Abstract
The two genetically similar severe acute respiratory syndrome coronaviruses, SARS-CoV-1 and SARS-CoV-2, have each been responsible for global epidemics of vastly different scales. Although both viruses arose from similar origins, they quickly diverged due to differences in their transmission dynamics and spectrum of clinical presentations. The potential involvement of multiple organs systems, including the respiratory, cardiac, gastrointestinal and neurological, during infection necessitates a comprehensive understanding of the clinical pathogenesis of each virus. The management of COVID-19, initially modelled after SARS and other respiratory illnesses, has continued to evolve as we accumulate more knowledge and experience during the pandemic, as well as develop new therapeutics and vaccines. The impact of these two coronaviruses has been profound for our health care and public health systems, and we hope that the lessons learned will not only bring the current pandemic under control, but also prevent and reduce the impact of future pandemics.
Keywords: COVID-19, SARS, Coronavirus: Infectious disease, Public health
Introduction
The coronavirus is no new guest to humans, and neither are humans a new host. The first strain of coronavirus was identified in 1965 as a significant cause of the common cold [1]. Since then, a total of seven human-infecting strains have been discovered of which four generally cause mild symptoms, and three cause potentially severe disease. The latter includes the 2002 Severe Acute Respiratory Syndrome (SARS), 2012 Middle East Respiratory Syndrome (MERS), and the 2019 Coronavirus Disease (COVID-19) [2]. The global SARS epidemic, caused by SARS-CoV-1, redirected significant attention to the Coronaviridae family [3]. Years after the disruptive SARS epidemic, there were many warnings about the possibility of the re-emergence of SARS and other novel viruses. One report in 2007 described the large reservoir of SARS-CoV-like viruses in bats, coupled with the trade of wild animals in China, to be a “time bomb” [4]. Likewise, after the MERS outbreak, a multidisciplinary group of scientists re-iterated the potential risk of SARS-CoV's from bat populations [5]. Years later, SARS-CoV-2, responsible for the current COVID-19 pandemic, emerged to affirm the prior warnings. The disease followed a similar pattern to SARS, starting in China around a wet market selling wild animals and spreading to over 200 countries [6], [7]. Both diseases have similar clinical manifestations, predominantly causing a respiratory illness [8]. Ongoing research on SARS-CoV-2 has revealed unique features about its genome, pathogenesis, clinical course, transmissibility, and control measures. By comparing and contrasting SARS-CoV-2 to SARS-CoV-1, we aim to understand the current pandemic and its future trajectory. Herein, we present a comprehensive comparative review of both viruses to provide a resource for clinicians, scientists, and public health experts managing the COVID-19 pandemic and guide future efforts to deal with subsequent coronavirus outbreaks. We used multiple search terms including “SARS”, “SARS-CoV”, 2019 novel coronavirus” and “COVID-19” to identify published articles and preprints on PubMed and Google Scholar, as well as additional search terms specific to each subtopic, and information from public health agency websites including the World Health Organization and the Centers for Disease Control and Prevention as of March 24, 2021.
Background and epidemiology
The two viruses responsible for SARS and COVID-19 outbreaks mirror one another in many different ways; not only are they genetically related, but the stories behind their emergence are strikingly similar. Both SARS and COVID-19 began as outbreaks of atypical pneumonia located around wet markets [9]. Both outbreaks were reported to the World Health Organization (WHO) when excess cases of pneumonia were detected. Although SARS was reported almost four months after the outbreak had begun, COVID-19 was reported after only one month [10], [11]. The etiological agents of SARS and COVID-19 are believed to have originated from bats and transmitted to humans via intermediate hosts [12]. The intermediate host for SARS-CoV-1 is thought to be the Asian palm civet, but the identity of the COVID-19 intermediary is still not confirmed. Some studies suggest pangolins may have played a role in the evolution of SARS-CoV-2 but this remains an active area of investigation [12], [13], [14].
The SARS outbreak started as atypical pneumonia associated with wet markets in Guangdong, China, in mid-November 2002 [10]. By February of the following year, China reported to the WHO that the disease had affected 305 individuals, 5 of whom had died [15]. China became ground zero for the international outbreak after a 65-year-old physician from Guangzhou, who had been treating patients with what was thought to be atypical pneumonia in China, arrived at a hotel in Hong Kong on 21 February 2003 [16]. Similarly, the outbreak of COVID-19 was believed to have originated around a wet market in Wuhan, China, in November 2019 [17]. Consequently, the market was closed on 1st of January 2020. On 7th of January, the etiological agent of the outbreak was identified as a novel betacoronavirus, and by 21 January, a total of 270 cases were reported in Wuhan [18].
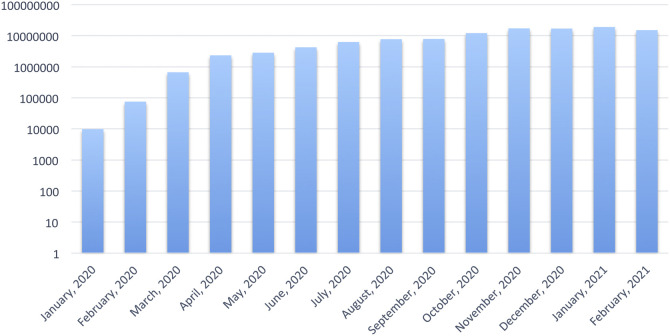
The global spread of both diseases was primarily mediated by commercial travel. Much of the worldwide spread of SARS could be traced back to the pivotal transmission event in the Hong Kong hotel where many international visitors were also staying, bringing the infection back to their respective countries, including Vietnam, Canada, Singapore, USA, Philippines, and Australia [16]. The SARS epidemic lasted approximately nine months, spreading to 29 countries, infecting 8096 people and resulting in 774 deaths [19]. In contrast to 2003, China's connectivity to the rest of the world in 2020 was far more significant, and thus COVID-19 was able to spread much faster. This was compounded by the Chinese New Year celebrations’ coincidental timing that involved millions of people traveling within and outside China [7], [20]. In a period of 1 year, the world witnessed an exponential rise in cases, with approximately 120 million people infected and over 2 million fatalities [21]. The initial increase in the period from December 2019 to mid-March 2020 reflected the rise and wane of the outbreak within China, followed by outbreaks in the rest of the world, most notably in the Americas, Europe, and Asia. (Fig. 1 ). More recently, the emergence of new genetic variants of SARS-CoV-2 have contributed to further spread of the virus [22].
Fig. 1.
Global incidence of new COVID-19 cases [22].
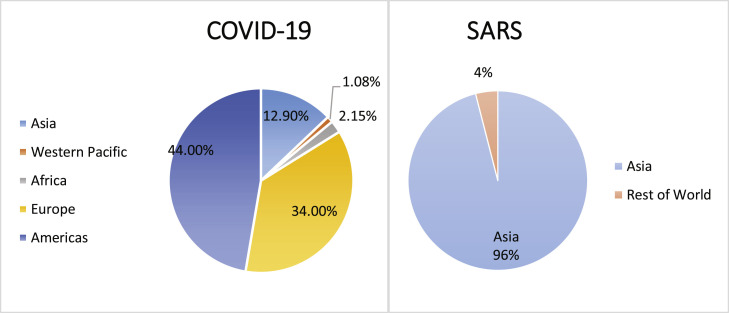
From a geographical perspective, the SARS outbreak was concentrated mainly in Asia, accounting for 96% of all cases globally (Fig. 2 ), with 7775 cases and 729 deaths in Asia compared to 321 cases and 45 deaths in the rest of the world [19]. In contrast, the widespread and sustained transmission of COVID-19 is occurring on all continents. After the initial outbreak in China, the disease's incidence was concentrated in Europe and the Americas. The Americas currently account for almost half of the total COVID-19 cases worldwide (44%) (Fig. 2) [21].
Fig. 2.
Geographic distribution of COVID-19 and SARS cases (March 14, 2021).
Virology
Structure and genome
Coronaviruses are taxonomically classified under the subfamily Orthocoronavirinae, in the family Coronaviridae, and the order Nidovirales [23]. They are enveloped viruses, composed of a single-stranded, positive-sense RNA genome [3], [24]. In contrast to other RNA viruses, coronaviruses are uniquely characterized by their long, non-segmented genome [25]. The large genome, varying from 26 to 32 kb, allows coronaviruses to acquire more genetic diversity and thus greater capacity for cross-species transmission [26]. Currently, there are four known genera (alpha, beta, gamma, and delta) with all human coronaviruses falling under the genus betacoronavirus [23]. The only coronaviruses documented to have caused human epidemics have been SARS-CoV-1, MERS-CoV, and SARS-CoV-2, but there is evidence suggesting that HCoV-OC43 may have caused a pandemic at the end of the 19th century [27], [28]. All human coronaviruses are speculated to have arisen through recombination and other mutational events in their original hosts, bats and rodents, as well as through intermediary animals [29].
