Abstract
Background
Autoimmune atrophic gastritis (AAG) is a chronic disease that can progress to gastric cancer (GC). To better understand AAG pathology, this proteomics study investigated gastric proteins whose expression levels are altered in this disease and also in GC.
Methods
Using two-dimensional difference gel electrophoresis (2D-DIGE), we compared protein maps of gastric corpus biopsies from AAG patients and controls. Differentially abundant spots (|fold change|≥ 1.5, P < 0.01) were selected and identified by LC–MS/MS. The spots were further assessed in gastric antrum biopsies from AAG patients (without and with Helicobacter pylori infection) and from GC patients and unaffected first-degree relatives of GC patients.
Results
2D-DIGE identified 67 differentially abundant spots, with 28 more and 39 less abundant in AAG-corpus than controls. LC–MS/MS identified these as 53 distinct proteins. The most significant (adjusted P < 0.01) biological process associated with the less abundant proteins was “tricarboxylic acid cycle”. Of the 67 spots, 57 were similarly differentially abundant in AAG-antrum biopsies irrespective of H. pylori infection status. The differential abundance was also observed in GC biopsies for 14 of 28 more abundant and 35 of 39 less abundant spots, and in normal gastric biopsies of relatives of GC patients for 6 and 25 spots, respectively. Immunoblotting confirmed the different expression levels of two more abundant proteins (PDIA3, GSTP gene products) and four less abundant proteins (ATP5F1A, PGA3, SDHB, PGC).
Conclusion
This study identified a proteomics signature of AAG. Many differential proteins were shared by GC and may be involved in the progression of AAG to GC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10120-020-01148-3.
Keywords: Autoimmune disease, Biological markers, Gastric cancer, Gastritis, Proteomics
Introduction
Autoimmune atrophic gastritis (AAG) is a T-cell-driven disease characterized by inflammation, loss of the oxyntic mucosa of the gastric corpus and fundus, and ultimately complete atrophy of the stomach mucosa [1, 2]. Patients with AAG have autoantibodies against H + K + ATPase, a parietal cell protein [3]. Antibody-mediated destruction of parietal cells reduces the production of intrinsic factor [4] and leads to the progressive loss of acid secretion, resulting in hypochlorhydria, hypergastrinemia, and hyperplasia of both G cells [5] and enterochromaffin-like cells [6]. Oxyntic atrophy leads to a loss of zymogenic chief cells, reducing pepsinogen I release [7]. AAG is diagnosed by endoscopy and serological detection of autoantibodies against parietal cells [8] and intrinsic factor [9]. Serological testing for gastrin-17 and pepsinogen I and II helps in the diagnosis: high gastrin-17 levels or a low pepsinogen I/II ratio suggests AAG [10, 11].
AAG is also known as type A gastritis. It is distinguished from type B gastritis, which is caused by Helicobacter pylori infection, environmental factors or diet, and which has an antrum-predominant anatomic distribution [12, 13]. However, many patients with AAG of the gastric corpus have H. pylori infection [14]. In a late stage of atrophy, inflammation decreases, the oxyntic mucosa is replaced by metaplastic epithelium, and the hyperplasia of enterochromaffin-like cells may progress to type I gastric carcinoid or adenocarcinoma [15–18]. Gastric atrophy and intestinal metaplasia are preneoplastic conditions. Patients with severe atrophy and intestinal metaplasia have an increased risk of intestinal-type gastric cancer (GC), and the risk increases with lesion severity [17, 19, 20].
Little is known about AAG pathogenesis because: (i) the disease has low prevalence, (ii) patients may have concomitant H. pylori infection, (iii) the disease causes few symptoms in early stages, and (iv) it is detectable only by endoscopy [21, 22]. Therefore, molecular characterization of AAG tissue should provide insight into the pathogenesis of this disease and into the process by which it progresses to neoplasia. One approach now widely used to molecularly characterize tissue specimens is provided by proteomics. In particular, the method called difference gel electrophoresis (DIGE) has been successfully used to characterize many human cancers (reviewed in Ref. [23]), proving useful in the discovery of tissue markers for GC [24], colorectal cancer [25], ulcerative colitis and Crohn's disease [26], but not yet for AAG.
We used DIGE in a quantitative proteomics study of gastric corpus biopsies from patients with AAG and patients in whom AAG was excluded, allowing us to identify a set of 53 differentially abundant proteins (67 spots). To test the specificity of these findings, we examined the abundance of these proteins in additional biopsies, including from the antrum of AAG patients, without or with H. pylori infection. Moreover, to shed light on the neoplastic progression of AAG, we examined their abundance in biopsies from GC patients and from first-degree relatives of GC patients, who are at risk of GC development [27].
Methods
Study design
The study enrolled consecutive patients undergoing esophagogastroduodenoscopy and gastric biopsy at the Unit of Oncological Gastroenterology of Centro di Riferimento Oncologico di Aviano (CRO) and Fondazione IRCCS San Matteo Hospital between April 2014 and December 2016. Patients whose tissues were analyzed gave written informed consent. The study was approved by CRO Institutional Review Board (decision no. 14) and San Matteo Hospital Institutional Ethics Committee (protocol no. 20170000832).
The study was organized in three parts. In Part I, gastric corpus biopsy specimens were examined from nine patients subsequently diagnosed with AAG (called “AAG-corpus”) and from nine patients in whom gastric disease was histologically excluded (controls); these samples had been collected at the first endoscopy examination, at San Matteo Hospital. Proteins that were differentially abundant between the groups were identified. In Part II, these differentially abundant proteins were examined in additional sets of biopsy specimens from patients recruited at CRO: (1) nine gastric antrum biopsies from AAG patients (AAG-antrum) without H. pylori infection; (2) nine gastric antrum biopsies from AAG patients with H. pylori infection (AAG-antrum-HP); (3) six gastric biopsies from first-degree relatives of GC patients (FDR-GC); and (4) six tumor biopsies from GC patients. The differential abundance of the proteins in these biopsy sets was compared to the controls of Part I. In Part III, validation analyses for seven spots were done by: (i) examining additional FDR-GC and GC proteome maps (six each), and (ii) immunoblotting pooled samples from Parts I and II.
Diagnostic procedures
All study subjects underwent esophagogastroduodenoscopy with gastric sampling according to the updated Sydney system [28]. At least two biopsies from the antrum, one from the incisura angularis, two from the corpus and two from each visible lesion were taken.
According to international criteria [1, 28], AAG was diagnosed when any grade of atrophy of the oxyntic mucosa was present, sparing the antrum. A diagnosis of AAG was excluded in subjects (controls) who had esophagogastroduodenoscopy for dyspepsia or other upper gastrointestinal complaints, when the examination and histopathological assessment were unremarkable and antiparietal cell antibodies were absent. Gastric cancer was diagnosed histologically based on Lauren’s criteria [29]. First-degree relatives of GC patients underwent esophagogastroduodenoscopy and biopsy for GC surveillance, and their gastric tissue was included when free of neoplasia and other gastric abnormalities.
AAG was also diagnosed serologically according to the presence of antiparietal cell antibodies [30]. Antibody levels were estimated by indirect immunofluorescence using a kit from Euroimmun (Lübeck, Germany). H. pylori infection was diagnosed on tissue sections using hematoxylin and eosin and Giemsa stain.
Sample preparation
Biopsy specimens were transported at 4 °C and stored at − 80 °C. Soluble proteins were extracted from frozen biopsies. Briefly, each frozen sample was homogenized in 200 µL 30 mM Tris HCl pH 8.5, 4% (w/v) CHAPS, 7 M urea, 2 M thiourea, and protease inhibitor cocktail (cOmplete EDTA-free tablets; Roche) on ice using Sample Grinding Kits (Cytiva—formerly GE Healthcare Life Sciences). The samples were lysed (4 °C, 15 min) and then centrifuged (12,000 g, 4 °C, 5 min). Supernatants were treated with 2-D Clean-Up Kit (Cytiva) and centrifuged. The resulting pellets were resuspended in 60 µL rehydration buffer (7 M urea, 2 M thiourea, 4% (w/v) CHAPS). Protein concentration was determined with Bradford reagent (Bio-Rad). Samples were stored at − 80 °C until 2D-DIGE.
