Abstract
Background: We previously reported that cannabidiol (CBD), a cannabinoid with a low toxicity profile, downregulated the expression of the prometastatic gene inhibitor of DNA binding 1 (ID1) in cancer cells, leading to inhibition of tumor progression in vivo. While CBD is broadly used, including in the self-medication of cancer patients, and CBD-based therapies are undergoing clinical evaluation for cancer treatment, its mechanisms of action are still poorly understood.
Methods: In this study, using microarray analysis and Western blot analysis for validation, we attempted to identify the full spectrum of genes regulated by CBD across various aggressive cancer cell lines, including the breast, brain, head and neck, and prostate.
Results: We confirmed that ID1 was a major target downregulated by CBD and also discovered that CBD inhibited FOXM1 (Forkhead box M1), a transcriptional activator involved in cell proliferation, while simultaneously upregulating GDF15 (growth differentiation factor 15), a cytokine associated with tissue differentiation.
Conclusion: Our results suggest that, by modulating expression of shared key cancer-driving genes, CBD could represent a promising nontoxic therapeutic for treating tumors of various origins.
Keywords: microarray analysis, transcription, cannabinoid, ID1, FOXM1, GDF15, Western blot, short hairpin RNA, metastasis
Introduction
Inhibitor of DNA binding (ID) helix-loop-helix (HLH) proteins are negative regulators of basic HLH (bHLH) transcription factors, and many reports now show that the ID family of HLH proteins controls cellular processes related to tumor progression.1 We found that reducing ID1 using antisense technology led to significant reductions in cancer cell proliferation and invasiveness in culture and metastasis in vivo in various types of cancer.2–5 Furthermore, ID1 overexpression in breast cancer cells was also found to be one of the most significant genes within a gene signature set that is correlated with the propensity of primary human breast cancer cells to metastasize to the lung.6 Reducing ID1 expression could provide a rational therapeutic strategy for the treatment of aggressive human cancers. It is not possible at this point, however, to use antisense technology in human patients. In our search for a nontoxic exogenous compound that could inhibit ID1 expression, a potential candidate agent, cannabidiol (CBD), was found.7
The endocannabinoid system was discovered through research focusing on the primary psychoactive active component of Cannabis sativa, Δ9-tetrahydrocannabinol (Δ9-THC), and other synthetic cannabinoids.8,9 Δ9-THC and additional cannabinoid agonists have been shown to interact with two G-protein coupled receptors, CB1 and CB2.8,9 More studies demonstrate that CB1 and CB2 receptor agonists show promise as tumor inhibitors.10 However, the psychotropic effects of Δ9-THC and additional cannabinoid agonists, mediated through the CB1 receptor, limit their clinical utility.9 In addition to Δ9-THC, CBD is also present in appreciable quantities in C. sativa and does not have appreciable affinity for CB1 or CB2 receptors or psychotropic activities.11 However, GPR55, a putative receptor for CBD, has been characterized.12
Even though we have shown that CBD inhibits cancer cell aggressiveness in various types of cancer through the downregulation of ID1 gene expression, no in-depth transcriptomic analysis has been published to date. Beyond the decrease in ID1 gene expression, there is no clear mechanism(s) that could explain the antitumor activity of CBD in so many types of cancer cells. Using an unbiased microarray analysis, we identified a CBD-induced gene expression profile consistent across four different cancer types, which include key regulators of tumor cell proliferation and invasion. We not only confirm downregulation of ID1 gene expression as a common mechanism of CBD action across various types of cancer but also identify additional genes, such as the transcription factor Forkhead box M1 (FOXM1), as a mediator of CBD antiproliferative effects. Even though CBD is broadly used for various diseases including cancer, its mechanisms of action are still poorly understood. We suggest that regulation of a specific set of genes by CBD leads to inhibition of human cancer cell proliferation, migration, and invasiveness, thus providing some mechanisms for the antimetastatic activity of the compound.