SARS-CoV-2 was given its name because its genetic sequence was more than 80% identical to SARS-CoV-1 [30]. The RNA genome has several open reading frames that encode for several structural and non-structural proteins [26]. The viral transmission capacity and cellular tropism is mainly determined by the spike protein which also gives these viruses their distinctive crown-like appearance under electron microscopy [31]. Both viruses use the spike protein to gain entry to cells via angiotensin-converting enzyme 2 (ACE2) [32]. The sequence of the receptor binding domain (RBD) of the SARS-CoV-2 spike protein is similar to that of SARS-CoV-1 [33]. However, the RBD of the spike protein of SARS-CoV-2 exhibits greater affinity for binding to ACE2 than the SARS-CoV-1 RBD, which may explain the higher intra-host replication and inter-host transmission efficacy of SARS-CoV-2 [32].
Transmission
The primary mode of transmission for both SARS-CoV-1 and SARS-CoV-2 is via direct or indirect contact with infectious droplets, released when coughing, sneezing, or even speaking [34], [35]. Both viruses can also remain viable in aerosols and on inanimate surfaces, which allows for indirect transmission [36]. The stability of each virus is similar to one another in various environmental conditions, so differences in transmissibility between SARS-CoV-1 and SARS-CoV-2 are mostly due to differences in receptor affinity and replication efficiency. Additionally, the superspreading effect, when an individual transmits the virus to a disproportionate number of contacts, has been an important driver in both outbreaks [37], [38]. WHO estimates of the basic reproductive number (R0) for SARS-CoV-2 range from 1.4 to 6.9 with a mean of 3.28, while the R0 estimates for SARS-CoV-1 range from 2 to 4 [34], [39]. However, the R0 estimates of both viruses are not reflective of the difficulty in controlling the spread of each virus. Patients with SARS were maximally infectious during the second week of illness, whereas COVID-19 patients are most infectious in the pre-symptomatic and early symptomatic phase of illness. The control of SARS-CoV-2 is further complicated by a population of infectious individuals who are asymptomatic at the time of transmission, both from pre-symptomatic individuals and individuals who remain asymptomatic throughout the course of infection. As a result, significantly greater effort has been required to reduce the effective reproductive number (Re) of SARS-CoV-2 compared to SARS-CoV-1 [34], [40], [41]. Other routes of transmission, including fecal, transplacental, and sexual have also been documented but their overall importance has been limited to specific scenarios for both COVID-19 and SARS [42], [43], [44], [45], [46]. Additionally, nosocomial transmission of both viruses played an important role in amplifying each outbreak, especially in the early stages, when insufficient protection was used by health care workers (HCWs) attending to infected patients, compounded by transmission amongst HCWs themselves [47], [48].
Clinical course
Clinical manifestation
The incubation period of SARS-CoV-2 is between 2 and 14 days, with anecdotal reports of periods up to 24 days. The virus produces a spectrum of disease severities, ranging from asymptomatic, to mild, moderate, and critical, life-threatening infection [49]. In comparison, the incubation period of SARS-CoV-1 is slightly shorter, at 2–10 days [50]. Patients over the age of 60 and those with comorbidities, such as diabetes, cardiovascular disease, and hypertension, are at higher risk of developing severe disease with both viruses [51], [52]. For COVID-19, symptomatic individuals can be classified into three major groups based on severity. In a Chinese study of 55,924 confirmed cases, approximately 80% of patients had mild to moderate illness, 13.8% had severe disease, and 6.1% were critically ill [53]. Symptoms of mild to moderate disease include fever, cough, sore throat, and mild pneumonia. Symptoms of severe disease include severe pneumonia (SpO2 < 93%), shortness of breath, and respiratory distress [24], [54]. Critical cases are defined as patients with respiratory failure, multi-organ dysfunction, sepsis, or shock [24]. The case severity of SARS was much reportedly higher, with 50–85% of SARS patients needing oxygen supplementation, while intensive care unit (ICU) admission rates ranged from 19% to 32% [55], [56], [57]. The overall case fatality rate (CFR) for SARS is 14–15%, which is significantly higher than the CFR estimate of approximately 2% for COVID-19 (Table 1 ) [21], [58].
Table 1.
Comparative summary between SARS and COVID-19.
| SARS (SARS-CoV-1) |
COVID-19 (SARS-CoV-2) |
||
|---|---|---|---|
| Epidemiology | Source of infection | Zoonotic | |
| WHO classification | Epidemic | Pandemic | |
| Duration | November 1, 2002–July 31, 2003 | Decemeber 2019 - ongoing | |
| Number of cases | 8096 [19] | 119,512,530[22] (March 14, 2021) |
|
| Number of deaths | 774 [19] | 2,642,612 [22] (March 14, 2021) |
|
| Total countries affected | 29 [19] | 200+ [21] | |
| Transmission | Primary mode of transmission | Infectious droplets, aerosols | |
| Viability on various surfaces | Similar viability | ||
| Basic reproductive number (R0) | 2–4 [34] | 1.4–6.9 [39] | |
| Additional modes of transmission | Faecal | Faecal, and potentially vertical, and sexual [42], [43], [44], [45], [46] | |
| Nosocomial transmission | Yes | ||
| Peak of contagiousness | Symptomatic phase [34] | Asymptomatic, presymptomatic, and early symptomatic phases [40] | |
| Clinical Course & Complications | Incubation period | 2–10 days [50] | 2–14 days [49] |
| Case Fatality Rate (As per confirmed cases and deaths) |
14–15% [58] | 2% [21] | |
| Most common cause of death | Acute respiratory distress syndrome (ARDS) |
||
| Extra-pulmonary disease | Cardiovascular diseases, renal impairment, coagulopathy | Cardiovascular diseases, renal impairment, coagulopathy | |
| Cytokine storm | Associated with ARDS | Associated with ARDS and other complications, more so than in SARS | |
| Therapeutics and Vaccines | Antiviral drugs | No approved drugs | Remdesivir (emergency use authorisation) [136] |
| Other treatments | No evidence of benefit from corticosteroid therapy No anticoagulant protocols needed |
Corticosteroid therapy e.g., dexamethasone [140] Anticoagulant protocols crucial [126] |
|
| Convalescent plasma therapy may have benefit | |||
| Vaccines | Approximately 30 vaccines initially developed with no clinical trials conducted [154] | 150+ vaccines developed; more than 30 candidates have reported results from clinical trials [153] As of March 14, 2021, 6 vaccines have been deployed globally |
|
| Prevention | Public health measures | Isolation of cases Quarantine of close contacts Closure of businesses and schools Restrictions on mass gatherings Cordon sanitaire of high-risk areas |
|
| Infection control precautions | Standard Contact Droplet Airborne |
||
A significant proportion of COVID-19 patients are asymptomatic, with estimates ranging from 4% to 41% [59]. In contrast, asymptomatic patients were not as evident during the SARS outbreak, but subsequently, asymptomatic seropositive HCWs have been identified [60], [61]. Symptoms that overlap between the two infections include fever, cough, headache, and shortness of breath, with fever being the most common symptom for both infections [62], [63]. According to two meta-analysis studies, the incidence of respiratory failure or acute respiratory distress syndrome (ARDS) in hospitalized COVID-19 patients ranged from 14.8% to 19.5%, with two studies reporting a prevalence of around 32% to 42%. As for SARS, there is a paucity of data, but studies reported the prevalence of ARDS to range from 20% to 49% [64], [65], [66], [67], [68], [69]. Gastrointestinal (GI) symptoms have been reported in COVID-19 patients, with diarrhea being the most common manifestation, followed by vomiting and GI bleeding. Some patients reported GI symptoms without any respiratory symptoms. COVID-19 patients with GI symptoms had a longer incubation period, higher liver enzymes, and prolonged coagulation times [70], [71]. Neurological manifestations have also been reported in COVID-19. The most common neurological symptoms were asthenia, headache, anosmia followed by encephalopathy, seizures, and stroke. In severe cases, encephalitis and coma have been reported [72], [73], [74]. In a systematic review, headaches were reported in 7.5% of COVID-19 patients, while 6.1% reported dizziness [75]. Other less common neurologic manifestations include cerebral haemorrhage and cerebral venous thrombosis (0.5%) [76]. In comparison, the most common neurological manifestation in SARS were neuromuscular. Other reported complications included axonopathic polyneuropathy, myopathy, and large artery ischemic stroke [77], [78]. Skin lesions have been widely reported as a potential manifestation of COVID-19, but not SARS [79]. The finding of orchitis remains fairly unique to SARS patients, but its prevalence is still being studied in COVID-19 patients [80].
