Abstract
Reports of the novel and more contagious strains of SARS-CoV-2 originating in different countries have further aggravated the pandemic situation. The recent substitutions in spike protein may be critical for the virus to evade the host's immune system and therapeutics that have already been developed. Thus, this study has employed an immunoinformatics pipeline to target the spike protein of this novel strain to construct an immunogenic epitope (CTL, HTL, and B cell) vaccine against the new variant. Our investigation revealed that 12 different epitopes imparted a critical role in immune response induction. This was validated by an exploration of physiochemical properties and experimental feasibility. In silico and host immune simulation confirmed the expression and induction of both primary and secondary immune factors such as IL, cytokines, and antibodies. The current study warrants further lab experiments to demonstrate its efficacy and safety.
Keywords: SARS-CoV-2, New variant, Vaccine designing, In silico cloning, Immune simulation
Graphical abstract
1. Introduction
A novel SARS-CoV-2 was reported as the etiological agent of COVID-19 in December 2019 and transmitted around the world with unprecedented speed. Due to this prolific spread, a pandemic was declared by the World Health Organization. The pathogen and the disease it causes have devastated the world's health systems and profoundly affected many countries' economies. As such, SARS-CoV-2 has not only jeopardized prevailing health systems but also human safety [1]. It has also placed a substantial economic burden on the global population due to lack of job availability as a result of social distancing. The latest updates, as of April 12, 2021, reported 136 million infected persons and 2,930,000 deaths. Symptoms such as coughing, fever, shortness of breath, myalgia, and dyspnea are associated with COVID-19 patients, although some asymptomatic cases have also surfaced [[2], [3], [4]].
Within the subfamily, Orthocoronavirinae of the family Coronaviridae forms four different genera (α, β, γ, δ), which accommodate many types of coronaviruses (CoV) [5,6]. Mammals (α and β) and birds (γ and δ) are hosts of CoV [7,8], with β-CoVs responsible for the 2003, 2012, and 2019 epidemics. The decedents of β-CoVs poses a serious threat to human safety and also highlights the vulnerability of existing medical systems to outbreaks from them [9,10]. These coronaviruses are particularly dangerous because they transmit from animals to humans and from humans to humans [11]. Case fatality ratios of 10%, 35%, and 5% have been reported for SARS-CoV, MERS, and SARS-CoV-2, respectively [12]. The continuous spread of SARS-CoV-2 and the emergence of novel, more contagious strains have exposed more people to this deadly pathogen. Methods such as the development of vaccines and novel therapies have been adopted to control COVID-19 [13,14].
The SARS-CoV-2 genome (30 kb) consists of regions that encode structural, nonstructural, and accessory proteins [15]. The envelope, spike protein (S protein), nucleocapsid, and membrane proteins are the structural proteins [16]. Unlike other coronaviruses, SARS-CoV and SARS-CoV-2 encode an additional spike protein that has similarities with acetyl esterase and hemagglutination [17]. The SARS-CoV-2 virus infects host cells by binding to the host's cellular ACE2 receptors using spike protein positioned on the surface of the virus that is composed of S1 and S2 subunits, of which the receptor-binding domain (RBD) of the S1 subunit facilitates binding to ACE2, whereas S2 is involved in membrane fusion. After the membranes fuse, the viral genome enters into the cytoplasm [18].
The spike protein is multifunctional, and the study conducted by Walls et al. addressed its structure, function, and immunogenicity [19]. The main focus of anti-CoV therapies is to block the interaction of the virus with receptors and stop replication [20]. The SARS virus binds to the ACE2 receptor [21,22]. The binding of the RBD in the spike protein with the ACE2 receptor triggers the host immune system to produce antibodies targeting the RBD, leading to immunization. As such, immunization targeting the RBD would be instrumental in blocking the binding of the virus to the host cell and thus controlling viral invasion and infection. Therefore, the RBD is a significant target in the design of anti-CoV therapies [18]. The RBD of the SARS-CoV-2 virus has 75% structural identity and 83% sequence similarity with SARS-CoV. However, 59% similarity has been reported among the residues present at the receptor-binding motif (RBM) in the RBD (26). The substitution of critical residues in the RBD of SARS-CoV-2 has played a vital role in spreading COVID-19. Moreover, the sequence disparity in RBM between SARS-CoV and SARS-CoV-2 has generated different antigenicity and antibodies against the RBD that are not effective against the other [19]. For instance, TLR4 has been reported to be strongly associated with the pathophysiology of COVID-19. Robust interaction of the spike protein and TLR4 has been reported, and blocking TLR4 has reduced the viral infection load hence suggest a possible mechanism in which the virus may be binding to and activating TLR4 to increase expression of ACE2, which promotes viral entry [[23], [24], [25], [26]]. This is possible due to the strongest binding of spike protein with TLR4 which increases the expression of ACE2 receptor thus facilitating SARS-CoV-2 entry. Consequently, it is important to design spike protein based antigenic vaccine that is recognized by TLR4 and hence curtail the binding of viral spike protein to maintain the optimal ACE2 expression. In this regard, blocking of TLR4 would reduce the viral infection as reported previously [27].
Reports of the novel and more contagious strains of SARS-CoV-2 in England and other countries, amid a second wave of the virus in the country in December 2020, aggravated the situation further. Mutations of the novel strains may escape the host's immune system so that vaccines that have already been developed may become useless [28]. Therefore, in the present study, an immunoinformatics approach was employed to target the spike protein of this novel strain to construct an immunogenic epitope (CTL, HTL, and B cell) vaccine using the spike protein from SARS-CoV-2. This has been the most widely used approach for designing vaccines against several other pathogens and also against earlier strains of SARS-CoV-2 [[29], [30], [31], [32], [33]]. Furthermore, molecular docking studies were conducted to dock a vaccine construct with TLR4 receptor and check the stability of complex through molecular dynamics (MD) simulations. These efforts were made to search for and develop an effective vaccine against COVID-19 caused by the novel strain of SARS-CoV-2.
2. Methodology
2.1. Data retrieval and variants modeling
The spike protein sequence of the novel SARS-CoV-2 strain was obtained for this study from UniProt (https://www.uniprot.org/) under the accession number P0DTC2 [34]. The latest reported sequence was retrieved and checked for new mutations and deleted regions. Finally, the sequence was submitted to the SWISS-MODEL server (https://swissmodel.expasy.org/) for variant modelling [35]. A comparative modelling approach was used to model the structure using the wild structure of the spike protein. The structure of the wild type spike protein was retrieved from RCSB (https://www.rcsb.org/) using accession ID (6VSB) and superimposed on the mutant modelled spike protein to compare the RMSD differences [36]. The antigenicity of the wild type spike protein and the new variants spike protein was predicted and compared prior to further analyses.
2.2. Data processing
2.2.1. Prediction of immunogenic peptide vaccine sequences
Spike protein sequence was used to obtain highly immunogenic and antigenic epitopes to design a novel antigenic vaccine candidate against the novel strains. To achieve the desired objectives, cytotoxic T lymphocyte (CTL) epitopes were predicted with the help of NetCTL 1.2 (http://www.cbs.dtu.dk/services/NetCTL/) based on a combined score [37]. The cut-off value used to predict CTL epitopes was set at 0.75. Helper T lymphocyte (HTL) epitopes (15mer) were obtained by IEDB server (http://tools.iedb.org/mhcii/) that showed good affinity for human MHC molecules (HLA-DRB1*01:02, HLA-DRB1*01:01, HLA-DRB1*01:04, HLA-DRB1*01:03, HLA-DRB1*01:05) [38]. IC50 score was used for sorting the HTL epitopes and those with IC50 value < 50 nM were considered good binders. Percentile ranking is inversely proportional to epitopes’ binding affinity and implies that a lower percentile rank would depict higher binding affinity. B cell epitopes are key in generating protective host antibody response. ABCPred (http://crdd.osdd.net/raghava/abcpred/) was used to predict linear B cell epitopes [39]. To filter the best predictions of these, a cut off score of 0.8 was defined in the process. To select the best combination of epitopes that passes all experimental principles, multiple analyses including toxicity, virulence, solubility, synthesis, and purification index were determined. The toxicity and virulence of each peptide were checked through ToxinPred (http://crdd.osdd.net/raghava/toxinpred/) [40] and VirulentPred (http://bioinfo.icgeb.res.in/virulent/) [41], respectively. For solubility analysis, the CamSol Intrinsic (http://www-vendruscolo.ch.cam.ac.uk/camsolmethod.html) [42] method of Vendruscolo lab software was used, while Thermofisher Scientific-Peptide Synthesis and Prototypic Peptide Analyzing Tool was used for synthesis and purification of the peptides [43]. Together, the T cell, HTL, and B cell epitopes were filtered out further to construct the final multi-epitope vaccine candidate (MEVC).
