Abstract
After the emergence of the pandemic, repurposed drugs have been considered as a quicker way of finding potential antiviral agents. SARS-CoV-2 3CLpro is essential for processing the viral polyproteins into mature non-structural proteins, making it an attractive target for developing antiviral agents. Here we show that Vitamin K3 screened from the FDA-Approved Drug Library containing an array of 1,018 compounds has potent inhibitory activity against SARS-CoV-2 3CLpro with the IC50 value of 4.78 ± 1.03 μM, rather than Vitamin K1, K2 and K4. Next, the time-dependent inhibitory experiment was carried out to confirm that Vitamin K3 could form the covalent bond with SARS-CoV-2 3CLpro. Then we analyzed the structure-activity relationship of Vitamin K3 analogues and identified 5,8-dihydroxy-1,4-naphthoquinone with 9.8 times higher inhibitory activity than Vitamin K3. Further mass spectrometric analysis and molecular docking study verified the covalent binding between Vitamin K3 or 5,8-dihydroxy-1,4-naphthoquinone and SARS-CoV-2 3CLpro. Thus, our findings provide valuable information for further optimization and design of novel inhibitors based on Vitamin K3 and its analogues, which may have the potential to fight against SARS-CoV-2.
Keywords: SARS-CoV-2 3CLpro, Vitamin K3, Covalent inhibitors
1. Introduction
COVID-19 (Coronavirus disease 2019) is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1,2], which is widely spread among people and highly contagious [3]. The clinical manifestations of COVID-19 patients are fever, dry cough, lymphopenia and disseminated intravascular coagulation, and so on [[4], [5], [6]]. In severe cases, SARS-CoV-2 leads to acute respiratory syndrome, renal failure, and eventually death [7]. Although the vaccines have been approved in several countries, the high mutation rate of SARS-CoV-2 [8] and the lack of data regarding more complete safety [9] and durable clinical efficacy of these vaccines are significant concerns [10,11]. Besides, vaccines are only given to healthy people, there are currently no effective drugs used for the treatment of patients with COVID-19 or blocking the transmission of the virus. Therefore, there is an urgent need to discover and develop new antiviral agents for COVID-19 treatment.
SARS-CoV-2 is classified as β-coronavirus, whose genomic sequence is highly identical to that of SARS-CoV (79.6%), and the amino acid sequences of the seven conserved domains of the replicase in ORF1ab of SARS-CoV-2 share 94.4% identity to SARS-CoV, indicating that they belong to the same species, SARSr-CoV [12]. Some key proteins in a variety of processes of the viral life cycle including invasion, replication, budding and release of SARS-CoV-2 have been considered as the potential targets for antiviral agents. During the processes, 3-chymotrypsin-like protease (3CLpro, or main protease, Mpro), is essential for the transcription and replication of SARS-CoV-2 [13], making it an attractive therapeutic target of COVID-19. SARS-CoV-2 3CLpro together with papain-like protease (PLpro) can cleavage polyproteins into functional proteins, including structural proteins, accessory proteins [14], and non-structural proteins (NSPs) [15]. This proteolysis plays an important role in the SARS-CoV-2 life cycle, as it produces the helicase and RNA-dependent RNA polymerase that is required for viral transcription and replication [16]. Because of the high sequence conservatism of 3CLpro of SARS-CoV-2 and SARS-CoV (~96%) [17], broad-spectrum antiviral inhibitors like N3, Compound 11a and 11b, baicalin and baicalein and α-ketoamide inhibitor 13b, have been developed based on this target (Fig. S1) [13,16,18,19]. Although several types of SARS-CoV-2 3CLpro inhibitors have been reported, none of them have moved into clinical trials yet. Thus, searching for broad-spectrum anti-coronavirus candidates targeting 3CLpro holds much promise for preventing the infection of SARS-CoV-2 [20,21].
In such emergence of the pandemic, repurposed drugs have been considered as a quicker way of finding potential antiviral agents. In the present study, we selected SARS-CoV-2 3CLpro as the potential antiviral target to screen the L1300 Selleck FDA-Approved Drug Library containing an array of 1018 compounds by FRET (Fluorescence resonance energy transfer) and 6 drugs were found to have the inhibitory activities against the protease. Among of them, Vitamin K3 was chosen for further study because it had stronger inhibitory activity than other compounds. As one of the synthetic forms of Vitamin K, Vitamin K3 also is known as Menadione, which is a fat-soluble substance and can be used as a clotting drug and Vitamin supplement [22,23]. A study has found that Vitamin K3 had good antibacterial activity against multi-resistant bacterial strain P. aeruginosa 03 and showed a significant synergistic antibacterial effect when combined with the aminoglycoside class of antibiotics by cell membrane permeabilization mechanism [24]. The combination of Vitamin K3 and ultraviolet light A as photosensitizer can inactivate Staphylococcus aureus, Pseudomonas aeruginosa, Staphylococcus epidermidis, Klebsiella pneumoniae, Bacillus cereus and Escherichia coli [25]. In addition, Vitamin K3 exhibited a spectrum of anticancer activities and effects against various cancer cells [[26], [27], [28]]. Recent research has also found that Vitamin K3 can inhibit the activity of SARS-CoV-2 3CLpro and serve as a potential lead molecule for further antiviral studies to combat COVID-19 [29]. However, the mechanism of action of Vitamin K3 and the binding mode with SARS-CoV-2 3CLpro remain largely unknown.
Here we found that Vitamin K3 showed time-dependent inhibition of SARS-CoV-2 3CLpro by a 4.4-fold decrease in the IC50 value (from 20.96 to 4.78 μM) in 30 min. Then we analyzed the structure-activity relationship of Vitamin K3 analogues and identified a Vitamin K3 analogue 5,8-dihydroxy-1,4-naphthoquinone with 9.8 times higher inhibitory activity than that of Vitamin K3. Further research found that the two compounds could efficiently block the enzymatic activities of SARS-CoV 3CLpro. Finally, mass spectrometric analysis and molecular docking study verified the covalent binding between SARS-CoV-2 3CLpro and Vitamin K3 or 5,8-dihydroxy-1,4-naphthoquinone. Thus, our findings provide valuable information for further optimization and design of novel inhibitors based on the structures of Vitamin K3 and its analogues, which may have the potential to fight against SARS-CoV-2.
2. Materials and methods
2.1. Expression and purification of SARS-CoV-2 3CLpro
Construction of the expression vector of SARS-CoV-2 3CLpro for producing N terminal tag-cleavable fusion proteins in E. coli BL21 (DE3) was accomplished according to reported procedures with modification [30]. Briefly, different from modified GST fusion protein expression vector (pGSTM), pET29a(+) was used to create the recombinant expression plasmid of SARS-CoV-2 3CLpro with ubiquitin-like protein Smt3 and the five amino acids SAVLQ at the N-terminus followed by a modified HRV 3C protease cleavage site (SGVTFQ↓GP) connected to a His6-tag at the C-terminus by homologous recombination, eventually producing the eight amino acids GPHHHHHH at the C-terminus of SARS-CoV-2 3CLpro. The plasmid DNA was transformed into E. coli BL21 (DE3) to express SARS-CoV-2 3CLpro by the auto-induction method as described previously [31]. The cells were lysed by sonication in ice and the lysate was centrifuged at 4 °C for 30 min at 18000 rpm. The supernatant was loaded onto 2 mL Ni-NTA agarose (GE Healthcare), eluted with 300 mM imidazole and further purified through Superdex 200 10/300 GL column (GE Healthcare). The protein of interest was concentrated by centrifugation using a 10 kDa molecular weight cut-off (MWCO) concentrator and stored in a solution (25 mM HEPES, 150 mM NaCl, 1 mM DTT, pH 7.4) for enzymatic inhibition assay.