Complications
ARDS is a common sequela in COVID-19 and SARS patients, and is the commonest cause of death in COVID-19 patients [81], [82], [83], [84]. Pulmonary inflammation and extensive lung damage in SARS patients were associated with high levels of pro-inflammatory cytokines. Recent studies of COVID-19 showed similar findings (e.g., elevated levels of IL1B, IL6, IL12, IFNγ, IP10, and MCP 1) [84]. Multiple studies show a correlation between higher concentrations of inflammatory mediators and disease severity [84], [85]. The same immune reaction brought about by the cytokine storm can also rapidly progress to multiple organ failure [55]. In addition, COVID-19 patients can also die of cardiovascular and renal complications. This has been linked to either direct viral injury due to the expression of the ACE2 receptor in these organs, or indirectly via the cytokine storm [86].
Serious respiratory infections are well-recognized triggers for cardiovascular complications, especially in patients with underlying cardiovascular disease [87], [88]. Cardiovascular involvement is highly associated with mortality in COVID-19 patients. Mortality rates were reported to be significantly higher in those with elevated plasma troponin T (TnT) and high-sensitivity cardiac troponin I (hs-cTnI) levels compared to patients with normal plasma levels [89]. While cardiovascular events were documented to occur post SARS infection, cardiovascular complications such as heart failure (23%), acute cardiac injury (17%), and arrhythmia (16.7%) have been widely reported in COVID-19 patients, with a higher prevalence among non-survivors [90], [91]. A study of cardiovascular complications of SARS showed that the majority were either self-limiting or reversible, with the most common being tachycardia [92].
Kidney damage has been reported with both coronavirus infections but less commonly in SARS. The pathogenesis of kidney damage includes direct viral injury, indirect damage through deposition of immune complexes or via the cytokine storm [93], [94]. Additionally, thrombotic complications, such as pulmonary embolism and deep vein thrombosis, have been documented in SARS and COVID-19 patients, but are significantly more common in COVID-19. These complications are thought to be associated with the exaggerated inflammatory immune response, hypoxia, endothelial injury, or diffuse intravascular coagulation similar to that seen in sepsis [95], [96]. An inflammatory shock similar to atypical Kawasaki disease and Kawasaki disease shock syndrome, termed multisystem inflammatory syndrome in children (MIS-C), has also been reported in children with COVID-19, but was not observed in SARS [97].
Investigation and diagnosis
The diagnostic techniques used to identify the presence of SARS-CoV-1 and SARS-CoV-2 are similar. For the diagnosis of infection with SARS-CoV-1, the patient must present with indicative signs and symptoms combined with a positive laboratory test [98]. As for SARS-CoV-2, the diagnostic criteria include epidemiological evidence with indicative clinical signs and symptoms, and a positive laboratory test [99]. In both diseases, reverse transcription-polymerase chain reaction (RT-PCR) is performed primarily on upper respiratory tract samples, such as nasopharyngeal swabs or saliva, to detect the virus. Other sample types include lower respiratory tract samples, such as bronchoalveolar lavages, as well as stool, which may have greater sensitivity than upper respiratory tract samples depending on the clinical presentation and stage of infection. RT-PCR requires the extraction of viral RNA from a sample and uses reverse transcriptase to convert the RNA to cDNA, which is then amplified by PCR targeting genetic targets specific to each virus [98], [100]. One study of 213 confirmed COVID-19 patients showed that sputum samples had the highest positivity rate in both mild and severe cases (82.2% and 88.9%), followed by nasal swabs (73.3% and 72.1%) and finally throat swabs (60.0% and 61.3%) [101]. Another study similarly found higher SARS-CoV-2 viral loads in nasopharyngeal swabs compared to oral throat swabs [102]. As for SARS-CoV-1, tracheal aspirates were reported to yield higher positivity rates (66.7%) compared to nasopharyngeal swabs (29.7–40.0%) as most SARS patients had lower respiratory tract infections at the time of diagnosis [103]. It is important to note that the diagnostic yield of the test is affected by the quality of the sample, as well as the timing of the sample collection in relation to the stage of illness. The addition of a lower respiratory tract sample can be useful when an upper respiratory sample is negative for SARS-CoV-2 in cases that are highly suspicious for COVID-19 [104].
Serological testing is an essential tool for both epidemiological studies and vaccine evaluation, as well as to identify patients with higher antibody titers for hyperimmune plasma donation [105]. Furthermore, serological tests are typically less expensive and easier to perform than molecular tests, making them a better option for population level surveillance, especially for the identification of individuals who may have had asymptomatic or subclinical infection [106]. The use of serological testing for SARS-CoV-2 is less clear for the diagnosis of acute infection, as reliable detection of antibodies does not occur until at least 2 weeks post onset. For the detection of recent infection, serology would need to be complemented with viral testing (such as RT-PCR), which may be useful to enhance contact tracing efforts [107]. Serological testing should not be used as definitive evidence of immunity until further studies identify which antibodies and epitopes are most predictive of immunity and how long that immunity lasts [108]. Both the IgM and IgG responses in COVID-19 are detectable earlier than in SARS. For SARS-CoV-1, enzyme-linked immunosorbent assays (ELISAs) can detect serum antibodies (IgM and IgG) with 80–85% sensitivity by the end of the second week, and 100% by the third week [98]. Various commercial tests for SARS-CoV-1 serology have sensitivities ranging from 72% to 100%, and specificities ranging from 98% to 100% [106].
SARS-CoV-1 and SARS-CoV-2 have similar radiological findings in those who have lower respiratory tract infection [98], [109]. The most common chest x-ray findings are consolidation and ground glass opacification [110]. In cases where chest x-ray findings are not diagnostic, a CT scan may be indicated. The use of CT to diagnose both infections is crucial in cases that present early in the course of the disease when lung abnormalities are not yet clear on chest X-ray [111]. This will allow for earlier isolation and treatment of patients, especially when viral testing is not available or may be falsely negative. CT scan can also be used to assess the changes during clinical follow-up [98]. The CT findings most commonly associated with both diseases are bilateral pulmonary ground-glass opacities and consolidation. These findings are most prominent 10 days after the onset of symptoms in patients with SARS-CoV-1 and SARS-CoV-2 [109], [112]. These CT scan findings are also seen in some asymptomatic SARS-CoV-2 patients, in which unilateral opacifications progress to bilateral consolidations in 7–21 days [109]. At initial presentation in SARS patients, 86% of the cases in one case series presented with a unilateral focal opacity. In severe cases, the virus led to diffuse consolidation, a sign of the development of ARDS. The lesions were mostly located in the lower lung lobes [98].
COVID-19 can affect multiple organs and tissues at the histopathological level. In the lungs, findings include interstitial pneumonia, alveolar hyperplasia, and pulmonary fibrosis [113]. As for the kidneys, one post-mortem study reported that acute kidney injury developed in 94% of COVID-19 deaths, with the most common pathological finding being acute tubular injury. Other findings related to the kidneys included hypertensive arterionephrosclerosis and diabetic glomerulosclerosis. Focal thrombi were present in 14% of autopsies [114]. In the brain, spongiosis and edema were the most common histopathological findings in COVID-19 [115]. Similar to COVID-19, the lungs of SARS patients also showed alveolar damage, edema, and pulmonary fibrosis. The kidneys of SARS patients also showed acute tubular injury [116]. In SARS, neuronal edema and degeneration were the two most common findings in the brain. Damage to the heart was also reported in SARS with evidence of myocardial fiber edema and atrophy [116]. Likewise, in COVID-19, acute myocyte necrosis along with the presence of inflammatory infiltrates and apoptotic bodies have been reported in autopsies [117]. As for dermatoses related to COVID-19, several features have been identified, including necrotic keratinocytes and acantholytic clefts [117]. This has not been commonly reported in SARS [116].
Management
Patients with SARS and COVID-19 infection with mild symptoms and no risk factors for severe disease development can be managed at home while maintaining infection control precautions [11], [118], [119]. Patients who are managed at home are advised to continue infection control precautions for 10 days following resolution of fever in the case of SARS and for variable durations in the case of COVID-19 depending on the guidelines of different countries, which may also include clinical or laboratory criteria [11], [118], [120].
A limited set of therapeutics, including remdesivir, corticosteroids, and monoclonal antibodies, have been indicated for use in critical cases of COVID-19 or cases where there is a high risk of disease progression [121]. Similar therapeutic guidelines have not been developed for SARS due to more limited extent of the outbreak and challenges with demonstrating clinical efficacy; however, similar approaches were used and investigated [122].
