Abstract
Hypoxia-inducible factor-3α (HIF-3α), a member of HIF family, can mediate adaptive responses to low oxygen and ischemia. It is believed that HIF plays crucial roles in stroke-related diseases. However, there are no reports on the association between HIF-3α genetic variants and ischemic stroke (IS) susceptibility. Therefore, we examined the association between HIF-3α gene polymorphisms (rs3826795, rs2235095, and rs3764609) and IS risk. The study population included 302 controls and 310 patients with ischemic stroke. Three polymorphisms in HIF-3α (rs3826795, rs2235095, and rs3764609) were genotyped using SNPscan technique. Our study showed a strong association of rs3826795 in HIF-3α with the risk of IS. The genotype and allele frequencies were shown to differ between the two groups. The rs3826795 in an intron of HIF-3α was related to a prominent increased IS risk (AA vs GG adjusted odd ratio [OR], 2.21; 95% confidence intervals [95% CI], 1.10–4.44; P = 0.03; AA vs AG/GG OR = 1.74, 95% CI, 1.02–2.97, P = 0.04; A vs G OR = 1.48, 95% CI, 1.05–2.07, P = 0.02). Logistic regression analysis suggested that rs3826795 posed a risk factor for IS in addition to common factors. Furthermore, when compared to controls, increased levels of homocysteic acid and level of non-esterified fatty acid were found in the cases (P < 0.01). However, no significant association was found between rs2235095 or rs3264609 and IS risk. These findings indicated that the rs3826795 polymorphism may be a potential target for predicting the risk of IS.
Keywords: Ischemic stroke, Hypoxia-inducible factor-3α, Gene, Polymorphism
Introduction
Stroke is considered to be the main cause of death and permanent disability worldwide, causing serious economic and social impacts (Amp and Wilkins 2017; Brand et al. 2009; Susan a. Randolph 2016). Likewise, both the life quality and expectancy of life in patients are seriously influenced. Ischemic stroke (IS) is a more common type, and it accounts for approximately 85% of total cases (Rosamond et al. 2007). Previously, atherosclerosis has been found to play a major role in the IS process (Amarenco et al. 2004). Moreover, the occurrence and development of IS are also affected by a host of risk factors, including environmental and genetic risk factors (Feigin et al. 2009; Fonseca and Ferro 2015; Guzik and Bushnell 2017; Hankey 2014).
The hypoxia-inducible factor-α (HIF-α) family has 3 different members (Duan 2016). However, it is well known more about HIF-1α and HIF-2α than HIF-3α due to multiple splicing variants of HIF-3α. Hara and his team discovered human HIF-3α (hHIF-3α) in 2001 (Hara et al. 2001). The HIF-3α gene is located in 19q13.32 with a full length of 43 kb, and contains 8 alternative splicing and 19 introns (Pasanen et al. 2010). It is thought that HIF-3α is a negative regulator by competing with HIF-1α/HIF-2α for common HIF-β (Duan 2016). HIF participates actively in angiogenesis, erythropoiesis, cell cycle regulation, metabolic reprograming, and tumorigenic changes (Greer et al. 2012; Mylonis and Simos 2019; Shay and Celeste Simon 2012). Some HIF-3α target genes have been identified and contribute to various diseases, such as idiopathic pulmonary fibrosis (Aquino-Galvez et al. 2019).
Single nucleotide polymorphism (SNP) is a single base transversion or substitution, which occurs in more than 1% of the entire population. The SNP in the coding region may cause changes in the amino acid sequence, and these changes may remarkably alter the activity and biological characteristics of the encoded protein. Several reports have shown that polymorphisms of HIF gene can contribute to a variety of diseases. Huang et al. (Huang et al. 2018) found that HIF-1α C1772T TT genotype and HIF-1α G1790A AA genotype were involved in renal cell carcinoma susceptibility. Guo et al. (2015) showed that rs2057482 polymorphism in HIF-1α was associated with clinical outcome of Chinese aggressive hepatocellular carcinoma patients. In a Moscow population specifically, the IVS9-675C > A was prominently associated with the stroke risk (Tupitsyna et al. 2006). HIF-3α and HIF-1α have similar structures in the same family. We speculated that the polymorphisms of HIF-3α may be related to IS. However, no literature has reported the association between SNP of HIF-3α and IS risk. In order to verify our hypothesis, we conducted the case-control research to investigate whether these three SNP (rs3826795, rs3765609, rs2235095) in the HIF-3α gene were linked to IS risk in a Chinese population.
Methods
Study Population
The study subjects consisted of 310 IS patients and 302 controls. The patients with IS were continuously recruited from neurological department in Affiliated Hospital of Youjiang Medical University for Nationalities, Guangxi, China, from August 2016 to October 2019. The IS patients were ultimately diagnosed according to clinical simple symptom, examinations of nervous system, magnetic resonance imaging, or/and cranial computed tomography scan. These patients suffering from brain tumors, severe inflammatory diseases, and hereditary diseases were absolutely excluded from subjects of IS. The exclusion criteria for controls were as follows: history of stroke, tumorous, autoimmune diseases, genetic diseases, and cardiovascular diseases. A total of 302 gender- and age-matched healthy volunteers were eventually recruited after physical examinations in the same hospital during the same period. We collected the clinical data from medical record review of hospital, such as gender, age, smoking status, triglyceride (TG), total cholesterol (TC), apolipsprotein A1 (Apo-A1), apolipoprotein B (Apo-B) and high-density lipoprotein cholesterol (HDL-C), homocysteic acid (Hcy), and non-esterified fatty acid (NEFA). The study has been approved by our hospital ethics committee. Likewise, all subjects were unrelated Chinese, and they were selected continuously from the same geographical area.
