Abstract
Brucellosis, caused by several species of the genus Brucella, is a zoonotic disease that affects humans and animal species worldwide. Information on the Brucella species circulating in different hosts in Kenya is largely unknown, thus limiting the adoption of targeted control strategies. This study was conducted in multi-host livestock populations in Kenya to detect the circulating Brucella species and assess evidence of host–pathogen associations. Serum samples were collected from 228 cattle, 162 goats, 158 sheep, 49 camels, and 257 humans from Narok and Marsabit counties in Kenya. Information on age, location and history of abortion or retained placenta were obtained for sampled livestock. Data on age, gender and location of residence were also collected for human participants. All samples were tested using genus level real-time PCR assays with primers specific for IS711 and bcsp31 targets for the detection of Brucella. All genus positive samples (positive for both targets) were further tested with a speciation assay for AlkB and BMEI1162 targets, specific for B. abortus and B. melitensis, respectively. Samples with adequate quantities aggregating to 577 were also tested with the Rose Bengal Test (RBT). A total of 199 (33.3%) livestock and 99 (38.5%) human samples tested positive for genus Brucella. Animal Brucella PCR positive status was positively predicted by RBT positive results (OR = 8.3, 95% CI 4.0–17.1). Humans aged 21–40 years had higher odds (OR = 2.8, 95% CI 1.2–6.6) of being Brucella PCR positive compared to the other age categories. The data on detection of different Brucella species indicates that B. abortus was detected more often in cattle (OR = 2.3, 95% CI 1.1–4.6) and camels (OR = 2.9, 95% CI 1.3–6.3), while B. melitensis was detected more in sheep (OR = 3.6, 95% CI 2.0–6.7) and goats (OR = 1.7, 95% CI 1.0–3.1). Both B. abortus and B. melitensis DNA were detected in humans and in multiple livestock host species, suggesting cross-transmission of these species among the different hosts. The detection of these two zoonotic Brucella species in humans further underpins the importance of One Health prevention strategies that target multiple host species, especially in the multi-host livestock populations.
Subject terms: Policy and public health in microbiology, Bacterial infection
Introduction
The genus Brucella consists of several species of gram negative, facultative bacteria, causing brucellosis in humans, livestock, and wildlife hosts worldwide1. Brucella abortus, B. melitensis and B. suis are the most reported zoonotic species of Brucella2. Brucellosis causes reproductive disorders such as abortion, infertility, reduced milk yield and retained placenta in livestock, while humans suffer from a long and debilitating illness that is characterized by undulating fever3. Brucellosis in humans is largely due to transmission from infected animals through consumption of raw contaminated livestock products, particularly milk or through contact with secretions from infected animals, especially during parturition4. Brucella bacteria are highly infectious, thus posing an occupational hazard to persons involved in handling products from infected animals, including laboratory personnel, veterinarians, slaughter personnel and farmers who assist animals when giving birth1.
Brucellosis remains endemic or is a re-emerging neglected zoonosis in many parts of the world, especially Africa and Asia5. Brucellosis is emerging as a growing problem in intensive, peri-urban, small holder dairy production systems6, 7. Brucellosis has been successfully controlled in several developed countries using host species-specific livestock vaccines or test and slaughter policies in animals with no history of vaccination8. In Kenya, seroprevalences ranging from 0.1 to 46.5% in humans9–11 and 1.0–38.0% in livestock have been reported, with higher prevalence recorded in pastoral areas9, 12, 13.
While several serological studies conducted in animals and humans in many parts of sub-Saharan Africa revealed that Brucella antibodies are widespread, serology tests are unable to indicate which Brucella species were responsible for inducing antibodies in the host14, 15. A modelling framework that integrated serological data sets from northern Tanzania to determine the source of human infection, hypothesized that sheep and goats are the main source of human infection16. In Kenya, a strong association between human and animal Brucella seropositivity has also been reported9. However, information on the Brucella species circulating in the different hosts remains limited. This knowledge is key to understanding the epidemiology of brucellosis and could form the basis for the development of targeted control programs. Therefore, this study was conducted to identify the species of Brucella circulating in livestock and humans in the mixed livestock-human ecosystems in Narok and Marsabit in Kenya.
Methods
Study area
The study was conducted in pastoral areas of Narok and Marsabit counties in Kenya (Fig. 1). The two sites were purposefully selected due to high numbers of livestock kept in close contact with humans under a nomadic pastoralism system. Marsabit County has the highest reported Brucella spp. seroprevalence in humans and camels in Kenya8, 9, 13, while several serological studies have also demonstrated the exposure to Brucella spp. in Narok County12, 13, 17.
Figure 1.
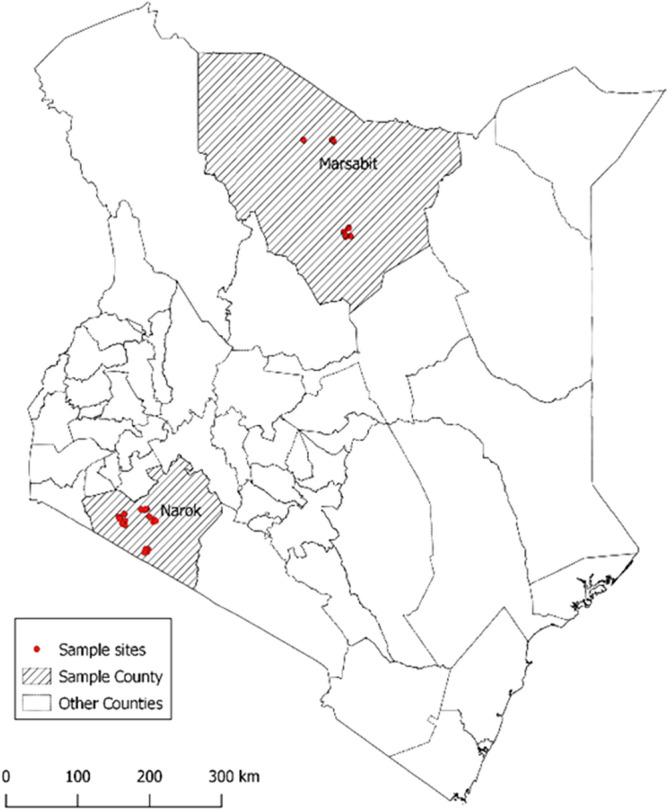
Map of Kenya showing the location of the study areas in Narok and Marsabit. The map was drawn by Fred Otieno using QGIS software, version 3.4.14–1 (http://www.gisagmaps.com/qgis-download/)19.
Narok County is located on the southern border of Kenya and Tanzania with an altitude of 1700–3000 m above sea level. It is a predominantly pastoral area with cattle, sheep and goats being kept in proximity with wildlife; Marsabit is the largest, most arid, and sparsely populated County in Kenya. Marsabit mostly comprises a vast lowland scrub desert ranging from 400 to 700 m above the sea level. Nomadic pastoralism that involves mixed herds of camels, cattle, sheep and goats is the main economic activity in Marsabit, practiced by up to 80% of the occupants18.