2D-DIGE and image analysis
For 2D-DIGE, an internal standard was prepared by pooling equal amounts of protein extracts including all samples used in this study. This standard was labeled with Cy2 dye while individual samples were labeled, in equal numbers, with either Cy3 or Cy5 (CyDye DIGE Fluor Minimal Labeling Kit; Cytiva). Then, internal standard, one Cy3-labeled sample and one Cy5-labeled sample (25 µg each) were combined and separated by isoelectrofocusing (IEF) on Immobiline Drystrip gels with a nonlinear pH gradient (pH 3–10 NL IPG, Cytiva) for 30 kVh on a Protean IEF Cell system (Bio-Rad). After IEF, strips were frozen at − 80 °C. For the second dimension, strips were defrosted, incubated 15 min in equilibrating buffer (4 M urea, 2 M thiourea, 50 mM Tris HCl pH 8.9, 30% glycerol, 2% SDS) containing 65 mM DTT and 15 min in equilibrating buffer containing 135 mM iodoacetamide, and subjected to electrophoresis on 8–16% polyacrylamide gels (Criterion TGX, Bio-Rad) in running buffer (25 mM Tris pH 8.3, 192 mM glycine, 0.1% SDS) at 120 V for 20 min and 150 V for 50 min.
Gel images were acquired using an Amersham Typhoon Scanner (Cytiva) at 100 µm. Images were analyzed with DeCyder image analysis software v6.5 (Cytiva). The DyCyder Differential In-gel Analysis module detected spots in each gel and normalized their staining intensity to the same spot of the internal standard (standardized abundance). The Biological Variation Analysis module compared the standardized abundance of spots across gels and calculated an average ratio (fold change) between the experimental and control groups. Differentially abundant spots were those that: (i) were present on > 70% of spot maps per group; (ii) had a standardized abundance average ratio (fold change) of ≥ 1.50 or ≤ − 1.50; and (iii) had P < 0.01 on Student’s t test. The differentially abundant spots were then analyzed using the Extended Data Analysis module for principal component analysis and hierarchical clustering. Multiple group comparison of standardized spot abundance was performed with one-way ANOVA with a significance level of P < 0.01.
Protein identification by MS
To identify the proteins of the differentially abundant spots, a 2D-DIGE pick gel was prepared and separated. After Coomassie blue staining, spots of interest were excised with the Screen Picker (Proteomics Consult), destained overnight with 25 mM ammonium bicarbonate in 50% acetonitrile, dehydrated with 100% acetonitrile, dried, and digested overnight with trypsin (200 ng/spot; T6567 Sigma-Aldrich) in 40 mM ammonium bicarbonate, 9% acetonitrile at 37 °C. The trypsin digest was extracted with 1% trifluoroacetic acid (Sigma-Aldrich), lyophilized and stored at − 80 °C until shipping.
Samples were analyzed at the Proteomics Facility of CEINGE-Biotecnologie Avanzate (Naples, Italy). LC–MS/MS was done on a Proxeon EASY nano liquid chromatography system coupled with an LTQ Orbitrap XL mass spectrometer with ETD (Thermo Fisher Scientific, Massachusetts, USA). CEINGE provided raw MS data in.mgf format.
Mascot Server v2.3 (Matrix Science, Boston, USA) was used to search for matches between the MS data and proteins in the NCBI nr and Swiss-Prot databases, selected for human taxonomy.
Protein functional annotation
Functional annotation of the differentially abundant proteins between AAG-corpus and control samples was done with DAVID 6.8 [31]. Gene ontology (GO) biological processes, molecular functions and cellular components associated with the proteins were reported together with P values (Fisher’s exact test) and adjusted P values (Benjamini–Hochberg procedure). Strongly enriched annotation categories (adjusted P < 0.05) were considered.
Immunoblotting
Differentially abundant proteins of interest were examined by immunoblotting of pools of protein extracts from all experimental groups and controls from Parts I and II. Protein (10 µg per pool) was fractionated on 12% Criterion TGX Stain-Free gels (Bio-Rad). Gel images were acquired with the Chemidoc system (Bio-Rad) to document equal protein loading among samples. Then, proteins were electrotransferred onto nitrocellulose membranes and probed with following primary antibodies: anti-ATP5F1A (1:1000; #ab151229, AbCam), anti-SDHB [21A11AE7] (1:500; #ab14714, AbCam), anti-PGA3 (1:1000; #PA5-49728, Thermo Fisher Scientific), anti-pepsinogen II/PGC (1:500; #ab135862, AbCam), anti-PDIA3 (internal region; 1:1000; #ABIN3187755, Antibodies-online; Aachen, Germany), anti-GSTP1 (1:1000; #PA5-29558, Thermo Fisher Scientific), and anti-PSME1 (1:1000; #ab14714, AbCam). Primary antibodies were detected with HRP-conjugated goat anti-rabbit and anti-mouse IgG Fc fragment antibodies (1:10,000 dilution; Bethyl Laboratories, Montgomery (TX), USA) and Clarity Western ECL Substrate (Bio-Rad). Blots were imaged with the Chemidoc system.
Results
Differentially abundant proteins in AAG
In Part I of this study, protein abundance in the gastric corpus was compared between nine AAG patients (AAG-corpus) and nine controls without AAG (Table 1). Six of nine AAG patients had intestinal metaplasia. No study subject had H. pylori infection, by study design.
Table 1.
Clinicopathological characteristics of the study groups, by study design
| Group | Gastric biopsy (n) | Cases (n) | Female (n) | H. pylori infection (n) | Age (years)a |
|---|---|---|---|---|---|
| Part I | |||||
| AAG-corpusb | Corpus (9) | 9 | 6 | 0 | 51 (7) |
| Controls | Corpus (9) | 9 | 3 | 0 | 47 (13) |
| Part II | |||||
| AAG-antrumb | Antrum (9) | 9 | 6 | 0 | 50 (14) |
| AAG-antrum-HPb | Antrum (9) | 9 | 5 | 9 | 45 (11) |
| FDR-GC | Antrum (5), corpus (1) | 6 | 4 | 0d | 44 (6) |
| GCc | Antrum (1), corpus (4), fundus (1) | 6 | 3 | 0d | 63 (14) |
| Part III | |||||
| FDR-GC | Antrum (6) | 6 | 4 | 0d | 62 (12) |
| GCc | Antrum (3), corpus (3) | 6 | 2 | 4d | 64 (13) |
AAG autoimmune atrophic gastritis, FDR-GC first-degree relative of a patient with gastric cancer, GC gastric cancer, HP H. pylori infection, ND not determined
aAge at biopsy, mean (SD)
bSix patients had intestinal metaplasia
cLauren’s histological classification: intestinal type (n = 1), diffuse type (n = 2), indeterminate (n = 2); data missing for one case
dData missing for two cases
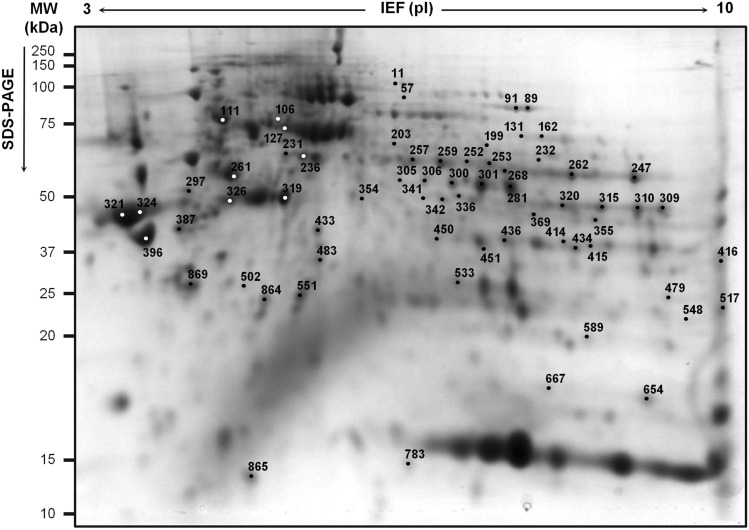
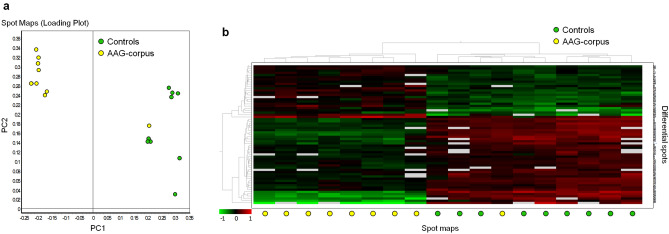
Each gastric biopsy was processed by 2D-DIGE to generate a proteome map (Fig. 1). Comparison of proteome maps between the two groups identified 67 spots as differentially abundant (P < 0.01, Student’s t test). In particular, 28 spots were more abundant in the AAG-corpus group and 39 spots were less abundant, compared to controls. At principal component analysis of the 67 spots, the nine spot maps in each group grouped together with the exception of one AAG-corpus sample that grouped with controls (Fig. 2a). Similar results were obtained with hierarchical clustering (Fig. 2b). No particular clinical or histological findings explained the misclassification of the one AAG-corpus sample.