In-patient care for SARS and COVID-19 is mainly supportive, focusing on managing the complications of infection such as pneumonia, respiratory failure, and sepsis, as well as complications from prolonged hospital stay, including secondary bacterial infections and thromboembolism [123], [124], [125]. Because of the importance of endothelial dysfunction in COVID-19, many guidelines recommend prophylactic doses of anticoagulant or antiplatelet therapies for all in-patient cases, including full therapeutic doses for those at higher risk of developing thromboembolic disease. Low-molecular weight heparins have been specifically recommended for their downregulation of IL-6 and blocking of the SARS-CoV-2 spike protein [126], [127], [128]. Recent studies suggest that therapeutic anticoagulation doses in COVID-19 patients with critical illness may worsen mortality outcomes due to the increased risk of bleeding, but rather, prophylactic dosages may be more suitable, unless otherwise indicated by a hypercoagulable state [129], [130]. The need for long-term antithrombotic therapy remains unclear. Similar antithrombotic protocols were not recommended for the treatment of SARS.
Treatment and vaccination
Treatment
Other than supportive care and infection control measures, there are no specific or highly effective treatments for SARS-CoV-1 and SARS-CoV-2 [3], [11]. Empirical therapy for community acquired pneumonia should be administered in patients suspected to have SARS [3], [131]. However, bacterial superinfection has not been a prominent feature of COVID-19, so empiric antibiotic therapy has not been broadly recommended [132].
As many therapeutic and vaccine development projects were not successfully carried to completion during the SARS outbreak, there exist gaps in knowledge to guide the development of vaccines and treatments against COVID-19 [133]. Several antiviral agents have nonetheless been used experimentally for the treatment of both diseases. Remdesivir is a nucleoside analogue that was originally developed against Ebola, but later shown to inhibit SARS-CoV-1 in animal models [134], [135]. Remdesivir gained an emergency use authorization (EUA) for the treatment of COVID-19 by the FDA, as clinical trials showed modest benefits in critically ill patients [136]. Preliminary evidence suggested that hydroxychloroquine and chloroquine may have antiviral effects against both viruses, but EUA for the use of both drugs against COVID-19 was revoked by the FDA following findings showing a lack of benefit and concerns of serious side effects [137], [138].
Furthermore, various immunomodulators have been used to improve the prognostic outcome of SARS and COVID-19 patients. Corticosteroids were used during the SARS outbreak, but did not seem to be effective [139]. However, the use of dexamethasone in a clinical trial has shown a significant decrease in the mortality of COVID-19 patients requiring respiratory support [140]. In terms of monoclonal antibodies, tocilizumab (IL-6 inhibitor) has been recommended along with dexamethasone in hospitalised COVID-19 cases presenting with rapidly deteriorating respiratory function [141]. Other neutralizing antibodies, such as bamlanivimab and etesevimab, have been considered for outpatients with mild to moderate COVID-19 disease who are at increased risk of disease progression [142]. In the case of SARS, although some SARS-CoV-1 antibodies have been developed, they have not been sufficiently studied [122], [143]. Finally, the use of convalescent plasma has shown promise for seriously ill patients for both viruses. However, the FDA has recommended further evidence from controlled clinical studies to determine the efficacy and safety of this approach [144], [145].
Vaccination
There have been many efforts to develop effective vaccines against SARS-CoV-1 and SARS-CoV-2 [146], [147], [148], [149], [150], [151], [152], [153]. The fact that affected individuals produce high titers of neutralizing antibodies against SARS-CoV-1 suggests that the induction of neutralizing antibodies could be an effective strategy to prevent infection [146]. Studies have explored the use of different classes of vaccines, such as nucleic-acid based, protein-based, and inactivated whole virus vaccines, many of which have successfully demonstrated the induction of neutralizing antibodies [147], [148], [149], [150], [151], [154]. Although the use of SARS-CoV-1 whole virus vaccine was effective in animals, its use in humans raised serious safety concerns due to the risk of inflammatory responses and potential for SARS-like disease. Therefore, none of the vaccines developed for SARS have been evaluated in clinical trials [152], [154]. Conversely, there are currently over 100 vaccine candidates from 8 classes of vaccines being developed and tested in clinical trials against SARS-CoV-2, including whole virus vaccines, viral vectors, and nucleic acid and protein-based vaccines. This accelerated effort has produced several efficacious vaccines, which are now being deployed globally to mitigate the COVID-19 pandemic. Additionally, other efforts are ongoing to study the immunomodulatory effect of existing vaccines, such as the Bacillus Calmette-Guerin (BCG) vaccine for tuberculosis, which may have a protective effect against severe illness in COVID-19 [153], [155], [156].
Prevention
Prevention in the community
In addition to the current emergency approval and distribution of COVID-19 vaccines, public health containment measures are the only effective strategies to prevent or slow down human transmission in the community [20]. Hand hygiene is strongly advised in the setting of coronaviruses, as these viruses are efficiently inactivated via handwashing with soap and water or alcohol-based sanitizers [157]. Good respiratory hygiene, social distancing, and the universal use of facemasks are also fundamental elements of controlling these viral outbreaks [158], [159]. The detection and isolation of confirmed cases, along with the tracking and quarantine of close contacts, is essential to prevent community transmission. These methods resulted in the elimination of SARS from the human population and remain important tools for controlling the COVID-19 pandemic [20], [160], [161]. Restrictions on movement have also been imposed worldwide, both within countries as well as between countries, with individuals from high-incidence regions prevented from traveling or having to go through additional screening or quarantine on arrival [162], [163], [164].
Prevention in hospitals
SARS and COVID-19 have been shown to spread in the hospital setting [165], [166]. Appropriate infection prevention methods such as standard, contact, and droplet precautions must be practiced by HCWs (Table 1). Additionally, the use of N95 respirators in the setting of aerosol-generating procedures has been greatly emphasized [167], [168]. Disinfection of hospital areas and patients’ items has been shown to be useful in preventing hospital-acquired infection [169]. Due to the magnitude of the COVID-19 outbreak, some countries resorted to building new temporary hospitals, and converting public venues such as stadiums and parking lots into shelter hospitals for treatment and isolation. These methods have been shown to be effective in preventing the nosocomial spread of disease while maintaining health care services for non-COVID-19 patients [170].
Preventing future zoonotic outbreaks
The SARS and COVID-19 outbreaks have emphasized the need to be able to predict and prevent future outbreaks. Regulatory oversight of wet markets and the wild animal trade must be one key component of strategies to prevent future zoonotic outbreaks [171]. Although these markets are an essential source of food and income for many parts of the world, additional safety measures need to be implemented along with enforcement of these rules. Surveillance efforts to monitor and characterize animal viruses in the environment must continue to provide early warnings for potential zoonotic pathogens. The capacity of the health care system, including clinical laboratory testing and stockpiles of resources such as masks, to deal with emergencies such as a pandemic must be maintained [172]. In addition, it is essential that communicable disease prevention and control programs and agencies continue to be supported even when global outbreaks are not in the headlines [173]. The current pandemic has also demonstrated the successful incorporation of new technology for public health, with the use of mobile applications to assist with contact tracing. One study in Spain has shown that the use of digital contact tracing was almost twice as effective as manual contact tracing techniques [174].
Limitations of comparative reviews
It is worth noting that the amount of literature available on COVID-19 is much larger than that on SARS. In fact, a PubMed search using the term “COVID-19” provides over 100,000 results from the period of 2019 to early-2021 alone. In contrast, a search using the term “SARS” between 2002 and 2019 provides a list of 9,000 results. Therefore, a comparative discussion using the available literature is prone to selection and/or reporting bias. Additionally, since the SARS outbreak was more concentrated in Asia compared to the global distribution of COVID-19, the findings related to SARS may not be representative of the potential spectrum of clinical disease, complications and public health implications, had it spread globally to more diverse populations. Finally, a larger dedicated international effort has been directed towards studying COVID-19 compared to the SARS epidemic. Again, this highlights the need for a cautious approach when drawing parallels between SARS and COVID-19, despite the obvious and significant similarities between both viruses.
Future perspective
The past is the present's gift for the future; if we choose to accept it, we may be carving the path for well-informed, proactive measures for the management of future health crises. These measures cannot focus only on health, but must involve a collaborative effort that takes into consideration global and community-level inequalities. It is evident, as the scientific and clinical community tackles the COVID-19 pandemic with incomplete knowledge, that previous epidemics, such as SARS, can guide us to better understand the trajectory of the outbreak, the expected pathogenesis, and potential complications of the disease, both clinically and at the population level. Global health issues of this magnitude require multidisciplinary efforts from governments, research and scientific bodies, health care systems, and international entities. As evident from the current situation and past experiences, it is imperative that strict measures for virus source control are implemented, including the banning, or at least regulation, of the wild animal trade, and global monitoring and surveillance efforts for zoonotic viral reservoirs and significant viral mutations. Without significant changes in the global approach to zoonotic diseases, another coronavirus outbreak in the next few years would not be unthinkable. Viral outbreaks and infectious diseases have plagued humanity for centuries, and it is inevitable that they will continue to do so. However, we hope that lasting changes will result from this unprecedented modern-day outbreak, and that the next comparative review on the topic will be discussing the proactive infection prevention measures, successful containment, efficient global response, and rapid activation of public health emergency systems in the effort to reduce the magnitude of damage from the next pandemic.
