Abstract
Adult mammalian astrocytes are sensitive to inflammatory stimuli in the context of neuropathology or mechanical injury, thereby affecting functional outcomes of the central nervous system (CNS). In contrast, glial cells residing in the spinal cord of regenerative vertebrates exhibit a weak astroglial reaction similar to those of mammals in embryonic stages. Macrophage migration inhibitory factor (MIF) participates in multiple neurological disorders by activation of glial and immune cells. However, the mechanism of astrocytes from regenerative species, such as gecko astrocytes (gAS), in resistance to MIF-mediated inflammation in the severed cords remains unclear. Here, we compared neural stem cell markers among gAS, as well as adult (rAS) and embryonic (eAS) rat astrocytes. We observed that gAS retained an immature phenotype resembling rat eAS. Proinflammatory activation of gAS with gecko (gMIF) or rat (rMIF) recombinant protein was unable to induce the production of inflammatory cytokines, despite its interaction with membrane CD74 receptor. Using cross-species screening of inflammation-related mediators from models of gMIF- and rMIF-induced gAS and rAS, we identified Vav1 as a key regulator in suppressing the inflammatory activation of gAS. The gAS with Vav1 deficiency displayed significantly restored sensitivity to inflammatory stimuli. Meanwhile, gMIF acts to promote the migration of gAS through regulation of CXCL8 following cord lesion. Taken together, our results suggest that Vav1 contributes to the regulation of astrocyte-mediated inflammation, which might be beneficial for the therapeutic development of neurological diseases.
Keywords: MIF, astrocyte, neuroinflammation, immunosuppression, cell migration, cytokine, spinal cord, Vav1, reptile
Abbreviations: 4-IPP, 4-iodo-6-phenylpyrimidine; ANOVA, analysis of variance; DAMPs, damage-associated molecular pattern molecules; DMEM, Dulbecco's modified eagle's medium; eAS, embryonic rat astrocytes; ELISA, enzyme-linked immunosorbent assay; gAS, adult gecko astrocytes; GFAP, glial fibrillary acid protein; IPA, Ingenuity Pathway Analysis Software; IPTG, isopropyl-b-D-thiogalactopyranoside; MAPK, mitogen-activated protein kinase; MIF, macrophage migration inhibitory factor; NODs, oligomerization domain proteins; NSC, neural stem cell; PBS, phosphate-buffered saline; Q-PCR, quantitative real-time polymerase chain reaction; rAS, adult rat astrocytes; SCI, spinal cord injury; TLRs, Toll-like receptors
Astrocytes are the most abundant cell population in the central nervous system (CNS) that provide nutrients, recycle neurotransmitters, and maintain other structural and functional homeostasis (1). Insults to the CNS will result in reactivity of astrocytes, which undergo morphological and functional changes relevant to the clinical outcomes (2). Aside from glial scarring, astrocytes also act as immune-competent cells by sensing danger signals, secreting cytokines and chemokines, and subsequently activating innate and adaptive immune responses (3, 4, 5, 6). Therefore, astrocytes, together with microglia, have widely been shown to mediate CNS inflammation and neuropathogenesis. Interference of astrocyte-mediated inflammation has been shown to reduce infiltration of leukocytes into the CNS and ameliorate neurological disorders (7, 8, 9, 10). Constitutively, membrane of astrocyte expresses a profile of pattern-recognition receptors (PRRs) similar to that of microglia, such as the Toll-like receptors (TLRs), nucleotide-binding oligomerization domain proteins (NODs), and scavenger receptors (SRs) (2, 3, 11). In the context of neuropathology, ligation of the damage-associated molecular pattern molecules (DAMPs) to these receptors will drive a complex inflammatory network and effector events of astrocytes (2, 12).
Astrocytes, however, are essential for axonal growth, neuronal survival, and formation of synapse during the development of CNS (13). Inspired by these amazing features, fetal (immature) astrocytes are tested in therapeutic applications in the damaged CNS through transplantation. Indeed, the immature astrocytes are observed to benefit for neuroprotection and axonal growth, rather than deteriorate the lesion environment such as forming glial scar or triggering innate immunity (14, 15, 16, 17). The outcomes of transplantation are tightly associated with the embryonic stages of astrocyte acquisition. Transcriptome analysis of astrocytes isolated from developing and mature CNS identifies an entirely different molecular repertoire, which governs distinct morphology and function (16, 18, 19). However, the mechanism of the astroglial reaction weakening in immature astrocytes is yet to be elucidated.
Several vertebrates such as fish and some amphibians can successfully regenerate their spinal cord following injury. The CNS of these regenerative models is thought to lack “real” astrocytes. Instead, a population of glial cells builds a “glial bridge” (or tube in amphibian) favoring for the axonal elongation at the lesion sites (20, 21, 22). Comparatively, glial cells of the regenerative spinal cord can hardly form glial scar or activate excessive inflammation (23), which in turn decrease GFAP and elevate protein levels of neural stem cell (NSC)-related factors including fibroblast growth factor and Nestin after spinal cord injury (20, 24). These physiological features are very similar to those of mammalian immature astrocytes in the developing spinal cord. Given the immunosuppressive activity manifested by the glial cells or immature astrocytes, uncovering the underlying mechanism may provide a therapeutic clue for the inflammatory neuropathology mediated by the adult counterparts of mammals.
Macrophage migration inhibitory factor (MIF) is a potent proinflammatory cytokine that is expressed by a wide spectrum of cell types in multiple tissues (25). As a critical mediator of innate and adaptive immunity, MIF is implicated in the pathogenesis of many inflammatory and autoimmune diseases, such as sepsis, rheumatoid arthritis, colitis, asthma, pancreatitis, and cancer (25, 26). In the pathological CNS, MIF is highly induced within astrocytes, neurons, microglia, ependymal cells, and epithelial cells of the choroid plexus, thereby associated with exacerbation of neurological disorders or severed cord (27, 28, 29). MIF has been found to promote the production of inflammatory cytokines and chemokines from rodent astrocytes, in association with activation of excessive inflammation in the CNS (10, 30). Supposing that the glial cells or immature astrocytes are resistant to the activation of proinflammatory MIF, and the intrinsic mechanism might be conservative for other mediators, it will be extremely informative for controlling astrocyte-induced inflammation in the neurological disorder if the intricate regulatory mechanism has been delineated. In the present study, amniotic geckos were selected as an animal model for their ability in circumventing excessive inflammation and evoking less astrocytic responses in the severed spinal cord (31, 32). Transcriptome analysis of astrocytes from adult geckos (gAS), adult (rAS) and embryonic rats (E18, eAS) demonstrated that gAS expressed high abundance of NSC markers, a feature of immature astrocytes. Proinflammatory MIF was unable to induce the inflammatory responses of gAS, rather than rAS. Integration of inflammation-related signal cascades in MIF-stimulated gAS and rAS identified that Vav1 protein was a key regulator responsible for suppressing inflammation in the gAS, and a deficient expression of the protein could restore the cellular sensitivity to the inflammatory stimuli. Our results identified a novel player that mediated inflammation of astrocytes, which might be beneficial for the therapeutic development of neurological diseases.
Results
Adult gecko astrocytes display a profile of NSC markers similar to those of embryonic rat astrocytes
Though reptiles have not retained the same regenerative ability as shown in anamniotes following spinal cord injury, several lizards and turtles still exhibit anatomical and functional recovery of the lumbar and thoracic cord after complete transection (33, 34). To shed light on the physiological property of adult astrocytes isolated from gecko spinal cord, we screened the profile of NSC markers by transcriptome sequencing and made a comparison with those of eAS and rAS. The purity of the isolated astrocytes reached above 95% prior to sequencing, as determined by immunostaining of GFAP. The morphology of gAS appeared to be fibrous type with many long fiber-like processes, and the astrocytic responses to various stimuli were less sensitive than those of the mammals (32). A total of 18 NSC markers were examined on the basis of definition by Kuegler et al. (35). Results revealed that 13 out of 18 NSC marker genes were abundantly expressed in the gAS in comparison with those of rAS (RPKM >2-fold). Meanwhile, their expression abundance was much closer between gAS and eAS (Table 1, Table S1). Several typical NSC markers such as Fabp7, Sox2, Pax3, and Pax6 were expressed in the gAS at a ratio of 87- to 60,587-fold comparing with those of the rAS. The molecular profile of gAS reflects an immature cell trait, suggesting a distinct physiological function between the gAS and rAS.
Table 1.
