Abstract
To date, the COVID-19 pandemic has claimed over 1 million human lives, infected another 50 million individuals and wreaked havoc on the global economy. The crisis has spurred the ongoing development of drugs targeting its etiological agent, the SARS-CoV-2. Targeting relevant protein-protein interaction interfaces (PPIIs) is a viable paradigm for the design of antiviral drugs and enriches the targetable chemical space by providing alternative targets for drug discovery. In this review, we will provide a comprehensive overview of the theory, methods and applications of PPII-targeted drug development towards COVID-19 based on recent literature. We will also highlight novel developments, such as the successful use of non-native protein-protein interactions as targets for antiviral drug screening. We hope that this review may serve as an entry point for those interested in applying PPIIs towards COVID-19 drug discovery and speed up drug development against the pandemic.
Keywords: COVID-19, PPIIs, Antiviral strategy, Drug discovery, SARS-CoV-2
1. Introduction
COVID-19 has become one of the most formidable public health crises of this century. As of now, over 50 million people have been infected with a death toll of over 1 million individuals worldwide [1]. It has also caused global social and economic disruption, with losses amounting to over US$ 8.5 trillion according to data from the United Nations [2]. COVID-19 is caused by a novel coronavirus named SARS-CoV-2 [3]. Given the urgency imposed by the crisis, several initiatives were quickly set up to gain a better understanding about the virus and to utilize that knowledge to pursue potential therapies and vaccines against the disease [4]. SARS-CoV-2 belongs to the beta-coronavirus subfamily and shares considerable similarity with SARS-CoV and MERS-CoV at the protein level [5]. The viral architecture is essentially identical to that of SARS-CoV and MERS-CoV, comprised of a phospholipid envelope, the structural proteins N (nucleocapsid), M (membrane), S (spike) and E (envelope), and non-structural proteins such as the 3C-like protease and the RNA-dependent RNA polymerase (RdRP). In a fortunate turn of events, the high similarity between SARS-CoV-2 and other coronaviruses allowed researchers to leverage knowledge from past research to speed up development of potential therapies. For example, the interaction between the SARS-CoV-2 S protein and the human angiotensin-converting enzyme 2 (ACE2) receptor, one of the most promising drug targets, was quickly elucidated based on what was known about its SARS-CoV counterpart [6], [7].
The battle against COVID-19 has been waged in several forms, but vaccine development and drug design remain two of the most important ones. Vaccines provide the means for large-scale immunization of the population which is essential for a return to normal life. Ye et al. provide an excellent summary of current preclinical efforts and technologies for vaccine development against SARS-CoV-2 [8]. However, given the scale of the pandemic, it may be necessary to vaccinate a large percentage of the global population before life can return to normal. Hence, drugs designed to combat COVID-19 will be required to “hold the fort” during the vaccine development/production/distribution time window. Even after successful implementation of a vaccination program, there is still a need for anti-COVID-19 drugs to deal with sporadic cases.
Several approaches have been applied towards drug development against COVID-19. Gil et al. provide a comprehensive overview of potential drug targets and therapeutic options against SARS-CoV-2 infections [9]. The best-known approach is to inhibit key enzymes in SARS-CoV-2 such as the RNA-dependent RNA polymerase (RdRp) and 3C-like protease (3CLpro). Remdesivir, a drug which has been in the spotlight, is a prime example because it is a nucleotide analogue which inhibits RdRp and has received emergency use authorization against COVID-19 in several countries [10], [11]. Although highly effective, this approach relies on targeting the catalytic site, which limits the chemical space that may be explored and may reduce the chances of identifying successful hits. A second popular approach is to disrupt the interaction between proteins which are essential for viral processes [12], [13]. In this review, we shall focus on the second approach and discuss various aspects of targeting protein-protein interaction interfaces (PPIIs) for anti-COVID-19 drug discovery.
2. Role of PPIIs in drug development
Nearly all biological processes involve some type of protein-protein interaction (PPI). In human cells, it is estimated that more than 300,000 PPIs participate in processes such as immunity, signal transduction, molecules transportation, and maintenance of cellular organization [14]. Aberrations in these interactions are correlated with many human disorders, including cancer, infectious diseases, autoimmune diseases and neurodegeneration [15], [16], [17], [18], [19], [20]. Other organisms, including pathogens, also have their own set of essential PPIs. The ability to target and manipulate these interactions may thus be a viable strategy for drug discovery [21] (Table 1). Paclitaxel, more commonly known as Taxol®, is an anticancer agent extracted from the Pacific yew Taxus brevifolia [22]. Paclitaxel arrests the cell cycle in the mitotic phase by inhibiting microtubule disassembly [23]. Binding of paclitaxel to a hydrophobic patch in β-tubulin stabilizes the interdimer contacts between β-tubulin molecules of adjacent protofilaments, resulting in microtubule stabilization [24], [25], [26]. Colchicine, which has been used to treat gout since 1961, utilizes a mechanism opposite to paclitaxel [27], [28]. Colchicine inhibits the assembly of microtubules by disrupting the interaction between α- and β-tubulin [29], [30], [31]. Although the mechanism of action of the two drugs were elucidated long after they were approved for medical use, recent studies have proven the possibility of targeting PPIs a priori for development of new treatments against refractory diseases [32], [33], [34].
Table 1.
Examples of PPII modulators described in the present review.
| Name | Protein complex | PPI modulators | PDB code | EC50 / IC50 (μM) | Ref |
|---|---|---|---|---|---|
| Paclitaxel | microtubules | Allosteric stabilizer | 1JFF | 1.41 ± 0.32 (IC50) | [26] |
| Colchicine | α- / β-tubulin | Orthosteric inhibitor | 1SA0 | 3.2 (IC50) | [31] |
| BIO8898 | CD40L/CD40 | Allosteric inhibitor | 3LKJ | 25 (IC50) | [40] |
| Compound 2/4 | gp120–CD4 | Orthosteric inhibitor | N/A | 22/9 (EC50) | [40] |
| FK506 | FKBP12/ calcineurin | Orthosteric stabilizer | 1TCO | 0.047 (IC50) | [44] |
| Rapamycin | FKBP12/mTOR | Orthosteric stabilizer | 2RSE | 0.002 (EC50) | [45] |
| Nucleozin | Influenza nucleoprotein | Orthosteric stabilizer | 3RO5 | 0.17 (IC50) | [54] |
PPIs usually involve a large and flat interaction interface between the proteins. These protein-protein interaction interfaces (PPIIs) typically have an area of 1500–3000 Å2 and are often complementary in shape and electrostatic properties [35], [36], [37]. Although the interface is usually composed of several amino acid residues, only a small subset of these residues, called “hot spots”, contributes significantly to the binding free energy [38], [39]. These hot-spots are prime targets for small-molecule orthosteric PPII disruptors, such as colchicine, which act directly on the PPII. Disruption of PPIIs may also be achieved by allosteric means where the small-molecules can bind to sites that are topologically distinct from PPIIs (Fig. 1). BIO8898 is a small-molecule inhibitor of the trimeric cytokine CD40-ligand (CD40L) developed by the company Biogen Idec targeted at autoimmune diseases [40]. The inhibitor intercalates at the trimer interface of CD40L and allosterically disrupts the CD40L/CD40 interaction associated with several types of autoimmune diseases. Caporuscio et al. reported two novel inhibitors against HIV-1 that target the Phe43 pocket in the gp120-CD4 interaction through molecular dynamics simulations, and Zhan et al. summarized the potential compounds against a variety of PPIs in HIV infection [41], [42]. Similarly, PPII stabilizers may also act in an orthosteric or allosteric fashion (Fig. 1). For example, FK506 and rapamycin inhibit calcineurin and mTOR kinase activity, respectively, by stabilizing the corresponding interactions between FKBP12 and calcineurin or mTOR [43], [44], [45]. Both FK506 and rapamycin bind first to FKBP12 and form part of the interaction interface with calcineurin and mTOR, respectively, and both are considered orthosteric PPII stabilizers [46]. On the other hand, paclitaxel is believed to exert allosteric stabilization in addition to its orthosteric effect in microtubules because the interface between α- and β-tubulin within the same protofilament, which is not part of the paclitaxel binding site, is also affected by paclitaxel binding [47], [48].