Funding
No funding sources.
Competing interests
None declared.
Ethical approval
Not required.
References
- 1.Tyrrell D., Bynoe M. Cultivation of viruses from a high proportion of patients with colds. Lancet. 1966;1:76–77. doi: 10.1016/s0140-6736(66)92364-6. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and, Prevention, Human coronavirus types. https://www.cdc.gov/coronavirus/types.html [accessed 24.03.21].
- 3.Froude S., Hughes H. In: Oxford textbook of medicine. 6th ed. Firth J., Conlon C., Cox T., editors. Oxford University Press; Oxford: 2020. Newly discovered viruses. [Google Scholar]
- 4.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menachery V.D., Yount B.L., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization Coronavirus disease 2019 (COVID-19) situation report – 84. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200413-sitrep-84-covid-19.pdf?.sfvrsn=44f511ab_2 [accessed 24.03.21].
- 7.Chen S., Yang J., Yang W., Wang C., Bärnighausen T. COVID-19 control in China during mass population movements at New Year. Lancet. 2020;395:764–766. doi: 10.1016/S0140-6736(20)30421-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G., et al. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun. 2020;109:102434. doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung S.M., Jung S.M., Kinoshita R., Thompson R.N., Linton N.M., Yang Y., Akhmetzhanov A.R., et al. Epidemiological identification of a novel pathogen in real time: analysis of the atypical pneumonia outbreak in Wuhan China, 2019–2020. J Clin Med. 2020;9:637. doi: 10.3390/jcm9030637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control Prevention. CDC SARS response timeline, https://www.cdc.gov/about/history/sars/timeline.htm; [accessed 24.03.21].
- 11.World Health Organization. Pneumonia of unknown cause – China. https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/ [accessed 24.03.21].
- 12.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. 2020;30:1578. doi: 10.1016/j.cub.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 14.Liu P., Jiang J.Z., Wan X.F., Hua Y., Li L., Zhou J., et al. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020;16:e1008421. doi: 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization. Severe acute respiratory syndrome (SARS) – multi-country outbreak – update 27. https://www.who.int/csr/don/2003_04_11/en/ [accessed 24.03.21].
- 16.Heymann D.L., Mackenzie J.S., Peiris M. SARS legacy: outbreak reporting is expected and respected. Lancet. 2013;381:779–781. doi: 10.1016/S0140-6736(13)60185-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020 doi: 10.1056/2020NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization. Novel coronavirus (2019-nCoV) Situation Report – 2; 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200122-sitrep-2-2019-ncov.pdf?.sfvrsn=4d5bcbca_2 [accessed 24.03.21].
- 19.World Health Organization. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. https://www.who.int/csr/sars/country/table2004_04_21/en/ [accessed 24.03.21].
- 20.Wilder-Smith A., Chiew C.J., Lee V.J. Can we contain the COVID-19 outbreak with the same measures as for SARS? Lancet Infect Dis. 2020;20(5):102–107. doi: 10.1016/S1473-3099(20)30129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Health Organization. Weekly epidemiological update on COVID-19; 16 March 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update---16-march-2021 [accessed 24.03.21].
- 22.World Health Organization. Coronavirus disease (COVID-2019) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports [accessed 24.03.21].
- 23.Fahmi M., Kubota Y., Ito M. Nonstructural proteins NS7b and NS8 are likely to be phylogenetically associated with evolution of 2019-nCoV. Infect Genet Evol. 2020;81:104272. doi: 10.1016/j.meegid.2020.104272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls Publishing; Treasure Island (FL): 2021. Features, evaluation and treatment coronavirus (COVID-19) StatPearls [internet] [PubMed] [Google Scholar]
- 25.Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo P.C., Huang Y., Lau S.K., Yuen K.-Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vijgen L., Keyaerts E., Moës E., Thoelen I., Wollants E., Lemey P., et al. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. 2005;79(3):1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J.M., Chung Y.S., Jo H.J., Lee N.J., Kim M.S., Woo S.H., et al. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res Perspect. 2020;11(1):3. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Nature. 2020;17(6):613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization. Consensus document on the epidemiology of severe acute respiratory syndrome (SARS). https://www.who.int/csr/sars/en/WHOconsensus.pdf [accessed 24.03.21].
- 35.World Health Organization. Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations; 2020. https://apps.who.int/iris/bitstream/handle/10665/331601/WHO-2019-nCoV-Sci_Brief-Transmission_modes-20201-eng.pdf [accessed 24.03.21].
- 36.Ather A., Patel B., Ruparel N.B., Diogenes A., Hargreaves K.M. Coronavirus disease 19 (COVID-19): implications for clinical dental care. J. Endod. 2020;46(5):584–595. doi: 10.1016/j.joen.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong G., Liu W., Liu Y., Zhou B., Bi Y., Gao G.F. MERS SARS, and Ebola: the role of super-spreaders in infectious disease. Cell Host Microbe. 2015;18(4):398–401. doi: 10.1016/j.chom.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwok K.O., Chan H.H., Huang Y., Hui D.S., Tambyah P.A., Wei W.I., et al. Inferring super-spreading from transmission clusters of COVID-19 in Hong Kong Japan and Singapore. J Hosp Infect. 2020;105(4):682–685. doi: 10.1016/j.jhin.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020;27(2) doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.World Health Organization. Coronavirus disease 2019 (COVID-19) situation report – 73. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200402-sitrep-73-covid-19.pdf?sfvrsn=5ae25bc7_2 [accessed 24.03.21].
- 41.Chowdhury R., Heng K., Shawon M.S., Goh G., Okonofua D., Ochoa-Rosales C., et al. Dynamic interventions to control COVID-19 pandemic: a multivariate prediction modelling study comparing 16 worldwide countries. Eur J Epidemiol. 2020;35(5):389–399. doi: 10.1007/s10654-020-00649-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amirian E.S. Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int J Infect Dis. 2020;95:363–370. doi: 10.1016/j.ijid.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alzamora M.C., Paredes T., Caceres D., Webb C.M., Valdez L.M., La Rosa M. Severe COVID-19 during pregnancy and possible vertical transmission. Am J Perinatol. 2020;37(8):861. doi: 10.1055/s-0040-1710050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karimi-Zarchi M., Neamatzadeh H., Dastgheib S.A., Abbasi H., Mirjalili S.R., Behforouz A., et al. Vertical transmission of coronavirus disease 19 (COVID-19) from infected pregnant mothers to neonates: a review. Fetal Pediatr Pathol. 2020;39(3):246–250. doi: 10.1080/15513815.2020.1747120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patrì A., Gallo L., Guarino M., Fabbrocini G. Sexual transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a new possible route of infection? JAAD. 2020;82(6) doi: 10.1016/j.jaad.2020.03.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li A., Ng P. Severe acute respiratory syndrome (SARS) in neonates and children. Arch Dis Fetal Neonat Ed. 2005;90(6):F461–F465. doi: 10.1136/adc.2005.075309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borras-Bermejo B., Martínez-Gómez X., San Miguel M.G., Esperalba J., Antón A., Martin E., et al. Early release-asymptomatic SARS-CoV-2 infection in nursing homes, Barcelona, Spain April 2020. Emerg Infect Dis. 2020:26. doi: 10.3201/eid2609.202603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lau J.T., Fung K.S., Wong T.W., Kim J.H., Wong E., Chung S., et al. SARS transmission among hospital workers in Hong Kong. Emerg Infect Dis. 2004;10(2):280. doi: 10.3201/eid1002.030534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leung C. The difference in the incubation period of 2019 novel coronavirus (SARS-CoV-2) infection between travelers to Hubei and nontravelers: the need for a longer quarantine period. Infect Control Hosp Epidemiol. 2020;41(5):594–596. doi: 10.1017/ice.2020.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lessler J., Reich N.G., Brookmeyer R., Perl T.M., Nelson K.E., Cummings D.A. Incubation periods of acute respiratory viral infections: a systematic review. Lancet Infect Dis. 2009;9(5):291–300. doi: 10.1016/S1473-3099(09)70069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng Z., Peng F., Xu B., Zhao J., Liu H., Peng J., et al. Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect. 2020;81(2):e16–e25. doi: 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan J.W., Ng C.K., Chan Y.H., Mok T.Y., Lee S., Chu S.Y., et al. Short term outcome and risk factors for adverse clinical outcomes in adults with severe acute respiratory syndrome (SARS) Thorax. 2003;58(8):686–689. doi: 10.1136/thorax.58.8.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.World Health Organization. Report of the WHO-China joint mission on coronavirus disease 2019 (COVID-19). https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf [accessed 24.03.21].
