Abstract
Bacterial biofilms are a major cause of delayed wound healing. Consequently, the study of wound biofilms, particularly in host-relevant conditions, has gained importance. Most in vitro studies employ refined laboratory media to study biofilms, representing conditions that are not relevant to the infection state. To mimic the wound milieu, in vitro biofilm studies often incorporate serum or plasma in growth conditions, or employ clot or matrix-based biofilm models. While incorporating serum or plasma alone is a minimalistic approach, the more complex in vitro wound models are technically demanding, and poorly compatible with standard biofilm assays. Based on previous reports of clinical wound fluid composition, we have developed an in vitro wound milieu (IVWM) that includes, in addition to serum (to recapitulate wound fluid), matrix elements and biochemical factors. With Luria-Bertani broth and Fetal Bovine Serum (FBS) for comparison, the IVWM was used to study planktonic growth, biofilm features, and interspecies interactions, of common wound pathogens, Staphylococcus aureus and Pseudomonas aeruginosa. We demonstrate that the IVWM recapitulates widely reported in vivo biofilm features such as biomass formation, metabolic activity, increased antibiotic tolerance, 3D structure, and interspecies interactions for monospecies and mixed-species biofilms. Further, the IVWM is simple to formulate, uses laboratory-grade components, and is compatible with standard biofilm assays. Given this, it holds potential as a tractable approach to study wound biofilms under host-relevant conditions.
Keywords: Wounds, Biofilms, Milieu, Staphylococcus aureus, Pseudomonas aeruginosa, Mixed-species, Interspecies interactions
Introduction
Wound healing is mediated by several host factors, including inflammatory, immune and biochemical components [1,2]. Following injury, a protein-rich fluid leaks into the wound, which along with cellular and matrix elements, results in a characteristic wound milieu [3,4]. Interplay across various factors is reflected in this milieu, which is known to influence progression and outcome of the wound state [[5], [6], [7], [8]]. Microbial infections are the single-most-important cause of delayed wound healing [2,9]. In wounds, diverse bacterial species are known to form biofilms, observed as microscopic bacterial aggregates enmeshed in a self-produced matrix [[10], [11], [12]]. Previous work has reported that biofilm aggregates of different bacterial species exist in close approximation with each other, albeit occupying spatially-distinct niches in the wound bed [[13], [14], [15], [16]]. In the infected wound bed, these biofilm aggregates interface with host cellular and matrix elements [13,[17], [18], [19], [20]], in the presence of the composite wound milieu.
Hitherto, the study of biofilms in wounds has typically relied on in vivo animal systems or in vitro laboratory studies [17,19]. In vivo systems pose scientific, technical and ethical challenges, and are also limited by availability and accessibility. On the other hand, the majority of in vitro biofilm studies employ laboratory media [13,18] (such as refined protein broths) to grow biofilms and analyze the effects of antimicrobial treatments. However, the composition of laboratory media is not relevant in the context of the wound infection state. Recognizing this, recent studies have incorporated serum or plasma into in vitro growth conditions, to more closely represent the host milieu [[20], [21], [22], [23], [24], [25]]. This is relevant given that wound fluid has been shown to resemble the biochemical and nutrient profile of serum [20]. However, the wound milieu is more complex and includes additional host factors and matrix elements [26]. To recapitulate this, clot and matrix-based in vitro wound biofilm models have been developed that more closely mimic in vivo conditions [14,20,25,27,28]. However, these models are technically demanding, low-throughput, and poorly compatible with standard biofilm assays.