DNA Isolation and Genotyping
Genomic DNA was extracted from peripheral blood samples of each participant using a commercial kit (DP318, Qiangen, China). The primers of rs3826795, rs3764609, and rs2235095 were designed and synthesized by Shanghai Sangon Corporation. Three SNP sites were genotyped on the ABI 3500 Genetic Analyzer133 (CA, USA) using the custom-by-design 48-Plex SNPscan kit (G0104KS, Shanghai, China). Primer information is listed in Table 1. Moreover, a total of 10% of these samples were chosen at random to be examined by Sanger sequencing. Finally, we got a 100% consistent result.
Table 1.
Primer information for genotyping assay of the HIF-3α gene
| SNP | Length | Primer sequences(5′-3′) |
|---|---|---|
| Rs3764609 | ||
| F | 20 | TCTGCCTTGTACCCCAGACA |
| R | 20 | TCTGCCTTGTACCCCAGACG |
| Rs3826795 | ||
| F | 24 | GGAGACCCCTGAGCTGGATTGGTA |
| R | 24 | GGAGACCCCTGAGCTGGATTGATG |
| Rs2235095 | ||
| F | 36 | AGCTCAATCAATTAACGTTAACATCAATAAAACCTA |
| R | 36 | AGCTCAATCAATTAACGTTAACATCAATAAAACTTG |
HIF-3α hypoxia-inducible factor-3α, F forward, R reverse
Single Nucleotide Polymorphism Selection
We used the following criteria to select SNP: (1) tagSNPs in HIF-3α gene; (2) silico analysis predicted the potential functional SNP of promoter region in HIF-3α gene; (3) the frequency of secondary allele > 5% in Chinese Han population or candidate SNP sites previously reported in the literature. Eventually, three SNP were ascertained, including rs3826795, rs3764609, and rs2235095.
Statistical Analysis
Continuous data were presented as mean ± standard deviation (SD) and compared using Student’s t test. Comparisons of categorical variables and Hardy-Weinberg equilibrium (HWE) were analyzed by χ2 test. Both odds ratios (ORs) and 95% confidence interval (CI) were calculated after adjustments for TG, TC, HDL-C, Apo-1, Apo-B, Hcy, and NEFA using logistic regression. Haplotype analysis and analysis of linkage disequilibrium (LD) were conducted on online SHEsis software (http://analysis.bio-x.cn/myAnalysis.php) (Yong and HE 2005). All statistical analysis is achieved through SPSS25 (version 25.0; SPSS, IL, USA). P < 0.05 was regarded really significant.
Results
Features of the Study Population
These clinical baseline characteristics are displayed in Table 2. The IS and control groups were not significantly different in sex, age, and smoking status (P = 0.68, 0.13, and 0.11, respectively). On the contrary, TG, TC, Apo-B, NEFA, and Hcy levels were significantly higher in IS patients, while HDL-C and Apo-A1 levels were lower in control groups (P all < 0.001).
Table 2.
Clinical characteristics of the patients group and the control group
| Variables | Controls, n = 302 | Patients with IS, n = 310 | P value |
|---|---|---|---|
| Age, year (mean ± SD) | 62.2 ± 11.8 | 62.0 ± 11.3 | 0.68 |
| Male/female | 187 /115 | 210 /100 | 0.13 |
| Smoking, yes/no | 88/214 | 109/201 | 0.11 |
| TC, mmol/L | 4.62 ± 1.17 | 4.88 ± 0.94 | 0.001 |
| TG, mmol/L | 1.44 ± 1.37 | 1.82 ± 1.58 | < 0.001 |
| HDL-C, mmol/L | 2.11 ± 10.45 | 1.13 ± 0.32 | < 0.001 |