- 54.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stainsby B., Howitt S., Porr J. Neuromusculoskeletal disorders following SARS: a case series. JCCA. 2011;55(1):32. [PMC free article] [PubMed] [Google Scholar]
- 56.Xu J., Qi L., Chi X., Yang J., Wei X., Gong E., et al. Orchitis: a complication of severe acute respiratory syndrome (SARS) Biol Reprod. 2006;74(2):410–416. doi: 10.1095/biolreprod.105.044776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yam L.Y., Chen R.C., Zhong N.S. SARS: ventilatory and intensive care. Respirology. 2003;8:S31–S35. doi: 10.1046/j.1440-1843.2003.00521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.World Health Organization. Update 49 – SARS case fatality ratio, incubation period. https://www.who.int/csr/sars/archive/2003_05_07a/en/ [accessed 24.03.21].
- 59.Byambasuren O., Cardona M., Bell K., Clark J., McLaws M.L., Glasziou P. Estimating the extent of true asymptomatic COVID-19 and its potential for community transmission: systematic review and meta-analysis. JAMMI. 2020;5(4):223–234. doi: 10.3138/jammi-2020-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee H.K., Tso E.Y., Chau T., Tsang O.T. Asymptomatic severe acute respiratory syndrome-associated coronavirus infection. Emerg Infect Dis. 2003;9(11):1491. doi: 10.3201/eid0911.030401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leung G.M., Hedley A.J., Ho L.-M., Chau P., Wong I.O., Thach T.Q., et al. The epidemiology of severe acute respiratory syndrome in the 2003 Hong Kong epidemic: an analysis of all 1755 patients. Ann Intern Med. 2004;141(9):662–673. doi: 10.7326/0003-4819-141-9-200411020-00006. [DOI] [PubMed] [Google Scholar]
- 62.World Health Organization. SARS (severe acute respiratory syndrome) international travel and health. https://www.who.int/ith/diseases/sars/en/ [accessed 24.03.21].
- 63.Centers for Disease Control Prevention. Symptoms of coronavirus. https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html [accessed 24.03.21].
- 64.Zhu J., Ji P., Pang J., Zhong Z., Li H., He C., et al. Clinical characteristics of 3,062 COVID-19 patients: a meta-analysis. J Med Virol. 2020;22(10):1902–1914. doi: 10.1002/jmv.258841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun P., Qie S., Liu Z., Ren J., Li K., Xi J. Clinical characteristics of hospitalized patients with SARS-CoV-2 infection: a single arm meta-analysis. J Med Virol. 2020;92(6):612–617. doi: 10.1002/jmv.25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodriguez-Morales A.J., Cardona-Ospina J.A., Gutiérrez-Ocampo E., Villamizar-Peña R., Holguin-Rivera Y., Escalera-Antezana J.P., et al. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Travel Med Infect Dis. 2020;34:101623. doi: 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu C., Chen X., Cai Y., Zhou X., Xu S., Huang H., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan China. JAMA Intern Med. 2020;180(7):925–1036. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen C.-Y., Lee C.-H., Liu C.-Y., Wang J.-H., Wang L.-M., Perng R.-P. Clinical features and outcomes of severe acute respiratory syndrome and predictive factors for acute respiratory distress syndrome. J China Med Assoc. 2005;68(1):4–10. doi: 10.1016/S1726-4901(09)70124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peiris J.S.M., Chu C.-M., Cheng V.C.-C., Chan K.S., Hung I.F., Poon L.L., et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hajifathalian K., Mahadev S., Schwartz R.E., Shah S., Sampath K., Schnoll-Sussman F., et al. SARS-COV-2 infection (coronavirus disease 2019) for the gastrointestinal consultant. World J Gastroenterol. 2020;26(14):1546. doi: 10.3748/wjg.v26.i14.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian Y., Rong L., Nian W., He Y. gastrointestinal features in COVID-19 and the possibility of faecal transmission. Aliment Pharm Ther. 2020;51(9):843–851. doi: 10.1111/apt.15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zubair A.S., McAlpine L.S., Gardin T., Farhadian S., Kuruvilla D.E., Spudich S. Neuropathogenesis and neurologic manifestations of the coronaviruses in the age of coronavirus disease 2019: a review. JAMA Neurol. 2020;77(8):1018–1027. doi: 10.1001/jamaneurol.2020.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Payus A.O., Lin C.L., Noh M.M., Jeffree M.S., Ali R.A. SARS-CoV-2 infection of the nervous system: a review of the literature on neurological involvement in novel coronavirus disease-(COVID-19) Bosnian J Basic Med Sci. 2020;20(3):283. doi: 10.17305/bjbms.2020.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Di Carlo D.T., Montemurro N., Petrella G., Siciliano G., Ceravolo R., Perrini P. Exploring the clinical association between neurological symptoms and COVID-19 pandemic outbreak: a systematic review of current literature. J Neurol. 2020:1–9. doi: 10.1007/s00415-020-09978-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fisicaro F., Di Napoli M., Liberto A., Fanella M., Di Stasio F., Pennisi M., et al. Neurological sequelae in patients with covid-19: a histopathological perspective. Int J Environ Res Public Health. 2021;18(4):1415. doi: 10.3390/ijerph18041415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsai L.K., Hsieh S.T., Chao C.C., Chen Y.C., Lin Y.H., Chang S.C., et al. Neuromuscular disorders in severe acute respiratory syndrome. Arch Neurol. 2004;61(11):1669–1673. doi: 10.1001/archneur.61.11.1669. [DOI] [PubMed] [Google Scholar]
- 78.Umapathi T., Kor A.C., Venketasubramanian N., Lim C.T., Pang B.C., Yeo T.T., et al. Large artery ischaemic stroke in severe acute respiratory syndrome (SARS) J Neurol. 2004;251(10):1227–1231. doi: 10.1007/s00415-004-0519-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao Q., Fang X., Pang Z., Zhang B., Liu H., Zhang F. COVID-19 and cutaneous manifestations: a systematic review. J Eur Acad Dermatol. 2020;34(11) doi: 10.1111/jdv.16778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pan F., Xiao X., Guo J., Li H., Patel D. No evidence of SARS-CoV-2 in semen of males recovering from COVID-19. Fertil Steril. 2020;113(6):1135–1139. doi: 10.1016/j.fertnstert.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mehta P., Mcauley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet (London, England) 2020;395(10229):1033. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan China. Intensive Care Med. 2020;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu Z., Shi L., Wang Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang C., Wang Y., Li X., Zhao J., Hu Y., Zhang L., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cameron M.J., Bermejo-Martin J.F., Danesh A., Muller M.P., Kelvin D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133(1):13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang T., Du Z., Zhu F., Cao Z., An Y., Gao Y., et al. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet. 2020;395(10228):e52. doi: 10.1016/S0140-6736(20)30558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Madjid M., Safavi-Naeini P., Solomon S.D., Vardeny O. Potential effects of coronaviruses on the cardiovascular system: a review. JAMA Cardiol. 2020;5(7):831–840. doi: 10.1001/jamacardio.2020.1286. [DOI] [PubMed] [Google Scholar]
- 88.Hamming I., Timens W., Bulthuis M., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. JPTLAS. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Atallah B., Mallah S.I., Abdelwareth L., AlMahmeed W., Fonarow G.C. A marker of systemic inflammation or direct cardiac injury: should cardiac troponin levels be monitored in COVID-19 patients? Eur Heart J. 2020;6(3):204–207. doi: 10.1093/ehjqccoqcaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult in-patients with COVID-19 in Wuhan China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu C., Wong R.S., Wu E., Kong S.L., Wong J., Yip G.W., et al. Cardiovascular complications of severe acute respiratory syndrome. Postgrad Med J. 2006;82(964):140–144. doi: 10.1136/pgmj.2005.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng Y., Luo R., Wang K., Zhang M., Wang Z., Dong L., et al. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020;97(5):829–838. doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chu K.H., Tsang W.K., Tang C.S., Zhang M., Wang Z., Dong L., et al. Acute renal impairment in coronavirus-associated severe acute respiratory syndrome. Kidney Int. 2005;67(2):698–705. doi: 10.1111/j.1523-1755.2005.67130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Klok F., Kruip M., Van Der Meer N., Arbous M.S., Gommers D.A., Kant K.M., et al. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiang-Hua Y., Le-Min W., Ai-Bin L., Zhu G., Riquan L., Xu-You Z., et al. Severe acute respiratory syndrome and venous thromboembolism in multiple organs. Am J Respir Crit Care Med. 2010;182(3):436–437. doi: 10.1164/ajrccm.182.3.436. [DOI] [PubMed] [Google Scholar]
- 97.Riphagen S., Gomez X., Gonzalez-Martinez C., Wilkinson N., Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395(10237):1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leung C., Chiu W. Clinical picture, diagnosis, treatment and outcome of severe acute respiratory syndrome (SARS) in children. Paediatr Respir Rev. 2004;5(4):275–288. doi: 10.1016/j.prrv.2004.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang Y.-Y., Jin Y.-H., Ren X.-Q., Yi-Rong L., Xiao-Chun Z., Xian-Tao Z., et al. Updating the diagnostic criteria of COVID-19 “suspected case” and “confirmed case” is necessary. Mil Med Res. 2020;7(1):1–3. doi: 10.1186/s40779-020-00245-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. AJR. 2020;215(1):87–93. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 101.Yang Y., Yang M., Shen C., Wang F., Wang Z., Li J., et al. Laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. Innovation. 2020;1(3):100061. doi: 10.1016/j.xinn.2020.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chan P.K., To W.-K., Ng K.-C., Lam R.K., Ng T.K., Chan R.C., et al. Laboratory diagnosis of SARS. Emerg Infect Dis. 2004;10(5):825–831. doi: 10.3201/eid1005.030682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hase R., Kurita T., Muranaka E., Sasazawa H., Mito H., Yano Y. A case of imported COVID-19 diagnosed by PCR-positive lower respiratory specimen but with PCR-negative throat swabs. J Infect. 2020;52(6):423–426. doi: 10.1080/23744235.2020.1744711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., et al. Assay techniques and test development for COVID-19 diagnosis. ACS. 2020;6(5):591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rashid Z.Z., Othman S.N., Samat M.N.A., Ali U.K., Wong K.K. Diagnostic performance of COVID-19 serology assays. Malays J Pathol. 2020;42(1):13–21. [PubMed] [Google Scholar]
- 107.Centers for Disease Control and Prevention. COVID-19 Serology Surveillance Strategy, https://www.cdc.gov/coronavirus/2019-ncov/covid-data/serology-surveillance/index.html; [accessed 24 March 2021].
