Abstract
Objectives
Seroprevalence surveys provide crucial information on cumulative severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) exposure. This Slovenian nationwide population study is the first longitudinal 6-month serosurvey using probability-based samples across all age categories.
Methods
Each participant supplied two blood samples: 1316 samples in April 2020 (first round) and 1211 in October/November 2020 (second round). The first-round sera were tested using Euroimmun Anti-SARS-CoV-2 ELISA IgG (ELISA) and, because of uncertain estimates, were retested using Elecsys Anti-SARS-CoV-2 (Elecsys-N) and Elecsys Anti-SARS-CoV-2 S (Elecsys-S). The second-round sera were concomitantly tested using Elecsys-N/Elecsys-S.
Results
The populations of both rounds matched the overall population (n = 3000), with minor settlement type and age differences. The first-round seroprevalence corrected for the ELISA manufacturer's specificity was 2.78% (95% highest density interval [HDI] 1.81%–3.80%), corrected using pooled ELISA specificity calculated from published data 0.93% (95% CI 0.00%–2.65%), and based on Elecsys-N/Elecsys-S results 0.87% (95% HDI 0.40%–1.38%). The second-round unadjusted lower limit of seroprevalence on 11 November 2020 was 4.06% (95% HDI 2.97%–5.16%) and on 3 October 2020, unadjusted upper limit was 4.29% (95% HDI 3.18%–5.47%).
Conclusions
SARS-CoV-2 seroprevalence in Slovenia increased four-fold from late April to October/November 2020, mainly due to a devastating second wave. Significant logistic/methodological challenges accompanied both rounds. The main lessons learned were a need for caution when relying on manufacturer-generated assay evaluation data, the importance of multiple manufacturer-independent assay performance assessments, the need for concomitant use of highly-specific serological assays targeting different SARS-CoV-2 proteins in serosurveys conducted in low-prevalence settings or during epidemic exponential growth and the usefulness of a Bayesian approach for overcoming complex methodological challenges.
Keywords: Coronavirus disease 2019, Probability-based sample, Seroprevalence, Severe acute respiratory syndrome coronavirus 2
Introduction
The coronavirus disease 2019 (COVID-19) pandemic has already affected over 120 million people, with over 2.6 million COVID-19-related deaths as of 16 March 2021. Detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in nasopharyngeal swabs is a COVID-19 reference diagnostic standard [1,2]. However, because of a significant number of asymptomatic/mild infections, demanding implementation and scale-up of molecular testing and frequent changes in testing strategies, the SARS-CoV-2 RNA positives identified represent only the tip of the pandemic iceberg [[2], [3], [4]]. As a result, seroprevalence surveys measuring the population immune response to SARS-CoV-2 remain integral for understanding cumulative population exposure and insight into pandemic dynamics during the COVID-19 pre-vaccination era [4,5]. As far as we know, and as summarized in Table 1 and the Supplementary material (Table S6), all population-based seroprevalence surveys published in peer-reviewed literature until 31 January 2021 were conducted during or just following the pandemic's first wave [4,[6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17]]. The most recent systematic review and meta-analysis, also assessing non-peer-reviewed seroprevalence surveys deposited in open-access preprint repositories or posted at websites, showed a general lack of peer-reviewed population-based studies from much of the world and significant data heterogeneity, and called for longitudinal surveys to continually monitor seroprevalence around the globe [2].
Table 1.
SARS-CoV-2 population-based prevalence studies on a probability-based sample with results published in peer-reviewed literature until 31 January 2021
| Country | SARS-CoV-2 serology assay | Period of sampling | Study sample size (no. tested) | Age range (years) | Seroprevalence estimate (% seropositive, 95% CI) | Reference |
|---|---|---|---|---|---|---|
| Slovenia | Euroimmun ELISA; Roche Elecsys-N and Elecsys-S ECLIA |
20 April to 1 May (first round); 17 October to 10 November 2020 (second round) | 1316 (first round) 1211 (second round) |
0–99 | First round (April 2020): 0.87% (95% HDI 0.40%–1.38%); Second round (October/November 2020): unadjusted lower limit of seroprevalence on 11 November 2020: 4.06% (95% HDI 2.97%–5.16%) and unadjusted upper limit of seroprevalence on 3 October 2020: 4.29% (95% HDI 3.18%–5.47%) |
Maver and Oštrbenk Valenčak et al., 2020 [6] |
| Iceland | Roche Elecsys-N ECLIA; Wantai ELISA |
3 April to 8 July 2020 | 30 576: a) 1237 recovered b) 4222 quarantined c) 23 452 unknown exposure |