Comparison of neural stem cell markers in gAS, rAS, and eAS
| Gene | gAS | rAS | eAS | gAS/rAS | gAS/eAS |
|---|---|---|---|---|---|
| Fabp7 | 60.588 | 0.001 | 127.600 | 60587.571 | 0.475 |
| Sox2 | 13.354 | 0.027 | 12.749 | 493.570 | 1.047 |
| Pax3 | 0.211 | 0.001 | 0.337 | 210.761 | 0.626 |
| Sox11 | 5.967 | 0.031 | 1.803 | 193.311 | 3.310 |
| Cdh2 | 90.030 | 0.643 | 29.294 | 140.028 | 3.073 |
| Cxcr4 | 4.826 | 0.036 | 6.445 | 132.528 | 0.749 |
| Sema5b | 1.617 | 0.017 | 1.903 | 94.986 | 0.850 |
| Pax6 | 2.496 | 0.029 | 1.558 | 87.025 | 1.602 |
| Rtn1 | 1.621 | 0.162 | 5.987 | 10.033 | 0.271 |
| Chrdl1 | 3.269 | 0.387 | 1.189 | 8.446 | 2.750 |
| Prkcz | 1.132 | 0.165 | 0.502 | 6.849 | 2.253 |
| Notch1 | 4.662 | 1.370 | 4.437 | 3.403 | 1.051 |
| Metrn | 61.767 | 21.460 | 80.300 | 2.878 | 0.769 |
| Dbx2 | 0.334 | 0.282 | 0.001 | 1.186 | 333.807 |
| Mbnl1 | 1.639 | 8.450 | 3.642 | 0.194 | 0.450 |
| Gata2 | 0.050 | 0.451 | 0.083 | 0.110 | 0.598 |
| Zic1 | 7.834 | 94.134 | 29.275 | 0.083 | 0.268 |
| Fgfr2 | 0.595 | 26.097 | 6.926 | 0.023 | 0.086 |
Astrocytes of gecko spinal cord express CD74 receptor of MIF
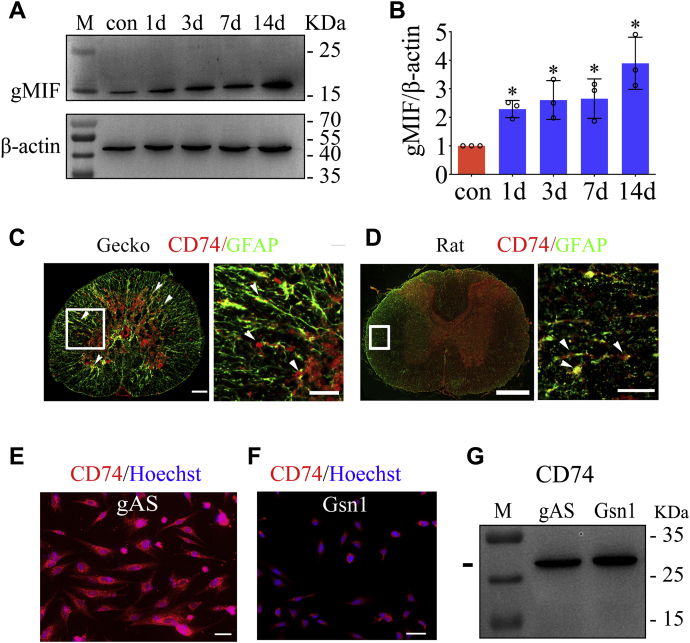
Proinflammatory MIF has been shown to be inducibly expressed at the severed cord to mediate neuropathology by binding with CD74 receptor (10, 29, 30). Injury of gecko spinal cord, however, can induce little production of inflammatory cytokines following tail amputation, though some DAMPs are actively emitted at the lesion sites (31, 36, 37). These indicate that DAMPs are inefficient in activating inflammation of gecko glial cells. Western blot showed that the protein level of gMIF temporally increased at the injured cord following gecko tail amputation (Fig. 1, A and B). Immunostaining demonstrated that the CD74 receptor of MIF was distributed in the GFAP-positive astrocytes of gecko spinal cord, a similar pattern to those of the rat (Fig. 1, C and D). Further examination on gecko primary astrocytes or astrocyte cell line Gsn1 recapitulated the receptor localization in the gAS (Fig. 1, E–G). The data indicate that elevation of gMIF protein levels in the injured cord can potentially mediate cell events of gecko astrocytes via binding to CD74 receptor.
Figure 1.
Temporal analysis of gMIF expression in the severed cord and localization of CD74 receptor in the gAS. A, western blot analysis of gMIF in the 0.5 cm cord segments at injured sites following tail amputation (n = 10) at 1 day, 3 days, 1 week, and 2 weeks, respectively. B, statistical analysis of (A). C, colocalization of CD74 receptor with GFAP-positive cells detected by immunohistochemistry in the spinal cord of gecko (C) and rat (D). E and F, analysis of CD74 receptor in the primary cultured astrocytes (E) and gecko astrocyte cell line Gsn1 (F). Arrowheads indicated the CD74-positive signals. G, western blots analysis of CD74 in the gAS and Gsn1. Error bars represent the standard deviation (p < 0.05). Scale bars, 100 μm in (C) and 50 μm in the magnification, 500 μm in (D) and 50 μm in the magnification, 50 μm in (E) and (F).
Gecko astrocytes are insensitive to proinflammatory stimulation of gMIF
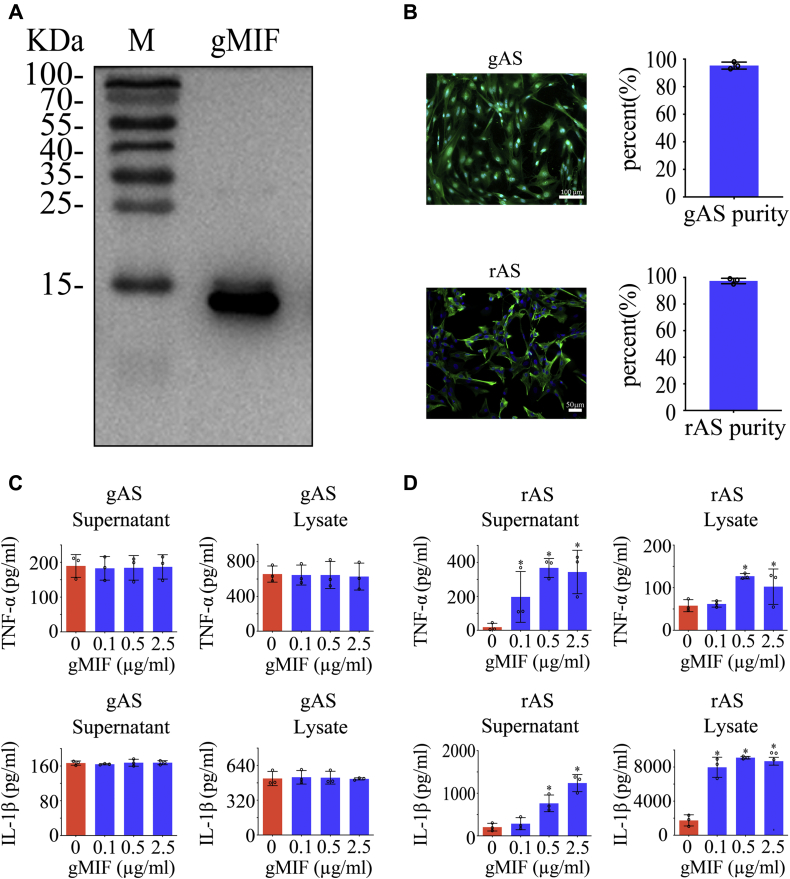
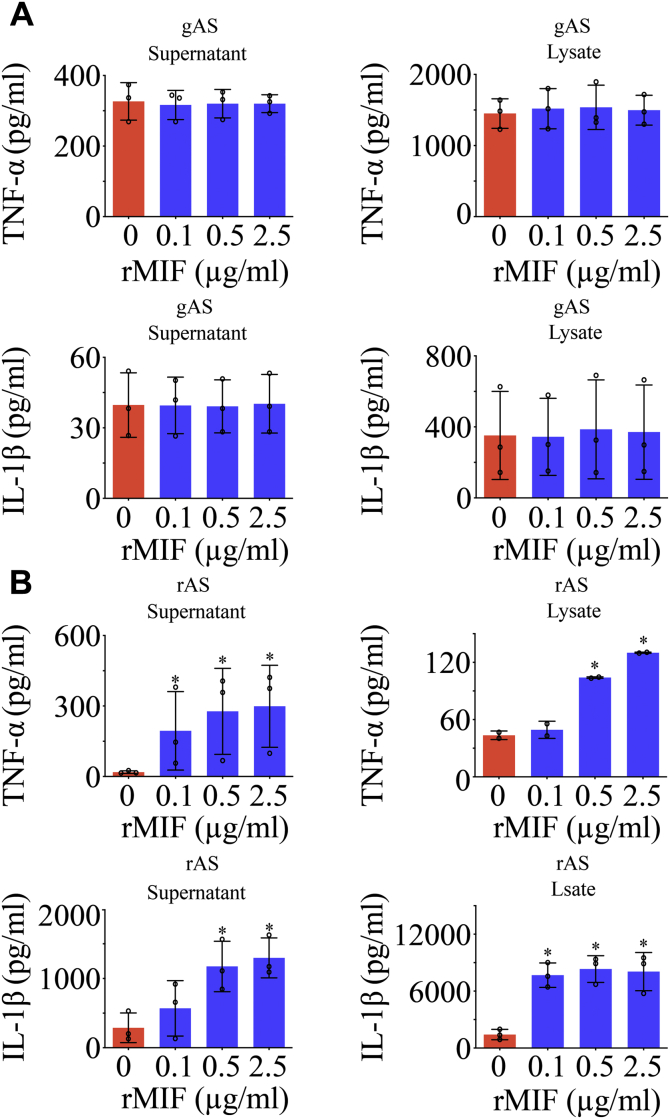
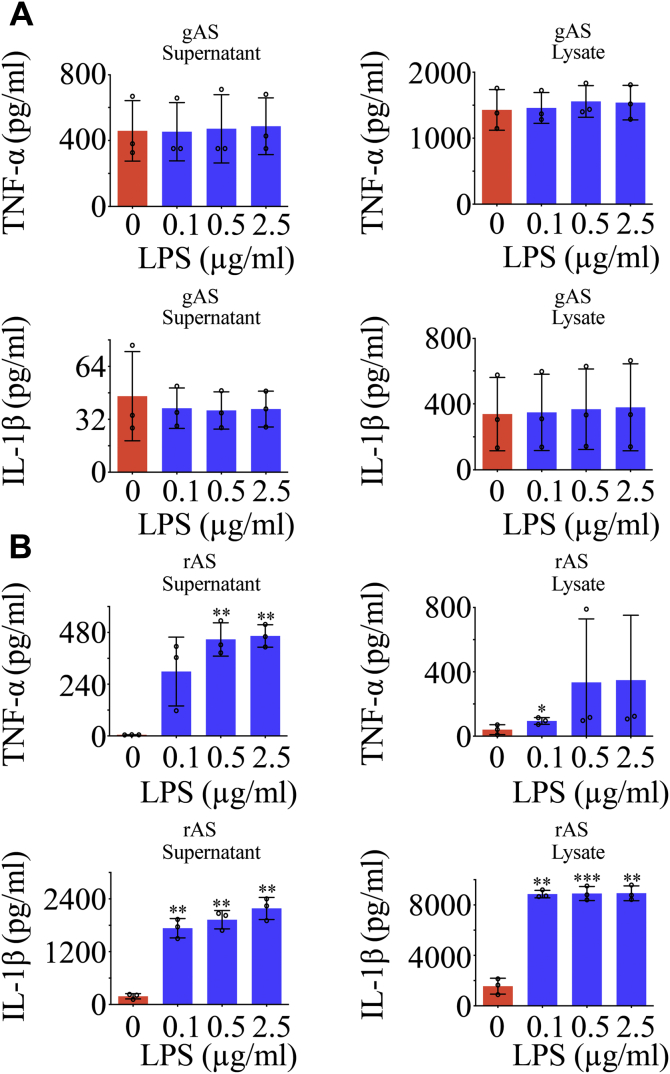
To clarify the potential effects of gMIF on the inflammatory activation of gAS, the recombinant gMIF protein was prepared and applied to stimulate the primary cultured gAS or rAS (purity > 92%) at concentration of 0 to 2.5 μg/ml (Fig. 2, A and B). ELISA assays demonstrated that gMIF at various concentration was unable to induce the production of TNF-α and IL-1β from gAS. On the contrary, gMIF significantly promoted the expression of TNF-α and IL-1β protein in rAS (Fig. 2, C and D). To validate that the insensitivity of gMIF in the activation of inflammation results from the unique property of gAS, recombinant rMIF protein was employed to stimulate gAS or rAS at the same concentration as gMIF. Similarly, rMIF was sufficient in inducing the production of TNF-α and IL-1β in rAS, rather than in gAS (Fig. 3, A and B). Even a substitution with the robust proinflammatory cytokine LPS also failed in activating gAS inflammation (Fig. 4, A and B). The results indicate that gAS are insensitive to the proinflammatory stimulation though they express the receptors of DAMPs.
