Abstract
Introduction: In mammals, sn-1-diacylglycerol lipases (DAGL) generate 2-arachidonoylglycerol (2-AG) that, as the major endocannabinoid, modulates synaptic neurotransmission by acting on CB1 cannabinoid receptors (CB1R). Even though the insect genome codes for inaE, which is a DAGL ortholog (dDAGL), its products and their functions remain unknown particularly because insects lack chordate-type cannabinoid receptors.
Materials and Methods: Gain-of-function and loss-of-function genetic manipulations were carried out in Drosophila melanogaster, including the generation of both dDAGL-deficient and mammalian CB1R-overexpressing flies. Neuroanatomy, dietary manipulations coupled with targeted mass spectrometry determination of arachidonic acid and 2-linoleoyl glycerol (2-LG) production, behavioral assays, and signal transduction profiling for Akt and Erk kinases were employed. Findings from Drosophilae were validated by a CB1R-binding assay for 2-LG in mammalian cortical homogenates with functionality confirmed in neurons using high-throughput real-time imaging in vitro.
Results: In this study, we show that dDAGL is primarily expressed in the brain and nerve cord of Drosophila during larval development and in adult with 2-LG being its chief product as defined by dietary precursor availability. Overexpression of the human CB1R in the ventral nerve cord compromised the mobility of adult Drosophilae. The causality of 2-LG signaling to CB1R-induced behavioral impairments was shown by inaE inactivation normalizing defunct motor coordination. The 2-LG-induced activation of transgenic CB1Rs affected both Akt and Erk kinase cascades by paradoxical signaling. Data from Drosophila models were substantiated by showing 2-LG-mediated displacement of [3H]CP 55,940 in mouse cortical homogenates and reduced neurite extension and growth cone collapsing responses in cultured mouse neurons.
Conclusions: Overall, these results suggest that 2-LG is an endocannabinoid-like signal lipid produced by dDAGL in Drosophila.
Keywords: 2-monoacylglycerol, behavior, development, diacylglycerol lipase, inaE
Introduction
Ligand-receptor coevolution is a critical driving force of phylogenetic selection to increase fitness and survival by gain of function.1 Evolutionarily, Hydra vulgaris (Cnidaria) first shows an ancestral “endocannabinoid system” (i.e., 2-arachidonoylglycerol [2-AG] and anandamide (AEA) content and the existence of SR141716-sensitive binding sites).2 Similarly, cannabinoid (ant-)agonist-binding sites were broadly found in mollusks, annelids, as well as in some crustaceans,1,3,4 with the first orthologs of the CB1 cannabinoid receptor (CB1R) described in nematodes (such as the NPR-19 receptor in C. elegans5). However, classical CB1Rs appear unique to chordates. Insects, which likely emerged from a crustacean-like ancestor,6,7 uniquely lack arachidonic acid (AA)-derived endocannabinoids, cannabinoid receptors, and specific cannabinoid-binding sites,8 suggesting that the lack of the endocannabinoid system could contribute to safeguarding the species' survival (e.g., by “secondary loss” and evolutionary selection to protect against the toxicity of phytocannabinoids).9
Nevertheless, the genome of the fruit fly (Drosophila melanogaster) contains inaE (“inactivation no afterpotential E,” CG33174 gene, X chromosome), an ancestral form of sn-1-diacylglycerol lipases (DAGLs10,11), which is essential for phototransduction by acting downstream from phospholipase C and producing signal lipids that act as potential excitatory messengers at TRP/TRPL channels.11 Accordingly, Drosophila with defective inaE do not generate physiologically meaningful photoreceptor responses.11 Notably, inaE duplication and mutations gave rise to vertebrate DAGLα and β isoforms10,12 (Fig. 1A), which produce the endocannabinoid 2-AG. Because plants, unless genetically modified, do not contain long-chain polyunsaturated fatty acids (PUFAs), including AA,13 inaE, if functionally conserved in frugivore insects (e.g., Drosophila), might instead generate alternative, nonarachidonate-containing endocannabinoid-like molecules (Fig. 1A).
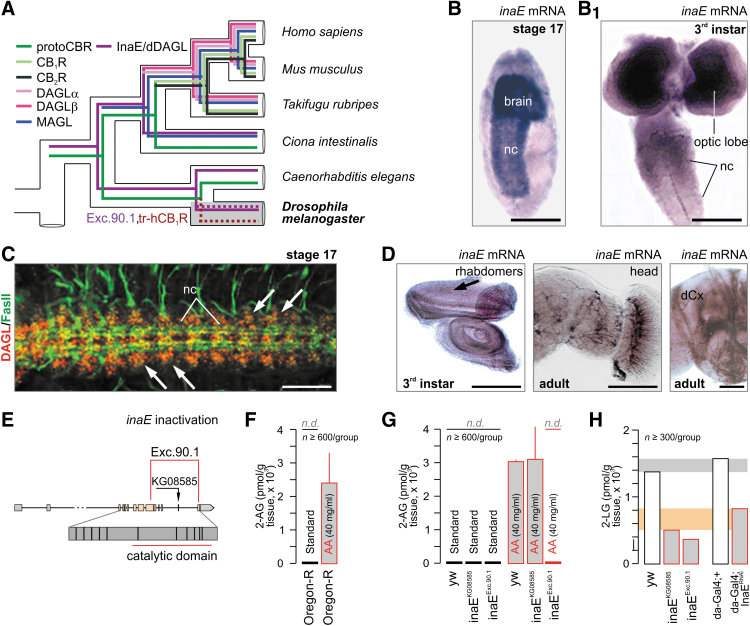
FIG. 1.
InaE/dDAGL function and ligands in Drosophila melanogaster. (A) In silico reconstruction of the evolution of endocannabinoid signaling. Data were modified from Refs.,1,3,8 and expanded by highlighting our experimental strategy: transgenic hCB1R (tr-hCB1R) expression in fruit fly. Note that classical cannabinoid receptors are restricted to chordates. In this study, “protoCBR” refers to ancestral cannabinoid receptor-like binding sites that are molecularly distinct from CB1Rs (and CB2Rs). (B, B1) inaE mRNA and (C) dDAGL protein localized to brain and nerve cord (nc) in Drosophila embryos and larvae as shown by in situ hybridization and immunohistochemistry, respectively. Fasciclin II (FasII) was used as an axonal marker in Drosophila embryos. (D) inaE localization by two in situ hybridization probes: eye imaginal disc, head, and the dendritic field of the calyx of the mushroom bodies (dCx) are shown. (E) InaE inactivation strategy showing the excision 90.1 (Exc.90.1) line. (F, G) Dietary AA supplementation allows 2-AG synthesis by dDAGL in third instar larvae. Note that dDAGL catalytic inactivity (inaEExc.90.1) precludes 2-AG synthesis. (H) 2-LG may be an endogenous dDAGL-derived signal lipid in Drosophila. Accordingly, dDAGL inactivation by two independent strategies reduces 2-LG content. Gray and orange underlays highlight biological variation between the genetic strategies. Horizontal gray bar: 2-LG content in yw and daughterless-GAL4;+ controls. Orange bar: residual 2-LG after genetic manipulation. Example from n=2 qualitatively identical studies. Note that we followed a population strategy with each sample comprising n≥300 larvae. Scale bars=50 μm (B, B1, C), 110 μm (D). AA, arachidonic acid; 2-AG, 2-arachidonoylglycerol; CB1R, CB1 cannabinoid receptor; hCB1R, human CB1R; 2-LG, 2-linoleoyl glycerol; MAGL, monoacylglycerol lipase; yw, yellow-white.
Previous targeted lipidomics identified a library of putative signaling lipids (both 2-acyl glycerols and N-acyl amides), most prominently 2-linoleoyl glycerol (2-LG), in Drosophila larvae,14 setting them significantly apart from vertebrates. In addition, the use of T1117, a lipophilic fluorescent cannabinoid, was shown to accumulate in Drosophila larvae, particularly the fat body, upon its gastrointestinal availability,14 suggesting that dietary precursors efficiently shape the cannabinoid-like insect lipidome. However, the identity of endocannabinoid-like signal lipids and if they could qualify as ancestral endocannabinoids remains unknown.
From an evolutionary perspective, an archetypical endocannabinoid precursor in insects might be expected to have lower efficacy than the potent vertebrate endocannabinoids, probably as partial agonists.15 Thus, and even if they remain present in the mammalian brain, they could be functionally inferior to either 2-AG or AEA with their effects partially or completely masked. This assumption is compatible with the notion of the “entourage effect,”16–18 which was initially defined as inactive (or marginally active) glycerol esters enhancing 2-AG activity at CB1R.16
In this study, we show that Drosophila with mutant InaE (termed, Drosophila-DAGL, dDAGL) lacking catalytic activity have a diminished axonal innervation of skeletal muscles. If fed by artificial AA-containing diets, Drosophila can generate 2-AG, although lethal by the third instar larval stage. However, 2-LG is the bioactive lipid present in Drosophila. Based on our InaE data, we have generated Drosophila ectopically expressing CB1Rs predominantly in cholinergic neurons targeting motor neurons and find this gain-of-function mutation to compromise motor coordination. Once exposed to 2-LG, CB1Rs in Drosophila signal through Erk and Akt alike their mammalian counterparts.19–21 Even though 2-LG has been identified as a partial agonist at CB1Rs in a recombinant cell system,15 in-depth insights in its physiological effects, if any, remain unknown. In this study, we demonstrate 2-LG to bind to mammalian CB1Rs and to displace CP 55,940 in mouse cortical homogenates. Alike 2-AG,22 2-LG dose dependently inhibits neuronal differentiation, including a change in the morphology of growth cones, a major subcellular domain where CB1Rs accumulate. Cumulatively, these data suggest that 2-LG is a low-efficacy CB1R ligand with archetypical biological activity that, in mammals, is superseded by 2-AG and AEA.