Fig. 1.
Two-dimensional proteome map of pooled proteins (450 µg) of gastric corpus biopsy specimens from patients with AAG-corpus and controls. Proteins were resolved on an immobilized pH 3–10 gradient, followed by SDS-PAGE (8–16% acrylamide). The numbers indicate the 67 differentially abundant spots between AAG-corpus and control groups (i.e. present on > 70% of spot maps, |fold change|≥ 1.5, Student's t test P < 0.01). IEF, isoelectric focusing
Fig. 2.
Grouping of differentially abundant spots on 2D-DIGE between AAG-corpus and control groups. a Principal component analysis. Each circle represents an individual spot map. b Hierarchical clustering. The dendrogram on the left orders the spots so that similar data are displayed next to each other. The dendrogram on the top orders the samples by similarity
The 67 differentially abundant spots were identified by LC–MS/MS as 53 distinct proteins (Table 2). In particular, 25 proteins were more abundant in the AAG-corpus group, with a fold change from 1.5 to 4.4 with respect to controls. Furthermore, 28 proteins were less abundant, compared to controls, with a fold change from − 1.5 to − 13.0.
Table 2.
Differentially abundant proteins in gastric corpus tissue between patients with autoimmune atrophic gastritis (AAG) and controls
| Spot no.a | MW (pI)b | Database | Accession | Gene | Protein | Score | Matches | Seq | Seq. cov. % | Fold changec | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| More abundant in AAG (n = 28) | |||||||||||
| 783* | 16,102 (6.8) | SwissProt | HBB_HUMAN | HBB | Hemoglobin subunit beta | 177 | 8 | 6 | 42 | 4.4 | 1.71E–04 |
| 865*° | 10,161 (5.3) | NCBInr | gi|20,664,042 | S100A6 | Chain A, crystal structure of calcium-free (or Apo) human S100a6 | 155 | 22 | 5 | 32 | 3.3 | 1.64E–06 |
| 11,826 (5.2) | SwissProt | S10AE_HUMAN | S100A14 | Protein S100-A14 | 103 | 3 | 3 | 33 | – | – | |
| 231* | 54,541 (5.6) | NCBInr | gi|220,702,506 | PDIA3d | Chain A, protein disulfide-isomerase A3 erp57 | 808 | 50 | 33 | 54 | 2.6 | 6.41E–04 |
| 236 | 54,541 (5.6) | NCBInr | gi|220,702,506 | PDIA3d | Chain A, protein disulfide-isomerase A3 erp57 | 1380 | 93 | 39 | 56 | 2.4 | 8.16E-05 |
| 551*° | 23,583 (5.4) | NCBInr | gi|726,098 | GSTP1-1 | Glutathione S-transferase P1-1 class | 7083 | 261 | 12 | 64 | 2.2 | 3.55E–06 |
| 436° | 38,808 (7.6) | SwissProt | ANXA2_HUMAN | ANXA2 | Annexin A2 | 673 | 56 | 27 | 57 | 2.1 | 3.75E–04 |
| 297 | 53,676 (5.1) | SwissProt | VIME_HUMAN | VIM | Vimentin | 1255 | 79 | 34 | 53 | 2.1 | 4.22E-03 |
| 548 | 22,653 (8.9) | SwissProt | TAGL_HUMAN | TAGLN2 | Transgelin | 404 | 28 | 16 | 74 | 2.1 | 5.14E-03 |
| 415 | 38,808 (7.6) | SwissProt | ANXA2_HUMAN | ANXA2 | Annexin A2 | 522 | 37 | 25 | 56 | 2.0 | 7.37E–03 |
| 434*° | 38,808 (7.6) | SwissProt | ANXA2_HUMAN | ANXA2 | Annexin A2 | 912 | 77 | 27 | 58 | 1.9 | 2.09E-05 |
| 667* | 18,719 (8.2) | SwissProt | COF1_HUMAN | CFL1 | Cofilin-1 | 151 | 23 | 11 | 39 | 1.8 | 1.09–03 |
| 354 | 42,829 (5.9) | SwissProt | ILEU_HUMAN | SERPINB1 | Leukocyte elastase inhibitor | 635 | 33 | 22 | 48 | 1.8 | 3.49E–03 |
| 131* | 61,305 (7.3) | NCBInr | gi|237,823,914 | FGA | Chain A, crystal Structure of human fibrinogen | 479 | 29 | 20 | 37 | 1.8 | 2.94E–03 |
| 479*° | 26,193 (8.6) | SwissProt | LEG3_HUMAN | LGALS3 | Galectin-3 | 712 | 53 | 11 | 36 | 1.6 | 1.30E–03 |
| 57 | 74,380 (6.6) | SwissProt | LMNA_HUMAN | LMNA | Prelamin-A/C | 621 | 42 | 32 | 37 | 1.7 | 1.32E–03 |
| 96,590 (6.1) | SwissProt | PDC6I_HUMAN | PDCD6IP | Programmed cell death 6-interacting protein | 616 | 47 | 33 | 29 | – | – | |
| 387 | 33,027 (4.6) | NCBInr | gi|47,519,616 | TPM2 | Tropomyosin beta chain isoform 2 | 1171 | 93 | 39 | 78 | 1.7 | 9.30E–04 |
| 326* | 44,079 (5.0) | SwissProt | K1C19_HUMAN | KRT19 | Keratin, type I cytoskeletal 19 | 4978 | 280 | 38 | 73 | 1.7 | 1.86E–04 |
| 589* | 22,324 (8.3) | SwissProt | PRDX1_HUMAN | PRDX1 | Peroxiredoxin-1 | 807 | 81 | 15 | 56 | 1.6 | 6.91E–06 |
| 654* | 18,719 (8.2) | SwissProt | COF1_HUMAN | CFL1 | Cofilin-1 | 643 | 80 | 14 | 45 | 1.6 | 2.65E–04 |
| 232* | 51,926 (8.1) | NCBInr | gi|5,453,595 | CAP1 | Adenylyl cyclase-associated protein | 984 | 46 | 19 | 45 | 1.5 | 4.34E-04 |
| 11 | 74,380 (6.6) | SwissProt | LMNA_HUMAN | LMNA | Prelamin-A/C | 657 | 39 | 31 | 35 | 1.5 | 6.52E–03 |
| 117,059 (6.4) | SwissProt | ODO1_HUMAN | OGDH | 2-oxoglutarate dehydrogenase, mitochondrial | 479 | 45 | 28 | 22 | – | – | |
| 533*° | 28,909 (6.6) | SwissProt | CAH1_HUMAN | CA1 | Carbonic anhydrase 1 | 700 | 50 | 11 | 54 | 1.5 | 9.17E–03 |
| 203 | 61,066 (6.1) | SwissProt | TCPG_HUMAN | CCT3 | T-complex protein 1 subunit gamma | 917 | 54 | 29 | 43 | 1.5 | 7.36E–03 |
| 869 | 27,899 (4.7) | SwissProt | 1433Z_HUMAN | YWHAZ | 14–3-3 protein zeta/delta (protein kinase C inhibitor protein 1) | 1472 | 173 | 23 | 60 | 1.5 | 4.31E–06 |
| 483 | 28,876 (5.8) | SwissProt | PSME1_HUMAN | PSME1 | Proteasome activator complex subunit 1 | 961 | 95 | 53 | 58 | 1.5 | 1.41E–03 |
| 127* | 70,294 (5.5) | SwissProt | HS71A_HUMAN | HSPA1A | Heat shock 70 kDa protein 1A | 1441 | 94 | 38 | 46 | 1.5 | 9.55E–03 |
| 414 | 38,808 (7.6) | SwissProt | ANXA2_HUMAN | ANXA2 | Annexin A2 | 592 | 36 | 24 | 56 | 1.5 | 1.23E–04 |
| 502 | 23,031 (5.1) | SwissProt | GDIR2_HUMAN | ARHGDIB | Rho GDP-dissociation inhibitor 2 | 599 | 26 | 9 | 40 | 1.5 | 5.53E–05 |
| Less abundant in AAG (n = 39) | |||||||||||
| 321*° | 42,349 (4.2) | SwissProt | PEPA3_HUMAN | PGA3 | Pepsinogen-3 (pepsin A-3) | 713 | 112 | 9 | 11 | − 13.0 | 4.9E–05 |
| 301*° | 45,380 (6.8) | SwissProt | LIPG_HUMAN | LIPF | Gastric triacylglycerol lipase | 3625 | 154 | 10 | 22 | − 5.3 | 4.2E–06 |
| 319*° | 42,617 (5.3) | SwissProt | KCRB_HUMAN | CKB | Creatine kinase B-type | 11,761 | 455 | 23 | 56 | − 4.8 | 4.5E–06 |
| 396* | 42,798 (4.4) | SwissProt | PEPC_HUMAN | PGC | Gastricsin (pepsinogen C) | 2384 | 64 | 4 | 8 | − 4.5 | 2.6E–05 |
| 324*° | 42,349 (4.2) | SwissProt | PEPA3_HUMAN | PGA3 | Pepsinogen-3 (pepsin A-3) | 168 | 66 | 10 | 12 | − 4.1 | 3.1E–04 |