2.2. Enzymatic inhibition assay of SARS-CoV-2 3CLpro or SARS-CoV 3CLpro by FRET
Dabcyl-KNSTLQSGLRKE-Edans (Sangon Biotech, Shanghai, China) was synthesized as a substrate to measure the protease activity of SARS-CoV-2 3CLpro. For the inhibition assay of SARS-CoV-2 3CLpro, 4 μg/mL protease was incubated with the indicated concentrations of tested compounds in reaction buffer (0.1 M PBS, 1 mM EDTA, pH 7.4) for 30 min at 37 °C. The fluorogenic substrate at a final concentration of 20 μM was added to initiate the reaction. The fluorescence intensity change was measured immediately every 2 min for 20 min at 340 nm (excitation) / 490 nm (emission) using Spectramax® ID3 (Molecular Devices, California, USA) plate reader. The inhibition ratios of the protease with compounds added at various concentrations were calculated compared to the reaction including the solvent control. An FDA-Approved Drug Library containing an array of 1,018 compounds was obtained from Selleck Chemicals (# L1300) and used for screening the inhibitors by a FRET assay against SARS-CoV-2 3CLpro. Vitamin K3 analogues were purchased from MCE. The IC50 values of Vitamin K3 and its analogues were calculated by fitting the curve of normalized inhibition ratio with the concentration of the test compounds. Similarly, the inhibitory activities of SARS-CoV 3CLpro by Vitamin K3 and its analogues were also determined according to the previously published method [32,33].
2.3. Analysis of the inhibitory mechanism of Vitamin K3 and 5,8-dihydroxy-1,4-naphthoquinone
The time-dependent inhibition (TDI) assay of Vitamin K3 or 5,8-dihydroxy-1,4-naphthoquinone was carried out according to the above conditions of IC50 determination, except that the incubation time was 3 min and 33 min respectively. To obtain the inhibition constant (K I) and the rate of enzyme inactivation (k inact) values of the two compounds, SARS-CoV-2 3CLpro was incubated with the indicated concentrations of tested compounds in reaction buffer (0.1 M PBS, 1 mM EDTA, pH 7.4) for 5, 10, 20, 30, 40 min at 37 °C, then allocated 10 μL protease (4 μg/mL final concentration), added 20 μM fluorogenic substrate to initiate the reaction. K I and k inact values of the compounds were calculated by nonlinear regression using GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, USA) according to the equation kobs = (k inact·[I]) / (K I + [I]) [34], in which kobs is the inactivation rate constant, [I] is the initial inhibitor concentration, k inact is the maximal inactivation rate constant and K I is the inhibitor concentration that produces 50% the maximal rate of inactivation [35].
2.4. Molecular weight analysis by mass spectrometry
Molecular weight of SARS-CoV-2 3CLpro with or without compounds was analyzed through Agilent 1290 Infinity II UPLC and Q-TOF 6530 mass spectrometry. The detailed experimental procedure was performed as follows. 8.4 μM SARS-CoV-2 3CLpro with 90 μM Vitamin K3 or 5,8-dihydroxy-1,4-naphthoquinone was incubated in reaction buffer (0.1 M PBS, 1 mM EDTA, pH 7.4) for 30 min at 37 °C. 10 μL protein samples were separated on a Waters ACQUITY UPLC Protein BEH C4 column (2.1 × 50 mm, 1.7 μm, 300 Å). The mobile phase A is 0.1% formic acid in ddH2O and mobile phase B is 0.1% formic acid in acetonitrile. The column temperature is 60 °C and the flow rate is 0.3 mL/min. The gradient is 10% MPB (mobile phase B) from 0 to 1 min, then 10% to 50% MPB from 1 min to 6 min, 50% to 90% MPB from 6 min to 7.5 min and keep 90% MPB for 0.5 min. The mass spectrometry parameters were shown as follows. Positive scan mode on ESI source; Scan mass range: 100–3200 m/z; Gas temperature: 300 °C; Drying gas: 8 L/min; Nebulizer: 35 psi; Sheath gas temperature: 350 °C; Drying gas: 8 L/min; Nebulizer: 35 psi; Sheath gas temperature: 350 °C; Sheath gas flow: 12 L/min. The acquired data is processed with Agilent BioConfirm software (version 10.0).
2.5. Catalytic site flexibility of SARS-CoV-2 3CLpro
To incorporate the dynamics and the possible induced fit-effect of the 3CLpro catalytic site in the modeling process and generate a suitable protein model, 84 SARS-CoV-2 3CLpro crystal structures from PDB were superimposed using chain A of 6LU7 [12] as a reference structure. PyMOL (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC) was used to read and align the structures. If multiple asymmetric chains exist, then each chain will be extracted and aligned to the reference. Residues within 3.5 Å of the peptidomimetic inhibitor in chain A of 6LU7 was defined as the catalytic site. Flexibility detection was based on the superposition of the crystal structures of SARS-CoV-2 3CLpro. Flexibility of catalytic residues were determined by visual check of the unicity of the spatial distribution of the corresponding atoms.
2.6. The similarity between Vitamin K3 and the SARS-CoV-2 3CLpro binders in PDB
All structures (259 compounds) of co-crystallized SARS-CoV-2 3CLpro binders were downloaded from PDB, which were provided as a CSV file in the supplementary data. As similar compounds often share a similar binding mode, the 2D similarity between Vitamin K3 and the co-crystallized SARS-CoV-2 3CLpro binders was calculated using DataWarrior [36].
2.7. Covalent docking using AutoDockFR
2.7.1. Ligand preparation
To use AutoDockFR for covalent docking, the receptor and covalent ligand were prepared according to the method described previously [37]. The ligand must be in the product form (Fig. S2), and the receptor and covalent ligand need to share 3 atoms. In this case, the atoms (all sidechain atoms and backbone carbon atoms) of cysteine should be included as part of the ligand structure to allow the conformational sampling of the sidechain of C145 during docking. The 2D structure of the product was converted to 3D structure by RDkit (version: 2020.3) [38] and PDBQT format by OpenBabel (version 3.0) [39], respectively. To meet the requirement of AutoDockFR, the atoms in the root section in PDBQT format must correspond to those atoms of backbone carbon, alpha carbon and beta carbon of cysteine, and this operation was achieved by using the MGLTools script, prepare_ligand4.py, with the “-R” option.
2.7.2. Protein model preparation
Crystal structure 6YNQ [40] was processed using PyMOL and Autodock/Vina plugin [41] and was saved in PDBQT format. The center of docking grid was defined as the center of mass of the ligand in 6YNQ, and a cube with length of 22.5 Å was defined as the grid box. Residues of N142 and M165 were defined as flexible residues, and C145 was defined as the covalent residue. The detected flexible residues were chosen to be modeled explicitly during docking. The atom index of backbone carbon, alpha carbon and beta carbon of C145 was specifically defined as overlapping atoms for the ligand. Program “agfr” was used to generate the grid file.