Sample size determination
The sample size was calculated using the formula for detecting presence or absence of a disease20. Prevalence estimates from a previous study were assumed as 15.3% for humans, 3.3% for cattle, 3.6% for small ruminants (sheep and goats) in Narok and 46.5% for humans, 11.2% cattle, 16.1% small ruminants and 11.1% for camels in Marsabit9. The sample size calculated for each host was then adjusted to cater for potential clustering of Brucella species at the herd level using inter-cluster correlation coefficient (icc) of 0.4 and a sample size of 2.5 (1–5 animals) collected from each animal species at the herd level, which then translated into a design effect of 1.6. Therefore, a minimum of 228, 160, 40, 183 samples, from humans, small ruminants (sheep and goats), camels, and cattle, was estimated to be the total samples needed from the two sites.
Study design and sampling strategy for animals
Data were collected through a cross-sectional study that targeted herds with current or previous clinical signs suggestive of brucellosis. The targeted study design was adopted to increase chances of getting animals with circulating Brucella DNA for subsequent species identification.
Animal health providers and public health officers working in Narok and Marsabit were consulted to identify four locations from each of the two sites with reported cases of illness in animals and or people with clinical signs suggestive of brucellosis. In each location, animal herds with suspected brucellosis or reported history of brucellosis within the past two years were identified and visited. A maximum of five adult animals were randomly selected for sampling from each species within the herd when present (cattle, sheep, goats, and camels). Blood was collected via jugular venipuncture into a plain 10 ml vacutainer tube. The samples were then labeled and transported in a cool box and ice packs (~ 4 °C) to the field laboratory, where they were centrifuged on the same day of collection at 5000 rpm for five minutes to obtain serum. The serum samples were transported at 4 °C and stored at − 20 °C until testing at the Biosciences East and Central Africa (BecA) laboratories, hosted at the International Livestock Research Institute (ILRI), Nairobi, Kenya.
Sampling strategy for humans
The health care facilities visited by people living in and around the sampled livestock populations were identified. Three dispensaries in Narok (Oloolaimutia, Talek and Aitong) were visited and included in the study. The Marsabit County hospital, serving the entire county and one dispensary in North Horr Sub-County were included in the Marsabit study site. Patients referred to the laboratory at each facility for brucellosis testing, based on clinical suspicion, were approached, and consented for inclusion in this study. Venous blood was drawn from participants into a 6 ml plain vacutainer tube by a laboratory technologist working in the study health facilities, upon receiving written informed consent. The blood samples were kept upright at room temperature for not less than 10 min to facilitate clotting, then centrifuged at 5000 rpm for five minutes to obtain serum for routine serological testing of brucellosis at the hospital. A volume of approximately 1 ml extracted serum from each human subject was aliquoted into cryotubes for this study. All human sera were also transported at 4 °C and stored at − 20 °C until further molecular and serological testing at the BecA laboratory facility at ILRI, Nairobi, Kenya.
Questionnaire administration
A brief structured questionnaire was administered to the head of each household visited for animal sampling to capture information on sex, location, history of abortion and retained placenta in livestock. For human sampling, information on the age, gender, and location of residence of each sampled human subject was recorded.
Sample selection and testing rationale
This study is focused on the molecular epidemiology of Brucella. Therefore, the lab procedure begun with DNA extraction from all the samples for PCR testing to detect the genus Brucella and to identify Brucella spp. Samples with sufficient quantities were later selected for RBT testing to assess if RBT positive status could be used as a predictor for PCR positivity.
DNA extraction and purification
Genomic DNA was extracted from all serum samples using QIAamp DNA mini kit, (QIAGEN Germany) as per manufacturer’s guidelines. Briefly, 20 µl of proteinase K was added to 200 µl serum, mixed with 200 µl of lysis buffer and left to digest at room temperature for 2 h. The lysate was then loaded into the spin columns (provided with the kit), and manufacturers guidelines followed, before eluting the genomic DNA in 50 µl of elution buffer. The DNA quality and quantity for each of the extracts derived from the original sera were assessed using a NanoDrop 2000c Spectrophotometer (ThermoFisher Scientific, USA) before being stored at − 20 °C until PCR was done.
Real-time PCR
The PCR assays were first optimized to our local environmental conditions with reference to published standards (supplementary material S1). Each DNA extract was tested using primers targeting the Brucella-specific insertion sequence IS711 to detect the genus Brucella as previously published21. A second assay was also run independently on all the samples with the bcsp31target, as adopted from a previous study to detect Brucella genus22. See supplementary table S2 for primer and probe details. Both quantitative PCR assays were performed using an ABI 7500 thermocycler machine (Applied Biosystems, Life Technologies, Singapore). All reaction mixtures (20 µl) were prepared by mixing 4 µl of DNA template with 0.25 µM of fluorescent probe, 0.5 µM of each of the primers and 10 µl of the Luna Universal Probe qPCR master mix (404 with UDG; New England BioLabs, MA, USA). All test runs were performed with the following PCR conditions; pretreatment with UDG 50 °C for 2 min, then polymerase activation and DNA denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s of amplification step, then 1 min of annealing at 57 °C. The positive controls (DNA extracts from Brucella strains, B. melitensis 16M and B. abortus 544) used in this study were both sourced from the Friedrich-Loeffler-Institute Brucella Reference Laboratory in Germany). Positive controls, no template control and samples were all loaded in duplicates before sealing the plate in every run. A run was only considered valid if all negative/no-template controls did not amplify, and positive controls amplified within agreeable range of the standard curve equivalent. A test run was classified as valid when amplification was observed for positive controls and no amplification was observed for the negative controls.
All samples that showed amplification and a cycle threshold (Ct) value < 40 in one or both duplicate wells were considered assay positive. Samples positive in both the IS711 and bcsp31 assays were classified as Brucella genus PCR positive. Samples classified as Brucella spp. positive were then subjected to a multiplex speciation assay22 with oligonucleotide primers and probes detecting specific IS711 insertions downstream of the alkB gene for B. abortus and downstream of the BMEI1162 locus for B. melitensis. Speciation assays were run using identical reaction volumes and conditions as described above on an ABI 7500 thermocycler. All samples, no template control, B. abortus and B. melitensis positive controls were tested in duplicates in all the runs. Any sample with amplification and a cycle threshold (Ct) value < 40 with the respective target was classified as positive for B. abortus or B. melitensis.
Rose Bengal Test (RBT)
Animal and human serum samples were tested for antibodies against Brucella spp. using the RBT. The test was conducted, with the rapid slide agglutination suspension of B. abortus biovar 1 Weybridge strain No. 99 antigen (RSA-RB, IDVet, France). Briefly, the antigen and serum samples were brought to room temperature in a biosafety cabinet and vortexed. Thereafter, 25 µl of each serum sample (for cattle and camels) or 30 µl (for goat, sheep, and human sera) were mixed with an equal volume of antigen on the glossy side of a white tile using a clean, wooden splint. The tile was then gently rocked at room temperature for four minutes before observing agglutination under natural light. Any sample that had visible agglutination after four minutes was considered as RBT positive23.