| Apo-A1,g/L | 1.76 ± 1.13 | 1.23 ± 0.26 | < 0.001 |
| Apo-B, g/L | 0.75 ± 0.31 | 1.00 ± 0.31 | < 0.001 |
| Hcy, umol/L | 13.6 ± 3.91 | 15.0 ± 3.84 | < 0.001 |
| NEFA, mmol/L | 0.53 ± 0.28 | 0.71 ± 0.32 | < 0.001 |
IS, ischemic stroke; SD, standard deviation; TG, triglycerides, HDL, high-density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride; Apo-A1, apolipsprotein A1; Apo-B, apolipoprotein B; Hcy, homocysteic acid; NEFA, non-esterified fatty acid
Main Influence of HIF-3α Polymorphisms on IS Risk
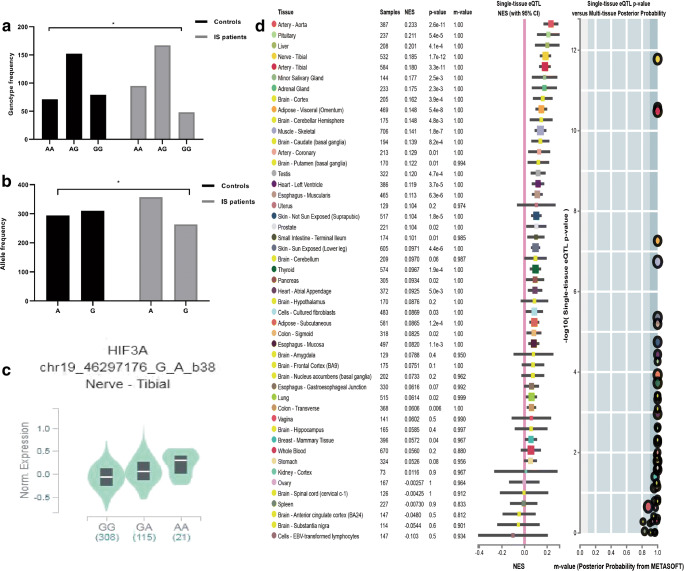
Both genotypes and allele frequencies of the three SNP (rs3826795, rs2235095, and rs3764609) between two groups are displayed in Table 3. In the case and control group, genotype distribution did not deviate from HWE. The allele and genotype distribution of rs3826795 in two groups is presented in Fig. 2a and b. The AA genotype frequency of rs3826795 was 30.6% in the cases and 23.5% in the control groups (OR = 2.21, 95% CI, 1.10–4.44, P = 0.03). Moreover, rs3826795 AA genotype increased IS risk in AA vs GG+GA model analysis (OR 1.74, 95% CI = 1.02–2.97, P = 0.04). Moreover, the frequency of the rs3826795 A allele was 57.6% in IS and 42.4% in controls (OR = 1.48; 95% CI, 1.05–2.07, P = 0.02). By exploring the impact of 3 polymorphisms on IS risk, we found that the rs3826795 polymorphism significantly increased risk of IS. However, rs2235095 and rs3764609 sites were not different in the comparison between two groups.
Table 3.
Association between HIF-3α polymorphisms and risk of IS
| Polymorphisms | Controls, n = 302 (%) | IS, n = 310 (%) | Adjusted OR (95% CI)a | P value |
|---|---|---|---|---|
| Rs3826795 | ||||
| GG | 79 (21.2) | 48 (15.3) | 1.00 | |
| AG | 152 (50.3) | 167 (53.4) | 1.40 (0.75–2.61) | 0.29 |
| AA | 71 (23.5) | 95 (31.3) | 2.21 (1.10–4.44) | 0.03 |
| AA+AG vs GG | 223 (73.8) | 262 (84.7) | 1.64 (0.86–1.72) | 0.10 |
| AA vs AG + GG | 1.74 (1.02–2.97) | 0.04 | ||
| G | 310 (51.3) | 263 (48.7) | 1.00 | |
| A | 294 (42.4) | 357 (57.6) | 1.48 (1.05–2.07) | 0.02 |
| Rs3764609 | ||||
| GG | 45 (14.9) | 44 (14.2) | 1.00 | |
| AG | 160 (53.0) | 150 (48.4) | 0.91 (0.45–1.86) | 0.80 |
| AA | 97 (32.1) | 116 (37.4) | 1.34 (0.64–2.79) | 0.43 |
| AA+AG vs GG | 1.08 (0.55–2.12) | 0.82 | ||
| AA vs AG+GG | 1.43 (0.87–2.35) | 0.15 | ||
| G | 250 (41.4) | 238 (38.4) | 1.00 | |
| A | 354 (58.6) | 382 (61.6) | 1.22 (0.86–1.72) | 0.26 |
| Rs2235095 | ||||
| GG | 104 (34.4) | 107 (34.5) | 1.00 | |
| AG | 150 (49.7) | 156 (50.3) | 1.05 (0.62–1.78) | 0.84 |
| AA | 48 (15.9) | 47 (15.2) | 1.06 (0.50–2.22) | 0.89 |
| AA+AG vs GG | 1.06 (0.64–1.74) | 0.84 | ||
| AA vs AG+GG | 1.02 (0.52–2.01) | 0.95 | ||
| G | 358 (59.3) | 370 (59.7) | 1.00 | |
| A | 246 (40.7) | 250 (40.3) | 1.03 (0.73–1.46) | 0.86 |
CI, confidence interval; IS, ischemic stroke; OR, odds ratio; HIF-3α, hypoxia-inducible factor-3α
aAdjusted by total cholesterol, triglyceride, high-density lipoprotein cholesterol, apolipoprotein A1, apolipoprotein B, homocysteic acid, and non-esterified fatty acid
Fig. 2.
