoV-2 by a PCR-Based Assay. A Diagnostic Validity Study. 2020 doi: 10.1016/j.cca.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamote K., Janssens E., Schillebeeckx E., Lapperre T.S., De Winter B.Y., van Meerbeeck J.P. The scent of COVID-19: viral (semi-)volatiles as fast diagnostic biomarkers? J. Breath Res. 2020;14(4) doi: 10.1088/1752-7163/aba105. [DOI] [PubMed] [Google Scholar]
- 5.Phillips M., Herrera J., Krishnan S., Zain M., Greenberg J., Cataneo R.N. Variation in volatile organic compounds in the breath of normal humans, Journal of chromatography. B, Biomed. Sci. Appl. 1999;729(1–2):75–88. doi: 10.1016/s0378-4347(99)00127-9. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Aguilar M., Diaz de Leon-Martinez L., Gorocica-Rosete P., Padilla R.P., Thirion-Romero I., Ornelas-Rebolledo O., Flores-Ramirez R. Identification of breath-prints for the COPD detection associated with smoking and household air pollution by electronic nose. Respir. Med. 2020;163 doi: 10.1016/j.rmed.2020.105901. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Aguilar M., Ramirez-Garcia S., Ilizaliturri-Hernandez C., Gomez-Gomez A., Van-Brussel E., Diaz-Barriga F., Medellin-Garibay S., Flores-Ramirez R. Ultrafast gas chromatography coupled to electronic nose to identify volatile biomarkers in exhaled breath from chronic obstructive pulmonary disease patients: A pilot study. Biomed. Chromatogr. 2019;33(12) doi: 10.1002/bmc.4684. [DOI] [PubMed] [Google Scholar]
- 8.Mendez-Rodriguez K.B., Figueroa-Vega N., Ilizaliturri Hernandez C.A., Cardona-Alvarado M., Borjas Garcia J.A., Kornhauser C., Malacara J.M., Flores-Ramirez R., Perez-Vazquez F.J. Identification of metabolic markers in patients with type 2 Diabetes Mellitus by Ultrafast gas chromatography coupled to electronic nose. A pilot study. Biomed. Chromatogr. 2020 doi: 10.1002/bmc.4956. [DOI] [PubMed] [Google Scholar]
- 9.L.-M. Lorena Díaz de, R.-A. Maribel, G.-R. Patricia, R. Carlos Alberto Domínguez, B. Verónica Martínez, T.-T. Juan Alberto, O.-R. Omar, R. José Alfonso Cruz, B.-S. Berenice, F.-R. Rogelio, Identification of profiles of volatile organic compounds in exhaled breath by means of an electronic nose as a proposal for a screening method for breast cancer: a case-control study, J. Breath Res. (2020). [DOI] [PubMed]
- 10.Gandhi P., Bafna R., Arabale G., Engineer S., Phadke S. Olfactory device for large scale pre-screening for COVID-19. Trans. Indian National Acad. Eng. 2020:1–4. doi: 10.1007/s41403-020-00126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schivo M., Aksenov A.A., Linderholm A.L., McCartney M.M., Simmons J., Harper R.W., Davis C.E. Volatile emanations from in vitro airway cells infected with human rhinovirus. J. Breath Res. 2014;8(3) doi: 10.1088/1752-7155/8/3/037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aksenov A.A., Sandrock C.E., Zhao W., Sankaran S., Schivo M., Harper R., Cardona C.J., Xing Z., Davis C.E. Cellular scent of influenza virus infection. ChemBioChem. 2014;15(7):1040–1048. doi: 10.1002/cbic.201300695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.J. Cohen, Statistical power analysis for the behavioral sciences, Academic Press, 2013.
- 14.W.T. Anderson MJ, Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology., Ecology 84 (2003) 511–525.