2.2.2. Construction of multi-epitope peptide vaccine
The final vaccine candidate was composed of adjuvant, CTL, HTL, and B cell epitopes joined together by AAY, GPGPG, and KK linkers, respectively [44,45], while human beta defensin-2 (hBD-2) was used as an adjuvant at the N-terminal of the vaccine sequence to reinforce stability and enhance immunogenic response [46]. Human-defensin 2 plays a critical function in innate antiviral immunity and can potentiate antigen-specific immunity initiation. Previously it has been used experimentally (in vitro and in vivo) along with antigen designed from the spike protein of the Middle East respiratory syndrome-coronavirus (MERS-CoV). It was observed that when the hBD-2 was attached the host immune response was robustly higher than without hBD-2. Upon the injection several immune response factors such as IFN-β, TNF-α, IFN-γ, IL-6 and IL-1β were significantly higher. This shows that the closely related specie favourable increases the immune response when hBD-2 is attached. Hence, using it here would significantly trigger a robust immune response [47].
2.2.3. Physiochemical properties, structure prediction, and validation
The vaccine construct needed to be antigenic for eliciting the proper immune response. For this purpose, the VexiJen server (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) was employed to predict the vaccine's antigenicity while keeping the threshold at the default 0.4 [48]. Another important parameter, allergenicity, was predicted with the help of AlgPred server (http://crdd.osdd.net/raghava/algpred/) at an accuracy of around 85% [49]. Allergenic sequence can be identified when there is a score greater than the threshold (>0.4).
Physiochemical properties such as amino acid composition, molecular weight, theoretical pI, in vivo and in vitro half-life, instability index, aliphatic index, and grand average of hydropathicity (GRAVY) for experimental processing parameters were employed to verify the vaccine. Therefore, to unveil these properties for the vaccine construct, an online webserver, ProtParam (https://web.expasy.org/protparam/), was used [50].
The secondary and tertiary structure of the vaccine, was predicted using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/), a freely available online tool [51], and Robetta webserver (https://robetta.bakerlab.org/) [52]. Upon submission, the submitted sequences were deconstructed into recognized domains for a forecast structure. In the next step, de novo homology modelling was used to model 3D structure. The choice of process depends on the availability of templates in the database. If templates are matched, then comparative modelling is used; otherwise, de novo modelling would be performed.
Vaccine 3D structure was subjected to refinement using Galaxy Refine to improve the local and global structure quality of the vaccine [53]. During the process, the protein side-chain was reconstructed and repacked using CASP10 refinement and then relaxed through MD simulation. The quality of the vaccine 3D model was tested using online tools such as ERRAT [54] and ProSA-web (https://saves.mbi.ucla.edu/) [55]. The latter is commonly used in protein structure validation to investigate input 3D model quality, assessed by means of a quality score. In cases when the score of the input structure is within the range of experimentally determined structure scores, the submitted structure is most likely to have no errors. The problematic part of the 3D molecule can be viewed in the ProSA-web results, thus making it easy for users to improve the overall structure quality. ERRAT is mainly deployed to find and fix non-bonded interactions present in a given structure.
2.3. Validation of the final vaccine construct
2.3.1. Molecular docking of the peptide vaccines and Vaccine-TLR4
In order to check the interaction of the peptides considered for the construction of the final vaccine candidate, T cell epitopes were docked against their respective major histocompatibility complex (MHC) molecules. Based on the lowest percentile rank, HLA-A*01:01, HLA-A*26:01, HLA-A*01:01, and HLA-A*01:01 molecules were used for docking. The 3D structures of the peptides were modelled using PepFOLD3 webserver [56]. Furthermore, to dock the MEVC (multi-epitopes vaccine construct) with TLR4 (PDB ID: 3FXI), we used ClusPro webserver [57]. ClusPro uses the fast Fourier transform correlation approach to perform rigid-body docking. The server then predicts how global energy can be split into atomic contact energy, hydrogen bonding energy, and Van der Waals (vdW) interaction energy, both attractive and repulsive.
2.3.2. Molecular dynamics simulation of the Vaccine-TLR4 complex
The stability of the complex was checked by MD simulation performed on Amber20 [58] using FF19SB force field. The system solvation was performed in a rectangular water box with TIP3P water molecules, and the system was neutralized with the addition of counter ions [59]. Energy minimization protocol was used for the removal of bad clashes in the system. The steepest descent algorithm [60] and conjugate gradient algorithm were used for 6000 and 3000 cycles, respectively [61]. After heating up to 300K, the system was equilibrated at a constant pressure of 1 atm with weak restraint and then equilibrated without restraint. Finally, the production step was run for 50 ns. Long-range electrostatic interaction was treated with particle mesh Ewald algorithm [62] with a cut-off distance of 10.0 Å. SHAKE algorithm was used to treat a covalent bond [63]. The MD simulation production step was performed on PMEMD.CUDA and trajectories were processed using the Amber20 CPPTRAJ package [64].
2.3.3. Binding free energy calculations
To estimate the real binding energy of the MEVC and TLR4, the MMGBSA approach was used. This is the best method to estimate the real binding energy of different biological complexes such as protein-ligand, protein-protein, and protein-DNA/RNA [12,[65], [66], [67], [68], [69], [70]]. MMGBSA.py [71] script was used to estimate the total binding free energy of the MEVC-TLR4 complex. Each energy term, such as vdW, electrostatic, GB, and SA, was calculated as a part of the total binding energy.
2.3.4. In silico cloning and host immune simulation
Codon optimization, expression, and reverse translation were performed using the Java codon adaptation tool (JCat) [72]. Optimization is important for the expression of vaccine structure in a host, E. coli (strain K12). Rho-independent transcription, restriction cleavage sites, and prokaryotic ribosome binding site were optimized through the consideration of three extra options. JCat provided the output in terms of codon adaptation index (CAI) and %GC content in order to confirm a high level of protein expression. For cloning the final vaccine in E. coli pET-28a (+), vector modification N and C terminal with XhoI and NdeI restriction sites were performed, respectively. Finally, for expression, the prepared optimized sequence, along with the restriction sites, was ligated in the pET-28a (+) vector utilizing the SnapGene tool. C-ImmSim server was used to simulate the host immune system in response to the vaccine antigen [73]. The PSSM method was used to estimate immune system response against the antigen. This server is used as an agent-based modelling approach to understand the dynamics of the host immune system [73]. Production of various immune substances such as antibodies, interferon, and cytokines upon vaccine administration is estimated. Additionally, the webserver predicted Th1 and Th2 responses and plotted a Simpson Index D value at default parameters. All the parameters were used as default while stimulating the host immune system in response to the vaccine.