ND | Overall seroprevalence estimate for Iceland: 0.9% (95% CI 0.8%–0.9%) a) recovered persons: 91.1% (95% CI 89.4%–92.6%) b) quarantined persons: 2.3% (95% CI 1.9%–2.8%) c) persons with unknown exposure: 0.3% (95% CI 0.2%–0.4%) |
Gudbjartsson et al., 2020 [7] |
| Spain | Orient Gene Biotech POCT; Abbott IgG CLIA |
27 April to 11 May 2020 | 51 958 | <1 to >90 | POCT: 5.0% (95% CI 4.7%–5.4%); Abbott: 4.6% (95% CI 4.3%–5.0%), with a specificity–sensitivity range of 3.7% (95% CI 3.3%–4.0%; both tests positive) to 6.2% (95% CI 5.8%–6.6%; either test positive) | Pollán et al., 2020 [8] |
| USA (Indiana) | CLIA (not specifically defined) | 25–29 April 2020 | 3629 | ≥12 | 1.01% (95% CI 0.76%–1.45%); estimated overall population SARS-CoV-2 prevalence of active or current infection: 2.79% (95% CI 2.02%–3.70%) | Menachemi et al., 2020 [9] |
| USA (Los Angeles County, CA) | Premier Biotech POCT | 10–14 April 2020 | 863 | ≥18 | 4.06% (exact binomial CI, 2.84%–5.60%); adjusted unweighted and weighted seroprevalence: 4.34% (bootstrap CI, 2.76%–6.07%) and 4.65% (bootstrap CI, 2.52%–7.07%) | Sood et al., 2020 [10] |
| Switzerland (Geneva) | Euroimmun ELISA | 6 April to 9 May 2020 | 2766 (1339 households in 5 weeks) | ≥5 | 1st to 5th week, respectively: 4.8% (95% CI 2.4%–8.0%, n = 341); 8.5% (95% CI 5.9%–11.4%, n = 469); 10.9% (95% CI 7.9%–14.4%, n = 577); 6.6% (95% CI 4.3%–9.4%, n = 604); 10.8% (95% CI 8.2%–13.9%, n = 775) | Stringhini et al., 2020 [4] |
| India | Kavach ELISA; Euroimmun ELISA | 11 May to 4 June 2020 | 28 000 | ≥18 | 0.73% after adjusting for test performance (95% CI 0.34%–1.13%) | Muhrekar et al., 2020 [11] |
| Faroe Islands | Wantai ELISA | 27 April to 1 May 2020 | 1075 | 0–100 | 0.6% (exact binomial 95% CI 0.2%–1.2%); 0.7% (bootstrap 95% CI 0.3%–1.3%) after adjustment for test sensitivity and specificity |
Petersen et al., 2020 [12] |
| Brazil | Wondfo POCT | 14–21 May 2020; 4–7 June 2020 | 25 025 and 31 165 | ≥1 | Corrected prevalence estimates: 1.6% (95% CI 1.4%–1.8%) in the first survey and 2.8% (95% CI 2.5%–3.1%) in the second; city-level prevalence ranged from 0 to 25.4% in both surveys | Hallal et al., 2020 [13] |
| Iran | Pishtaz Teb ELISA | 17 April to 2 June 2020 | 3530 from the general population, 5372 from a high-risk group | ND | Overall population weight-adjusted and test performance-adjusted seroprevalence: 17.1% (95% CI 14.6%–19.5%); in the high-risk population: 20.0% (95% CI 18.5%–21.7%) | Poustchi et al., 2020 [14] |
| Netherlands | Laboratory-developed immunoassay | 31 March to 11 May 2020 | 3207 | 2–90 | Overall weighted seroprevalence: 2.8% (95% CI 2.1%–3.7%) | Vos et al., 2020 [15] |
| Italy (northeast region) | Abbott Architect CLIA | 5–15 May 2020 | 6075 | >10 | 23.1% (95% CI 22.0%–24.1%) | Stefanelli et al., 2020 [16] |
| China | Innovita colloidal gold detection kit | 6 March to 3 May 2020 | 63 107: Hubei 49 257; other 29 provinces 13 850 | 17–63 | All of China: 0.74%; Wuhan: 1.68%; Hubei province without Wuhan: 0.59%; other 29 provinces: 0.38% | Duan et al., 2021 [17] |
Abbreviations: CLIA, chemiluminescent microparticle immunoassay; ECLIA, electrochemiluminescence assay; ELISA, enzyme-linked immunosorbent assay; HDI, high-density interval; ND, no data; POCT, point-of-care test; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Non-peer-reviewed seroprevalence surveys with results deposited in open-access preprint repositories or posted at websites and those performed retrospectively on residual serum samples were not considered eligible. A more detailed summary of each study is provided in the Supplementary material (Table S6).
Here we present SARS-CoV-2 seroprevalence estimates for Slovenia assessed in the same participants in two rounds of a nationwide population study on a probability-based sample: at the end of the first epidemic wave (April 2020) and during exponential growth of the second epidemic wave (October/November 2020). Both study rounds were accompanied by significant logistical and methodological challenges. Although performed only 6 months apart, each round had a unique set of challenges, with several important lessons learned.
Materials and methods
Study design and population
The Slovenian nationwide population study is the first using a probability-based sample representative of the entire country and across all age categories, combined with a longitudinal follow up of the entire cohort over the next 6 months [6]. The study design and results of baseline SARS-CoV-2 RNA testing were detailed previously [6]. Briefly, a sample of 3000 residents was selected from the Central Population Register and PCR testing of nasopharyngeal swabs (n = 1366) performed in late April 2020 showed SARS-CoV-2 RNA prevalence of 0.15% (posterior mean 0.18%, 95% CI 0.03%–0.47%) [6].
