Abstract
Spodoptera frugiperda (J. E. Smith) is a highly adaptable polyphagous migratory pest in tropical and subtropical regions. Small heat shock proteins (sHsps) are molecular chaperones that play important roles in the adaptation to various environment stressors. The present study aimed to clarify the response mechanisms of S. frugiperda to various environmental stressors. We obtained five S. furcifera sHsp genes (SfsHsp21.3, SfsHsp20, SfsHsp20.1, SfsHsp19.3, and SfsHsp29) via cloning. The putative proteins encoded by these genes contained a typical α-crystallin domain. The expression patterns of these genes during different developmental stages, in various tissues of male and female adults, as well as in response to extreme temperatures and UV-A stress were studied via real-time quantitative polymerase chain reaction. The results showed that the expression levels of all five SfsHsp genes differed among the developmental stages as well as among the different tissues of male and female adults. The expression levels of most SfsHsp genes under extreme temperatures and UV-A-induced stress were significantly upregulated in both male and female adults. In contrast, those of SfsHsp20.1 and SfsHsp19.3 were significantly downregulated under cold stress in male adults. Therefore, the different SfsHsp genes of S. frugiperda play unique regulatory roles during development as well as in response to various environmental stressors.
Keywords: Spodoptera frugiperda, Small heat shock proteins, Gene cloning, Expression analysis, Environmental stressors
Introduction
Heat shock proteins (Hsps) are a class of highly conserved proteins that are synthesized by cells or organisms in response to stress from environmental factors and harmful stimuli. As molecular chaperones, Hsps respond to various biological stressors, such as bacteria, viruses, fungi, and insects, as well as to non-biological hazards, such as ultraviolet (UV) light, heavy metals, high/low temperatures, moisture, various agents, and salt; as a result, they improve the organism’s ability to resist stress (Parsell and Lindquist 1993; Zhao and Jones 2012; Hu et al. 2018). Based on their molecular weight, Hsps are divided into various families: Hsp100, Hsp90, Hsp70, Hsp60, Hsp40, and small Hsps (sHsps) (Feder and Hofmann 1999). sHsps have molecular masses ranging from approximately 12 to 43 kDa and generally have a large N-terminal region, conserved α-crystallin domain, and extended C-terminal features (Haslbeck et al. 2005). As a molecular chaperone, sHsps help in maintaining the initial cytoskeleton, participate in redox metabolism, maintain growth and development, and enhance resistance to environmental stress. Furthermore, they are closely related to the dormancy and diapause of organisms (Van Montfort et al. 2001; Garrido et al. 2012; Wettstein et al. 2012). In addition, sHsps play important roles in the growth and development of insects as well as in responses to various environmental stressors (Michaud et al. 2002; Takahashi et al. 2010; King and Macrae 2015).
In nature, insects have strong adaptability toward changing and adverse environmental conditions (Grubor-Lajsic et al. 1992; Bale and Hayward 2010). The growth, development, survival, and geographic distribution of insects are affected by various environmental factors, including temperature variations and UV irradiation exposure (Bale et al. 2002; Wang et al. 2014). Several studies have reported that sHsps participate in the regulation of the tolerance of insects to temperature fluctuations. For example, in response to cold and heat stresses, the expression levels of multiple sHsp genes were significantly upregulated in Liriomyza trifolii, Choristoneura fumiferana, Bactrocera dorsalis, and Lasioderma serricorne (Dou et al. 2017; Quan et al. 2018; Chang et al. 2019; Yang et al. 2019). Hsp20.1, Hsp21.2, Hsp21.4, and Hsp22 in Laodelphax striatellus were silenced by RNAi, following which the mortality of the insect significantly increased at 42°C (Wang et al. 2019). UV-A (315–400 nm) is widely used as a light trap for insect pests to control different kinds of insect pest populations, including Lepidopteron (Antignus 2000; Steinbauer 2003). UV-A irradiation, as another important environmental stressor, can induce the production of large amounts of reactive oxygen species by insects, resulting in oxidative stress; this can adversely affect normal physiological functions (Meyer-Rochow et al. 2002; Meng et al. 2009; Ali et al. 2017). However, insect sHsps, such as Apis cerana Hsp27.6 and Tribolium castaneum Hsp27 and Hsp21.8b, as molecular chaperones, are significantly upregulated to ensure the correct folding of newly synthesized proteins as well as to protect functional proteins from degeneration and aggregation due to UV-A stress (Liu et al. 2012; Sang et al. 2012; Xie et al. 2018). In addition, the expression levels of insect sHsps vary at different developmental stages and across different tissues (Michaud et al. 2002; Yang et al. 2019; Lu et al. 2014; Chen and Zhang 2015; Pan et al. 2018). Furthermore, the expression patterns of sHsps differ between sexes. For example, the expression levels of the Hsp23 and Hsp27 genes are significantly higher in the adult male Plutella xylostella than in adult females (Xiao et al. 2013). Moreover, the expression levels of five sHsp genes of Chilo suppressalis also differ according to sex (Lu et al. 2014). A similar phenomenon has also been reported in Bombyx mori (Li et al. 2009).
Spodoptera frugiperda (J. E. Smith) (Lepidoptera: Noctuidae) is a major migratory pest native to and widely distributed in the tropical and subtropical regions of America (Luginbill 1928; Sparks 1979). S. frugiperda has a very mixed diet, and its larvae can harm a variety of crops, including corn, rice, wheat, sorghum, cotton, and various vegetables (Montezano et al. 2018). In 2016, S. frugiperda invaded Africa and many Asian countries, resulting in major crop losses, particularly corn (Day et al. 2017). The ability of S. frugiperda to migrate to many areas and quickly settle is related to its strong adaptability to various environmental factors. However, there have been relatively few studies on the mechanisms underlying the ability of S. frugiperda to adapt to environmental stressors. In the present study, the full-length complementary DNAs (cDNAs) of five S. frugiperda sHsp genes were cloned and their expression patterns were examined in different tissues of male and female adults at different developmental stages to assess their roles in the adaptation to high and low temperatures as well as in response to UV-A irradiation. These findings will expand our current knowledge of the molecular mechanisms underlying the ability of S. frugiperda to adapt to environmental stressors.
Methods
Insect and sample preparation
The S. frugiperda specimens used in this study were originally collected from corn fields in Guizhou Province, China, in July 2019 and reared on young corn leaves in an artificial climate chamber at a temperature of 27°C ± 1°C, relative humidity of 70–80%, and 14:10-h light:dark photoperiod.
Samples were collected at different developmental stages: eggs, 1st–6th instar larva, 3-day-old pupae, and 3-day-old adults (females and males). A total of 100 eggs, i.e., 50 1st instar; 30 2nd instar; 20 3rd instar; 15 4th instar; and 10 5th–6th instar larvae, 3-day-old pupae, and 3-day-old adults, were collected per biological replicate. For the tissue-specific experiments, the head, thorax, abdomen, antenna, compound eye, foot, wing, midgut, and ovary/testis of male and female adults were placed in 1.5-mL centrifuge tubes containing RNA storage reagent (Sangon Biotech Co. Ltd., Shanghai, China). The head, thorax, abdomen, and wings of 15 male and female adults and the antenna, compound eye, foot, midgut, and ovary/testis tissues of 20 male and female adults were collected. All samples were immediately frozen in liquid nitrogen and stored at −80°C until further analysis.