2.7.3. Docking
Covalent docking was carried with default options using program “adfr”, and the result was visualized with PyMOL and Autodock/Vina plugin. We firstly conducted re-docking. The re-docking was done using the SARS-CoV-2 3CLpro structure model (PDB: 6YNQ) with the same protocol (allowing M165, C145 and N142 to be flexible) as that of Vitamin K3 docking. As shown in Fig. S3, the PDB ligand 2-Methyl-1-tetralone docking pose (cyan stick model) was almost identical to that (gray stick model) in the crystal structure.
3. Results and discussion
3.1. Inhibitor screening against SARS-CoV-2 3CLpro by FRET
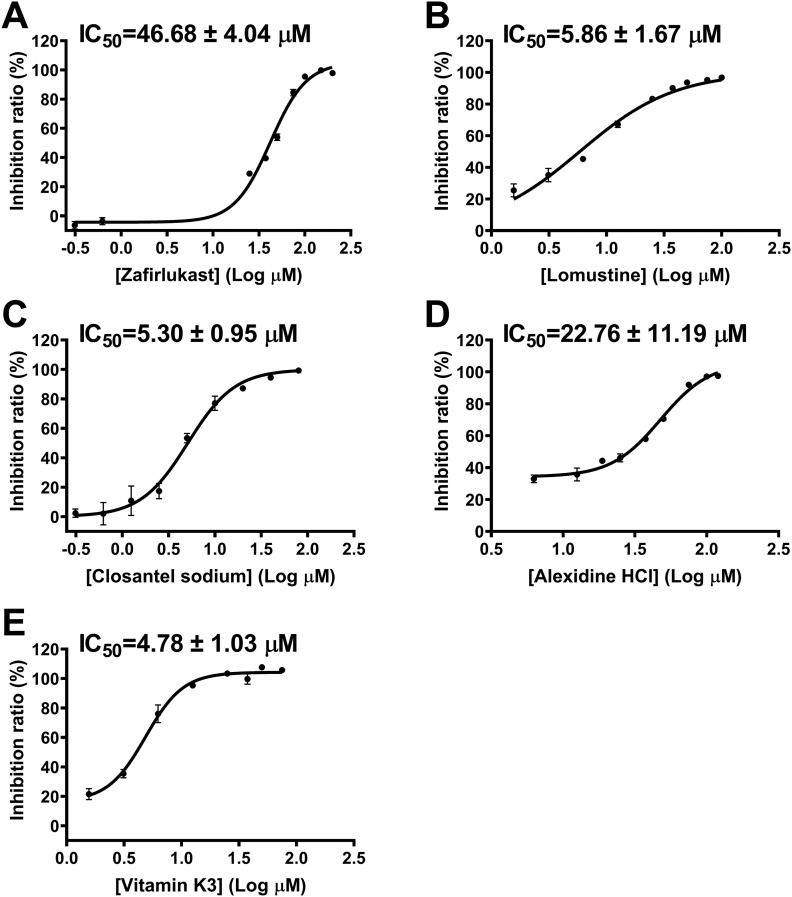
According to the above method of the expression and purification of SARS-CoV-2 3CLpro, the protein solution was concentrated to 1 mg/mL and diluted to 4 μg/mL in the reaction buffer. Pilot screening for SARS-CoV-2 3CLpro inhibitors was carried out by FRET-based assay using the L1300 Selleck FDA-Approved Drug Library containing an array of 1,018 compounds. Among these compounds, 6 hits (Nafamostat Mesylate, Zafirlukast, Alexidine HCl, Closantel sodium, Lomustine and Vitamin K3, Fig. 1A, B) were selected for secondary screening using four concentrations (0, 1, 10, 100 μM). As shown in Fig. 1C, these compounds showed dose-dependent inhibition of SARS-CoV-2 3CLpro. However, Nafamostat Mesylate presented the least inhibitory activity compared to other compounds because the residual activity of the protease was about 60% at a concentration of 100 μM. We thus next determined IC50 values of 5 compounds except for Nafamostat Mesylate. 3 compounds (Closantel sodium, Lomustine and Vitamin K3) displayed better inhibition of SARS-CoV-2 3CLpro with IC50 values of 5.30, 5.86, and 4.78 μM, while Zafirlukast and Alexidine HCl showed less inhibitory activities against the protease with IC50 values of 46.68, and 22.76 μM, respectively (Fig. 2 ). Among them, Vitamin K3, also known as menadione, was chosen for further study because its inhibitory activity is stronger than that of other compounds.
Fig. 1.
Screening of drug molecules for SARS-CoV-2 3CLpro protease activity inhibition. (A) 6 drugs were screened as the potent SARS-CoV-2 3CLpro inhibitors from the FDA-Approved Drug Library containing an array of 1,018 compounds. (B) The chemical structure of 6 drugs. (C) Effects of different concentrations of 6 drugs on the residual activity of SARS-CoV-2 3CLpro.
Fig. 2.
Inhibition of SARS-CoV-2 3CLpro by 5 drugs. (A) Representative inhibition curves for Zafirlukast, (B) Lomustine, (C) Closantel sodium (D)Alexidine HCl, and (E)Vitamin K3 against SARS-CoV-2 3CLpro.
3.2. Time-dependent inhibitory effects of Vitamin K3 on SARS-CoV-2 3CLpro
Due to Vitamin K3 covalently binding to QsrR dimer at the Cys-5 site [42], we tried to determine whether Vitamin K3 could also form a covalent bond with SARS-CoV-2 3CLpro. To confirm the covalent bond formation, the covalent binding between the compound and the targeted protein was confirmed by time-dependent inhibition and mass spectrometry [43]. The time dependence of the inhibition of the SARS-CoV-2 3CLpro proteolysis by Vitamin K3 was subsequently performed. The inhibition ratios of SARS-CoV-2 3CLpro were measured when different concentrations of Vitamin K3 incubated with the protease at 3 min and 33 min, respectively. To our great delight, Vitamin K3 exhibited a time-dependent inhibition in enzyme activity by a 4.4-fold decrease in the IC50 value from 20.96 to 4.78 μM in 30 min (Fig. 3A). The result suggested that Vitamin K3 could covalently bind to and inhibit SARS-CoV-2 3CLpro in a time-dependent manner. Since K I and k inact are useful to characterize the inhibitory potential to irreversible or mechanism-based inhibitors (MBIs) [44], the remaining activities of SARS-CoV-2 3CLpro in 40 min were measured with increasing concentrations of Vitamin K3 ranging from 5 to 80 μM. The result demonstrated that Vitamin K3 inhibited the activity of SARS-CoV-2 3CLpro in a time and dose-dependent manner (Fig. 3B) with K I and k inact values of 112.2 μM and 0.305 min−1, respectively (Fig. 3C). These results suggested that Vitamin K3 was an irreversible inhibitor of SARS-CoV-2 3CLpro.
Fig. 3.
Inhibition of SARS-CoV-2 3CLpro by Vitamin K3. (A) The inhibition ratio curves of SARS-CoV-2 3CLpro under the action of Vitamin K3 for 3 min and 33 min. (B) Time course of inhibition of SARS-CoV-2 3CLpro in the presence of Vitamin K3 at different concentrations. (C) Dependence of the values for Kobs on the concentrations of Vitamin K3.