Figure 2.
Analysis of gMIF proinflammatory effects on the gAS and rAS.A, western blot analysis of prepared gMIF recombinant protein using anti-His tag antibody. M, marker. B, primary cultured gAS and rAS stained with GFAP and Hoechst 33342 with purity over 92%. C and D, ELISA assay of TNF-α and IL-1β production in gAS (C) and rAS (D) stimulated with gradient gMIF for 24 h. All experiments were carried out in triplicate. Error bars represent the standard deviation (p < 0.05). Scale bars, 100 μm in gAS; 50 μm in rAS.
Figure 3.
Analysis of rMIF proinflammatory effects on the gAS and rAS. ELISA assay of TNF-α and IL-1β production in gAS (A) and rAS (B) stimulated with gradient rMIF for 24 h. All experiments were carried out in triplicate. Error bars represent the standard deviation (p < 0.05).
Figure 4.
Analysis of LPS effects on the gAS and rAS. ELISA assay of TNF-α and IL-1β production in gAS (A) and rAS (B) stimulated with gradient LPS for 24 h. All experiments were carried out in triplicate. Error bars represent the standard deviation (p < 0.05).
Interaction of gMIF with CD74 receptor is unable to activate intracellular inflammation-related signaling
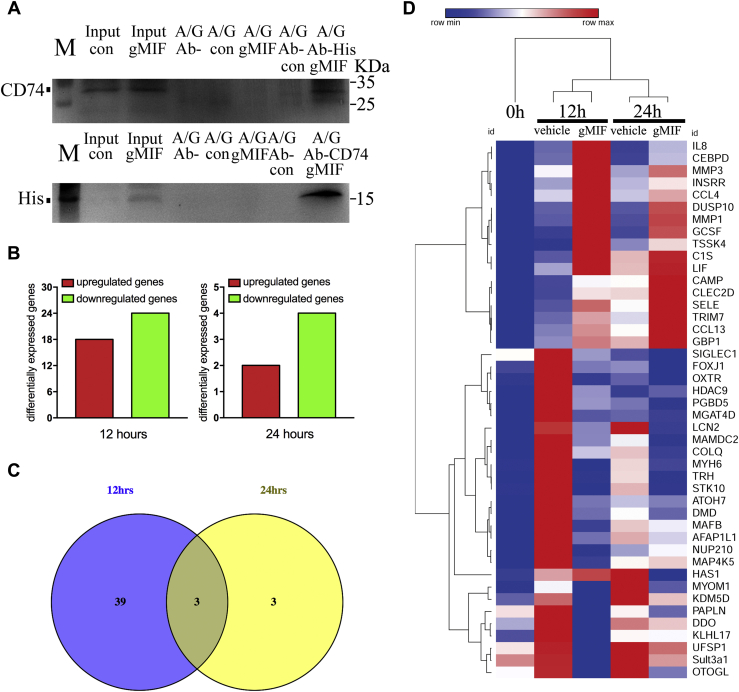
To exclude the possibility of gMIF failure in binding to the CD74 receptor, immunoprecipitation assays were performed using anti-His or -CD74 antibody to examine the potential interaction of gMIF with CD74 receptor. Addition of gMIF to the gAS culture resulted in the coprecipitation with CD74, as detected by anti-His or -CD74 antibody (Fig. 5A). The result suggests that gMIF/CD74 regulatory axis remains active in the gAS.
Figure 5.
Transcriptome sequencing of gAS following stimulation with or without 2.5 μg/ml gMIF for 12 h and 24 h.A, binding assay of gMIF with CD74 receptor in the gAS using anti-His or -CD74 antibody immunoprecipitation. A/G, protein A plus G-Sepharose beads; Ab-His, anti-His antibody; Ab-CD74, anti-mouse CD74 antibody. B, the bar graph showing the number of upregulated (red) genes and downregulated genes in the gAS following stimulation with 2.5 μg/ml gMIF for 12 h and 24 h. C, integration of DEGs following 2.5 μg/ml gMIF treatment of gAS at 12 h and 24 h, respectively. D, heatmap of integrated DEGs in response to stimulation of gMIF. The color scale shown at the top illustrates the relative expression level of the indicated mRNA across all samples: red denotes expression >0 and blue denotes expression <0.
Transcriptome sequencing was then carried out to unveil gMIF-mediated intracellular signal cascades of gAS, among which the clues in association with modulating inflammation might be deposited. The gAS were incubated in the culture medium for 12 h and 24 h in the presence or absence of 2.5 μg/ml gMIF. A total of 42 and 6 differentially expressed genes (DEGs) were found to be dynamically regulated by gMIF at 12 h and 24 h, respectively (Fig. 5, B–D, Table S2). However, GO analysis for biological process demonstrated that these 45 DEGs were irrelevant to regulating inflammation of gAS (data not shown). The data indicate that gMIF/CD74 regulatory axis is unable to activate inflammation of gAS.
Vav1 is identified as a key mediator in suppressing gMIF-induced inflammation of gAS
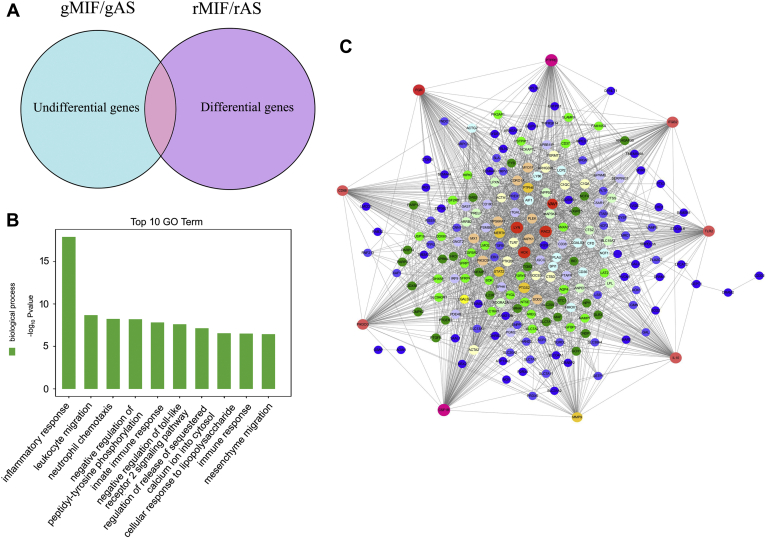
To uncover the underlying mechanism of MIF-mediated distinct inflammatory effects between gAS and rAS, a cross-species screening for the key mediator(s) in suppressing gMIF-induced inflammation in gAS was then performed. Genes with unaltered expression in gAS after gMIF stimulation were integrated to the DEGs of rAS challenged by rMIF (Table S3) (10), followed by GO annotation of the intersected inflammation-related genes (Fig. 6, A and B). A total of 234 genes were thus obtained from the integration (Table S4), and GO analysis demonstrated that inflammatory response was among top ten enriched GO terms under biological process (Fig. 6B). A gene regulatory network was further constructed using the Ingenuity Pathway Analysis (IPA) Software on the basis of these inflammation-related genes to identify the key mediator(s), highlighting that Vav1, Rac2, and C1QC potentially regulated the differential inflammatory responses between gAS and rAS (Fig. 6C).
Figure 6.
Cross-species screening for the key mediators in suppressing gMIF-induced inflammation in gAS.A, integration of the unaltered genes in gAS following 2.5 μg/ml gMIF stimulation for 12 h and 24 h and the DEGs in rAS stimulated with 2.0 μg/ml rMIF for 12 h and 24 h, respectively. B, GO annotation of the integrated inflammation-related genes. C, a reconstructed gene network was created using IPA on the basis of the integrated genes involved in inflammatory responses.
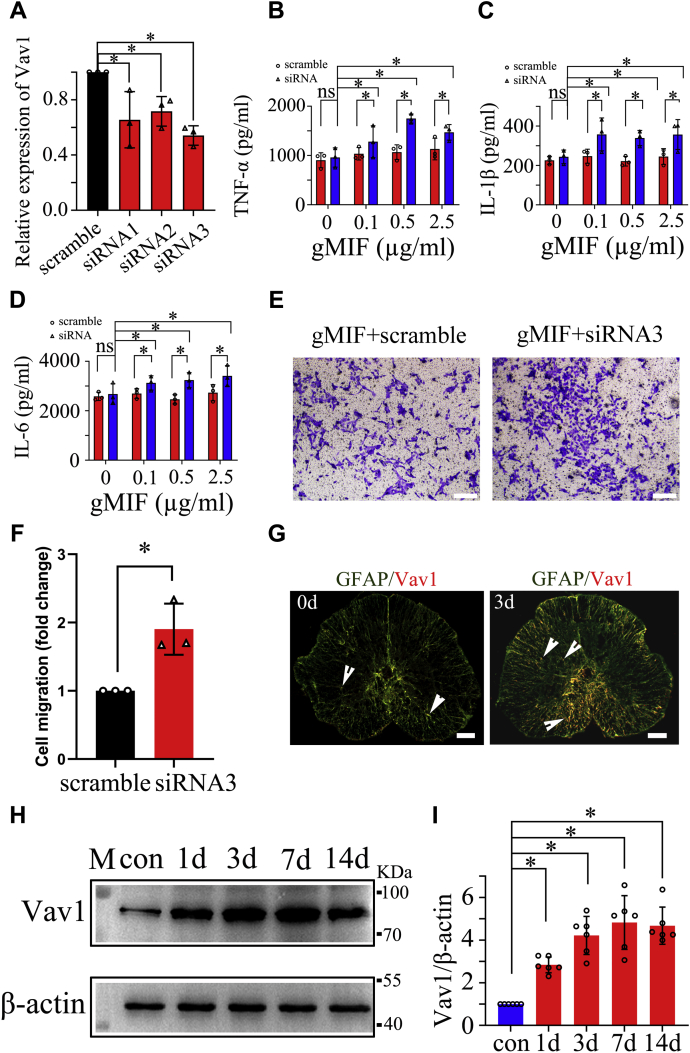
To validate the deduced function based on bioinformatics, siRNAs of Vav1, Rac2, and C1QC were synthesized and transfected to gAS respectively, for observation of the effects on the production of inflammatory cytokines. Results demonstrated that knockdown of the Vav1, rather than Rac2 and C1QC, was efficient in promoting proinflammatory stimuli-mediated expression of TNF-α, IL-1β, and IL-6 in gAS (Fig. 7, A–D, Fig. S1). Vav1 deficiency in gAS also increased the sensitivity to inflammatory stimuli of rMIF or LPS (Fig. S2), indicating that Vav1 is implicated in the inflammatory inactivation of gAS.