Materials and Methods
Cell culture and treatments
We used the human breast cancer cell lines MDA-MB231 and MDA-MB436 (ATCC), human glioblastoma (GBM) U251 and SF126 (ATCC), human oral squamous cell carcinoma SAS and human salivary gland cancer cell line ACCM (Japanese Collection of Research Bioresources), and human prostate cancer cell lines PC3 and DU145 (ATCC). In all experiments, the different cell populations were first cultured in RPMI-1640 media containing 10% fetal bovine serum (FBS). On the first day of treatment, the media were replaced with vehicle control (Veh) (ethanol) or CBD (2 μM) in RPMI and 0.1% FBS as previously reported.7,9 The media with the appropriate compounds were replaced after 24 h, for a total of 2-day treatment, a time corresponding to significant effects of CBD as previously reported in time-course experiments by our laboratory.7 CBD was obtained from the National Institutes of Health (NIH) through the National Institute of Drug Abuse.
Microarray analysis
To evaluate genome-wide changes in gene expression, cancer cell lines were obtained from breast, GBM, head and neck (HN), and prostate (as described previously). Total RNA was extracted from these cell lines using TRIzol (Invitrogen, Carlsbad, CA) and hybridized as technical replicates to Agilent Human Whole Genome OneArray™ Microarrays by Phalanx Biotech Group, Inc. (Hsinchu, Taiwan).13 A total of 12,712 genes with a minimum detection p-value of <0.05 in at least one sample were retained for further analysis. Given the inherent gene expression variation between cell lines, we normalized gene expression changes within a cell line between treatment and vehicle. To normalize each gene expression value, the log2 mean of each treatment and control sample was subtracted from individual values, resulting in positive and negative log2-folds. Normalized log2-fold changes for all treatment and control samples were then compared using an empirical Bayes moderated t-test in AltAnalyze version 2.0.814 between all CBD-treated and vehicle-treated lines. Genes with a false discovery rate-adjusted p-value of <0.05 and fold change >2 were analyzed using the GO-Elite module15 in AltAnalyze to identify and visualize enriched biological pathways, ontologies, and gene-sets. Clustering was performed in AltAnalyze using the HOPACH algorithm via remote connection to R.16
Knockdown experiments
Human PC3 prostate cancer cells were infected with control short hairpin RNA (Ctl shRNA) or with shRNA against growth differentiation factor 15 (GDF15 shRNA) purchased as lentiviral particles (sc-39798-V; Santa Cruz Biotechnology), selected with puromycin and pooled.
Western analysis
Cells were scraped, lysed, and the protein extracts were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis using 10% acrylamide gels and blotted onto nitrocellulose membranes. Spectra Multicolor Broad Range Protein Ladder was from Thermo Scientific and included 10 recombinant proteins (10, 17, 26, 34, 42, 52, 72, 95, 140, and 260 kDa). After blocking, blots were hybridized with rabbit monoclonal anti-ID1 (BCH-1#195–14; Biocheck), mouse monoclonal anti-FOXM1 (A-11/sc-271746; Santa Cruz Biotechnology), goat polyclonal anti-GDF15 (AF957; R&D Systems), as well as anti-actin (Chemicon), diluted in Tris–HCl buffer saline pH 7.6–0.1% Tween 20+10% nonfat dry milk. Membranes were incubated with horseradish peroxidase-labeled secondary antibodies, and the signal was detected using enhanced chemiluminescence. Band intensity values were obtained directly from the blot using AlphaeaseFC software (San Leandro, CA) or from film using the ImageJ (NIH, MD).
Results
CBD downregulates ID1 gene expression in various types of cancer
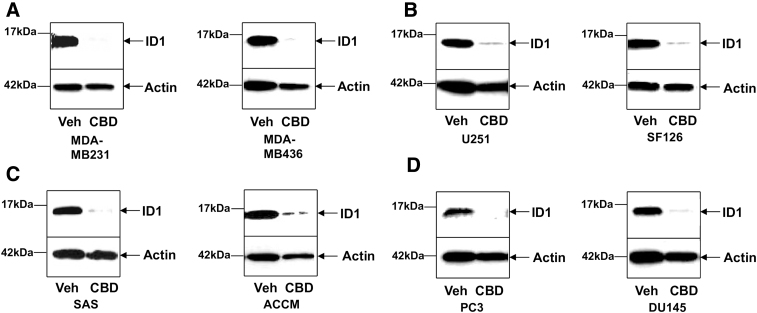
We previously demonstrated that CBD inhibited tumor cancer cell proliferation and invasion in various types of cancer.17 Even though the decrease in ID1 expression seemed to be necessary for the majority of effects of CBD on metastatic progression,7,18 we could not rule out that other important transcriptional regulators of tumor aggressiveness would be modulated by CBD. Therefore, we used eight different human cancer cells lines, two breast cancer cell lines (MDA-MB231 and MDA-MB436), two GBM lines (U251 and SF126), two HN cancer lines (oral squamous cell carcinoma SAS and salivary gland cancer cell line ACCM), and two prostate cancer lines (PC3 and DU145). Following a 48-h CBD treatment, RNA and protein were harvested from the same plate for all cell lines. Western blot analysis of ID1 was performed to confirm the downregulation of protein expression by CBD (Fig. 1A–D). RNA samples were collected for microarray analysis as described below.