3.3. The inhibitory activities of Vitamin K3 and its analogues against SARS-CoV-2 3CLpro
Since Vitamin K3 showed strong inhibitory activity, in order to further understand the structure-activity relationship between SARS-CoV-2 3CLpro and its analogues, we firstly bought Vitamin K3 analogues and examined the inhibitory activities of Vitamin K family, including Vitamin K1, K2, K3 and K4 (Fig. 4A). Experimental results showed that Vitamin K3 was the only one capable of inhibiting SARS-CoV-2 3CLpro, while other compounds had no inhibition at the high and middle concentration (Fig. 4B). Based on the result, a series of Vitamin K3 analogues were further evaluated for their inhibitory activities against SARS-CoV-2 3CLpro. As shown in Fig. 5 , the inhibitory activities of these Vitamin K3 analogues were determined at different concentrations (0, 1, 10, 100 μM). The result showed that 6 out of 8 compounds had inhibitory activities and their IC50 values were measured. As indicated in Table 1 , 6 analogues had better inhibitory activities than Vitamin K3 against SARS-CoV-2 3CLpro with IC50 values from 0.49 to 4.11 μM, and 5,8-dihydroxy-1,4-naphthoquinone showed the strongest inhibitory activity (IC50 = 0.49 ± 0.11 μM), which is 9.8 times more potent than Vitamin K3 against SARS-CoV-2 3CLpro.
Fig. 4.

Inhibitory activities of Vitamins K1, K2, K3 and K4 against SARS-CoV-2 3CLpro. (A) The chemical structures of Vitamins K1, K2, K3 and K4. (B) Effects of different concentrations of Vitamins K1, K2, K3 and K4 against SARS-CoV-2 3CLpro.
Fig. 5.
Inhibitory activities of different concentrations of Vitamin K3 analogues against SARS-CoV-2 3CLpro.
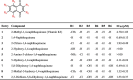
Table 1.
Structure of Vitamin K3 analogues and their IC50 values.
 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Entry | Compound | R1 | R2 | R3 | R4 | R5 | R6 | IC50 (μM) |
| 1 | 2-Methyl-1,4-naphthoquinone (vitamin K3) | -CH3 | -H | -H | -H | -H | -H | 4.78 ± 1.03 |
| 2 | 1,4-Naphthoquinone | -H | -H | -H | -H | -H | -H | 0.69 ± 0.13 |
| 3 | 2-Chloro-1,4-naphthoquinone | -Cl | -H | -H | -H | -H | -H | 1.92 ± 0.49 |
| 4 | 2-Hydroxy-1,4-naphthoquinone | -OH | -H | -H | -H | -H | -H | >100 |
| 5 | 2-Amino-3-chloro-1,4-naphthoquinone | -NH2 | -Cl | -H | -H | -H | -H | >100 |
| 6 | 5-Hydroxy-1,4-naphthoquinone | -H | -H | -OH | -H | -H | -H | 4.11 ± 0.24 |
| 7 | 5,8-Dihydroxy-1,4-naphthoquinone | -H | -H | -OH | -H | -H | -OH | 0.49 ± 0.11 |
| 8 | 2-Methyl-5-hydroxy-1,4-naphthoquinone | -CH3 | -H | -OH | -H | -H | -H | 1.32 ± 0.33 |
| 9 | 2,3-Dichloro-5,8-dihydroxy-1,4-naphthoquinone | -Cl | -Cl | -OH | -H | -H | -OH | 1.53 ± 0.37 |
It can be seen from above results that the inhibitory activities of Vitamin K1 and K2 were decreased by introducing a long carbon chain at the quinone-core of Vitamin K3. Meanwhile, change the naphthoquinone scaffold (Vitamin K4) also lost its inhibitory activity against SARS-CoV-2 3CLpro.
For 1,4-naphthoquinone (2), removal of the methyl at R1 position significantly increases the inhibitory activity with the IC50 value of 0.69 ± 0.13 μM, compared with Vitamin K3, namely, 2-methyl-1,4-naphthoquinone (1, IC50: 4.78 ± 1.03 μM). Similarly, for 2-chloro-1,4-naphthoquinone (3), the introduction of chlorine at the R1 position also augments the inhibitory activity. However, other Vitamin K3 analogues as possessing hydroxyl group and amino at R1 position, such as 2-hydroxy-1,4-naphthoquinone (4) and 2-amino-3-chloro-1,4-naphthoquinone (5), almost lose their inhibitory activities. These results implied that introduction groups into R1- or/and R2-position are detrimental to inhibitory activity.
Additionally, we turned our attention to other positions of Vitamin K3 to confirm whether change at other positions could provide more potent SARS-CoV-2 3CLpro inhibitors. Further introduction of hydroxyl groups at the R3 and R6 positions results in the increase of inhibitory activity for 5,8-dihydroxy-1,4-naphthoquinone (7). In the same way, the inhibitory activity of 2,3-dichloro-5,8-dihydroxy-1,4-naphthoquinone (9) is decreased when highly polar functional group Cl atom is introduced at R1 and R2 position in comparison to 5,8-dihydroxy-1,4-naphthoquinone (7). We found that the compound with one hydroxyl at R3, 5-hydroxy-1,4-naphthoquinone (6) lowers its inhibitory activity compared to 1,4-naphthoquinone (2). On this basis, the inhibition activity of 2-methyl-5-hydroxy-1,4-naphthoquinone (8) is increased when introducing the hydroxyl at R3 position compared with Vitamin K3.
As thiol group of cysteine of SARS-CoV-2 3CLpro attacks the C3 position of Vitamin K3 to form the covalent bond, any bulky substitution at C3 will block the reaction. Analogues, such as Vitamin K1 and K2, have long alkyl chain substituents at C3 position, which do not form the covalent bond due to the steric hindrance. Vitamin K4 can't form the covalent bond due to the lack of naphthoquinone unit as the reactive group. Other Vitamin K3 analogues with no substitution at C3 position inhibit the protease with varying IC50. Meanwhile, electrophilicity alteration caused by substituents may have impact on the binding activity and the covalent reaction between SARS-CoV-2 3CLpro and the compounds. Therefore, we speculate that removal of methyl at R1 position may improve the inhibitory activity of Vitamin K3, while Vitamin K3 loses the inhibitory activity upon the introduction of long carbon chain or amino and hydroxyl group at R1 position.
Due to 5,8-dihydroxy-1,4-naphthoquinone (Fig. 6A) having the strongest inhibitory activity against SARS-CoV-2 3CLpro, we selected the compound for further study. The time-dependent inhibition assay was performed to study the inhibitory mechanism of the compound. By comparing the IC50 values at 3 min and 33 min (Fig. 6B), the results showed that IC50 values of 5,8-dihydroxy-1,4-naphthoquinone decreased from 0.79 to 0.49 μM with the increase of time. From time course of inhibition of SARS-CoV-2 3CLpro in the presence of 5,8-dihydroxy-1,4-naphthoquinone at different concentrations, the results revealed that the protease activity reduced greatly (Fig. 6C), and K I and k inact values were 3.05 μM and 0. 05 min−1, respectively (Fig. 6D). These results indicate that Vitamin K3 analogue 5,8-dihydroxy-1,4-naphthoquinone had stronger inhibitory activity and can also covalently bind to SARS-CoV-2 3CLpro as an irreversible inhibitor.
Fig. 6.