Data management and analysis
All the data were merged and cleaned in MS Excel version 2018 (IBM, California). Further analyses were performed using R statistical software version 3.6.324. Descriptive analyses were done using the aggregated data from both sites. Analyses of associations between Brucella spp. PCR test status in animal and human populations were performed using mixed effects logistic regression models created with the function glmer in the package lme425 with sampling location included as a random effect, and the data being specified as having binomial distribution. Variables evaluated in the human model to assess association with PCR status included age category, gender and RBT results. For livestock, variable history of abortion, RBT status, animal spp. and history of retained placenta were included in the model as potential predictors for Brucella PCR status. In this model male ruminants and camels were excluded due to absence of appropriate clinical history (abortion and retained placenta) and low numbers of observations of the assessed variables, respectively.
For analysis of B. abortus and B. melitensis outcomes data from humans and livestock were combined to evaluate the influence of host species on Brucella species detection. The percentage positive for each test by species and site was calculated and plotted, with binomial exact confidence intervals. Both models were fitted with sampling location included as a random effect, and the data being specified as having binomial distribution.
The maximal models were simplified using a likelihood ratio tests, with a p-value of ≤ 0.05 being considered statistically significant to get the final models. The intra-cluster correlation coefficients (icc) for within-location clustering of brucellosis for both livestock and humans were calculated from the variance components of the final multivariable models using the icc function in sjstats package26.
Ethics approval and consent to participate
The research approval for this study was granted by the National Commission for Science, Technology, and Innovation (Ref. no. NACOSTI/P/19/81438/29438). Additional approvals for both livestock and human components of the study were provided by ILRI’s Institutional Animal Care and Use Committee (Ref. no. ILRI-IACUC2018-16) and Research Ethics Committee (Ref. no. ILRI-IREC2018-14), respectively. ILRI is recognized by the National Commission for Science and Technology in Kenya (NACOSTI/NBC/AC/01813) and internationally by the Federation wide assurance (number FWA00026536) to review and approve research studies. The research was performed in accordance with the relevant guidelines and regulations prescribed by the above research regulatory committees. For human subjects, a written informed consent was sought and obtained from all the adult participants (18 years and above), while parent’s consent was obtained for participants below the age of 12 years and both the parent or guardian consent together with the subject’s assent obtained for those between 13–17 years of age. The livestock owners also provided informed consent before inclusion into the study.
Results
Livestock population summary, RBT and Brucella spp. PCR results
The demographic characteristics of the sampled livestock are summarized in Table 1. A total of 597 livestock including 228 cattle, 162 goats, 158 sheep were sampled from Narok and Marsabit Counties combined. Camels (n = 49) were sampled in Marsabit County only, as these animals were not kept in Narok County. Most samples were collected from female animals (92%) (Table 1). Both IS711 and bcsp31 targets detected Brucella spp. in our samples (supplementary table S3). The results of the RBT and the genus-specific Brucella spp. PCR are shown in Table 1. The overall RBT positivity in livestock was 23.3% in cattle; 8.3% in goats; 5.7% in sheep; and 9.3% in camels (Table 1). The overall PCR positivity of Brucella spp. detected by PCR in livestock was 18.9% in cattle; 38.3% in goats; 38% in sheep; and 69.4% in camels (Table 1). The proportion of animals with a history of retained placenta was 2% and those with previous abortion was 12% (Table 1).
Table 1.
Summary of livestock population composition, descriptive characteristics, RBT results and Brucella spp. PCR results.
| Variable | Category | Rose Bengal results | PCR results | ||||
|---|---|---|---|---|---|---|---|
| Total | No RBT positive | % RBT positive (95% CI) | Total | Number PCR positive | % Brucella spp. PCR positive (95% CI) | ||
| Species | Cattle | 228 | 55 | 24.1 (18.7–30.2) | 228 | 43 | 18.9 (14.0–24.6) |
| Goats | 158 | 38 | 8.2 (4.5–13.7) | 162 | 62 | 38.3 (30.8–46.2) | |
| Sheep | 138 | 8 | 5.4 (2.3–10.4) | 158 | 60 | 38.0 (30.4–46.0) | |
| Camels | 41 | 4 | 9.8 (2.7–23.1) | 49 | 34 | 69.4 (54.6–81.7) | |
| Sex | Male | 45 | 19 | 13.3 (5.1–26.8) | 45 | 19 | 42.2 (27.7–57.8) |
| Female | 530 | 74 | 14.0 (11.1–17.2) | 552 | 180 | 32.6 (28.7–36.7) | |
| Abortion | No | 508 | 63 | 12.4 (9.7–15.6) | 527 | 164 | 31.1 (21.2–35.3) |
| Yes | 67 | 17 | 25.4 (15.5–37.5) | 70 | 35 | 50.0 (37.8–62.2) | |
| Retained placenta | No | 568 | 78 | 16.0 (12.9–19.6) | 585 | 192 | 33.3 (29.5–37.3) |
| Yes | 10 | 2 | 25.0 (3.2–65.1) | 12 | 7 | 63.6 (30.8–89.0) | |
| Total | Total samples | 575 | 80 | 13.9 (11.1–17.0) | 597 | 199 | 33.3 (29.6–37.3) |
PCR polymerase chain reaction, CI confidence interval, RBT Rose Bengal Test.
Human population summary, RBT and Brucella spp. PCR results
A total of 257 humans were sampled (110 in Marsabit and 147 in Narok). Majority of samples were collected from female participants (57.6%). The age of human participants ranged from 3 to 96 years with the mean age of 32.6 years. Participant age was converted into a categorical variable with 3 levels for analysis: ≤ 20 years, 21–40 and > 40 years.
The overall PCR positivity of Brucella spp. in humans was 40.1% (95% CI 32.5–44.8), while the RBT positivity was 19.3% (95% CI 14.5–24.9, n = 238) (Table 2).
Table 2.
Summary of human population descriptive, RBT results and Brucella spp. PCR results among suspected brucellosis patients referred for testing at the sampled medical facilities.
| Variable | Category | Narok | Marsabit | Combined human data | ||||
|---|---|---|---|---|---|---|---|---|
| Total tested | Number PCR positive | Total tested | Number PCR positive | Total tested | Number PCR positive | % Brucella spp. PCR positive (95% CI) | ||
| Age category (years) | ≤ 20 | 39 | 13 | 18 | 2 | 58 | 16 | 27.6 (16.7–40.9) |
| 21–40 | 65 | 36 | 54 | 24 | 99 | 52 | 52.5 (42.2–62.7) | |
| > 40 | 23 | 8 | 10 | 3 | 54 | 17 | 31.5 (19.5–45.6) | |
| RBT result | Negative | 116 | 31 | 76 | 25 | 192 | 58 | 30.2 (26.2–34.4) |
| Positive | 48 | 12 | 15 | 7 | 46 | 20 | 43.5 (28.9–58.9) | |
| Gender | Male | 55 | 22 | 54 | 18 | 109 | 40 | 36.7 (27.7–46.4) |
| Female | 92 | 38 | 56 | 21 | 148 | 59 | 39.9 (31.9–48.2) | |
| Total (site) | Total samples | 147 | 60 | 110 | 39 | 257 | 99 | 38.5 (32.5–44.8) |
PCR polymerase chain reaction, CI confidence interval, RBT Rose Bengal Test.