In this study, we have developed a simple in vitro wound milieu (IVWM) that includes, in addition to serum (to recapitulate wound fluid), matrix elements such as collagen, fibrinogen and fibronectin, and host factors such as lactoferrin and lactic acid. The formulation of the milieu is based on the composition of clinical wound fluid, as reported across previous studies [29,30]. We employ this composite milieu to study planktonic growth, biofilm features, and interspecies interactions of Staphylococcus aureus and Pseudomonas aeruginosa, two of the most common bacterial pathogens isolated from wound infections [31]. Using laboratory media (Luria-Bertani broth) and fetal bovine serum (FBS) for comparison, we demonstrate that the in vitro wound milieu recapitulates key in vivo biofilm features such as biomass formation, metabolic activity, antibiotic tolerance, three-dimensional structure, and interspecies interactions. While expectedly different from laboratory media, we find that these features are distinct from those observed with serum alone. Notably, the impact of the IVWM on mixed-species biofilm growth differs from that in serum, and similar to in vivo conditions appears to provide an advantage to P. aeruginosa [17,19]. Further, the IVWM is easy to formulate, high-throughput, and is compatible with standard biofilm assays.
Materials and Methods
Bacterial strains and growth conditions
All experiments were carried out using fluorescently tagged strains of Pseudomonas aeruginosa (PAO1-pUCP18, mCherry [32]) and Staphylococcus aureus (Strain AH 133-pAH13, GFP [33]). These strains were a gift from Dr. Kendra Rumbaugh (Texas Tech University Health Science Center, Lubbock, TX). Selection for SA-GFP was done with 10 μg/mL erythromycin (Himedia) and for PAO1-mCherry with 100 μg/mL ampicillin (Himedia) on Luria-Bertani (LB) agar (Sigma-Aldrich, USA, L2897-250G) plates and in overnight LB broth (Sigma-Aldrich, USA, L3022-250G) cultures. Strains were streaked onto LB agar and incubated overnight at 37 °C. For overnight cultures, isolated colonies were grown in LB broth under shaking conditions at 37 °C unless otherwise stated.
Preparation of the in vitro wound milieu
An in vitro wound milieu (IVWM) was prepared with sterile fetal bovine serum (FBS) (Thermofisher Scientific, Brazil, 10270106) as the base component. Other components added included sterile rat tail collagen (Sigma, USA, 122–20, 50 μg/mL), lactoferrin (Sigma, USA, L4040, 2 mg/mL stock prepared by dissolving in 1X PBS (pH 7.4, ThermoFisher Scientific, USA, 20012027) and filter sterilized), fibronectin (Sigma, USA, F4759, 1 mg/mL stock solution prepared using autoclaved distilled water), fibrinogen (Sigma, USA, F3879, 0.9% NaCl (prewarmed at 37 °C) was used to prepare a stock solution of 10 mg/mL and filter sterilized) and lactic acid (Sigma, USA, W261114, 11.4 M stock concentration). As per manufacturers’ instructions, collagen and lactoferrin were stored at 4 °C, FBS, fibronectin and fibrinogen were stored at −20 °C and lactic acid was stored at room temperature. Components were either purchased sterile or filter sterilized using a 0.22 μm syringe filter (Millex, Ireland, SLGV033RS). The components were combined in concentrations given in Table 1 to result in the final IVWM. The IVWM was freshly prepared each time and was used immediately after use (not stored). The pH of the IVWM was measured using a pH probe (Hanna Instruments, Romania, HI2210) and pH strip (Qualigens). The specific gravity of IVWM was calculated as the relative weight of IVWM compared to the weight of an equal volume of distilled water in an analytical balance (Mettler-Toledo, India, ME204) [34].
Table 1.