Figure 7.
Effects of Vav1 on suppression of gMIF-mediated inflammation in gAS. A, interference efficiency of three siRNA oligonucleotides for gecko Vav1 was measured by RT-PCR, and siRNA3 was used for the knockdown experiments. B–D, ELISA assay of TNF-α (B), IL-1β (C) and IL-6 (D) production in the gAS following siRNA3 knockdown for 24 h, then treatment with different concentration of gMIF for 24 h. E, transwell determination of gAS migration following siRNA3 knockdown for 24 h, followed by stimulation with 2.5 μg/ml gMIF for 24 h. F, statistical analysis of (E). G, colocalization of Vav1 with GFAP-positive astrocytes in the spinal cord following gecko tail amputation at 0 day and 3 days, respectively. H, western blot analysis of Vav1 in the 0.5 cm cord segments at injured sites following tail amputation (n = 10) at 1 day, 3 days, 1 week, and 2 weeks, respectively. I, statistical analysis of (H). Error bars represent the standard deviation (p < 0.05). Scale bars, 100 μm in (E) and (G).
Vav1 has been shown to mediate cell migration in a cell-type-specific manner. For example, Vav1−/− dendritic cells (DCs) increase rates of migration, while an active form of Vav1 induces the migration of mammary epithelial cells (38, 39). To examine whether Vav1 also affects the migration of gAS while blocking inflammation, Vav1 was knocked down for 24 h, followed by migration for another 24 h in the presence of 2.5 μg/ml gMIF. Transwell assays revealed that knockdown of Vav1 remarkably promoted the migration of gAS in comparison with the scramble (Fig. 7, E and F). These data indicate that Vav1 negatively regulates the migration of gAS in addition to suppressing inflammatory activation of these cells.
We next sought to address the role of Vav1 in the regenerating spinal cord of gecko. The expression changes of Vav1 were examined by immunochemistry following gecko tail amputation at 0 day, 1 day, 3 days, 7 days, and 14 days, respectively. Results displayed that the protein levels of Vav1 significantly increased at the lesion sites of spinal cords, with a colocalization with GFAP-positive astrocytes (Fig. 7, G–I). The data demonstrate that Vav1 is a key mediator in suppressing gMIF-induced inflammation of gAS.
gMIF promotes the migration of gAS through regulation of CXCL8
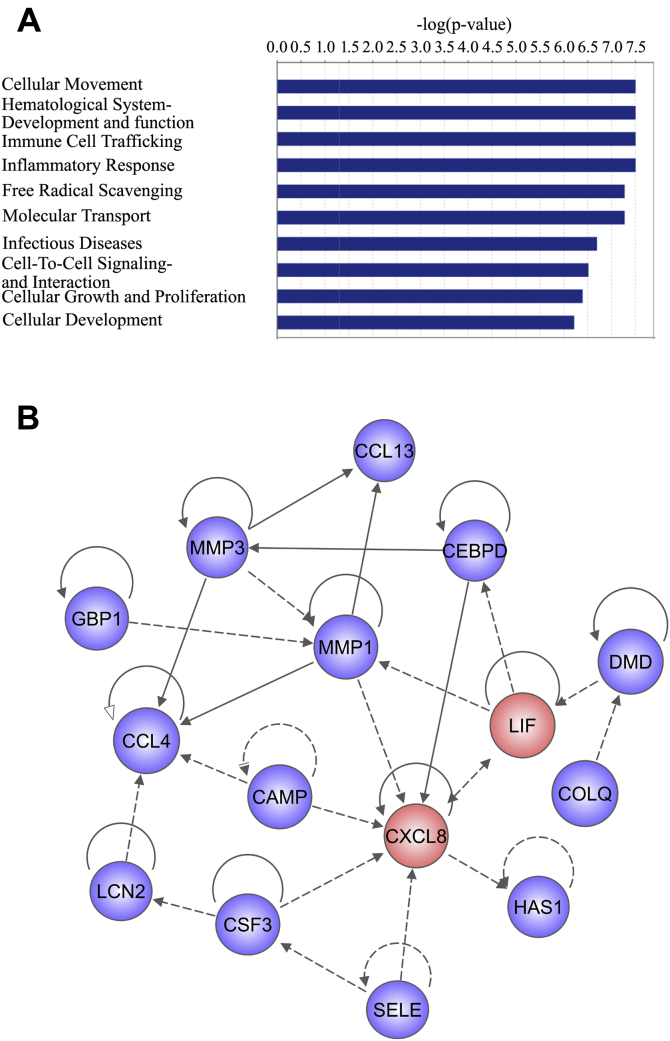
To elucidate the exact roles of gMIF on gAS, the 45 DEGs of gMIF-induced gAS were re-examined and subjected to GO enrichment analysis. Among the top ten GO terms, cellular movement was included as the most significant functional annotation, indicating that gMIF possibly acts to promote the migration of gAS (Fig. 8A). To identify the key regulator(s) modulated by gMIF, the gene regulatory network was thus established using IPA, which highlighted the neutrophil chemoattractant CXCL8/IL-8 as the important effector with the highest weight value (Fig. 8B).
Figure 8.
Analysis of gMIF-mediated regulators in the gAS based on trancriptome sequencing. A, GO terms of DEGs in gAS following stimulation with or without 2.5 μg/ml gMIF for 12 h and 24 h, respectively. B, a reconstructed gene network was created using IPA on the basis of integrated DEGs.
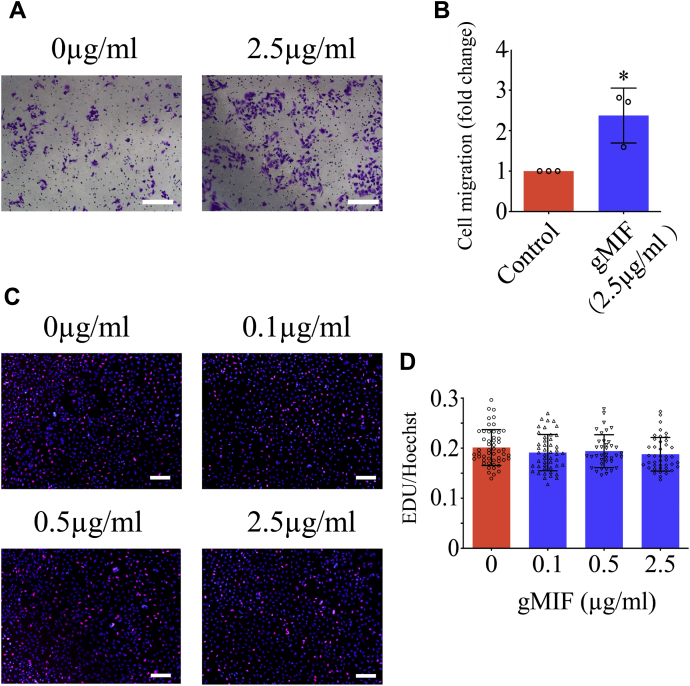
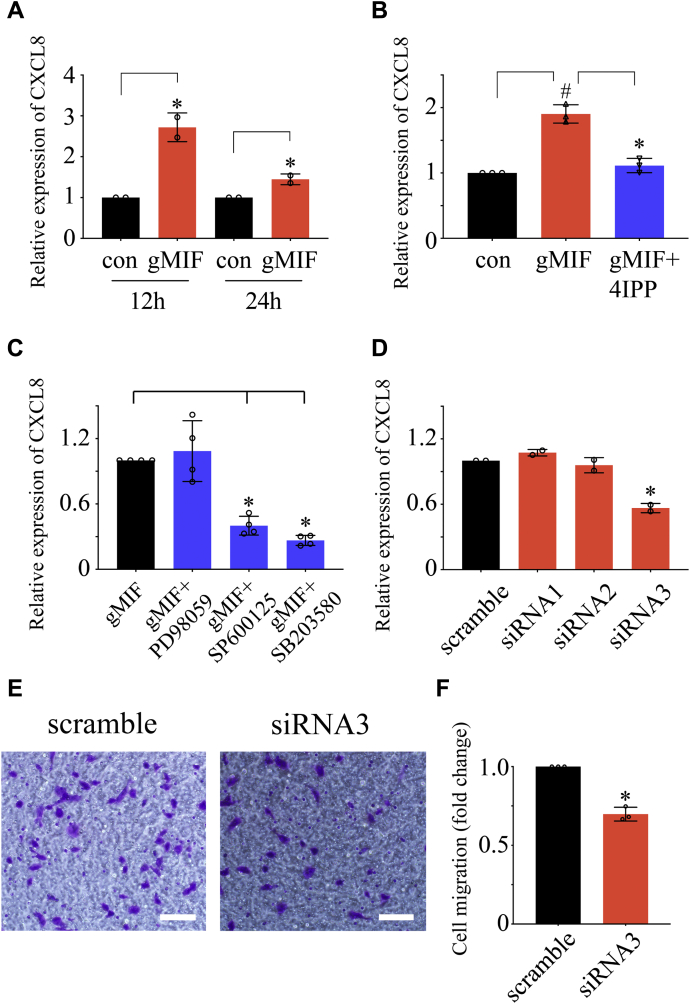
To verify inference drawn from transcriptome analysis, transwell assays were performed to examine the effects of gMIF on the migration of gAS. Stimulation of 2.5 μg/ml gMIF for 24 h significantly facilitated the migration of the cells in comparison with control (Fig. 9, A and B). However, gMIF was found to have no influence on the proliferation of the cells, as detected by EdU assay following cell treatment with 0 to 2.5 μg/ml recombinant gMIF for 24 h (Fig. 9, C and D). Given that CXCL8 potentially influenced such cellular events under regulation of gMIF, CXCL8 expression was examined by RT-PCR following exposure of gAS to 2.5 μg/ml gMIF for 12 h and 24 h. As was expected, the transcripts of CXCL8 were significantly increased in response to the challenges of gMIF (Fig. 10A). However, addition of 50 μM 4-IPP, the inhibitor of MIF, attenuated such effects (Fig. 10B). When the cells were treated with 10 μM SP600125, 10 μM SB203580, or 10 μM PD98059, the inhibitor of JNK, p38, and ERK, respectively, in the presence of 2.5 μg/ml gMIF for 12 h, CXCL8 expression was remarkably decreased by the SP600125 and SB203580 (Fig. 10C). The data indicate that gMIF regulates CXCL8 through activating JNK and p38 kinases.