Temperature and UV-A experiments
The experimental insect treatments were as follows: (1) high- and low-temperature treatment: 3-day-old male and female adults were exposed to temperatures of 42°C and 4°C for 0 (control), 30, 60, 90, 120, and 150 min. Ten samples were collected per biological replicate (insects). (2) UV-A treatment: 3-day-old male and female adults were irradiated with 315–400 nm UVA light (NanJing HuaQiang Electronic Engineering Co., Ltd., Nanjing, China) at a frequency of 300 μW/cm2. To eliminate the influence of other light sources, after adapting to a 2-h scotophase period at 27°C±1°C, 3-day-old female adults were exposed to UVA for 0 (control), 30, 60, 90, and 120 min. Ten female adults were collected per biological replicate (insects). All samples were immediately frozen in liquid nitrogen and stored at −80°C until analysis.
RNA extraction and cDNA synthesis
Total RNA was extracted from each sample using the Eastep® Super Total RNA Extraction Kit (Shanghai Promega Biological Products, Ltd., Shanghai, China) and processed in a spin column with DNase I to remove genomic DNA. RNA concentration and purity were measured using the NanoDrop 2000C spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). RNA integrity was verified via 1% agarose gel electrophoresis. The cDNA for cloning and quantitative real-time polymerase chain reaction (qRT-PCR) was synthesized from 1 μg of RNA from each sample using the HiFiScript cDNA Synthesis Kit (CoWin Biosciences, Beijing, China).
Identification and sequencing of SfsHsp cDNAs
The sequences of the homologous insect genes Hsp21.3, Hsp20.0, Hsp20.1, Hsp19.3, and Hsp29 were retrieved from the database of the National Center for Biotechnology Information (NCBI) and amplified using simple primer pairs targeting the conserved regions (Table 1). The full-length open reading frames (ORFs) of the five genes were confirmed by PCR. The PCR reactions were conducted under the following parameters: an initial denaturation step at 95°C for 3 min, followed by 35 cycles at 95°C for 30 s, 55–65°C (depending on the gene simple primer) for 30 s, and 72°C for 1 min and a final extension step at 72°C for 10 min. The PCR products were separated via 1% agarose gel electrophoresis and subcloned into the pMDTM 19-T vector (TaKaRa Biotechnology (Dalian) Co., Ltd., Dalian, China) for sequencing.
Table 1.
Primers used in this study
| Application of primers | Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|
| Full-length confirmation | Hsp21.3 | GCGTAGCGACACTGTGATTC | CTCTGCTCAATGCAGGTTGTG |
| Hsp20.0 | CCCGCCGGCAAAACATTCA | TCACTTAGCTGTTCCGTTGGC | |
| Hsp20.1 | CATTCAACTGAACGCGACACT | AGCAAGCTCAGCTCGACAG | |
| Hsp19.3 | CGCGAATACAACAACACAACAA | CATACAAACTTAACACAATTAAGGA | |
| Hsp29 | GCAGCCGGCATAGTACATTA | ACGCTTTAATGACTGTCGGT | |
| qPCR analysis | Hsp21.3 | TCGACACTGAATTCTCCAGCA | TCTGCCAACTGTCTGCTGTC |
| Hsp20.0 | GACAGCTGATGGCTACGTGA | GCGATGACTGTCAAGACACC | |
| Hsp20.1 | CAGCCGCGACTACTACAGAC | TCCTCGTGCTTAGCTTCCAC | |
| Hsp19.3 | TGGACCAGAACTTCGGACTG | ACATCCACGTTGATCTGCCAT | |
| Hsp29 | TTCATCACCACCGTAGCCTG | TGATGAGTTGACTCCACGGC | |
| RPL27 | GAAGCCAGGTAAAGTGGTGCT | GTGTCCGTAGGGCTTGTCTG | |
| β-actin | GATCTGGCACCACACCTTCT | GGCGTGTTGAAGGTCTCGAA |
Bioinformatics and phylogenetic analyses
The ORFs were predicted using the ORF finder graphical analysis tool (https://www.ncbi.nlm.nih.gov/orffinder/). Multiple sequence alignments were generated using the DNAMAN 8.0 sequence analysis software (Lynnon Biosoft, San Ramon, CA, USA). Sequence similarities and the presence of conserved domains were determined using the Basic Local Alignment Search Tool algorithm-based programs available on the NCBI website (http://www.blast.ncbi.nlm.nih.gov/Blast). Molecular weight, isoelectric point, and instability index were predicted using the ExPASy ProtParam tool (http://us.expasy.org/tools/protparam.html). The domains were predicted using the ExPASy PROSITE tool (https://prosite.expasy.org/prosite.html). Phylogenetic analysis was performed using the MEGA 5.0 software with the neighbor-joining (1000 replicates) method.
qRT-PCR
qRT-PCR reactions were conducted in a 20-μL reaction mixture containing 10 μL of TB Green® Premix Ex Taq II (TaKaRa), 1 μL of cDNA as a template, 1 μL of gene-specific primers, and 7 μL of nuclease-free water using the CFX-96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). All primers used for qRT-PCR were designed online using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) available on the NCBI website (Table 1). The qRT-PCR reactions were conducted under the following conditions: an initial denaturation step at 95°C for 30 s, followed by 40 cycles at 95°C for 5 s and 60°C for 30 s. All reactions were analyzed using melting curves from 60 to 95°C to ensure the specificity and consistency of the products amplified. All experiments were performed in triplicate and each experiment was repeated three times. The ribosomal protein L27 (RPL27) and β-actin genes were used as internal reference genes. The relative transcript levels were determined using the 2−ΔΔCT method. The geometric mean of two selected internal control genes was used for normalization (Livak and Schmittgen 2001).
Data analysis
All data were analyzed via one-way analysis of variance (ANOVA) using IBM SPSS Statistics for Windows, version 19.0 (IBM Corporation, Armonk, NY, USA). Multiple comparisons and analyses were performed using Tukey’s method. Prior to ANOVA, the homogeneity of the data was tested. A probability (p) value of <0.05 was considered statistically significant.