Composition of the in vitro wound milieu (IVWM) and rationale for inclusion of components.
| Components | Final concentration in IVWM | Rationale | References |
|---|---|---|---|
| FBS | 70% | Major component and base of IVWM; at this concentration, FBS accounts for several components in wound fluid | [29,30,38,40] |
| Lactic acid | 11–12 mM | Host biochemical factor released in response to tissue damage; based on levels detected in wounds immediately following injury | [[41], [42], [43]] |
| Lactoferrin | 20–30 μg/mL | Host biochemical factor increased in the presence of microbes | [[44], [45], [46], [47], [48], [49], [50], [51], [52]] |
| Fibrinogen | 200–400 μg/mL | Host matrix protein, notably absent from serum, present in the wound milieu | [[54], [55], [56]] |
| Fibronectin | 30–60 μg/mL | Host matrix protein, present in lower concentrations in the wound milieu | [55,[57], [58], [59], [60], [61], [62]] |
| Collagen | 10–12 μg/mL | Host matrix protein, critical component of the wound bed | [20,21,25,28,[63], [64], [65], [66]] |
Planktonic growth
For growth curves in LB
Overnight cultures of S. aureus and P. aeruginosa were grown in LB broth under shaking conditions at 37 °C. The next day, cultures were quantified by measuring optical density (O.D.) at 600 nm with a multimode microplate reader (Tecan, Austria, Infinite M Plex), and this value was converted to colony forming units (CFU)/mL. To set up growth curves, overnight cultures were diluted in sterile LB broth (to result in ~106 cells/mL) and 100 μL of the diluted culture (consisting of ~105 cells) was added per well, in replicates of three, to a sterile, transparent, round bottom, untreated 96-well polystyrene plate (Corning, USA, 3788). A similar protocol was used throughout this study to quantify overnight cultures and use the appropriate seeding density in each experiment. Uninoculated LB broth was used as a control. Plates were incubated in the multimode microplate reader (Tecan, Austria, Infinite M Plex) at 37 °C with shaking in orbital mode (2 mm amplitude), and O.D. was measured at 600 nm every 30 min for 12–14 h.
For growth curves in FBS and IVWM
Growth curves in FBS and IVWM were done using overnight cultures set up in FBS. Briefly, each bacterial colony was inoculated in FBS and incubated overnight at 37 °C under shaking conditions. The next day, cultures were quantified by measuring O.D. at 600 nm with a multimode microplate reader (Tecan, Austria, Infinite M Plex). To set up growth curves, each culture was diluted in sterile FBS or IVWM (to result in ~106 cells/mL) and 100 μL of this diluted culture (consisting of ~105 cells) was added per well, in replicates of three, to a sterile, transparent, round bottom, untreated 96-well polystyrene plate (Corning, USA, 3788). Uninoculated FBS or IVWM were used as controls. The plate reader was set to 37 °C with shaking in orbital mode (2 mm amplitude), and absorbance was measured at 600 nm every 30 min for 12–14 h.
For all growth curves, O.D. versus time was plotted and growth rates (doubling times) were calculated.
Colony forming units (CFUs) for viability of planktonic cultures
In order to quantify the proportion of living cells in the planktonic cultures, in monospecies and mixed-species conditions, colony count assays were carried out. Overnight cultures were set up as done for planktonic growth curves (as described above). The next day, cultures were diluted to ~106 cells/mL in respective media. From these diluted cultures, 100 μL (containing ~105 cells) was added to 100 μL of the respective media (LB, FBS, IVWM) in a fresh tube (for monospecies cultures). For mixed-species cultures, 100 μL of each of the diluted cultures of S. aureus and P. aeruginosa (in LB, FBS, IVWM) were added together in a single fresh tube. All conditions were set up in at least 3 replicates. Cultures were incubated under shaking conditions at 37 °C for 7 h (till stationary phase). Dilutions were plated on selective media such as Pseudomonas isolation agar BioVeg (SRL Chemicals, India, 36124) and Staphylococcus Medium 110 (SRL Chemicals, India, 39726). Plates were incubated overnight for 24 h at 37 °C. Based on protocols using Staphylococcus Medium 110, these plates required incubation for up to 36–48 h to obtain visible colonies.