Factors associated with Brucella spp. PCR status in humans
The final model fitted for human Brucella spp. PCR status only had age category as a significant variable positively associated with Brucella PCR status, LRT χ2 = 9.8 (Table 3). Individuals aged 21–40 were more likely to be PCR positive than individuals with an age category of less or equal to 20 years (Table 3). Sex and RBT status were not significantly associated with PCR status, therefore, we dropped the variables from the final model. The icc for within-location clustering of human brucellosis estimated for this model was < 0.001 (0.0–0.9).
Table 3.
Summary of the final mixed-effects logistic regression models run to assess associations between variables and Brucella spp. PCR status in human.
| Variables | Category (years) | Odds ratio (95% CI) | Odds ratio P-value |
LRT χ2 |
LRT p-value |
Df |
|---|---|---|---|---|---|---|
| Age category | ≤ 20 | 1 (baseline) | 9.81 | 0.03 | 2 | |
| 21–40 | 2.8 (1.2–6.6) | 0.016 | ||||
| > 40 | 1.2 (0.4–3.1) | 0.771 |
CI confidence interval, LRT χ2 Likelihood Ratio Test Chi-square value, P-value according to Pearson Chi-square test, Df degrees of freedom.
Factors predicting Brucella spp. PCR positivity in livestock
There was a positive and significant relationship between RBT status and Brucella PCR status, with RBT seropositive animals having elevated odds (OR = 8.3, 95% CI 4.0–17.1) of testing positive by PCR. Goats and sheep had higher odds (OR = 2.6, 95% CI 1.3–5.2) and (OR = 2.2, 95% CI 1.1–4.6) respectively of being PCR positive for Brucella spp. compared to cattle (Table 4).
Table 4.
Summary of multivariable mixed-effects logistic regression models run to predict Brucella spp. PCR status in livestock.
| Variables | Category (years) | Odds ratio (95% CI) | Odds ratio P-value |
LRT χ2 |
LRT p-value |
Df |
|---|---|---|---|---|---|---|
| Animal species | Cattle | 1 (baseline) | 7.99 | 0.018 | 2 | |
| Goats | 2.6 (1.3–5.2) | 0.005 | ||||
| Sheep | 2.2 (1.1–4.6) | 0.036 | ||||
| RBT | Positive | 8.3 (4.0–17.1) | < 0.001 | 35.98 | < 0.001 | 1 |
Location icc = 0.4.
CI confidence interval, P-value according to Pearson Chi-square test, Df degrees of freedom, RBT Rose Bengal Test, icc intra-cluster correlation coefficients.
Detection of Brucella abortus and Brucella melitensis
Of 298 Brucella spp. PCR positive livestock and human samples, 117 (39.3%) were positive for the B. abortus specific target, 111 (37.2%) were positive for the B. melitensis target and 68 (22.8%) did not amplify with either B. abortus or B. melitensis primer targets. No samples amplified with more than one target. The distribution of Brucella spp. differed in the two study sites, with B. abortus detected in most typed samples from Marsabit 50 (67.7.0%, 95% CI 54.8–77.1), whereas B. melitensis was detected in majority of typed samples from Narok 54 (65.9%, 95% CI 54.6–76.0).
Brucella abortus was detected in all species with the highest positivity in camels (64.7%), followed by cattle (60.5%), humans (44.4%), goats (24.2%), and sheep (20.0%). Sheep and goats had a significant reduced odds of association OR = 0.2, 95% CI 0.1–0.5 and OR = 0.2, 95% CI 0.0.6, with B. abortus compared to cattle, while the lower odds observed in camels and humans were not significant when compared with cattle (Table 5).
Table 5.
Summary of mixed-effects logistic regression models of Brucella abortus status in different host species.
| Level | Positivity for targeted Brucella spp. | Univariable mixed-effects logistic regression | |||||
|---|---|---|---|---|---|---|---|
| Host | n/N | % positivity and (95% CI) | Odds Ratio 95% CI |
Odds ratio P-value |
LRT χ2 |
LRT p-value |
LRT df |
| Cattle | 26/43 | 60.5 (44.4–75.0) | Baseline | 16.71 | 0.002 | 4 | |
| Goats | 15/62 | 24.2 (14.2–36.7) | 0.2 (0.1–0.6) | 0.003 | |||
| Sheep | 12/60 | 20.0 (10.8–32.3) | 0.2 (0.1–0.5) | 0.001 | |||
| Camel | 22/34 | 64.7 (46.4–80.3) | 0.9 (0.3–2.7) | 0.916 | |||
| Human | 44/99 | 44.4 (34.5–54.8) | 0.5 (0.2–1.0) | 0.055 | |||
Location icc = 0.10.
CI confidence interval, LRT χ2 Likelihood Ratio Test Chi-square value, P-value according to Pearson Chi-square test, n number of positives, N total number tested.
Brucella melitensis was found in the highest proportion in sheep (63.3%), followed by goats (50.0%), humans (29.3%), camel (22.2%), and cattle (16.3%). The mixed-effects model found that sheep had higher odds of being associated with B. melitensis OR = 1.8, 95% CI 0.8–3.8, while cattle, camels and humans had lower odds OR = 0.2, 95% CI 0.1–0.6, OR = 0.3, 95% CI 0.1–0.9, and OR = 0.5, 95% CI 0.3–1.2 compared to goats respectively (Table 6).
Table 6.
Summary of mixed-effects logistic regression models of Brucella melitensis status in different host species.
| Level | Positivity for targeted Brucella spp. | Univariable mixed-effects logistic regression | |||||
|---|---|---|---|---|---|---|---|
| Host | n/N | % positivity and (95% CI) | Odds Ratio 95% CI |
Odds ratio P-value |
LRT χ2 |
LRT p-value |
LRT df |
| Goats | 31/62 | 50.0 (37.0–63.0) | Baseline | 22.98 | < 0.001 | 4 | |
| Cattle | 7/43 | 16.3 (06.8–30.7) | 0.2 (0.1–0.6) | 0.003 | |||
| Sheep | 38/60 | 63.3 (49.9–75.4) | 1.8 (0.8–3.8) | 0.134 | |||
| Camel | 6/34 | 17.6 (06.8–34.5) | 0.3 (0.1–0.9) | 0.037 | |||
| Human | 29/99 | 29.3 (20.6–39.3) | 0.5 (0.3–1.2) | 0.177 | |||
Location icc = 0.05.