All participants provided written informed consent before enrolment and for individuals younger than 18 years a parent or legal guardian provided consent. The Slovenian National Medical Ethics Committee (consent number 0120-199/2020/19) approved the study protocol. The study is registered with ClinicalTrials.gov (NCT04376996).
To assess SARS-CoV-2 seroprevalence, participants provided two blood samples: the first-round sample in April 2020 and the second-round sample in October/November 2020.
Serological testing
First-round sera were tested in early May 2020 using the enzyme-linked immunosorbent assay anti-SARS-CoV-2 ELISA IgG (ELISA; Euroimmun, Lübeck, Germany) targeting the S1-domain of the spike (S) protein of SARS-CoV-2. The manufacturer's evaluation for ELISA showed 99.6% specificity and 99.4% sensitivity, and our internal evaluation showed 100% specificity (see Supplementary material, item S2). ELISA specificity in other evaluations varied significantly (Supplementary material, item S3). Due to uncertain seroprevalence estimates based on ELISA results, anonymized first-round sera were retested in early November 2020 using two electrochemiluminescence double-antigen sandwich immunoassays concomitantly: Elecsys Anti-SARS-CoV-2 (Elecsys-N) and Elecsys Anti-SARS-CoV-2 S (Elecsys-S) (Roche Diagnostics, Mannheim, Germany) on a cobas e411 analyser following the manufacturer's instructions, using cut-off values for positive results of ≥1.0 and ≥ 0.8 U/mL, respectively. Second-round sera were tested in early November 2020 using the same Elecsys-N/Elecsys-S combined approach. Additionally, all second-round sera positive by either Elecsys-N or Elecsys-S as well as 50 anti-SARS-CoV-2-negative samples were tested further by neutralization test.
Elecsys-N is an assay for qualitative detection of total anti-SARS-CoV-2 antibodies against SARS-CoV-2 nucleoprotein (N). The manufacturer's extensive evaluation showed 99.80% specificity and 99.5% sensitivity, and manufacturer-independent evaluations even with values spanning manufacturer claims [3,5,[18], [19], [20], [21], [22], [23], [24]] (see Supplementary material, item S5).
Elecsys-S is an assay for quantitative detection of total anti-SARS-CoV-2 antibodies against the S1 receptor binding domain. The manufacturer's extensive evaluation showed 99.98% specificity and 98.8% sensitivity, and our internal unpublished Elecsys-N/Elecsys-S head-to-head comparison showed overall agreement of both assays of 98.68% [20] (see Supplementary material, item S5).
A neutralization test (NT) was performed in a BSL-3 laboratory using SARS-CoV-2 (strain Slovenia/SI-4265/20, D614G; EVA-GLOBAL-Ref-SKU: 005V-03961). NT was based on the standardized protocol [25]. Briefly, Vero E6 cells were seeded in a concentration of 105/well (96-well plate) 1 day before NT was performed. Triplicate serial dilutions of heat-inactivated plasma samples (56°C, 30 min) were incubated with 100 TCID50 of SARS-CoV-2 for 1 h at 37°C. The plasma–virus mixture was added to the cells and incubated at 37°C with 5% CO2 for 4 days. The neutralization end-point titre was determined as the end-point plasma dilution that inhibited the SARS-CoV-2-induced cytopathic effect. For the presence of neutralizing anti-SARS-CoV-2 antibodies the NT titre ≥1:20 was considered positive if inhibition occurred in at least two out of three parallels; and NT titre ≥1:10 was considered positive if inhibition occurred in all three parallels.
Statistical analysis
We used three different but related models to estimate seroprevalence, as detailed in the Supplementary material (item S1). Model 1 is the baseline model for estimating seroprevalence from test results where the assay's sensitivity or specificity is not 100%. Model 2 is an extension of Model 1, where two tests are performed simultaneously. Model 3 is an extension of Model 1 that allows for inclusion of more than one source of estimates of specificity. We also corrected the estimates for seroprevalence for non-response bias using post-stratification on age group, sex, region and settlement type (urban/rural, size). However, because no significant differences were observed between estimates, post-stratification results are not shown. To provide the most reliable seroprevalence assessment for the challenging second-round period, two estimates of seroprevalence for October/November 2020 were calculated: (a) an estimate of the lower bound of the seroprevalence at the end of the sample collection period and (b) an upper bound of seroprevalence 14 days before the first samples were collected (see Supplementary material, item S6).
The analysis was performed in R and we implemented the models in the Stan probabilistic programming language and used the built-in Hamiltonian Monte Carlo-based Markov Chain Monte Carlo sampler [26]. We set the number of sampling iterations to a number sufficient for the sampling-based approximation error to be lower than the number of decimal places reported. The results are reported as the posterior mean and a 95% highest density interval (95% HDI).