The rs3826795 AA genotype was associated with increased levels of HIF-3α compared to GG, P < 0.05 (C). Allele and genotype frequencies distribution of rs3826795 in control and IS patients (A, B), expression quantitative trait loci (eQTL) analysis of rs3826795 with gene expression in single tissue (D). *There were significant differences in genotype and allele frequencies of rs3826795 between two groups in Guangxi population (P < 0.01)
Haplotype Analysis
SHEsis was chosen to perform LD measurement and haplotype analysis, and the possible eight haplotypes are enumerated in Table 4. We observed that AGA was the main haplotype in IS and GGA was the main haplotype in controls (22.3% and 21.9%, respectively). Furthermore, we observed that the AGA haplotype may be associated with an increased risk of IS (OR 2.64, 95% CI 1.90–3.67, P < 0.001), while the GGA haplotype may be related to a reduced risk of IS (P < 0.05).
Table 4.
Haplotype analysis in the patients with IS and the controls
| IS (%) | Controls (%) | OR (95% CI) | P value | |
|---|---|---|---|---|
| AAA | 81 (13.1) | 49 (8.2) | 1.68 (1.16–2.44) | 0.01 |
| AAG | 57 (9.2) | 63 (10.6) | 0.86 (0.59–1.26) | 0.44 |
| AGA | 138 (22.3) | 59 (9.8) | 2.64 (1.90–3.67) | < 0.001 |
| AGG | 80 (12.9) | 120 (20.0) | 0.59 (0.43–0.80) | < 0.001 |
| GAA | 61 (9.8) | 111 (18.6) | 0.48 (0.34–0.67) | < 0.001 |
| GAG | 50 (8.1) | 20 (3.3) | 2.59 (1.52–4.41) | < 0.001 |
| GGA | 101 (16.3) | 131 (21.9) | 0.70 (0.52–0.93) | 0.01 |
| GGG | 51 (8.2) | 46 (7.6) | 1.08 (0.71–1.64) | 0.72 |
IS, ischemic stroke; OR, odds ratio; 95% confidence interval
Multiple Logistic Regression Analysis
IS risk factors were analyzed by logistic regression analysis. The results are shown in Table 5; the risk factors contained TG (OR = 1.02; 95% CI = 0.86–1.20, P = 0.85), TC (OR = 1.37; 95% CI = 1.08–1.73), HDL-C (OR = 1.00; 95% CI = 0.93–1.08), Apo-A1 (OR = 0.00; 95% CI = 0.00–0.01), Apo-B (OR = 52.76; 95% CI = 18.90–147.21), Hcy (OR = 1.10; 95% CI = 1.03–1.18), and NEFA (OR = 8.86; 95% CI = 3.52–22.32) (P all < 0.05). TC, Apo-A1, Apo-B, Hcy, and NEFA were still connected with IS risk after logistic regression analysis. However, after correction by comparisons, TG and HDL-C had no statistical significance. Therefore, further researches are necessary to confirm our results in larger sample sizes.
Table 5.
Logistic regression analysis for identifying risk factors of IS
| Variables | B | OR (95%CI) | P value |
|---|---|---|---|
| TG | 0.02 | 1.02 (0.86–1.20) | 0.85 |
| TC | 0.31 | 1.37 (1.08–1.73) | 0.01 |
| HDL-C | 0.01 | 1.00 (0.93–1.08) | 0.94 |
| APO-A1 | − 5.68 | 0.00 (0.00–0.01) | < 0.001 |
| APO-B | 3.97 | 52.76 (18.9–147.21) | < 0.001 |
| Hcy | 0.09 | 1.10 (1.03–1.18) | 0.01 |
| NEFA | 2.18 | 8.86 (3.52–22.32) | < 0.001 |
IS, ischemic stroke; TG, triglycerides; TC, total cholesterol; HDL, high-density lipoprotein cholesterol; Apo-A1, apolipsprotein A1; Apo-B, apolipoprotein B; Hcy, homocysteic acid; NEFA, non-esterified fatty acid
Association Between rs3826795 Polymorphism and Serum Hcy and NEFA Levels
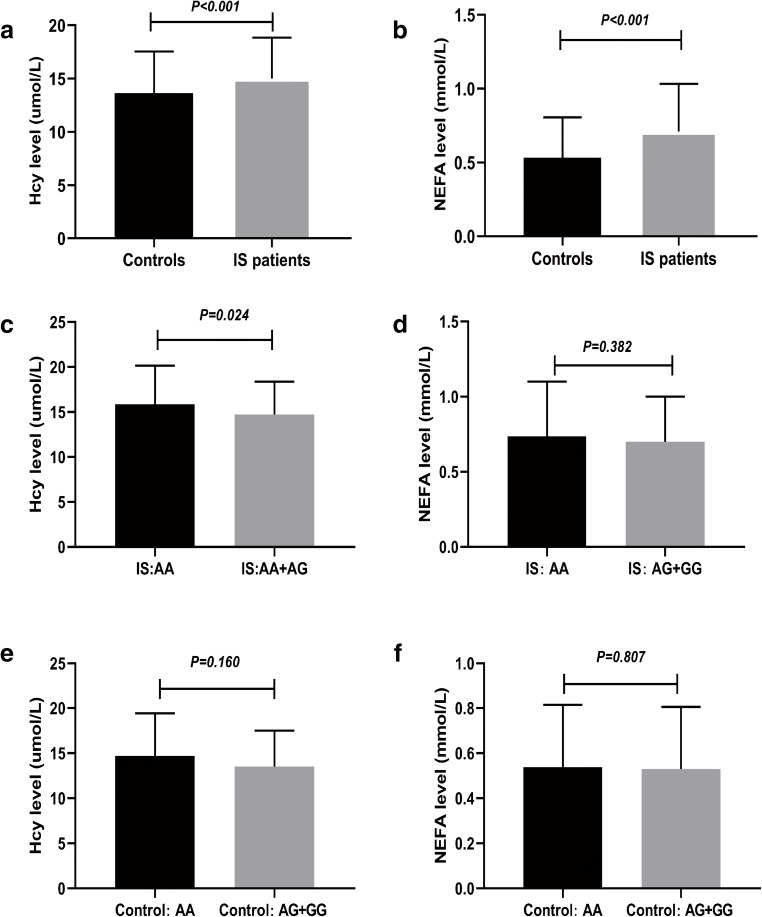
We explored the association between serum levels of Hcy and NEFA and IS. As shown in Fig. 1a and b, compared to the control group, Hcy and NEFA levels in IS patients were significantly upregulated. Even using logistic regression analysis to adjust for common risks, such as the age, sex, smoking, TG, TC, HDL-C, Apo-A1, and Apo-B, Hcy and NEFA levels were still related to an increased risk of IS (Table 5) (P < 0.05). Additionally, we explored the association between polymorphism of HIF-3α rs3826795 and the levels of Hcy and NEFA. We found that patients with the rs3826795 AA genotype had higher levels of Hcy than those with AG+GG genotypes (P < 0.05). However, individuals with the AA genotype of rs3826795 in the control group had no significant difference in Hcy levels compared with the AG+GG control group (P = 0.160). Whether in the IS group or the control group, the NEFA levels of rs826795AA individuals were not statistically different from those of rs3826795 AG+GG individuals (P > 0.05). (Fig. 1c–f).