Inhibition of SARS-CoV-2 3CLpro by 5,8-dihydroxy-1,4-naphthoquinone. (A) Chemical structure of 5,8-dihydroxy-1,4-naphthoquinone. (B) The inhibition ratio curves of SARS-CoV-2 3CLpro under the action of 5,8-dihydroxy-1,4-naphthoquinone for 3 min and 33 min. (C) Time course of inhibition of SARS-CoV-2 3CLpro in the presence of 5,8-dihydroxy-1,4-naphthoquinone at different concentrations. (D) Dependence of the values for Kobs on the concentrations of 5,8-dihydroxy-1,4-naphthoquinone.
Due to 3CL proteases of SARS-CoV-2 and SARS-CoV share significant homology, we also determined the inhibitory activities of Vitamin K3 and 5,8-dihydroxy-1,4-naphthoquinone against SARS-CoV 3CLpro. As shown in Fig. S4A and S4B, the two inhibitors exhibited dose-dependent inhibition of SARS-CoV 3CLpro activity, with IC50 values of 10.21 μM for Vitamin K3 and 1.75 μM for 5,8-dihydroxy-1,4-naphthoquinone, respectively. To show the selectivity of Vitamin K3 and 5,8-dihydroxy-1,4-naphthoquinone, in vitro selectivity assessment of the two compounds with human chymotrypsin-C (CTRC) was performed. The results demonstrate that Vitamin K3 and 5,8-dihydroxy-1,4-naphthoquinone can selectively inhibit SARS-CoV-2 and SARS-CoV 3CLpro, rather than human CTRC (Fig. S5).
3.4. Characterization of covalent binding of Vitamin K3 and 5,8-dihydroxy-1,4-naphthoquinone
To confirm if Vitamin K3 or 5,8-dihydroxy-1,4-naphthoquinone is covalently bound to native SARS-CoV-2 3CLpro, an intact protein molecular weight analysis was processed by mass spectrometry after incubation with the two compounds for 30 min at 37 °C, respectively. The protein sample without compound incubation was as control. The sequence of SARS-CoV-2 3CLpro is SGFRKMAFPSGKVEGCMVQVTCGTTTLNGLWLDDVVYCPRHVICTSEDMLNPNYEDLLIRKSNHNFLVQAGNVQLRVIGHSMQNCVLKLKVDTANPKTPKYKFVRIQPGQTFSVLACYNGSPSGVYQCAMRPNFTIKGSFLNGSCGSVGFNIDYDCVSFCYMHHMELPTGVHAGTDLEGNFYGPFVDRQTAQAAGTDTTITVNVLAWLYAAVINGDRWFLNRFTTTLNDFNLVAMKYNYEPLTQDHVDILGPLSAQTGIAVLDMCASLKELLQNGMNGRTILGSALLEDEFTPFDVVRQCSGVTFQGPHHHHHH. As shown in Fig. 7 , the molecular weight of SARS-CoV-2 3CLpro was detected as 34,773.6 Da ~ 34,774.2 Da, which was consistent with its theoretical value (34,773.66 Da). After incubation with Vitamin K3, about 70% of proteins were covalently bound, of which 55% of proteins were bound with one compound and about 15% of proteins were bound with two compounds. Similarly, after incubation with 5,8-dihydroxy-1,4-naphthoquinone, about 90% of proteins were covalently bound, of which 80% of proteins were bound with one compound and about 10% of proteins were bound with two compounds. The covalent binding extent is highly dependent on the incubation time, temperature and compound concentration [45,46]. The adduct peaks corresponding to the covalent binding of more than one compound indicate that the compound can react with protein at multiple residues. There are 12 cysteine residues in SARS-CoV-2 3CLpro protein, so it is possible that multiple cysteine residues are captured at high concentration of the compound. Indeed, a thorough analysis of the available crystal structures of SARS-CoV-2 3CLpro revealed that more than one cysteine residue can be captured by various compounds. For example, 4 cysteine residues (44, 145, 156, 300) are labelled by SU3327 (halicin) in the structure of PDB 7NBY, (Fig. S6). These results suggested that the two compounds form the covalent binding to SARS-CoV-2 3CLpro in two states at this reaction condition. 5,8-Dihydroxy-1,4-naphthoquinone had a stronger binding affinity, which was consistent with their IC50 values.
Fig. 7.
Mass spectra of native SARS-CoV-2 3CLpro (8.4 μM) after incubation with 90 μM Vitamin K3 or 5,8-dihydroxy-1,4-naphthoquinone. All measurements were processed on the UHPLC-MS system with positive ESI source.
3.5. Binding mode of Vitamin K3 or 5,8-dihydroxy-1,4-naphthoquinone with SARS-CoV-2 3CLpro
As Vitamin K3 forms an adduct with SARS-CoV-2 3CLpro, we further investigated its binding mode by molecular modeling. To use AutoDockFR for covalent docking, we firstly prepared the ligand, ie, the product form of Vitamin K3 after reaction with cysteine under aerobic condition (Fig. 8A). To choose a suitable protein model, all available SARS-CoV-2 3CLpro crystal structures from PDB were superimposed. As demonstrated in Fig. 8B, the backbone atoms of the catalytic site residues show low positional variance, while the sidechain atoms of N142, M165 and Q189 are flexible. Thus N142, M165 and Q189 were defined as flexible residues during the docking study. On the other hand, by 2D similarity comparison between Vitamin K3 and all co-crystallized 3CLpro binders, ligand in 6YNQ, 2-Methyl-1-tetralone, is most similar to Vitamin K3 (FragFP similarity = 0.55) (Fig. 8C). Same as Vitamin K3, 2-Methyl-1-tetralone forms a covalent bond with the Cys145 [40], thus the protein structure of 6YNQ is chosen for docking study.
Fig. 8.
(A) The product form of Vitamin K3 after reaction with cysteine under aerobic condition. (B) Flexible residues in the catalytic active site of SARS-CoV-2 3CLpro. (C) Ligands in available SARS-CoV-2 3CLpro co-crystal structures are most similar to Vitamin K3. (D) Potential reaction between Vitamin K3 and Cys145 of SARS-CoV-2 3CLpro. (E) The binding mode between Vitamin K3 and SARS-CoV-2 3CLpro was predicted by covalent docking using AutoDockFR. Docking pose of Vitamin K3 and side chain of N142 and M165 are shown as stick and colored in megantas, SARS-CoV-2 3CLpro surface is colored in gray, H-bond between Vitamin K3 and protein is rendered as yellow dash line. (F) Schematic plot to highlight the positions that are not in close contact with protein, and the surrounding gray line indicated the protein surface. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
When quinone of Vitamin K3 reacts with cysteine as an electrophile, there could be multiple routes that lead to different products [42,47,48]. As the mass shift of Vitamin K3 and SARS-CoV-2 3CLpro adduct is 170 Da, and the incubation is under aerobic condition, the reaction [49] between the quinone like Vitamin K3 and the thiol moiety should reduce Vitamin K3 to the dihydroquinone form, then the dihydroquinone is rapidly oxidized back to the quinone form (Fig. 8D). Similarly, 5,8-dihydroxy-1,4-naphthoquinone has the same binding mode due to the oxidized reaction between the quinone and the thiol moiety of SARS-CoV-2 3CLpro, which forms the irreversible covalent adduct at the cysteine residue of the enzyme [50].