3. Results and discussion
3.1. Data retrieval and variants modeling
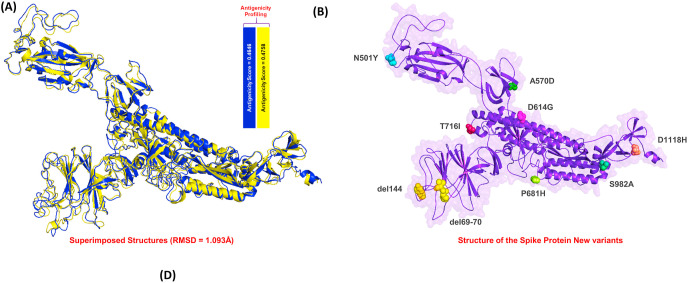
The new variants of SARS-CoV-2 that has emerged in the U.K. and other parts of the world have created an alarming situation. It consists of numerous mutations compared with the strain that emerged in Wuhan, China, at the end of 2019. The new strain consists of seven mutations presented on the spike protein (K17 N/T, E484K, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H) and three deletions (residues 69–70, 144) [74]. Furthermore, the spike protein mutations can further exacerbate SARS-CoV-2 infectivity and severity and potentially hinder the effectiveness of vaccines. Therefore, in the current study, computational modelling and integrated immunoinformatics were used to develop a MEVC to help in the development of a vaccine against the new variant.
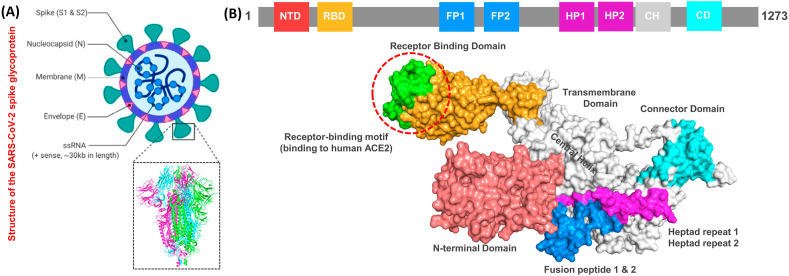
The spike protein is made up of RBD, N-terminal domain (NTD), Fusion peptide 1 and 2 (FP1 and FP2), hepated peptide 1 and 2 (HP1 and HP2), connector domain (CD), transmembrane, and central helix. The perfusion topology of the spike protein is depicted in Fig. 1 A, while Fig. 1B shows the mapping of the different domains of the spike protein. The new variant sequence of the spike protein was modelled using the SWISS-MODEL server.
Fig. 1.
The figure shows the general structure of the spike protein and its different domains. (A) represent the basic structure of the SARS-CoV-2 and the crystallographically solved structure of the spike protein. (B) shows the domain organization and different motifs present on the surface of the spike protein.
Superimposition of the wild type (WT) and the new variant structure revealed a1.093 Å RMSD difference between the two structures, indicating that residue deletion and mutations had induced some conformational changes required for the SARS-CoV-2 new variant's evolution (Fig. 2 A). Furthermore, the antigenicity analysis revealed a score of 0.4646 WT and 0.4758 for the new variant (Fig. 2B) indicates that the new variant is more antigenic than the WT, and thus presents the possibility of requiring alternate therapeutic opportunities. Therefore, the focus of this study was to design novel vaccine candidates from the spike protein of the new variants. This was used to map the antigenic peptides and join them with linkers to design a more potent MEVC.
Fig. 2.
The figure shows the superimposed structures and the reported mutations on the spike protein. (A) shows the wild type (Blue) and the new variant (deletions and mutations) (yellow) structures superimposed. The RMSD reported for these two structure was 1.093 Å while the antigenicity scale for the wild and mutant spike proteins is also given. (B) shows the representation of the deleted regions and mutated residues on the spike protein.
3.2. Immunogenic epitope prediction
Adaptive or acquired immune responses are exceedingly synchronized and systematic in removing and neutralization an invading pathogen [75]. The memory B cells created during acquired immunity will recognize the target pathogen on subsequent encounters after early identification. These memory cells memorize the antibodies against a distinct pathogen, which forms the basis of vaccination [76]. The adaptive immune system's B and T cells develop humoral and cell-mediated immunity against a particular pathogen [75]. In this study, the new variants' spike protein was used in the crucial examination of B and T cell epitope mapping.
For this variant, 42 B cell epitope targets were selected based on criteria of cut-off (≥0.5) and sequence length (≥16-mer). Of the 42 epitopes, 4 exhibited the highest score for vaccine development: TTRTQLPPAYTNSFTRGVYY19-39 (0.986), ALHRSYLTPGDSSSGWTAGA240-260 (0.888), NNKSWMESEFRVYSSANNCT145-165 (0.881), and DQLTPTWRVYSTGSNVFQTR624-644 (0.841) (Table 1 ). B cell epitope prediction is essential because the antibodies that bind to these could lead to the activation of five protection mechanisms, namely neutralization (obstructing bacterial adhesion potential to the mucosa), agglutination (reducing an infectious unit of the invader), complement activation (for cell demolition and infection), cell-mediated cytotoxicity directed by antibody coating (macrophages, natural killer cells and eosinophils causing the annihilation of the target cell appended by an antibody), and opsonization (to augment phagocytosis by tagging antigen with antibody to augment phagocytosis) [77].
Table 1.
Predicted and shortlisted T cell, HTL and B cell epitopes with their residues position, and the predicted scores and percentile ranks are also tabulated. The respective MHC molecules, allergenicity and antigenicity indexes are given.
| CTL Epitopes | |||||
|---|---|---|---|---|---|
| Molecules | Peptide Sequence | Peptide Position | Combined Score | Allergenicity | Antigenicity |
| HLA-A*01:01 | LTDEMIAQY | 862–870 | 3.66 | Non-allergenic | 0.675 |
| HLA-A*26:01 | WTAGAAAYY | 255–263 | 3.11 | Non-allergenic | 0.658 |
| HLA-A*01:01 | TSNQVAVLY | 601–608 | 3.07 | Non-allergenic | 0.667 |
| HLA-A*01:01 |
GAEHVNNSY |
649–657 |
1.99 |
Non-allergenic |
0.657 |
| HTL Epitopes | |||||
|
Molecules |
Peptide Sequence |
Peptide Position |
Percentile Rank |
Allergenicity |
Antigenicity |
| HLA-DRB-3*01:01 | HTPINLVRDLPQGFS | 207–221 | 0.51 | Non-allergenic | 0.617 |
| HLA-DRB-5*01:01 | GINITRFQTLLALHR | 232–246 | 0.52 | Non-allergenic | 0.664 |
| HLA-DRB-5*01:01 | TRFASVYAWNRKRIS | 345–359 | 0.52 | Non-allergenic | 0.666 |
| HLA-DRB-4*01:01 |
LQIPFAMQMAYRFNG |
894–908 |
0.73 |
Non-allergenic |
0.660 |
| B Cell Epitopes | |||||
|
Peptide Sequence |
Peptide Position |
Score |
Allergenicity |
Antigenicity |
|
| TTRTQLPPAYTNSFTRGVYY | 19–39 | 0.986 | Non-allergenic | 0.657 | |
| ALHRSYLTPGDSSSGWTAGA | 240–260 | 0.888 | Non-allergenic | 0.514 | |
| NNKSWMESEFRVYSSANNCT | 145–165 | 0.881 | Non-allergenic | 0.655 | |
| DQLTPTWRVYSTGSNVFQTR | 624–644 | 0.841 | Non-allergenic | 0.694 | |
Every B cell epitope helps in the mapping of T cell epitopes in order to predict MHC I and MHC II binding sites. The MHC I glycoproteins present on the surface of all nucleated cells, and the foremost function of the class I MHC gene product is to present the peptide antigens to cytotoxic T cells. The molecules present on the nucleated cell surface have a crucial function in presenting peptides from intracellular to cytotoxic T cells and exogenous proteins in order to generate a prompt immune reaction to completely eradicate the cell [78]. On the other hand, MHC I glycoproteins are expressed on antigen-presenting cells (APCs), such as B cells, monocytes, thyme epithelial cells, and dendritic cells [79]. The antigenic peptides are confined on the surface, regulating the helper T cells, restraining infection, conscripting phagocytes, and ultimately leading to full-force antibody reaction [80].