CI confidence interval, LRT χ2 Likelihood Ratio Test Chi-square value, P-value according to Pearson Chi-square test, n number of positives, N total number tested.
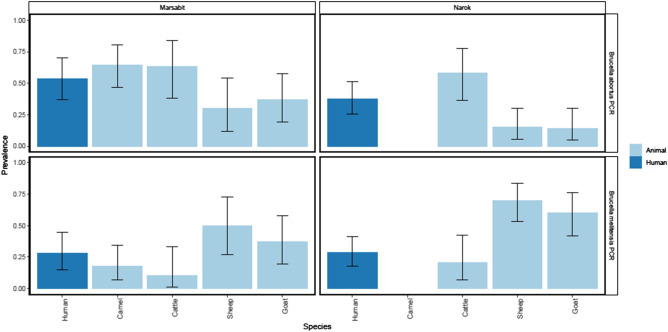
Analysis of Brucella spp. detected in different animal hosts in Marsabit and Narok gave a similar distribution trend that is comparable to those observed in the combined data (Fig. 2).
Figure 2.
Graphical presentation of the proportion of each host species testing positive for B. abortus and B. melitensis in Narok and Marsabit. The four panels show data for each site and Brucella species respectively. In each panel the bars indicate the proportion of each species testing positive. The error bars indicate exact binomial confidence intervals.
Discussion
This molecular study on the epidemiology of Brucella spp. in various host species has highlighted some key findings: we detected Brucella DNA in serum from humans and all the livestock species studied. This study also identified RBT positive status as a significant predictor for Brucella PCR positivity in livestock, while age category was associated with Brucella PCR positive status in humans. Finally, both B. abortus and B. melitensis were detected in humans and all the livestock species included in the study.
The detection of Brucella in four different animal hosts indicates a complex epidemiology. In this study, we have demonstrated that camels have a higher proportion of Brucella PCR positives than other species. This could partly be attributed to the high frequencies of migration of camel herds27 that enhance sharing of grazing areas and watering points or direct interactions between herds, thereby increasing their exposure to infectious pathogens such as Brucella. Detection of zoonotic species Brucella (B. abortus and B. melitensis) in camels could present a public health problem, given the steady rise in camel populations in Kenya28, primarily for milk production29–31. Consumption of raw milk9, and the significant increase in production of camel milk29, 32 and the extensive reach of camel milk value chains further highlight the need for more investigations and intervention programs to control the potential role of camels in transmission of brucellosis to humans, and other possible zoonotic diseases.
People within the age of 21–40 years were more likely to get positive Brucella PCR status. Earlier studies have also reported higher prevalence rate in humans within the similar age category33, 34. Based on observation made during field data collection, the high positivity in the age group 21–40 years could be attributed to their primary responsibility of herding, milking, and helping animals during parturition. Thus, having the highest exposure compared to the younger population (below 21 years) that are either school going or have less contact when taking care of animals. Those above 40 years of age tend to take more leadership roles as they reduce their active involvement in taking care of animals, hence reducing their risk of acquiring zoonotic infection through direct contact with the animals or contaminated animal products. This finding is comparable to earlier studies35 that reported high prevalence of brucellosis in the same age category and associated this to their occupational roles with livestock. The distribution of human cases across all age groups points to consumption of contaminated animal products as an alternative transmission route as observed by other studies within the region36, 37.
Our study found out that the odds of Brucella PCR positive status were eight times higher in animals with RBT positive results compared with those with negative results. This finding agrees with earlier recommendation that RBT is an appropriate test to be used for diagnosis of brucellosis and may be a useful screening test in low resource regions where PCR testing is constrained38. However, these two tests detect different markers of infection and even though complementary, they give overlapping but different information39. The low sensitivity of RBT observed in this study, and previous studies40–42 makes it necessary for it to be considered alongside other serological tests such as ELISA, slow Agglutination Tests (SAT) the Fluorescence Polarization Assay (FPA) and Complement Fixation Tests (CFT)43–45. This study however, exploited quantitative real-time PCR assays to detect Brucella DNA extracted from serum samples of the various hosts. Real-time PCR assays have been recommended as rapid and sensitive tests for detection of Brucella spp.21, 46. In this study, we tested DNA extracted from serum samples for Brucella positivity. Serum has been previously shown to be a good sample type for Brucella DNA detection in livestock and humans47. The exploitation of the same sample type for both serological testing (to advise Brucella exposure), and direct nucleic acid detection is a growing field of brucellosis diagnosis47–49. The added convenience of bypassing the need for tedious and dangerous culture procedures makes PCR a powerful brucellosis surveillance tool. Future studies should focus on generating more data to explore the agreement between serology and molecular approaches, and how each of the approaches informs about infection status in animals and humans.
Location was used as a random effect in our analysis to account for the potential effect of clustering of Brucella PCR positive cases in the models. The icc for location was estimated to be 0.4 for the livestock dataset. This is an indication that brucellosis infection in livestock tends to cluster within locations, which may be due to close or frequent interaction of herds within the same location. However, the human PCR positive cases were not clustered by locations (icc = 0). This may be attributed to the fact that humans may not only get exposed to Brucella infection within their residential location but could also acquire infection in other locations through consumption of contaminated animal products and contact with infected animals or their products. The broader distribution of milk and other dairy products in Kenya50 may also contribute to the lack of clustering of brucellosis cases in humans.
Both B. abortus and B. melitensis were detected in all livestock species. Our findings of both species of the pathogen across all hosts is consistent with earlier reports that Brucella spp. are not host specific and that cross transmission of Brucella spp. from one livestock host to the other could be occurring in areas with close interaction between different animal species14. Congregation of animals around communal watering points, keeping of mixed herds, and sharing of grazing sites have been reported to increase chances of brucellosis transmission10. Therefore, this could have contributed to the cross transmission of Brucella spp. observed in the two pastoral areas studied. We identified B. abortus most commonly in cattle and camels, while infection in sheep and goats were mainly associated with B. melitensis. This finding is consistent with existing knowledge on the preferential nature of Brucella spp. in the different animal hosts51.
Brucellosis infection in sheep and goats with B. melitensis is endemic in East Africa with earlier studies in Kenya in 197252 and 197653, in Tanzania 196754, and 199655 reporting presence of B. melitensis in the region. Earlier studies in East Africa also reported circulation of B. abortus in cattle in Kenya 200256, in Tanzania57, 58 and in Uganda59. This study is among the first reports of Brucella spp. circulating in camels in Kenya, although infection of camels with B. abortus has also been reported in Sudan60 and Pakistan61.
Our study found that B. abortus and B. melitensis are detectable in humans with clinical suspicion of brucellosis in Narok and Marsabit. Previous findings in the wider region, also reported the presence of B. abortus and B. melitensis in the human population62, 63. Human infection with Brucella spp. may be transmitted from multiple livestock species, given that B. abortus and B. melitensis detected in humans were also found in cattle, sheep, goats, and camels kept in the same locality. Brucellosis control programs aimed at reducing human brucellosis should therefore target all livestock hosts studied.