Results
Cloning, characterization, and phylogenetic analysis of five SfsHsp genes from S. frugiperda
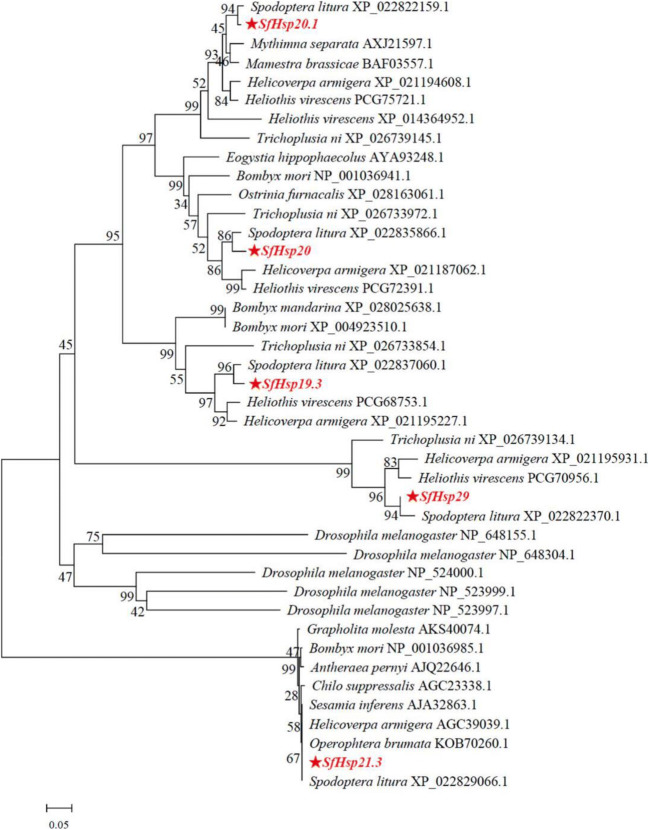
Five SfsHsp genes (SfsHsp21.3, SfsHsp20, SfsHsp20.1, SfsHsp19.3, and SfsHsp29) from S. frugiperda (GenBank accession numbers: MN842800, MN842801, MN842802, MN842803, and MN842804, respectively) were cloned. Sequencing revealed that the five SfsHsp genes contained ORFs of 564, 546, 531, 516, and 765 bp, respectively, encoding 187, 181, 176, 171, and 254 amino acids. The predicted molecular weights of the SfsHsp genes were 19.3–29.0 kDa, theoretical isoelectric points were 5.79–6.34, and instability indices were 39.45–64.02 (Table 2). The deduced amino acid sequences all contained a typical α-crystal domain with lengths of 98, 99, 110, 99, and 114 amino acids, respectively (Fig. 1). To analyze the correlation among these five SfsHsps and sHsps of other insect species, a phylogenetic tree was constructed using the neighbor-joining method with the whole sequences of the five SfsHsps. The results showed that the five SfsHsps were clustered into five branches separately from other homologous species, indicating relatively high conservation of the five SfsHsps (Fig. 2). All the five genes had the highest homology with the sHsps of S. litura.
Table 2.
Characteristics of the mRNAs of sHsp of Spodoptera frugiperda
| Gene | ORF (bp) | Protein length (aa) | Molecular weight (kDa) | Isoelectric point (IP) | Instability index (II) | GenBank accession number |
|---|---|---|---|---|---|---|
| Hsp21.3 | 564 | 187 | 21.3 | 5.79 | 49.18 | MN842800 |
| Hsp20.0 | 546 | 181 | 20.0 | 5.96 | 39.45 | MN842801 |
| Hsp20.1 | 531 | 176 | 20.1 | 6.07 | 64.02 | MN842802 |
| Hsp19.3 | 516 | 171 | 19.3 | 6.10 | 39.97 | MN842803 |
| Hsp29 | 765 | 254 | 29.0 | 6.34 | 43.85 | MN842804 |
Fig. 1.
Nucleotide sequences and the predicted amino acid sequences of five small heat shock proteins (sHSPs) of Spodoptera frugiperda. The highly conserved α-crystallin domain is underlined. The asterisk indicates a translational termination codon
Fig. 2.
Phylogenetic analysis of the small heat shock proteins (sHsps) of Spodoptera frugiperda and other insects. The phylogenetic tree was generated using the MEGA 6.06 software using the neighbor-joining method. Numbers on the branches are the bootstrap values obtained from 1000 replicates. Sequence labels are indicated by the species name and GenBank accession number. The asterisk denotes the sequences of the sHsps of S. frugiperda
Expression levels of the five SfsHsp genes at different developmental stages
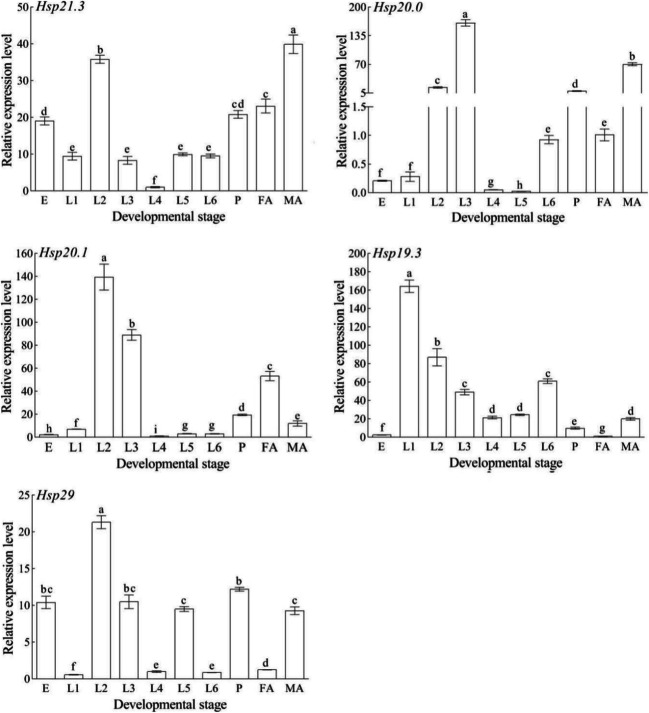
The mRNA expression levels of the five SfsHsp genes at different developmental stages were determined via qRT-PCR (Fig. 3). The SfsHsp genes were expressed at all developmental stages; however, they had different expression patterns. The expression level of SfsHsp21.3 was highest in male adults and lowest in 1st instar larva. Furthermore, the expression level of SfsHsp20 expression was highest in 3rd instar larva and lowest in 5th instar larva. In addition, the expression levels SfsHsp20.1 and SfsHsp29 were highest in 2nd instar larva and lowest in 4th and 1st instar larva, respectively. The expression level of SfsHsp19.3 was highest in 1st instar larva and lowest in female adults.
Fig. 3.