FIG. 1.
CBD inhibits ID1 gene expression in human cancer cell lines from various origins. Proteins from (A) two breast cancer cell lines MDA-MB231 and MDA-MB436, (B) two GBM U251 and SF126, (C) two tumor types in the HN region (oral squamous cell carcinoma SAS and salivary gland cancer cell line ACCM), and (D) two prostate cancer cell lines PC3 and DU145 were extracted and analyzed for ID1 by Western blot analysis as described in the Materials and Methods section. Cells were treated either with Veh or with CBD. Normalization was carried out by stripping the blots and re-probing with an anti-actin antibody. For each cell line, a representative experiment from three to five independent experiments is shown, and some of the bands were cropped from the original blots. CBD, cannabidiol; GBM, glioblastoma; HN, head and neck; Veh, vehicle control.
CBD treatment results in a common gene expression response across multiple cancers
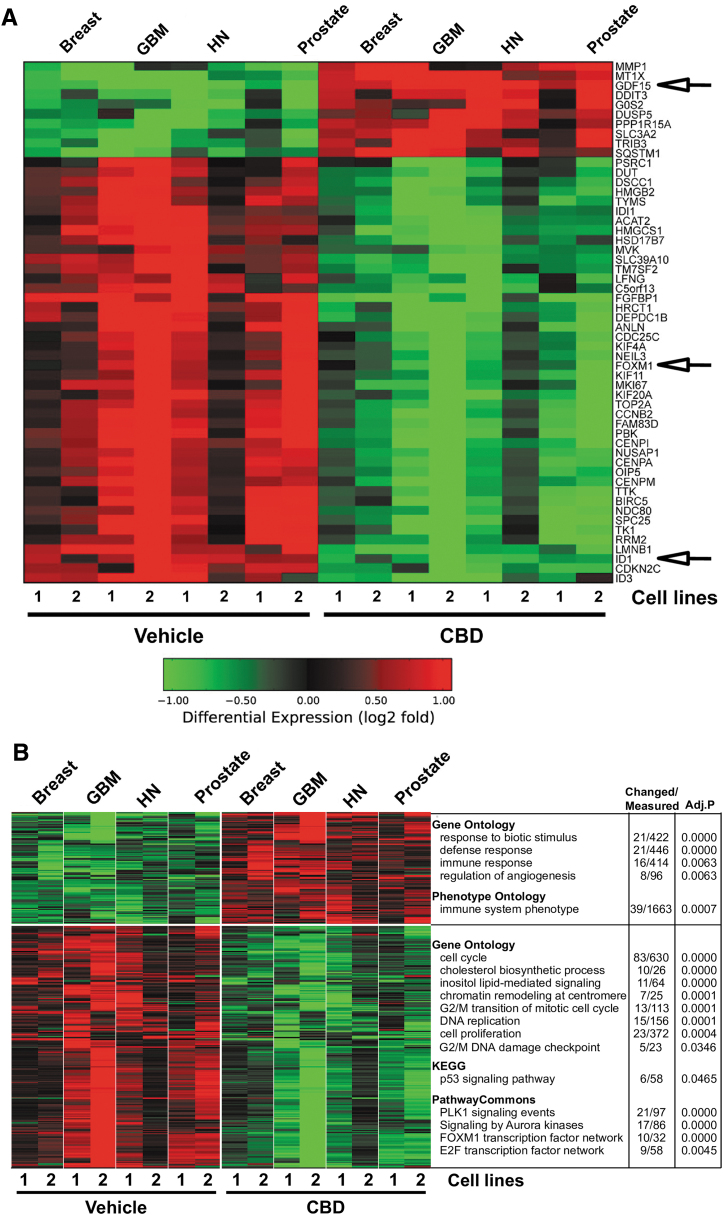
To identify an optimal set of CBD-responsive genes and pathways, we performed genome-wide expression profiling on the eight aggressive cancer cell lines (breast, GBM, HN, and prostate) with and without CBD treatment. When treatments and controls within each cell line were normalized to themselves, the gene expression differences between CBD and vehicle were largely consistent between tumors of distinct origins. The majority of regulated genes were decreased upon CBD treatment following stringent filtering of these data. Importantly, only a few transcriptional regulators were found among the top 44 most significantly downregulated genes, including ID1, FOXM1, and ID3 (Fig. 2A). As previously mentioned above, the ID family of HLH proteins controls cellular processes related to tumor progression, including migration and invasion.1 FOXM1 is exclusively expressed in proliferating cells, controls cell cycle, and is overexpressed in many types of cancer.19 We also examined the Gene Ontology pathways that were modulated upon CBD treatment. Downregulated genes were largely composed of factors regulating cell cycle, cell proliferation, DNA replication, DNA damage, p53 signaling, cholesterol biosynthesis, Aurora kinase signaling, and transcriptional regulation by FOXM1 and E2F (Fig. 2B).
