Graphical abstract
Keywords: SARS-CoV-2, COVID-19, Diagnostic testing, Real time-PCR, Multiplex
Abstract
Objective
The aim of this study was to evaluate the QIAstat-Dx® Respiratory SARS-CoV-2 Panel (QIAstat-SARS-CoV-2), which is a closed, fully automated, multiplex polymerase chain reaction (PCR) assay that detects severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and 21 other pathogens that cause respiratory disease.
Methods
Nasopharyngeal swabs from patients with or suspected of having coronavirus disease 2019 were collected and tested at Bichat–Claude Bernard Hospital, Paris, France. Using the World Health Organisation-approved real-time-PCR assay developed by the Charité Institute of Virology as the reference, positive percent agreement (PPA) and negative percent agreement (NPA) were calculated.
Results
In total, 189 negative and 88 positive samples were analyzed. QIAstat-SARS-CoV-2 had an NPA of 90.48% (95% confidence interval (CI), 85.37%, 94.26%) and a PPA of 94.32% (95% CI, 87.24%, 98.13%). Co-infections were detected by QIAstat-SARS-CoV-2 in 4/277 specimens. The methods exhibited comparable failure rates (23/307 [7.5%] vs. 6/298 [2.0%] for QIAstat-SARS-CoV-2 and reference methods, respectively). The turnaround time was shorter for QIAstat-SARS-CoV-2 compared with the reference method (difference in mean –14:30 h [standard error, 0:03:23; 95% CI, –14:37, –14:24]; P < 0.001).
Conclusions
QIAstat-SARS-CoV-2 shows good agreement with the reference assay, providing faster and accurate results for detecting SARS-CoV-2.
Introduction
Early identification of patients with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection enables rapid isolation to prevent transmission (European Centre for Disease Prevention and Control, 2020, National Institutes of Health, 2020, The World Health Organization, 2020). Symptoms of coronavirus disease 2019 (COVID-19) are common to a range of respiratory pathogens; therefore, accurate diagnostics are required for differential diagnosis.
The QIAstat-Dx® Respiratory SARS-CoV-2 Panel (QIAstat-SARS-CoV-2) is a closed, fully automated, multiplex assay that detects SARS-CoV-2 and 21 other respiratory pathogens. Prior independent validation studies of the SARS-CoV-2 assay demonstrated comparable performance with a WHO-recommended reverse-transcription polymerase chain reaction (RT-PCR) assay (Visseaux et al., 2020). The remaining 21 targets in the panel have been previously validated (Boers et al., 2020).
This study aimed to evaluate the SARS-CoV-2 assay performance characteristics in QIAstat-SARS-CoV-2 against the WHO-recommended reference method (WHO-Charité) (Corman et al., 2020).
Methods
This was an observational, retrospective study of specimens from patients with or suspected of having COVID-19. The primary objective was to compare the performance of QIAstat-SARS-CoV-2 with the WHO-Charité reference method (Corman et al., 2020), which was the standard-of-care test at the investigation site (acceptance criteria: positive percent agreement [PPA] and negative percent agreement [NPA] ≥90%). Secondary objectives were to evaluate failure rates and differences between methods in mean cycle threshold (Ct) values and time-to-results.
Collection of nasopharyngeal swab specimens and analysis was performed at Bichat–Claude Bernard Hospital, Paris, France. De-identified, residual samples were tested, and only transport medium liquid samples were included. Further methodological details are provided in the Supplementary data.
The study was conducted following the ethical principles of the Declaration of Helsinki and regulations regarding Good Clinical Practice.