Relative expression levels of five SfsHsp genes at different the developmental stages of Spodoptera frugiperda. E, egg; L1–L6, 1st–6th instar larva; P, pupa; FA, female adult; and MA, male adult. Different letters above the bars indicate significant differences among the developmental stages based on one-way analysis of variance followed by the least significant difference test (p < 0.05)
Expression levels of the five SfsHsp genes in different tissues
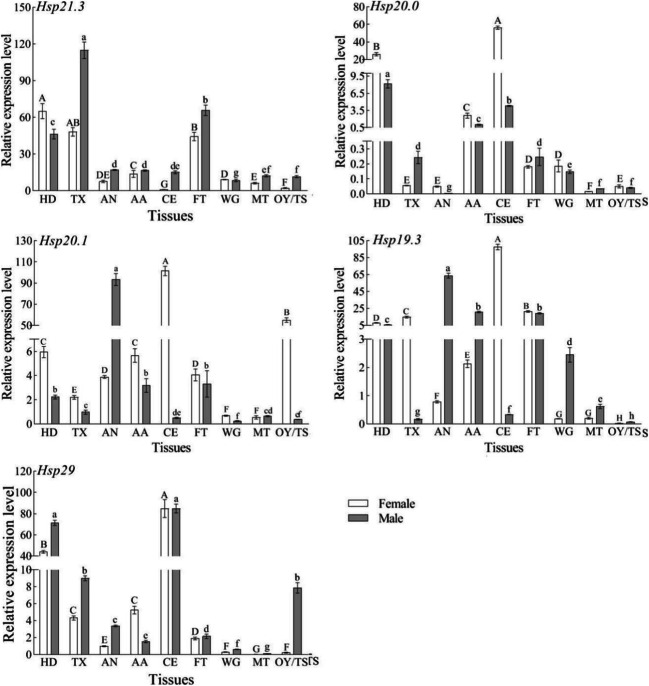
The five S. frugiperda SfsHsp genes were expressed in nine tissue types of male and female adults. However, the expression patterns of these genes significantly varied among the different tissue types and between male and female adults (Fig. 4). In female adults, the expression level of SfsHsp21.3 was highest in the head and lowest in the compound eyes. The expression levels of SfsHsp20, SfsHsp20.1, SfsHsp19.3, and SfsHsp29 were highest in the eyes and lowest in the midgut, midgut, ovary, and midgut, respectively. In male adults, the expression levels of SfsHsp20.1 and SfsHsp19.3 were highest in the abdomen, and those of SfsHsp21.3, SfsHsp20, and SfsHsp29 were highest in the chest, head, and compound eyes, respectively. In contrast, the expression levels of SfsHsp21.3 and SfsHsp20.1 were lowest in the wings. Lastly, the expression levels of SfsHsp20 and SfsHsp29 were lowest in the midgut, and the expression level of SfsHsp19.3 was the lowest in the testis.
Fig. 4.
Relative expression levels of five SfsHsp genes in the different tissues of Spodoptera frugiperda male and female adults. HD, head; TX, thorax; AN, abdomen; AA, antenna; CE, compound eye; FT, foot; WG, wing; MT, midgut; OY/TS, ovary/testis. Different letters above the bars indicate significant differences among tissues based on one-way analysis of variance followed by the least significant difference test (p < 0.05)
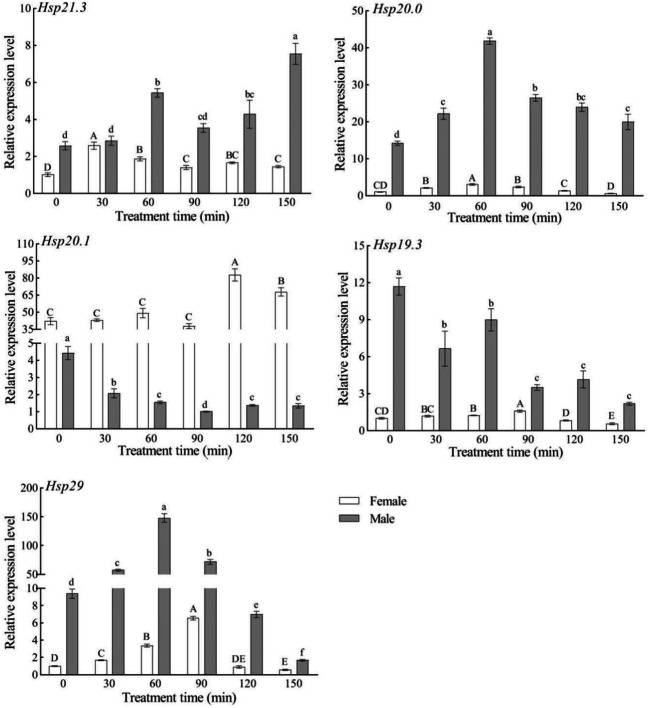
Expression levels of the five SfsHsp genes in response to high-temperature (42°C) stress
In response to high-temperature (42°C) stress, the expression patterns of the five SfsHsp genes widely varied (Fig. 5). In female adults, the expression levels of SfsHsp21.3, SfsHsp20, SfsHsp20.1, SfsHsp19.3, and SfsHsp29 first increased and then decreased with time, reaching peaks at 120, 120, 90, 120, and 120 min, respectively. They were significantly higher than those in the control (0 min) (p < 0.05) by 1.9-, 6.7-, 1.6-, 4.2-, and 5.1-fold. In male adults, the expression level of SfsHsp21.3 was found to be significantly higher than that in the control (0 min) over time. The expression levels of SfsHsp20 and SfsHsp20.1 initially increased and then decreased with time, with both peaking at 60 min. Likewise, the expression levels of SfsHsp19 and SfsHsp29 increased with time.
Fig. 5.
Effect of high-temperature (42°C) stress on the expression levels of the five SfsHsp genes of Spodoptera frugiperda. Different letters above the bars indicate significant differences based on one-way analysis of variance followed by the least significant difference test (p < 0.05)
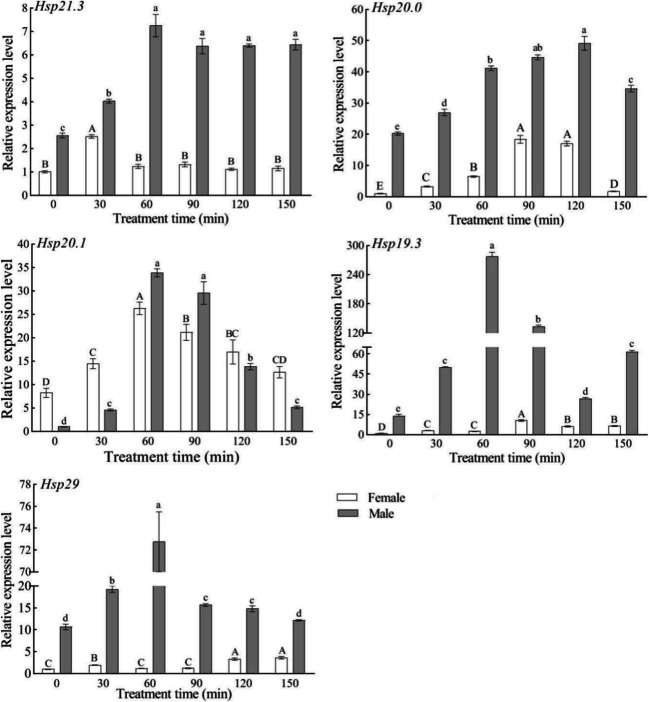
Expression levels of the five SfsHsp genes in response to low-temperature (4°C) stress
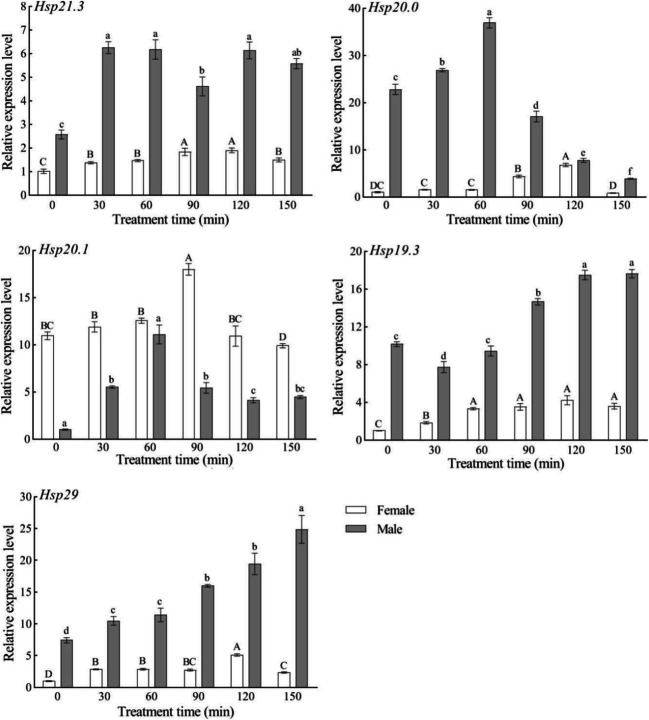
In response to low-temperature (4°C) stress, the expression levels of the five SfsHsp genes increased with time in female adults, reaching maximum expression at 30, 60, 120, 90, and 90 min, respectively; however, the expression levels then decreased. In male adults, the expression levels of SfsHsp21.3, SfsHsp20, and SfsHsp29 increased with time, reaching maximum expression at 150, 60, and 60 min, respectively, whereas the expression levels of SfsHsp20.1 and SfsHsp19.3 decreased with time (Fig. 6).