| 320*° | 51,333 (8.9) | SwissProt | IDHP_HUMAN | IDH2 | Isocitrate dehydrogenase, mitochondrial | 1220 | 75 | 24 | 40 | − 4.1 | 2.2E–05 |
| 315*° | 51,333 (8.9) | SwissProt | IDHP_HUMAN | IDH2 | Isocitrate dehydrogenase, mitochondrial | 1592 | 111 | 26 | 45 | − 3.4 | 2.1E–05 |
| 355*° | 45,456 (9.0) | SwissProt | THIL_HUMAN | ACAT1 | Acetyl-CoA acetyltransferase, mitochondrial | 784 | 41 | 23 | 44 | − 3.1 | 5.8E–05 |
| 300* | 45,380 (6.8) | SwissProt | LIPG_HUMAN | LIPF | Gastric triacylglycerol lipase | 531 | 57 | 7 | 15 | − 3.1 | 6.0E–04 |
| 433*° | 36,900 (5.7) | SwissProt | LDHB_HUMAN | LDHB | L-lactate dehydrogenase B chain | 1772 | 145 | 21 | 45 | − 3.0 | 6.3E–06 |
| 89*° | 86,113 (7.4) | SwissProt | ACON_HUMAN | ACO2 | Aconitate hydratase, mitochondrial | 912 | 57 | 27 | 31 | − 2.9 | 9.0E–06 |
| 281*° | 45,380 (6.8) | SwissProt | LIPG_HUMAN | LIPF | Gastric triacylglycerol lipase | 1492 | 82 | 9 | 22 | − 2.9 | 2.0E–05 |
| 309*° | 51,333 (8.9) | SwissProt | IDHP_HUMAN | IDH2 | Isocitrate dehydrogenase | 1082 | 78 | 25 | 42 | − 2.8 | 1.9E-05 |
| 310*° | 51,333 (8.9) | SwissProt | IDHP_HUMAN | IDH2 | Isocitrate dehydrogenase | 1527 | 112 | 24 | 39 | − 2.6 | 1.5E–05 |
| 91*° | 86,113 (7.4) | SwissProt | ACON_HUMAN | ACO2 | Aconitate hydratase, mitochondrial | 827 | 52 | 30 | 33 | − 2.5 | 2.9E–05 |
| 306*° | 47,481 (7.01) | SwissProt | ENOA_HUMAN | ENO1 | Alpha-enolase | 386 | 32 | 19 | 35 | − 2.3 | 5.6E–03 |
| 451* | 37,571 (6.50) | NCBInr | gi|4,104,867 | AFAR | Aflatoxin B1-aldehyde reductase | 985 | 48 | 14 | 36 | − 2.3 | 2.3E–04 |
| 341*° | 49,852 (7.26) | SwissProt | EFTU_HUMAN | TUFM | Elongation factor Tu, mitochondrial | 145 | 7 | 7 | 13 | − 2.2 | 4.3E–04 |
| 369* | 45,456 (9.0) | SwissProt | THIL_HUMAN | ACAT1 | Acetyl-CoA acetyltransferase, mitochondrial | 822 | 42 | 22 | 42 | − 2.2 | 2.0E–04 |
| 303*° | 54,773 (8.9) | SwissProt | FUMH_HUMAN | FH | Fumarate hydratase, mitochondrial | 583 | 38 | 24 | 41 | − 2.1 | 1.7E–05 |
| 342* | 46,915 (6.5) | SwissProt | IDHC_HUMAN | IDH1 | Isocitrate dehydrogenase [NADP] cytoplasmic | 940 | 53 | 25 | 45 | − 2.0 | 9.0E–04 |
| 416* | 38,837 (9.2) | SwissProt | ROA1_HUMAN | HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A | 478 | 34 | 16 | 37 | − 2.0 | 4.4E–04 |
| 35,937 (9.0) | SwissProt | MDHM_HUMAN | MDH2 | Malate dehydrogenase, mitochondrial | 383 | 28 | 17 | 42 | – | – | |
| 247*° | 59,828 (9.2) | SwissProt | ATPA_HUMAN | ATP5F1A | ATP synthase subunit alpha, mitochondrial | 3866 | 218 | 32 | 42 | − 2.0 | 5.3E–05 |
| 259*° | 55,454 (6.3) | SwissProt | AL1A1_HUMAN | ALDH1A1 | Retinal dehydrogenase I | 2546 | 184 | 29 | 48 | − 1.9 | 3.9E–05 |
| 864 | 28,061 (5.3) | NCBInr | NP_000030.1 | APOA1 | Apolipoprotein A-I isoform 1 preproprotein | 707 | 102 | 26 | 74 | − 1.8 | 9.3E–05 |
| 199* | 59,947 (6.9) | SwissProt | CATA_HUMAN | CAT | Catalase | 612 | 41 | 27 | 37 | − 1.8 | 1.1E–03 |
| 305*° | 44,101 (6.1) | SwissProt | PA2G4_HUMAN | PA2G4 | Proliferation-associated protein 2G4 | 488 | 32 | 20 | 45 | − 1.7 | 5.1E–03 |
| 517* | 26,325 (8.5) | NCBInr | gi|119,615,235 | SDHB | Succinate dehydrogenase complex, subunit B, iron sulfur (Ip), isoform CRA_b | 261 | 31 | 15 | 47 | − 1.7 | 2.0E–05 |
| 253° | 52,908 (7.1) | NCBInr | gi|237,823,915 | FGB | Chain B, crystal structure off human fibrinogen | 894 | 35 | 20 | 45 | − 1.7 | 9.6E–04 |
| 268 | 59,828 (9.2) | SwissProt | ATPA_HUMAN | ATP5F1A | ATP synthase subunit alpha, mitochondrial | 824 | 59 | 29 | 41 | − 1.7 | 1.0E–03 |
| 56,315 (6.7) | NCBInr | gi|20,151,189 | GLUD1 | Chain A, Structure of human glutamate dehydrogenase-apo form | 565 | 51 | 26 | 44 | – | – | |
| 261*° | 56,525 (5.3) | SwissProt | ATPB_HUMAN | ATP5F1B | ATP synthase subunit beta, mitochondrial | 6610 | 353 | 23 | 50 | − 1.6 | 9.6E–03 |
| 336* | 49,852 (7.3) | SwissProt | EFTU_HUMAN | TUFM | Elongation factor Tu, mitochondrial | 827 | 58 | 22 | 46 | − 1.6 | 4.4E–03 |
| 106*° | 73,920 (5.9) | SwissProt | GRP75_HUMAN | HSPA9 | Stress-70 protein, mitochondrial | 1042 | 55 | 34 | 43 | − 1.6 | 4.9E–04 |
| 450* | 42,902 (5.3) | SwissProt | KCRB_HUMAN | CKB | Creatine kinase B-type | 1627 | 79 | 59 | 49 | − 1.6 | 4.9E–03 |
| 262 | 59,828 (9.2) | SwissProt | ATPA_HUMAN | ATP5F1A | ATP synthase subunit alpha, mitochondrial | 1864 | 123 | 25 | 43 | − 1.6 | 3.8E–03 |
| 257* | 55,454 (10.9) | SwissProt | AL1A1_HUMAN | ALDH1A1 | Retinal dehydrogenase 1 | 1093 | 62 | 24 | 41 | − 1.6 | 5.9E–03 |
| 252*° | 54,143 (8.0) | SwissProt | DLDH_HUMAN | DLD | Dihydrolipoyl dehydrogenase, mitochondrial | 299 | 29 | 17 | 21 | − 1.6 | 4.7E–04 |
| 162*° | 67,144 (9.0) | SwissProt | AIFM1_HUMAN | AIFM1 | Apoptosis-inducing factor 1, mitochondrial | 784 | 33 | 20 | 30 | − 1.6 | 2.1E–04 |
| 111*° | 72,402 (5.1) | SwissProt | GRP78_HUMAN | HSPA5 | 78 kDa glucose-regulated protein (endoplasmic reticulum chaperone BiP) | 10,489 | 469 | 52 | 62 | − 1.5 | 4.1E–05 |
Proteins are sorted by fold change
aSpot numbers refer to Fig. 1. Five spots (11, 57, 268, 416, and 865) contained two proteins
bMolecular weight in Daltons
cCalled “average ratio” by DeCyder
dAlso ERP57
*Spots also differential in GC vs. controls
°Spots also differential in FDR-GC vs. control
According to DAVID Bioinformatics Resources, proteins more abundant in AAG-corpus were involved (adjusted P < 0.05) in the molecular functions “structural molecule activity” (GO: 0005198; including FGA, KRT19, KRT18, LMNA, VIM), “cadherin binding involved in cell–cell adhesion” (GO:0098641; including ANXA2, HSPA1A, KRT18, PRDX1, YWHAZ) (DAVID_raw data, sheet 1), and “protein binding” (GO:0005515; including CA1, CFL1, VIM, ARHGDIB, PRXD1). Regarding biological processes, proteins less abundant in AAG-corpus than controls were involved in “tricarboxylic acid cycle” (GO:0006099; including ACO2, DLD, FH, IDH1, IDH2, MDH2, SDHB), followed by “isocitrate metabolic process” (GO:0006102; including ACO2, IDH1, IDH2) and “malate metabolic process” (GO:0006108; including FH, LIPF, MDH2) (DAVID_raw data, sheet 2).