Biofilm formation
Overnight cultures of S. aureus and P. aeruginosa (in LB media) were each diluted to ~106 cells/mL in LB, FBS and IVWM. This was done by diluting 1 μL of overnight culture in 99 μL of respective media (or larger volumes but in the same proportion), to make a 1:100 dilution. Therefore, given the 1:100 dilution, there is minimal residual LB contaminating the FBS and IVWM media conditions. From these diluted cultures, 50 μL (containing ~105 cells) was added in multiple replicates (at least three) to a transparent, round bottom, untreated 96-well polystyrene plate. To these wells, 50 μL of the media in which biofilm formation was to be tested was added, to maintain a constant volume of 100 μL. For mixed-species biofilms, individual cultures of P. aeruginosa and S. aureus were diluted to ~106 cells/mL (as above). From these diluted cultures, 50 μL of each strain (containing ~105 cells) was added to wells in at least three replicates to make a total volume of 100 μL. Biofilms were allowed to grow under static conditions at 37 °C for 24 h. These pre-formed biofilms were used to measure metabolic activity (XTT assay, see below), determine viable colony counts (CFU assay, see below) and to visualize the biofilm (confocal microscopy, see below).
Colony forming units (CFUs) of biofilms
In order to quantify the proportion of living cells in the biofilms grown under different conditions, overnight cultures of S. aureus and P. aeruginosa were each diluted to ~106 cells/mL in LB, FBS and IVWM, and biofilm growth was set up as described in the above section. Briefly, for monospecies biofilms, 50 μL of the diluted cultures (containing ~105 cells) were added to 50 μL of the respective media and allowed to incubate at 37 °C for 24 h under static conditions. For the mixed-species biofilms, 50 μL (containing ~105 cells) of each of the diluted cultures (in a 1:1 ratio) were added and allowed to incubate at 37 °C for 24 h under static conditions. The next day, the supernatant in the wells was gently removed, and wells were washed once with 150 μL of LB broth to remove planktonic cells. Each biofilm (monospecies or mixed-species) was dislodged by scraping with a sterile 200 μL pipette (50 times in circular motion) in 100 μL of LB media, following which the suspended cells were mixed 50 times with a 20–200 μL pipette set at 100 μL. From this suspension of cells, ten-fold serial dilutions were made in LB media, and dilutions were plated on Pseudomonas isolation agar BioVeg (SRL Chemicals, India, 36124) and Staphylococcus Medium 110 (SRL Chemicals, India, 39726) to identify colony counts of each strain. The mixed-species biofilms were plated on both sets of media. Plates were incubated for 24 h at 37 °C. Based on protocols using Staphylococcus Medium 110, these plates required incubation for up to 36–48 h to obtain visible colonies. The next day, colony forming units (CFUs) were counted, and back calculations were done to obtain CFU/mL.
XTT assay for biofilm metabolic activity
Pre-formed biofilms of S. aureus and P. aeruginosa were grown for 24 h in different media (as described above) under monospecies and mixed-species conditions. The next day, biofilms were washed once with 150 μL of respective media (LB, FBS or IVWM), after removing suspended media. Menadione (SRL, India, 61495) solution (7 mg/mL) was diluted 1:100 in sterile distilled water. A mixture of LB:XTT:Menadione in 79:20:1 ratio was freshly prepared [35] (XTT, Invitrogen, USA, X6493), and 150 μL of this mixture was added to each well (including media only controls). The plates were covered in aluminum foil and incubated for 4 h at 37 °C under static conditions. From each well, 100 μL was transferred to a new 96-well plate and absorbance was measured at 492 nm. For each set of biofilm conditions, metabolic activity was normalized to log10 (CFU) of the biofilm, which was calculated from replicate biofilms that had been set up simultaneously. For the mixed-species biofilms, metabolic activity was normalized to the log10 (total CFU) of both species in the biofilm.