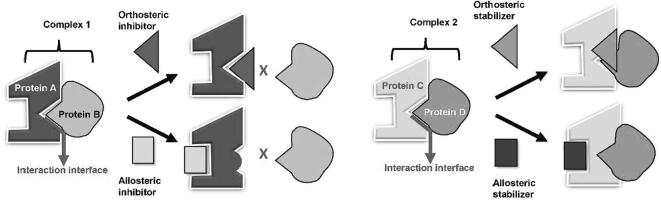
Fig. 1.
Different strategies for designing PPI modulators. Modulation of PPIs can be achieved by using inhibitors or stabilizers to target the orthosteric or allosteric sites of the protein-protein complex. (Right) graphic expression of PPI inhibitors. Protein A binds to Protein B to form Complex 1. Orthosteric inhibitors bind directly to the PPII, which hinders Protein B from binding with Protein A. On the other hand, allosteric inhibitors bind to a region distal from the PPII on Protein A, which induces a conformational change to obstruct Protein B from binding with Protein A. (Left) graphic expression of PPI stabilizers. Protein C interacts with Protein D to form Complex 2. Orthosteric stabilizers bind directly to the PPII, which enhances the binding affinity between Protein C and D. Allosteric stabilizers bind to a region distal to the PPII on protein C, which induces a conformational change to enhance the binding affinity between Protein C and Protein D.
Orthosteric PPII inhibitors and stabilizers are easier to design as long as there is enough information about the interaction interface as a guide [49]. In contrast, the design of allosteric modulators faces difficulties in identifying a suitable target site [49], [50]. However, it may sometimes be difficult to design orthosteric PPII modulators, e.g. when the interface is particularly flat and few hits are available, and allosteric compounds may provide alternative choices for drug design.
Non-native contact modulators comprise a particularly interesting category. Nucleozin induces aggregation of influenza A nucleoproteins in the nucleus of the host cell, which stops viral replication [51], [52], [53]. The structure of influenza A nucleoprotein in complex with nucleozin revealed that nucleozin exerted its antiviral activity by stabilizing a non-native interface between two adjacent nucleoprotein trimers, leading to abnormal viral nucleoprotein oligomerization and suppression of the influenza virus [51], [54].
3. Strategies for the design of PPII modulators
PPIIs have long been ignored as drug targets because of their flat interacting surfaces, which is not conducive towards conventional methods of drug discovery. Fortunately, technological advancements have made it possible to design and screen potential modulators of PPIIs with therapeutic potential. Several excellent and detailed reviews have been written on the subject [55], [56]. For the sake of brevity, we only highlight a few of the strategies:
3.1. Structural studies
Structural biology is arguably one of the most important tools for the design of PPII modulators. Unlike conventional methods where one may not require structural information for screening of potential therapeutic molecules, targeting PPIIs requires intimate knowledge of the interaction interface, which can only be provided by its detailed atomic structure. Energetics analysis of the structural data may provide clues about potential interaction hot spots, which may then be experimentally validated. In recent years, a combination of X-ray crystallography and alanine scanning mutagenesis has become the main strategy for this purpose [34], [39]. Even when a lead has been identified, elucidation of the structure of the proteins in complex with the lead molecule is still desirable, since it may yield additional information useful in the optimization process [57], [58], [59]. In addition, structural elucidation of the protein-modulator complex is currently the only way to identify allosteric PPII modulators.
3.2. Structure-based drug design
Once the structure of the interface is known, two different strategies may be employed to design PPII modulators. First, novel compounds may be generated through bioisosterism and de novo design based on the structure surrounding the hot spots. However, because PPIIs are often flat, a larger interaction surface between the compound and the interface may be desired. This leads to the second approach, peptidomimetic design, which employs small molecules or short peptide derivatives that mimic a binding peptide [60], [61], [62]. Human knowledge plays a key role in either approach since interpretation of the structure has to be carried out manually before the design phase.
3.3. High-throughput screening
To accelerate the development of PPII modulators, screening methods which are amenable to a high degree of automation may be employed. High-throughput screening (HTS) experiments and virtual screening are widely used approaches in traditional drug discovery [63], [64]. In theory, HTS does not even require the elucidation of the protein structures as long as a suitable validation assay targeting the PPII is available. Virtual screening, on the other hand, requires that the structures of the interacting proteins are known before hand, but has the advantage of high speed and low cost because the screening is carried out in silico. However, both HTS and virtual screening are limited by the chemical space represented by their compound libraries, and conventional libraries, which were not developed for this use case, may be less effective at screening PPII modulators. Virtual screening further suffers from an interface degeneracy problem, because there may be multiple potential PPIIs, but only one or a few of them are actually physiologically relevant. Although these issues are not trivial, both strategies have been successfully applied towards the identification of compounds regulating PPIs in recent years [65], [66], [67].
3.4. Fragment-based drug discovery
The hot spots on the PPII are often scattered at the interface and do not form a continuous surface. This is often a problem because compounds targeting a single hot spot may not bind to the protein tightly enough. Fragment-based drug discovery (FBDD) is an alternative approach that may be employed to solve this problem. FBDD starts from the identification of small fragments (~200 Da) that target a single site at the PPII. Once fragments for several sites have been identified, they may be linked into a single molecule to obtain a ‘lead’ which has a much higher binding affinity towards the target [68], [69]. Compared to HTS, FBDD allows for the initial screening of a smaller library, because N hits could be combined to produce N × N leads for further screening (assuming two screening sites), thus increasing the combinatorial space. However, optimization of the linker between the fragments by organic methods still requires the structure of the lead-target complex to be solved first, which is not absolutely necessary for HTS.
3.5. Computational tools
In silico strategies offer flexibility and insights which may not be accessible with experimental approaches. Advances in computational and systems biology, combined with the increasing amount of structural knowledge available has ushered several computational tools that may assist in the design of PPII modulators. Databases of small-molecule PPI inhibitors, such as TIMABL (http://www-cryst.bioc.cam.ac.uk/databases/timbal), 2P2I (http://2p2idb.cnrs-mrs-fr) or iPPI-DB (http://www.ippidb.cdithem.fr) contain three-dimensional structures of several protein-protein and protein-inhibitor complexes. These databases may serve as starting points for molecule design, or as resources to explore possible interaction “rules” at the protein-protein interface. Another aspect is the collaborative development of new theoretical and computational modeling approaches, such as OpenMM and other open force-field initiatives [70]. The increasing use of machine learning algorithms, such as those employed on the Alpha Fold system for protein fold prediction [71], may also provide more accurate predictions of binding residues at PPIIs. On the small-molecule side, novel computational methods may help in optimizing desired pharmacophore properties (such as oral bioavailability) and/or expanding the chemical space beyond the “rule of five” at the initial stages of the design process [72].
4. Application to COVID-19 drug discovery
4.1. Potential candidate targets
CoVs share several proteins that are essential in the viral life cycle. Many of these proteins form PPIs with each other or host proteins, making them attractive targets for the design of PPII modulators (Fig. 2). A comprehensive list of potential targets is listed below (Table 2).