Fig. 6.
Effect of low-temperature (4°C) stress on the expression levels of the five SfsHsp genes of Spodoptera frugiperda. Different letters above the bars indicate significant differences based on one-way analysis of variance followed by the least significant difference test (p < 0.05)
Expression levels of the five SfsHsp genes in response to UV-A-induced stress
In response to exposure to UV-A irradiation, the expression levels of the five SfsHsp genes in male and female adults increased with time, with maximum expression in female adults at 30, 90, 60, 90, and 150 min, respectively. The maximum expression of the five genes (SfsHsp21.3, SfsHsp20, SfsHsp20.1, SfsHsp19.3, and SfsHsp29) was reached in male adults at 60, 120, 60, 60, and 60 min, respectively (Fig. 7).
Fig. 7.
Effect of UV-A-induced stress on the expression levels of the five SfsHsp genes of Spodoptera frugiperda. Different letters above the bars indicate significant differences based on one-way analysis of variance followed by the least significant difference test (p < 0.05)
Discussion
Insects have developed strategies to adapt to adverse environmental conditions during long-term evolution, including extreme temperatures and UV-A irradiation. sHsps play important roles in insect growth and development and in responses to various environmental stressors (Michaud et al. 2002; Takahashi et al. 2010; King and Macrae 2015). However, in the long-term evolution of different insects, the types, numbers, structures, and functions of sHsps differ among species (Chen and Zhang 2015; Xiao et al. 2013; Haslbeck et al. 2008; Wang et al. 2019). For example, 12 sHsp genes in Drosophila melanogaster had different subcellular localization. Hsp22 and Hsp27 were located in the mitochondrial matrix and nucleus, respectively. The other sHsps were located in the cytoplasm (Morrow and Tanguay 2015). Moutaoufik and Tanguay (2021) predicted that there are 13 sHsps in S. frigiperda. In the present study, five new sHsp genes (SfsHsp21.3, SfsHsp20, SfsHsp20.1, SfsHsp19.3, and SfsHsp29) of S. frugiperda were identified and cloned. The predicted protein products of all five SfsHsp contained the characteristic α-crystallin domain, which is consistent with the findings of previous studies of sHsps in other insect species (Yang et al. 2019; Wang et al. 2019). Phylogenetic analysis revealed that the five SfsHsps were clustered into five branches separately from other homologous species. Insect sHsps can be divided into two categories (specific and homologous) according to the clustering methods used to construct the phylogenetic trees (Lu et al. 2014; Xiao et al. 2013; Shen et al. 2011). The five SfsHsps identified in the present study were homologous. This clustering mode has also been applied in previous studies, which have indicated that the evolution of sHsps is complicated (Quan et al. 2018; Chang et al. 2019; Franck et al. 2004; Martín-Folgar et al. 2015). Nevertheless, all five SfsHsps had the highest homology with the sHsps of S. litura. Ye et al. (2019) conducted a comparative genomics analysis of Lepidopteran insects and reported that S. frugiperda and S. litura are closely related.
Insect sHsps play important roles in developmental regulation. For example, the expression levels of Hsp19.8, Hsp21.4, Hsp21.5, and Hsp21.7b in C. suppressalis were highest in the adult stage, whereas those of Hsp21.7a were highest in the 1st instar stage. The expression levels of Hsp19.7, Hsp20, and Hsp20.7 in S. litura and those of Hsp19.8 in C. pomonella were also highest in the adult stage (Lu et al. 2014; Shen et al. 2011; Garczynski et al. 2011). However, the expression levels of three sHsp genes (Hsp19.5, Hsp20.8, and Hsp21.7) in S. litura and those of Hsp20.4 in Liriomyza sativa were lowest in the adult stage, whereas the expression level of Hsp24 in Lucilia cuprina was lowest in 1st instar larvae (Shen et al. 2011; Huang et al. 2009; Concha et al. 2012). In the present study, the five SfsHsp genes were expressed in all tested developmental stages; however, they had different expression patterns. The expression levels of SfsHsp20.1, SfsHsp20, SfsHsp19.3, and SfsHsp29 were all highest in young larval stages (1st–3rd instar larva), whereas the expression level of SfsHsp21.3 was highest in the adult stage. All five sHsps exhibited lower expression at the 4th instar in S. frugiperda. However, Hsp19.7, Hsp20, Hsp20.4, Hsp20.7, and Hsp20.8 exhibited lower expression at the 3rd instar in S. litura; this difference may be related to the growth and development of insects at different instars (Shen et al. 2011; Jagla et al. 2018). In addition, the expression levels of four SfsHsp genes significantly differed between male and female adults, with a similar phenomenon in the sHsps of C. suppressalis (Lu et al. 2014). This result may be due to the different roles of sHsps in female and male worms (Jagla et al. 2018). Nevertheless, the exact underlying mechanism needs to be further investigated. Overall, the multiple expression patterns indicate that sHsp genes play different roles in insect development.
The expression patterns of insect sHsps are tissue-specific. For example, the expression levels of Hsp21.4 and Hsp21.7a in C. suppressalis and those of Hsp21.4 in Sesamia inferens were highest in the head, similar to the expression levels of Hsp19.1 and Hsp22.6 in B. mori. In A. cerana, the expression level of Hsp22.6 was the highest in the midgut, whereas the expression levels of Hsp24.2 and Hsp23 differed among the tissues (Lu et al. 2014; Xiao et al. 2013; Sun et al. 2014; Zhang et al. 2014). In the present study, the five SfsHsp genes of S. frugiperda were expressed in nine tissue types of male and female adults. However, the expression patterns of these genes significantly varied among the different tissue types and between male and female adults. The expression levels of the five SfsHsp genes were the highest in the head and compound eyes of female adults, whereas those of SfsHsp20 and SfsHsp29 were the highest in the head and compound eyes of male adults; this may be because these tissues are the center of insect perception of changes in the external environment. However, in male adults, the expression level of SfsHsp19.3 was the highest in the abdomen, whereas that of SfsHsp21.3 was the highest in the chest. In addition, the high expression of SfsHsp20.1 in the female ovary and male abdomen suggests that it is related to the reproduction of S. frugiperda. In Drosophila, Hsp26 and Hsp27 showed high transcript levels in the testis and ovaries (Jagla et al. 2018). The expression patterns of insect sHsp genes widely vary across tissues, indicating large functional differences among species.