A total of 1242 T cell epitopes were predicted, among which 37 were MHC binders, while the rest were not. An allergenicity check was applied on these 37 epitopes using AlgPred to remove allergenic epitopes which could possibly cause autoimmune reactions [76]. This analysis revealed that only 11 epitopes were non-allergenic while the rest possessed allergenic properties. To further screen the remaining epitopes, percentile rank was predicted, and only the lowest percentile rank peptides were selected for the final vaccine construct. NetMHCpan EL 4.1 score was also considered for the final epitope selections, with the best binder having the highest score. Of the 11 epitopes, four were selected; of these, LTDEMIAQY862-870, which binds to the HLA-A*01:01 molecule, was antigenic and non-allergenic and had the strongest percentile rank score of 0.01. The NetMHCpan EL score for this peptide was predicted to be 0.997. Among the others. WTAGAAAYY255-263, with a combined score of 3.11, was also classified as antigenic and non-allergenic. This epitope significantly binds to the HLA-A*26:01 based on percentile rank, which was reported to be 0.03, while its predicted NetMHCpan EL value was forecast to be 0.847. The NetMHCpan EL score and the percentile rank for the peptide TSNQVAVLY601-608 (antigenic, non-allergenic) were reported to be 0.917 and 0.03, respectively. The HLA-A*01:01 was reported to be the target binder for this peptide. GAEHVNNSY649-657, which was classified as non-allergenic and antigenic, also binds to the HLA-A*01:01 molecule. The predicted NetMHCpan EL score was 0.698, while the percentile rank was predicted to be 0.1. Thus, the epitopes with the highest NetMHCpan EL score and lowest percentile rank strongly interacted with the MHC molecule to produce robust immune response; therefore, these four epitopes were selected for the final vaccine construct. The selected top T cell epitopes are given in Table 1.
On the other hand, for acquired or adaptive immunity, helper T cells are essential factors that not only help the B cell produce and release antibodies but also help the CTL destroy infected cells. Upon activation and presentation on the surface of APCs, they act as effectors and thus mature into a specific type of HTL required for a specific immune response [81]. Seeing this important role of HTL epitopes herein, we also predicted helper T cell epitopes that would aid the B and T cell epitopes to trigger a robust immune response. The HTL epitopes were mapped on the spike protein using IEDB. Multiple HTL epitopes were predicted against the given list of molecules. Following similar criteria, four HTL epitopes that were non-allergenic and in the lowest percentile rank were shortlisted. 15-mers HTL epitopes HTPINLVRDLPQGFS (percentile rank: 0.51) (HLA-DRB-3*01:01), GINITRFQTLLALHR (percentile rank: 0.52) (HLA-DRB-5*01:01), TRFASVYAWNRKRIS (percentile rank: 0.52) (HLA-DRB-5*01:01), and LQIPFAMQMAYRFNG (percentile rank: 0.76) (HLA-DRB-4*01:01) were classified as active and non-allergenic (Table 1).
The next vital step was to check the toxicity of the epitopes that had been predicted through ToxinPred. All the selected epitopes should be nontoxic in nature, so that when the vaccine construct enters the body, it will not initiate a toxic reaction. All the non-allergenic epitopes showed no toxicity, and their virulent nature was re-validated through VirulentPred. All epitopes that are used to design vaccine constructs should be virulent in nature so that the immune defense is triggered against them. All the non-allergenic and nontoxic epitopes were found to be virulent in nature. Another crucial step was to check the intrinsic solubility of the peptides using the CamSol Intrinsic method of Vendruscolo lab software. All of the peptides showing good solubility values were selected, and all the peptides showed positive solubility values. The solubility of the peptides is very important so that they can interact fully with the solvent. Further, the synthesis and purification of the peptides were checked using Thermo Fisher Scientific Peptide Synthesis and Prototypic Peptide Analyzing Tool [43]. All the peptides showing favourable intrinsic solubility values also showed easy synthesis and purification except for GAEHVNNSY, which had an acceptable stability problem. Thus, these analyses further confirmed the validity of our shortlisted epitopes for MEVC.
3.3. Construction of MEVC
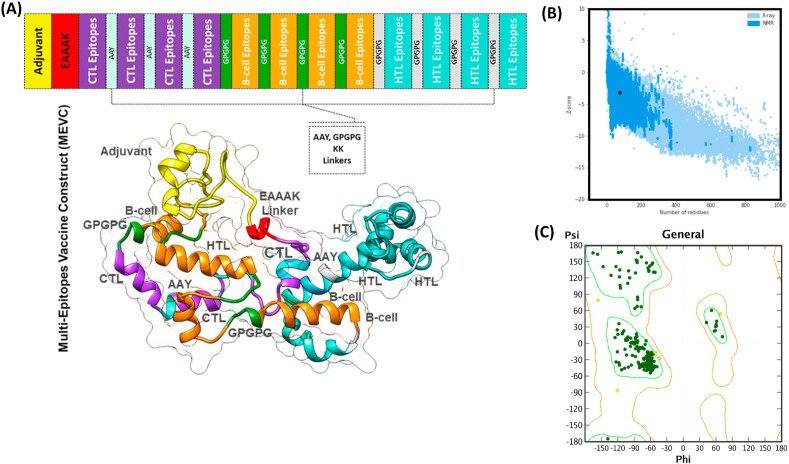
Mortality and morbidity owing to infectious diseases have been significantly reduced due to vaccines based on huge proteins and whole organisms [77]. However, due to the high antigenic burden of such vaccines, imprecise immunological responses are generated that are associated with reactogenic reactions [82]. The proposed peptide vaccines are suitable substitutes as they are cost-effective, can be easily produced, trigger specific immune responses, are easy to manufacture and use clinically, lower the risk of antigen-induced anaphylaxis, and are flexible to changes in antigen [118]. However, peptides cannot be used alone because they are feebly immunogenic and require appropriate adjuvants [77]. Thus, multi-epitope peptides containing overlying epitopes offer a solution to the weak immunogenicity of individual antigenic peptides [83]. Thus, a MEVC was designed from the shortlisted B cell, T cell, and HTL epitopes (Table 1) for a more robust immune response. Different types of linkers such as EAAK, GPGPG, AAY and KK were used to join these small peptides together and design a full-length MEVC. These linkers were used for constructing the MEVC because they help to facilitate the epitope display, hence allowing effectual immune processes [84]. Further, these linkers also help to keep the epitopes separate and prevent them from folding [85]. The addition of an adjuvant can further completely enhance the immunogenicity of a multi-epitope peptide vaccine [86]. Once the peptide was modelled, a nontoxic human beta defensin-2 (hBD-2) expressing in mammalian cells was attached at the N-terminal as an adjuvant, and EAAAK linker was also added. This adjuvant has the capability to express its own self and produce a robust immune response against the antigen attached to it [83]. A 263 amino acids long MEV was designed, and its antigenic and allergenic properties were calculated again to avoid any immune reaction in experimental conditions. The final MEVC was classified as a protective antigen with an antigenicity score of 0.449 and a non-allergenic with a score of −0.476. Hence, this systematic analysis confirmed that our designed vaccine construct is able to provoke an immune response without any allergenic reaction, and thus it should be considered for further validations and analyses. The topographical organization of the MEVC, including the adjuvant position, CTL, HTL, B cell epitopes, and the respective linkers, is given in Fig. (3A) . Furthermore, the 3D structure of the final vaccine construct is also shown in Fig. (3A), whereby each component of the vaccine is mapped and coloured differently.
Fig. 3.