Figure 9.
Effects of gMIF on the migration and proliferation of gAS.A, transwell determination of gAS migration stimulated with 2.5 μg/ml gMIF for 24 h. B, statistical analysis of (A). C, EdU assays of gMIF effects on proliferation of gAS stimulated with 0 to 2.5 μg/ml gMIF for 24 h. D, quantification data as shown in (C). Error bars represent the standard deviation. Scale bars, 100 μm
Figure 10.
Determination of promoting effects of CXCL8 on gAS migration under regulation of gMIF.A, RT-PCR analysis of CXCL8 expression in gAS following 2.5 μg/ml gMIF stimulation for 12 h and 24 h, respectively. B, RT-PCR analysis of CXCL8 expression in gAS following 2.5 μg/ml gMIF stimulation for 24 h in the presence or absence of 50 μM 4-IPP, the inhibitor of MIF. C, expression analysis of CXCL8 following 2.5 μg/ml gMIF stimulation for 12 h in the presence of 10 μM SP600125, 10 μM SB203580, or 10 μM PD98059, the inhibitor of JNK, P38, and ERK, respectively. D, interference efficiency of three siRNA oligonucleotides for gecko CXCL8 was measured by RT-PCR. E, transwell determination of gAS migration after CXCL8 siRNA3 knockdown for 24 h, followed by stimulation with 2.5 μg/ml gMIF for 24 h. F, quantification data as shown in (E). Error bars represent the standard deviation. Scale bars, 100 μm.
To substantiate that gMIF-mediated migratory effect of gAS is influenced by CXCL8, gAS were transfected with CXCL8 siRNA3 for 24 h, followed by stimulation with 2.5 μg/ml gMIF for 24 h. Transwell assay revealed that knockdown of CXCL8 markedly attenuated the promigratory effects of gMIF (Fig. 10, D–F). The results indicate that gMIF promotes the migration of gAS through regulation of CXCL8.
Discussion
Spinal cord regeneration does not exclusively occur in some lower vertebrates, but appears in the immature mammals such as newborn opossums or fetal rats (22, 24, 40). They share the common features of spinal cord regeneration, including permissive microenvironment, intrinsic capacity for axonal regrowth, and limited astrocytic gliosis (41). Mammalian astrocytes are a population of cells sensitive to the injury of CNS, and their impacts on the functional outcomes of CNS largely depend on the developmental stage. In stark contrast to the reactive astrocytes in the adult CNS, immature astrocytes are amenable in supporting axonal growth and modifying the inhibitory environment. A transplantation of type-1 astrocytes derived from embryonic glial-restricted precursors to the transected spinal cord of the adult rat significantly promotes axonal growth and restoration of locomotor function (42). Interestingly, these transplanted cells occupy the lesion sites, forming a rostral-caudal connection similar to the glial cell “bridge” occurring in the regenerative vertebrates (20, 21, 22), suggesting a conserved orchestration of these cells in repairing the served spinal cord. Phylogenetically, reptiles represent the first vertebrate group that appears typical astrocytes in the CNS (43, 44). To date, the property of reptile astrocytes, especially their responses to the CNS injury, is still limited. In the present study, by the characterization of NSC markers, we have demonstrated that gAS are akin to the mammalian immature astrocytes at the molecular level, which partly explain their beneficial function in the spinal cord regeneration.
Damage to the gecko spinal cord is incapable of triggering excessive inflammation, but can indeed facilitate recruitments of leukocytes at lesion sites (31, 36). Several DAMPs have been observed to be inducibly increased by nerve injury, but the accumulating microglia/macrophages have circumvented excessive activation of inflammation through either invalid interaction with the ligands or intracellular signal blocking by negative regulators (31, 37). However, the mechanism of gAS in resistance to the various inflammatory stimuli is yet to be elucidated, though the membranes of these cells express to some extent the profile of inflammatory receptors. As a potent proinflammatory cytokine, MIF is abundantly expressed in the pathological CNS of mammals and associated with clinical worsening of multiple sclerosis (MS), Alzheimer's disease (AD), and traumatic spinal cord (10, 30, 45, 46). Astrocytes, together with microglia, are certainly the primary target cells of MIF and the active players that contribute to the inflammatory pathology (10, 47). In the present study, gAS have been shown to be insensitive to the proinflammatory stimuli of gMIF, suggesting a unique inflammation-suppressive function of the immature adult astrocytes. The similar character has been observed in the astrocytes from fetal rat spinal cord, which produces less glial scar and inflammation in response to cordotomy (48). These properties of gAS or immature astrocytes have provided a perfect opportunity for unveiling the mechanisms of astrocyte-mediated neuropathology.
The guanine nucleotide exchange factor Vav1 is originally detected and functionally studied in the hematopoietic system, playing a critical role in the development and activation of B and T cells (49). Deregulation of Vav1 has been reported to contribute to various hematologic malignancies and solid tumors such as neuroblastoma, lung cancer, and breast carcinomas (50). Vav1 GEF activity is associated with tyrosine phosphorylation of Tyr174, by which creates an “open” active conformation, thereby interacts with Rho/RacGTPase to facilitate cytoskeletal rearrangement and cell migration (51). So, the protein has been found to be required for phagocytosis of immune cells (52, 53). The physiological importance of Vav1 in the CNS, including negative regulation of motor circuit activity, maintenance of effector T cell susceptibility to neuroinflammation, has been increasingly concerned (54, 55). Here, we revealed that Vav1 was abundantly expressed in the astrocytes of gecko to sustain the insensitivity to inflammatory stimuli during the spontaneous spinal cord regeneration. A consistent role of Vav1 is observed in the fungi-mediated infection, in which Vav1 controls NFκB activation and proinflammatory gene transcription through cooperating downstream Dectin-1/2 and Mincle to engage Card9 (56). Also, Vav1 is shown in macrophages to form a nuclear DNA-binding complex with HSF1 at the HSE2 region of the IL-6 promoter to suppress IL-6 gene transcription (57). Our finding of Vav1 in suppressing inflammation of astrocytes has provided a potential therapeutical target for neuropathology.
CXCL8, also known as IL-8, belongs to CXC family that is one of the first chemokines identified in human brain (58). It can induce chemotaxis in neutrophils, basophils, B and T lymphocytes. Moreover, CXCL8 can promote angiogenesis by inducing the migration of endothelial cells (59). In the CNS, CXCL8 is produced from a variety of cell types including microglia, astrocytes, and endothelial cells (58, 60). Inflammatory stimuli to these cells will result in nuclear localization of several transcription factors such as NFκB and AP-1 and subsequent binding to the CXCL8 promoter for the transcriptional expression (61). Astrocytes per se, together with other cell types, express CXCL8 receptors CXCR1 and CXCR2 (62). So, it is possible that CXCL8 regulates the migration of gAS in a manner of autocrine. However, the mechanism of gMIF-mediated expression of CXCL8 as well as its promigratory pathways in gAS deserves further study.
In conclusion, adult gecko astrocytes have the similar molecular and functional features to those of immature rat astrocytes. The gAS are resistant to gMIF-induced inflammatory activation through a sustaining expression of Vav1 protein. gMIF turns to promote the migration of gAS by upregulating CXCL8 expression, which might be beneficial for the spontaneous spinal cord regeneration.
Experimental procedures
Animals
Adult Gekko japonicus geckos were obtained from the Experimental Animal Center of Nantong University. They were fed mealworms ad libitum and were housed in an air-conditioned room with a controlled temperature (22–25 °C) and saturated humidity. Anesthesia was induced by cooling the animals on ice prior to tail amputation. Amputation was performed at the sixth caudal vertebra, identified based on the special tissue structure present at that position (63), by placing a slipknot of nylon thread and pulling gently until the tail was detached, thus mimicking the process of natural defense.
All experiments were conducted in accordance with the guidelines of the NIH (Guide for the Care and Use of Laboratory Animals: 1985) and the Guidelines for the Use of Animals in Neuroscience Research by the Society for Neuroscience. The experiments were approved according to the Animal Care and Use Committee of Nantong University and the Jiangsu Province Animal Care Ethics Committee. All geckos were anaesthetized on ice prior to sacrifice.
Cell culture and treatment
The culture of primary astrocytes referred to previous description (32). Briefly, the spinal cords removed from adult and E18 rats, as well as from adult geckos, were enzymatically dissociated. The dispersed cells were seeded on a 6-cm dish with a density of 1 × 105 cells/ml in Dulbecco's Modified Eagle's Medium (DMEM)/F-12 medium with 10% fetal bovine serum (FBS) and 1% P-S (Penicillin 100 units/ml, Streptomycin 100 μg/ml). The osmotic pressure of the culture media for rats was 300 to 350 mmol/kg, but for the geckos was 200 to 260 mmol/kg. Cultures were incubated with 5% CO2 at 37 °C for rats or at 30 °C for geckos. Astrocytes were synchronized by serum deprivation for 48 h, followed by passage 3 before using. The purity of the astrocytes was determined with GFAP immunostaining. Cells were stimulated with different concentration of recombinant MIF in the presence or absence of 40 μM 4-IPP or following siRNA interference of Vav1 and CXCL8.