Materials and Methods
Drosophila lines and husbandry
Wild-type D. melanogaster (Oregon-RS, w1118 and yw), as well as the cha-Gal4, UAS-GFP, elav-Gal4, and inaEKG08585 lines were obtained from the Bloomington Drosophila Stock Center (BDSC, University of Indiana), whereas inaERNAi flies were purchased from the Vienna Drosophila RNAi Center (VDRC). Daughterless-Gal4 (da-Gal4) flies were a kind gift from Dr. A. Wredenberg (Karolinska Institutet). Flies were raised at 25°C with a 12-h light/12-h dark cycle, and reared on standard medium (Nutrifly Bloomington formulation; Genesee Scientific) unless specified otherwise.14
Cloning of dDAGL and characterization of transcripts
Two vertebrate DAGLs exist with a canonical structure that consists of a four transmembrane domain, followed by a catalytic domain and a long (DAGLα) or short (DAGLβ) tail.10 Drosophila has a single DAGL (dDAGL) located on the X chromosome at position 13677782 (version FB2010_07) and denoted as CG33174. Based on intron/exon boundary analysis, CG33174 can be classified as the DAGLα homolog, with DAGLβ arising from a gene duplication event (G.W., unpublished).
Based on homology with murine DAGL, we designed primers (5′/3′ GCGATCAATGCGCGAAACAGA/CAACAGCAGCCAAAGCAT) to generate a 500 base pair genomic fragment of the Drosophila DAGL by PCR and used this to probe a Drosophila embryonic cDNA library (Stratagene) according to the manufacturer's instructions. We isolated a splice form of dDAGL consisting of 3951 bp that utilizes exons 3–14 of the gene and encodes a protein of 708 amino acid residues with an intact catalytic domain. We denoted this splice variant CG33174-RC, with exons 1 and 2 encoding the 5′ UTR. Two other splice variants have been reported,11 corresponding to CG33174-RA and CG33174-RD. RA shares exons 3–13 with RC and encodes a truncated 644 residue protein, which is catalytically dead as it terminates before a critical histidine, which constitutes the third member of the lipase catalytic triad (G.W., unpublished). RD shares exons 3–13 and part of exon 14 with RC, but differs in being spliced into an additional exon (exon 15). This encodes a 739 reside protein with a longer tail than RC, but importantly shares a full catalytic domain with the protein encoded by RC.
Generation of dDAGL mutant strains
We obtained the inaEKG08585 line from BDSC and confirmed that a P-element was inserted 21,746 bp downstream of the gene origin (start at×13677782, CG33174, http://flybase.org/cgi-bin/gbrowse/dmel), in the 13th intron of the dDAGL gene, within the exons encoding the catalytic domain (exons 8–14). We generated 500 excision lines using standard genetic techniques23 and characterized these using primers designed to recognize sequences in the catalytic domain. Genomic DNA of adult flies was PCR amplified according to http://engels.genetics.wisc.edu/flyDNA.html. Primers 5′-TTGGCCGTAACGCGGATTA-3′ and 5′-CTCATCGAAGTCCGTGGAAT-3′, which bind 557 bp upstream (f) and 1661 bp downstream (r) of the P-element insertion site, were used to examine the excision lines. Only those excision lines lacked the dDAGL fragment, confirmed with the above primers, were processed further.
PCR was performed further with primers designed to bind sequences 5′ and 3′ into the dDAGL gene. We selected three overlapping excisions that totally or partially delete the catalytic domain of the enzyme. We amplified and sequenced their limits using primers GACACGCACATCAATCAG (5′f) and GCAAGATGTGGCATTTGG (5′f), located 1294 bp and 2258 bp upstream of the P-element insertion site, respectively, and GGACAAAGCCACATAAACAAA (3′r), 1970 bp downstream of the insertion site. The three mutant lines were homozygous viable, although with a delayed development with respect to wild type and, thus, better maintained as a stock over a balancer chromosome. Phenotypes were assessed on homozygous lines. For rescue experiments, we used the inaEExc.90.1 containing two copies of UAS-dDAGL on the second chromosome. UAS-dDAGL flies were generated by PCR amplification from CG33174-RC from the start (ATG) to the end of exon 14 (TGA) with a proofreading polymerase. We sequenced the blunt-end product and cloned it into a pENTR/D-TOPO vector (Invitrogen) according to the manufacturer's instructions. We used Gateway technology to insert the product into a UASt destination vector with 3 FLAG tags (The Drosophila Gateway™ Vector Collection, Department of Embryology, Carnegie Institution of Washington). Positive clones were sequenced and sent for generation of transgenic flies (www.thebestgene.com). The phenotype was analyzed in male embryos, which contained the 90.1 excision on the X chromosome, and were homozygous for UAS-DAGL (second X chromosome) and heterozygous for elav-Gal4 (third X chromosome).
Generation of anti-dDAGL antibodies
Antibodies were made by Eurogentec under its double XP program using the peptide antigens EP041911 (H2N-CRHHPKPDEQKYDSGW-CONH2) and EP041912 (H2N-NYKRSNSMNRNWRQRC-CONH2). Two rabbits were immunized with each peptide and the antibodies were affinity purified against separate antigens. Both antibodies were tested and titrated in whole-mount embryos, cryosectioned, and stained (see section Commercial antibodies, immunohistochemistry, and confocal microscopy). EP041912 gave the best signal-to-noise ratio and was used for all further experiments.
In situ hybridization
Embryos (third instar larval) and adult brains were collected and processed according to published protocols.24 Tissues were immersion fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) (0.1 M, pH7.4), embedded in paraffin, and sectioned at 6 μm. RNA probes were generated using our full-length CG33,174-PC product, inserted in Bluescript SK(−), and linearized with EcoRI for T7 polymerase (antisense probes) and XhoI for T3 polymerase (sense control probes; not shown). Probes for the dDAGL catalytic domain region were generated with PCR of CG33,174-PC (primer pairs (5′/3′): CCGGAACAGCGGCCATACTCGCCATTCTCC/CGGGCTTGGGATGATGTCGCAC), which recognized all three cloned dDAGL transcripts. Probes were tagged using the Roche Digoxigenin Labeling Kit and hybridization and signal detection were performed following published protocols.24
Commercial antibodies, immunohistochemistry, and confocal microscopy
Collection of stage 17 embryos, larval dissection, immunohistochemical staining, and confocal microscopy were performed according to published protocols.25 Tissues were immersion fixed as above, stained as whole mounts with cocktails of select primary antibodies at 4°C overnight: mouse anti-Fasciclin II (FasII, 1D4 IgG), mouse anti-MHC (DSHB, Iowa), goat anti-CB1R antibodies (1:200; provided by Dr. M. Watanabe), and rabbit anti-dDAGL (see section Generation of anti-dDAGL antibodies). Subsequently, tissues were exposed to Alexa Fluor 488 or 594-conjugated or DyLight 549-tagged secondary antibodies (1: 200; Jackson ImmunoResearch) in phosphate buffer (0.1 M, pH 7.4) containing 0.3% Triton X-100 and 1% bovine serum albumin (Sigma-Aldrich). Tyramine signal amplification (TSA) protocol (PerkinElmer) was used to enhance anti-dDAGL immunosignal when necessary. Larval abdominal muscles were labeled with Alexa Fluor 546-conjugated phalloidin (1:50, 30 min at 22–24°C; Invitrogen). Embryos were mounted in ProLongGold Antifade Reagent (Invitrogen). Larval preparations were mounted in vectashield (Vector Laboratories), and imaged on either a Zeiss LSM700 laser-scanning microscope or a Zeiss Axioplan 2 HXP120C microscope equipped with a ProgRes C14 camera (Jenoptik). All images were minimally and linearly processed with Adobe Photoshop for contrast enhancement.
UAS-human CB1R expression
We generated a Drosophila strain for the conditional expression (“inducible gain-of-function”) of the human CB1R (hCB1R). Using the Gal4 binary system, we subcloned the entire open reading frame of the hCB1R into a pUAST vector.26 Transgenic lines were generated by standard P-element-mediated transgenesis (GenetiVision, Houston), followed by selection of transgenic flies and balancing transgene insertion.
Lipid extraction and LC-MS/MS analysis
For mass spectrometry (MS), third instar larvae (n≥300/sample) were collected, rinsed twice in PBS (50 mM), flash frozen in liquid N2, and stored at −80°C for lipid extraction. Measurements were performed as published14,27 Tissue samples and AEA-d4 standards were homogenized in acetonitrile, centrifuged, and lipids extracted using a solid-phase extraction procedure on a Cerex SPE positive-pressure (N2) manifold (Crawford Scientific) with Strata-X 33 μm polymeric reversed-phase cartridges (flow rate: 1 mL/min; Phenomenex). Analytes were eluted by methanol, dried under N2, and residues resuspended in 50 μL of mobile phase before injection (10 μL) onto the chromatograph. AEA, 2-AG (Tocris) and 2-LG (Cayman Chemicals) standards were dissolved in dimethylsulfoxide. A Surveyor LC system coupled to a TSQ Quantum, triple quadrupole mass spectrometer (Thermo Scientific) was used for the analysis. The LC column was a 150×2.1 mm ACE 5 μm C8 with precolumn (Hichrom), maintained at 30°C with a mobile-phase flow rate of 200 μL/min. Isocratic elution was achieved with a mobile phase consisting of 15% water/85% methanol (both containing 0.5% formic acid). MS analysis was performed using electrospray ionization in positive ion mode using source conditions optimized for the transmission of the relevant parent ions by flow injection analysis. The spray voltage was 3500 V, sheath gas 40 (N2, arbitrary units), auxiliary gas 10 (N2, arbitrary units), and capillary temperature 375°C. Quantification was undertaken using single reaction monitoring on the dominant product ion determined for each compound using collision energy 16 V and a collision gas pressure of 1.3 mTorr (argon). The parent ion—product ion transitions monitored were AEA m/z 348.2–m/z 62.2, AEA-d4 m/z 352.2–m/z 66.2, 2-AG m/z 352.2–m/z 287.2, and 2-LG m/z 355.2–m/z 263.2. Under these conditions the retention time for AEA and AEA-d4 was 7.3 min. Two peaks were present in the chromatogram for 2-AG and 2-LG, at retention times 7.66/8.10 and 7.72/8.24 min, respectively (Supplementary Fig. S1A, A1). This is explained by the presence of biologically inactive 1(3)-isomers, which exist in equilibrium with 2-AG and 2-LG. The areas of both peaks were combined to determine 2-AG and 2-LG concentrations. System control, data collection, and all subsequent quantitative analyses were carried out using Xcalibur 2.0.6 software (Thermo Scientific). Weighted linear regression analysis curves were constructed using the ratio of analyte/internal standard to concentration of analyte in the calibration standard, and tissue extract concentration was determined from this curve. The assayed concentration was then divided by the tissue weight and the results expressed as nmol/g tissue.