Results
From 1368 and 1245 participants in the first and second rounds, respectively, 1316 and 1211 blood samples were collected between 20 April and 1 May 2020 and 17 October and 10 November 2020, respectively. As detailed in the Supplementary material (item S7), the populations of both rounds matched the overall population well (n = 3000) without observed statistically significant mismatches in sex (p = 0.187 and p = 0.216) and region (p = 0.43 and p = 0.44). Minor differences were observed in settlement type, where individuals from a rural settlement with <2000 inhabitants were more likely to participate (p = 0.011), and in the age structure, where children in the age groups 0–10 years were less likely to be included and adults in the age group 51–60 years more likely (both p = 0.000).
As detailed in the Supplementary material (items S1 and S4), after testing 1316 blood samples, four different estimates of SARS-CoV-2 seroprevalence in Slovenia for late April 2020 were calculated: three based on ELISA results and one based on concomitant Elecsys-N/Elecsys-S testing results. Uncorrected crude seroprevalence based on ELISA results was estimated at 3.11% (posterior mean 3.15%, 95% HDI 2.23%–4.10%), seroprevalence corrected for the manufacturer's estimate of ELISA specificity at 2.78% (95% HDI 1.81%–3.80%) and seroprevalence corrected for the estimate of ELISA specificity based on our meta-analysis (see Supplementary material, item S3) at 0.93% (95% CI 0.00%–2.65%). The unadjusted lower limit of anti-SARS-CoV-2 seroprevalence estimate for the first round based on the Elecsys-N/Elecsys-S results corrected for the manufacturers' estimates of the specificities of both assays was 0.87% (95% HDI 0.40%–1.38%).
For the second round, based on Elecsys-N/Elecsys-S concomitant testing of 1211 blood samples and as detailed in the Supplementary material (item S6), the unadjusted lower limit of anti-SARS-CoV-2 seroprevalence in Slovenia on 11 November 2020 was 4.06% (95% HDI 2.97%–5.16%) and on 3 October 2020 the unadjusted upper limit of seroprevalence was 4.29% (95% HDI 3.18%–5.47%). Unadjusted age-specific anti-SARS-CoV-2 seroprevalence estimates for four age categories (0–20, 21–40, 41–60 and > 60 years) are presented in the Supplementary material (Table S4).
All 50 anti-SARS-CoV-2-negative samples tested negative by NT and three-quarters of second-round anti-SARS-CoV-2-positive samples contained measurable levels of anti-SARS-CoV-2 neutralizing antibodies. The unadjusted lower limits of prevalence of anti-SARS-CoV-2 neutralizing antibodies in Slovenia on 11 November 2020, as well as the unadjusted upper limit of prevalence on 3 October 2020, were 2.97% (95% HDI 2.06%–3.87%).
Discussion
Seroprevalence surveys on a probability-based sample provide crucial information on cumulative SARS-CoV-2 exposure in communities. Such serosurveys are particularly useful in settings where limited SARS-CoV-2 RNA testing and/or contact tracing capacity prevents reliable assessment of the COVID-19 burden through cumulative incidence data from official notification systems, which is an unfortunate reality in most of the world. To address this knowledge gap, in March 2020 the WHO recommended nationwide population-based, age-stratified epidemiological surveys and designed a flexible investigation study protocol to facilitate the collection and sharing of COVID-19 epidemiological data in a standardized format [27]. As a result, numerous SARS-CoV-2 seroprevalence surveys on a probability-based sample were performed, but so far the majority have resulted in reports deposited in open-access preprint repositories or posted on websites [2], and only a dozen have had results published in peer-reviewed literature [4,[6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17]]. Comparing the results of published seroprevalence surveys is complicated because serological assay combinations with substantially different performance characteristics and different approaches regarding estimate calculations and interpretation of the results were used and, despite being labelled as probability-based sample surveys, the great majority excluded children or adolescents, or included only individuals seeking various health-care services, resulting in underrepresentation of younger cohorts. In addition, with the start of large-scale vaccination against SARS-CoV-2, the value and purpose of population-based seroprevalence surveys will shift from past/current aims toward the duration and dynamic of protective immunity after vaccination to dictate the future course of public health measures and mitigating strategies [28].
As of 31 January 2021, all published pre-vaccination SARS-CoV-2 serosurveys were conducted during or just following the first wave of the COVID-19 pandemic (Table 1 and Supplementary material Table S6) [4,[6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17]]. Our study estimated the SARS-CoV-2 seroprevalence in Slovenia in two time periods: at the end of the relatively mild first epidemic wave (April 2020) and during the devastating exponential growth phase of the second epidemic wave (October/November 2020). Due to considerably different circumstances, each study round dealt with distinct logistical and methodological challenges, resulting in several practical and informative lessons learned.