Protein abundance in AAG vs. other gastric conditions
In Part II, we examined the levels of the 67 spots (53 differentially abundant proteins) in four other sets of specimens (Table 1). Two sets were gastric antrum biopsies from AAG patients without and with confirmed H. pylori infection (called AAG-antrum and AAG-antrum-HP, respectively); two out of three patients in each group had intestinal metaplasia. Two additional sets were gastric biopsies from first-degree relatives of GC patients (FDR-GC) and from GC patients.
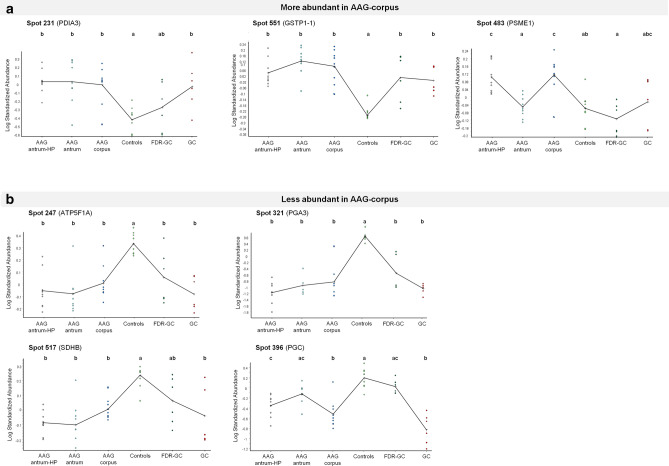
Examination of proteome maps from the AAG-antrum and AAG-antrum-HP groups revealed that, of the 67 spots identified in Part I, 57 were also differentially abundant (vs. controls) in these groups (P < 0.01) with the same directions of effect, and one spot (548) was differentially abundant but with the opposite direction. The 57 concordant spots corresponded to 43 unique proteins. Of the remaining nine spots, two (127 and 415) had similar levels as controls, two (111 and 305) were differentially abundant only in AAG-antrum, and five (131, 203, 232, 483 and 783) were differentially abundant only in AAG-antrum-HP. Log10 standardized abundance values for seven spots are shown in Fig. 3, and for the remaining 60 spots in Fig. S1.
Fig. 3.
Abundances of seven differentially abundant spots in the five experimental groups and controls, expressed as log standardized abundance from 2D-DIGE. Dots mark individual samples, and the line connects group mean values. Different letters indicate significantly different groups (P < 0.01, one-way ANOVA). a Three selected spots more abundant in AAG-corpus than controls; b Four selected spots less abundant in AAG-corpus than controls. Data for the remaining 60 spots are shown in Figure S1
Examination of GC proteome maps revealed that 14 of the 28 spots more abundant in AAG-corpus (than controls) were also more abundant in GC (see asterisks in Table 2). Furthermore, 35 of the 39 spots less abundant in AAG-corpus (than controls) were also less abundant in GC. Altogether, 30 unique proteins that were differentially abundant in AAG-corpus were also differentially abundant in GC. Examination of FDR-GC proteome maps revealed that 6 of the 28 spots more abundant in AAG-corpus were also more abundant in FDR-GC (see circles in Table 2), and 25 of the 39 spots less abundant were also less abundant in FDR-GC, for a total of 23 unique proteins maintaining their differential abundance status.
Validation of differentially abundant proteins
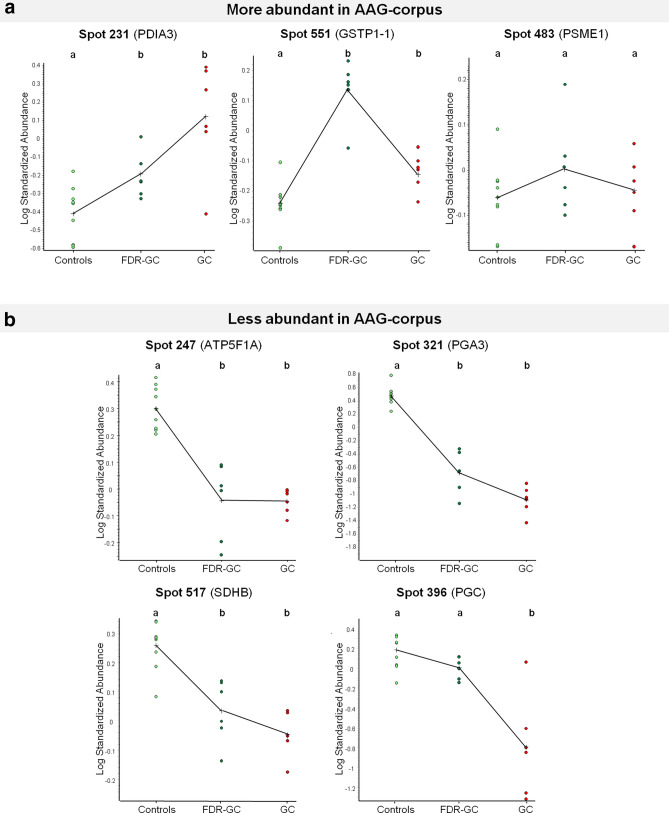
In Part III, seven spots (seven distinct proteins) identified in Part I were chosen for validation (Fig. 3). These spots included three that were more abundant (Fig. 3a) and four that were less abundant (Fig. 3b) in AAG-corpus than controls. Two spots (231 and 551) were selected because their abundance was significantly higher in all AAG groups and GC than in controls. The spots were identified as chain A, protein disulfide-isomerase A3 erp57 (PDIA3 or ERP57; spot 231), and glutathione S-transferase P1-1 class (GSTP; spot 551); this latter spot was also more abundant in FDR-GC. Spot 483 was more abundant in AAG-corpus and AAG-antrum-HP than in controls, while its abundance in GC was similar to that of controls; this spot was identified as proteasome activator complex subunit 1 (PSME1). Three spots (247, 321 and 517) were selected because their abundance in all AAG groups and GC was significantly lower than in controls. Spots 247 and 321 were also significantly less abundant in FDR-GC, while spot 517 had similar levels between FDR-GC and controls. These three spots were identified as ATP synthase subunit alpha, mitochondrial (ATP5F1A; spot 247), pepsinogen-3 (PGA3; spot 321), and succinate dehydrogenase complex, subunit B, iron sulfur (Ip), isoform CRA_b (SDHB; spot 517). Finally, spot 396, identified as pepsinogen C (PGC), was selected for being more abundant in AAG-antrum than AAG-antrum-HP, AAG-corpus and GC, in accordance with the physiological production of pepsinogen C by the antrum. Spots 321 (pepsinogen-3) and 396 (pepsinogen C) were also selected for their association with gastric diseases.
First, 2D-DIGE was repeated in an attempt to validate the findings for the seven spots in additional FDR-GC and GC biopsy samples (Part III in Table 1). For the three spots more abundant in AAG-corpus (Fig. 4a), this analysis confirmed the higher abundance (vs. controls) of PDIA3 in GC, the higher abundance GSTP1-1 in both FDR-GC and GC, and the similar levels to controls for PSME1 in both groups. For the four spots less abundant in AAG-corpus (Fig. 4b), this analysis confirmed the lower abundance of all in GC, while for FDR-GC it confirmed the lower levels of ATP5F1A and PGA3 and the similar abundance of PGC.