Preparation of gecko MIF (gMIF) recombinant protein and polyclonal rabbit anti-gMIF antibody
The preparation of gMIF recombinant protein and polyclonal rabbit anti-gMIF antibody referred to the previous description (36). The open reading frame of gMIF was amplified from cDNA using NdeI- and XhoI-linked primers: anti-sense primer 5′- CTC GAG TTG CAA AGG TGG ATC CGT TCC AG -3′ and sense primer 5′- CAT ATG CCG ATG CTC GTG ATT AAC ACC A -3′. The antisense primer was designed to omit the stop codon and enable translation of the C-terminal His tag on the pET28a(+) vector. The pET28a(+) vector (Novagen) and PCR amplicon were digested with NdeI and XhoI, and ligated. E. coli BL21 (DE3) was transformed with pET28a(+)-gMIF expression vector and grown in LB broth. After inducing expression with 1.0 mmol/l IPTG for 6 h, the bacterial cells were harvested and disrupted in NTA binding buffer (20 mM Tris-HCl, 500 mM NaCl, and 10 mM imidazole, pH 7.9). The cell suspension was disrupted by sonication and centrifuged at 12,000g for 10 min at 4 °C. The supernatant was loaded onto a Ni2+-chelating column chromatography, and the C-terminal His-tagged recombinant fusion protein was eluted with the elution buffer (20 mM Tris-HCl, 500 mM NaCl, and 500 mM imidazole, pH 7.9) following the manufacturer’s instructions. The eluted fusion protein was collected and analyzed by anti-His tag mouse monoclonal antibody (GeneTex).
Polyclonal rabbit anti-gMIF antibody was commercially prepared using synthesized peptides (CSIGKIGGPQNKAYS/CLTQQLAKATGKPAQ) by GenScript Biotech Corp.
Western blot analysis
Protein was extracted from cells with a buffer containing 1% SDS, 100 mM Tris-HCl, 1 mM PMSF, and 0.1 mM β-mercaptoethanol. Alternatively, protein was extracted from 0.5 cm cord segments after gecko tail amputation at 1 day, 3 days, 1 week, and 2 weeks, respectively. After centrifugation at 13,000 r/min for 30 min at 4 °C, 20 μg of total protein of each sample was loaded into a 10% SDS-PAGE gel and transferred to PVDF membranes (Millipore Sigma). The membrane was then blocked with 5% nonfat dry milk in TBS containing 0.05% Tween-20 (TBS-T) for 1 h and incubated with polyclonal rabbit anti-gMIF (1:50; GenScript), polyclonal rabbit anti-Vav1 (1:1000; Proteintech Group Inc), or polyclonal rabbit anti-CD74 (1:1000; Biorbyte) primary antibodies. After reaction with the second antibody conjugated with goat anti-rabbit or goat anti-mouse HRP (1:5000; Santa Cruz Biotechnology) at room temperature for 2 h, the HRP activity was detected using enhanced chemiluminescence. The membrane was scanned with a ChemiDOC XRS+ Imager (Bio-Rad). The data were analyzed using PDQuest 7.2.0 software (Bio-Rad). β-actin (1:5000; Cell Signaling Technology) was used as an internal control.
Immunofluorescence assay
The spinal cord segments were harvested, postfixed, and sectioned. The sections were allowed to incubate with polyclonal rabbit anti-CD74 (1:100; Bioss), monoclonal mouse anti-GFAP antibody (1:500; Millipore Sigma), or polyclonal rabbit anti-Vav1 (1:100; Proteintech Group Inc) at 4 °C for 36 h. The sections were further reacted with the Cy3-labeled secondary antibody goat anti-rabbit IgG (1:400; Thermo Fisher Scientific) or the FITC-labeled secondary antibody donkey anti-mouse IgG (1:400; Thermo Fisher Scientific) at 4 °C overnight, followed by observation under a confocal laser scanning microscope (Leica).
ELISA
Primary gAS or rAS were stimulated with 0 to 2.5 μg/ml recombinant gMIF for 24 h. Cell supernatants were harvested, while the cells were lysed in the buffer that contained 1% SDS, 100 mM Tris-HCl, 1 mM PMSF, and 0.1 mM β-mercaptoethanol. The lysates were centrifuged at 12,000g for 15 min. The levels of TNF-α and IL-1β were assessed using the appropriate ELISA kits (BD Biosciences) according to the manufacturer’s directions. Plates were read using a 96-well plate reader (Biotek Synergy2; Bio Tek) at a 450 nm wavelength.
Sequencing of mRNA
Total RNA of gAS stimulated with 2.5 μg/ml recombinant gMIF for 0 h, 12 h, or 24 h was extracted using the mirVana miRNA Isolation Kit (Ambion) according to the manufacturer’s instructions. They were then selected by RNA Purification Beads (Illumina) and had undergone library construction and RNA-seq analysis. The library was constructed by using the Illumina TruSeq RNA sample Prep Kit v2 and sequenced by the Illumina HiSeq-2000 for 50 cycles. High-quality reads that passed the Illumina quality filters were kept for the sequence analysis. Using the SOAP program (BGI-Shenzhen) (64), clean reads were mapped to the reference genomes (Assembly Gekko_japonicus_V1.1 and RGSC Genome Assembly v6.0) and gene sequences. No more than five mismatches were allowed in the alignment. Proportions of the clean reads were further mapped back to the genome and genes, providing an overall assessment of the sequencing quality. An average read depth of 30× per sample was achieved, and all samples were analyzed in triplicate. The calculation of gene expression used the reads per kilobase of transcript per million reads mapped (RPKM) method.
Transcriptome analysis of rAS following stimulation of rat MIF recombinant protein (rMIF) referred to the method by Zhou et al. (10).
For comparison of gene expression among gAS, rAS, and eAS, transcriptome sequencing referred to the method by Gu et al. (32).
Bioinformatics analysis
Differentially expressed mRNA was designated in criteria of greater or less than twofold changes in comparison with control. Function of genes was annotated by Blastx against the NCBI database or the AGRIS database (http://arabidopsis.med.ohio-state.edu/downloads.html) with E-value threshold of 10−5. Gene ontology (GO) classification was obtained by WEGO (http://wego.genomics.org.cn/cgi-bin/wego/index.pl) via GO id annotated by Perl and R program. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were assigned to the sequences using KEGG Automatic Annotation Server (KAAS) online. For all heatmaps, genes were clustered by Jensen–Shannon Divergence.
The reconstructed gene networks were created using IPA on the basis of differentially expressed genes in gMIF-stimulated gAS, alternatively the inflammation-related genes integrated from gMIF- or rMIF-stimulated gAS or rAS, to investigate their regulatory relationships and cellular functions (65).
Quantitative real-time polymerase chain reaction (Q-PCR)
Total RNA was prepared with Trizol (Gibco) from cultured gAS stimulated with 2.5 μg/ml recombinant gMIF in the presence or absence of 40 μM 4-IPP at different time points, respectively. The first-strand cDNA was synthesized using an Omniscript Reverse Transcription Kit (QIAGEN) in a 20 μl reaction system that contained 2 μg total RNA, 0.2 U/μl M-MLV reverse transcriptase, 0.5 mM dNTP mix, and 1 μM Oligo-dT primer. The cDNA was diluted 1:5 before use in the Q-PCR assays. The sequence-specific primers of gecko CXCL8/IL-8 were designed and synthesized by Invitrogen and included forward primer 5'- CTT GGA TGG GAG GTG AAC TG -3' and reverse primer 5'- GCA CAC TTC TCT GCC ATC TT -3'. Q-PCR reactions were performed in a final volume of 20 μl (1 μl cDNA template and 19 μl Q-PCR reaction buffer containing 2.5 mmol/l MgCl2, 0.2 mmol/l dNTPs, antisense and sense primers 0.5 μmol/l, Taqman probe 0.4 μmol/l, DNA polymerase 0.2 μl and 1×DNA polymerase buffer). The Rotor-Gene 5 software (Corbett Research, Rotor-Gene) was used for real-time PCR analysis. The reactions were processed using one initial denaturation cycle at 94 °C for 5 min followed by 38 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. Fluorescence was recorded during each annealing step. At the end of each PCR run, data were automatically analyzed by the system and amplification plots were obtained. gMIF full-length plasmid was used to prepare standard curves and as a specificity control for real-time PCR. The expression levels of the CXCL8 cDNA were normalized to an endogenous EF-1α cDNA using forward primer 5'-CCT TCA AAT ATG CCT GGG T-3', reverse primer 5'-CAG CAC AGT CAG CTT GAG AG-3', and Taqman probe 5'-TTG GAC AAG CTG AAG GCA GAA CGT G-3'. In addition, a negative control without the first-strand cDNA was also performed.
Transwell migration assay
The migration of gAS was studied using 6.5 mm transwell chambers with 8 μm pores (Corning Incorporated) as previously described (31). A total of 100 μl of gAS (2 × 105 cells/ml) with or without siRNA knockdown for 24 h was resuspended in DMEM followed by transfer to the top chambers of each transwell and allowed to migrate at 37 °C in 5% CO2. Moreover, 2.5 μg/ml recombinant gMIF was added into the lower chambers. After migration for 24 h, the upper surface of each membrane was cleaned with a cotton swab at the indicated time point. Cells adhering to the bottom surface of each membrane were stained with 0.1% crystal violet, imaged, and counted using a DMR inverted microscope (Leica Microsystems). Assays were performed three times using triplicate wells.
Cell proliferation assay
Primary astrocytes were resuspended in fresh prewarmed (37 °C) complete medium, counted, and plated at a density of 2 × 105 cells/ml on 0.01% poly-L-lysine-coated 96-well plates. Following treatment with 0 to 2.5 μg/ml recombinant gMIF for 24 h, 50 mM EdU was applied to the cultures and the cells were grown for an additional 2 h. Finally, the cells were fixed with 4% formaldehyde in PBS for 30 min. After labeling, the cells were assayed using Cell-Light EdU DNA Cell Proliferation Kit (Ribobio) according to the manufacturer’s protocol. Analysis of astrocytes proliferation (ratio of EdU+ to all cells) was performed using images of randomly selected fields obtained on a DMR fluorescence microscope (Leica Microsystems). Assays were performed three times using triplicate wells.