- 15.Hajian-Tilaki K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Caspian J. Intern. Med. 2013;4(2):627–635. [PMC free article] [PubMed] [Google Scholar]
- 16.Armijos-Jaramillo V., Yeager J., Muslin C., Perez-Castillo Y. SARS-CoV-2, an evolutionary perspective of interaction with human ACE2 reveals undiscovered amino acids necessary for complex stability. Evol. Appl. 2020;13(9):2168–2178. doi: 10.1111/eva.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan B., Chu H., Yang D., Sze K.-H., Lai P.-M., Yuan S., Shuai H., Wang Y., Kao R.Y.-T., Chan J.F.-W., Yuen K.-Y. Characterization of the lipidomic profile of human coronavirus-infected cells: implications for lipid metabolism remodeling upon coronavirus replication. Viruses. 2019;11(1):73. doi: 10.3390/v11010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.B. Shen, X. Yi, Y. Sun, X. Bi, J. Du, C. Zhang, S. Quan, F. Zhang, R. Sun, L. Qian, W. Ge, W. Liu, S. Liang, H. Chen, Y. Zhang, J. Li, J. Xu, Z. He, B. Chen, J. Wang, H. Yan, Y. Zheng, D. Wang, J. Zhu, Z. Kong, Z. Kang, X. Liang, X. Ding, G. Ruan, N. Xiang, X. Cai, H. Gao, L. Li, S. Li, Q. Xiao, T. Lu, Y.J. Zhu, H. Liu, H. Chen, T. Guo, Proteomic and Metabolomic Characterization of COVID-19 Patient Sera, medRxiv (2020) 2020.04.07.20054585. [DOI] [PMC free article] [PubMed]
- 19.Bourgonje A.R., Abdulle A.E., Timens W., Hillebrands J.-L., Navis G.J., Gordijn S.J., Bolling M.C., Dijkstra G., Voors A.A., Osterhaus A.D., van der Voort P.H., Mulder D.J., van Goor H. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19) J. Pathol. 2020;251(3):228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grassin-Delyle S., Roquencourt C., Moine P., Saffroy G., Carn S., Heming N., Fleuriet J., Salvator H., Naline E., Couderc L.-J., Devillier P., Thévenot E.A., Annane D. Metabolomics of exhaled breath in critically ill COVID-19 patients: A pilot study. EBioMedicine. 2021;63 doi: 10.1016/j.ebiom.2020.103154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan B., Broza Y.Y., Li W., Wang Y., Wu S., Liu Z., Wang J., Gui S., Wang L., Zhang Z., Liu W., Zhou S., Jin W., Zhang Q., Hu D., Lin L., Zhang Q., Li W., Wang J., Liu H., Pan Y., Haick H. Multiplexed nanomaterial-based sensor array for detection of COVID-19 in exhaled breath. ACS Nano. 2020;14(9):12125–12132. doi: 10.1021/acsnano.0c05657. [DOI] [PubMed] [Google Scholar]
- 22.Wintjens A.G.W.E., Hintzen K.F.H., Engelen S.M.E., Lubbers T., Savelkoul P.H.M., Wesseling G., van der Palen J.A.M., Bouvy N.D. Applying the electronic nose for pre-operative SARS-CoV-2 screening. Surg. Endosc. 2020 doi: 10.1007/s00464-020-08169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R. de Vries, R.M. Vigeveno, S. Mulder, N. Farzan, D.R. Vintges, J.J. Goeman, S. Bruisten, B. van den Corput, J.J.M. Geelhoed, L.G. Visser, M. van der Lubben, P.J. Sterk, J.C.C.M. in ’t Veen, G.H. Groeneveld, Ruling out SARS-CoV-2 infection using exhaled breath analysis by electronic nose in a public health setting, medRxiv (2021) 2021.02.14.21251712.
- 24.Li Y., Ren B., Peng X., Hu T., Li J., Gong T., Tang B., Xu X., Zhou X. Saliva is a non-negligible factor in the spread of COVID-19. Mol. Oral Microbiology. 2020;35(4):141–145. doi: 10.1111/omi.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonnweber T., Sahanic S., Pizzini A., Luger A., Schwabl C., Sonnweber B., Kurz K., Koppelstätter S., Haschka D., Petzer V., Boehm A., Aichner M., Tymoszuk P., Lener D., Theurl M., Lorsbach-Köhler A., Tancevski A., Schapfl A., Schaber M., Hilbe R., Nairz M., Puchner B., Hüttenberger D., Tschurtschenthaler C., Aßhoff M., Peer A., Hartig F., Bellmann R., Joannidis M., Gollmann-Tepeköylü C., Holfeld J., Feuchtner G., Egger A., Hoermann G., Schroll A., Fritsche G., Wildner S., Bellmann-Weiler R., Kirchmair R., Helbok R., Prosch H., Rieder D., Trajanoski Z., Kronenberg F., Wöll E., Weiss G., Widmann G., Löffler-Ragg J., Tancevski I. Cardiopulmonary recovery after COVID-19 – an observational prospective multi-center trial. Eur. Respir. J. 2020;2003481 doi: 10.1183/13993003.03481-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Logue J.K., Franko N.M., McCulloch D.J., McDonald D., Magedson A., Wolf C.R., Chu H.Y. Sequelae in Adults at 6 Months After COVID-19 Infection. JAMA Network Open. 2021;4(2) doi: 10.1001/jamanetworkopen.2021.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meghji J., Mortimer K., Agusti A., Allwood B.W., Asher I., Bateman E.D., Bissell K., Bolton C.E., Bush A., Celli B., Chiang C.-Y., Cruz A.A., Dinh-Xuan A.-T., El Sony A., Fong K.M., Fujiwara P.I., Gaga M., Garcia-Marcos L., Halpin D.M.G., Hurst J.R., Jayasooriya S., Kumar A., Lopez-Varela M.V., Masekela R., Mbatchou Ngahane B.H., Montes de Oca M., Pearce N., Reddel H.K., Salvi S., Singh S.J., Varghese C., Vogelmeier C.F., Walker P., Zar H.J., Marks G.B. Improving lung health in low-income and middle-income countries: from challenges to solutions. The Lancet. 2021;397(10277):928–940. doi: 10.1016/S0140-6736(21)00458-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.