Fig. 4.
Validation analysis for seven differentially abundant spots in two additional sets of gastric biopsies vs. controls, expressed as log standardized abundance from 2D-DIGE. Dots mark individual samples, and the line connects group mean values. Different letters indicate significantly different groups (P < 0.01, one-way ANOVA). a Three selected spots more abundant in AAG-corpus than controls; b Four selected spots less abundant in AAG-corpus than controls
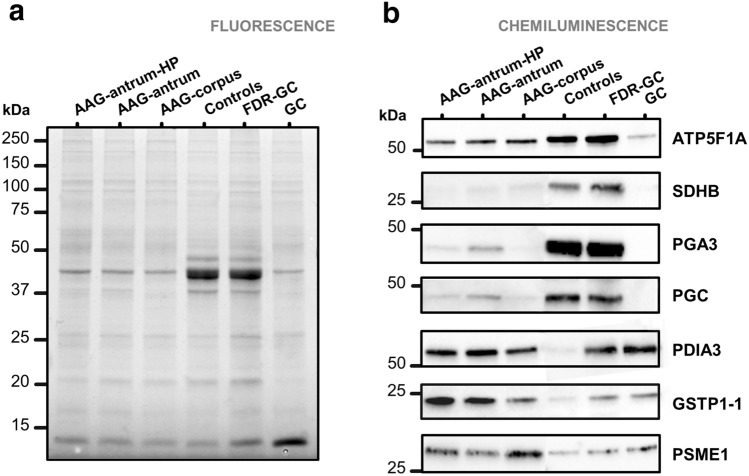
Finally, the abundance of the seven selected proteins was examined by immunoblotting of pooled proteins (Fig. 5). This analysis confirmed the lower levels of ATP synthase subunit alpha (ATP5F1A), succinate dehydrogenase complex subunit B (SDHB), pepsinogen-3 (PGA3), and pepsinogen C (PGC) in all three AAG groups and in GC than in controls. In FDR-GC, immunoblotting confirmed the similar abundances of PGC and PSME1, and the higher abundance of GSTP1-1, but did not confirm the similar abundance of SDHB and PDIA3 or the lower abundance of ATP5F1A or PGA3. The analysis also confirmed the higher levels of PDIA3 in AAG and GC than in controls. For GSTP1-1, immunoblotting confirmed the higher abundance in all AAG groups and in GC than in controls. Finally, for PSME1, the higher abundance in AAG-antrum-HP and AAG-corpus than in controls was confirmed.
Fig. 5.
Immunochemical detection of seven differentially abundant proteins, indicated by gene names on the right. Samples are pools of extracted protein from six gastric biopsy specimens per study group. a Image of the gel acquired with Chemidoc system before transfer to nitrocellulose membranes. b Western blots of the selected proteins
Discussion
This study identified a gastric tissue profile comprising 53 corpus proteins differentially abundant between AAG-corpus and controls. Of these proteins, 25 were more abundant and 28 were less abundant in AAG-corpus. These results were also obtained for 43 proteins in AAG-antrum biopsies, irrespective of H. pylori infection status. In GC biopsies and gastric biopsies of first-degree relatives of GC patients, 30 and 23 proteins, respectively, maintained their differential abundance status. At the individual protein level, the gastric tissue profile includes PDIA3, GSTP1-1, and PSME1, which were more abundant in AAG-corpus, and ATP5F1A, PGA3, PGC,, and SDHB, which were less abundant. These proteins were also differentially abundant in AAG-antrum, AAG-antrum-HP, and GC. Regarding FDR-GC, only PDIA3 and GSTP1-1 maintained the differential status.
The proteomics profile of AAG-corpus was partially observed in biopsies of the antrum (for 43 of 53 unique proteins), reflecting a common response of the gastric mucosa to chronic inflammation. However, ten proteins did not have the same changes in abundance, and some differences were found between antrum biopsies with and without H. pylori infection. The lack of complete overlap may reflect real pathophysiological differences, or may be an artifact due to small sample number and is worthy of further investigation.
According to DAVID, the 28 less abundant proteins were enriched in “tricarboxylic acid (TCA) cycle”, suggesting that the gastric corpus in AAG has TCA cycle impairments. The decreased abundance of proteins of the TCA cycle agrees with previous findings of decreased respiratory capacity and deficient mitochondrial respiratory complex I in corpus mucosal cells of patients with atrophic gastritis [32]. It is possible that, in AAG, loss of zymogenic mucosal cells and consequent reduction of zymogen granule secretion reduce energy demand. Moreover, energy metabolism (i.e. TCA cycle and oxidative phosphorylation) may be decreased by lower iron availability [33] due to achlorhydria [34].
We found that proteins involved in “structural molecule activity” and “cadherin binding involved in cell–cell adhesion” were more abundant in AAG tissues. In AAG, autoantibodies against parietal cells cause apoptosis and atrophy in the corpus mucosa [35]. An increased synthesis of intercellular adhesion molecules by oxyntic mucosa cells may reflect an attempt to counteract apoptosis and atrophy. Thus, the observed increase in abundance of some proteins with adhesive properties in AAG may be a reaction of the atrophic gastric mucosa to parietal cell loss and gastric cell phenotype changes.
This study has several limitations. First, the sample size is small and does not reflect the range of severity of AAG, from pre-atrophic to severe atrophic. In all AAG groups, two out of three patients had advanced atrophy with intestinal metaplasia, but how this condition affected the proteomics profiles could not be investigated. Intestinal metaplasia is a common finding in AAG; hence we included these patients as they are part of the spectrum of AAG histopathology and natural history. Another limitation is the heterogeneity of GC samples (intestinal, diffuse and indeterminate), which precluded identifying differences related to histology but made our protein signature more reflective of all GC types.
Possible roles of selected differentially abundant proteins
Protein disulfide isomerase A3 (PDIA3) was more abundant in all AAG groups and in GC than controls. It is a member of the protein disulfide isomerase-like family [36] and an endoplasmic reticulum chaperone implicated in oncogenesis, tumor progression, and immune response of carcinoma cells [37–39]. PDIA3 has been found increased in spasmolytic polypeptide-expressing metaplasia, a reparative tissue that develops in response to oxyntic atrophy [40] and H. pylori infection [41]. High levels of PDIA3 in gastric adenocarcinoma have been associated with better prognosis [37, 42].
GST protein P1-1 class (GSTP1-1) was also more abundant in all AAG groups and GC than controls. It belongs to the glutathione S-transferase family, which has various functions (e.g., detoxification of exogenous substances) [43, 44]. The increased abundance of GSTP1-1 in AAG may be a mechanism to counteract oxidative stress, which is known to occur in stomach disorders, including gastritis [45].
Proteasome activator complex subunit 1 (PSME1) was more abundant in AAG-corpus and AAG-antrum-HP than controls. It is a multicatalytic proteinase complex, involved in immunoproteasome assembly required for efficient antigen processing [46]. Its abundance in AAG-antrum-HP suggests a protective function against H. pylori infection. PSME1 content in GC was similar to that in controls.
ATP synthase subunit alpha (ATP5F1A) was less abundant in all AAG groups and GC than controls. It is a mitochondrial protein that generates ATP in the presence of an H+ gradient across membranes [47]. Reduced ATP synthase abundance in AAG-corpus may reflect a decreased energy demand by gastric tissues, consistent with the above-described TCA cycle impairment and lower SDHB abundance. Decreased ATP synthase in AAG-antrum, irrespective of H. pylori infection, and in GC may reflect reduced energy demand and respiratory activity in these diseases.
Pepsinogen 3 (PGA3) abundance was also less abundant in all AAG groups and GC than controls. It is an aspartic protease primarily secreted by gastric chief cells (zymogenic), and it is activated into the digestive enzyme pepsin when it comes in contact with acid produced by parietal cells. Aspartic proteinases are classified into two major groups: pepsinogen I or A, to which PGA3 belongs, and pepsinogen II or C [48]. Low pepsinogen content may reflect the loss of parietal cells. Our results confirm those of Kuipers et al. [49], who found at the gene level a loss of PGA3 expression in patients with atrophic gastritis or GC.
Pepsinogen C (PGC) was less abundant in AAG-corpus, AAG-antrum-HP, and GC than controls. These results agree with a previous study that showed progressively lower PGC expression, compared to normal gastric mucosa, in lesions of increasing severity, namely gastric erosions, atrophic gastritis, and GC (where only 2.4% of specimens had detectable levels) [50].