Based on the proposed product structure, the binding mode between Vitamin K3 and SARS-CoV-2 3CLpro was predicted by covalent docking using AutoDockFR. As shown in Fig. 8E, the two ketone oxygen atoms form two H bonds with sidechain of N142 and H163, the naphthalene ring patches the small pocket between the L141, N142, M165 and E166, and a covalent bond between Vitamin K3 and C145 is similar in orientation as that of 2-Methyl-1-tetralone in 6YN5. Based on the mass spectrometry and docking result, Vitamin K3 or 5,8-dihydroxy-1,4-naphthoquinone might covalently bind to Cys145 and lead to enzyme activity inhibition. Meanwhile, some other cysteine residues might be modified at the high concentration of the compound. According to the predicted binding mode of Vitamin K3 (Fig. 8F), modifications at positions 1, 8 and 9 will not clash with SARS-CoV-2 3CLpro and might be promising directions to improve the binding affinity of Vitamin K3.
4. Conclusion
Because of the indispensability of SARS-CoV-2 3CLpro to complete the life cycle of the coronavirus, it is one of the most promising drug targets for anti-viral agents. In the present study, 6 drugs were screened as the inhibitors of SARS-CoV-2 3CLpro (Fig. 1). Of these 6 drugs, Vitamin K3 was chosen for further study because it exhibited the strongest inhibitory effect on SARS-CoV-2 3CLpro. Based on the previous results, Vitamin K3 contains the structure of quinone, which is electrophilic and leads to covalent modification of the thiol of cysteine via Michael-type addition [48,51,52]. We further determined whether Vitamin K3 can form the covalent bond with SARS-CoV-2 3CLpro. The time-dependent inhibition assay was conducted to explore the inhibitory mechanism of Vitamin K3. The results showed that Vitamin K3 exhibited a time-dependent inhibition of enzyme activity by a 4.4-fold decrease in the IC50 value (from 20.96 to 4.78 μM) at 3 min and 33 min, with K I and k inact values of 112.2 μM and 0.305 min−1, respectively. Since Vitamin K as dietary supplements shares a similar structure with a 2-methyl-1,4 naphthoquinone ring and a variable aliphatic chain [53], in order to analyze the structure-activity relationship between SARS-CoV-2 3CLpro and its analogues, we examined the inhibitory activities Vitamin K family, including Vitamin K1, K2, K3 and K4 (Fig. 4.) and other analogues of Vitamin K3 (Fig. 5, Table 1). Finally, 5,8-dihydroxy-1,4-naphthoquinone was identified with 9.8 times higher inhibitory activity than Vitamin K3. To investigate the binding mode of Vitamin K3 and 5,8-dihydroxy-1,4-naphthoquinone, the mass spectrometric study was performed and revealed the covalent binding between the two compounds and SARS-CoV-2 3CLpro. Due to the high degree of structural homology of 3CLpro of SARS-CoV-2 and SARS-CoV, we determined the inhibitory activities of the two compounds against SARS-CoV 3CLpro. The result showed that they also inhibited SARS-CoV 3CLpro and might be used as broad-spectrum antiviral agents targeting 3CLpro of both SARS-CoV-2 and SARS-CoV.
To better understand the detailed binding site of Vitamin K3 and 5,8-dihydroxy-1,4-naphthoquinone with SARS-CoV-2 3CLpro, we performed molecular modeling to analyze the covalent binding site of the two compounds to SARS-CoV-2 3CLpro. The result demonstrated that two H bonds are formed between the two ketone oxygen atoms of Vitamin K3 and the sidechain of N142 and H163 of SARS-CoV-2 3CLpro and a covalent binding between Vitamin K3 and C145. The knowledge gained in this study provides valuable information for further optimization and design of novel inhibitors based on Vitamin K3 and its analogues. Although Vitamin K3 often had different effects on cell apoptosis in PC-12 cells depending on its concentrations, it is safe in clinical application [54]. Therefore, further modification of Vitamin K3 may have the potential to fight against SARS-CoV-2.
CRediT authorship contribution statement
Lili Chen, Hongzhuan Chen and Guangbo Ge conceived and designed the experiments. Qing Hu, Ruyu Wang, Haonan Wang, Guanghao Zhu, Mengge Wang, Yishu Zhao, Chunyu Li, and Yani Zhang carried out the experiments and data analysis. Ruyu Wang wrote the manuscript. Lili Chen, Qian Zhang, Hongzhuan Chen and Guangbo Ge critically revised the manuscript.
Acknowledgments
Acknowledgments
We thank Dr. Zhenting Gao for performing molecular modeling study and providing suggestions on experiment design. We also thank Dr. Yanyan Yu for her valuable comments and suggestions on mass spectrometry experiment. Supported by Scientific Research Project of Shanghai Municipal Health Commission on Traditional Chinese Medicine for Prevention and Treatment of COVID-19 (2020XGKY07), Emergency Scientific Research Program of Shanghai University of Traditional Chinese Medicine (2019YJ 06-01) and the National Key Research and Development Program of China (2020YFC0845400).
Declaration of competing interest
The authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2021.04.129.
Appendix A. Supplementary data
Supplementary materials 1: Methods and Results.
Supplementary material 2: Structures of 259 compounds
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/s0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hussein M., Toraih E., Elshazli R., Fawzy M., Houghton A., Tatum D., Killackey M., Kandil E., Duchesne J. Meta-analysis on serial intervals and reproductive rates for SARS-CoV-2. Ann. Surg. 2021;273(3):416–423. doi: 10.1097/sla.0000000000004400. [DOI] [PubMed] [Google Scholar]
- 4.Tsai S.C., Lu C.C., Bau D.T., Chiu Y.J., Yen Y.T., Hsu Y.M., Fu C.W., Kuo S.C., Lo Y.S., Chiu H.Y., Juan Y.N., Tsai F.J., Yang J.S. Approaches towards fighting the COVID-19 pandemic (review) Int. J. Mol. Med. 2021;47(1):3–22. doi: 10.3892/ijmm.2020.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iba T., Levy J.H., Connors J.M., Warkentin T.E., Thachil J., Levi M. The unique characteristics of COVID-19 coagulopathy. Crit. Care. 2020;24(1) doi: 10.1186/s13054-020-03077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.Q., Wang Q., Miao H. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct. Target Ther. 2020;5(1) doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas-Rüddel D., Winning J., Dickmann P., Ouart D., Kortgen A., Janssens U., Bauer M. Coronavirus disease 2019 (COVID-19): update for anesthesiologists and intensivists March 2020. Anaesthesist. 2020;69(4):225–235. doi: 10.1007/s00101-020-00758-x. [DOI] [PubMed] [Google Scholar]