Antibiotic susceptibility of pre-formed biofilms
Pre-formed biofilms of S. aureus and P. aeruginosa were grown as monospecies biofilms for 24 h as previously described above (in the section on biofilm formation). Briefly, overnight cultures S. aureus and P. aeruginosa were each diluted to ~106 cells/mL in LB, FBS and IVWM. From these diluted cultures, ~105 cells (100 μL) were added in replicates of three, to a transparent, round bottom 96-well polystyrene plate and the plate was incubated for 24 h at 37 °C. After 24 h, the suspended media was gently removed and the biofilms were washed once with LB. Antibiotics were diluted to varying concentrations 0–64 μg/mL for tobramycin (TCI, Japan, T2503) and 0–512 μg/mL for vancomycin (Himedia, India, CMS217) in the respective media in which biofilm susceptibility was to be tested (LB, FBS, IVWM) and 100 μL of the antibiotic-media solution was added into the wells. Wells with untreated biofilms (no antibiotic, only media added) were also included. Uninoculated media was used as a control. Plates were incubated at 37 °C for 24 h. After 24 h, the XTT assay was performed to quantify viability (as described above). Antibiotic susceptibility was quantified as the Minimum Biofilm Eradication Concentration (MBEC). The concentration resulting in an 80% reduction in biofilm metabolic activity (representing metabolically-active cells), compared to the untreated biofilms, was considered as the MBEC80 value for that particular antibiotic.
Biofilm visualization using confocal microscopy
To visualize in situ three-dimensional biofilm structure, 24-h old undisturbed biofilms were set up as previously described in LB, FBS or IVWM (as monospecies and mixed-species). Briefly, LB overnight cultures of P. aeruginosa and S. aureus were diluted in the respective media in which biofilm structure was to be observed (to obtain ~106 cells/mL in LB, FBS and IVWM), and mixed in a 1:1 ratio for mixed-species biofilms to obtain a total volume of 100 μL (50 μL (~105 cells) of the diluted culture of each strain). For monospecies biofilms, 50 μL (~105 cells) of the diluted culture was mixed with 50 μL of the respective media. After incubation at 37 °C for 24 h (static conditions), the biofilms were directly examined with confocal laser scanning microscopy (Leica, Germany, LASX TCS SP8). To enable this direct visualization, biofilms were grown in 96-well, black polystyrene tissue-culture treated flat-bottom plates with a transparent bottom (Corning, USA, 3603). These plate specifications allow visualization of the biofilm structure in the well and minimum interference of fluorescence signals from neighboring wells. The tissue-culture treatment imparts an overall negative charge to the surface and results in a hydrophilic surface, and this treatment is known to reduce the attachment of bacteria. In doing so, this enables the study of the role of the media conditions (host components in FBS and IVWM) in biofilm formation.
It is important to note that the wells were not rinsed prior to imaging. As seen in previous biofilm studies [36], rinsing or pipetting (even gently) would disturb the structure of the biofilm. Given this, the biomass imaged at the bottom of the well, would include biofilm, as well as attached or unattached single cells.
Wells were imaged using a 488 nm laser for excitation and a 496–551 nm emission filter for S. aureus-GFP, and a 561 nm laser for excitation and a 590–625 nm emission filter for P. aeruginosa-mCherry. To visualize the 3D architecture of the biofilms across the entire well, an 8x8 tile scan approach was used with an overlap of 30% and a Z-stack step size of 10 μm. The images were processed and reconstructed in the Leica Application Suite (LAS) software. Mean intensity measurements were carried out using the LAS software. Biofilm thickness was measured using ImageJ; for the mixed-species biofilms, this was done by exporting and measuring biofilms separately in each channel.
Statistical analysis
All analysis was performed using GraphPad Prism 8. A two-way ANOVA with Tukey’s multiple comparisons test was performed and a p-value of <0.05 was considered significant.