AA feeding assay
Female flies (3–10 days) of appropriate genotypes were allowed to lay eggs in vials containing standard food (Nutrifly formulation) with or without AA supplement (40 mg/mL; Cayman Chemicals) at 25°C for 24 h.14 Three days later, third instar larvae (n≥300/sample) were collected and processed to determine tissue AEA and 2-AG contents.
RNA analysis
Total RNA was isolated by organic extraction with phenolic Tri Reagent (Sigma-Aldrich) and further processed by solid-phase purification using RNeasy Plus columns (Qiagen). RNase-free DNase I (Qiagen) treatment was optionally performed to eliminate genomic DNA from the purified total RNA. For each sample, 1 μg of RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative real-time PCRs and data analysis were performed using an iCycler iQ real-time detection system (Bio-Rad). Each sample was run in triplicate, including appropriate controls. Ribosomal protein rp49 mRNA was used as internal control. The primer pairs: (rp49; 5′/3′ f) CCAAGGACTTCATCCGCCACC, (rp49, 5′/3′ r) GCGGGTGCGCTTGTTCGATCC, (hCB1R, 5′/3′ f) AAGGTGACATGGCATCCAAAT, and (hCB1R, 5′/3′ r) AGGACGAGAGAGACTTGTTGTAA were designed using Primer 3 (http://frodo.wi.mit.edu) and custom synthesized (DNA Technology A/S).
CB1R internalization
Fat bodies of da-Gal4;UAS-hCB1R larvae at the third instar stage were acutely dissected in PBS (50 mM) and intact tissues exposed to WIN 55,212-2 (1 μM; Tocris) or 2-LG (2 μM; Cayman Chemicals) for 5 or 15 min. Subsequently, fat bodies were immersion fixed in 4% PFA, and stained using a goat anti-CB1R antibody to visualize subcellular receptor localization. Hoechst 33,342 was used as nuclear counterstain. Images were acquired on a Zeiss LSM780 laser-scanning microscope.
Signal transduction
Tissues from n=25 adult elav-Gal4;+ or elav-Gal4;UAS-CB1R males (3–6 days old) were homogenized with a microtissue grinder in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid [EDTA], 1% NP40, 1 mM phenylmethanesulfonyl fluoride and a protease inhibitor cocktail [Complete; Roche]). Aliquots were exposed to 2-LG (2 μM; Cayman Chemicals) or AM251 (500 nM, CB1R antagonist; Tocris) alone or in combination for 1, 3, or 10 min at 37°C. Next, samples were centrifuged at 14,000 rpm at 4°C for 30 min, with supernatant denaturated in Laemmli buffer (5×) and boiled at 95°C for 5 min. Fifteen micrograms of each sample was separated by SDS-PAGE (10%) and transferred onto PVDF membranes. Protein samples were exposed to primary antibodies (phospho-Erk1/2, total Erk1/2, phospho-Drosophila Akt, total Akt; 1:100; Cell Signaling) at 4°C overnight, and detected using horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch) and chemiluminescent detection (ECL Prime, Amersham). E7 anti-β-tubulin monoclonal antibody (1:1000; Developmental Studies Hybridoma Bank) served as loading control. Experiments at 1 and 3 min were performed in duplicate, while 10-min treatments were performed in triplicate.
Locomotor activity assay
Locomotion activity was measured according to Feany and Bender.28 Briefly, groups of 10 males (6-day-old) were transferred into 140×20 mm vials. After a 10-min resting period, the flies were tapped to the bottom of the vials and the number of flies able to climb a vertical distance of 8 cm within 10 s were recorded (Supplementary Video S1).29,30 Results from n≥60 flies/group were expressed as percentages from n=3 independent experiments.
Ethics approval of rodent studies
Experiments on live animals conformed to the 2010/63/EU European Communities Council Directive and were approved by the Austrian Ministry of Science and Research (66.009/0145-WF/II/3b/2014, and 66.009/0277-WF/V/3b/2017). Particular effort was directed toward minimizing the number of animals used and their suffering during experiments.
Radioligand binding in mouse cortical membranes
Adult C57Bl6/J mouse cortices were dissected and homogenized with a glass/teflon Potter-type homogenizer in ice-cold 50 mM Tris-HCl buffer (pH 7.4) containing 3 mM EDTA. Total membranes were fractionated at 35,000 g for 10 min followed by a washing step, repeated centrifugation, and pellet resuspension in 50 mM Tris-HCl buffer. Intracellular constituents were removed from resuspended membrane preparations by incubation at 23°C for 2 h before suspensions were centrifuged, resuspended, aliquoted, and stored at −80°C. BSA at a concentration of 0.1% was used throughout the experiments to retain [3H]CP 55,940 (American Radiolabeled Chemicals) in solution and protect it from being precipitated on glass surfaces. MgCl2 (3 mM) was used to increase binding specificity. Binding experiments followed a displacement design with 0.5 nM [3H]CP 55,940 challenged by increasing concentrations of unlabeled 2-LG (0, 1, 5, 12.5, 25, 50 and 100 μM; Cayman Chemicals). Additionally, JZL 195 (100 nM; Sigma-Aldrich) was present to inhibit both monoacylglycerol lipase (MAGL) and fatty acid amide hydrolase activity. Nonspecific binding was determined for immoderately high-ligand concentrations. In a separate study, O-2050 was additionally used as CB1R antagonist. The composite was incubated at 23°C for 2 h before being rinsed over glass microfiber filters (Whatman GF/C, presoaked in 0.3% polyethylene) in a vacuum-driven Höfer filtration box. Filters were washed (4×) with 50 mM Tris-HCl (containing 0.1% BSA) buffer before being exposed to a scintillation cocktail (Rotiszint 11; Roth) for 20 min and measured in a Packard TRI-CARB 2100TR scintillation counter. Raw scintillation counts were expressed as the percentage of specifically bound radioligand. Protein concentrations of the samples were determined by the Bradford method (Bio-Rad). Experiments were performed in quadruplicate using n=3 biological replicates each.
Cultures of mouse neurons and IncuCyte imaging
Neurons were isolated from E14.5 C57Bl6/J mouse cortices and grown in Neurobasal A medium (GIBCO, Life Technologies) supplemented with l-glutamine (2 mM), B27 supplement (2%), and penicillin/streptomycin (1%). Cells were plated in either 96-well plates (25,000 cells/well for automated imaging) or in 24-well plates (25,000 cells/well for immunocytochemistry) pretreated with poly-d-lysine in 0.1 M borate buffer overnight. Drug challenges were initiated on the first day in vitro (DIV) with 2-LG replenished every 24 h. An IncuCyte live-cell imaging system (Essen Bioscience), placed in an incubator with stable 5% CO2 intake and temperature control at 37°C, was used for live cell (including neurite) imaging. Phase-contrast images were taken at hourly intervals (or every other hour). “The area of cell viability,” that is the surface area occupied by cell bodies (mm2/mm2), was taken as a measure of cell viability. The growth rate of neurites was obtained by using a “neurite length” mask (mm/mm2). All measures were optimized in preliminary experiments, including drug doses and treatment paradigms. Dose/response curves were constructed from duplicate experiments with n≥12 wells/condition in each experiment.
Neurons at low density on coverslips were fixed with 4% PFA in PBS on ice for 15 min before being processed for immunohistochemistry. Multiple immunofluorescence labeling was performed by applying cocktail of affinity-purified antibodies27: Alexa Fluor 555 phalloidin (1:500; Invitrogen) and mouse anti-β-III-tubulin (1:300; Millipore). Images were acquired on a Zeiss 880LSM confocal laser-scanning microscope. High-resolution images were acquired with optical zoom ranging from 1.5×to 3.0×at 40×primary magnification to limit signal detection. Emission spectra for each dye were limited as follows: Cy2 (505–530 nm), Cy3 (560–610 nm), and Cy5 (650–720 nm). Morphometric analysis of cultured neurons was aided by the ZEN2012 software, as well as ImageJ with Fiji applications and included: (1) filopodia number (n), (2) neurite branching (n), (3) length of the primary neurite (μm), (4) the surface area and number of growth cones, and (5) fluorescence intensity distribution of phalloidin/F-actin in growth cones and along neurite shafts. The longest process emanating from neuronal somata was considered as the prospective axon. The number of neurons analyzed (per cover-slip) was n=56 (control) and n=29 (2-LG, 1 μM). The number of growth cones analyzed was n=26 (control) and n=17 (2-LG, 1 μM).
Statistical analysis
Data were expressed as mean±s.e.m. Individual data points for mammalian neurons were shown as scatter plots. The comparison of independent groups was statistically performed using Student's t-test. Behavioral data were analyzed using multivariate ANOVA followed by Bonferroni's post hoc test. A p-value of <0.05 was considered statistically significant.