FIG. 2.
CBD-dependent gene expression signatures. (A) Principle component analysis performed in AltAnalyze on 12,712 genes following normalization in eight cell lines corresponding to four different types of cancers. The most significantly regulated genes between CBD and vehicle treatment were clustered in AltAnalyze using HOPACH (red, upregulation; green, downregulation). (B) 363 regulated genes (false discovery rate; p<0.05, fold >2) between CBD and vehicle treatment, clustered in AltAnalyze using HOPACH (red, upregulation; green, downregulation). The top unique GO-Elite results are displayed to the right of the corresponding gene clusters.
Examination of upregulated genes with CBD revealed increase in the expression of genes such as Tribbles Pseudokinase 3 (TRIB3) and GDF15 among the top 10 most upregulated genes (Fig. 2A) and, using Gene Ontology, sets of genes corresponding to inhibitors of angiogenesis and regulators immune response (Fig. 2B). TRIB3 is known to be activated as a result of cellular stress and ultimately leads to autophagy-mediated cell death, a process implicated in CB1 and CB2 receptor antitumor activity across cancers.10 The characterization of GDF15 as one of the most upregulated gene is particularly interesting since its expression has been linked to the inhibition of the development of some cancers, such as intestinal and prostate tumors.20 Even though various genes were found to be regulated upon CBD treatment, we opted to focus on the regulation of the major transcriptional regulators, ID1 and FOXM1 genes. We hypothesized that CBD treatment of cancer cells would lead to inhibition of FOXM1 and ID1 genes expression leading to inhibition of proliferation and invasion. The downregulation of these genes could be dependent on the upregulation of GDF15.
GDF15 and FOXM1/ID1 expression in cancer cells treated with CBD
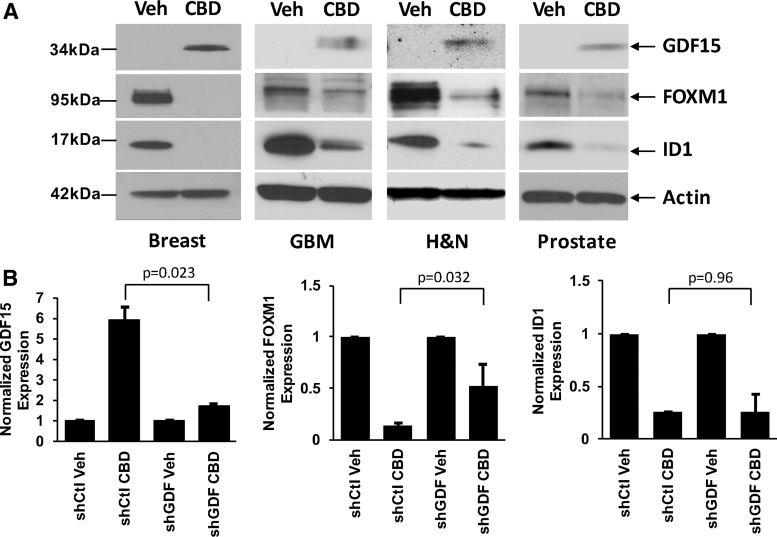
Using Western blot, we validated GDF15/FOXM1/ID1 protein expression changes in cells treated with CBD (at 2 μM for 48 h) vs vehicle control (Veh). All cell lines investigated (human breast cancer cells [MDA-MB231], human GBM cells [U251], human HN cancer cells [SAS], and human prostate cancer cells [PC3]) showed the same pattern of expression of these important genes (Fig. 3A). GDF15 protein expression was upregulated, whereas both FOXM1 and ID1 protein expressions were downregulated across all cancers. We next asked whether the increase in GDF15 was linked to the decrease in expression of FOXM1 and/or ID1. To do this, we used a cancer cell line that could be efficiently infected, PC3. We investigated the effect of GDF15 knockdown on expression of FOXM1 and ID1 in these prostate cancer cells infected with a control shRNA (shCtl) or with an shRNA against GDF15 (shGDF) and treated with either vehicle control (Veh) or 2 μM CBD (CBD). Using Western blot analysis from several independent experiments, we confirmed that GDF15 expression was reduced upon knockdown and treatment with CBD (Fig. 3B, left panel). We next determined that this reduction in GDF15 expression was linked to a reduced effect of CBD on FOXM1 downregulation (compare second and fourth samples in Fig. 3B, center panel). However, the difference between the decrease in ID1 expression by CBD was not significant when comparing shCtl and shGDF cells (second and fourth samples in Fig. 3B, right panel). These data suggest that the increase in GDF15 expression upon treatment with the cannabinoid compound is necessary for the downregulation of FOXM1 expression, but not for its effect on ID1 gene expression.
FIG. 3.