(A) represent the topographic structure of the multi-epitopes vaccine construct (MEVC) and the 3D homology model of MEVC. Each component of the MEVC is coloured differently and tagged. (B) & (C) shows the ProSA-web and Ramachandran plot analysis results.
3.4. Analyzing physicochemical properties
ProtParam server determined the MW and pI of the MEVC to be 29.0 kDa and 10.04, respectively. The pI values of the model show that our candidate protein is acidic in nature which shows the suitability of the designed vaccine candidate. Charge distribution in the residues revealed that 11 residues were negatively charged, whereas 37 were positively charged. Similarly, the extinction coefficient of the protein in water was found to be 54695 M− 1 cm− 1 at 280 nm. In vitro analysis of the half-life of the vaccine construct in mammalian reticulocytes was found to be 30h, whereas in vivo analysis in yeast and E. coli revealed the half-life to be by 20h and >10h, respectively. The proposed protein was found to be stable, with an instability index of 29.66. The aliphatic index and GRAVY value of the proposed vaccine were determined to be 57.98 and −0.565, respectively, which shows that temperature changes may not lead to instability in the proposed protein. Similarly, the negative GRAVY value indicates that the protein is hydrophilic and can lead to improved interactions with the nearby water molecules.
3.5. MEVC structure modeling and validation
Robetta predicted the 3D model of the candidate MEVC using the comparative modelling approach. The native spike protein was used as a template for comparative modelling, followed by the selection of one of the predicted models based on a confidence score of 0.02. In order to select the best model, ProSA-web, ERRAT, and Ramachandran plot evaluations were performed (Fig. 3B & 3C). The final analysis revealed that a model with a confidence score of 0.02 predicted by Robetta would be the best model and subject to further analysis.
The refined structure was subjected to potential errors and quality of the best model using ProSA-web. The Z-score of the preliminary input model was found to be −4.51, which endorses the range commonly found in native proteins with similar size. The structure was further subjected to ERRAT validation, which revealed an overall quality factor of 92.70. In addition, a Ramachandran plot analysis was carried out, which revealed that 94.977%, 4.566%, and 0.457% of the residues in the primary model were present in the favoured, allowed, and outlier regions. Thus the modelled MEVC is acceptable for further processes, such as molecular docking, simulation, and free energy calculations.
3.6. Validation of the final vaccine construct
3.6.1. Molecular docking of the peptide vaccines and Vaccine-TLR4
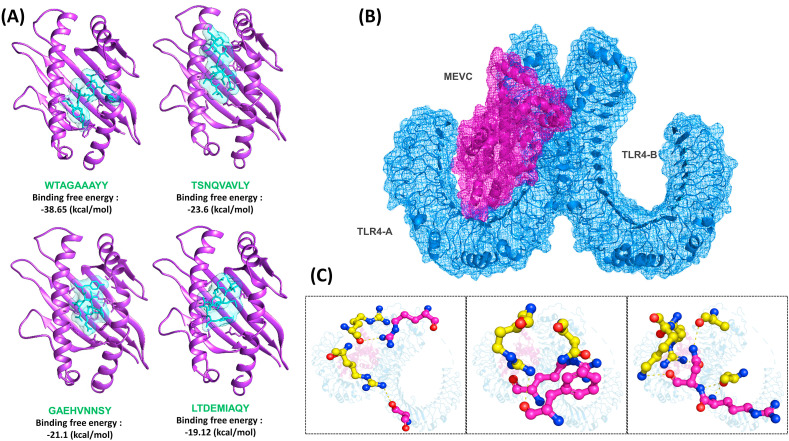
To stimulate a proper immune response, a vaccine should show a very good binding affinity with the host's immune receptors. To evaluate peptide-MHC binding, protein-peptide docking of the selected epitopes was carried out against the respective molecule. For interaction evaluation, binding energy was considered. The docking energy for LTDEMIAQY-HLA-A*01:01 was reported to be −2650.42 kcal/mol; for WTAGAAAYY-HLA-A*26:01, it was reported to be −2159.24 kcal/mol; for TSNQVAVLY-HLA-A*01:01, it was reported to be −2587.54 kcal/mol; while for GAEHVNNSY-HLA-A*01:01, the docking score was reported to be −2880.01 kcal/mol spike protein. The docked peptides against the specific molecule are shown in Fig. 4 A. Furthermore, after construction and refinement, the best model of MEV was docked against the TLR4 (Fig. 4B). TLR4 has been reported to be strongly associated with the pathophysiology of COVID-19. Robust interaction of the spike protein and TLR4 has been reported, and blocking TLR4 has reduced the viral infection load hence suggest a possible mechanism in which the virus may be binding to and activating TLR4 to increase expression of ACE2, which promotes viral entry [[23], [24], [25], [26]]. The protein-protein docking approach revealed a docking score of −256.02 kcal/mol. Since the TLR4 acts as a dimer, interactions with both the chains were evaluated. The MEVC formed 8 salt bridges with chain A of the TLR4 and 25 hydrogen bonds, while only three hydrogen bonds were reported with chain B. Among the hydrogen bonds formed by the MEVC were Glu603-Arg36, Glu605-Arg36, His456-Tyr10, Asp502-Arg12, Ser552-Arg38, Glu505-Lys8, Ser528-Lys8, Asn526-Lys8, Ser86-Arg142, Arg289-Ser94, Asp84-Arg142, Ser360-His101, Ser360-Arg108, Arg382-Arg108, Ser317-His90, Arg264-Ala85, Asn265-Lys152, Asn265-Als85, Asn265-Arg151, and Gln266-Arg151. On the other hand, chain B of TLR4 formed only three interactions with the MEVC. Among these, two bonds were formed by Glu439 and Ser438 with Gln112, while one bond was reported between Arg460 and Tyr10 of the vaccine (Fig. 4C). Overall, these results show that the MEVC significantly interacted with the immune receptor TLR4, and therefore would potentially activate the immune receptor.
Fig. 4.
Interaction of peptides vaccines with MHC molecules and MEVC with TLR4. (A) show the docked complexes of LTDEMIAQY-HLA-A*01:01, WTAGAAAYY-HLA-A*26:01, TSNQVAVLY-HLA-A*01:01 and GAEHVNNSY-HLA-A*01:01. (B) represent the interaction of key amino acids of the MEVC-TLR4 complex while (C) shows the specific interacting residues.
3.7. MD simulations
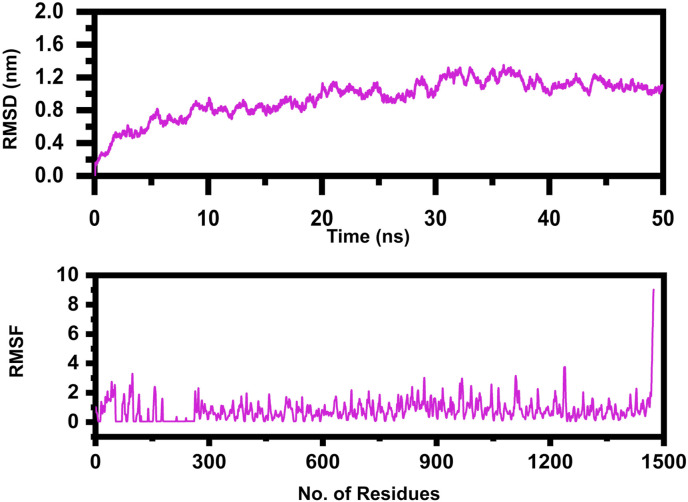
MD simulations for 50 ns of the MEVC-TLR4 complex were carried out to investigate system stability. There are different examples in which time-dependent MD simulations were applied on docked complexes to explore protein-ligand interactions, conformational fluctuations, structural and architectural changes, and dynamical shifts of the proteins. The MD simulations aid in understanding dynamic behavior of the complex and also highlight the critical residues that play a vital role in identifying and binding the ligand [87]. To shed light on biomolecular movements within a solvated environment, RMSD and RMSF were plotted as a function of time. Investigation of the MEVC-TLR4 complex led to an assessment of minor structural variations and atomic-level transitions. Deviation of the backbone Cα atoms was observed first for the complete production run. The average RMSD value calculated for the complex was 0.4 nm, with a maximum of 0.6 nm at 30th ns. No substantial structural movements were reported that elucidate complex stability. The average RMSF value for the complex was 2.0 Å. The regions illustrating higher fluctuation were loops that involved the regular conversion of sheets into helices and helices into sheets. The graph indicates that most of the residues of the active site remained stable. The highest peak of the graph indicates the loop region. The RMSD and RMSF graph is shown in Fig. 5 .