Abortions and retained placenta were rarely reported in our study, therefore our ability to assess their association with Brucella PCR status was limited by the low numbers of observations. The targeted study design limits the power of inference to population level prevalence but could have also contributed to the high PCR positivity reported in the study. The PCR speciation assay used only targeted B. abortus and melitensis. Therefore, 22.8% of the genus Brucella PCR positive samples that did not amplify with our two targets could not be identified, and other Brucella species circulating in the targeted population might have been missed. Future studies could focus on the known risk factors for positivity, more epidemiologically focused study designs and improved typing options using real-time PCR to address some of these prevailing challenges in the molecular epidemiology of brucellosis in the region.
Conclusion
This is among the first studies in the region to undertake a population level molecular study aimed at detecting circulating species of Brucella in several livestock hosts and humans. This study provided evidence of the presence of B. abortus and B. melitensis in multiple livestock species and humans in Kenya. Future studies should consider expanding the range of real-time PCR typing options to shed more light on all the Brucella species present in the targeted population. Our findings also confirmed that B. melitensis have a significant positive association with sheep and goats, while B. abortus is associated with cattle and camels. Cross transmission of different Brucella spp. between different animal hosts and the potential of human infection being caused by multiple livestock hosts was also highlighted. Brucellosis control programs in Kenya should use a One Health strategy targeting multiple host species.
Supplementary Information
Acknowledgements
We appreciate the support offered by the veterinary and medical departments in Narok and Marsabit Counties. We greatly acknowledge the contribution of the hospital staffs and patients attending the sampled health facilities, as well as pastoralists in our study sites. We are grateful to Prof. Heinrich Neubauer and Dr. Falk Melzer from Friedrich-Loeffler-Institute, for offering the controls for this study. We also appreciate the diagnostic training offered by Dr. Roland Ashford (from the Animal and Plant Health Agency (APHA), UK) that guided the development of aspects of laboratory protocols.
Author contributions
Concept development: J.A., C.O., E.S., G.S., R.K., B.B., Formal analysis: J.A., J.H., A.L., D.N., E.C., C.O., B.Bett, Funding acquisition: B.B., R.K., R.P., Investigation: J.A., C.O., R.P., D.O., BBett, E.C., Methodology: J.A., J.H., E.S., R.P., A.L., C.M., G.S., D.O., Supervision: C.O., G.S., E.S., R.P., E.F., Writing original draft: J.A., D.N., A.L., Writing review and editing: All Authors. All the authors have read and approved the manuscript.
Funding
This study was supported by the DELTAS Africa Initiative [Afrique One-ASPIRE/DEL-15-008] and the Africa Biosciences Challenge Fund (ABCF) fellowship. Afrique One-ASPIRE is funded by a consortium of donors including the African Academy of Sciences (AAS) Alliance for Accelerating Excellence in Science in Africa (AESA), the New Partnership for Africa’s Development Planning and Coordinating (NEPAD) Agency, the Wellcome Trust [107753/A/15/Z] and the UK government. And the ABCF program is funded by the Australian Department for Foreign Affairs and Trade (DFAT) through the BecA-CSIRO partnership; the Syngenta Foundation for Sustainable Agriculture (SFSA); the Bill & Melinda Gates Foundation (BMGF); the UK Department for International Development (DFID) and the Swedish International Development Cooperation Agency (Sida). J.A and B.Bett were also funded by the United States Defense Threat Reduction Agency (GRANT 12686246_R, to B.Bett). The funders had no role in the study design, data collection and analysis, writing or choice of the journal.
Data availability
All the data are included in this article and its supplementary files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88327-z.
References
- 1.D’Anastasio, R., Staniscia, T., Milia, M. L., Manzoli, L. & Capasso, L. Origin, evolution and paleoepidemiology of brucellosis. Epidemiol. Infect. [Internet]. 139(1), 149–156 (2011). http://www.ncbi.nlm.nih.gov/pubmed/20447329. [DOI] [PubMed]
- 2.Alton, L. Laboratory Techniques in Brucellosis, Vol. 91, 408–409 (Harefuah, 1976). [PubMed]
- 3.Megid J, Mathias LA, Robles CA. Clinical manifestations of brucellosis in domestic animals and humans. Open Vet. Sci. J. [Internet] 2010;4(1):119–126. doi: 10.2174/1874318801004010119. [DOI] [Google Scholar]
- 4.Goonaratna, C. Brucellosis in humans and animals. Ceylon Med. J. [Internet]. 52(2), 66 (2009). www.who.int/csr/resources/publications/Brucellosis.pdf.
- 5.Seleem MN, Boyle SM, Sriranganathan N. Brucellosis: A re-emerging zoonosis. Vet. Microbiol. 2010;140(3):392–398. doi: 10.1016/j.vetmic.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 6.Ducrotoy M, Ammary K, Lbacha HA, Zouagui Z, Mick V, Prevost L, et al. Narrative overview of animal and human brucellosis in Morocco: Intensification of livestock production as a driver for emergence ? Infect. Dis. Poverty [Internet] 2015 doi: 10.1186/s40249-015-0086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Njeru MF, et al. Human brucellosis in febrile patients seeking treatment at remote hospitals, Northeastern Kenya, 2014–2015. Emerg. Infect. Dis. 2016;2007(7):2014–2015. doi: 10.3201/eid2212.160285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDermott JJ, Arimi S. Brucellosis in sub-Saharan Africa: Epidemiology, control and impact. Vet. Microbiol. 2002;90(1):111–134. doi: 10.1016/S0378-1135(02)00249-3. [DOI] [PubMed] [Google Scholar]
- 9.Osoro EM, Munyua P, Omulo S, Ogola E, Ade F, Mbatha P, et al. Strong association between human and animal brucella seropositivity in a linked study in Kenya, 2012–2013. Am. J. Trop. Med. Hyg. 2015;93(2):224–231. doi: 10.4269/ajtmh.15-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kairu-Wanyoike S, Nyamwaya D, Wainaina M, Lindahl J, Ontiri E, Bukachi S, et al. Positive association between Brucella spp. Seroprevalences in livestock and humans from a cross-sectional study in Garissa and Tana River Counties, Kenya. PLoS Negl. Trop. Dis. [Internet] 2019;13(10):1–15. doi: 10.1371/journal.pntd.0007506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiambi SG, Fèvre EM, Omolo J, Oundo J, de Glanville WA. Risk factors for acute human brucellosis in Ijara, north-eastern Kenya. PLoS Negl. Trop. Dis. 2020;14(4):e0008108. doi: 10.1371/journal.pntd.0008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nthiwa D, Alonso S, Odongo D, Kenya E, Bett B. Zoonotic pathogen seroprevalence in cattle in a wildlife–livestock interface, Kenya. Ecohealth [Internet] 2019;16(4):712–725. doi: 10.1007/s10393-019-01453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Njeru J, Wareth G, Melzer F, Henning K, Pletz MW, Heller R, et al. Systematic review of brucellosis in Kenya: Disease frequency in humans and animals and risk factors for human infection. BMC Public Health [Internet] 2016;16(1):1–15. doi: 10.1186/s12889-016-3532-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godfroid J, Al Dahouk S, Pappas G, Roth F, Matope G, Muma J, et al. A “One Health” surveillance and control of brucellosis in developing countries: Moving away from improvisation. Comp. Immunol. Microbiol. Infect. Dis. [Internet] 2013;36(3):241–248. doi: 10.1016/j.cimid.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Godfroid J, Scholz HC, Barbier T, Nicolas C, Wattiau P, Fretin D, et al. Brucellosis at the animal/ecosystem/human interface at the beginning of the 21st century. Prev. Vet. Med. [Internet] 2011;102(2):118–131. doi: 10.1016/j.prevetmed.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Viana M, Shirima GM, John KS, Fitzpatrick J, Kazwala RR, Buza JJ, et al. Integrating serological and genetic data to quantify cross-species transmission: Brucellosis as a case study. Parasitology. 2016;143(7):821–834. doi: 10.1017/S0031182016000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey P, Doern C. Brucellosis in rural Narok County, Kenya: A retrospective review. Open Forum Infect. Dis. 2019;6:S614–S615. doi: 10.1093/ofid/ofz360.1543. [DOI] [Google Scholar]
- 18.Fratkin, E., Nathan, M. A. & Roth, E. A. Is settling good for pastoralists? The effects of pastoral sedentarization on children’s nutrition , growth , and health among Rendille and Ariaal of Marsabit District, Northern Kenya, 02111, 27–28 (2006). https://pdfs.semanticscholar.org/212f/08b904fbe600958ebf53369748354a28e5ae.pdf.