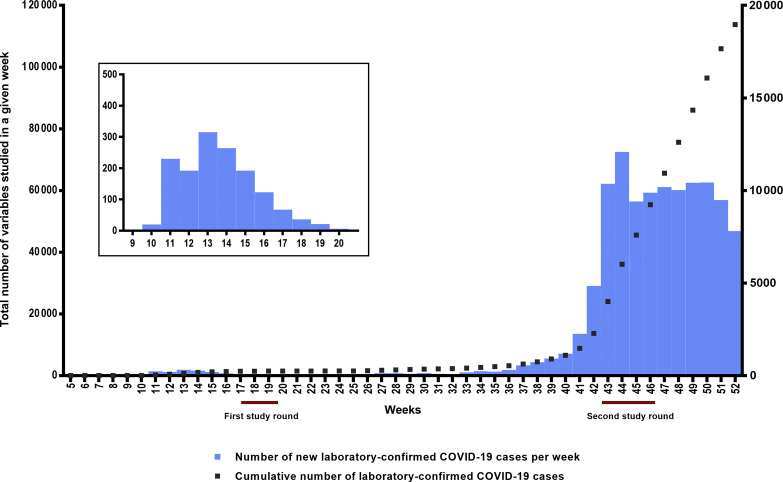
Slovenia was relatively spared during the first COVID-19 epidemic wave, with approximately 1400 cumulative laboratory-confirmed COVID-19 cases and 50 deaths/million reported (Fig. 1 ). After 2 months of draconian containment measures, based on favourable incidence data from the national official notification system and supported by low active COVID-19 prevalence estimates in a nationwide study on a probability-based sample [6], the restrictions in the country started to loosen in May 2020, ultimately leading to Slovenia being the first country in Europe to officially declare the end of the first wave of the epidemic as of 31 May 2020. However, because of an initially very conservative national diagnostic strategy (mainly due to a limited supply of reagents and consumables), it was predicted that the cumulative number of laboratory-confirmed COVID-19 cases captured through the official national notification system poorly reflected actual cumulative SARS-CoV-2 exposure during the first wave. When deciding in late April 2020 which anti-SARS-CoV-2 assay to use for testing the first-round samples, we and many others had no other choice than to use assay(s) with largely unproven performance, with very limited real-life experience in daily diagnostics and in population-based studies, heavily relying on the manufacturer's evaluation data and/or limited independent evaluation data, with the assay's performance assessed on a very limited number of samples and on a potentially biased sample population. As a result of extreme containment measures at that time (total lockdown, border closure), the only assay with relatively reliable performance data and with the assay quantities available to us for timely testing of all first-round samples was Euroimmun's ELISA. From today's perspective it is obvious that our initial estimate of SARS-CoV-2 seroprevalence in Slovenia for late April 2020 corrected for the manufacturer's estimate of ELISA specificity (2.78%) was an overestimation that was later corrected using two completely different approaches. First, after accumulation of published manufacturer-independent ELISA performance data, we innovatively corrected the first-round seroprevalence using pooled ‘meta-specificity’ (see Supplementary material, item S3). Second, when it became clear that concomitant use of highly-specific serological assays targeting different SARS-CoV-2 proteins offers reliable seroprevalence estimates in low-prevalence settings [3,5,18,19,24,29,30], all first-round samples were retested in November 2020 using an Elecsys-N/Elecsys-S combination. Interestingly, using these two different approaches—correction of suboptimal ELISA results using pooled ‘meta-specificity’ versus concomitant use of two highly specific assays—very similar seroprevalence estimates for late April 2020 were determined (0.93% versus 0.87%). Slovenian seroprevalence in late April 2020 was compatible with population-based seroprevalence estimates for Iceland [7], the Faroe Islands [12], India [11] and China [17], and substantially lower than those recorded in countries heavily affected by the first COVID-19 epidemic wave [4,8,10,[14], [15], [16]].
Fig. 1.
New laboratory-confirmed coronavirus disease 2019 (COVID-19) cases per week and cumulative number of laboratory-confirmed COVID-19 cases in Slovenia from week 5 to week 52 in 2020 captured though the national official notification system. New laboratory-confirmed COVID-19 cases per week are presented using blue bars, and the corresponding numbers are on the right y-axis. The cumulative number of laboratory-confirmed COVID-19 cases is presented by the dotted line, and corresponding numbers are on the left y-axis. For a nationwide seroprevalence population study on a probability-based sample, blood samples were collected between 20 April and 1 May 2020 (first study round) and 17 October and 10 November 2020 (second study round). National official notification system row data are available at https://www.nijz.si/sl/dnevno-spremljanje-okuzb-s-sars-cov-2-covid-19. Total data are presented from week 5 to week 52 in 2020; detailed data from week 9 to week 20 in 2020 are presented in the magnified window.