Fig. 1.
Association between rs3826795 polymorphism and levels of Hcy and NEFA. a An increased level of Hcy in IS patients compared to controls (P < 0.001). b Increased level of NEFA in IS patients compared to controls (P < 0.001). c Increased level of Hcy in IS patients carrying the rs3826795 AA compared to those carrying the rs3826795 AG+GG (P = 0.024). d–f The levels of Hcy and NEFA showed no significant differences among different genotype groups (P > 0.05). IS ischemic stroke, Hcy homocysteic acid, NEFA non-esterified fatty acid
Bioinformatics Analysis
We obtained HIF-3α gene polymorphisms and tissue-specific expression in the GTEX database (https://www.gtexportal.org/home/). GTEX studied autopsy samples from healthy human donors. The healthy individuals carrying the rs3826795AA genotype increased the expression of HIF-3α (Fig. 2c). The analysis of the expressed quantitative trait locus (eQTL) suggested that the rs3826795 polymorphism is related to HIF-3α expression level in a single tissue (Fig. 2d).
Discussion
In the current case-control study, we examined whether the SNP in HIF-3α affected IS risk. This is the first study to explore the relationship between rs3826795, rs3764609, and rs2235095 polymorphisms and IS risk. We found that the distributions of AA genotype and A allele of rs3826795 were significantly different between cases and controls. Further analysis found that AA genotype and A allele of rs3826795 were associated with an increased IS risk. Moreover, a dramatically increased level of Hcy and NEFA was found in IS patients compared with the control groups. Especially, IS patients carrying the rs3826795AA genotype had a higher level of Hcy than those with AG+GG genotypes. Our results indicate that rs3826795 in HIF-3α may be a risk biomarker related to the etiology of IS.
The HIF family has 3 distinct members from mammals to insects (HIF-1/2/3). HIF are involved in a wide range of biological processes, including vasomotor control, angiogenesis, energy metabolism, nucleotide metabolism, and cell proliferation and viability (Tekin et al. 2010). Numerous functional processes are linked to the pathogenesis of atherosclerosis. Genetic factors in the HIF gene may be associated with atherosclerosis (Jain et al. 2018) and inflammation, which are linked to the occurrence of IS (Cheng et al. 2014; Cole et al. 2008; Davis et al. 1987). Karshovska and his colleagues found that HIF-1α could increase atherosclerosis by necrotic core formation (Karshovska et al. 2020). Furthermore, other researchers found that in the process of atherosclerosis, HIF-1 has an effect on macrophages and vascular cells, which is used as a target for treating atherosclerosis (Jain et al. 2018). However, as a member of HIF family, the HIF-2 had a protective effect on atherosclerosis by inhibiting adipose, plasma ceramide, and plasma cholesterol levels (Zhang et al. 2019). Taken together, HIF- (1, 2, 3) belongs to same family. HIF-1α and HIF-2α are strongly linked to transcriptional regulation of HIF-3α. The evidence demonstrated that the HIF-3α may play a key role in IS, which may be a therapeutic target for this disease.