Results and discussion
Development of an in vitro wound milieu (IVWM) that mimics host conditions
Following injury, the wound bed is bathed in protein-rich exudate, which along with additional host elements, results in a characteristic wound milieu [1,3,4,37]. The composition of wound fluid has been widely reported to resemble that of serum [29,30,38], with several in vitro wound studies using serum to mimic wound conditions [20,25,28,39]. To develop the IVWM, we used FBS as the base component, to which relevant host matrix and biochemical factors were added. Components were chosen based on previous reports of biochemical analyses across clinical wound fluids [29,30], as well as their identified roles in the wound bed.
Based on analysis of wound fluid composition [29], we decided to use 70% FBS as the base component of the IVWM, since at this concentration the levels of multiple components in serum fit into the range of values for that component in wound fluid [29,40]. While the concentrations of several biochemical factors in serum and wound fluid are similar [29], the wound milieu is also characterized by the presence of additional host-derived biochemical factors. In the initial inflammatory state of wound repair, the wound milieu is characterized by increased levels of lactate and lactoferrin. Lactate in the wound is a result of local tissue damage, and increased concentrations are reflected in the local milieu [[41], [42], [43]]. While the levels of lactoferrin in the plasma and serum of healthy individuals are typically low [[44], [45], [46], [47]], in the presence of microbial infections [48], increased levels are reported in the wound milieu. In addition to possessing antimicrobial properties [[49], [50], [51]], lactoferrin has also been shown to be important for wound re-epithelialization [52]. Given this, lactate and lactoferrin were included in the IVWM at concentrations that mimicked host conditions (Table 1).
The IVWM composition also included relevant matrix components such as fibrinogen, fibronectin and collagen [53]. While fibrinogen is present in wound tissue [54,55] and plasma [56], it is notably absent from serum. On the other hand, the matrix protein fibronectin is typically present at high concentrations in serum or plasma [57], but undergoes degradation under inflammatory wound conditions [[58], [59], [60]]. To mimic this, fibronectin was added to the IVWM, to resemble lower concentrations as relevant to the wound milieu [55,[60], [61], [62]]. Given its critical role as an extracellular matrix protein in the wound bed [20,21,25,28,[63], [64], [65], [66]], collagen was also included in the IVWM (Table 1). At the concentration of collagen added, the IVWM was in liquid form (not a gel) resembling the wound fluid milieu.
The pH of the freshly formulated IVWM was determined to be 5.5–6.0 and specific gravity was 0.966, which is similar to that measured in wound fluid [8,30,67]. However, following overnight growth of P. aeruginosa and S. aureus biofilms, alone and together (with an initial seeding density of 105 cells/mL), the pH of the IVWM was determined to be 8.0–8.5 for P. aeruginosa, 6.5–7.0 for S. aureus and 8.0–8.5 for the mixed-species biofilm. This represents an increase in pH following biofilm growth for both pathogens, and a notable alkaline shift for P. aeruginosa, an effect that could result from the production of alkaline by-products of bacterial metabolism [68]. In chronic, non-healing wounds, the wound pH is reported to be elevated, likely the consequence of bacterial infections [69]. Previous studies have reported a range of pH values (ranging from 5 to 9) for wound fluid, with increasing pH correlating with infected wounds [67,70]. While in this study we aimed to develop a wound milieu that recapitulated the pH of the basal wound state (slightly acidic), this milieu can be further adapted to mimic variations observed in pH, such as the alkaline shift observed in the presence of bacterial infection and non-healing states. Given that the individual components of the IVWM are recommended to be stored at different temperatures, the IVWM was freshly prepared each time and used immediately after use (not stored). This is an important point to consider while replicating the milieu or using it with modifications.