In contrast to the first epidemic wave, Slovenia was one of the countries most affected in the second epidemic wave, with currently the third-worst COVID-19-related cumulative mortality globally (https://origin-coronavirus.jhu.edu/). With lessons learned during first-round testing, second-round specimens were also tested with the Elecsys-N/Elecsys-S combination, and all positives were tested further by NT. However, other methodological and logistic challenges arose. As shown in Fig. 1, second-round specimens were collected at the turning point of the second epidemic wave—during the collection period, the cumulative number of laboratory-confirmed COVID-19 cases in the country increased 3.6-fold, from 13 675 to 48 837. In addition, several study participants were either isolated or quarantined. All of these extended the sample collection period over 25 days and resulted in non-random sampling collection order (participants with COVID-19-compatible symptoms/signs were tested later because of logistical issues). Consequently, the seroprevalence estimate for the second round could not be calculated as point prevalence, but we had to present the results as the period (see Supplementary material, item S6). As a result, the seroprevalence estimates for October/November 2020 were presented as the upper bound of seroprevalence 14 days before the sample collection starting date and the lower bound of seroprevalence at the end of the sample collection period because: (a) the sensitivity of the serological assays correlates with the time that elapsed between exposure and testing, (b) sample collection was performed during epidemic exponential growth and (c) in contrast to the first round, the second round was not accompanied with concomitant SARS-CoV-2 RNA testing. In comparison to the first round, concomitant use of Elecsys-N/Elecsys-S was even more beneficial in the second round not only to secure high specificity but also to increase net sensitivity by capturing the participants with an anti-S-only or anti-N-only early convalescent response [[31], [32], [33]].
An important study limitation is that, as the result of stringent study protocol requirements, all first-round samples were anonymized after initial ELISA testing, making it impossible to individually link the Elecsys-N/Elecsys-S results from both rounds. However, the recent study that followed a large cohort of medical workers with the Elecsys-N/Elecsys-S combination showed the persistence of anti-SARS-CoV-2 antibodies in all participants up to 6 months [31]. We strongly believe that all first-round anti-SARS-CoV-2-positive participants were also captured by the Elecsys-N/Elecsys-S combination in their second-round samples and that a risk of 6-month ‘seroreversion’ when using the Elecsys-N/Elecsys-S combination is negligible. An additional important inherited limitation of all seroprevalence surveys is that some individuals remain seronegative post-infection, as evidenced by the detection of memory T-cell SARS-CoV-2 responses in the absence of antibodies [34,35].
In conclusion, the nationwide population study on a probability-based sample showed at least a four-fold increase in anti-SARS-CoV-2 prevalence from late April 2020 to October/November 2020. Significant logistical and methodological challenges accompanied both study rounds with several important lessons learned, including (but not limited to) the need for caution when relying on assays' evaluation data generated by manufacturers, the importance of manufacturer-independent assay performance assessments and early data sharing through peer-reviewed literature, the need for concomitant use of two highly specific serological assays targeting different SARS-CoV-2 proteins in seroprevalence surveys conducted in both low-prevalence settings and during epidemic exponential growth, and the power and flexibility of the Bayesian approach for overcoming methodological challenges arising in seroprevalence surveys of emerging pathogens.
Author contributions
MPO, AOV and ES contributed equally to this work. MPO, AOV, PMV and TAZ were responsible for the study concept and design. AOV, PMV, KRR, TAZ, MPE, MPO, NK, MK, BZ, JD, ES, SK, KS and VV were responsible for the acquisition, analysis or interpretation of data. MPO, AOV, ES and PMV drafted the manuscript, and NK, MK, KRR, BZ, JD, SK, KS and VV provided essential parts. BZ, JD, ES, SK and VV were responsible for statistical analysis. All authors approved the final version of the manuscript submitted to CMI. All authors had full access to the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Funding
This work was funded by the Government of the Republic of Slovenia, the Institute of Microbiology and Immunology, Faculty of Medicine, University of Ljubljana, Slovenia and the Slovenian Research Agency (grants: P5-0399, P2-0209, P5-0410, P5-0151 and P3-0083). All the authors declare no competing interests. The funders had no role in the study design, data collection, analysis, interpretation of the results, writing of the manuscript and decision to submit the work for publication.
Transparency declaration
The authors have no conflicts of interest to disclose.
Acknowledgements
We would like to thank all the participants who accepted the invitation and volunteered to participate in the study. This study would not have been possible without the dedicated work of many people: the colleagues responsible for first-contact surveying of the participants and organization of sample collection: Mateja Škamperle, Tina Triglav, Barbara Šoba Šparl, Tjaša Cerar Kišek, Sabina Islamović, Anja Šterbenc, Mateja Pirš, Miša Pavletič, Karin Kregar, Ivana Velimirović, Maja Lunar, Jana Mlakar, Grega Gimpelj Domjanič, Anja Erbežnik, Katarina Ogrin; the fieldwork coordinators: Mateja Pirš, Anja Šterbenc, Lara Hudej, Grega Gimpelj Domjanič, Mitja Gajski, Tadej Pliberšek, Petra Hrvat, Jasmina Livk, Petra Čamernik, Nataša Krošelj, Tanja Kozinc, Polona Pretnar, Danijela Petrović, Maja Accetto Kos, Katka Pohar, Jana Boben, Rok Tomazin, Veronika Pavlič, Aljoša Obreza, Anja Erbežnik, Katarina Ogrin; the entire COVID-19 diagnostic team: Mateja Jelovšek, Robert Krošelj, Blanka Kušar, Katja Strašek Smrdel, Maja Lunar, Marko Kolenc, Martin Sagadin, Monika Jevšnik Virant, Nina Žigon, Petra Markočič, Rok Kogoj, Tina Uršič, Urška Glinšek Biškup, Maja Accetto Kos, Tina Štamol, Lara Hudej; the computer science students and teaching assistants that rapidly developed a data collection platform: Rafael Frančišek Irgolič, Robert Cvitkovič, Andrej Čopar, Gregor Krmelj, Nejc Debevc, Andreja Kovačič, Jaka Kokošar, Nejc Hirci, Tomaž Hočevar, Matjaž Pančur, Ajda Pretnar; the questionnaire reviewers: Lea Korva, Sabina Islamović, Jasmina Zlotrg; the longitudinal follow-up medical student team: Sara Ručigaj, Joanna Prusnik, Andrej Lipužič, Tanja Adamlje, Tjaša Pugelj, Boris Podobnik, Manca Bregar, Katja Strouhal, Gjoko Markoski, Rok Gerbec, Veronika Pučnik, Martin Janakjiev, Ana Benedik; and IT support: May Doušak. In addition, we would like to thank all the employees of the Institute of Microbiology and Immunology, Faculty of Medicine, University of Ljubljana, as well as health-care workers of the Pacient service and employees of the Episcenter call centre. We also thank the Statistical Office of the Republic of Slovenia for their help with the sample selection.