Temperature is an important environmental factor that regulates the growth, development, reproductive capacity, population density, and distribution of insects. It plays a vital role in the growth and decline of populations and affects almost all ecological and physiological processes of insects (Bale et al. 2002; Willmer et al. 2005; Hoffmann et al. 2003). Natural fluctuations in environmental temperatures pose a certain threat to insect reproduction. To adapt to the environment, insects must be tolerant to increases in temperature. The differences in the ability of insects to adapt to temperature determine distribution and population dynamics (Hausmann et al. 2005; Samietz et al. 2005). Resistance to extremely high and low temperatures is related to the genetic makeup of insects, such as Hsps, which are generally considered temperature-tolerance proteins. In the present study, the expression levels of the five SfsHsp genes of S. frugiperda were significantly upregulated at 42°C and 4°C, whereas SfsHsp20.1 and SfsHsp19.3 of the adult males did not respond to exposure to 4°C. However, the expression patterns of the five SfsHsp genes significantly differed at 42°C and 4°C as well as between male and female adults. In response to extreme temperatures (42°C and 4°C), the expression levels of the five SfsHsp genes in male and female adults tended to initially increase and then decrease with time, but the peak of each SfsHsp gene differed among the time points and between male and female adults. For example, at 4°C, the expression levels of SfsHsp21.3, SfsHsp20, SfsHsp20.1, SfsHsp19.3, and SfsHsp29 in female adults peaked at 30, 60, 120, 90, and 90 min, respectively. At 42°C and 4°C, the expression level of the SfsHsp29 gene peaked in male and female adults at 120, 150, 90, and 60 min, respectively, which may have been due to the threshold of the protective effects of Hsps genes. When environmental stress exceeds a certain limit, the expression levels of SfsHsp genes will decrease. A similar phenomenon was also observed in response to heat stress in A. cerana (Hsp27.6 showed maximum expression at 120 min) and L. striatellus (Hsp20.1, Hsp21.2, Hsp21.4, Hsp21.5, and Hsp22 showed maximum expression at 2 h) (Liu et al. 2012; Wang et al. 2019). These findings indicate that SfsHsps play important roles in S. frugiperda’s response to environmental temperature stress.
As an important environmental stress factor, UV-A irradiation can result in the photo-oxidation of endogenous photosensitizers in insects, which produces large amounts of reactive oxygen species, including superoxide anions, hydroxyl radicals, and hydrogen peroxide, which destroy the functional activities of proteins involved in lipid peroxidation and DNA base pair modification, leading to mutations (Meyer-Rochow et al. 2002; Meng et al. 2009; Cadet et al. 2005). Previous studies have shown that insect sHsps, as molecular chaperones, can help to ensure the correct folding of newly synthesized proteins and prevent functional proteins from degeneration and aggregation due to UV-A stress (Johnston et al. 1998; Basha et al. 2012; Liu et al. 2012; Xie et al. 2018). In this study, the expression levels of five SfsHsp genes were significantly upregulated in response to UV-A irradiation, indicating that SfsHsps play important roles in the molecular mechanism underlying the response of S. frugiperda to UV-A-induced stress. In response to UV-A irradiation, the expression levels of five SfsHsp genes (SfsHsp21.3, SfsHsp20, SfsHsp20.1, SfsHsp19.3, and SfsHsp29) increased with time in adults, with maximum expression levels at 30, 90, 60, 90, and 150 min in female adults and at 60, 120, 60, 60, and 60 min in male adults, respectively. Similar results were observed with the sHsps of other insect species. For example, under different durations of UV-A-induced stress, the expression levels of Hsp22.6 and Hsp27.6 in A. cerana as well as those of Hsp27 and Hsp21.8b in T. castaneum peaked at 2 and 4 h, respectively (Liu et al. 2012; Sang et al. 2012; Xie et al. 2018; Zhang et al. 2014). In response to UV-A-induced stress, the changes in the expression levels of insect sHsps over time were somewhat similar to the responses to temperature stress, which may be due to the threshold phenomenon of the protective effect of insect sHsps in response to UV-A-induced stress. A similar phenomenon has also been reported in Hsp70 and Hsp90 in S. littoralis under UV-C exposure (Guz et al. 2020). The variation in the expression patterns suggests that the sHsp genes may have different roles in the responses of insects to UV-A-induced stress. In addition, in response to UV-A irradiation for different lengths of time, with the exception of SfsHsp20.1, the expression levels of the other four SfsHsp genes of S. frugiperda were higher in males than in females. These results suggest that S. frugiperda males are more resistant to UV-A irradiation than females. Similar results were reported for Hsp90 in S. frugiperda under UV-A stress (Zhou et al. 2020).
Conclusions
In summary, five sHsp genes of S. frugiperda were identified and cloned. The putative sHsps contained typical conserved domains. The expression levels of these sHsp genes varied among different developmental stages and tissues. Responses to extreme temperatures (42°C and 4°C) and UV-A irradiation indicate that these genes play important roles in the mechanisms underlying the ability of the insects to adapt to environmental stress. However, further studies are warranted to clarify the exact roles and physiological mechanisms of the sHsp genes of S. frugiperda.
Code availability
Not applicable
Funding
This research was funded by the National Key R&D Program of China (grant no. 2017YFD0200900), the National Natural Science Foundation of China (grant nos. 31401754, 31460483), and the Science and Technology Project of the China Tobacco Corporation (grant no. 201919).
Declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
Not applicable
Consent to participate
Not applicable
Consent for publication
Not applicable
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ali A, Rashid MA, Huang QY, Lei CL. Influence of UV-A radiation on oxidative stress and antioxidant enzymes in Mythimna separata (Lepidoptera: Noctuidae) Environ Sci Pollut Res. 2017;24:8392–8398. doi: 10.1007/s11356-017-8514-7. [DOI] [PubMed] [Google Scholar]
- Antignus Y. Manipulation of wavelength-dependent behaviour of insects: an IPM tool to impede insects and restrict epidemics of insect-borne viruses. Virus Res. 2000;71:213–220. doi: 10.1016/S0168-1702(00)00199-4. [DOI] [PubMed] [Google Scholar]
- Bale JS, Hayward SAL. Insect overwintering in a changing climate. J Exp Biol. 2010;213:980–994. doi: 10.1242/jeb.037911. [DOI] [PubMed] [Google Scholar]
- Bale JS, Masters GJ, Hodkinson ID, Awmack C, Bezemer TM, Brown VK, Butterfield J, Buse A, Coulson JC, Farrar J, Good JEG, Harrington R, Hartley S, Jones TH, Lindroth RL, Press MC, Symrnioudis I, Watt AD, Whittaker JB. Herbivory in global climate change research: direct effects of rising temperature on insect herbivores. Glob Chang Biol. 2002;8:1–16. doi: 10.1046/j.1365-2486.2002.00451.x. [DOI] [Google Scholar]
- Basha E, O’Neill H, Vierling E. Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 2012;37:106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J, Sage E, Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Chang YW, Zhang XX, Lu MX, Du YZ, Zhu-Salzman K. Molecular cloning and characterization of small heat shock protein genes in the invasive leaf miner fly, Liriomyza trifolii. Genes. 2019;10:775. doi: 10.3390/genes10100775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XE, Zhang YL. Identification of multiple small heat-shock protein genes in Plutella xylostella (L.) and their expression profiles in response to abiotic stresses. Cell Stress Chaperon. 2015;20:23–35. doi: 10.1007/s12192-014-0522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha C, Edman RM, Belikoff EJ, Schiemann AH, Carey B, Scott MJ. Organization and expression of the Australian sheep blowfly (Lucilia cuprina) hsp23, hsp24, hsp70 and hsp83 genes. Insect Mol Biol. 2012;21:169–180. doi: 10.1111/j.1365-2583.2011.01123.x. [DOI] [PubMed] [Google Scholar]
- Day R, Abrahams P, Bateman M, Beale T, Clottey V, Cock M, Colmenarez Y, Corniani N, Early R, Godwin J, Gomez J, Moreno PG, Murphy ST, Oppong-Mensah B, Phiri N, Pratt C, Silvestri S, Witt A. Fall armyworm: impacts and implications for Africa. Outlooks Pest Manag. 2017;28:196–201. doi: 10.1564/v28_oct_02. [DOI] [Google Scholar]
- Dou W, Tian Y, Liu H, Shi Y, Smagghe G, Wang JJ. Characteristics of six small heat shock protein genes from Bactrocera dorsalis: diverse expression under conditions of thermal stress and normal growth. Comp Biochem Physiol B. Biochem Mol Biol. 2017;213:8–16. doi: 10.1016/j.cbpb.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Franck E, Madsen O, Van-Rheede T, Ricard G, Huynen MA, de-Jong WW. Evolutionary diversity of vertebrate small heat shock proteins. J Mol Evol. 2004;59:792–805. doi: 10.1007/s00239-004-0013-z. [DOI] [PubMed] [Google Scholar]
- Garczynski SF, Unruh TR, Guédot C, Neven LG. Characterization of three transcripts encoding small heat shock proteins expressed in the codling moth, Cydia pomonella (Lepidoptera: Tortricidae) Insect Sci. 2011;18:473–483. doi: 10.1111/j.1744-7917.2010.01401.x. [DOI] [Google Scholar]
- Garrido C, Paul C, Seigneuric R, Kampinga HH. The small heat shock proteins family: the long forgotten chaperones. Int J Biochem Cell Biol. 2012;44:1588–1592. doi: 10.1016/j.biocel.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Grubor-Lajsic G, Block W, Worland R. Comparison of the cold hardiness of two larval Lepidoptera (Noctuidae) Physiol Entomol. 1992;17:148–152. doi: 10.1111/j.1365-3032.1992.tb01192.x. [DOI] [Google Scholar]
- Guz N, Dageri A, Altincicek B, Aksoy S. Molecular characterization and expression patterns of heat shock proteins in Spodoptera littoralis, heat shock or immune response? Cell Stress Chaperon. 2020;26:29–40. doi: 10.1007/s12192-020-01149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Kastenmüller A, Buchner J, Weinkauf S, Braun N. Structural dynamics of archaeal small heat shock proteins. J Mol Biol. 2008;378:362–374. doi: 10.1016/j.jmb.2008.01.095. [DOI] [PubMed] [Google Scholar]
- Hausmann C, Samietz J, Dorn S. Thermal orientation of Anthonomus pomorun (Coleoptera: Curculionidae) in early spring. Physiol Entomol. 2005;30:48–53. doi: 10.1111/j.0307-6962.2005.00427.x. [DOI] [Google Scholar]
- Hoffmann AA, Sørensen JG, Loeschcke V. Adaptation of Drosophila to temperature extremes: bringing together quantitative and molecular approaches. J Therm Biol. 2003;28:175–216. doi: 10.1016/S0306-4565(02)00057-8. [DOI] [Google Scholar]
- Hu F, Ye K, Tu XF, Lu YJ, Thakur K, Wei ZJ. Identification and expression analysis of four heat shock protein genes associated with thermal stress in rice weevil, Sitophilus oryzae. J Asia-Pac Entomol. 2018;21:872–879. doi: 10.1016/j.aspen.2018.06.009. [DOI] [Google Scholar]
- Huang LH, Wang CZ, Kang L. Cloning and expression of five heat shock protein genes in relation to cold hardening and development in the leaf miner, Liriomyza sativa. J Insect Physiol. 2009;55:279–285. doi: 10.1016/j.jinsphys.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Jagla T, Magiera MD, Poovathumkadavil P, Daczewska M, Jagla K. Developmental expression and functions of the small heat shock proteins in Drosophila. Int J Mol Sci. 2018;19:3441. doi: 10.3390/ijms19113441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AM, Macrae TH. Insect heat shock proteins during stress and diapause. Annu Rev Entomol. 2015;60:59–75. doi: 10.1146/annurev-ento-011613-162107. [DOI] [PubMed] [Google Scholar]
- Li ZW, Li X, Yu QY, Xiang ZH, Kishino H, Zhang Z. The small heat shock protein (shsp) genes in the silkworm, Bombyx mori, and comparative analysis with other insect shsp genes. BMC Evol Biol. 2009;9:215. doi: 10.1186/1471-2148-9-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xi D, Kang M, Guo X, Xu B. Molecular cloning and characterization of Hsp27.6: the first reported small heat shock protein from Apis cerana cerana. Cell Stress Chaperon. 2012;17:539–551. doi: 10.1007/s12192-012-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu MX, Hua J, Cui YD, Du YZ. Five small heat shock protein genes from Chilo suppressalis: characteristics of gene, genomic organization, structural analysis, and transcription profiles. Cell Stress Chaperon. 2014;19:91–104. doi: 10.1007/s12192-013-0437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luginbill P. The fall army worm, USDA Technol. Bull. 1928;34:91. [Google Scholar]