In the study, our experiment confirmed that HIF-3α polymorphism and IS risk were related. As for rs3826795, to our knowledge, the relationship between rs3826795 and IS was revealed for the first time. We observed that the rs3826795 AA genotype had a 2.21 times elevated IS risk. Meanwhile, our results indicated the A allele is closely related to an increased risk of IS. Additionally, the A-G-A haplotype may cause the susceptibility of IS. These differences were significant even after adjusting for potential confounding factors (i.e., TC, TG, HDL-C, Hcy, and NEFA). In addition to common factors, risk factors of IS included atherosclerosis as well as obesity (Aa and Samiee 2009; Bhupathiraju and Hu 2016; Carl et al. 2014). A research by Wang et al. (2017) showed that the HIF-3α rs3826795 polymorphism interacted with ALT of obesity, and ALT elevation was closely related to central adiposity and related features including hypertension and dyslipidemia. Huang et al. (2015) reported that DNA methylation of HIF-3α rs3826795 interacted with total B vitamins in relation to BMI changes. Rausch and his team (Johnson et al. 2012) found that obesity was relevant to increased HIF expression in male C57BL/6 J mice. The evident indicates that the HIF-3α may play a vital role in IS. Interestingly, rs3826795 is located in the first intron of HIF-3α, and is a noncoding region of HIF-3α. However, there is still an association between rs3826795 and IS. The possible reason is that introns can dramatically influence expression of gene, such as containing enhancer or increasing mRNA accumulation (Moabbi et al. 2012; Rose 2008). The structure of human HIF-3α has a forceful commonality with Zebrafish HIF-3α (Zhang et al. 2012). Hypoxia and ischemia significantly increase HIF-3α levels in the liver, heart, brain, and ovary. Yet, hypoxia did not elevate HIF-3α levels in the kidney, gill, and testis. Heidbreder et al. (2003) suggested that hypoxia increased HIF-3α but not HIF-1α and HIF-2α levels in the hippocampus and cerebral cortex, rat’s lung. Hence, HIF-3α mRNA expression regulated by hypoxia and ischemia is tissue-specific. Yan et al. (Jun Yan et al. 2014) reported that HIF-3α expression in whole blood cell was upregulated after acute IS. In addition, the GTEx database showed that the rs3826795 had differences in HIF-3α expression (Fig. 2d). We observed that the AA genotype frequency in IS was clearly higher than that in controls. The GTEx database also showed that subjects carrying the rs3826795 AA genotype had higher levels of HIF-3α expression (Fig. 2c). Further mechanism experiments are needed to verify the connection between rs3826795 SNP and HIF-3α expression. In a word, the important role of rs3826795 in HIF-3α may be considered as a novel target for treating IS.
In this present study, we also studied the relationship between Hcy or NEFA and IS risk. Compared with the controls, the Hcy and NEFA serum levels among IS patients were significantly upregulated. Serum NEFA and Hcy were associated with atherosclerosis and IS formation, and high levels of Hcy and NEFA were correlated to poor prognosis in IS patients (Jickling and Spence 2014; Wei et al. 2019). Wei (Wei et al. 2019) found that serum Hcy levels were upregulated in IS patients, especially those carrying rs2666433 AA genotype. Similar to Wei’s findings, further comparing the difference in Hcy and NEFA expression between rs3826795 AA and AG+ GG, we found that IS patients with rs3826795 AA had higher levels of Hcy. We speculated that rs3826795 AA may increase the risk of IS by upregulating the levels of Hcy. Further experiments are needed to explore the potential molecular mechanism of HIF-3α rs3826795 affecting the occurrence of IS.
The current research has several limitations. Hospital-based cases and controls cannot eliminate selection bias. The involvement of environmental factors in the development of IS has been widely accepted. Due to the lack of objective data, we cannot assess the impact of gene-environment interaction. The same polymorphism varies among different races. Therefore, the results we got do not directly apply to all races. Perhaps, once these limitations are eliminated, we can have a better and more comprehensive understanding of these SNPs in the development of IS.
Conclusion
In conclusion, we reported that rs3826795AA polymorphism in the HIF-3α may be associated with the susceptibility of IS for the first time. These findings suggest that the rs3826795 may be an underlying biomarker for the occurrence and development of IS. A larger sample size is required in various ethnic groups. Further researches are crucially important to understand biological function of the rs3826795 in the development of IS.
Acknowledgments
The authors want to thank the patients for their participation in the study.
Authors’ Contributions
Ye-Sheng Wei designed the study; Xi-Xi Gu drafted and revised the manuscript; Yong-Ling He, Zhuan-Zhi Tang, Zeng-Zhi Neng, Xiang Shi, and Yong-Chao Qiao acquired, analyzed, and interpreted the data. All authors read and approved the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81560552), the Natural Science Foundation of Guangxi, China (No. 2018GXNSFAA138120), and the Key Research Projects of Guangxi, China (No. 2018AB58018).
Data Availability
The raw datasets generated and/or analyzed during the current study are not publicly available in order to protect participant confidentiality.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no competing interests.
Ethics Approval and Consent to Participate
All participants signed informed consent.
Consent for Publication
Not applicable.
Code Availability
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xi-Xi Gu and Zhuan-Zhi Tang contributed equally to this work.
Contributor Information
Wu-Xiang Shi, Email: swx_56@126.com.