- 8.Forni G., Mantovani A. COVID-19 vaccines: where we stand and challenges ahead. Cell Death Differ. 2021;28(2):626–639. doi: 10.1038/s41418-020-00720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodgson S.H., Mansatta K., Mallett G., Harris V., Emary K.R.W., Pollard A.J. What defines an efficacious COVID-19 vaccine? A review of the challenges assessing the clinical efficacy of vaccines against SARS-CoV-2. Lancet Infect. Dis. 2021;21(2):e26–e35. doi: 10.1016/s1473-3099(20)30773-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bar-Zeev N., Inglesby T. COVID-19 vaccines: early success and remaining challenges. Lancet. 2020;396(10255):868–869. doi: 10.1016/s0140-6736(20)31867-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hossain M.S., Hami I., Sawrav M.S.S., Rabbi M.F., Saha O., Bahadur N.M., Rahaman M.M. Drug repurposing for prevention and treatment of COVID-19: a clinical landscape. Discoveries (Craiova) 2020;8(4) doi: 10.15190/d.2020.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu A., Wang Y., Zeng C., Huang X., Xu S., Su C., Wang M., Chen Y., Guo D. Prediction and biochemical analysis of putative cleavage sites of the 3C-like protease of Middle East respiratory syndrome coronavirus. Virus Res. 2015;208:56–65. doi: 10.1016/j.virusres.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar R., Verma H., Singhvi N., Sood U., Gupta V., Singh M., Kumari R., Hira P., Nagar S., Talwar C., Nayyar N., Anand S., Rawat C.D., Verma M., Negi R.K., Singh Y., Lal R. Comparative genomic analysis of rapidly evolving SARS-CoV-2 reveals mosaic pattern of phylogeographical distribution. mSystems. 2020;5(4) doi: 10.1128/mSystems.00505-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su H.X., Yao S., Zhao W.F., Li M.J., Liu J., Shang W.J., Xie H., Ke C.Q., Hu H.C., Gao M.N., Yu K.Q., Liu H., Shen J.S., Tang W., Zhang L.K., Xiao G.F., Ni L., Wang D.W., Zuo J.P., Jiang H.L., Bai F., Wu Y., Ye Y., Xu Y.C. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020;41(9):1167–1177. doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta Mol. basis Dis. 2020;1866(10) doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 20.Jukič M., Janežič D., Bren U. Ensemble docking coupled to linear interaction energy calculations for identification of coronavirus main protease (3CL(pro)) non-covalent small-molecule inhibitors. Molecules. 2020;25(24) doi: 10.3390/molecules25245808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cannalire R., Cerchia C., Beccari A.R., Di Leva F.S., Summa V. Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities. J. Med. Chem. 2020 doi: 10.1021/acs.jmedchem.0c01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki R., Suzuki Y., Yonezawa Y., Ota Y., Okamoto Y., Demizu Y., Huang P., Yoshida H., Sugimura K., Mizushina Y. DNA polymerase gamma inhibition by vitamin K3 induces mitochondria-mediated cytotoxicity in human cancer cells. Cancer Sci. 2008;99(5):1040–1048. doi: 10.1111/j.1349-7006.2008.00771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tintino S.R., Souza V.C.A., Silva J., Oliveira-Tintino C.D.M., Pereira P.S., Leal-Balbino T.C., Pereira-Neves A., Siqueira-Junior J.P., da Costa J.G.M., Rodrigues F.F.G., Menezes I.R.A., da Hora G.C.A., Lima M.C.P., Coutinho H.D.M., Balbino V.Q. Effect of vitamin K(3) inhibiting the function of NorA efflux pump and its gene expression on Staphylococcus aureus. Membranes (Basel) 2020;10(6) doi: 10.3390/membranes10060130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrade J.C., Morais Braga M.F., Guedes G.M., Tintino S.R., Freitas M.A., Quintans L.J., Jr., Menezes I.R., Coutinho H.D. Menadione (vitamin K) enhances the antibiotic activity of drugs by cell membrane permeabilization mechanism. Saudi J. Biol. Sci. 2017;24(1):59–64. doi: 10.1016/j.sjbs.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu F., Vostal J.G. Inactivation of bacteria via photosensitization of vitamin K3 by UV-A light. FEMS Microbiol. Lett. 2014;358(1):98–105. doi: 10.1111/1574-6968.12544. [DOI] [PubMed] [Google Scholar]
- 26.de Carvalho Scharf Santana N., Lima N.A., Desoti V.C., Bidóia D.L., de Souza Bonfim Mendonça P., Ratti B.A., Nakamura T.U., Nakamura C.V., Consolaro M.E., Ximenes V.F., de Oliveira Silva S. Vitamin K3 induces antiproliferative effect in cervical epithelial cells transformed by HPV 16 (SiHa cells) through the increase in reactive oxygen species production. Arch. Gynecol. Obstet. 2016;294(4):797–804. doi: 10.1007/s00404-016-4097-7. [DOI] [PubMed] [Google Scholar]
- 27.Kishore C., Sundaram S., Karunagaran D. Vitamin K3 (menadione) suppresses epithelial-mesenchymal-transition and Wnt signaling pathway in human colorectal cancer cells. Chem. Biol. Interact. 2019;309 doi: 10.1016/j.cbi.2019.108725. [DOI] [PubMed] [Google Scholar]
- 28.Nutter L.M., Cheng A.L., Hung H.L., Hsieh R.K., Ngo E.O., Liu T.W. Menadione: spectrum of anticancer activity and effects on nucleotide metabolism in human neoplastic cell lines. Biochem. Pharmacol. 1991;41(9):1283–1292. doi: 10.1016/0006-2952(91)90099-q. [DOI] [PubMed] [Google Scholar]
- 29.He Z., Zhao W., Niu W., Gao X., Gao X., Gong Y., Gao X. Molecules inhibit the enzyme activity of 3-chymotrypsin-like cysteine protease of SARS-CoV-2 virus: the experimental and theory studies. bioRxiv. 2020 doi: 10.1101/2020.05.28.120642. [DOI] [Google Scholar]
- 30.Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., Chen C., Jin Y., Bartlam M., Rao Z. Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366(3):965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Chen L., Li J., Luo C., Liu H., Xu W., Chen G., Liew O.W., Zhu W., Puah C.M., Shen X., Jiang H. Binding interaction of quercetin-3-beta-galactoside and its synthetic derivatives with SARS-CoV 3CL(pro): structure-activity relationship studies reveal salient pharmacophore features. Bioorg. Med. Chem. 2006;14(24):8295–8306. doi: 10.1016/j.bmc.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L., Chen S., Gui C., Shen J., Shen X., Jiang H. Discovering severe acute respiratory syndrome coronavirus 3CL protease inhibitors: virtual screening, surface plasmon resonance, and fluorescence resonance energy transfer assays. J. Biomol. Screen. 2006;11(8):915–921. doi: 10.1177/1087057106293295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrelson J.P., Stamper B.D., Chapman J.D., Goodlett D.R., Nelson S.D. Covalent modification and time-dependent inhibition of human CYP2E1 by the meta-isomer of acetaminophen. Drug Metab. Dispos. 2012;40(8):1460–1465. doi: 10.1124/dmd.112.045492. [DOI] [PubMed] [Google Scholar]
- 35.Ghanbari F., Rowland-Yeo K., Bloomer J.C., Clarke S.E., Lennard M.S., Tucker G.T., Rostami-Hodjegan A. A critical evaluation of the experimental design of studies of mechanism based enzyme inhibition, with implications for in vitro-in vivo extrapolation. Curr. Drug Metab. 2006;7(3):315–334. doi: 10.2174/138920006776359293. [DOI] [PubMed] [Google Scholar]