Immunoprecipitation
Primary astrocytes of gecko were washed twice with cold phosphate-buffered saline and then extracted with lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerolphosphate, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, and Roche Applied Science’s complete protease inhibitors). Whole-cell extracts were centrifuged at 14,000 rpm for 20 min to remove the debris. The proteins in the supernatant were measured using a Protein Assay Kit II (Bio-Rad). For the immunoprecipitation analysis, 500 μg of total cell lysates was precleared with protein A plus G-Sepharose before incubation with specific antibodies, followed by the addition of protein A plus G-Sepharose. The precipitated proteins were resolved in 2× SDS-PAGE sample buffer and separated by electrophoresis on 10 to 12% SDS-PAGE. Following transfer onto a PVDF membrane, they were incubated with anti-His or anti-CD74 antibody and further incubated with horseradish-peroxidase-conjugated secondary antibody (Santa Cruz).
Statistical analysis
The statistical significance of the differences between groups was analyzed by one-way analysis of variance (ANOVA) followed by Bonferroni’s post hoc comparison test with SPSS 15.0 (SPSS). The normality and homoscedasticity of the data were verified prior to statistical analysis using Levene’s test. Statistical significance was set at p < 0.05 and p < 0.01.
Data availability
All the data described are contained within the article and associated supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors have declared that no competing interests exist.
Acknowledgments
We are grateful to Prof. Mei Liu for providing the original data about the comparison of stem cell marker among gAS, eAS, and rAS. This study was supported by the National Natural Science Foundation of China (No. 31871211; 31702022) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author contributions
Yongjun Wang designed this work. Yongjun Wang wrote the paper. N. D., C. S., and B. H. performed the experiments. Yongjun Wang, Yingjie Wang, H. L., T. Y., and H. S. analyzed the data. All the authors have approved the present version of the article and have agreed to be accountable for all aspects of the work regarding questions related to the accuracy or integrity of any part of the work.
Edited by Peter Cresswell
Contributor Information
Yingjie Wang, Email: wyj2010@ntu.edu.cn.
Yongjun Wang, Email: wyjbs@ntu.edu.cn.
Supporting information
References
- 1.Pekny M., Pekna M. Astrocyte reactivity and reactive astrogliosis: Costs and benefits. Physiol. Rev. 2014;94:1077–1098. doi: 10.1152/physrev.00041.2013. [DOI] [PubMed] [Google Scholar]
- 2.Colombo E., Farina C. Astrocytes: Key regulators of neuroinflammation. Trends Immunol. 2016;37:608–620. doi: 10.1016/j.it.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Farina C., Aloisi F., Meinl E. Astrocytes are active players in cerebral innate immunity. Trends Immunol. 2007;28:138–145. doi: 10.1016/j.it.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Sofroniew M.V. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009;32:638–647. doi: 10.1016/j.tins.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson M.A., Burda J.E., Ren Y., Ao Y., O'Shea T.M., Kawaguchi R., Coppola G., Khakh B.S., Deming T.J., Sofroniew M.V. Astrocyte scar formation aids central nervous system axon regeneration. Nature. 2016;532:195–200. doi: 10.1038/nature17623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wheeler M.A., Jaronen M., Covacu R., Zandee S.E.J., Scalisi G., Rothhammer V., Tjon E.C., Chao C.C., Kenison J.E., Blain M., Rao V.T.S., Hewson P., Barroso A., Gutiérrez-Vázquez C., Prat A. Environmental control of astrocyte pathogenic activities in CNS inflammation. Cell. 2019;176:581–596.e518. doi: 10.1016/j.cell.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang Z., Altuntas C.Z., Gulen M.F., Liu C., Giltiay N., Qin H., Liu L., Qian W., Ransohoff R.M., Bergmann C., Stohlman S., Tuohy V.K., Li X. Astrocyte-restricted ablation of interleukin-17-induced Act1-mediated signaling ameliorates autoimmune encephalomyelitis. Immunity. 2010;32:414–425. doi: 10.1016/j.immuni.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang Z., Wang C., Zepp J., Wu L., Sun K., Zhao J., Chandrasekharan U., DiCorleto P.E., Trapp B.D., Ransohoff R.M., Li X. Act1 mediates IL-17-induced EAE pathogenesis selectively in NG2+ glial cells. Nat. Neurosci. 2013;16:1401–1408. doi: 10.1038/nn.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayo L., Trauger S.A., Blain M., Nadeau M., Patel B., Alvarez J.I., Mascanfroni I.D., Yeste A., Kivisäkk P., Kallas K., Ellezam B., Bakshi R., Prat A., Antel J.P., Weiner H.L. Regulation of astrocyte activation by glycolipids drives chronic CNS inflammation. Nat. Med. 2014;20:1147–1156. doi: 10.1038/nm.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y., Guo W., Zhu Z., Hu Y., Wang Y., Zhang X., Wang W., Du N., Song T., Yang K., Guan Z., Wang Y., Guo A. Macrophage migration inhibitory factor facilitates production of CCL5 in astrocytes following rat spinal cord injury. J. Neuroinflammation. 2018;15:253. doi: 10.1186/s12974-018-1297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liddelow S.A., Barres B.A. Reactive astrocytes: Production, function, and therapeutic potential. Immunity. 2017;46:957–967. doi: 10.1016/j.immuni.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Gorina R., Font-Nieves M., Marquez-Kisinousky L., Santalucia T., Planas A.M. Astrocyte TLR4 activation induces a proinflammatory environment through the interplay between MyD88-dependent NFkappaB signaling, MAPK, and Jak1/Stat1 pathways. Glia. 2011;59:242–255. doi: 10.1002/glia.21094. [DOI] [PubMed] [Google Scholar]
- 13.Sofroniew M.V., Vinters H.V. Astrocytes: Biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J.J., Chuah M.I., Yew D.T., Leung P.C., Tsang D.S. Effects of astrocyte implantation into the hemisected adult rat spinal cord. Neuroscience. 1995;65:973–981. doi: 10.1016/0306-4522(94)00519-b. [DOI] [PubMed] [Google Scholar]
- 15.Hill C.E., Proschel C., Noble M., Mayer-Proschel M., Gensel J.C., Beattie M.S., Bresnahan J.C. Acute transplantation of glial-restricted precursor cells into spinal cord contusion injuries: Survival, differentiation, and effects on lesion environment and axonal regeneration. Exp. Neurol. 2004;190:289–310. doi: 10.1016/j.expneurol.2004.05.043. [DOI] [PubMed] [Google Scholar]
- 16.Haas C., Neuhuber B., Yamagami T., Rao M., Fischer I. Phenotypic analysis of astrocytes derived from glial restricted precursors and their impact on axon regeneration. Exp. Neurol. 2012;233:717–732. doi: 10.1016/j.expneurol.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X., Chen M., Wang F., Windrem M., Wang S., Shanz S., Xu Q., Oberheim N.A., Bekar L., Betstadt S., Silva A.J., Takano T., Goldman S.A., Nedergaard M. Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem Cell. 2013;12:342–353. doi: 10.1016/j.stem.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A., Thompson W.J., Barres B.A. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S., Li G., Duncan J.A., 3rd, Cheshier S.H., Shuer L.M., Chang E.F., Grant G.A. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldshmit Y., Sztal T.E., Jusuf P.R., Hall T.E., Nguyen-Chi M., Currie P.D. Fgf-dependent glial cell bridges facilitate spinal cord regeneration in zebrafish. J. Neurosci. 2012;32:7477–7492. doi: 10.1523/JNEUROSCI.0758-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen J.P., Sagasti A. Learning to swim, again: Axon regeneration in fish. Exp. Neurol. 2017;287:318–330. doi: 10.1016/j.expneurol.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 22.Tazaki A., Tanaka E.M., Fei J.F. Salamander spinal cord regeneration: The ultimate positive control in vertebrate spinal cord regeneration. Dev. Biol. 2017;432:63–71. doi: 10.1016/j.ydbio.2017.09.034. [DOI] [PubMed] [Google Scholar]
- 23.Zukor K.A., Kent D.T., Odelberg S.J. Meningeal cells and glia establish a permissive environment for axon regeneration after spinal cord injury in newts. Neural Dev. 2011;6:1. doi: 10.1186/1749-8104-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diaz Quiroz J.F., Echeverri K. Spinal cord regeneration: Where fish, frogs and salamanders lead the way, can we follow? Biochem. J. 2013;451:353–364. doi: 10.1042/BJ20121807. [DOI] [PubMed] [Google Scholar]
- 25.Calandra T., Roger T. Macrophage migration inhibitory factor: A regulator of innate immunity. Nat. Rev. Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lue H., Kleemann R., Calandra T., Roger T., Bernhagen J. Macrophage migration inhibitory factor (MIF): Mechanisms of action and role in disease. Microbes Infect. 2002;4:449–460. doi: 10.1016/s1286-4579(02)01560-5. [DOI] [PubMed] [Google Scholar]
- 27.Koda M., Nishio Y., Hashimoto M., Kamada T., Koshizuka S., Yoshinaga K., Onodera S., Nishihira J., Moriya H., Yamazaki M. Up-regulation of macrophage migration-inhibitory factor expression after compression-induced spinal cord injury in rats. Acta Neuropathol. 2004;108:31–36. doi: 10.1007/s00401-004-0853-z. [DOI] [PubMed] [Google Scholar]
- 28.Nishio Y., Koda M., Hashimoto M., Kamada T., Koshizuka S., Yoshinaga K., Onodera S., Nishihira J., Okawa A., Yamazaki M. Deletion of macrophage migration inhibitory factor attenuates neuronal death and promotes functional recovery after compression-induced spinal cord injury in mice. Acta Neuropathol. 2009;117:321–328. doi: 10.1007/s00401-008-0476-x. [DOI] [PubMed] [Google Scholar]