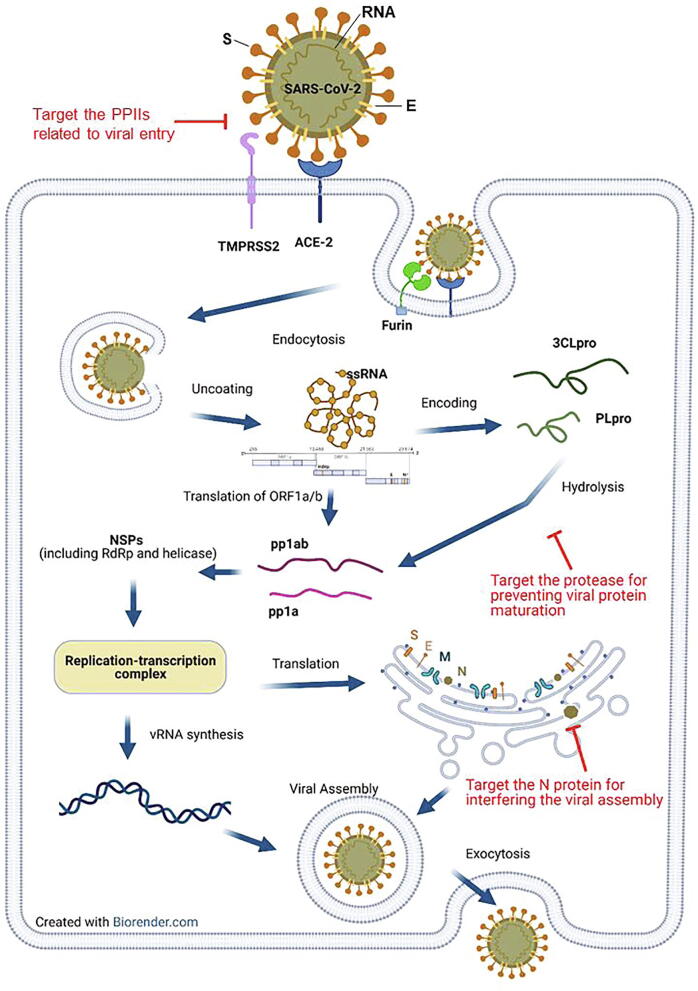
Fig. 2.
Critical PPIs and proteins involved in the virus life cycle. The PPIs suitable for inhibitor design are highlighted in red. Host proteins involved in CoV processing might be candidates that can be targeted through the PPII strategy: these include primary cellular receptors for CoV, such as ACE2 or DPP4, and host proteases, such as TMPRSS2 or furin. Host receptors are recognized by CoV spike proteins and the binding of receptor and S1 domain of S protein subsequently activates the conformational changes of S protein. For host proteases, the serine protease TMPRSS2 is responsible for two distinct functions during the CoV infection, including an alternative pathway for viral entry and activation of S protein for virus-cell fusion [128]. In parallel, the protease furin, which is predominantly expressed on the trans-Golgi network and intracellular vesicles, activates the S protein by cleaving at the S1/S2 cleavage site, thus facilitating membrane fusion [129], [130]. PPII inhibitors targeting host receptors and proteases may provide a potent way to prevent CoV from entering host cells during the early stages of infection. During later stages of infection, N protein dimerization and interaction with viral RNA is required for formation of RNP complexes and viral assembly. Targeting N dimers using a PPII strategy is a potential mechanism of inhibiting late steps of viral production. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Examples of PPII modulators against SARS-CoV/ SARS-CoV-2.
| Name | Virus | Type | Mechanism of inhibition | Targets | Ref |
|---|---|---|---|---|---|
| SSAA09E2 | SARS-CoV | Small molecule | Disturbing S-ACE2 interaction | PPII of S-ACE2 complex | [75] |
| VE607 | SARS-CoV | [76] | |||
| Methylene blue | SARS-CoV-2 | [77] | |||
| Diammonium Glycyrrhizinate | SARS-CoV-2 | [78] | |||
| corilagin | SARS-CoV-2 | [79] | |||
| CB6 | SARS-CoV-2 | Antibody | [80] | ||
| B38 | SARS-CoV-2 | [81] | |||
| 311mab-31B5 and 311mab-32D4 | SARS-CoV-2 | [85] | |||
| COVA2-15 | SARS-CoV-2 | [86] | |||
| IgG1 ab1 | SARS-CoV-2 | [87] | |||
| hrsACE2 | SARS-CoV-2 | Soluble peptide analogues of ACE2 | [83] | ||
| ACE2-Ig | SARS-CoV-2 | [88] | |||
| EK1C4 | SARS-CoV-2 | Lipopeptide | Disturbing 6-HB formation of S protein | S protein | [90] |
| Arbidol | SARS-CoV-2 | Small molecule | Modulating S protein trimerization | [96], [127] | |
| S471-503 | SARS-CoV | Soluble peptide analogues of S | Disturbing S-ACE2 interaction | host ACE2 | [97] |
| 438YKYRYL443 | SARS-CoV | host ACE2 | [98] | ||
| Chloroquine | SARS-CoV-2 | Small molecule | atypical PPI inhibition | [105] | |
| Octapeptide | SARS-CoV-2 | Peptide-based inhibitor | Disturbing intra-dimer of 3CLpro | 3C-like protease | [108], [109] |
| P3 | SARS-CoV-2 | Small molecule | Stabilizing a non-native dimer of N-NTD | Nucleocapsid protein | [113] |
4.1.1. PPIIs related to viral entry
Viral entry is initiated through the interaction between the receptor-binding domain of viral spike glycoprotein (S-RBD) and host receptors. Subsequently, host proteases such as TMPRSS2 or furin cleave the S protein and activates membrane fusion. The receptor for MERS-CoV S protein is dipeptidylpeptidase 4 (DPP4), whereas both SARS-CoV and SARS-CoV-2 S proteins bind to ACE2 [73], [74]. Being the first stage in viral infection, PPIIs involved in the viral entry process, such as those between S and host receptors/proteases, are considered to be one of the most promising drug development targets. Several PPII inhibitors targeting the interaction between S and host receptors have been identified. For example, Sarafianos et al. identified three compounds that block SARS-CoV entry from a chemical library containing 3000 compounds. One compound (designated SSAA09E2) was found to obstruct the binding of SARS-CoV S protein to ACE2 [75]. In addition, Kao et al. identified 104 compounds that inhibit SARS-CoV-induced cytopathic effects from a library of 50,240 small molecules. 18 compounds were found to block the S-ACE2-mediated entry of SARS-CoV, among which one compound, VE607, inhibited plaque formation of SARS-CoV at low micromolar range [76]. In light of these successful SARS-CoV studies, Bojadzic et al. initiated a screen to find compounds which interfere with the PPI between SARS-CoV-2 S protein and ACE2. They identified methylene blue as an inhibitor of SARS-CoV-2 entry with an IC50 of 3.5 μM [77]. Kalhor et al. conducted structure-based virtual screening with FDA approved drug databases to discover the PPI inhibitors against S-ACE2 complex. They identified 6 compounds can bind to the ACE2 binding pocket on SARS-CoV-2 S protein, from which they further proposed Diammonium Glycyrrhizinate as the most potent compound by MD simulation technique [78]. Hanson et al. also developed an AlphaLISA RBD − ACE2 platform to facilitate the screening of PPII inhibitors perturbing this host–pathogen interaction. They identified corilagin as a potential inhibitor against the ACE2 − RBD complex with an IC50 of 5.5 μM [79].