Yong-Chao Qiao, Email: qiaoyc@glmc.edu.cn.
Ye-Sheng Wei, Email: yeshengwei56@163.com.
References
- Aa M, Samiee R. Correlation of obesity and cardiovascular diseases risk factors in girls: Tehran Lipid and Glucose Study (TLGS) Int J Endocrinol Metab. 2009;2009:35–40. [Google Scholar]
- Amarenco P, Labreuche J, Lavallee P, Touboul P-J. Statins in stroke prevention and carotid atherosclerosis: systematic review and up-to-date meta-analysis. Stroke. 2004;35:2902–2909. doi: 10.1161/01.STR.0000147965.52712.fa. [DOI] [PubMed] [Google Scholar]
- Amp LW, Wilkins Correction to: Heart disease and stroke statistics—2018 update: a report from the American Heart Association. Circulation. 2017;137:e493. doi: 10.1161/CIR.0000000000000573. [DOI] [PubMed] [Google Scholar]
- Aquino-Galvez A, et al. Dysregulated expression of hypoxia-inducible factors augments myofibroblasts differentiation in idiopathic pulmonary fibrosis. Respir Res. 2019;20:130. doi: 10.1186/s12931-019-1100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhupathiraju SN, Hu FB. Epidemiology of obesity and diabetes and their cardiovascular complications. Circ Res. 2016;118:1723–1735. doi: 10.1161/CIRCRESAHA.115.306825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand HS, Mekenkamp WCG, Baart JA. Prevalence of carotid artery calcification on panoramic radiographs. Ned Tijdschr Tandheelkd. 2009;116:69–73. [PubMed] [Google Scholar]
- Carl, et al. Obesity and cardiovascular diseases: implications regarding fitness, fatness, and severity in the obesity paradox. J Am Coll Cardiol. 2014;63:1345–1354. doi: 10.1016/j.jacc.2014.01.022. [DOI] [PubMed] [Google Scholar]
- Cheng YC, Cole JW, Kittner SJ, Mitchell BD. Genetics of ischemic stroke in young adults. Circ Cardiovasc Genet. 2014;7:383–392. doi: 10.1161/circgenetics.113.000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JW, Brown DW, Giles WH, Stine OC, Kittner SJ. Ischemic stroke risk, smoking, and the genetics of inflammation in a biracial population: the stroke prevention in young women study. Thromb J. 2008;6:11. doi: 10.1186/1477-9560-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis PH, Dambrosia JM, Schoenberg BS, Schoenberg DG, Whisnant JP. Risk factors for ischemic stroke: a prospective study in Rochester, Minnesota. Ann Neurol. 1987;22:319–327. doi: 10.1002/ana.410220307. [DOI] [PubMed] [Google Scholar]
- Duan C. Hypoxia-inducible factor 3 biology: complexities and emerging themes. Am J Phys Cell Phys. 2016;310:C260–C269. doi: 10.1152/ajpcell.00315.2015. [DOI] [PubMed] [Google Scholar]
- Feigin VL, Lawes CMM, Bennett DA, Barker-Collo SL, Parag V. Worldwide stroke incidence and early case fatality reported in 56 population-based studies: a systematic review. Lancet Neurol. 2009;8:355–369. doi: 10.1016/S1474-4422(09)70025-0. [DOI] [PubMed] [Google Scholar]
- Fonseca AC, Ferro JM. Cryptogenic stroke. Eur J Neurol. 2015;22:618–623. doi: 10.1111/ene.12673. [DOI] [PubMed] [Google Scholar]
- Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31:2448–2460. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, et al. SNP rs2057482 in HIF1A gene predicts clinical outcome of aggressive hepatocellular carcinoma patients after surgery. Sci Rep. 2015;5:11846. doi: 10.1038/srep11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzik A, Bushnell C. Stroke epidemiology and risk factor management. Continuum (Minneap Minn) 2017;23:15–39. doi: 10.1212/CON.0000000000000416. [DOI] [PubMed] [Google Scholar]
- Hankey GJ. Secondary stroke prevention. Lancet Neurol. 2014;13:178–194. doi: 10.1016/S1474-4422(13)70255-2. [DOI] [PubMed] [Google Scholar]
- Hara HS, Kobayashi J, Kondo C, Imura Y. Expression and characterization of hypoxia-inducible factor (HIF)-3alpha in human kidney: suppression of HIF-mediated gene expression by HIF-3alpha. Biochem Biophys Res Commun. 2001;287:808–813. doi: 10.1006/bbrc.2001.5659. [DOI] [PubMed] [Google Scholar]
- Heidbreder M, Frohlich F, Johren O, Dendorfer A, Qadri F, Dominiak P. Hypoxia rapidly activates HIF-3alpha mRNA expression. FASEB J. 2003;17:1541–1543. doi: 10.1096/fj.02-0963fje. [DOI] [PubMed] [Google Scholar]
- Huang L, Li MQ, Ou C, Huang WC, Liu JF, Huang H. Association of hypoxia-inducible factor-1 alpha gene polymorphism with renal cell carcinoma susceptibility. J Cancer Res Ther. 2018;14:S1105–s1109. doi: 10.4103/0973-1482.199456. [DOI] [PubMed] [Google Scholar]