- 108.Infectious Diseases Society of America. IDSA COVID-19 Antibody Testing Primer, https://www.idsociety.org/globalassets/idsa/public-health/covid-19/idsa-covid-19-antibody-testing-primer.pdf; [accessed 24 March 2021].
- 109.Zhai P., Ding Y., Wu X., Long J., Zhong Y., Li Y. The epidemiology, diagnosis and treatment of COVID-19. Int J Antimicrob Agents. 2020;55(5):105955. doi: 10.1016/j.ijantimicag.2020.105955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wong H.Y.F., Lam H.Y.S., Fong A.H.-T., Leung S.T., Chin T.W., Lo C.S. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2020;296(2):201160. doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chan P.K., Ng K.-C., Chan R.C., Lam R.K., Chow V.C., Hui M., et al. Immunofluorescence assay for serologic diagnosis of SARS. Emerg Infect Dis. 2004;10(3):530–532. doi: 10.3201/eid1003.030493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ooi G.C., Daqing M. SARS: radiological features. Respirology. 2003;8:S15–S19. doi: 10.1046/j.1440-1843.2003.00519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mansueto G. COVID-19: brief check through the pathologist's eye (autopsy archive) Pathol Res Pract. 2020;216(11):153195. doi: 10.1016/j.prp.2020.153195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Santoriello D., Khairallah P., Bomback A.S., Xu K., Kudose S., Batal I., et al. Postmortem kidney pathology findings in patients with COVID-19. J Am Soc Nephrol. 2020;31(9):2158–2167. doi: 10.1681/ASN2020050744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Remmelink M., De Mendonça R., D’Haene N., De Clercq S., Verocq C., Lebrun L., et al. Unspecific post-mortem findings despite multiorgan viral spread in COVID-19 patients. Critical Care. 2020;24(1):1. doi: 10.1186/s13054-020-03218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gu J., Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. Am J Pathol. 2007;170(4):1136–1147. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Deshmukh V., Motwani R., Kumar A., Kumari C., Raza K. Histopathological observations in COVID-19: a systematic review. J Clin Pathol. 2021;74(2):76–83. doi: 10.1136/jclinpath-2020-206995. [DOI] [PubMed] [Google Scholar]
- 118.Centers for Disease Control and Prevention. SARS | Guidance | Infection Control | Care of Patients at Home, https://www.cdc.gov/sars/guidance/i-infection/patients-home.html; [accessed 24 March 2021].
- 119.World Health Organization. Considerations for quarantine of individuals in the context of containment for coronavirus disease (COVID-19). Interim guidance, https://www.who.int/publications/i/item/considerations-for-quarantine-of-individuals-in-the-context-of-containment-for-coronavirus-disease-(covid-19); [accessed 24 March 2021].
- 120.European Centre for Disease Prevention and Control. Guidance for discharge and ending isolation in the context of widespread community transmission of COVID-19–first update, https://www.ecdc.europa.eu/en/publications-data/covid-19-guidance-discharge-and-ending-isolation; [accessed 24 March 2021].
- 121.National Institute of Health. Therapeutic Management of Adults With COVID-19, https://www.covid19treatmentguidelines.nih.gov/therapeutic-management/; [accessed 5 April 2021].
- 122.Medscape. Severe Acute Respiratory Syndrome (SARS) Treatment & Management, https://emedicine.medscape.com/article/237755-treatment#d10; [accessed 5 April 2021].
- 123.Meng L., Qiu H., Wan L., Ai Y., Xue Z., Guo Q., et al. Intubation and Ventilation amid the COVID-19 OutbreakWuhan’s Experience. Anesthesiology. 2020;132(6):1317–1332. doi: 10.1097/ALN0000000000003296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cheung T.M., Yam L.Y., So L.K., Lau A.C., Poon E., Kong B.M., et al. Effectiveness of noninvasive positive pressure ventilation in the treatment of acute respiratory failure in severe acute respiratory syndrome. Chest. 2004;126(3):845–850. doi: 10.1378/chest.126.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ford N., Vitoria M., Rangaraj A., Norris S.L., Calmy A., Doherty M. Systematic review of the efficacy and safety of antiretroviral drugs against SARS MERS or COVID-19: initial assessment. J Int AIDS Soc. 2020;23(4):e25489. doi: 10.1002/jia2.25489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.National Institutes of Health. Antithrombotic Therapy in Patients with COVID-19, https://www.covid19treatmentguidelines.nih.gov/adjunctive-therapy/antithrombotic-therapy/; [accessed 24 March 2021].
- 127.Atallah B., Mallah S.I., Almahmeed W. Anticoagulation in COVID-19. EHJ-CVP. 2020;6(4):260–261. doi: 10.1093/ehjcvp/pvaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.European Society of Cardiology. ESC guidance for the diagnosis and management of CV disease during the COVID-19 pandemic. ESOCJ, https://www.escardio.org/Education/COVID-19-and-Cardiology/ESC-COVID-19-Guidance; [accessed 24 March 2021].
- 129.Cuker A., Tseng E.K., Nieuwlaat R., Angchaisuksiri P., Blair C., Dane K., et al. American Society of Hematology 2021 guidelines on the use of anticoagulation for thromboprophylaxis in patients with COVID-19. Blood advances. 2021;5(3):872–888. doi: 10.1182/bloodadvances.2020003763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.National Institute of Health. NIH ACTIV Trial of blood thinners pauses enrollment of critically ill COVID-19 patients, https://www.nih.gov/news-events/news-releases/nih-activ-trial-blood-thinners-pauses-enrollment-critically-ill-covid-19-patients; [accessed 5 April 2021].
- 131.Fujii T., Nakamura T., Iwamoto A. Current concepts in SARS treatment. J Infect Chemother. 2004;10(1):1–7. doi: 10.1007/s10156-003-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Clancy C.J., Nguyen M.H. COVID-19, superinfections and antimicrobial development: What can we expect? Clin. Infect Dis. 2020;71(10):2736–2743. doi: 10.1093/cid/ciaa524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem. 2020;21(5):730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Warren T., Jordan R., Lo M., Soloveva V., Ray A., Bannister R., et al. Nucleotide prodrug GS-5734 is a broad-spectrum filovirus inhibitor that provides complete therapeutic protection against the development of Ebola virus disease (EVD) in infected non-human primates. Open Forum Infect Dis. 2015;2(suppl_1) doi: 10.1093/ofid/ofv130.02. LB-2. [DOI] [Google Scholar]
- 135.Agostini M.L., Andres E.L., Sims A.C., Sheahan T.P., Lu X., Smith E.C. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9(2) doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., et al. Remdesivir for the treatment of Covid-19—preliminary report. N Engl J Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Biot C., Daher W., Chavain N., Fandeur T., Khalife J., Dive D., et al. Design and synthesis of hydroxyferroquine derivatives with antimalarial and antiviral activities. J. Med. Chem. 2006;49(9):2845–2849. doi: 10.1021/jm0601856. [DOI] [PubMed] [Google Scholar]
- 138.U.S. Food & Drug Administration. Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Chloroquine and Hydroxychloroquine, https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-chloroquine-and; [accessed 24 March 2021].