Results
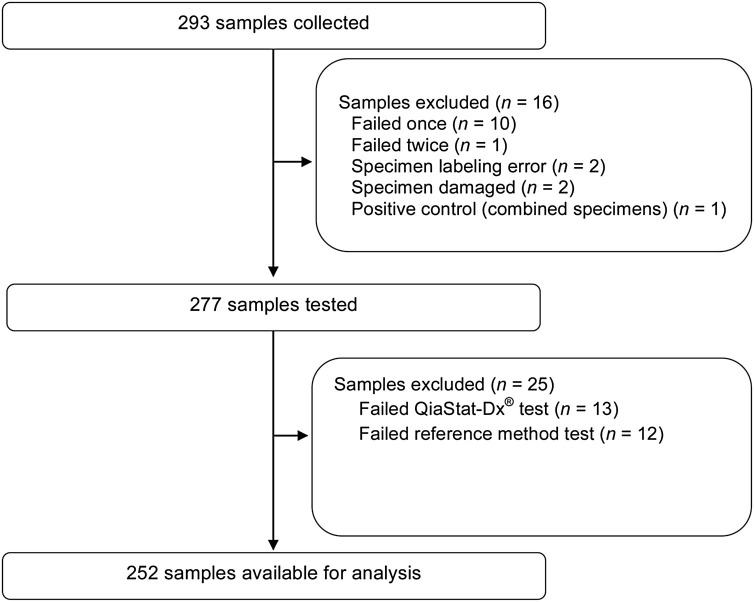
The analysis took place between March 23 and June 04, 2020. 293 specimens were collected, and 16 samples were excluded from the analysis (Figure 1 ). The remaining 277 samples were tested using both QIAstat-SARS-CoV-2 and the reference method. An additional 25 results were excluded based on a failed QIAstat-SARS-CoV-2 (n = 13) or reference method test (n = 12; Figure 1). Patient age at hospital admission was available for 237 subjects (mean 58.2 years [range, 1–97 years; standard deviation, 20.24 years]).
Figure 1.
Study flow diagram.
In total, 18/189 samples that were negative according to the reference method were positive using QIAstat-SARS-CoV-2 (NPA: 90.48% [95% confidence interval (CI), 85.37%, 94.26%]; Table 1 ). According to the reference method, five of 88 samples that were positive were negative according to QIAstat-SARS-CoV-2 (PPA: 94.32% [95% CI, 87.24%, 98.13%]). These 23 discordant results were investigated by reviewing patient medical source data. An alternative RT-PCR (Cobas or Altona) method on different samples obtained during the patients’ follow-up confirmed one of five QIAstat-SARS-CoV-2-negative reference assay-positive results to be positive. Seven of 18 QIAstat-SARS-CoV-2-positive reference assay-negative results were confirmed as positive. Both methods exhibited comparable failure rates; 23/307 (7.5%) QIAstat-SARS-CoV-2 tests failed compared with 6/298 (2.0%) reference method assays.
Table 1.
Performance of QIAstat-Dx® Respiratory SARS-CoV-2 Panel system in comparison with the reference method.
| QiaStat-Dx® | Reference method |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 83 | 18 | 101 |
| Negative | 5 | 171 | 176 |
| Total | 88 | 189 | 277 |
| PPA, % (95% CI) | 94.32 (87.24, 98.13) | ||
| NPA, % (95% CI) | 90.48 (85.37, 94.26) | ||
CI, confidence interval; NPA, negative percent agreement; PPA, positive percent agreement; SARS-CoV-2; severe acute respiratory syndrome coronavirus 2.
The mean Ct value for QIAstat-SARS-CoV-2 was significantly lower than for each component of the reference method (difference in mean: −6.747 [standard error (SE), 0.329; 95% CI, –7.398, –6.097]; P < 0.001 and –4.979 [SE, 0.271; 95% CI, –5.516, –4.443]; P < 0.001, for RNA-dependent RNA polymerase- and E gene-components, respectively). When plotting individual Ct values obtained with QIAstat-SARS-CoV-2 against those for each component of the reference method, a proportional shift was observed over the whole range of experimental Ct values (Supplementary Figure 1). Of the 277 QIAstat-SARS-CoV-2 results analyzed, four specimens were positive for multiple pathogens, including two co-infections of SARS-CoV-2 and another respiratory pathogen (Supplementary Table 1).
The mean time to result was significantly shorter for QIAstat-SARS-CoV-2 than the reference method (Δ –14:30 h [SE, 03:23; 95% CI, –14:37, –14:24]; P < 0.001).
Discussion
The QIAstat-SARS-CoV-2 panel demonstrated PPA and NPA with the WHO-recommended assay greater than 90%, both in this study and in another previous smaller study (Visseaux et al., 2020).