- 36.Sander T., Freyss J., von Korff M., Rufener C. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015;55(2):460–473. doi: 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- 37.Ravindranath P.A., Forli S., Goodsell D.S., Olson A.J., Sanner M.F. AutoDockFR: advances in protein-ligand docking with explicitly specified binding site flexibility. PLoS Comput. Biol. 2015;11(12) doi: 10.1371/journal.pcbi.1004586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riniker S., Landrum G.A. Better informed distance geometry: using what we know To improve conformation generation. J. Chem. Inf. Model. 2015;55(12):2562–2574. doi: 10.1021/acs.jcim.5b00654. [DOI] [PubMed] [Google Scholar]
- 39.O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open babel: an open chemical toolbox. J. Cheminform. 2011;3 doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Günther S., Reinke P.Y.A., Fernández-García Y., Lieske J., Lane T.J., Ginn H.M., Koua F.H.M., Ehrt C., Ewert W., Oberthuer D., Yefanov O., Meier S., Lorenzen K., Krichel B., Kopicki J.-D., Gelisio L., Brehm W., Dunkel I., Seychell B., Gieseler H., Norton-Baker B., Escudero-Pérez B., Domaracky M., Saouane S., Tolstikova A., White T.A., Hänle A., Groessler M., Fleckenstein H., Trost F., Galchenkova M., Gevorkov Y., Li C., Awel S., Peck A., Barthelmess M., Schlünzen F., Xavier P.L., Werner N., Andaleeb H., Ullah N., Falke S., Srinivasan V., Franca B.A., Schwinzer M., Brognaro H., Rogers C., Melo D., Zaitsev-Doyle J.J., Knoska J., Peña Murillo G.E., Mashhour A.R., Guicking F., Hennicke V., Fischer P., Hakanpää J., Meyer J., Gribbon P., Ellinger B., Kuzikov M., Wolf M., Beccari A.R., Bourenkov G., Stetten D.v., Pompidor G., Bento I., Panneerselvam S., Karpics I., Schneider T.R., Garcia Alai M.M., Niebling S., Günther C., Schmidt C., Schubert R., Han H., Boger J., Monteiro D.C.F., Zhang L., Sun X., Pletzer-Zelgert J., Wollenhaupt J., Feiler C.G., Weiss M.S., Schulz E.-C., Mehrabi P., Karničar K., Usenik A., Loboda J., Tidow H., Chari A., Hilgenfeld R., Uetrecht C., Cox R., Zaliani A., Beck T., Rarey M., Günther S., Turk D., Hinrichs W., Chapman H.N., Pearson A.R., Betzel C., Meents A. Inhibition of SARS-CoV-2 main protease by allosteric drug-binding. bioRxiv. 2020 doi: 10.1101/2020.11.12.378422. 2020.11.12.378422. [DOI] [Google Scholar]
- 41.Seeliger D., de Groot B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010;24(5):417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji Q., Zhang L., Jones M.B., Sun F., Deng X., Liang H., Cho H., Brugarolas P., Gao Y.N., Peterson S.N., Lan L., Bae T., He C. Molecular mechanism of quinone signaling mediated through S-quinonization of a YodB family repressor QsrR. Proc. Natl. Acad. Sci. U. S. A. 2013;110(13):5010–5015. doi: 10.1073/pnas.1219446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vazquez-Rodriguez S., Wright M., Rogers C.M., Cribbs A.P., Velupillai S., Philpott M., Lee H., Dunford J.E., Huber K.V.M., Robers M.B., Vasta J.D., Thezenas M.L., Bonham S., Kessler B., Bennett J., Fedorov O., Raynaud F., Donovan A., Blagg J., Bavetsias V., Oppermann U., Bountra C., Kawamura A., Brennan P.E. Design, synthesis and characterization of covalent KDM5 inhibitors. Angew. Chem. Int. Ed. Eng. 2019;58(2):515–519. doi: 10.1002/anie.201810179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krippendorff B.F., Neuhaus R., Lienau P., Reichel A., Huisinga W. Mechanism-based inhibition: deriving K(I) and k(inact) directly from time-dependent IC(50) values. J. Biomol. Screen. 2009;14(8):913–923. doi: 10.1177/1087057109336751. [DOI] [PubMed] [Google Scholar]
- 45.Goldman J.L., Koen Y.M., Rogers S.A., Li K., Leeder J.S., Hanzlik R.P. Bioactivation of trimethoprim to protein-reactive metabolites in human liver microsomes. Drug Metab. Dispos. 2016;44(10):1603–1607. doi: 10.1124/dmd.116.072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandrasekaran A., Shen L., Lockhead S., Oganesian A., Wang J., Scatina J. Reversible covalent binding of neratinib to human serum albumin in vitro. Drug Metab. Lett. 2010;4(4):220–227. doi: 10.2174/187231210792928206. [DOI] [PubMed] [Google Scholar]
- 47.Wissner A., Floyd M.B., Johnson B.D., Fraser H., Ingalls C., Nittoli T., Dushin R.G., Discafani C., Nilakantan R., Marini J., Ravi M., Cheung K., Tan X., Musto S., Annable T., Siegel M.M., Loganzo F. 2-(Quinazolin-4-ylamino)-[1,4]benzoquinones as covalent-binding, irreversible inhibitors of the kinase domain of vascular endothelial growth factor receptor-2. J. Med. Chem. 2005;48(24):7560–7581. doi: 10.1021/jm050559f. [DOI] [PubMed] [Google Scholar]
- 48.Klein P., Barthels F., Johe P., Wagner A., Tenzer S., Distler U., Le T.A., Schmid P., Engel V., Engels B., Hellmich U.A., Opatz T., Schirmeister T. Naphthoquinones as covalent reversible inhibitors of cysteine proteases-studies on inhibition mechanism and kinetics. Molecules. 2020;25(9) doi: 10.3390/molecules25092064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott G.K., Atsriku C., Kaminker P., Held J., Gibson B., Baldwin M.A., Benz C.C. Vitamin K3 (menadione)-induced oncosis associated with keratin 8 phosphorylation and histone H3 arylation. Mol. Pharmacol. 2005;68(3):606–615. doi: 10.1124/mol.105.013474. [DOI] [PubMed] [Google Scholar]
- 50.Duval R., Bui L.C., Mathieu C., Nian Q., Berthelet J., Xu X., Haddad I., Vinh J., Dupret J.M., Busi F., Guidez F., Chomienne C., Rodrigues-Lima F. Benzoquinone, a leukemogenic metabolite of benzene, catalytically inhibits the protein tyrosine phosphatase PTPN2 and alters STAT1 signaling. J. Biol. Chem. 2019;294(33):12483–12494. doi: 10.1074/jbc.RA119.008666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valente C., Moreira R., Guedes R.C., Iley J., Jaffar M., Douglas K.T. The 1,4-naphthoquinone scaffold in the design of cysteine protease inhibitors. Bioorg. Med. Chem. 2007;15(15):5340–5350. doi: 10.1016/j.bmc.2007.04.068. [DOI] [PubMed] [Google Scholar]
- 52.Fisher A.A., Labenski M.T., Malladi S., Gokhale V., Bowen M.E., Milleron R.S., Bratton S.B., Monks T.J., Lau S.S. Quinone electrophiles selectively adduct “electrophile binding motifs” within cytochrome c. Biochemistry. 2007;46(39):11090–11100. doi: 10.1021/bi700613w. [DOI] [PubMed] [Google Scholar]
- 53.Ivanova D., Zhelev Z., Getsov P., Nikolova B., Aoki I., Higashi T., Bakalova R. Vitamin K: redox-modulation, prevention of mitochondrial dysfunction and anticancer effect. Redox Biol. 2018;16:352–358. doi: 10.1016/j.redox.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J., Ke J.-Y., Lian Y.-L., Zhang Y., Huang J.-F., Wang L. Vitamin K3 regulates reactive oxygen species and extracellular-regulated protein kinase in differentiated PC-12 cells within a safe dose range. Reprod. Dev. Med. 2018;2(2):74. doi: 10.4103/2096-2924.242757. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials 1: Methods and Results.
Supplementary material 2: Structures of 259 compounds