Planktonic growth of P. aeruginosa and S. aureus in LB, FBS and IVWM
To determine the effects of the IVWM on the planktonic growth of P. aeruginosa and S. aureus (monospecies), we performed growth curves in the IVWM, and in LB medium and FBS for comparison (Fig. 1). LB broth is widely-used to study planktonic and biofilm states under in vitro conditions [18,35,[71], [72], [73]], however, its composition (refined yeast extract and tryptone) poorly mimics the wound infection state. To recapitulate factors relevant to the wound milieu, several media incorporate FBS, in varying concentrations, into growth conditions [20,24,74,75]. Given that the concentrations of FBS employed across different planktonic and biofilm wound pathogen studies range widely, we decided to use the maximum possible FBS concentration (100% FBS) for comparison, as opposed to 70% used in the IVWM [[76], [77], [78], [79], [80]]. Further, developing 70% FBS would require the addition of a diluent (to make up the 30%), which would itself influence bacterial dynamics in the media. This dilution would also have to be maintained in the IVWM (to maintain consistency across both host-relevant conditions) which would alter the composition and dilute the concentrations of relevant host and matrix factors in the IVWM. Finally, while this does not allow the parsing of the individual roles of the additional host components in the IVWM, this study is focused on developing and evaluating a composite milieu which represents host-relevant conditions.
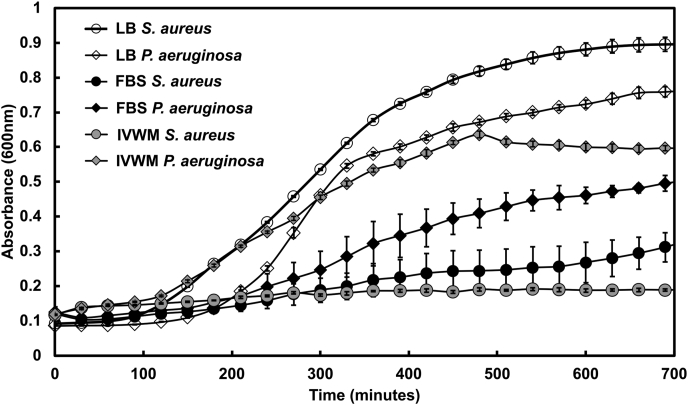
Fig. 1.
IVWM supports the planktonic growth of P. aeruginosa, but not of S. aureus. Planktonic growth curves of Pseudomonas aeruginosa (PAO1) and Staphylococcus aureus (AH133) were performed in Luria-Bertani broth (LB), Fetal Bovine Serum (FBS) and the in vitro wound milieu (IVWM). Optical density (OD600) was measured at intervals of 30 min for 12 h. Error bars represent SEM, n = 3 (biological replicates).
When examined in LB medium, P. aeruginosa and S. aureus showed typical growth curves, with doubling times of 30 ± 1 min and 28 ± 1 min respectively, in accordance with previous reports [81,82]. In 100% FBS, P. aeruginosa was observed to grow slower as compared to growth in LB medium, with a doubling time of 56 ± 7 min. On the other hand, S. aureus showed markedly impaired growth in FBS. In the IVWM, consisting of 70% FBS with additional matrix and host components, P. aeruginosa was observed to double every 43 ± 7 min, which is faster than that seen in FBS alone, and cultures were also observed to enter exponential phase earlier. Similar to that observed in FBS, S. aureus demonstrated impaired growth in the IVWM.
Taken together, as compared with LB media, the planktonic growth of P. aeruginosa and S. aureus is notably different under growth conditions that incorporate host factors. In the presence of FBS, both pathogens displayed markedly slower growth; components in serum are known to impair the growth of S. aureus [80,83,84]. However, when grown in IVWM, which contains serum at a concentration that recapitulates wound fluid (70% FBS), and additional matrix and biochemical factors, a greater difference in growth across the two species was observed. The IVWM was observed to better support the growth of P. aeruginosa (as compared with FBS alone), while S. aureus showed significantly impaired growth. This indicates that in the IVWM, P. aeruginosa has a distinct growth advantage in planktonic state, as compared with its co-pathogen S. aureus.