In addition to using the screening approach to find small-molecule candidates that block the S-ACE2 interaction, macromolecular PPI inhibitors have also been developed. In this regard, the most common approach is to mask the S-ACE2 interaction through monoclonal antibodies which recognize either the S protein or ACE2 [80], [81], [82]; or through the application of recombinant soluble proteins/peptides, which competes with normal proteins for the binding to their respective interacting partners [83], [84]. Currently, several antibodies recognizing the S protein have been reported. For example, Shi et al. isolated two specific human monoclonal antibodies from a convalescent COVID-19 patient that exhibited SARS-CoV-2 neutralization activity in vitro. One of these antibodies, termed CB6, further exhibited antiviral activity in a rhesus monkey model. The mechanism behind the antiviral activity of CB6 was further revealed by structural studies, which showed that CB6 interacted with the RBD of SARS-CoV-2 S protein in an orthosteric fashion and interfered with the virus–receptor interaction [80]. Yan et al. also isolated four antibodies from a convalescent patient which neutralized SARS-CoV-2. Two antibodies (B38 and H4) blocked the interaction between the S-RBD and ACE2 via binding to different epitopes on the RBD. Both antibodies were able to relieve the symptoms of infected animals in mouse model experiments. The crystal structure of the RBD-B38 complex revealed that the B38-binding surface on the RBD overlaps with its ACE2-binding interface (Fig. 3A). In addition, Chen et al. identified three SARS-CoV-2 antibodies from 26 recovered COVID-19 patients. Two of them, 311mab-31B5 and 311mab-32D4, exhibited neutralizing activities in host cells ectopically expressing hACE2. Enzyme-linked immunosorbent assays (ELISA) and flow cytometry-based blockade experiments proved that both antibodies specifically bind to SARS-CoV-2 RBD and may disrupt the PPI between RBD-ACE2 [85]. Brouwer et al. used an ELISA-based approach with SARS-CoV-2 stabilized prefusion S protein to isolate 19 neutralizing antibodies from blood samples of three convalescent COVID-19 patients. One of them, COVA2-15, showed picomolar neutralizing activity against infectious SARS-CoV-2. Single-particle negative-stain electron microscopy (EM) revealed that the COVA2-15 epitope partially overlapped with the binding region of ACE2 in S protein, suggesting that the antibody may block receptor engagement [86]. Li et al. also identified a potent monoclonal antibody (mAb), IgG1 ab1, from large antibody libraries. ELISA experiments revealed that IgG1 ab1 exhibited high-affinity to RBD and competed with ACE2 in vitro. More importantly, IgG1 ab1 showed high therapeutic efficacy in an animal experiment of SARS-CoV-2 infection [87].
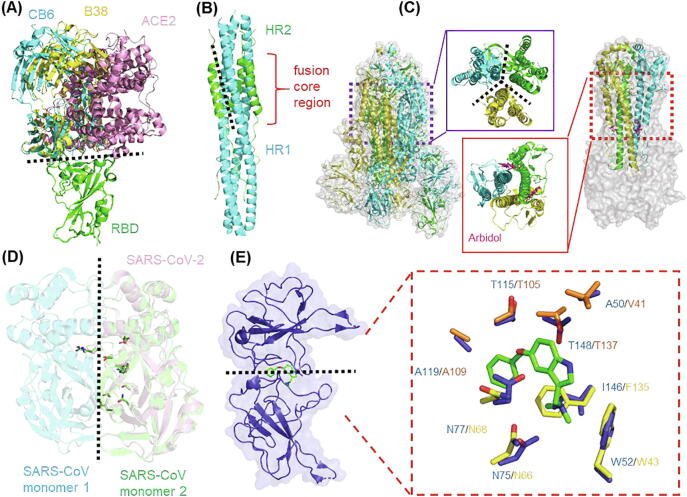
Fig. 3.
Main strategies for PPI modulator design against SARS-CoV-2. (A) Hot-spot for PPI inhibitor design against RBD-ACE2 complex. Structures of SARS-CoV-2 S protein RBD in complex with ACE2 (PDB: 6LZG), CB6 (PDB: 7C01) and B38 (PDB: 7BZ5) complex are shown in cartoon. The RBDs of each structure are aligned to show the PPI suitable for modulator design. (B) Hot-spot for PPI inhibitor design against fusion core region of S protein. SARS-CoV-2 6-HB structure is shown in cartoon with HR1 and HR2, colored in green and cyan, respectively (PDB: 6LXT). (C) Hot-spot for PPI modulator design against S protein trimerization. (Left) Structure of SARS-CoV-2 S protein trimer (PDB: 6VSB). The trimeric interface is enlarged in the middle. (Right) Structure of the influenza HA in complex with arbidol (PDB: 5T6N). The arbidol target site of the trimeric interface is enlarged in the middle. (D) Hot-spot for PPI inhibitor design against intra-dimer of 3CLpro. The structure of 3CLpro of SARS-CoV-2 (PDB:6Y2E) is aligned with that of SARS-CoV (PDB: 1UK4). The key residues involved in dimerization are shown in stick representation. (E) Hot-spot for PPI stabilizer design against N-NTD. The structure of SARS-CoV-2 N-NTD (PDB: 6M3M) is aligned to P3: MERS CoV N-NTD complex (PDB: 6KL6), the interacting residues are shown in sticks and highlighted in right box. The residues of SARS-CoV-2 and MERS CoV N-NTD are shown in orange and yellow, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Development of soluble peptide analogues of ACE2 is an alternative approach employed to compete with normal ACE2 for the binding to S protein. Monteil et al. have shown that human recombinant ACE2 (hrACE2) reduced the replication of SARS-CoV-2 by a factor 1000–5000 times in a cell model [83]. In addition, Hu et al. connected the extracellular domain of human ACE2 with human IgG‐Fc to generate a novel recombinant protein (ACE2-Ig). This chimeric recombinant protein displayed high affinity for binding to RBD of SARS-CoV2 and neutralized SARS-CoV-2 with potent efficacy in vitro [88]. These developments highlight the promise of using PPII inhibitors targeting the S-ACE2 complex as therapeutic agents against SARS-CoV-2.
Another indirect way of disrupting the interaction between the S protein and the host is by targeting regions of the protein involved in membrane fusion (Fig. 3B). The S protein forms a 6-helix bundle (6-HB) fusion core through two heptad repeats, HR1 and HR2, which brings the host and viral membranes close together for fusion and infection [89]. The inhibition of the formation of viral 6-HB could be a strategy to interfere the viral entry. For this purpose, Xia et al designed a series of lipopeptides against HR1 to disturb the formation of viral 6-HB. One of which, termed EK1C4, showed the capacity as a potent fusion inhibitor, which appeared to inhibit infection of several types of coronaviruses in cells, including SARS-CoV-2 [90], [91].
Based on the fact that S protein trimerization appeared to be the rate limiting step in other types of coronavirus infections [92], it has been suggested that manipulation of S protein trimerization may be another strategy to block the viral entry of SARS-CoV-2 (Fig. 3C). Kalathyia et al. employed molecular dynamics simulations to identify a highly conserved cavity within the S protein homotrimer of SARS-CoV-2 which may serve as a novel drug target for PPII inhibitor design [93]. Bongini et al. also performed molecular docking to identify eight available compounds targeting the trimer cavity which may interfere with trimerization of SARS-CoV-2 S protein [94]. Arbidol, a broad-spectrum antiviral drug against influenza, targets the trimerization interface of hemagglutinin (HA) and inhibits virus-host cell fusion by stabilizing the prefusion conformation of HA, which prevents further conformational rearrangements required for membrane fusion [95]. Since the role of HA in influenza is similar to S protein in SARS-CoV-2, arbidol is now being used in a clinical trial for treatment of SARS-CoV-2. By employing molecular dynamics and structural analysis, Vandakari suggested that arbidol may target the SARS-CoV-2 spike glycoprotein employing a mechanism similar to that of influenza virus [96].