Rapid testing for SARS-CoV-2 enables quick triage and the identification of patients requiring isolation (Brendish et al., 2020). In this study, QIAstat-SARS-CoV-2 was 14.5 h quicker than the reference method; this rapid testing with QIAstat-SARS-CoV-2 was consistent with previous results (Brendish et al., 2020, Visseaux et al., 2020).
Near-patient multiplex tests allow testing multiple pathogens in a single assay, simplifying testing workflows and assisting in the timely differential diagnosis of infectious diseases. The utility of near-patient multiplex testing using the QIAstat-Dx® Respiratory Panel to reduce time to diagnosis and improve patient management has previously been demonstrated (Bouzid et al., 2020).
In this study, co-infections were identified in four samples, including two samples positive for SARS-CoV-2. Co-infections with SARS-CoV-2, at rates in-line with this study, have previously been reported in the literature (Lai et al., 2020, Lansbury et al., 2020).
In conclusion, QIAstat-SARS-CoV-2 produces concordant results with the WHO-Charité reference method, but in a significantly shorter time and in a near-patient setting.
Funding
This study has been funded by QIAGEN Manchester Ltd., Manchester, UK.
Conflict of interest
BG is a contractor for QIAGEN. MCC, AE, JP, JL, and DM are employees of QIAGEN. DD has received personal fees from Viiv Healthcare, Gilead Sciences, and Janssen Cilag. BV has received grants from QIAGEN, personal fees from QIAGEN, BioMérieux, Hologic and Gilead, and non-financial support from QIAGEN and BioMérieux. SL, AS, QH, GAT, and NHF do not declare any conflicts of interest.
Acknowledgments
The authors would like to acknowledge Sarah Johnston, MBiolSci, of Ashfield MedComms, an Ashfield Health company, part of UDG Healthcare plc, for medical writing support that was funded by QIAGEN Manchester Ltd., Manchester, UK.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2021.04.066.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Boers S., Melchers W., Peters C., Toonen M., McHugh M., Templeton K., et al. Multicenter evaluation of QIAstat-Dx Respiratory Panel V2 for detection of viral and bacterial respiratory pathogens. J Clin Microbiol. 2020;58(6) doi: 10.1128/JCM.01793-19. e01793–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzid D., Lucet J.-C., Duval X., Houhou-Fidouh N., Casalino E., Visseaux B., et al. PCR implementation as point-of-care testing in a French emergency department. J Hosp Infect. 2020;105(June (2)):337–338. doi: 10.1016/j.jhin.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendish N., Poole S., Naidu V., Mansbridge C., Norton N., Wheeler H., et al. Clinical impact of molecular point-of-care testing for suspected COVID-19 in hospital (COV-19POC): a prospective, interventional, non-randomized, controlled study. Lancet Respir Med. 2020;8(December (12)):1192–1200. doi: 10.1016/S2213-2600(20)30454-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control . 2020. Diagnostic testing and screening for SARS-CoV-2. Available from: https://www.ecdc.europa.eu/en/covid-19/latest-evidence/diagnostic-testing. [Accessed October 2020] [Google Scholar]
- Lai C.-C., Wang C.-Y., Hseuh P.-R. Co-infections among patients with COVID19: the need for combination therapy with non-anti-SARS-CoV-2 agents? J Microbiol Immunol Infect. 2020;53(4):505–512. doi: 10.1016/j.jmii.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbury L., Lim B., Baskaran V., Lim W. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infection. 2020;81:266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health . 2020. COVID-19 treatment guidelines. Testing for SARS-CoV-2 infection. Available from: https://www.covid19treatmentguidelines.nih.gov/. [Accessed January 2021] [Google Scholar]
- Visseaux B., Le Hingrat Q., Collin G., Bouzid D., Lebourgeois S., Le Pluart D., et al. Evaluation of the QIAstat-Dx Respiratory SARS-CoV-2 Panel, the first rapid multiplex PCR commercial assay for SARS-CoV2 detection. J Clin Microbiol. 2020;58(8) doi: 10.1128/JCM.00630-20. e00630–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The World Health Organization . 2020. Diagnostic testing for SARS-CoV-2. Available from: https://www.who.int/publications/i/item/diagnostic-testing-for-sars-cov-2. [Accessed October 2020] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.