- 139.Russell C.D., Millar J.E., Baillie J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395(10223):473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Group R.C., Horby P., Lim W.S. Dexamethasone in hospitalized patients with COVID-19: preliminary report. NEJM. 2020;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.National Institute of Health. The COVID-19 Treatment Guidelines Panel's Statement on the Use of Tocilizumab for the Treatment of COVID-19, https://www.covid19treatmentguidelines.nih.gov/statement-on-tocilizumab/; [accessed 5 April 2021].
- 142.National Institute of Health. The COVID-19 Treatment Guidelines Panel's Statement on the Emergency Use Authorization of the Bamlanivimab Plus Etesevimab Combination for the Treatment of COVID-19 https://www.covid19treatmentguidelines.nih.gov/statement-on-bamlanivimab-plus-etesevimab-eua/; [accessed 5 April 2021].
- 143.Elshabrawy H.A., Coughlin M.M., Baker S.C., Prabhakar B.S. Human monoclonal antibodies against highly conserved HR1 and HR2 domains of the SARS-CoV spike protein are more broadly neutralizing. PloS one. 2012;7(11):e50366. doi: 10.1371/journal.pone.0050366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stockman L.J., Bellamy R., Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3(9):e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.U.S. Food & Drug Administration. Recommendations for investigational COVID-19 convalescent plasma, https://www.fda.gov/vaccines-blood-biologics/investigational-new-drug-ind-or-device-exemption-ide-process-cber/recommendations-investigational-covid-19-convalescent-plasma; [accessed 24 March 2021].
- 146.Yuchun N., Guangwen W., Xuanling S., Hong Z., Yan Q., Zhongping H., et al. Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J Infect Dis. 2004;190(6):1119. doi: 10.1086/423286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Marshall E., Enserink M. Caution urged on SARS vaccines. AAAS. 2004;303(5660):944–946. doi: 10.1126/science.303.5660.944. [DOI] [PubMed] [Google Scholar]
- 148.He Y., Zhou Y., Siddiqui P., Jiang S. Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem Biophys Res Commun. 2004;325(2):445–452. doi: 10.1016/j.bbrc.2004.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Takasuka N., Fujii H., Takahashi Y., Kasai M., Morikawa S., Itamura S., et al. A subcutaneously injected UV-inactivated SARS coronavirus vaccine elicits systemic humoral immunity in mice. Int Immunol. 2004;16(10):1423–1430. doi: 10.1093/intimm/dxh143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.tang L., Zhu Q., Qin E., Yu M., Ding Z., Shi H., et al. Inactivated SARS-CoV vaccine prepared from whole virus induces a high level of neutralizing antibodies in BALB/c mice. DNA Cell Biol. 2004;23(6):391–394. doi: 10.1089/104454904323145272. [DOI] [PubMed] [Google Scholar]
- 151.Xiong S., Wang Y.-F., Zhang M.-Y., Liu X.J., Zhang C.H., Liu S.S., et al. Immunogenicity of SARS inactivated vaccine in BALB/c mice. Immunol Lett. 2004;95(2):139–143. doi: 10.1016/j.imlet.2004.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang D., Lu J. Glycan arrays lead to the discovery of autoimmunogenic activity of SARS-CoV. Physiol Genomics. 2004;18(2):245–248. doi: 10.1152/physiolgenomics.00102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.World Health Organization. Draft landscape of COVID-19 candidate vaccines. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines [accessed 24.03.21].
- 154.World Health Organization. List of candidate vaccines developed against SARS-CoV. https://www.who.int/blueprint/priority-diseases/key-action/list-of-candidate-vaccines-developed-against-sars.pdf?ua=1 [accessed 24.03.21].
- 155.Callaway E. The race for coronavirus vaccines: a graphical guide. Nature. 2020;580(7805):576. doi: 10.1038/d41586-020-01221-y. [DOI] [PubMed] [Google Scholar]
- 156.O’neill L.A., Netea M. BCG-induced trained immunity: can it offer protection against COVID-19? Nat Rev Immunol. 2020;20(6):335–337. doi: 10.1038/s41577-020-0337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect. 2020;104(3):246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Centers for Disease Control and Prevention. Considerations for wearing cloth face coverings. https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/cloth-face-cover-guidance.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fprevent-getting-sick%2Fcloth-face-cover.html [accessed 24.03.21].
- 159.Mahase E. Covid-19: UK starts social distancing after new model points to 260 000 potential deaths. BMJ. 2020;368:m1089. doi: 10.1136/bmj.m1089. [DOI] [PubMed] [Google Scholar]
- 160.Mayr V., Nußbaumer-Streit B., Gartlehner G. Quarantine alone or in combination with other public health measures to control COVID-19: a rapid review. Cochrane Database Syst Rev. 2020;82(6):501–506. doi: 10.1055/a-1164-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Koo J.R., Cook A.R., Park M., Sun Y., Sun H., Lim J.T., et al. Interventions to mitigate early spread of SARS-CoV-2 in Singapore: a modelling study. Lancet Infect Dis. 2020;20(6):678–688. doi: 10.1016/S1473-3099(20)30162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gostin L.O., Wiley L.F. Governmental public health powers during the COVID-19 pandemic: stay-at-home orders, business closures, and travel restrictions. JAMA. 2020;323(21):2137–2138. doi: 10.1001/jama.2020.5460. [DOI] [PubMed] [Google Scholar]
- 163.World Health Organization. Updated WHO recommendations for international traffic in relation to COVID-19 outbreak. https://www.who.int/news-room/articles-detail/updated-who-recommendations-for-international-traffic-in-relation-to-covid-19-outbreak/ [accessed 24.03.21].
- 164.Cetron M., Maloney S., Koppaka R., Simone P. In: Isolation and quarantine: containment strategies for SARS. In et al. Knobler S., Mahmoud A., Lemon S., editors. National Academies Press (US); Institute of Medicine (US) Forum on Microbial Threats Washington DC: 2004. [Google Scholar]
- 165.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early transmission dynamics in Wuhan China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lee N., Sung J.J. Nosocomial transmission of SARS. Curr Infect Dis Rep. 2003;5(6):473–476. doi: 10.1007/s11908-003-0089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.World Health Organization. Infection prevention and control during health care when novel coronavirus (nCoV) infection is suspected. https://www.who.int/publications/i/item/infection-prevention-and-control-during-health-care-when-novel-coronavirus-(ncov)-infection-is-suspected-20200125 [accessed 24.03.21].
- 168.World Health Organization. Hospital infection control guidance for Severe Acute Respiratory Syndrome (SARS). https://www.who.int/ihr/lyon/surveillance/infectioncontrol/en/ [accessed 24.03.21].
- 169.Yang Y., Wang H., Chen K., Zhou J., Deng S., Wang Y. Shelter hospital mode: how do we prevent COVID-19 hospital-acquired infection? Infect Control Hosp Epidemiol. 2020;41(7):872–873. doi: 10.1017/ice.2020.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fang D., Pan S., Li Z., Yuan T., Jiang B., Gan D., et al. Large-scale public venues as medical emergency sites in disasters: lessons from COVID-19 and the use of Fangcang shelter hospitals in Wuhan China. BMJ Global Health. 2020;5(6):e002815. doi: 10.1136/bmjgh-2020-002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Peeri N.C., Shrestha N., Rahman M.S., Zaki R., Tan Z., Bibi S., et al. The SARS MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int J Epidemiol. 2020;49(3):717–726. doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Martin-Delgado J., Viteri E., Mula A., Serpa P., Pacheco G., Prada D., et al. Availability of personal protective equipment and diagnostic and treatment facilities for healthcare workers involved in COVID-19 care: A cross-sectional study in Brazil Colombia, and Ecuador. PLoS One. 2020;15(11):e0242185. doi: 10.1371/journal.pone.0242185. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Popescu S. Roadblocks to infection prevention efforts in health care: SARS-CoV-2/COVID-19 response. Disaster Med Public Health Prep. 2020;14(4):538–540. doi: 10.1017/dmp.2020.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rodríguez P., Graña S., Alvarez-León E.E., Battaglini M., Darias F.J., Hernán M.A., et al. A population-based controlled experiment assessing the epidemiological impact of digital contact tracing. Nat Commun. 2021;26(1):1–6. doi: 10.1038/s41467-020-20817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]