Succinate dehydrogenase complex, subunit B (SDHB) was also less abundant in all AAG groups and GC than controls. It is involved in the mitochondrial electron transport chain (complex II), a subpathway of the TCA cycle and is part of carbohydrate metabolism [51]. Decreased abundance of both SDHB and ATP5F1A in GC was recently reported [52].
Conclusion
This study identified a proteomics signature of stomach corpus in AAG, which includes decreased abundance of proteins involved in the TCA cycle and increased abundance of those in structural molecule activity and cadherin binding. Many of these AAG markers are shared with GC. These proteomics alterations may represent a link between AAG and GC and be part of the progression to gastric cancerogenesis. Our proteomic approach on tissue should be integrated with transcriptomic and biochemical data.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank the patients for donating tissue samples, Valerie Matarese for scientific editing on versions of the manuscript, and Dr. Simona Celentano from CEINGE-Biotecnologie Avanzate (Naples, Italy) for doing the LC–MS/MS analyses. Research reported in this publication was supported by the Italian Ministry of Health (Grant number RF-2016-02361525 to R. Cannizzaro) and Ministero della Salute-Ricerca Corrente. Manuscript editing was paid for by Centro di Riferimento Oncologico di Aviano (CRO), IRCCS.
Author contributions
OR, VDR, ADS and RC designed and supervised the study; OR developed methods for protein extraction, 2D-DIGE, western blotting, and drafted the manuscript; OR and VDR performed statistical analyses and data interpretation; AS and RC obtained funding; PG, MVL, RM, AS, ADS, MV and RC provided samples and clinical data; PG, MVL and MV critically revised the manuscript for intellectual content. All authors contributed to manuscript preparation.
Compliance with ethical standards
Conflict of interest
The authors disclose no competing interests.
Ethical standards
The collection and analysis of samples in this study was approved by the Institutional Review Board of Centro di Riferimento Oncologico di Aviano (CRO) and the Institutional Ethics Committee of San Matteo Hospital.
Informed consent
Informed consent was obtained from each subject.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ombretta Repetto and Valli De Re have contributed equally to this work.
Contributor Information
Ombretta Repetto, Email: orepetto@cro.it.
Valli De Re, Email: vdere@cro.it.
Paolo Giuffrida, Email: paolo.giuffrida01@universitadipavia.it.
Marco Vincenzo Lenti, Email: marco.lenti@unipv.it.
Raffaella Magris, Email: raffaella.magris@cro.it.
Marino Venerito, Email: m.venerito@med.ovgu.de.
Agostino Steffan, Email: asteffan@cro.it.
Antonio Di Sabatino, Email: a.disabatino@smatteo.pv.it.
Renato Cannizzaro, Email: rcannizzaro@cro.it.
References
- 1.Lenti MV, Rugge M, Lahner E, Miceli E, Toh BH, Genta RM, et al. Autoimmune gastritis. Nat Rev Dis Primers. 2020;6:56. doi: 10.1038/s41572-020-0187-8. [DOI] [PubMed] [Google Scholar]
- 2.Di Sabatino A, Lenti MV, Giuffrida P, Vanoli A, Corazza GR. New insights into immune mechanisms underlying autoimmune diseases of the gastrointestinal tract. Autoimmun Rev. 2015;14:1161–1169. doi: 10.1016/j.autrev.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Mårdh S, Song YH. Characterization of antigenic structures in auto-immune atrophic gastritis with pernicious anaemia. The parietal cell H, K-ATPase and the chief cell pepsinogen are the two major antigens. Acta Physiol Scand. 1989;136:581–587. doi: 10.1111/j.1748-1716.1989.tb08705.x. [DOI] [PubMed] [Google Scholar]
- 4.Lahner E, Norman GL, Severi C, Encabo S, Shums Z, Vannella L, et al. Reassessment of intrinsic factor and parietal cell autoantibodies in atrophic gastritis with respect to cobalamin deficiency. Am J Gastroenterol. 2009;104:2071–2079. doi: 10.1038/ajg.2009.231. [DOI] [PubMed] [Google Scholar]
- 5.Chlumska A, Boudova L, Benes Z, Zámecník M. Autoimmune gastritis. A clinicopathologic study of 25 cases. Cesk Patol. 2005;41(4):137–142. [PubMed] [Google Scholar]
- 6.Solcia E, Fiocca R, Villani L, Luinetti O, Capella C. Hyperplastic, dysplastic, and neoplastic enterochromaffin-like-cell proliferations of the gastric mucosa. Classification and histogenesis. Am J Surg Pathol. 1995;19:S1–S7. [PubMed] [Google Scholar]
- 7.Carmel R. Pepsinogens and other serum markers in pernicious anemia. Am J Clin Pathol. 1988;90:442–445. doi: 10.1093/ajcp/90.4.442. [DOI] [PubMed] [Google Scholar]
- 8.Rusak E, Chobot A, Krzywicka A, Wenzlau J. Anti-parietal cell antibodies—diagnostic significance. Adv Med Sci. 2016;61:175–179. doi: 10.1016/j.advms.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Khan S, Del-Duca C, Fenton E, Holding S, Hirst J, Doré PC, et al. Limited value of testing for intrinsic factor antibodies with negative gastric parietal cell antibodies in pernicious anaemia. J Clin Pathol. 2009;62:439–441. doi: 10.1136/jcp.2008.060509. [DOI] [PubMed] [Google Scholar]
- 10.Magris R, De Re V, Maiero S, Fornasarig M, Guarnieri G, Caggiari L, et al. Low pepsinogen I/II ratio and high gastrin-17 levels typify chronic atrophic autoimmune gastritis patients with gastric neuroendocrine tumors. Clin Transl Gastroenterol. 2020;11:e00238. doi: 10.14309/ctg.0000000000000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miceli E, Padula D, Lenti MV, Gallia A, Albertini R, Di Stefano M, et al. A laboratory score in the diagnosis of autoimmune atrophic gastritis: a prospective study. J Clin Gastroenterol. 2015;49:e1–e5. doi: 10.1097/MCG.0000000000000101. [DOI] [PubMed] [Google Scholar]
- 12.Strickland RG. Gastritis. In: Van der Reis L, editor. Immune disorders. Front Gastrointest Res. Basel: Karger; 1975. pp. 12–18. [DOI] [PubMed] [Google Scholar]
- 13.Strickland RG, Mackay IR. A reappraisal of the nature and significance of chronic atrophic gastritis. Am J Dig Dis. 1973;18:426–440. doi: 10.1007/BF01071995. [DOI] [PubMed] [Google Scholar]
- 14.Veijola LI, Oksanen AM, Sipponen PI, Rautelin HI. Association of autoimmune type atrophic corpus gastritis with Helicobacter pylori infection. World J Gastroenterol. 2010;16:83–88. doi: 10.3748/wjg.v16.i1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miceli E, Vanoli A, Lenti MV, Klersy C, Di Stefano M, Luinetti O. Natural history of autoimmune atrophic gastritis: a prospective, single centre, long-term experience. Aliment Pharmacol Ther. 2019;50:1172–1180. doi: 10.1111/apt.15540. [DOI] [PubMed] [Google Scholar]
- 16.Bizzaro N, Antico A, Villalta D. Autoimmunity and Gastric Cancer. Autoimmune gastritis has been associated with the development of two types of gastric neoplasms: intestinal type and type I gastric carcinoid. Int J Mol Sci. 2018;19(2):pii: E377. doi: 10.3390/ijms19020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lahner E, Esposito G, Galli G, Annibale B. Atrophic gastritis and pre-malignant gastric lesions. Transl Gastrointest Cancer. 2015;4:272–281. [Google Scholar]
- 18.Kokkola A, Sjoblom SM, Haapiainen R, Sipponen P, Puolakkainen P, Järvinen H. The risk of gastric carcinoma and carcinoid tumours in patients with pernicious anaemia. A prospective follow-up study. Scand J Gastroenterol. 1998;33:88–92. doi: 10.1080/00365529850166266. [DOI] [PubMed] [Google Scholar]
- 19.Kodama M, Murakami K, Okimoto T, Abe H, Sato R, Ogawa R, et al. Histological characteristics of gastric mucosa prior to Helicobacter pylori eradication may predict gastric cancer. Scand J Gastroenterol. 2013;48:1249–1256. doi: 10.3109/00365521.2013.838994. [DOI] [PubMed] [Google Scholar]