Fig. 5.
The figure represents the RMSD and RMSF of the MEVC-TLR4 complex after 50ns simulation.
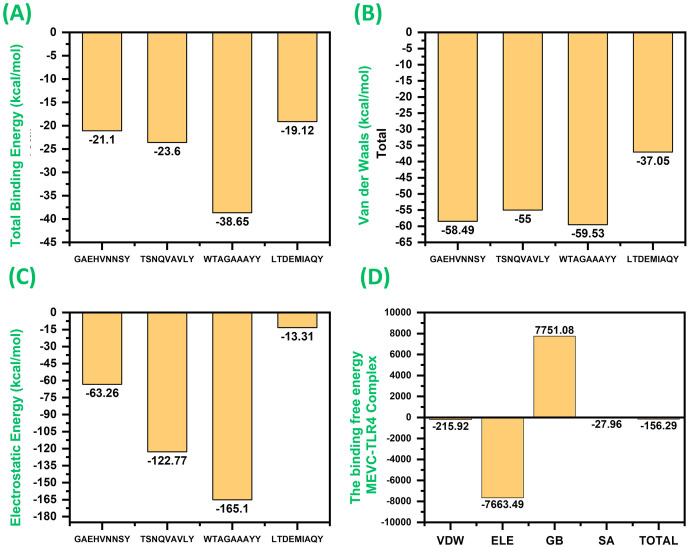
3.8. Binding free energy calculations
Amber20's MM-GBSA method, which combines molecular mechanical and continuum solvent approaches, was used to describe binding free energy of the system and molecular interactions within the TLR4-MHC. The entropy term is eliminated because of convergence problems in some cases, and it cannot be calculated. The formation of the complex led to highly favourable interactions, and the total binding energy for each complex revealed significant interaction energy. The total binding energy for each complex is given in Fig. 6 A. As given in Fig. 6, the total binding energy for LTDEMIAQY-HLA-A*01:01 was reported to be −21.1 kcal/mol; for WTAGAAAYY-HLA-A*26:01, ΔG was −23.6 kcal/mol; for TSNQVAVLY-HLA-A*01:01, ΔG was reported to be −38.65 kcal/mol; while for complex GAEHVNNSY-HLA-A*01:01, the total binding energy was −19.12 kcal/mol. The vdW energy for each of these complexes (Fig. 6B) was reported to be −58.49 kcal/mol, −55.0 kcal/mol, −59.53 kcal/mol, and −37.05 kcal/mol, respectively. The electrostatic contribution by each peptide-MHC complex was reported to be −63.26 kcal/mol, −122.77 kcal/mol, −165.1 kcal/mol, and −13.1 kcal/mol (Fig. 6C). In the case of the MEVC-TLR4 complex, the MM-GBSA results are shown in Fig. 6D. The total binding energy for the MEVC-TLR4 complex was reported to be −156.29 kcal/mol; the vdW was reported to be −215.92 kcal/mol, and the electrostatic energy was reported to be −7663.49 kcal/mol. These results show robust interaction of the peptides with MHC molecules and the MEVC-TLR4, which will significantly provoke the host immune response.
Fig. 6.
(A) represent the total binding energy of each LTDEMIAQY-HLA-A*01:01, WTAGAAAYY-HLA-A*26:01, TSNQVAVLY-HLA-A*01:01 and GAEHVNNSY-HLA-A*01:01 complex. (B) represent the vdW energy of each LTDEMIAQY-HLA-A*01:01, WTAGAAAYY-HLA-A*26:01, TSNQVAVLY-HLA-A*01:01 and GAEHVNNSY-HLA-A*01:01 complex. (C) represent the electrostatic energy of each LTDEMIAQY-HLA-A*01:01, WTAGAAAYY-HLA-A*26:01, TSNQVAVLY-HLA-A*01:01 and GAEHVNNSY-HLA-A*01:01 complex. (D) show the total binding energy, electrostatic, vdW, GB and SA for the MEVC-TLR4 complex.
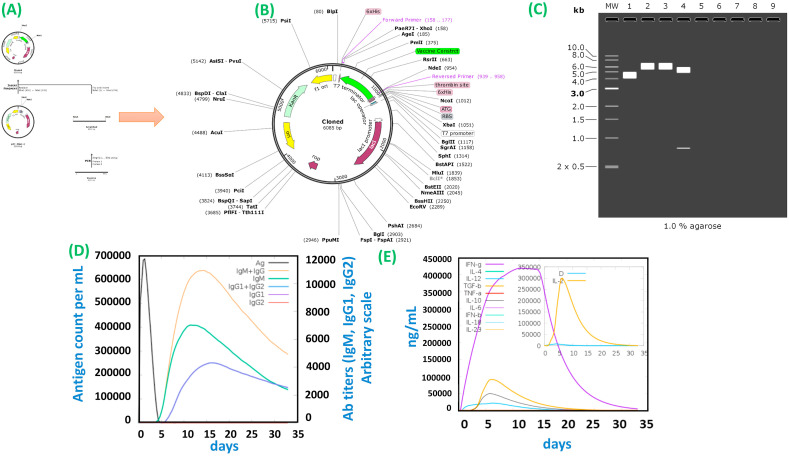
3.8.1. In silico Cloning and host immune simulation
For the maximum expression in the E. coli expression system, in silico cloning of the engineered construct was attained (Fig. 7 A and 7B). The CAI value of the vaccine construct came out to be 0.84, which is considered to be an ideal value. The vaccine construct has a GC content of 53.61%, suggesting the vaccine protein's high expression level in the E. coli system. To clone the vaccine gene for expression in E. coli into pET-28a (+) plasmid, XhoI and NdeI enzymes were selected, and restriction sites were inserted to the 3′ and 5′ ends of the sequence. The agarose gel electrophoresis to confirm the product is shown in Fig. 7C. The agarose 1% is an optimal experimental concentration of plasmids with a size more than 5 kB. Furthermore, host immune simulation was performed to determine the immune response upon the injection of the constructed antigen. Both secondary and primary immune responses appear to contribute significantly to the pathogens and possibly to the real immune response. The response of the in silico host immune system to the antigen is illustrated in Fig. 7D and 7E. High concentrations of IgM + IgG were described as a primary response, followed by IgM, IgG1 + IgG2, and IgG1 at the primary and secondary stages with antigen reduction concomitant. Strong cytokine and interleukin responses have also been observed. The IFN-gamma and IL-2 concentrations were significantly higher. All this demonstrates the effective immune response and acceptance of the MEVC after subsequent encounters.
Fig. 7.
(A) represent the in silico cloning of the MEVC in E. Coli. (B) The cloned MEVC construct. The CAI value of the vaccine construct came out to be 0.84, which is considered ideal value. The vaccine construct has a GC content equals to 53.61% suggesting the vaccine protein high expression level in the E. coli system. (C) 1% agarose gel electrophoresis of the MEVC and the vector. (D) (E) Show the immune response and host immune different factors secretion upon the injection of different doses at different intervals.