- Martín-Folgar R, de la Fuente M, Morcillo G, Martínez-Guitarte JL. Characterization of six small HSP genes from Chironomus riparius (Diptera, Chironomidae): differential expression under conditions of normal growth and heat-induced stress. Comp Biochem Physiol A. 2015;188:76–86. doi: 10.1016/j.cbpa.2015.06.023. [DOI] [PubMed] [Google Scholar]
- Meng JY, Zhang CY, Zhu F, Wang XP, Lei CL. Ultraviolet light-induced oxidative stress: effects on antioxidant response of Helicoverpa armigera adults. J Insect Physiol. 2009;55:588–592. doi: 10.1016/j.jinsphys.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Meyer-Rochow VB, Kashiwagi T, Eguchi E. Selective photoreceptor damage in four species of insects induced by experimental exposures to UV-irradiation. Micron. 2002;33:23–31. doi: 10.1016/s0968-4328(00)00073-1. [DOI] [PubMed] [Google Scholar]
- Michaud S, Morrow G, Marchand J, Tanguay RM. Drosophila small heat shock proteins: cell and organelle-specific chaperones? Prog Mol Subcell Biol. 2002;28:79–101. doi: 10.1007/978-3-642-56348-5_5. [DOI] [PubMed] [Google Scholar]
- Montezano DG, Specht A, Sosa-Gómez DR, Roque-Specht VF, Sousa-Silva JC, Paula-Moraes SV, Peterson JA, Hunt TE(H) Host plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. Afr Entomol. 2018;26:286–300. doi: 10.4001/003.026.0286. [DOI] [Google Scholar]
- Morrow G, Tanguay RM (2015) Drosophila small heat shock proteins: an update on their features and functions. In: Tanguay RM, Hightower LE (eds) The Big Book on Small Heat Shock Proteins, Heat Shock Proteins 8. Berlin, Germany, Chapter25, pp 579-606. 10.1007/978-3-319-16077-1_25
- Moutaoufik MT, Tanguay RM. Analysis of insect nuclear small heat shock proteins and interacting proteins. Cell Stress Chaperon. 2021;26:265–274. doi: 10.1007/s12192-020-01156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan DD, Lu MX, Li QY, Du YZ. Characteristics and expression of genes encoding two small heat shock protein genes lacking introns from Chilo suppressalis. Cell Stress Chaperon. 2018;23:55–64. doi: 10.1007/s12192-017-0823-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Quan GX, Duan J, Ladd T, Krell PJ. Identification and expression analysis of multiple small heat shock protein genes in spruce budworm, Choristoneura fumiferana (L.) Cell Stress Chaperon. 2018;23:141–154. doi: 10.1007/s12192-017-0832-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samietz J, Salser MA, Dingle H. Altiudinal variation in behavioural thermoregulation: local adaptation vs. plasticity in California grasshoppers. J Evol Biol. 2005;118:1087–1096. doi: 10.1111/j.1420-9101.2005.00893.x. [DOI] [PubMed] [Google Scholar]
- Sang W, Ma WH, Qiu L, Zhu ZH, Lei CL. The involvement of heat shock protein and cytochrome P450 genes in response to UV-A exposure in the beetle Tribolium castaneum. J Insect Physiol. 2012;58:830–836. doi: 10.1016/j.jinsphys.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Shen Y, Gu J, Huang LH, Zheng SC, Liu L, Xu WH, Feng QL, Kang L. Cloning and expression analysis of six small heat shock protein genes in the common cutworm, Spodoptera litura. J Insect Physiol. 2011;57:908–914. doi: 10.1016/j.jinsphys.2011.03.026. [DOI] [PubMed] [Google Scholar]
- Sparks AN. A review of the biology of the fall armyworm. Fla Entomol. 1979;62:82–87. doi: 10.2307/3494083. [DOI] [Google Scholar]
- Steinbauer MJ. Using ultra-violet light traps to monitor autumn gum moth, Mnesampela private (Lepidoptera: Geometridae), in south-east Australia. Aust For. 2003;66:279–286. doi: 10.1080/00049158.2003.10674922. [DOI] [Google Scholar]
- Sun M, Lu MX, Tang XT, Du YZ. Characterization and expression of genes encoding three small heat shock proteins in Sesamia inferens (Lepidoptera: Noctuidae) Int J Mol Sci. 2014;15:23196–23211. doi: 10.3390/ijms151223196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi KH, Rako L, Takano-Shimizu T, Hoffmann AA, Lee SF. Effects of small Hsp genes on developmental stability and microenvironmental canalization. BMC Evol Biol. 2010;10:284. doi: 10.1186/1471-2148-10-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Montfort RV, Slingsby C, Vierling E. Structure and function of the small heat shock protein/alpha-crystallin family of molecular chaperones. Adv Protein. Chem. 2001;59:105–156. doi: 10.1016/s0065-3233(01)59004-x. [DOI] [PubMed] [Google Scholar]
- Wang LH, Zhang YL, Pan L, Wang Q, Han YC, Niu HT, Shan D, Hoffmann A, Fang JC. Induced expression of small heat shock proteins is associated with thermotolerance in female Laodelphax striatellus planthoppers. Cell Stress Chaperon. 2019;24:115–123. doi: 10.1007/s12192-018-0947-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LJ, Zhou LJ, Zhu ZH, Ma WH, Lei CL. Differential temporal expression profiles of heat shock protein genes in Drosophila melanogaster (Diptera: Drosophilidae) under ultraviolet A radiation stress. Environ Entomol. 2014;43:1427–1434. doi: 10.1603/EN13240. [DOI] [PubMed] [Google Scholar]
- Wettstein G, Bellaye PS, Micheau O, Bonniaud P. Small heat shock proteins and the cytoskeleton: an essential interplay for cell integrity? Int J Biochem Cell Biol. 2012;44:1680–1686. doi: 10.1016/j.biocel.2012.05.024. [DOI] [PubMed] [Google Scholar]
- Willmer PC, Stone G, Johnston IA. Environmental physiology of animals. 2. Oxford: Wiley-Blackwell; 2005. p. e1333. [Google Scholar]
- Xiao XF, Lin HL, Zheng DD, Yang G, You MS. Identification and expression patterns of heat shock protein genes in the diamondback moth, Plutella xylostella (Lepidoptera: Yponomeutidae) Acta Entomol Sin. 2013;56:457–464. [Google Scholar]
- Xie J, Xiong W, Hu X, Gu S, Zhang S, Gao S, Song X, Bi J, Li B. Characterization and functional analysis of hsp21.8b: an orthologous small heat shock protein gene in Tribolium castaneum. J Appl Entomol. 2018;142:654–666. doi: 10.1111/jen.12519. [DOI] [Google Scholar]
- Yang WJ, Xu KK, Cao Y, Meng YL, Liu Y, Li C. Identification and expression analysis of four small heat shock protein genes in cigarette beetle, Lasioderma serricorne (Fabricius) Insects. 2019;10:139. doi: 10.3390/insects10050139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye XH, Yang Y, Mei Y, Xiao HM, Li F. The genome annotation and comparative genomics analysis of Spodoptera frugiperda. J Appl Entomol. 2019;41:706–717. doi: 10.3969/j.issn.1674-0858.2019.04.4. [DOI] [Google Scholar]
- Zhang YY, Liu YL, Guo XL, Li YL, Gao HG, Guo XQ, Xu B. sHsp22.6, an intronless small heat shock protein gene, is involved in stress defence and development in Apis cerana cerana. Insect Biochem Mol Biol. 2014;53:1–12. doi: 10.1016/j.ibmb.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Zhao L, Jones WA. Expression of heat shock protein genes in insect stress responses. Invert Surviv J. 2012;9:93–101. doi: 10.1155/2012/484919. [DOI] [Google Scholar]
- Zhou L, Meng JY, Yang CL, Li J, Hu CX, Zhang CY. Cloning of heat shock protein gene SfHsp90 and its expression under high and low temperature and UV-A stresses in Spodoptera frugiperda (Lepidoptera: Noctuidae) Acta Entomol Sin. 2020;63:533–544. doi: 10.16380/j.kcxb.2020.05.002. [DOI] [Google Scholar]