GDF15 and FOXM1/ID1 expression in cancer cells. (A) Using Western blotting, GDF15/FOXM1/ID1 expression was compared in cancer cell lines treated (CBD) or not (Veh) for 2 days. Results are presented in human breast cancer cells (MDA-MB231), human GBM cells (U251), human HN cancer cells (SAS), and human prostate cancer cells (PC3). For each cell line, a representative experiment from three independent experiments is shown, and some of the bands were cropped from the original blots. (B) The effects of GDF15 knockdown were determined on the expression of FOXM1 and ID1 in PC3 prostate cancer cells infected with a control shRNA (shCtl), or in cells infected with an shRNA against GDF15 (shGDF), and treated either with Veh or with CBD. Densitometry readings of the blots were taken from independent experiments, and the percentage relative expression was calculated for GDF15, FOXM1, or ID1. Data were compared using the Student's t-test. Error bars, ±SEM. p-Values indicate statistically significant or nonsignificant differences. FOXM1, Forkhead box M1; GDF15, growth differentiation factor 15; shRNA, short hairpin RNA.
Discussion
In this study, we investigated the expression of genes that are regulated by the cannabinoid compound CBD across cancers, focusing on four types of solid cancers from very different origins. Metastasis is the final and fatal step in the progression of many types of solid tumors. Currently available therapeutic strategies at this stage of cancer progression are often nonspecific, have only marginal efficacy, and are highly toxic. Therapeutic approaches targeting only specific mechanisms involved in the development of aggressive cancers are urgently needed.
It was previously demonstrated that the HLH protein ID1, an inhibitor of bHLH transcription factors, played a crucial role during cancer progression.1,21 It was suggested that targeting its expression may be highly effective and safe in advanced cancer patients, given (1) the relationship between high ID1 expression levels and aggressive cancer cell behaviors; (2) partial reduction in the ID1 activity could achieve significant outcomes; and (3) ID1 expression was found to be low in normal adult tissues, thereby eliminating unwanted toxicities generally associated with currently available therapeutic modalities. We reported that CBD could act as an effective inhibitor of ID1 expression, leading to inhibition of genotypic and phenotypic changes that allow aggressive cancers to invade and metastasize.17 In this study, an unbiased study of gene expression profiling across tumors of distinct origins confirms that, upon CBD treatment, ID1 was indeed the most strongly inhibited transcriptional regulator along with FOXM1. In addition, upregulation of TRIB3, an upstream indicator of autophagy-mediated cell death, was conserved across cancer types. TRIB3 has previously been implicated in cannabinoid control of tumor progression.8
Interestingly, even though there are about 2,500 transcriptional regulators in human cells, we only found three of them (ID1, ID3, and FOXM1) among the most downregulated genes. Interestingly, we did not detect any change in the expression of other ID members, ID2 and ID4. In the case of ID4, our results are consistent with previous observations that this HLH protein possesses unique roles compared with other ID proteins.22 This indicates a high specificity of CBD regulation of these transcriptional regulators, across cancer types. FOXM1 is a transcription factor exclusively expressed in proliferating cells. It is a protein of the Forkhead box transcription factor family, critical for cell cycle progression. FOXM1 activity is mediated by increased expression of Skp2 and Cks1, which are important for the processes that drive the cell cycle from G1 to S phase.19 In normal tissues, FOXM1 is only expressed in tissues that are regenerating or in progenitor cells. Studies have shown that FOXM1 is overexpressed in many human malignancies and in all human carcinomas. Overexpression of FOXM1 has been documented in GBM and breast cancer. In the latter, it has also been shown to correlate with a poor prognosis.19 Interestingly, the downregulation of FOXM1 expression by CBD appeared to be, at least partly, dependent on the upregulation of GDF15 expression.