Results
dDAGL expression during Drosophila development
InaE, the DAGLα homolog (dDAGL) based on the evolutionary conservation of intron/exon boundaries, was previously implicated in phototransduction in D. melanogaster.11 By PCR probing a Drosophila cDNA library, we isolated a dDAGL splice form that spans exons 3–14 of the inaE gene (3951 bp), and encodes an INAE protein of 708 residues (CG33174-RC). Recently, two other C-terminally truncated inaE splice variants (CG33174-RA/RD) were reported11 that contain a lipase-3 domain and exhibit sn-1 selectivity and DAGL-like activity in vitro. However, available knowledge is fragmented on the developmentally regulated and adult body-wide localization of dDAGL/INAE proteins in Drosophila. In this study, we used a hybridization probe recognizing all dDAGL mRNA transcripts to show that InaE mRNA is expressed in the developing Drosophila brain and ventral nerve cord by stage 17 (Fig. 1B), which persists throughout larval development (third instar, Fig. 1B1). By means of a dDAGL-specific antibody that we have generated (see Materials and Methods section), we then localized dDAGL/INAE protein to intersegmental neuroblast/neuron clusters along the ventral nerve cord (Fig. 1C), consistent with mRNA distribution. As in vertebrates, expression is dynamic during development and colocalizes with neuronal markers. Postembryonically, the protein is found in the larval brain and eye discs, and was particularly localized to photoreceptor rhabdomeres (Fig. 1D) and the dendritic field of the calyx region of the mushroom bodies (Fig. 1D). These data link dDAGL to neuronal differentiation and suggest evolutionarily conserved functions for dDAGL in for example, axonal growth and guidance.31
Dietary precursors drive 2-LG versus 2-AG production by dDAGL
Next, we used the Drosophila inaEKG08585 line to excise a P-element inserted at nucleotide (nt) 21,746 from the transcription start in intron 13, within the dDAGL/INAE catalytic domain. We characterized 500 excisions and recovered one that deletes most of the catalytic domain with only the terminal histidine of the catalytic triad remaining (Exc.90.1; nt 20004/exon 9–nt 22869/exon 14 henceforth the line termed as inaEExc.90.1; Fig. 1E), rendering dDAGL inactive. We then combined metabolomics and genetic tools to identify dDAGL products.
Drosophila do not express Δ5 and Δ6 desaturases,32 precluding the synthesis of C20/C22 PUFAs (including AA), suggesting the production of diunsaturated 2-acylglycerols instead. Indeed, targeted lipidomics revealed 2-linoleoylglycerol (2-LG) but not 2-AG in wild-type14 and mutant Drosophila (Fig. 1F, G, and Supplementary Fig. S1A, A1). Moreover, dDAGL hypomorphism or catalytic inactivation significantly reduced 2-LG content in third instar Drosophila larvae (Fig. 1H). Similarly, ubiquitous inaE RNAi knockdown by daughterless-Gal4 (da-Gal4, Supplementary Fig. S1B) reduced bodily 2-LG content in larvae (Fig. 1H). Since any 2-LG present in standard feeding formulations will be metabolized to triglycerides before delivery to tissues, where triglycerides act as precursors for monoacylglycerols, we interpret the presence of residual 2-LG in inaEKG08585, inaEExc.90.1, and inaERNAi as experimental confounds derived from undigested food. Thereby, 2-LG represents an evolutionary alternative to 2-AG in Drosophila.
We then fed third instar Drosophila larvae an AA-supplemented diet to show that both Oregon-R (Fig. 1F) wild-type and yellow-white (yw) control flies (Fig. 1G) can generate 2-AG, reaching concentrations found in mammalian brain. AEA was not detected (not shown). A similar dependence on exogenous AA was reported for the biosynthesis of 2-AG, but not AEA, in the lone star tick (Amblyomma americanum).33 The ability to utilize dietary AA in a pathway that leads to the generation of 2-AG was not obviously affected in inaEKG08585 flies, most probably because this is a hypomorphic line rather than a knockout. However, this pathway was completely lost in the inaEExc.90.1 line demonstrating an enzymatic requirement for dDAGL activity for this function (Fig. 1G). Targeted lipidomics has revealed 2-LG in the fruit fly on a standard diet.14 In this study, we found substantial (∼50–60%) reductions in 2-LG levels in the inaEExc.90.1 third instar Drosophila larvae. Similarly, ubiquitous InaERNAi knockdown by daughterless-Gal4 (Supplementary Fig. S1B) reduced bodily 2-LG content in larvae by a similar amount. Cumulatively, these data suggest that 2-LG is the primary dDAGL product in Drosophila with the substrate specificity of dDAGL tightly controlled by dietary choices and restrictions.
dDAGL loss of function inhibits skeletal muscle innervation
Mammalian endocannabinoids are developmentally significant since they participate in the establishment of correct axonal connectivity in the nervous system.31,34,35 Muscle innervation by motor axons in the developing Drosophila embryo is a well-characterized pathway (Fig. 2A, B), where a number of adhesion molecules, in particular Drosophila L1 (neuroglian, nrg), are known to function.36 Since dDAGL is localized to the ventral nerve cord that in adult Drosophila orchestrates motor output and its activity is required for L1 responses,37 we reasoned that dDAGL loss of function in Drosophila could adversely impact the development of muscle innervation. Axonal growth and guidance were impaired in Drosophila mutants with examples from the InaEExc.90.1 line shown (Fig. 2C–F). Three types of defects manifested in InaEExc.90.1 mutants: (1) stalling, whereby axons stop at variable points toward their targets in the muscle field or are delayed in their advance when compared with a wild-type embryo of the same age; (2) misrouting, which includes entering the muscle field at a wrong choice point or entering at the right point but extending in the wrong direction, and (3) defasciculation, which means that axons that should extend apposed to each other are running separate (Fig. 2G).
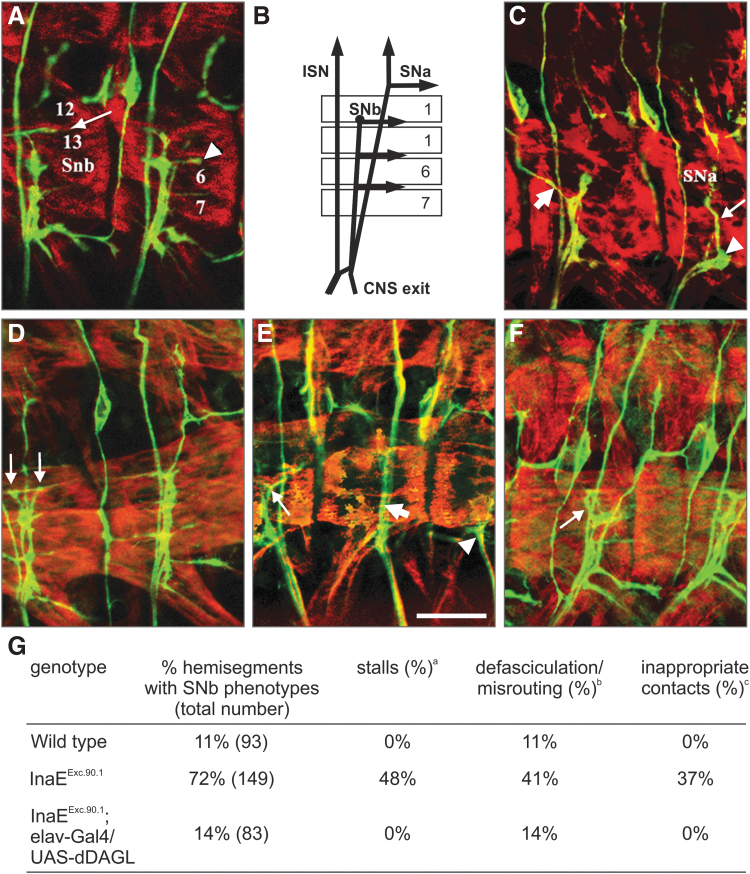
FIG. 2.
Mutations in dDAGL impair axonal outgrowth and pathfinding for muscle innervation by motor neurons in the SNb pathway, which are corrected by the targeted expression of CG33174-PC. (A, C–F) Apotome optical sections of three abdominal hemisegments in late-stage 16 filleted embryos stained for myosin heavy chain (MHC; red) and Fasciclin II (FasII, axonal marker; green). Anterior is left and dorsal up in all panels. (A) An overlay of five 500-μm optical sections showing the trajectory of the SNb as it grows toward and innervates the ventral muscle field. In the left hemisegment, arrow points to axonal extension in the 12–13 cleft. Arrowhead in the right hemisegment points to axons innervating the 6–7 cleft. (B) Schema depicting the three main motor axon pathways. ISN denotes the intersegmental nerve, with SNa and SNb denoting the segmental nerve A and B, respectively. (C, D, E) Abdominal hemisegments of inaEExc.90.1 embryos. In (C), focus is at the level of SNb entry into the muscle field (lateral transverse muscle 21 and 22). In the right hemisegment, SNb stalled at the entry point (arrowhead) but some fascicles proceeded to join SNa fascicles (thin arrow). In the left hemisegment, SNb stalls at the 6–13 cleft. There are some axons that misroute in the left hemisegment (thick arrow). In (D), axons can be seen to grow in two opposite directions past the 6–13 cleft (thin arrows) in the left hemisegment. (E) Another example points to SNb fascicles (thin arrow) following the top border of muscle 26. In the next hemisegment to the right, the thick arrow points to SNb fasciculating with the ISN instead running separately. In the right hemisegment, the SNb stalls before entering the ventral muscle field (arrowhead). (F) Hemisections from an inaEExc.90.1; elav-GAL4/UAS-dDAGL embryo are shown and axonal stalling is corrected with SNb fascicles reaching the 12–13 cleft. However, some misrouting can still be observed (thin arrow). (G) SNb projections in inaEExc.90.1 mutants as compared with wild-type control and the rescue by using UAS-DAGL. The three types of defects observed can be present alone or in combination in a given hemisegment. The percentage of hemisegments with phenotype refers to the total number irrespective of the severity of the defects. aSNb growth cones stop at any point on or after entering the ventral muscle field or are delayed in their advance at least one muscle width with respect to SNb growth cones of a similar age in control embryos. bSNb growth cones fail to defasciculate from ISN at the choice point (on entering the ventral muscle field) or they defasciculate from the main SNb branch at the wrong point and show abnormal trajectories. cSNb growth cones that enter the muscle field at the right point fail to make contact with their target muscle or show abnormal trajectories. Scale bar = 20 μm (E).