- 29.Leyton-Jaimes M.F., Kahn J., Israelson A. Macrophage migration inhibitory factor: A multifaceted cytokine implicated in multiple neurological diseases. Exp. Neurol. 2018;301:83–91. doi: 10.1016/j.expneurol.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Zhou Y., Chen S., Hu Y., Zhu Z., Wang Y., Du N., Song T., Yang Y., Guo A., Wang Y. Macrophage migration inhibitory factor facilitates prostaglandin E2 production of astrocytes to tune inflammatory milieu following spinal cord injury. J. Neuroinflammation. 2019;16:85. doi: 10.1186/s12974-019-1468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong Y., Gu Y., Huan Y., Wang Y., Liu Y., Liu M., Ding F., Gu X., Wang Y. HMGB1 protein does not mediate the inflammatory response in spontaneous spinal cord regeneration: A hint for CNS regeneration. J. Biol. Chem. 2013;288:18204–18218. doi: 10.1074/jbc.M113.463810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu Y., Yang J., Chen H., Li J., Xu M., Hua J., Yao J., Wang Y., Liu Y., Liu M. Different astrocytic activation between adult Gekko japonicus and rats during wound healing in vitro. PLoS One. 2015;10 doi: 10.1371/journal.pone.0127663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rehermann M.I., Marichal N., Russo R.E., Trujillo-Cenoz O. Neural reconnection in the transected spinal cord of the freshwater turtle Trachemys dorbignyi. J. Comp. Neurol. 2009;515:197–214. doi: 10.1002/cne.22061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alibardi L. Observations on the recovering lumbar spinal cord of lizards show multiple origins of the cells forming the bridge region including immune cells. J. Morphol. 2020;281:95–109. doi: 10.1002/jmor.21082. [DOI] [PubMed] [Google Scholar]
- 35.Kuegler P.B., Zimmer B., Waldmann T., Baudis B., Ilmjärv S., Hescheler J., Gaughwin P., Brundin P., Mundy W., Bal-Price A.K., Schrattenholz A., Krause K.H., van Thriel C., Rao M.S., Kadereit S. Markers of murine embryonic and neural stem cells, neurons and astrocytes: Reference points for developmental neurotoxicity testing. ALTEX. 2010;27:17–42. doi: 10.14573/altex.2010.1.16. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y., Wei S., Song H., Zhang X., Wang W., Du N., Song T., Liang H., Chen X., Wang Y. Macrophage migration inhibitory factor derived from spinal cord is involved in activation of macrophages following gecko tail amputation. FASEB J. 2019;33:14798–14810. doi: 10.1096/fj.201801966RRR. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X., He B., Li H., Wang Y., Zhou Y., Wang W., Song T., Du N., Gu X., Luo Y., Wang Y. SOCS3 attenuates GM-CSF/IFN-γ-mediated inflammation during spontaneous spinal cord regeneration. Neurosci. Bull. 2020;36:778–792. doi: 10.1007/s12264-020-00493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spurrell D.R., Luckashenak N.A., Minney D.C., Chaplin A., Penninger J.M., Liwski R.S., Clements J.L., West K.A. Vav1 regulates the migration and adhesion of dendritic cells. J. Immunol. 2009;183:310–318. doi: 10.4049/jimmunol.0802096. [DOI] [PubMed] [Google Scholar]
- 39.Wilsbacher J.L., Moores S.L., Brugge J.S. An active form of Vav1 induces migration of mammary epithelial cells by stimulating secretion of an epidermal growth factor receptor ligand. Cell Commun. Signal. 2006;4:5. doi: 10.1186/1478-811X-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicholls J., Saunders N. Regeneration of immature mammalian spinal cord after injury. Trends Neurosci. 1996;19:229–234. doi: 10.1016/0166-2236(96)10021-7. [DOI] [PubMed] [Google Scholar]
- 41.Bradbury E.J., Burnside E.R. Moving beyond the glial scar for spinal cord repair. Nat. Commun. 2019;10:3879. doi: 10.1038/s41467-019-11707-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davies J.E., Huang C., Proschel C., Noble M., Mayer-Proschel M., Davies S.J. Astrocytes derived from glial-restricted precursors promote spinal cord repair. J. Biol. 2006;5:7. doi: 10.1186/jbiol35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monzon-Mayor M., Yanes C., Ghandour M.S., de Barry J., Gombos G. Glial fibrillary acidic protein and vimentin immunohistochemistry in the developing and adult midbrain of the lizard Gallotia galloti. J. Comp. Neurol. 1990;295:569–579. doi: 10.1002/cne.902950406. [DOI] [PubMed] [Google Scholar]
- 44.Ahboucha S., Laalaoui A., Didier-Bazes M., Montange M., Cooper H.M., Gamrani H. Differential patterns of glial fibrillary acidic protein-immunolabeling in the brain of adult lizards. J. Comp. Neurol. 2003;464:159–171. doi: 10.1002/cne.10781. [DOI] [PubMed] [Google Scholar]
- 45.Cox G.M., Kithcart A.P., Pitt D., Guan Z., Alexander J., Williams J.L., Shawler T., Dagia N.M., Popovich P.G., Satoskar A.R., Whitacre C.C. Macrophage migration inhibitory factor potentiates autoimmune-mediated neuroinflammation. J. Immunol. 2013;191:1043–1054. doi: 10.4049/jimmunol.1200485. [DOI] [PubMed] [Google Scholar]
- 46.Oikonomidi A., Tautvydaitė D., Gholamrezaee M., Henry H., Bacher M., Popp J. Macrophage migration inhibitory factor is associated with biomarkers of Alzheimer's disease pathology and predicts cognitive decline in mild cognitive impairment and mild dementia. J. Alzheimers Dis. 2017;60:273–281. doi: 10.3233/JAD-170335. [DOI] [PubMed] [Google Scholar]
- 47.Vandenbark A.A., Meza-Romero R., Benedek G., Offner H. A novel neurotherapeutic for multiple sclerosis, ischemic injury, methamphetamine addiction, and traumatic brain injury. J. Neuroinflammation. 2019;16:14. doi: 10.1186/s12974-018-1393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujimoto Y., Yamasaki T., Tanaka N., Mochizuki Y., Kajihara H., Ikuta Y., Ochi M. Differential activation of astrocytes and microglia after spinal cord injury in the fetal rat. Eur. Spine J. 2006;15:223–233. doi: 10.1007/s00586-005-0933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarakhovsky A., Turner M., Schaal S., Mee P.J., Duddy L.P., Rajewsky K., Tybulewicz V.L. Defective antigen receptor-mediated proliferation of B and T cells in the absence of Vav. Nature. 1995;374:467–470. doi: 10.1038/374467a0. [DOI] [PubMed] [Google Scholar]
- 50.Farago M., Yarnitzky T., Shalom B., Katzav S. Vav1 mutations: What makes them oncogenic? Cell. Signal. 2020;65:109438. doi: 10.1016/j.cellsig.2019.109438. [DOI] [PubMed] [Google Scholar]
- 51.Turner M., Billadeau D. VAV proteins as signal integrators for multi-subunit immune-recognition receptors. Nat. Rev. Immunol. 2002;2:476–486. doi: 10.1038/nri840. [DOI] [PubMed] [Google Scholar]
- 52.Grommes C., Lee C.Y., Wilkinson B.L., Jiang Q., Koenigsknecht-Talboo J.L., Varnum B., Landreth G.E. Regulation of microglial phagocytosis and inflammatory gene expression by Gas6 acting on the Axl/Mer family of tyrosine kinases. J. Neuroimmune Pharmacol. 2008;3:130–140. doi: 10.1007/s11481-007-9090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah V.B., Ozment-Skelton T.R., Williams D.L., Keshvara L. Vav1 and PI3K are required for phagocytosis of beta-glucan and subsequent superoxide generation by microglia. Mol. Immunol. 2009;46:1845–1853. doi: 10.1016/j.molimm.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 54.Fry A., Laboy J., Norman K. VAV-1 acts in a single interneuron to inhibit motor circuit activity in Caenorhabditis elegans. Nat. Commun. 2014;5:5579. doi: 10.1038/ncomms6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kassem S., Gaud G., Bernard I., Benamar M., Dejean A.S., Liblau R., Fournié G.J., Colacios C., Malissen B., Saoudi A. A natural variant of the T cell receptor-signaling molecule Vav1 reduces both effector T cell functions and susceptibility to neuroinflammation. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roth S., Bergmann H., Jaeger M., Yeroslaviz A., Neumann K., Koenig P.A., Prazeres da Costa C., Vanes L., Kumar V., Johnson M., Menacho-Márquez M., Habermann B., Tybulewicz V.L., Netea M., Bustelo X.R. Vav proteins are key regulators of Card9 signaling for innate antifungal immunity. Cell Rep. 2016;17:2572–2583. doi: 10.1016/j.celrep.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zenker S., Panteleev-Ivlev J., Wirtz S., Kishimoto T., Waldner M.J., Ksionda O., Tybulewicz V.L.J., Neurath M.F., Atreya I. A key regulatory role for Vav1 in controlling lipopolysaccharide endotoxemia via macrophage-derived IL-6. J. Immunol. 2014;192:2830–2836. doi: 10.4049/jimmunol.1300157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Semple B.D., Kossmann T., Morganti-Kossmann M.C. Role of chemokines in CNS health and pathology: A focus on the CCL2/CCR2 and CXCL8/CXCR2 networks. J. Cereb. Blood Flow Metab. 2010;30:459–473. doi: 10.1038/jcbfm.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mamik M., Ghorpade A. CXCL8 as a potential therapeutic target for HIV-associated neurocognitive disorders. Curr. Drug Targets. 2016;17:111–121. doi: 10.2174/1389450116666150626124544. [DOI] [PubMed] [Google Scholar]
- 60.Ramjeesingh R., Leung R., Siu C. Interleukin-8 secreted by endothelial cells induces chemotaxis of melanoma cells through the chemokine receptor CXCR1. FASEB J. 2003;17:1292–1294. doi: 10.1096/fj.02-0560fje. [DOI] [PubMed] [Google Scholar]
- 61.Khalaf H., Jass J., Olsson P.E. Differential cytokine regulation by NF-kappaB and AP-1 in Jurkat T-cells. BMC Immunol. 2010;11:26. doi: 10.1186/1471-2172-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flynn G., Maru S., Loughlin J., Romero I.A., Male D. Regulation of chemokine receptor expression in human microglia and astrocytes. J. Neuroimmunol. 2003;136:84–93. doi: 10.1016/s0165-5728(03)00009-2. [DOI] [PubMed] [Google Scholar]
- 63.McLean K.E., Vickaryous M.K. A novel amniote model of epimorphic regeneration: The leopard gecko, Eublepharis macularius. BMC Dev. Biol. 2011;11:50. doi: 10.1186/1471-213X-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., Wang J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 65.Kramer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data described are contained within the article and associated supporting information.