GDF15 is part of the transformation growth factor beta superfamily. Its expression is increased in response to injuries in various organs such as the kidney and lung.23 Unlike ID1 and FOXM1, GDF15 has been linked to tumor suppression. In vivo studies on transgenic mice with prostate and intestinal cancer showed that increased expression of GDF15 inhibited cancer progression, and several tumor suppressors, such as EGR-1, GSK-3b, and p53, all upregulate GDF15 expression.20 Moreover, when cancer cells were treated with a variety of different antitumor agents, retinoids or resveratrol, all showed increased expression of GDF15, suggesting its role as part of a tumor suppressing response.24
Two studies investigated some transcriptomic analyses of CBD in encephalitogenic cells.25,26 The cells used in the investigations were nontransformed and were from a different tissue lineage in comparison to the metastatic cancer cells presented in our study. However, the results presented in these two studies showed specific effects of CBD on gene expression through alteration of histone methylation and provides some mechanisms by which CBD exerts its anti-inflammatory effects. In our studies, CBD could also modify chromatin remodeling, leading to repression of specific genes. The modulation of High Mobility Group Box 2 and Lamin B1 upon CBD treatment may strengthen this hypothesis.
Overall, we discovered that key genes modulated by CBD are conserved across cancer cells from various origins. We confirmed that inhibition of ID genes and upregulation of TRIB3 are indeed pathways impacted by CBD treatment in many types of cancer. In addition, we discovered that the upregulation of GDF15 triggers reduction in FOXM1 expression, and this may be a key pathway conserved across cancers, which underlies CBD-dependent inhibition of cell proliferation. The net effect of targeting these multiple pathways by a single molecule such as CBD could lead to the inhibition of tumor progression.
Abbreviations Used
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported by grants from the National Institutes of Health–National Cancer Institute (CA082548, CA135281) and the Susan Komen Breast Cancer Research Foundation (KG090385), and by funds from the Research and Education Leadership Committee of the CPMC Foundation.
Cite this article as: Desprez P-Y, Murase R, Limbad C, Woo RWL, Adrados I, Weitenthaler K, Soroceanu L, Salomonis N, McAllister SD (2021) Cannabidiol treatment results in a common gene expression response across aggressive cancer cells from various origins, Cannabis and Cannabinoid Research 6:2, 148–155, DOI: 10.1089/can.2019.0081.