We then went on to address the capacity of the CG33174-RC isoform of dDAGL to rescue the axonal defects. We used the UAS-Gal4 system to construct transgenic flies containing this isoform cloned into a UAS vector and used elav-Gal4 to drive dDAGL-RC in neurons during embryonic stages in the InaEExc.90.1 line. We observed an almost complete rescue of the stalling defects (Fig. 2F, G), but only a mild rescue of the defasciculation and misrouting defects. The remaining percentage of stalling, only slightly higher than the observed in wild-type embryos, and the poor rescue of the misrouting and defasciculation defects could reflect a diminished or absent modulation of UAS-dDAGL since dDAGL is probably not provided in the dynamic pattern observed for the endogenous protein. Nonetheless, the rescue experiment suggests that the phenotype in the dDAGL flies does indeed reflect a loss of function as opposed to toxic gain of function due to aberrant transcripts. These data suggest that, alike in mammalian organisms,10,12,15,37 dDAGL is involved in the control of axonal growth and guidance.
Transgenic expression of the mammalian CB1R reveals signaling potential of 2-LG
The above results show that DAGL activity can regulate 2-AG and 2-LG levels in Drosophila, but in the case of 2-AG this is likely to be a diet-related epiphenomenon. The possibility of 2-LG being a signaling lipid is difficult to test in the absence of precise knowledge on the nature of putative responsive receptor(s) or its signal-competent metabolites. Instead, we reasoned that 2-LG's signaling potential might be detected by using a CB1R as surrogate based on the coevolution of the DAGLs and cannabinoid receptors1 (Fig. 1A).
To test this hypothesis, we generated transgenic flies containing the full-length hCB1R and drove expression using a cha-Gal4 vector. This was chosen because it efficiently targets intersegmental cholinergic neurons, including upstream excitatory neurons for motor neurons that orchestrate axial movement, and even a subset of motor neurons.38 Alternatively, the transgene was more broadly expressed in the CNS by means of an elav-Gal4 vector. Reverse transcription PCR confirmed the expression of the hCB1R in transgenic flies (Fig. 3A and Supplementary Fig. S1C). Ectopically expressed hCB1Rs retained their axonal targeting in cha-Gal4/GFP+ neurons,39 as shown by high-resolution laser-scanning reconstruction with coexpressed GFP used as localization signal. In contrast, histochemistry for hCB1R failed to reveal a specific pattern of immunoreactivity in flies where binary Gal4::UAS expression of the hCB1R was not active (Fig. 3B–B3).
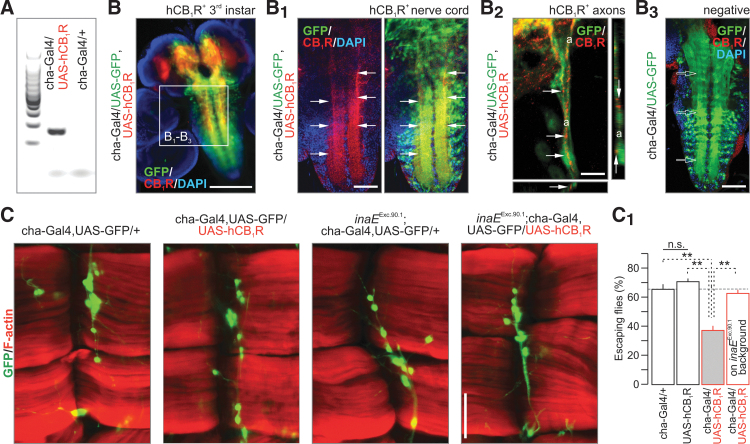
FIG. 3.
hCB1R expression disrupts locomotion in Drosophila. (A) Expression of hCB1R in fruit fly (mRNA). (B–B2) hCB1R immunolocalization upon expression by using a cha-Gal4 vector (primarily for excitatory intersegmental interneurons; GFP). Arrows point to hCB1R-dense cellular or subcellular compartments, including axons (a; B2). (B3) Histochemical control in nonoverexpressing flies. Open arrows show the lack of hCB1R labeling in GFP+ territories. DAPI was used as nuclear counterstain. (C) hCB1R expression affected neither the position nor the population size of intersegmental cholinergic neurons. Similarly, catalytic inactivation of inaE/dDAGL left cholinergic interneurons unaffected. (C1) hCB1R expression induced hypolocomotion [F(3, 187)=48.17, p<0.001]. See also Supplementary Video S1. In contrast, ectopic hCB1R expression on inaEExc.90.1 background rescued this phenotype. **p<0.01 (Bonferroni's post hoc test); n>60 animals/group; n.s., nonsignificant. Scale bar=50 μm (C).
This transgenic and cell type-specific model of hCB1R expression offers an opportunity to test if a tissue-endogeneous dDAGL product, likely 2-LG, activates hCB1Rs. Expressing hCB1R in cha-Gal4+ neurons is advantageous since optogenetic interrogation of these neurons was shown to affect locomotion.38 We find that hCB1R gain of function did not affect cha-Gal4/GFP+ neuron localization or numbers, suggesting that innervation of ventral longitudinal abdominal muscles (muscles 6,7) in third instar transgenic larvae remained normal (Fig. 3C). Subsequently, we exploited a locomotion (climbing) assay widely used to assess Drosophila behaviors upon transgene expression. The expression of hCB1R impaired motor control and locomotor activity (Fig. 3C1 and Supplementary Video S1). This inhibition was not seen when hCB1Rs were expressed in inaEExc.90.1 mutants (Fig. 3C1), neither when inaE was knocked down by RNAi (Supplementary Fig. S1E), suggesting that dDAGL produces an endogenous ligand that activates ectopically expressed hCB1Rs. These observations are also compatible with an inhibitory 2-LG action at ectopic CB1Rs. Moreover, they suggest that the loss of cannabinoid receptors in insects1 may be an evolutionary response to otherwise impaired neural functions limiting survival, rather than to the lack of endogenous ligands.
2-LG signaling at hCB1Rs in Drosophila adipocytes
Besides their native dDAGL expression, adipocytes of the fat body also expressed hCB1R levels and targeted the receptor correctly to their membrane surfaces (Fig. 4A). While 2-LG is considered as a participant in the “entourage effect”16 of endocannabinoid-like compounds (i.e., increased affinity of 2-AG at CB1Rs in the presence of 2-LG) with more recent data15 identifying 2-LG as a low-affinity partial CB1R agonist, we hypothesized that hCB1R in Drosophila might have increased ligand sensitivity and signal transduction efficacy in the absence of an endogenous “endocannabinoid tone,” thus being amenable to dissect 2-LG signal competence.
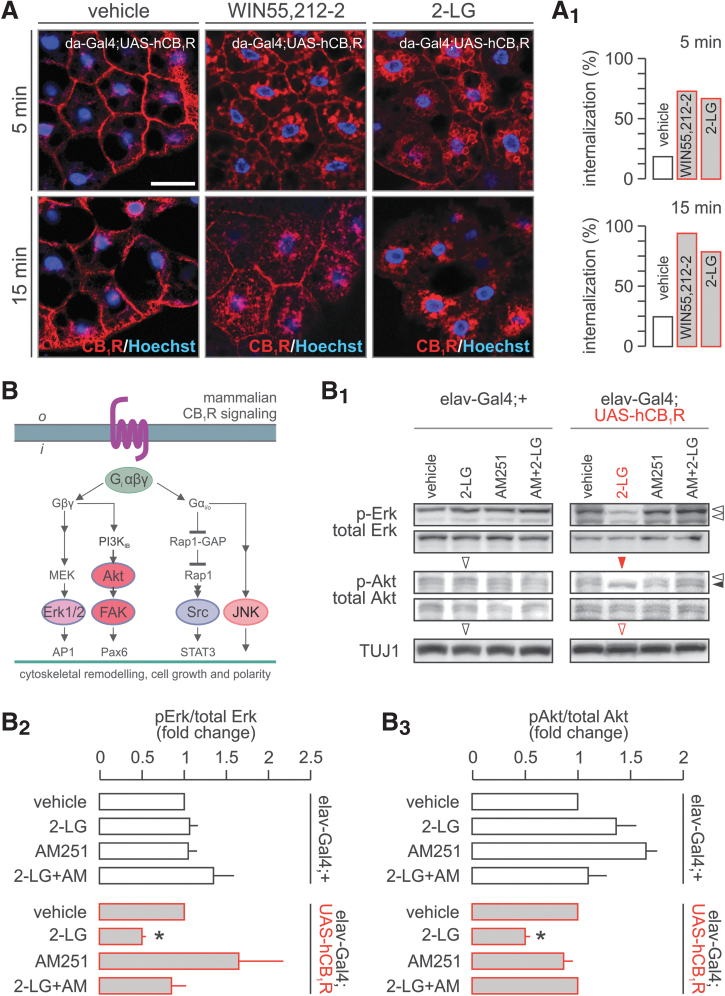
FIG. 4.
Paradoxical signaling by hCB1Rs in Drosophila. (A) WIN 55,212-2 and 2-LG induced rapid hCB1R internalization in Drosophila adipocytes. The percentage of cells with internalized hCB1Rs is shown in (A1). (B) Canonical signal transduction pathways activated by the CB1R in mammals. In Drosophila, 2-LG induced the inhibition of both extracellular signal-regulated protein kinase 1/2 (Erk1/2) and protein kinase B (Akt) pathways in an AM 251-dependent fashion. (B1) Representative experiment. (B2, B3) Data from triplicate experiments expressed as mean±s.e.m.; *p<0.05 (Student's t-test). Scale bar = 15 μm (A).