Host proteins such as ACE2, DPP4, TMPRSS2 or furin are also considered targets for inhibition of viral entry. Liu et al. generated a library comprising of peptides derived from S protein of SARS-CoV for identifying the epitopes of SARS-CoV to target the ACE2 receptors. They found one peptide, S471-503, which specifically interfered with the interaction between the S-RBD and ACE2, and inhibited SARS-CoV entrance in vitro [97]. By a similar approach, Meyer et al. synthesized one hexapeptide (438YKYRYL443), derived from SARS-CoV S-RBD, to bind to ACE2 and inhibit viral entry [98] https://elsevier.proofcentral.com/en-us/landing-page.html?token=c0d27m66064a67522d0470b27d0966 [99]. Several substrate analogues have been proposed to target either furin or TMRRSS2 for the inhibition of influenza. Although the exact mechanism has not been elucidated, these substrate analogues may inhibit viral entry by inhibiting the interaction between furin [100], [101] or TMPRSS2 [102] and their respective substrates. Since the host targets exhibit lower mutation rates, these results from previous studies may provide an important basis to develop PPII inhibitors against SARS-CoV-2.
It is worth noting that chloroquine, a repurposed anti-malarial drug that has gained lots of attention in COVID-19 treatment [103], [104], [105], has been proposed to reduce the affinity of SARS-CoV S protein to ACE2 [106] by increasing endosomal pH. Hence, chloroquine appears to be one atypical example for PPI inhibition that does not directly involve PPIIs.
4.1.2. 3C-like protease (3CLpro)
The active form of 3CLpro is a dimer which cleaves the peptide bond between a glutamine and a small amino acid (serine, alanine or glycine). It is essential for processing the coronavirus polyprotein into its functional constituents [107]. In addition to the active site, the intra-dimer PPII is also a valid target for drug development (Fig. 3D). As a proof of concept, an octapeptide derived from the N-terminus of SARS-CoV 3CLpro has been shown to disrupt protease dimerization and inhibit viral replication [108], [109]. The important residues involved in dimerization of SARS-CoV 3CLpro, Arg4, Ser10, Gly11, Glu14, Asn28, Ser139, Phe140, Ser147, Glu290, Arg298, all are conserved in SARS-CoV-2. It is conceivable that the octapeptide may also be active against SARS-CoV-2, and similar stratagems for drug development against COVID-19 may be gleaned from this example [110].
4.1.3. Nucleocapsid (N) protein
The N protein is a dimer which self-assembles with viral RNA to form the ribonucleoprotein (RNP) particle [111], [112]. Our group has recently identified a novel non-native PPII between N protein dimers of MERS-CoV [113]. Formation of the PPII inactivates the N protein by occluding its essential RNA-binding site but requires the presence of a “glue” molecule to stabilize the non-native contacts. The shape of the non-native PPII is highly conserved among other coronaviruses, making it a potential target for broad-spectrum antivirals that may also be effective against SARS-CoV-2 (Fig. 3E). Preliminary in vitro studies showed that at least one compound was effective across MERS-CoV, SARS-CoV and mouse hepatitis virus, and early studies assessing its efficacy against SARS-CoV-2 appear to yield promising results. Elucidation of other non-native PPIs among coronaviral proteins may contribute additional non-canonical targets for drug development.
4.1.4. PPIIs involving other proteins
The proteins listed above represent only a fraction of the possible targets for anti-COVID-19 drug development. For example, the viral membrane (M) protein has long been known to interact with the N protein and is also essential for virion assembly [114], [115], implying that the PPII between N-M may be another possible target. On a broader scale, Gordon et al used affinity-purification mass spectrometry to discover 332 protein-protein interactions between SARS-CoV-2 and humans [116]. However, these examples lack the structural characterization of the interaction interface, thus limiting their potential for PPII-based drug development.
4.2. In-silico exploration of potential PPII modulators against COVID-19
Several computational studies have been carried out since the early days of the COVID-19 outbreak. These can be broken down into the following categories:
4.2.1. Identification of PPI networks
PPI networks provide a wealth of information about possible pathogenesis mechanisms and drug interactions that may not be evident using conventional approaches. For example, disease mechanisms were revealed through comparative analyses of various host-coronavirus protein interaction networks [117], [118]. These PPI networks may also provide insights into possible drug repurposing [119], [120], [121]. In addition, open PPI network databases such as STRING-covid (https://string-db.org/cgi/covid.pl) and IMEx coronavirus interactome [122] may provide novel potential targets for the design of PPII modulators.
4.2.2. Modeling the interaction between potential PPII modulators and binding proteins
Virtual screening through docking is generally cheap but does not provide the free-energy information required for drug binding affinity estimation. However, accurate simulations that do provide the necessary energetic parameters for binding are usually time-consuming and computationally (and monetary) expensive. The Anton supercomputer developed by David Shaw and coworkers promises to vastly reduce the computational time required to conduct such calculations [123]. The Anton has been used to model the structure of several SARS-CoV-2 proteins, including the binding of drug molecules to the trimeric S protein, and all the trajectories are openly accessible and free of charge (for details, see https://www.deshawresearch.com/downloads/download_trajectory_sarscov2.cgi/). Modeling using less esoteric hardware have been carried out for the binding of remdesivir, favilavir, and ribavirin to SARS-CoV-2 RdRp [124]. The interaction between remdesivir, chloroquine, ciclesonide and niclosamide to ACE2 have also been modelled via autodock simulations [125].
5. Future perspectives
The variety of approaches towards COVID-19 drug discovery afforded by targeting PPIIs share a common goal: to find molecular entities that can stop viral activity in its track. In fact, using a combination of drugs targeting different PPIIs and conventional viral targets in a “cocktail” formulation may provide the best chance of inhibiting viral activity. From this perspective, the inherent variability of PPIIs provides an opportunity to diversify the chemical space of COVID-19 drugs and may help avoid drug resistance problems arising from the usage of drugs targeting a single mechanism. Another advantage to targeting PPIIs is the possibility to develop broad-spectrum antivirals which may be useful against other coronaviruses [113]. It is telling that three of the most important emerging diseases of the century (SARS, MERS, and COVID-19) are all caused by coronaviruses and having a coronavirus-specific broad-spectrum drug may help avert the next coronavirus health crisis.
One of the major obstacles to targeting PPIIs for drug discovery is the lack of a starting scaffold for further development [126]. However, the case for COVID-19 is very different. Thanks to a research intensity which has never been seen before in the history of drug development, there are now several candidate molecules available for repurposing tests or as leads for further development. The main issue today is a lack of experimental validation of these molecules. With the number of potential PPII-targeting compounds and biologics on the rise, there is an immediate need for increased validation capacity among laboratories worldwide.
We believe that targeting PPIIs for drug development against COVID-19 is a viable strategy that warrants further consideration from the scientific community. This is especially true in the current crisis, which unfortunately does not appear to be abating any time soon. Development of PPII-targeting drugs may provide an additional piece in the arsenal of anti-coronaviral treatments, and we sincerely hope that further studies in this direction will one day help find a cure for COVID-19.
CRediT authorship contribution statement
Chung-ke Chang: Writing - original draft, Writing - review & editing. Shan-Meng Lin: Writing - original draft, Writing - review & editing. Roshan Satange: Writing - review & editing. Shih-Chao Lin: Writing - review & editing. Sin-Cih Sun: Writing - original draft. Hung-Yi Wu: Resources. Kylene Kehn-Hall: Writing - review & editing. Ming-Hon Hou: Conceptualization, Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by the MOST 109-2327-B-005 -005 and 109ST001C from ENABLE Center, NCHU.
References
- 1.Organization WH (2020) WHO. World Health Organization Situation Report.
- 2.Nations U (2020) COVID-19 to slash global economic output by $8.5 trillion over next two years.