- 19.Vitalis, S., Arroyo Ohori, K. & Stoter, J. CityJSON in QGIS: Development of an open-source plugin. Trans GIS (2020). [DOI] [PMC free article] [PubMed]
- 20.Dohoo, I. R., Martin, S. W. & Stryhn, H. Methods in Epidemiologic Research (VER Inc., 2012). [Internet].
- 21.Matero P, Hemmilä H, Tomaso H, Piiparinen H, Rantakokko-Jalava K, Nuotio L, et al. Rapid field detection assays for Bacillusanthracis, Brucella spp., Francisellatularensis and Yersiniapestis. Clin. Microbiol. Infect. [Internet] 2011 doi: 10.1111/j.1469-0691.2010.03178.x. [DOI] [PubMed] [Google Scholar]
- 22.Probert W, Schrader Kimmi N, Khuong NY, Bystrom SL, Graves MH. Real-time multiplex PCR assay for detection of Brucella spp. J. Clin. Microbiol. 2004;42(3):1290–1293. doi: 10.1128/JCM.42.3.1290-1293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization. Brucellosis in humans and animals: WHO guidance [Internet]. WHO Library (World Health Organization, 2006). http://www.who.int/csr/resources/publications/deliberate/WHO_CDS_EPR_2006_7/en/.
- 24.R Core Team. An Introduction to dplR. Ind. Commer. Train [Internet] 10(1), 11–18 (2008). https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computing.
- 25.Bates D, Mächler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2014 doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 26.Brecht, A. et al.. Package ‘prevalence’ R [Internet] (2015). http://cran.r-project.org/web/packages/prevalence/prevalence.pdf.
- 27.O’Connor DA, Butt B, Foufopoulos JB. Mapping the ecological footprint of large livestock overlapping with wildlife in Kenyan pastoralist landscapes. Afr. J. Ecol. 2016;54:114–117. doi: 10.1111/aje.12241. [DOI] [Google Scholar]
- 28.FAO. Bovine brucellosis in Sub-Saharan Africa: Estimation of sero-prevalence and impact on meat and milk offtake potential (2002). ftp://ftp.fao.org/docrep/fao/009/ag274e/ag274e00.pdf.
- 29.Masinga, M., Kimenya, D. & Kivolonzi, P. The camel milk industry in Kenya: Results of a study commissioned by SNV to explore the potential of camel milk from Isiolo District (2008).
- 30.Watson EE, Kochore HH, Dabasso BH. Camels and climate resilience: Adaptation in northern Kenya. Hum. Ecol. Interdiscip. J. 2016;44:701–713. doi: 10.1007/s10745-016-9858-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muloi D, Alarcon P, Ombui J, Ngeiywa KJ, Abdullahi B, Muinde P, et al. Value chain analysis and sanitary risks of the camel milk system supplying Nairobi city, Kenya. Prev. Vet. Med. 2018;159:203–210. doi: 10.1016/j.prevetmed.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noor IM, Bebe BO, Guliye AY. Analysis of an emerging peri-urban camel production in Isiolo County, Northern Kenya. J. Camelid Sci. 2012;2012:41–61. [Google Scholar]
- 33.Alkahtani AM, Assiry MM, Chandramoorthy HC, Al-Hakami AM, Hamid ME. Sero-prevalence and risk factors of brucellosis among suspected febrile patients attending a referral hospital in southern Saudi Arabia (2014–2018) BMC Infect. Dis. 2020;20(1):1–8. doi: 10.1186/s12879-020-4763-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muloki HN, Erume J, Owiny DO, Kungu JM, Nakavuma J, Ogeng D, et al. Prevalence and risk factors for brucellosis in prolonged fever patients in post-conflict Northern Uganda. Afr. Health Sci. 2018;18(1):22–28. doi: 10.4314/ahs.v18i1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Assafi M, Al-berfkani MI. The seroprevalence of human brucellosis in different age groups patients and other associated risk factors in Duhok, Iraq. Innovaciencia. 2019;7:1–7. [Google Scholar]
- 36.Bodenham RF, Lukambagire AS, Ashford RT, Buza JJ, Rubach MP, Sakasaka P, et al. Prevalence and speciation of brucellosis in febrile patients from a pastoralist community of Tanzania. Sci. Rep. [Internet] 2020 doi: 10.1038/s41598-020-62849-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Migisha R, Nyehangane D, Boum Y, Page AL, Zúñiga-Ripa A, Conde-Álvarez R, et al. Prevalence and risk factors of brucellosis among febrile patients attending a community hospital in south western Uganda. Sci. Rep. 2018;8:15465. doi: 10.1038/s41598-018-33915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zakaria AM. Comparative assessment of sensitivity and specificity of rose bengal test and modified in-house ELISA by using IS711 TaqMan Real Time PCR assay as a gold standard for the diagnosis of bovine brucellosis. Biomed. Pharmacol. J. 2018;11(2):951–957. doi: 10.13005/bpj/1453. [DOI] [Google Scholar]
- 39.Al Dahouk S, Tomaso H, Nöckler K, Neubauer H, Frangoulidis D. Laboratory-based diagnosis of brucellosis—A review of the literature Part I: Techniques for direct detection and identification of Brucella spp. Clin. Lab. 2003;49:487–505. [PubMed] [Google Scholar]
- 40.Ahasan MS, Rahman MS, Rahman AKMA, Berkvens D. Bovine and Caprine Brucellosis in Bangladesh: Bayesian evaluation of four serological tests, true prevalence, and associated risk factors in household animals. Trop. Anim. Health Prod. 2017;49:1–11. doi: 10.1007/s11250-016-1151-1. [DOI] [PubMed] [Google Scholar]
- 41.Muma JB, Toft N, Oloya J, Lund A, Nielsen K, Samui K, et al. Evaluation of three serological tests for brucellosis in naturally infected cattle using latent class analysis. Vet. Microbiol. 2007;125:187–192. doi: 10.1016/j.vetmic.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 42.Sanogo M, Fretin D, Thys E, Saegerman C. Exploring the diversity of field strains of Brucellaabortus biovar 3 isolated in West Africa. Front. Microbiol. 2017;8:1–8. doi: 10.3389/fmicb.2017.01232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalleshamurthy T, Skariah S, Rathore Y, Ramanjinappa KD, Nagaraj C, Shome BR, et al. Comparative evaluation of fluorescence polarization assay and competitive ELISA for the diagnosis of bovine brucellosis vis-a-vis sero-monitoring. J. Microbiol. Methods. 2020;170:105858. doi: 10.1016/j.mimet.2020.105858. [DOI] [PubMed] [Google Scholar]