- 20.Vannella L, Lahner E, Osborn J, Bordi C, Miglione M, Delle Fave G, et al. Risk factors for progression to gastric neoplastic lesions in patients with atrophic gastritis. Aliment Pharmacol Ther. 2010;31:1042–1050. doi: 10.1111/j.1365-2036.2010.04268.x. [DOI] [PubMed] [Google Scholar]
- 21.Minalyan A, Benhammou JN, Artashesyan A, Lewis MS, Pisegna JR. Autoimmune atrophic gastritis: current perspectives. Clin Exp Gastroenterol. 2017;10:19–27. doi: 10.2147/CEG.S109123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toh BH. Diagnosis and classification of autoimmune gastritis. Autoimmun Rev. 2014;13:459–462. doi: 10.1016/j.autrev.2014.01.048. [DOI] [PubMed] [Google Scholar]
- 23.Kondo T. Cancer biomarker development and two-dimensional difference gel electrophoresis (2D-DIGE) Biochim Biophys Acta Proteins Proteom. 2019;1867:2–8. doi: 10.1016/j.bbapap.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Wu JY, Cheng CC, Wang JY, Wu DC, Hsieh JS, Lee SC, et al. Discovery of tumor markers for gastric cancer by proteomics. PLoS ONE. 2014;9:e84158. doi: 10.1371/journal.pone.0084158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Song MQ, Zhu JS, Zhou Z, Xu ZP, Chen WX, et al. Identification of differentially-expressed proteins between early submucosal non-invasive and invasive colorectal cancer using 2D-DIGE and mass spectrometry. Int J Immunopathol Pharmacol. 2011;24:849–859. doi: 10.1177/039463201102400404. [DOI] [PubMed] [Google Scholar]
- 26.Moriggi M, Pastorelli L, Torretta E, Tontini GE, Capitanio D, Bogetto SF, et al. Contribution of extracellular matrix and signal mechanotransduction to epithelial cell damage in inflammatory bowel disease patients: a proteomic study. Proteomics. 2017;17. [DOI] [PubMed]
- 27.Song M, Camargo MC, Weinstein SJ, Best AF, Männistö S, Albanes D, Rabkin CS. Family history of cancer in first-degree relatives and risk of gastric cancer and its precursors in a Western population. Gastric Cancer. 2018;21:729–737. doi: 10.1007/s10120-018-0807-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 29.Lauren P. The two histologic main types of gastric carcinoma: diffuse and so-called intestinal type carcinoma. An attempt at a histo-clinical classification. Acta Parhol Microbid Scan. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 30.Di Sabatino A, Biagi F, Lenzi M, Frulloni L, Lenti MV, Giuffrida P, et al. Clinical usefulness of serum antibodies as biomarkers of gastrointestinal and liver diseases. Dig Liver Dis. 2017;49:947–956. doi: 10.1016/j.dld.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 31.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Gruno M, Peet N, Tein A, Salupere R, Sirotkina M, Valle J, et al. Atrophic gastritis: deficient complex I of the respiratory chain in the mitochondria of corpus mucosal cells. J Gastroenterol. 2008;43:780–788. doi: 10.1007/s00535-008-2231-4. [DOI] [PubMed] [Google Scholar]
- 33.Oexle H, Gnaiger E, Weiss G. Iron-dependent changes in cellular energy metabolism: influence on citric acid cycle and oxidative phosphorylation. Biochim Biophys Acta. 1999;1413:99–107. doi: 10.1016/s0005-2728(99)00088-2. [DOI] [PubMed] [Google Scholar]
- 34.Betesh AL, Santa Ana CA, Cole JA, Fordtran JS. Is achlorhydria a cause of iron deficiency anemia? Am J Clin Nutr. 2015;102:9–19. doi: 10.3945/ajcn.114.097394. [DOI] [PubMed] [Google Scholar]
- 35.Bergman MP, Vandenbroucke-Grauls CM, Appelmelk BJ, D'Elios MM, Amedei A, Azzurri A, et al. The story so far: Helicobacter pylori and gastric autoimmunity. Int Rev Immunol. 2005;24:63–91. doi: 10.1080/08830180590884648. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Baig E, Williams DB. Functions of ERp57 in the folding and assembly of major histocompatibility complex class I molecules. J Biol Chem. 2006;281:14622–14631. doi: 10.1074/jbc.M512073200. [DOI] [PubMed] [Google Scholar]
- 37.Shimoda T, Wada R, Kure S, Ishino K, Kudo M, Ohashi R, et al. Expression of protein disulfide isomerase A3 and its clinicopathological association in gastric cancer. Oncol Rep. 2019;41:2265–2272. doi: 10.3892/or.2019.6999. [DOI] [PubMed] [Google Scholar]
- 38.Coe H, Michalak M. ERp57, a multifunctional endoplasmic reticulum resident oxidoreductase. Int J Biochem Cell Biol. 2010;42:796–799. doi: 10.1016/j.biocel.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Cicchillitti L, Di Michele M, Urbani A, Ferlini C, Donat MB, Scambia G, et al. Comparative proteomic analysis of paclitaxel sensitive A2780 epithelial ovarian cancer cell line and its resistant counterpart A2780TC1 by 2D-DIGE: the role of ERp57. J Proteome Res. 2009;8:1902–1912. doi: 10.1021/pr800856b. [DOI] [PubMed] [Google Scholar]
- 40.Nomura S, Yamaguchi H, Ogawa M, Wang TC, Lee JR, Goldenring JR. Alterations in gastric mucosal lineages induced by acute oxyntic atrophy in wild-type and gastrin-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2005;288:G362–G375. doi: 10.1152/ajpgi.00160.2004. [DOI] [PubMed] [Google Scholar]
- 41.Lee JR, Baxter TM, Yamaguchi H, Wang TC, Goldenring JR, Anderson MG. Differential protein analysis of spasomolytic polypeptide expressing metaplasia using laser capture microdissection and two-dimensional difference gel electrophoresis. Appl Immunohistochem Mol Morphol. 2003;11:188–193. doi: 10.1097/00129039-200306000-00018. [DOI] [PubMed] [Google Scholar]
- 42.Leys CM, Nomura S, LaFleur BJ, Ferrone S, Kaminishi M, Montgomery E, et al. Expression and prognostic significance of prothymosin-alpha and ERp57 in human gastric cancer. Surgery. 2007;141:41–50. doi: 10.1016/j.surg.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Singh S. Cytoprotective and regulatory functions of glutathione S-transferases in cancer cell proliferation and cell death. Cancer Chemoth Pharm. 2015;75:1–15. doi: 10.1007/s00280-014-2566-x. [DOI] [PubMed] [Google Scholar]
- 44.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki H, Nishizawa T, Tsugawa H, Mogami S, Hibi T. Roles of oxidative stress in stomach disorders. J Clin Biochem Nutr. 2012;50:35–39. doi: 10.3164/jcbn.11-115SR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vigneron N, van den Eynde BJ. Proteasome subtypes and regulators in the processing of antigenic peptides presented by class I molecules of the major histocompatibility complex. Biomolecules. 2014;4:994–1025. doi: 10.3390/biom4040994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Junge W, Nelson N. ATP synthase. Annu Rev Biochem. 2015;84:631–657. doi: 10.1146/annurev-biochem-060614-034124. [DOI] [PubMed] [Google Scholar]
- 48.Gritti I, Banfi G, Roi GS. Pepsinogens: physiology, pharmacology pathophysiology and exercise. Pharmacol Res. 2000;41:265–281. doi: 10.1006/phrs.1999.0586. [DOI] [PubMed] [Google Scholar]
- 49.Kuipers EJ, Peña AS, Crusius JB, Defize J, van der Stoop P, Meuwissen SG, Pals G. Absence of pepsinogen A3 gene expression in the gastric mucosa of patients with gastric cancer. J Clin Pathol. 1995;48:376–379. doi: 10.1136/jcp.48.4.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ning PF, Liu HJ, Yuan Y. Dynamic expression of pepsinogen C in gastric cancer, precancerous lesions and Helicobacter pylori associated gastric diseases. World J Gastroenterol. 2005;11:2545–2548. doi: 10.3748/wjg.v11.i17.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rutter J, Winge DR, Schiffman JD. Succinate dehydrogenase—assembly, regulation and role in human disease. Mitochondrion. 2010;10:393–401. doi: 10.1016/j.mito.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernández-Coto DL, Gil J, Hernández A, Herrera-Goepfert R, Castro-Romero I, Hernández-Márquez E, et al. Quantitative proteomics reveals proteins involved in the progression from non-cancerous lesions to gastric cancer. J Proteomics. 2018;186:15–27. doi: 10.1016/j.jprot.2018.07.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.