- 3.Coronaviridae Study Group of the International Committee on Taxonomy of V The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous COVID-19 therapies and vaccine landscape. Nat Mater. 2020;19(8):809. doi: 10.1038/s41563-020-0758-9. [DOI] [PubMed] [Google Scholar]
- 5.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ali A., Vijayan R. Dynamics of the ACE2–SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Sci Rep. 2020;10(1):14214. doi: 10.1038/s41598-020-71188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W., Choe H., Farzan M. Insights from the association of SARS-CoV S-protein with its receptor, ACE2. Adv Exp Med Biol. 2006;581:209–218. doi: 10.1007/978-0-387-33012-9_36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye T., Zhong Z., García‐Sastre A., Schotsaert M., De Geest B.G. Current status of COVID-19 (Pre)clinical vaccine development. Angew Chem Int Ed. 2020;59(43):18885–18897. doi: 10.1002/anie.202008319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gil C., Ginex T., Maestro I., Nozal V., Barrado-Gil L., Cuesta-Geijo M.Á. COVID-19: drug targets and potential treatments. J Med Chem. 2020;63(21):12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 10.Choy K.-T., Wong A.-L., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aschenbrenner D.S. Remdesivir receives emergency use authorization for severely Ill patients with COVID-19. AJN Am J Nurs. 2020;120(7):26. doi: 10.1097/01.NAJ.0000688196.83625.b1. [DOI] [PubMed] [Google Scholar]
- 12.Yang J., Petitjean S.J.L., Koehler M., Zhang Q., Dumitru A.C., Chen W. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiu S., Dick A., Ju H., Mirzaie S., Abdi F., Cocklin S. Inhibitors of SARS-CoV-2 entry: current and future opportunities. J Med Chem. 2020;63(21):12256–12274. doi: 10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun P., Gingras A.-C. History of protein-protein interactions: from egg-white to complex networks. Proteomics. 2012;12(10):1478–1498. doi: 10.1002/pmic.201100563. [DOI] [PubMed] [Google Scholar]
- 15.Petta I., Lievens S., Libert C., Tavernier J., De Bosscher K. Modulation of protein-protein interactions for the development of novel therapeutics. Mol Ther. 2016;24(4):707–718. doi: 10.1038/mt.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan D., Matthews J. Protein-protein interactions in human disease. Curr Opin Struct Biol. 2005;15(4):441–446. doi: 10.1016/j.sbi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Villoutreix B.O., Miteva M.A. Discoidin domains as emerging therapeutic targets. Trends Pharmacol Sci. 2016;37(8):641–659. doi: 10.1016/j.tips.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 18.White A.W., Westwell A.D., Brahemi G. Protein-protein interactions as targets for small-molecule therapeutics in cancer. Expert Rev Mol Med. 2008;10:1–14. doi: 10.1017/S1462399408000641. [DOI] [PubMed] [Google Scholar]
- 19.Blazer L.L., Neubig R.R. Small molecule protein-protein interaction inhibitors as CNS therapeutic agents: current progress and future hurdles. Neuropsychopharmacology. 2009;34(1):126–141. doi: 10.1038/npp.2008.151. [DOI] [PubMed] [Google Scholar]
- 20.Rosell M., Fernández-Recio J. Hot-spot analysis for drug discovery targeting protein-protein interactions. Expert Opin Drug Discov. 2018;13(4):327–338. doi: 10.1080/17460441.2018.1430763. [DOI] [PubMed] [Google Scholar]
- 21.Milroy L.-G., Grossmann T.N., Hennig S., Brunsveld L., Ottmann C. Modulators of protein-protein interactions. Chem Rev. 2014;114(9):4695–4748. doi: 10.1021/cr400698c. [DOI] [PubMed] [Google Scholar]
- 22.Downing K.H. Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu Rev Cell Dev Biol. 2000;16(1):89–111. doi: 10.1146/annurev.cellbio.16.1.89. [DOI] [PubMed] [Google Scholar]
- 23.Nogales E. Structural insights into microtubule function. Annu Rev Biophys Biomol Struct. 2001;30(1):397–420. doi: 10.1146/annurev.biophys.30.1.397. [DOI] [PubMed] [Google Scholar]
- 24.Xiao H., Verdier-Pinard P., Fernandez-Fuentes N., Burd B., Angeletti R., Fiser A. Insights into the mechanism of microtubule stabilization by Taxol. Proc Natl Acad Sci. 2006;103(27):10166–10173. doi: 10.1073/pnas.0603704103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang C.H., Horwitz S.B. Taxol®: the first microtubule stabilizing agent. Int J Mol Sci. 2017;18(8):1–11. doi: 10.3390/ijms18081733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Löwe J., Li H., Downing K.H., Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313(5):1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 27.Tangutur A.D., Kumar D., Krishna K.V., Kantevari S. Microtubule targeting agents as cancer chemotherapeutics: an overview of molecular hybrids as stabilizing and destabilizing agents. Curr Top Med Chem. 2017;17(22):2523–2537. doi: 10.2174/1568026617666170104145640. [DOI] [PubMed] [Google Scholar]
- 28.Fanale D., Bronte G., Passiglia F., Calò V., Castiglia M., Di Piazza F. Stabilizing versus destabilizing the microtubules: a double-edge sword for an effective cancer treatment option? Anal Cell Pathol. 2015;2015:1–19. doi: 10.1155/2015/690916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sternlicht H., Ringel I. Colchicine inhibition of microtubule assembly via copolymer formation. J Biol Chem. 1979;254(20):10540–10550. [PubMed] [Google Scholar]
- 30.Margolis R.L., Wilson L. Addition of colchicine–tubulin complex to microtubule ends: the mechanism of substoichiometric colchicine poisoning. PNAS. 1977;74(8):3466–3470. doi: 10.1073/pnas.74.8.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravelli R.B.G., Gigant B., Curmi P.A., Jourdain I., Lachkar S., Sobel A. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428(6979):198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 32.Nero T.L., Morton C.J., Holien J.K., Wielens J., Parker M.W. Oncogenic protein interfaces: small molecules, big challenges. Nat Rev Cancer. 2014;14(4):248–262. doi: 10.1038/nrc3690. [DOI] [PubMed] [Google Scholar]
- 33.Scott D.E., Bayly A.R., Abell C., Skidmore J. Small molecules, big targets: drug discovery faces the protein-protein interaction challenge. Nat Rev Drug Discov. 2016;15(8):533–550. doi: 10.1038/nrd.2016.29. [DOI] [PubMed] [Google Scholar]
- 34.Wells J.A., McClendon C.L. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450(7172):1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 35.Tsuchiya Y., Kinoshita K., Nakamura H. Analyses of homo-oligomer interfaces of proteins from the complementarity of molecular surface, electrostatic potential and hydrophobicity. Protein Eng Design Select PEDS. 2006;19(9):421–429. doi: 10.1093/protein/gzl026. [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Zhang X., Cao D. The role of shape complementarity in the protein-protein interactions. Sci Rep. 2013;3(1):3271. doi: 10.1038/srep03271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stites W.E. Protein−Protein interactions: interface structure, binding thermodynamics, and mutational analysis. Chem Rev. 1997;97(5):1233–1250. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]
- 38.Geppert T., Hoy B., Wessler S., Schneider G. Context-based identification of protein-protein interfaces and “hot-spot” residues. Chem Biol. 2011;18(3):344–353. doi: 10.1016/j.chembiol.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Moreira I.S., Fernandes P.A., Ramos M.J. Hot spots–a review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68(4):803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 40.Silvian L.F., Friedman J.E., Strauch K., Cachero T.G., Day E.S., Qian F. Small molecule inhibition of the TNF family cytokine CD40 ligand through a subunit fracture mechanism. ACS Chem Biol. 2011;6(6):636–647. doi: 10.1021/cb2000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporuscio F., Tafi A., González E., Manetti F., Esté J.A., Botta M. A dynamic target-based pharmacophoric model mapping the CD4 binding site on HIV-1 gp120 to identify new inhibitors of gp120–CD4 protein–protein interactions. Bioorg Med Chem Lett. 2009;19(21):6087–6091. doi: 10.1016/j.bmcl.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 42.Zhan P., Li W., Chen H., Liu X. Targeting protein-protein interactions: a promising avenue of anti-HIV drug discovery. Curr Med Chem. 2010;17(29):3393–3409. doi: 10.2174/092986710793176357. [DOI] [PubMed] [Google Scholar]