4. Conclusions
This research provided insight into vaccine design for a new variant of SARS-CoV-2 by taking advantage of immunoinformatics. The spike protein was screened to determine immune-dominant epitopes and subsequently develop an effective, safe, and highly specific MEVC. The proposed vaccine construct could offer long-lasting immunity to global populations against SARS-CoV-2. This study has opened the way for further experimental research of SARS-CoV-2 vaccine production. As the current research is based on an integrated computational pipeline, its only limitation is the requirement for further lab experiments to demonstrate the efficacy and safety of the proposed vaccine candidate.
Funding
Dong-Qing Wei is supported by grants from the Key Research Area Grant 2016YFA0501703 of the Ministry of Science and Technology of China, the National Science Foundation of China (Grant No. 32070662, 61832019, 32030063), the Science and Technology Commission of Shanghai Municipality (Grant No.: 19430750600), the Natural Science Foundation of Henan Province (162300410060), as well as SJTU JiRLMDS Joint Research Fund and Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2017ZD14). The computations were partially performed at the Pengcheng Lab. and the Center for High-Performance Computing, Shanghai Jiao Tong University.
Declaration of competing interest
Declared None.
Acknowledgments
The computations were partially performed at the Center for High-Performance Computing, Shanghai Jiao Tong University. We acknowledge their help.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., Liu L., Shan H., Lei C.-l., Hui D.S. MedRxiv; 2020. Clinical Characteristics of 2019 Novel Coronavirus Infection in China. [Google Scholar]
- 6.Weiss S.R., Leibowitz J.L. Advances in Virus Research. Elsevier; 2011. Coronavirus pathogenesis; pp. 85–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.-Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang Q., Song Y., Shi M., Cheng Y., Zhang W., Xia X.-Q. Inferring the hosts of coronavirus using dual statistical models based on nucleotide composition. Sci. Rep. 2015;5:17155. doi: 10.1038/srep17155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan A., Khan M., Saleem S., Babar Z., Ali A., Khan A.A., Sardar Z., Hamayun F., Ali S.S., Wei D.-Q. Phylogenetic analysis and structural perspectives of RNA-dependent RNA-polymerase inhibition from SARs-CoV-2 with natural products. Interdiscipl. Sci. Comput. Life Sci. 2020;12:335–348. doi: 10.1007/s12539-020-00381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan A., Ali S.S., Khan M.T., Saleem S., Ali A., Suleman M., Babar Z., Shafiq A., Khan M., Wei D.-Q. Combined drug repurposing and virtual screening strategies with molecular dynamics simulation identified potent inhibitors for SARS-CoV-2 main protease (3CLpro) J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1779128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koulgi S., Jani V., Uppuladinne M., Sonavane U., Nath A.K., Darbari H., Joshi R., Kumar A., Darbari H. ChemRxiv; 2020. Drug Repurposing Studies Targeting SARS-nCoV2: an Ensemble Docking Approach on Drug Target 3C-like Protease (3CLpro) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A. Khan, W. Heng, Y. Wang, J. Qiu, X. Wei, S. Peng, S. Saleem, M. Khan, S.S. Ali, D.-Q. Wei, In Silico and in Vitro Evaluation of Kaempferol as a Potential Inhibitor of the SARS-CoV-2 Main Protease (3CLpro), Phytotherapy research: PTR. [DOI] [PMC free article] [PubMed]
- 15.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Hu Y., Song Z.-G., Tao Z.-W., Tian J.-H., Pei Y.-Y. bioRxiv; China: 2020. Complete Genome Characterisation of a Novel Coronavirus Associated with Severe Human Respiratory Disease in Wuhan. [Google Scholar]
- 16.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 19.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Cell; 2020. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosch B.J., Van der Zee R., De Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS‐CoV‐2 spike glycoprotein with ACE‐2 receptor homologs and human TLRs. J. Med. Virol. 2020;92:2105–2113. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890. doi: 10.1016/j.chom.2020.04.017. e882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziegler C.G., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035. doi: 10.1016/j.cell.2020.04.035. e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandão S.C.S., Ramos J.d.O.X., Dompieri L.T., Godoi E.T.A.M., Figueiredo J.L., Sarinho E.S.C., Chelvanambi S., Aikawa M. Is Toll-like receptor 4 involved in the severity of COVID-19 pathology in patients with cardiometabolic comorbidities? Cytokine Growth Factor Rev. 2020:S1359–S6101. doi: 10.1016/j.cytogfr.2020.09.002. (1320)30205-30207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aboudounya M.M., Heads R.J. COVID-19 and toll-like receptor 4 (TLR4): SARS-CoV-2 may bind and activate TLR4 to increase ACE2 expression, facilitating entry and causing hyperinflammation. Mediat. Inflamm. 2021;2021:8874339. doi: 10.1155/2021/8874339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.-Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021 doi: 10.1002/jcp.30367. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D., Mai J., Zhou W., Yu W., Zhan Y., Wang N., Epstein N.D., Yang Y. Immunoinformatic analysis of T- and B-cell epitopes for SARS-CoV-2 vaccine design. Vaccines. 2020;8:355. doi: 10.3390/vaccines8030355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ismail S., Ahmad S., Azam S.S. Immunoinformatics characterization of SARS-CoV-2 spike glycoprotein for prioritization of epitope based multivalent peptide vaccine. J. Mol. Liq. 2020;314:113612. doi: 10.1016/j.molliq.2020.113612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C., Lee S.-S., Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J. Med. Virol. 2020;92:618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai Y., Chen H., Zhuang S., Feng X., Fang Y., Tang H., Dai R., Tang L., Liu J., Ma T., Zhong G. Immunodominant regions prediction of nucleocapsid protein for SARS-CoV-2 early diagnosis: a bioinformatics and immunoinformatics study. Pathog. Glob. Health. 2020;114:463–470. doi: 10.1080/20477724.2020.1838190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saha R., Ghosh P., Burra V.L.S.P. Designing a next generation multi-epitope based peptide vaccine candidate against SARS-CoV-2 using computational approaches, 3. Biotech. 2021;11:47. doi: 10.1007/s13205-020-02574-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magrane M. 2011. UniProt Knowledgebase: a Hub of Integrated Protein Data; p. 2011. Database. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwede T., Kopp J., Guex N., Peitsch M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larsen M.V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinf. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha S., Raghava G.P.S. Prediction of continuous B‐cell epitopes in an antigen using recurrent neural network. Proteins: Struct. Funct. Bioinf. 2006;65:40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 40.Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R., Raghava G.P. Peptide toxicity prediction. Comput. Peptidology. 2015:143–157. doi: 10.1007/978-1-4939-2285-7_7. Springer. [DOI] [PubMed] [Google Scholar]
- 41.Garg A., Gupta D. VirulentPred: a SVM based prediction method for virulent proteins in bacterial pathogens. BMC Bioinf. 2008;9:1–12. doi: 10.1186/1471-2105-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sormanni P., Vendruscolo M. Protein solubility predictions using the CamSol method in the study of protein homeostasis. Cold Spring Harb. Perspect. Biol. 2019;11:a033845. doi: 10.1101/cshperspect.a033845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scientific T.F. ThermoFisher Scientific; 2016. Thermo Fisher scientific.www.thermofisher.com [En línea]. Available: [Último acceso: 27 November 2018] [Google Scholar]
- 44.Negahdaripour M., Nezafat N., Eslami M., Ghoshoon M.B., Shoolian E., Najafipour S., Morowvat M.H., Dehshahri A., Erfani N., Ghasemi Y. Structural vaccinology considerations for in silico designing of a multi-epitope vaccine, Infection. Genet. Evol. 2018;58:96–109. doi: 10.1016/j.meegid.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 45.Parvizpour S., Pourseif M.M., Razmara J., Rafi M.A., Omidi Y. Epitope-based vaccine design: a comprehensive overview of bioinformatics approaches. Drug Discov. Today. 2020 doi: 10.1016/j.drudis.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 46.Schröder J.-M., Harder J. Human beta-defensin-2. Int. J. Biochem. Cell Biol. 1999;31:645–651. doi: 10.1016/s1357-2725(99)00013-8. [DOI] [PubMed] [Google Scholar]