- Huang T, Zheng Y, Qi Q, Xu M, Ley SH, Li Y, Kang JH, Wiggs J, Pasquale LR, Chan AT, Rimm EB, Hunter DJ, Manson JAE, Willett WC, Hu FB, Qi L. DNA methylation variants at HIF3A locus, B-vitamin intake, and long-term weight change: gene-diet interactions in two U.S. cohorts. Diabetes. 2015;64:3146–3154. doi: 10.2337/db15-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain T, Nikolopoulou EA, Xu Q, Qu A. Hypoxia inducible factor as a therapeutic target for atherosclerosis. Pharmacol Ther. 2018;183:22–33. doi: 10.1016/j.pharmthera.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Jickling GC, Spence JD. Free fatty acids to predict recurrent ischemic stroke. Neurology. 2014;82:1110–1111. doi: 10.1212/WNL.0000000000000275. [DOI] [PubMed] [Google Scholar]
- Johnson AR, Milner JJ, Makowski L. The inflammation highway: metabolism accelerates inflammatory traffic in obesity. Immunol Rev. 2012;249:218–238. doi: 10.1111/j.1600-065X.2012.01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun Yan JL, Greer JM, McCombe PA. Increased expression of the hypoxia-related genes in peripheral blood leukocytes of human subjects with acute ischemic stroke. Clin Exp Neuroimmunol. 2014;5:216–226. doi: 10.1111/cen3. [DOI] [Google Scholar]
- Karshovska E, et al. HIF-1alpha (hypoxia-inducible factor-1alpha) promotes macrophage necroptosis by regulating miR-210 and miR-383. Arterioscler Thromb Vasc Biol. 2020;40:583–596. doi: 10.1161/atvbaha.119.313290. [DOI] [PubMed] [Google Scholar]
- Moabbi AM, Agarwal N, El Kaderi B, Ansari A. Role for gene looping in intron-mediated enhancement of transcription. Proc Natl Acad Sci U S A. 2012;109:8505–8510. doi: 10.1073/pnas.1112400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonis I, Simos G. Hypoxia-inducible factors and the regulation of lipid metabolism. Cells. 2019;8:E214. doi: 10.3390/cells8030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasanen A, Heikkila M, Rautavuoma K, Hirsila M, Kivirikko KI, Myllyharju J. Hypoxia-inducible factor (HIF)-3alpha is subject to extensive alternative splicing in human tissues and cancer cells and is regulated by HIF-1 but not HIF-2. Int J Biochem Cell Biol. 2010;42:1189–1200. doi: 10.1016/j.biocel.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Rosamond W, et al. Heart disease and stroke statistics. Circulation. 2007;121:e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- Rose AB. Intron-mediated regulation of gene expression. Curr Top Microbiol Immunol. 2008;326:277–290. doi: 10.1007/978-3-540-76776-3_15. [DOI] [PubMed] [Google Scholar]
- Shay JE, Celeste Simon M. Hypoxia-inducible factors: crosstalk between inflammation and metabolism. Semin Cell Dev Biol. 2012;23:389–394. doi: 10.1016/j.semcdb.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Randolph SA. Ischemic stroke. Workplace Health Saf. 2016;64:444. doi: 10.1177/2165079916665400. [DOI] [PubMed] [Google Scholar]
- Tekin D, Dursun AD, Xi L. Hypoxia inducible factor 1 (HIF-1) and cardioprotection. Acta Pharmacol Sin. 2010;31:1085–1094. doi: 10.1038/aps.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupitsyna TV, Slominskii PA, Shadrina MI, Shetova IM, Skvortsova VI, Limborskaia SA. Association of the IVS9-675C > A polymorphism of the HIF-1alpha gene with acute ischemic stroke in the Moscow population. Genetika. 2006;42:858–861. [PubMed] [Google Scholar]
- Wang S, Song J, Yang Y, Zhang Y, Chawla NV, Ma J, Wang H. Interaction between obesity and the hypoxia inducible factor 3 alpha subunit rs3826795 polymorphism in relation with plasma alanine aminotransferase. BMC Med Genet. 2017;18:80. doi: 10.1186/s12881-017-0437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei GJ, et al. A genetic variant of miR-34a contributes to susceptibility of ischemic stroke among Chinese population front. Physiol. 2019;10:432. doi: 10.3389/fphys.2019.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong Y, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- Zhang P, Lu L, Yao Q, Li Y, Zhou J, Liu Y, Duan C. Molecular, functional, and gene expression analysis of zebrafish hypoxia-inducible factor-3alpha. Am J Phys Regul Integr Comp Phys. 2012;303:R1165–R1174. doi: 10.1152/ajpregu.00340.2012. [DOI] [PubMed] [Google Scholar]
- Zhang X, et al. Adipocyte hypoxia-inducible factor 2alpha suppresses atherosclerosis by promoting adipose ceramide catabolism. Cell Metab. 2019;30:937–951.e935. doi: 10.1016/j.cmet.2019.09.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw datasets generated and/or analyzed during the current study are not publicly available in order to protect participant confidentiality.