- 43.Harding M.W., Galat A., Uehling D.E., Schreiber S.L. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature. 1989;341(6244):758–760. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- 44.Griffith J.P., Kim J.L., Kim E.E., Sintchak M.D., Thomson J.A., Fitzgibbon M.J. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell. 1995;82(3):507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 45.Kobashigawa Y., Saio T., Ushio M., Sekiguchi M., Yokochi M., Ogura K. Convenient method for resolving degeneracies due to symmetry of the magnetic susceptibility tensor and its application to pseudo contact shift-based protein-protein complex structure determination. J Biomol NMR. 2012;53(1):53–63. doi: 10.1007/s10858-012-9623-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giordanetto F., Schäfer A., Ottmann C. Stabilization of protein–protein interactions by small molecules. Drug Discov Today. 2014;19(11):1812–1821. doi: 10.1016/j.drudis.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 47.Nogales E. Structural insights into microtubule function. Annu Rev Biochem. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- 48.Jordan M. Mechanism of action of antitumor drugs that interact with microtubules and tubulin. Curr Med Chem Anticancer Agents. 2002;2:1–17. doi: 10.2174/1568011023354290. [DOI] [PubMed] [Google Scholar]
- 49.Modell A.E., Blosser S.L., Arora P.S. Systematic targeting of protein-protein interactions. Trends Pharmacol Sci. 2016;37(8):702–713. doi: 10.1016/j.tips.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang N., Lodge J.M., Fierke C.A., Mapp A.K. Dissecting allosteric effects of activator-coactivator complexes using a covalent small molecule ligand. PNAS. 2014;111(33):12061–12066. doi: 10.1073/pnas.1406033111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pang B.o., Cheung N.N., Zhang W., Dai J., Kao R.Y., Zhang H. Structural characterization of H1N1 nucleoprotein-nucleozin binding sites. Sci Rep. 2016;6(1) doi: 10.1038/srep29684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kao R.Y., Yang D., Lau L.-S., Tsui W.H.W., Hu L., Dai J. Identification of influenza A nucleoprotein as an antiviral target. Nat Biotechnol. 2010;28(6):600–605. doi: 10.1038/nbt.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kukol A., Hughes D.J. Large-scale analysis of influenza A virus nucleoprotein sequence conservation reveals potential drug-target sites. Virology. 2014;454-455:40–47. doi: 10.1016/j.virol.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 54.Gerritz S.W., Cianci C., Kim S., Pearce B.C., Deminie C., Discotto L. Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers. Proc Natl Acad Sci. 2011;108(37):15366–15371. doi: 10.1073/pnas.1107906108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arkin M.R., Wells J.A. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3(4):301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 56.Arkin M., Tang Y., Wells J. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol. 2014;21(9):1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffer L., Muller C., Roche P., Morelli X. Chemistry-driven hit-to-lead optimization guided by structure-based approaches. Mol Inf. 2018;37(9-10):1800059. doi: 10.1002/minf.v37.9-1010.1002/minf.201800059. [DOI] [PubMed] [Google Scholar]
- 58.Curreli F., Belov D.S., Kwon Y.D., Ramesh R., Furimsky A.M., O'Loughlin K. Structure-based lead optimization to improve antiviral potency and ADMET properties of phenyl-1H-pyrrole-carboxamide entry inhibitors targeted to HIV-1 gp120. Eur J Med Chem. 2018;154:367–391. doi: 10.1016/j.ejmech.2018.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson A.C. The process of structure-based drug design. Chem Biol. 2003;10(9):787–797. doi: 10.1016/j.chembiol.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 60.Robertson N.S., Spring D.R. Using Peptidomimetics and Constrained Peptides as Valuable Tools for Inhibiting Protein-Protein Interactions. Molecules (Basel, Switzerland) 2018;23(4):959. doi: 10.3390/molecules23040959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang G., Andersen J., Gerona-Navarro G. Peptidomimetics targeting protein-protein interactions for therapeutic development. Protein Pept Lett. 2018;25(12):1076–1089. doi: 10.2174/0929866525666181101100842. [DOI] [PubMed] [Google Scholar]
- 62.Mason J.M. Design and development of peptides and peptide mimetics as antagonists for therapeutic intervention. Future Med Chem. 2010;2(12):1813–1822. doi: 10.4155/fmc.10.259. [DOI] [PubMed] [Google Scholar]
- 63.Macarron R., Banks M.N., Bojanic D., Burns D.J., Cirovic D.A., Garyantes T. Impact of high-throughput screening in biomedical research. Nat Rev Drug Discovery. 2011;10(3):188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 64.Kontoyianni M. Docking and virtual screening in drug discovery. Methods Mol Biol (Clifton NJ) 2017;1647:255–266. doi: 10.1007/978-1-4939-7201-2_18. [DOI] [PubMed] [Google Scholar]
- 65.Vassilev L.T. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 66.Allen J.G., Bourbeau M.P., Wohlhieter G.E., Bartberger M.D., Michelsen K., Hungate R. Discovery and optimization of chromenotriazolopyrimidines as potent inhibitors of the mouse double minute 2-tumor protein 53 protein-protein interaction. J Med Chem. 2009;52(22):7044–7053. doi: 10.1021/jm900681h. [DOI] [PubMed] [Google Scholar]
- 67.Tian W., Han X., Yan M., Xu Y., Duggineni S., Lin N. Structure-based discovery of a novel inhibitor targeting the β-catenin/Tcf4 interaction. Biochemistry. 2012;51(2):724–731. doi: 10.1021/bi201428h. [DOI] [PubMed] [Google Scholar]
- 68.Erlanson D.A., Fesik S.W., Hubbard R.E., Jahnke W., Jhoti H. Twenty years on: the impact of fragments on drug discovery. Nat Rev Drug Discov. 2016;15(9):605–619. doi: 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- 69.Robson-Tull J. Biophysical screening in fragment-based drug design: a brief overview. Biosci Horizons Int J Stud Res. 2019;11:1–12. [Google Scholar]
- 70.Eastman P., Swails J., Chodera J.D., McGibbon R.T., Zhao Y., Beauchamp K.A. OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comput Biol. 2017;13(7):e1005659. doi: 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577(7792):706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 72.DeGoey D.A., Chen H.-J., Cox P.B., Wendt M.D. Beyond the rule of 5: lessons learned from AbbVie’s drugs and compound collection. J Med Chem. 2018;61(7):2636–2651. doi: 10.1021/acs.jmedchem.7b00717. [DOI] [PubMed] [Google Scholar]
- 73.Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adedeji A.O., Severson W., Jonsson C., Singh K., Weiss S.R., Sarafianos S.G. Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J Virol. 2013;87(14):8017–8028. doi: 10.1128/JVI.00998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kao R.Y., Tsui W.H.W., Lee T.S.W., Tanner J.A., Watt R.M., Huang J.-D. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem Biol. 2004;11(9):1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bojadzic D., Alcazar O., Buchwald P. Methylene blue inhibits the SARS-CoV-2 Spike-ACE2 protein-protein interaction-a mechanism that can contribute to its antiviral activity against COVID-19. Front Pharmacol. 2020;11 doi: 10.3389/fphar.2020.600372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kalhor H., Sadeghi S., Abolhasani H., Kalhor R., Rahimi H. Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches. J Biomol Struct Dyn. 2020;1–16 doi: 10.1080/07391102.2020.1824816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hanson Q.M., Wilson K.M., Shen M., Itkin Z., Eastman R.T., Shinn P. Targeting ACE2–RBD interaction as a platform for COVID-19 therapeutics: development and drug-repurposing screen of an AlphaLISA proximity assay. ACS Pharmacol Trans Sci. 2020;3(6):1352–1360. doi: 10.1021/acsptsci.0c00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 81.Wu Y. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368(6496):1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barnes C.O., West A.P., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R. Structures of Human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182(4):828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Monteil V. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905–913.e907. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen X., Li R., Pan Z., Qian C., Yang Y., You R. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol Immunol. 2020;17(6):647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li W., Chen C., Drelich A., Martinez D.R., Gralinski L.E., Sun Z. Rapid identification of a human antibody with high prophylactic and therapeutic efficacy in three animal models of SARS-CoV-2 infection. Proc Natl Acad Sci. 2020;117(47):29832–29838. doi: 10.1073/pnas.2010197117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lei C, et al. (2020) Potent neutralization of 2019 novel coronavirus by recombinant ACE2-Ig. bioRxiv:929976.