Hence, we first expressed hCB1R ubiquitously, using the da-Gal4 driver, and tested hCB1R internalization as a functional read-out of G protein-coupled receptor activation. By superfusing WIN 55,212-2 (1 μM, cannabinoid receptor agonist) or 2-LG (2 μM) over the fat body, we find rapid hCB1R translocation. Both ligands induced hCB1R internalization, affecting >80% of adipocytes by 15 min after acute stimulation (Fig. 4A, A1). This response prompted us to test whether ectopic hCB1R activation could initiate kinase signaling triggering cytoskeletal reorganization (for mammalian candidates see Fig. 4B, B1) in tissues of 3–6-day-old flies (∼300/sample with tissues lysed, cleared, and proteins extracted). Neither 2-LG nor AM251, a CB1R antagonist, induced kinase activation in tissue homogenates of elav-Gal4;+ (control) flies, suggesting that there is little if any basal activation of the receptors in the absence of a stimulus. In contrast, 2-LG (2 μM, 10 min) inhibited (p<0.05) the phosphorylation of both extracellular signal-regulated kinases 1/2 (Erk1/2) and Akt upon hCB1R expression in postmitotic neurons of 3–6-day-old fruit flies (elav-Gal4;UAS-hCB1R; Fig. 4B1), while being ineffective at phosphorylating c-Jun N-terminal kinases or Src (not shown). Inhibition of Erk1/2 and Akt phosphorylation was rapid (Supplementary Fig. S1D), and commenced without transient activation. This, together with AM 251 antagonism of these responses (Fig. 4B–B3), supports the supposition that 2-LG is a bona fide partial agonist15 inducing paradoxical signaling, rather than desensitization, at hCB1R expressed ectopically in Drosophila.
2-LG binds to CB1Rs in mouse cortical homogenates
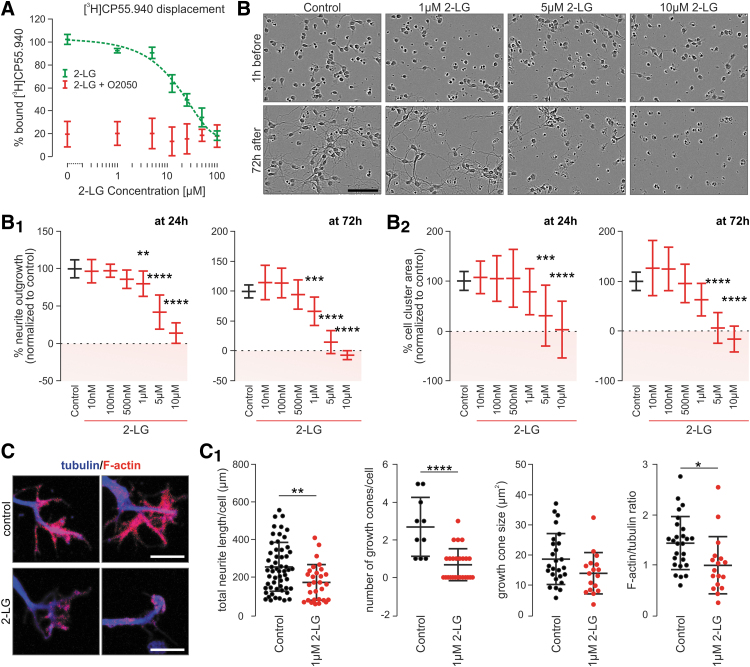
Recently, a recombinant CB1R-transcription factor interaction assay showed that, in mammalian cells, 2-LG also acts as a partial agonist of the CB1R.15 However, data from native mammalian systems on 2-LG binding to the cannabinoid receptor is still lacking. Therefore, we performed a classical radioligand-binding assay to test if 2-LG can replace [3H]CP 55,940, a high-affinity CB1R agonist,40 from its binding site in membrane fractions prepared from cerebral cortices of adult mice. We used JZL 195 (100 nM), a dual MAGL and fatty-acid amide hydrolase inhibitor,41 to inhibit potential 2-LG degradation in situ. JZL195 alone did not influence [3H]CP 55,940 binding (data not shown). Under these conditions, 2-LG replaced [3H]CP 55,940 in a dose-dependent fashion with a calculated IC50 of 22.82 μM (Fig. 5A). Notably, O-2050 (100 nM, used as a CB1R antagonist) coapplication completely eliminated both [3H]CP 55,940 binding and 2-LG modulation at all concentrations used (Fig. 5A). These results suggest that 2-LG has the ability to modulate mammalian CB1Rs, even if at concentrations a magnitude higher than those for 2-AG and AEA.
FIG. 5.
2-LG-induced growth phenotypes in mammalian neurons. (A) Displacement of [3H]CP 55,940 binding by increasing 2-LG concentrations in mouse cortical membranes. O-2050, a silent CB1R antagonist, was used as control. (B) Phase-contrast images of cultured mouse neurons before and after exposure to increasing 2-LG concentrations. Images are from an IncuCyte Zoom automated imaging platform. Quantitative analysis of the rate of neurite outgrowth (B1) and cell area coverage (a surrogate of cell survival, B2) 1 h before and 72 h after 2-LG treatment in vitro. Boxes in pink demonstrate neurite retraction (B1) and cell death (B2). (C, C1) 2-LG induces growth cone collapse and reduces the F-actin/β-III-tubulin ratio. Solid circles correspond to individual data points. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 from quadruplicate (A) and duplicate (B) experiments. Scale bars=10 μm (C), 100 μm (B).
2-LG reduces neurite outgrowth and induces growth cone collapse in mouse neurons
Next, we asked if 2-LG exposure of fetal mouse neurons affects their morphological differentiation, recapitulating developmental phenotypes imposed by endocannabinoids and synthetic CB1R agonists.27,34,42 First, we have measured if 2-LG affects neurite outgrowth using an automated IncuCyte imaging system with 2-LG replenished every 24 h during a period of 3 days in vitro (Fig. 5B). Indeed, when used in concentrations >500 nM, 2-LG significantly reduced neurite outgrowth (Fig. 5B1,) with 10 μM 2-LG inducing near-complete inhibition. As such, 2-LG at concentrations ≥5 μM significantly reduced the surface area occupied by neurons (Fig. 5B2) demonstrating that reduced cell survival was a significant confound in measuring decreased neurite occupancy. These data cumulatively show that 2-LG reduces neurite outgrowth in a pharmacological window of 500 nM–5 μM without significant cytotoxicity.
Finally, we used single-cell morphometry to test if 2-LG affects neurite growth cone morphology, an efficient means to reduce the rate of neurite growth among others by endocannabinoids.27,43,44 2-LG (1 μM) significantly reduced the length and branching of neurites per cell (Fig. 5C, C1). Moreover, 2-LG significantly reduced the number of growth cones, as well as the F-actin/β-III-tubulin ratio in individual growth cones (Fig. 5C, C1), defining subcellular sites and morphological correlates for its action. In sum, these data demonstrate that 2-LG is efficacious in modulating neuronal differentiation, at least in vitro, in a manner similar to classical endocannabinoids.42,43,45
Discussion
During evolution of the animal kingdom, the vertebrate DAGLs have likely coevolved with cannabinoid receptors.1 This hypothesis is logical because these enzymes are directly responsible for the “on-demand” synthesis of 2-AG in vertebrates,10,12,21 the signal lipid that is now emerging as the major endocannabinoid with ubiquitous functions in the developing and adult nervous systems.12,46 We also note that DAGL activity directly affects lipid metabolism, which, both in Drosophila and mammals, can impinge upon the degradation of glycerophospholipids as membrane components and triacylglycerols as energy sources (for reviews see Refs.47,48). Beyond the modulation of synaptic neurotransmission, Nomura et al.49 highlighted a key role for DAGLs in conjunction with MAGL to drive inflammatory responses by hydrolysis of 2-AG to AA.49 In this context, our data show that dietary supplementation of Drosophila with AA is lethal at late larval development, likely because of an overt and body-wide inflammatory response.
A highly conserved ortholog of DAGLα is found in Drosophila (inaE/dDAGL), but its function must differ as flies do not normally synthesize 2-AG and have no cannabinoid receptors in their genome (see Introduction section). We posed four related questions as a first step toward understanding the function of the dDAGL: (1) its pattern of expression, (2) its ability to regulate the level of alternative 2-monoacylglycols, (3) the ability of dDAGL-derived lipids to act as signaling molecules, and (4) a role for dDAGL in the regulation of axonal growth and guidance.
The first aim of this study was to gain insights into potential function of dDAGL in the nervous system by characterizing expression patterns in the embryonic and adult nervous system. Our results show widespread and dynamic patterns of dDAGL expression as revealed by in situ hybridization, and confirmed by immunocytochemistry. We have performed in situ hybridization for alternative dDAGL transcripts that differ only in the amino terminal tail region, and we report differences in their expression patterns, particularly in the adult nervous system. In the mammalian brain, the primary structural difference between DAGLα and DAGLβ is the presence of a long amino-terminal tail in DAGLα that is likely responsible for targeting it to dendritic spines.12 Interestingly, expression of the long-tail splice form of dDAGL is enriched over the short form in areas of the Drosophila nervous system (brain and nerve cord), where postsynaptic dendritic arborizations are present, for example, in the calyx of the mushroom bodies. We would speculate that differences in the tail region of dDAGL isoforms might allow for the localization to different cellular compartments. However, this can only be tested directly when specific antibodies will become available. Nonetheless, these initial studies are consistent with widespread functions for dDAGLs in the embryonic and adult nervous system.