References
- 1. Ke J, Wu R, Chen Y, et al. Inhibitor of DNA binding proteins: implications in human cancer progression and metastasis. Am J Transl Res. 2018;10:3887–3910 [PMC free article] [PubMed] [Google Scholar]
- 2. Murase R, Sumida T, Kawamura R, et al. Suppression of invasion and metastasis in aggressive salivary cancer cells through targeted inhibition of ID1 gene expression. Cancer Lett. 2016;377:11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fong S, Itahana Y, Sumida T, et al. Id-1 as a molecular target in therapy for breast cancer cell invasion and metastasis. Proc Natl Acad Sci U S A. 2003;100:13543–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soroceanu L, Murase R, Limbad C, et al. , Id-1 is a key transcriptional regulator of glioblastoma aggressiveness and a novel therapeutic target. Cancer Res. 2013;73:1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coppe JP, Itahana Y, Moore DH, et al. , Id-1 and Id-2 proteins as molecular markers for human prostate cancer progression. Clin Cancer Res. 2004;10:2044–2051 [DOI] [PubMed] [Google Scholar]
- 6. Minn AJ, Gupta GP, Siegel PM, et al. , Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McAllister SD, Christian RT, Horowitz MP, et al. , Cannabidiol as a novel inhibitor of Id-1 gene expression in aggressive breast cancer cells. Mol Cancer Ther. 2007;6:2921–2927 [DOI] [PubMed] [Google Scholar]
- 8. Pertwee RG. Pharmacology of cannabinoid CB1 and CB2 receptors. Pharmacological Therapeutics. 1997;74(November):129–180 [DOI] [PubMed] [Google Scholar]
- 9. McAllister SD, Murase R, Christian RT, et al. Pathways mediating the effects of cannabidiol on the reduction of breast cancer cell proliferation, invasion, and metastasis. Breast Cancer Res Treat. 2011;129:37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Velasco G, Sanchez C, Guzman M. Towards the use of cannabinoids as antitumour agents. Nat Rev Cancer. 2012;12:436–444 [DOI] [PubMed] [Google Scholar]
- 11. McPartland JM, Russo EB. Cannabis and cannabis extract: greater than the sum of the parts? J. Cannabis Therapeut. 2001;1:103–132 [Google Scholar]
- 12. Sharir H, Abood ME. Pharmacological characterization of GPR55, a putative cannabinoid receptor. Pharmacol Ther. 2010;126:301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang LH, Huang HS, Wu PT, et al. Role of macrophage CCAAT/enhancer binding protein delta in the pathogenesis of rheumatoid arthritis in collagen-induced arthritic mice. PLoS One. 2012;7:e45378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Emig D, Salomonis N, Baumbach J, et al. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 2010;38(Web Server issue):1W755–W762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zambon AC, Gaj S, Ho I, et al. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28:2209–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salomonis N, Cotte N, Zambon AC, et al. Identifying genetic networks underlying myometrial transition to labor. Genome Biol. 2005;6:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McAllister SD. Soroceanu L, Desprez PY. The antitumor activity of plantderived non-psychoactive cannabinoids. J Neuroimmune Pharmacol. 2015;10:255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Murase R, Kawamura R, Singer E, et al. Targeting multiple cannabinoid anti-tumour pathways with a resorcinol derivative leads to inhibition of advanced stages of breast cancer. Br J Pharmacol. 2014;171:4464–4477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raychaudhuri P, Park HJ. FoxM1: a master regulator of tumor metastasis. Cancer Res. 2011;71:4329–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X, Baek SJ, Eling TE. The diverse roles of nonsteroidal anti-inflammatory drug activated gene (NAG-1/GDF15) in cancer. Biochem Pharmacol. 2013;85:597–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nair R, Teo WS, Mittal V, et al. ID proteins regulate diverse aspects of cancer progression and provide novel therapeutic opportunities. Mol Ther. 2014;22:1407–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patel D, Morton DJ, Carey J, et al. Inhibitor of differentiation 4 (ID4): from development to cancer. Biochim Biophys Acta. 2015;1855:92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zimmers TA, Jin X, Hsiao EC, et al. Growth differentiation factor-15/macrophage inhibitory cytokine-1 induction after kidney and lung injury. Shock. 2005;23:543–548 [PubMed] [Google Scholar]
- 24. Martinez JM, Sali T, Okazaki R, et al. Drug-induced expression of nonsteroidal anti-inflammatory drug-activated gene/macrophage inhibitory cytokine-1/prostate-derived factor, a putative tumor suppressor, inhibits tumor growth. J Pharmacol Exp Ther. 2006;318:899–906 [DOI] [PubMed] [Google Scholar]
- 25. Kozela E, Juknat A, Gao F, et al. Pathways and gene networks mediating the regulatory effects of cannabidiol, a nonpsychoactive cannabinoid, in autoimmune T cells. J Neuroinflammation. 2016;13:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang X, Bam M, Nagarkatti PS, et al. Cannabidiol regulates gene expression in encephalitogenic T cells using histone methylation and noncoding RNA during experimental autoimmune encephalomyelitis. Sci Rep. 2019;9:15780. [DOI] [PMC free article] [PubMed] [Google Scholar]