Editor: L. Leibovici
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.03.009.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lai C.C., Wang J.H., Hsueh P.R. Population-based seroprevalence surveys of anti-SARS-CoV-2 antibody: an up-to-date review. Int J Infect Dis. 2020;101:314–322. doi: 10.1016/j.ijid.2020.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rostami A., Sepidarkish M., Leeflang M.M.G., Riahi S.M., Nourollahpour Shiadeh M., Esfandyari S. SARS-CoV-2 seroprevalence worldwide: a systematic review and meta-analysis. Clin Microbiol Infect. 2021;27:331–340. doi: 10.1016/j.cmi.2020.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mekonnen D., Mengist H.M., Derbie A., Nibret E., Munshea A., He H. Diagnostic accuracy of serological tests and kinetics of severe acute respiratory syndrome coronavirus 2 antibody: a systematic review and meta-analysis. Rev Med Virol. 2020 doi: 10.1002/rmv.2181. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 4.Stringhini S., Wisniak A., Piumatti G., Azman A.S., Lauer S.A., Baysson H. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study. Lancet. 2020;396:313–319. doi: 10.1016/S0140-6736(20)31304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turbett S.E., Anahtar M., Dighe A.S., Garcia Beltran W., Miller T., Scott H. Evaluation of three commercial SARS-CoV-2 serologic assays and their performance in two-test algorithms. J Clin Microbiol. 2020;59 doi: 10.1128/JCM.01892-20. e01892-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maver Vodičar P., Oštrbenk Valenčak A., Zupan B., Avšič Županc T., Kurdija S., Korva M. Low prevalence of active COVID-19 in Slovenia: a nationwide population study of a probability-based sample. Clin Microbiol Infect. 2020;26:1514–1519. doi: 10.1016/j.cmi.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollán M., Pérez-Gómez B., Pastor-Barriuso R., Oteo J., Hernán M.A., Pérez-Olmeda M. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020;396:535–544. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menachemi N., Yiannoutsos C.T., Dixon B.E., Duszynski T.J., Fadel W.F., Wools-Kaloustian K.K. Population point prevalence of SARSCoV-2 infection based on a statewide random sample – Indiana, April 25–29, 2020. Morb Mortal Wkly Rep. 2020;69:960–964. doi: 10.15585/mmwr.mm6929e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sood N., Simon P., Ebner P., Eichner D., Reynolds J., Bendavid E. Seroprevalence of SARS-CoV-2-specific antibodies among adults in Los Angeles county, California, on April 10–11, 2020. JAMA. 2020;323:2425–2427. doi: 10.1001/jama.2020.8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murhekar M.V., Bhatnagar T., Selvaraju S., Rade K., Saravanakumar V., Thangaraj J.W.V. Prevalence of SARS-CoV-2 infection in India: findings from the national serosurvey, May–June 2020. Indian J Med Res. 2020;152:48–60. doi: 10.4103/ijmr.IJMR_3290_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen M.S., Strøm M., Christiansen D.H., Fjallsbak J.P., Hansen Eliasen E., Johansen M. Seroprevalence of SARS-CoV-2-specific antibodies, Faroe Islands. Emerg Infect Dis. 2020;26:2761–2763. doi: 10.3201/eid2611.202736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallal P.C., Hartwig F.P., Horta B.L., Silveira M.F., Struchiner C.J., Vidaletti L.P. SARS-CoV-2 antibody prevalence in Brazil: results from two successive nationwide serological household surveys. Lancet Glob Health. 2020;8:e1390–e1398. doi: 10.1016/S2214-109X(20)30387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poustchi H., Darvishian M., Mohammadi Z., Shayanrad A., Delavari A., Bahadorimonfared A. SARS-CoV-2 antibody seroprevalence in the general population and high-risk occupational groups across 18 cities in Iran: a population-based cross-sectional study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30858-6. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vos E.R.A., den Hartog G., Schepp R.M., Kaaijk P., van Vliet J., Helm K. Nationwide seroprevalence of SARS-CoV-2 and identification of risk factors in the general population of The Netherlands during the first epidemic wave. J Epidemiol Community Health. 2020 doi: 10.1136/jech-2020-215678. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stefanelli P., Bella A., Fedele G., Pancheri S., Leone P., Vacca P. Prevalence of SARS-CoV-2 IgG antibodies in an area of northeastern Italy with a high incidence of COVID-19 cases: a population-based study. Clin Microbiol Infect. 2021;27:633.e1–633.e7. doi: 10.1016/j.cmi.2020.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan S., Zhou M., Zhang W., Shen J., Qi R., Qin X. Seroprevalence and asymptomatic carrier status of SARS-CoV-2 in Wuhan City and other places of China. PLoS Negl Trop Dis. 2021;15 doi: 10.1371/journal.pntd.