- 89.Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.-T. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019;5(4):eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Delmas B., Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J Virol. 1990;64(11):5367–5375. doi: 10.1128/jvi.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kalathiya U., Padariya M., Mayordomo M., Lisowska M., Nicholson J., Singh A. Highly conserved homotrimer cavity formed by the SARS-CoV-2 spike glycoprotein: a novel binding site. J Clin Med. 2020;9(5):1473. doi: 10.3390/jcm9051473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bongini P., Trezza A., Bianchini M., Spiga O., Niccolai N. A possible strategy to fight COVID-19: Interfering with spike glycoprotein trimerization. Biochem Biophys Res Commun. 2020;528(1):35–38. doi: 10.1016/j.bbrc.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kadam R.U., Wilson I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. PNAS. 2017;114(2):206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vankadari N. Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int J Antimicrob Agents. 2020;56(2):105998. doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hu H., Li L.i., Kao R.Y., Kou B., Wang Z., Zhang L. Screening and identification of linear B-Cell epitopes and entry-blocking peptide of severe acute respiratory syndrome (SARS)-associated coronavirus using synthetic overlapping peptide library. J Comb Chem. 2005;7(5):648–656. doi: 10.1021/cc0500607. [DOI] [PubMed] [Google Scholar]
- 98.Struck A.-W., Axmann M., Pfefferle S., Drosten C., Meyer B. A hexapeptide of the receptor-binding domain of SARS corona virus spike protein blocks viral entry into host cells via the human receptor ACE2. Antiviral Res. 2012;94(3):288–296. doi: 10.1016/j.antiviral.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Khan A.A., Khan Z. Comparative host–pathogen protein–protein interaction analysis of recent coronavirus outbreaks and important host targets identification. Briefings in Bioinformatics. 2021;22(2):1206–1214. doi: 10.1093/bib/bbaa207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shiryaev S.A., Remacle A.G., Ratnikov B.I., Nelson N.A., Savinov A.Y., Wei G.e. Targeting host cell furin proprotein convertases as a therapeutic strategy against bacterial toxins and viral pathogens. J Biol Chem. 2007;282(29):20847–20853. doi: 10.1074/jbc.M703847200. [DOI] [PubMed] [Google Scholar]
- 101.Ivanova T., Hardes K., Kallis S., Dahms S.O., Than M.E., Künzel S. Optimization of substrate-analogue furin inhibitors. ChemMedChem. 2017;12(23):1953–1968. doi: 10.1002/cmdc.201700596. [DOI] [PubMed] [Google Scholar]
- 102.Meyer D. Identification of the first synthetic inhibitors of the type II transmembrane serine protease TMPRSS2 suitable for inhibition of influenza virus activation. Biochem J. 2013;452(2):331–343. doi: 10.1042/BJ20130101. [DOI] [PubMed] [Google Scholar]
- 103.Gao J., Tian Z., Yang X.u. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 104.Borba M.G.S., Val F.F.A., Sampaio V.S., Alexandre M.A.A., Melo G.C., Brito M. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: a randomized clinical trial. JAMA Netw Open. 2020;3(4):e208857. doi: 10.1001/jamanetworkopen.2020.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vincent M.J. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2(1):69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ye G., Deng F., Shen Z., Luo R., Zhao L., Xiao S. Structural basis for the dimerization and substrate recognition specificity of porcine epidemic diarrhea virus 3C-like protease. Virology. 2016;494:225–235. doi: 10.1016/j.virol.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gan Y.-R. Synthesis and activity of an octapeptide inhibitor designed for SARS coronavirus main proteinase. Peptides. 2006;27(4):622–625. doi: 10.1016/j.peptides.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wei P., Fan K., Chen H., Ma L., Huang C., Tan L. The N-terminal octapeptide acts as a dimerization inhibitor of SARS coronavirus 3C-like proteinase. Biochem Biophys Res Commun. 2006;339(3):865–872. doi: 10.1016/j.bbrc.2005.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goyal B., Goyal D. Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb Sci. 2020;22(6):297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 111.Narayanan K., Kim K.H., Makino S. Characterization of N protein self-association in coronavirus ribonucleoprotein complexes. Virus Res. 2003;98(2):131–140. doi: 10.1016/j.virusres.2003.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin S.-M. Structure-based stabilization of non-native protein-protein interactions of coronavirus nucleocapsid proteins in antiviral drug design. J Med Chem. 2020;63(6):3131–3141. doi: 10.1021/acs.jmedchem.9b01913. [DOI] [PubMed] [Google Scholar]
- 114.Hsin W.-C. Nucleocapsid protein-dependent assembly of the RNA packaging signal of Middle East respiratory syndrome coronavirus. J Biomed Sci. 2018;25(1):47. doi: 10.1186/s12929-018-0449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J Virol. 2000;74(17):8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Messina F. COVID-19: viral-host interactome analyzed by network based-approach model to study pathogenesis of SARS-CoV-2 infection. J Transl Med. 2020;18(1):233. doi: 10.1186/s12967-020-02405-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gordon D.E. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370(6521) doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gordon DE, et al. (2020) A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature:459-468 [DOI] [PMC free article] [PubMed]
- 120.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6(14) doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Perišić O. Recognition of potential COVID-19 drug treatments through the study of existing protein-drug and protein-protein structures: an analysis of kinetically active residues. Biomolecules. 2020;10(9):1346. doi: 10.3390/biom10091346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Perfetto L, et al. (2020) The IMEx coronavirus interactome: an evolving map of Coronaviridae-host molecular interactions. Database (Oxford) 2020. [DOI] [PMC free article] [PubMed]
- 123.Shaw D.E., Deneroff M.M., Dror R.O., Kuskin J.S., Larson R.H., Salmon J.K. Anton, a special-purpose machine for molecular dynamics simulation. Commun ACM. 2008;51(7):91–97. [Google Scholar]
- 124.Byléhn F., Menéndez C.A., Perez-Lemus G.R., Alvarado W., de Pablo J.J. Modeling the binding mechanism of remdesivir, favilavir, and ribavirin to SARS-CoV-2 RNA-dependent RNA polymerase. ACS Cent Sci. 2021;7(1):164–174. doi: 10.1021/acscentsci.0c01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang X.Y. Biological, clinical and epidemiological features of COVID-19, SARS and MERS and AutoDock simulation of ACE2. Infectious Diseases of Poverty. 2020;9(1):99. doi: 10.1186/s40249-020-00691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mabonga L., Kappo A.P. Protein-protein interaction modulators: advances, successes and remaining challenges. Biophys Rev. 2019;11(4):559–581. doi: 10.1007/s12551-019-00570-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Herod MR, et al. (2019) The broad-spectrum antiviral drug arbidol inhibits foot-and-mouth disease virus genome replication. J Gen Virol 100(9):1293-1302. [DOI] [PubMed]
- 128.Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]