Given the likely functions in the nervous system, we turned our attention to the question of 2-monoacylglycerols that might be regulated by dDAGL. If AA is available in the diet, flies will accumulate 2-AG in a dDAGL-dependent manner. However, this is unlikely to be of physiological relevance as AA is not an essential dietary supplement. Therefore, dDAGL most likely regulates the expression of alternative 2-monoacylglycerols with 2-LG being a major candidate as the vertebrate DAGLs can synthesize this lipid,10 and basal levels are readily detectable in Drosophila.14 Our results show substantial reductions (50–60%) in 2-LG levels in the inaEKG08585 hypomorph as well as the inaEExc.90.1 and inaERNAi loss-of-function mutant lines. Notably, 2-LG of plant origin is present in standard feeding formulations and this might account in part for the residual 2-LG in inaEKG08585, inaEExc.90.1, and inaERNAi larvae. Irrespective of this confound, our results provide substantial evidence for inaE/dDAGL regulating 2-LG levels.
2-LG might be an important signaling lipid in Drosophila, but the absence of an identified receptor makes this a difficult hypothesis to test. Discovering a cognate “2-LG” receptor specific to Drosophila would necessitate the use of a chemical 2-LG-probe (whether radioactive or fluorescent) as bait with body-wide screens of its binding and subsequent molecular identification, as performed commonly for receptors and enzymes.50,51 To circumvent this problem and to specifically test if 2-LG can activate hCB1Rs, we have expressed the hCB1R in Drosophila and asked if 2-LG can activate ectopically expressed hCB1R as a proof of principle for a potential signaling function. The choice of hCB1R as a “surrogate” is based on the coevolution of the DAGLs and cannabinoid receptors.1 Although Drosophila are considered to have lost a cannabinoid receptor from their genome,8 the retention of dDAGL might still allow for the synthesis of ancestral endocannabinoids. Indeed, it has been proposed that cannabinoid receptors first recognized “a fatty acid ester ligand (akin to 2-AG) in ancestral metazoans.”1 We demonstrate that 2-LG can trigger rapid (within 5 min) hCB1R internalization in adipocytes and modulate kinase cascades in a hCB1R-dependent manner in Drosophila neurons. Both responses are inhibited by a selective CB1R antagonist, and this at face value identifies 2-LG as a novel endocannabinoid. The combination of our transgenic data in Drosophila and ligand binding in mouse cortical homogenates together with recent evidence from a transcriptional activator-based Tango assay,15 supersede previous studies suggesting that 2-LG does not obviously bind to or activate CB1R-dependent responses in a mammalian cell line yet enhance 2-AG binding through an “entourage” effect.16 A reason for this is that the cell type-specific and conditional expression of the hCB1R can significantly increase receptor sensitivity, revealing primary ligand properties. If this is the case, then perhaps 2-LG is an ancestral endocannabinoid, with 2-AG evolving to take its place with the appearance of the Δ5, Δ6 desaturases that allowed for the synthesis of arachidonate-containing lipids, and/or the emergence of AA as an essential dietary lipid.
We reasoned that if the fly does contain mammalian-type endocannabinoids, then the expression of the hCB1R might be all that is required to reconstitute an endocannabinoid signaling pathway, although one that would most likely disturb normal function. If there are no endocannabinoids, one might not expect to see any gain of function following expression of the hCB1R either. Locomotion is a complex integrated physiological response widely used to assess Drosophila behaviors upon transgene expression.29 In this study, we used cha-Gal4 as a driver to overexpress hCB1R because the ensuing transgene expression pattern was reminiscent of that seen upon dDAGL localization. Coincident hCB1R expression by cha-Gal438 in both intersegmental cholinergic neurons that excite motor neurons and some motor neurons themselves also helped our behavioral readouts by ensuring hCB1R gain of function in key cellular components of locomotor circuits. The transgenic expression of the hCB1R substantially impaired locomotor activity. Remarkably, the gain-of-function response was absent when there was no dDAGL activity in the fly. This clearly supports the hypothesis that dDAGL produces an endogenous ligand that activates ectopically expressed hCB1Rs to modulate an integrated physiological response. Further detailed studies will be required to dissect out the precise molecular basis for this response, however, we did not see any obvious perturbation of the gross anatomy of this particular circuit (unpublished observation) suggesting that the phenotype might be related to an inhibitory action of 2-LG (or another dDAGL product) at hCB1Rs.
Given that the above results all support the hypothesis that the dDAGL can synthesize an endocannabinoid-like ligand, the question arises as to natural function in the nervous system. Muscle innervation by motor axons in the developing Drosophila embryo is a well-characterized pathway, where a number of adhesion molecules, and in particular the Drosophila L1, are known to function,36 and in vertebrates DAGL activity is required for L1 responses.37 We show that mutant flies lacking dDAGL activity have axonal growth and guidance defects in this pathway, similar to those found in L1/nrg mutants, and that these defects can be partially rescued by transgenic expression of dDAGL in neurons. These findings are also akin to one of the functions of the mammalian DAGLα ortholog with 2-LG-induced slowing of neurite extension and axonal growth cone collapse in mouse neurons in vitro. We conclude that some aspects of DAGL function are conserved between the fly and mammals; however, different receptors must be used to respond to the signaling lipids generated by (d)DAGL enzymes. Our signaling and behavioral studies in transgenic fruit fly highlight the presence novel dDAGL generated in endocannabinoid-like signal lipids and counterpart receptors, the identification of which might yield new insights into DAGL function in vertebrates.
Conclusions
Taken together, a new in vivo model system in Drosophila is used in this study for the discovery of novel endocannabinoid ligands, receptor interactions, and signaling properties. Molecular pharmacology complemented with genetic and behavioral tools demonstrates that (d)DAGLs are promiscuous toward lipid precursors, with the identity of ensuing 2-monoacylglycerols dictated by dietary constraints. Thus, 2-AG as a major endocannabinoid is an evolutionary bypass, whose utilization coevolved with that of Δ5, Δ6 desaturases,10,12,14 as well as with preference for AA-containing diets. Thus, 2-LG in insects and lower invertebrates may be considered as an ancestral endocannabinoid-like ligand. Our signaling and behavioral studies in transgenic fruit flies highlight that as yet undescribed endocannabinoid-like signal lipids may exist even in organisms that lack both cannabinoid receptor orthologs and cannabinoid binding sites. Such endocannabinoid-like signal lipids could impact homeostatic control exerted by CB1Rs in peripheral organs, as well as at central synapses. Moreover, we suggest that the cell type-specific and conditional expression of hCB1Rs can significantly increase receptor sensitivity, revealing ligand properties otherwise masked by homeostatic control (such as the “entourage effect” for 2-LG in rodents16–18 or receptor modulation by interacting proteins). Whatever the exact molecular components and mechanisms are, the concept of reconstructing signaling networks in Drosophila appear appropriate to untangle intermolecular interactions and devise high-throughput screens of endocannabinoid-like bioactive compounds without the usual mammalian complexity and constraints. Moreover, this model uncovers that insects might have lost the CB1Rs because it could have represented a hindrance of biological fitness (reducing mobility), thus compromising the species' survival.
Author Contributions
P.D. and T.H. conceived the general concept of this study (DAGL function and hCB1R overexpression, respectively); G.T., E.K., V.D.M., P.D., and T.H. designed experiments; S.R., M.J.W., and K.M. contributed tools and test systems; G.T., J.B., S.R., G.W., G.A.C., and E.K. performed experiments; G.T., J.B., S.R., G.W., P.D., E.K., and T.H. analyzed data, G.T. and T.H. wrote the article. All authors commented on the article and approved its submission.
Supplementary Material
Acknowledgments
The authors thank O.K. Penz for her assistance with the maintenance of Drosophila stocks, M. Watanabe for anti-CB1 receptor antibodies, and T. Hummel for discussions and critical feedback on this article. GW Pharmaceuticals (United Kingdom) are acknowledged for providing access to an IncuCyte Zoom (Essen Bioscience) live-cell imaging platform.
Abbreviations Used
- 2-AG
2-arachidonoylglycerol
- 2-LG
2-linoleoyl glycerol
- AA
arachidonic acid
- BDSC
Bloomington Drosophila Stock Center
- CB1R
CB1 cannabinoid receptors
- DAGL
sn-1-diacylglycerol lipases)
- DIV
days in vitro
- EDTA
ethylenediaminetetraacetic acid
- hCB1R
human CB1R
- MAGL
monoacylglycerol lipase
- MS
mass spectrometry
- PFA
paraformaldehyde
- PUFAs
polyunsaturated fatty acids
- TSA
tyramine signal amplification
- VDRC
Vienna Drosophila RNAi Center
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported by the EMBO Young Investigator Program (T.H.), Swedish Research Council (T.H.); Novo Nordisk Foundation (T.H.); Hjärnfonden (T.H.); European Research Council (SECRET-CELLS, ERC-2015-AdG-695136; T.H.), intramural funds of the Medical University of Vienna (T.H.); and the Wellcome Trust (P.D.)
Supplementary Material
Cite this article as: Tortoriello G, Beiersdorf J, Romani S, Williams G, Cameron GA, Mackie K, Williams MJ, Di Marzo V, Keimpema E, Doherty P, Harkany T (2021) Genetic manipulation of sn-1-diacylglycerol lipase and CB1 cannabinoid receptor gain-of-function uncover neuronal 2-linoleoyl glycerol signaling in Drosophila melanogaster, Cannabis and Cannabinoid Research 6:2, 119–136, DOI: 10.1089/can.2020.0010.