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theel E.S., Slev P., Wheeler S., Couturier M.R., Wong S.J., Kadkhoda K. The role of antibody testing for SARS-CoV-2: is there one? J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00797-20. e00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coste A.T., Jaton K., Papadimitriou-Olivgeris M., Greub G., Croxatto A. Comparison of SARS-CoV-2 serological tests with different antigen targets. J Clin Virol. 2020;134:104690. doi: 10.1016/j.jcv.2020.104690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poljak M., Oštrbenk Valenčak A., Štamol T., Seme K. Head-to-head comparison of two rapid high-throughput automated electrochemiluminescence immunoassays targeting total antibodies to the SARS-CoV-2 nucleoprotein and spike protein receptor binding domain. J Clin Virol. 2021;137:104784. doi: 10.1016/j.jcv.2021.104784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manthei D.M., Whalen J.F., Schroeder L.F., Sinay A.M., Li S.H., Valdez R. Differences in performance characteristics among four high-throughput assays for the detection of antibodies against SARS-CoV-2 using a common set of patient samples. Am J Clin Pathol. 2021;155:267–279. doi: 10.1093/ajcp/aqaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan S.S., Saw S., Chew K.L., Huak C.Y., Khoo C., Pajarillaga A. Head-to-head evaluation on diagnostic accuracies of six SARS-CoV-2 serological assays. Pathology. 2020;52:770–777. doi: 10.1016/j.pathol.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zilla M., Wheeler B.J., Keetch C., Mitchell G., McBreen J., Wells A. Variable performance in 6 commercial SARS-CoV-2 antibody assays may affect convalescent plasma and seroprevalence screening. Am J Clin Pathol. 2021;155:343–353. doi: 10.1093/ajcp/aqaa228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lisboa Bastos M., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.P., Johnston J.C. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rijkers G., Murk J.L., Wintermans B., van Looy B., van den Berge M., Veenemans J. Differences in antibody kinetics and functionality between severe and mild severe acute respiratory syndrome coronavirus 2 infections. J Infect Dis. 2020;222:1265–1269. doi: 10.1093/infdis/jiaa463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stan Development Team . 2019. RStan: the R interface to Stan. R package version 2.19.1.http://mc-stan.org Available at: [Google Scholar]
- 27.WHO . 2020. Population-based age-stratified seroepidemiological investigation protocol for COVID-19 virus infection.https://apps.who.int/iris/bitstream/handle/10665/331656/WHO-2019-nCoV-Seroepidemiology-2020.1-eng.pdf Available at: [Google Scholar]
- 28.Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the post-pandemic period. Science. 2020;368:860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ripperger T.J., Uhrlaub J.L., Watanabe M., Wong R., Castaneda Y., Pizzato H.A. Orthogonal SARS-CoV-2 serological assays enable surveillance of low-prevalence communities and reveal durable humoral immunity. Immunity. 2020;53:925–933.e4. doi: 10.1016/j.immuni.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harley K., Gunsolus I.L. Comparison of the clinical performances of the Abbott Alinity IgG, Abbott Architect IgM, and Roche Elecsys total SARS-CoV-2 antibody assays. J Clin Microbiol. 2020;59:e02104–e02120. doi: 10.1128/JCM.02104-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.L’Huillier A.G., Meyer B., Andrey D.O., Arm-Vernez I., Baggio S., Didierlaurent A. Antibody persistence in the first six months following SARS-CoV-2 infection among hospital workers: a prospective longitudinal study. Clin Microbiol Infect. 2021;27:784.e1–784.e8. doi: 10.1016/j.cmi.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lumley S.F., O’Donnell D., Stoesser N.E., Matthews P.C., Howarth A., Hatch S.B. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med. 2021;384:533–540. doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lumley S.F., Wei J., O’Donnell D., Stoesser N.E., Matthews P.C., Howarth A. The duration, dynamics and determinants of SARS-CoV-2 antibody responses in individual healthcare workers. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab004. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarzkopf S., Krawczyk A., Knop D., Klump H., Heinold A., Heinemann F.M. Cellular immunity in COVID-19 convalescents with PCR-confirmed infection but with undetectable SARS-CoV-2-specific IgG. Emerg Infect Dis. 2021;27:122–129. doi: 10.3201/2701.203772. [DOI] [PubMed] [Google Scholar]
- 35.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.B., Olsson A. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.