- 44.Chisi SL, Marageni Y, Naidoo P, Zulu G, Akol GW, van Heerden H. An evaluation of serological tests in the diagnosis of bovine brucellosis in naturally infected cattle in KwaZulu-Natal province in South Africa. J. S. Afr. Vet. Assoc. 2017 doi: 10.4102/jsava.v88i0.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Díaz-Aparicio, E. et al.. Evaluation of serological tests for diagnosis of Brucella melitensis infection of goats [Internet]. J. Clin. Microbiol. 32, 1159–1165 (1994). http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=263630&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed]
- 46.Al Dahouk S, Nöckler K, Scholz HC, Pfeffer M, Neubauer H, Tomaso H. Evaluation of genus-specific and species-specific real-time PCR assays for the identification of Brucella spp. Clin. Chem. Lab. Med. 2007;45:1464–1470. doi: 10.1515/CCLM.2007.305. [DOI] [PubMed] [Google Scholar]
- 47.Zerva L, Bourantas K, Mitka S, Kansouzidou A, Legakis NJ. Serum is the preferred clinical specimen for diagnosis of human brucellosis by PCR. J. Clin. Microbiol. 2001;39:1661–1664. doi: 10.1128/JCM.39.4.1661-1664.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akoko J, Pelle R, Kivali V, Schelling E, Shirima G, Machuka EM, et al. Serological and molecular evidence of Brucella species in the rapidly growing pig sector in Kenya. BMC Vet. Res. [Internet] 2020 doi: 10.1186/s12917-020-02346-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al Dahouk S, Tomaso H, Nöckler K, Neubauer H. The detection of Brucella spp. using PCR-ELISA and real-time PCR assays. Clin. Lab. 2004;50:387–394. [PubMed] [Google Scholar]
- 50.Kiambi S, Alarcon P, Rushton J, Murungi MK, Muinde P, Akoko J, et al. Mapping Nairobi’s dairy food system: An essential analysis for policy, industry and research. Agric Syst. 2018;167:47–60. doi: 10.1016/j.agsy.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.OIE. Brucellosis (B. abortus, B. melitensis and B. suis) (2016). https://www.oie.int/en/animal-health-in-the-world/animal-diseases/Brucellosis/.
- 52.Philpott M, Auko O. Caprine brucellosis in Kenya. Br. Vet. J. [Internet] 1972;128(12):642–651. doi: 10.1016/S0007-1935(17)36637-X. [DOI] [PubMed] [Google Scholar]
- 53.Oomen, L. J. Human brucellosis in Kenya. Trop. Geogr. Med. [Internet] 28(1), 45–33 (1976). http://europepmc.org/abstract/MED/941242. [PubMed]
- 54.Mahlau EA, Hamond JA. Brucellosis survey of the western areas of Tanganyika. Bull. Epizoot. Dis. Afr. 1962;10:511–512. [Google Scholar]
- 55.Tekle M, Legesse M, Edao BM, Ameni G, Mamo G. Isolation and identification of Brucellamelitensis using bacteriological and molecular tools from aborted goats in the Afar region of north-eastern Ethiopia. BMC Microbiol. 2019;19:1–6. doi: 10.1186/s12866-019-1474-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muendo EN, Mbatha PM, Macharia J, Abdoel TH, Janszen PV, Pastoor R, et al. Infection of cattle in Kenya with Brucellaabortus biovar 3 and Brucella melitensis biovar 1 genotypes. Trop. Anim. Health Prod. 2012;44(1):17–20. doi: 10.1007/s11250-011-9899-9. [DOI] [PubMed] [Google Scholar]
- 57.Mathew, C. et al.. First isolation, identification, phenotypic and genotypic characterization of Brucellaabortus biovar 3 from dairy cattle in Tanzania. BMC Vet. Res. [Internet]. 11, 156 (2015). http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4508816&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed]
- 58.Assenga JA, Matemba LE, Muller SK, Malakalinga JJ, Kazwala RR. Epidemiology of Brucella infection in the human, livestock and wildlife interface in the Katavi-Rukwa ecosystem, Tanzania. BMC Vet. Res. [Internet] 2015;11(1):189. doi: 10.1186/s12917-015-0504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mugizi, D. R. et al.. Isolation and molecular characterization of Brucella isolates in cattle milk in Uganda. Biomed. Res. Int. [Internet] 2015, 1–9 (2015). http://www.hindawi.com/journals/bmri/2015/720413/. [DOI] [PMC free article] [PubMed]
- 60.Damir HA, Tageldin MH, Kenyon SJ, Idris OF. Isolation of Brucella abortus from experimentally infected dromedary camels in Sudan: A preliminary report. Vet. Res. Commun. 1989;13:403–406. doi: 10.1007/BF00402559. [DOI] [PubMed] [Google Scholar]
- 61.Fatima S, Khan I, Nasir A, Younus M. Serological, molecular detection and potential risk factors associated with camel brucellosis in Pakistan. Trop. Anim. Health Prod. [Internet] 2016;48:1711–1718. doi: 10.1007/s11250-016-1148-9. [DOI] [PubMed] [Google Scholar]
- 62.Bodenham RF, Lukambagire AHS, Ashford RT, Buza JJ, Cash-Goldwasser S, Crump JA, et al. Prevalence and speciation of brucellosis in febrile patients from a pastoralist community of Tanzania. Sci. Rep. [Internet] 2020;10(1):1–11. doi: 10.1038/s41598-020-62849-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Njeru J, Melzer F, Wareth G, El-adawy H, Henning K, Pletz MW, et al. Human brucellosis in febrile patients. Emerg. Infect Dis. 2016;22(12):2014–2015. doi: 10.3201/eid2212.160285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data are included in this article and its supplementary files.