References
- 1. McPartland JM, Norris RW, Kilpatrick CW. Coevolution between cannabinoid receptors and endocannabinoid ligands. Gene. 2007;397:126–135 [DOI] [PubMed] [Google Scholar]
- 2. De Petrocellis L, Melck D, Bisogno T, et al. Finding of the endocannabinoid signalling system in Hydra, a very primitive organism: possible role in the feeding response. Neuroscience. 1999;92:377–387 [DOI] [PubMed] [Google Scholar]
- 3. McPartland JM, Agraval J, Gleeson D, et al. Cannabinoid receptors in invertebrates. J Evol Biol. 2006;19:366–373 [DOI] [PubMed] [Google Scholar]
- 4. Elphick MR, Egertová M. The neurobiology and evolution of cannabinoid signalling. Philos Trans R Soc Lond B Biol Sci. 2001;356:381–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oakes MD, Law WJ, Clark T, et al. Cannabinoids activate monoaminergic signaling to modulate key C. elegans behaviors. J Neurosci Off J Soc Neurosci. 2017;37:2859–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Averof M, Akam M. Hox genes and the diversification of insect and crustacean body plans. Nature. 1995;376:420–423 [DOI] [PubMed] [Google Scholar]
- 7. Regier JC, Shultz JW, Zwick A, et al. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature. 2010;463:1079–1083 [DOI] [PubMed] [Google Scholar]
- 8. McPartland J, Di Marzo V, De Petrocellis L, et al. Cannabinoid receptors are absent in insects. J Comp Neurol. 2001;436:423–429 [DOI] [PubMed] [Google Scholar]
- 9. McPartland JM. Phylogenomic and chemotaxonomic analysis of the endocannabinoid system. Brain Res Brain Res Rev. 2004;45:18–29 [DOI] [PubMed] [Google Scholar]
- 10. Bisogno T, Howell F, Williams G, et al. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J Cell Biol. 2003;163:463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leung H-T, Tseng-Crank J, Kim E, et al. DAG lipase activity is necessary for TRP channel regulation in Drosophila photoreceptors. Neuron. 2008;58:884–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reisenberg M, Singh PK, Williams G, et al. The diacylglycerol lipases: structure, regulation and roles in and beyond endocannabinoid signalling. Philos Trans R Soc B Biol Sci. 2012;367:3264–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drexler H, Spiekermann P, Meyer A, et al. Metabolic engineering of fatty acids for breeding of new oilseed crops: strategies, problems and first results. J Plant Physiol. 2003;160:779–802 [DOI] [PubMed] [Google Scholar]
- 14. Tortoriello G, Rhodes BP, Takacs SM, et al. Targeted lipidomics in Drosophila melanogaster identifies novel 2-monoacylglycerols and N-acyl amides. PLoS One. 2013;8:e67865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu L, Williams G, Doherty P. 2-Linoleoylglycerol is a partial agonist of the human cannabinoid type 1 receptor that can suppress 2-arachidonolyglycerol and anandamide activity. Cannabis Cannabinoid Res. 2019;4:231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ben-Shabat S, Fride E, Sheskin T, et al. An entourage effect: inactive endogenous fatty acid glycerol esters enhance 2-arachidonoyl-glycerol cannabinoid activity. Eur J Pharmacol. 1998;353:23–31 [DOI] [PubMed] [Google Scholar]
- 17. Murataeva N, Dhopeshwarkar A, Yin D, et al. Where's my entourage? The curious case of 2-oleoylglycerol, 2-linolenoylglycerol, and 2-palmitoylglycerol. Pharmacol Res. 2016;110:173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blasco-Benito S, Seijo-Vila M, Caro-Villalobos M, et al. Appraising the ‘entourage effect’: antitumor action of a pure cannabinoid versus a botanical drug preparation in preclinical models of breast cancer. Biochem Pharmacol. 2018;157:285–293 [DOI] [PubMed] [Google Scholar]
- 19. Keimpema E, Mackie K, Harkany T. Molecular model of cannabis sensitivity in developing neuronal circuits. Trends Pharmacol Sci. 2011;32:551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dalton GD, Howlett AC. Cannabinoid CB1 receptors transactivate multiple receptor tyrosine kinases and regulate serine/threonine kinases to activate ERK in neuronal cells. Br J Pharmacol. 2012;165:2497–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Díaz-Alonso J, Guzmán M, Galve-Roperh I. Endocannabinoids via CB1 receptors act as neurogenic niche cues during cortical development. Philos Trans R Soc B Biol Sci. 2012;367:3229–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berghuis P, Dobszay MB, Wang X, et al. Endocannabinoids regulate interneuron migration and morphogenesis by transactivating the TrkB receptor. Proc Natl Acad Sci U S A. 2005;102:19115–19120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bellen HJ, Levis RW, Liao G, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nagaso H, Murata T, Day N, et al. Simultaneous detection of RNA and protein by in situ hybridization and immunological staining. J Histochem Cytochem. 2001;49:1177–1182 [DOI] [PubMed] [Google Scholar]
- 25. Tortoriello G, de Celis JF, Furia M. Linking pseudouridine synthases to growth, development and cell competition. FEBS J. 2010;277:3249–3263 [DOI] [PubMed] [Google Scholar]
- 26. Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Dev Camb Engl. 1993;118:401–415 [DOI] [PubMed] [Google Scholar]
- 27. Keimpema E, Barabas K, Morozov YM, et al. Differential subcellular recruitment of monoacylglycerol lipase generates spatial specificity of 2-arachidonoyl glycerol signaling during axonal pathfinding. J Neurosci. 2010;30:13992–14007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Feany MB, Bender WW. A Drosophila model of Parkinson's disease. Nature. 2000;404:394–398 [DOI] [PubMed] [Google Scholar]
- 29. Crowther DC, Kinghorn KJ, Miranda E, et al. Intraneuronal Abeta, non-amyloid aggregates and neurodegeneration in a Drosophila model of Alzheimer's disease. Neuroscience. 2005;132:123–135 [DOI] [PubMed] [Google Scholar]
- 30. Frydman-Marom A, Levin A, Farfara D, et al. Orally administrated cinnamon extract reduces β-amyloid oligomerization and corrects cognitive impairment in Alzheimer's disease animal models. PLoS One. 2011;6:e16564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maccarrone M, Guzmán M, Mackie K, et al. Programming of neural cells by (endo)cannabinoids: from physiological rules to emerging therapies. Nat Rev Neurosci. 2014;15:786–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shen LR, Lai CQ, Feng X, et al. Drosophila lacks C20 and C22 PUFAs. J Lipid Res. 2010;51:2985–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fezza F, Dillwith JW, Bisogno T, et al. Endocannabinoids and related fatty acid amides, and their regulation, in the salivary glands of the lone star tick. Biochim Biophys Acta. 2003;1633:61–67 [DOI] [PubMed] [Google Scholar]
- 34. Alpár A, Tortoriello G, Calvigioni D, et al. Endocannabinoids modulate cortical development by configuring Slit2/Robo1 signalling. Nat Commun. 2014;5:4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berghuis P, Rajnicek AM, Morozov YM, et al. Hardwiring the brain: endocannabinoids shape neuronal connectivity. Science. 2007;316:1212–1216 [DOI] [PubMed] [Google Scholar]
- 36. Hall SG, Bieber AJ. Mutations in the Drosophila neuroglian cell adhesion molecule affect motor neuron pathfinding and peripheral nervous system patterning. J Neurobiol. 1997;32:325–340 [PubMed] [Google Scholar]
- 37. Williams E-J, Walsh FS, Doherty P. The FGF receptor uses the endocannabinoid signaling system to couple to an axonal growth response. J Cell Biol. 2003;160:481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Inada K, Kohsaka H, Takasu E, et al. Optical dissection of neural circuits responsible for Drosophila larval locomotion with halorhodopsin. PLoS One. 2011;6:e29019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Salvaterra PM, Kitamoto T. Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Brain Res Gene Expr Patterns. 2001;1:73–82 [DOI] [PubMed] [Google Scholar]
- 40. White R, Robin Hiley C. The actions of some cannabinoid receptor ligands in the rat isolated mesenteric artery. Br J Pharmacol. 1998;125:533–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Long JZ, Nomura DK, Vann RE, et al. Dual blockade of FAAH and MAGL identifies behavioral processes regulated by endocannabinoid crosstalk in vivo. Proc Natl Acad Sci U S A. 2009;106:20270–20275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mulder J, Aguado T, Keimpema E, et al. Endocannabinoid signaling controls pyramidal cell specification and long-range axon patterning. Proc Natl Acad Sci U S A. 2008;105:8760–8765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Keimpema E, Tortoriello G, Alpár A, et al. Nerve growth factor scales endocannabinoid signaling by regulating monoacylglycerol lipase turnover in developing cholinergic neurons. Proc Natl Acad Sci U S A. 2013;110:1935–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jiang W, Zhang Y, Xiao L, et al. Cannabinoids promote embryonic and adult hippocampus neurogenesis and produce anxiolytic- and antidepressant-like effects. J Clin Invest. 2005;115:3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tortoriello G, Morris CV, Alpar A, et al. Miswiring the brain: Δ9-tetrahydrocannabinol disrupts cortical development by inducing an SCG10/stathmin-2 degradation pathway. EMBO J. 2014;33:668–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oudin MJ, Hobbs C, Doherty P. DAGL-dependent endocannabinoid signalling: roles in axonal pathfinding, synaptic plasticity and adult neurogenesis. Eur J Neurosci. 2011;34:1634–1646 [DOI] [PubMed] [Google Scholar]
- 47. Shin M, Ware TB, Hsu K-L. DAGL-Beta Functions as a PUFA-Specific Triacylglycerol Lipase in Macrophages. Cell Chem Biol. 2020. DOI: 10.1016/j.chembiol.2020.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ogasawara D, Deng H, Viader A, et al. Rapid and profound rewiring of brain lipid signaling networks by acute diacylglycerol lipase inhibition. Proc Natl Acad Sci U S A. 2016;113:26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nomura DK, Morrison BE, Blankman JL, et al. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science. 2011;334:809–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schreiber SL, Kotz JD, Li M, et al. Advancing biological understanding and therapeutics discovery with small-molecule probes. Cell. 2015;161:1252–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Federico S, Lassiani L, Spalluto G. Chemical probes for the adenosine receptors. Pharmaceuticals (Basel). 2019;12:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.