- 47.Kim J., Yang Y.L., Jang S.-H., Jang Y.-S. Human β-defensin 2 plays a regulatory role in innate antiviral immunity and is capable of potentiating the induction of antigen-specific immunity. Virol. J. 2018;15:124. doi: 10.1186/s12985-018-1035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaharieva N., Dimitrov I., Flower D., Doytchinova I. Current computer-aided drug design; 2019. VaxiJen Dataset of Bacterial Immunogens: an Update. [DOI] [PubMed] [Google Scholar]
- 49.Saha S., Raghava G. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006;34:W202–W209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garg V.K., Avashthi H., Tiwari A., Jain P.A., Ramkete P.W., Kayastha A.M., Singh V.K. MFPPI–multi FASTA ProtParam interface. Bioinformation. 2016;12:74. doi: 10.6026/97320630012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 52.Chivian D., Kim D.E., Malmström L., Bradley P., Robertson T., Murphy P., Strauss C.E., Bonneau R., Rohl C.A., Baker D. Automated prediction of CASP‐5 structures using the Robetta server. Proteins: Struct. Funct. Bioinf. 2003;53:524–533. doi: 10.1002/prot.10529. [DOI] [PubMed] [Google Scholar]
- 53.Ko J., Park H., Heo L., Seok C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012;40:W294–W297. doi: 10.1093/nar/gks493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lengths M., Angles M. Limitations of structure evaluation tools errat. Quick Guideline Comput. Drug Des. 2018;16:75. [Google Scholar]
- 55.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamiable A., Thévenet P., Rey J., Vavrusa M., Derreumaux P., Tufféry P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016;44:W449–W454. doi: 10.1093/nar/gkw329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017;12:255. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salomon‐Ferrer R., Case D.A., Walker R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscipl. Rev.: Comput. Mol. Sci. 2013;3:198–210. [Google Scholar]
- 59.Price D.J., Brooks C.L., III A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004;121:10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 60.Meza J.C. Steepest descent. Wiley Interdiscipl. Rev.: Comput. Stat. 2010;2:719–722. [Google Scholar]
- 61.Watowich S.J., Meyer E.S., Hagstrom R., Josephs R. A stable, rapidly converging conjugate gradient method for energy minimization. J. Comput. Chem. 1988;9:650–661. [Google Scholar]
- 62.Salomon-Ferrer R., Götz A.W., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theor. Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 63.Kräutler V., Van Gunsteren W.F., Hünenberger P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001;22:501–508. [Google Scholar]
- 64.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 65.Khan A., Junaid M., Kaushik A.C., Ali A., Ali S.S., Mehmood A., Wei D.-Q. Computational identification, characterization and validation of potential antigenic peptide vaccines from hrHPVs E6 proteins using immunoinformatics and computational systems biology approaches. PloS One. 2018;13 doi: 10.1371/journal.pone.0196484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khan A., Junaid M., Li C.-D., Saleem S., Humayun F., Shamas S., Ali S.S., Babar Z., Wei D.-Q. Dynamics insights into the gain of flexibility by Helix-12 in ESR1 as a mechanism of resistance to drugs in breast cancer cell lines. Front. Mol. Biosci. 2020;6:159. doi: 10.3389/fmolb.2019.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ali A., Khan A., Kaushik A.C., Wang Y., Ali S.S., Junaid M., Saleem S., Cho W.C., Mao X., Wei D.-Q. Immunoinformatic and systems biology approaches to predict and validate peptide vaccines against Epstein–Barr virus (EBV) Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-018-37070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khan A., Khan M., Saleem S., Babar Z., Ali A., Khan A.A., Sardar Z., Hamayun F., Ali S.S., Wei D.-Q. Phylogenetic analysis and structural perspectives of RNA-dependent RNA-polymerase inhibition from SARs-CoV-2 with natural products. Interdiscipl. Sci. Comput. Life Sci. 2020:1–14. doi: 10.1007/s12539-020-00381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khan M.T., Khan A., Rehman A.U., Wang Y., Akhtar K., Malik S.I., Wei D.-Q. Structural and free energy landscape of novel mutations in ribosomal protein S1 (rpsA) associated with pyrazinamide resistance. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-44013-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khan A., Khan M.T., Saleem S., Junaid M., Ali A., Ali S.S., Khan M., Wei D.-Q. Structural Insights into the mechanism of RNA recognition by the N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein. Comput. Struct. Biotechnol. J. 2020 doi: 10.1016/j.csbj.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grote A., Hiller K., Scheer M., Münch R., Nörtemann B., Hempel D.C., Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rapin N., Lund O., Castiglione F. Immune system simulation online. Bioinformatics. 2011;27:2013–2014. doi: 10.1093/bioinformatics/btr335. [DOI] [PubMed] [Google Scholar]
- 74.Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir. Med. 2021;9:e20–e21. doi: 10.1016/S2213-2600(21)00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bonilla F.A., Oettgen H.C. Adaptive immunity. J. Allergy Clin. Immunol. 2010;125:S33–S40. doi: 10.1016/j.jaci.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 76.Natoli G., Ostuni R. Adaptation and memory in immune responses. Nat. Immunol. 2019;20:783–792. doi: 10.1038/s41590-019-0399-9. [DOI] [PubMed] [Google Scholar]
- 77.Sanchez-Trincado J.L., Gomez-Perosanz M., Reche P.A. Fundamentals and methods for T-and B-cell epitope prediction. J. Immunol. Res. 2017:2017. doi: 10.1155/2017/2680160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jomaa M., Yuste J., Paton J.C., Jones C., Dougan G., Brown J.S. Antibodies to the iron uptake ABC transporter lipoproteins PiaA and PiuA promote opsonophagocytosis of Streptococcus pneumoniae. Infect. Immun. 2005;73:6852–6859. doi: 10.1128/IAI.73.10.6852-6859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Speziale P., Rindi S., Pietrocola G. Antibody-based agents in the management of antibiotic-resistant Staphylococcus aureus diseases. Microorganisms. 2018;6:25. doi: 10.3390/microorganisms6010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rao V.S., Srinivas K., Sujini G., Kumar G. Protein-protein interaction detection: methods and analysis. Int. J. Proteonomics. 2014:2014. doi: 10.1155/2014/147648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alberts B., Johnson A., Lewis J., Morgan D., Raff M., Keith Roberts P.W. 2018. Molecular Biology of the Cell. [Google Scholar]
- 82.Roche P.A., Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 2015;15:203–216. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tian Y., da Silva Antunes R., Sidney J., Lindestam Arlehamn C.S., Grifoni A., Dhanda S.K., Paul S., Peters B., Weiskopf D., Sette A. A review on T cell epitopes identified using prediction and cell-mediated immune models for Mycobacterium tuberculosis and Bordetella pertussis. Front. Immunol. 2018;9:2778. doi: 10.3389/fimmu.2018.02778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reche P., Flower D.R., Fridkis-Hareli M., Hoshino Y. Peptide-based immunotherapeutics and vaccines 2017. J. Immunol. Res. 2018:2018. doi: 10.1155/2018/4568239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baldauf K.J., Royal J.M., Hamorsky K.T., Matoba N. Cholera toxin B: one subunit with many pharmaceutical applications. Toxins. 2015;7:974–996. doi: 10.3390/toxins7030974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nezafat N., Ghasemi Y., Javadi G., Khoshnoud M.J., Omidinia E. A novel multi-epitope peptide vaccine against cancer: an in silico approach. J. Theor. Biol. 2014;349:121–134. doi: 10.1016/j.jtbi.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 87.Maiorov V.N., Crippen G.M. 1994. Significance of Root-Mean-Square Deviation in Comparing Three-Dimensional Structures of Globular Proteins. [DOI] [PubMed] [Google Scholar]