Interspecies interactions between planktonic P. aeruginosa and S. aureus under different conditions
To understand the effects of the IVWM, in comparison with LB and FBS, on interspecies interactions between the two co-pathogens under planktonic conditions, P. aeruginosa and S. aureus were co-cultured in LB, FBS and IVWM (with a starting ratio of 1:1, with ~105 CFU of each strain), and after 7 h were plated on selective media to obtain viable counts of each bacterial species. In LB media, S. aureus showed a significant decrease in viable cells under mixed-species conditions (Fig. 2A and B); the percentage of viable cells recovered for S. aureus was ~3% of viable cells obtained when S. aureus was grown alone (Fig. 2B). This is in accordance with previously published reports of P. aeruginosa actively killing and outcompeting S. aureus under planktonic conditions in LB medium [13,14,18,76].
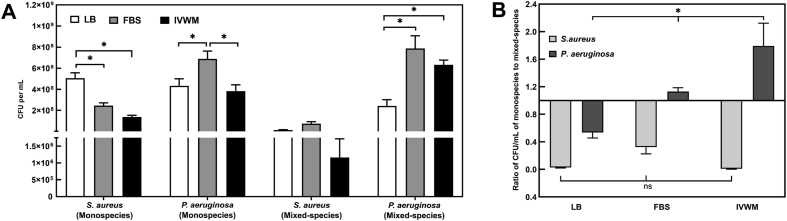
Fig. 2.
In the IVWM, interspecies interactions between planktonic P. aeruginosa and S. aureus result in P. aeruginosa significantly outcompeting S. aureus. Viable counts of planktonic P. aeruginosa and S. aureus grown under mixed-species conditions in LB, FBS and IVWM were quantified by measuring Colony Forming Units (CFU). (A) CFUs of planktonic monospecies and mixed-species cultures of Pseudomonas aeruginosa and Staphylococcus aureus in LB, FBS and IVWM (B) Ratio of CFU/mL of the colony counts in monospecies to that in mixed-species planktonic cultures of Pseudomonas aeruginosa and Staphylococcus aureus in LB, FBS and IVWM. Error bars represent SEM, n = 4 (biological replicates). A p-value of <0.05 was considered significant (∗).
In FBS and IVWM, S. aureus demonstrated the same trend, with reduced recovery of viable cells under mixed-species conditions (Fig. 2A and B). Notably, under mixed-species conditions in the IVWM, viable S. aureus cells recovered from planktonic co-cultures were very few, representing ~0.2% of the total viable cells (of both species). On the other hand, the IVWM supported the growth and recovery of P. aeruginosa under mixed-species conditions, comparable to that observed under monospecies conditions. It is important to note that this effect observed in the IVWM is similar to that seen under in vivo conditions, where in spite of co-existence between the two common wound pathogens, P. aeruginosa outcompetes S. aureus in wound infections [14,17,76,[85], [86], [87]].
Biofilm formation, metabolic activity and coexistence of P. aeruginosa and S. aureus biofilms under different conditions
To understand the effects of the IVWM on biofilm formation, metabolic activity and interspecies interactions, P. aeruginosa and S. aureus biofilms were grown in microtiter plates under monospecies and mixed-species conditions, and assayed for metabolic activity (by the XTT assay) and viable cells (CFUs). For comparison, monospecies and mixed-species biofilms were grown in LB and FBS, and assayed in a similar manner. For each set of biofilm conditions, metabolic activity was normalized to log10 (CFU) of the biofilm.
As previously reported across several studies [18,71], in LB broth, P. aeruginosa and S. aureus formed biofilms that displayed the presence of metabolic activity (Fig. 3A). Notably, in FBS and IVWM, P. aeruginosa and S. aureus biofilms displayed increased metabolic activity as compared to that in LB medium (Fig. 3A). Several host components, including serum, plasma, and matrix factors such as collagen, fibrinogen, fibronectin, have been shown to support the formation of P. aeruginosa [[88], [89], [90]] and S. aureus biofilms [78,[91], [92], [93], [94], [95], [96], [97], [98]].
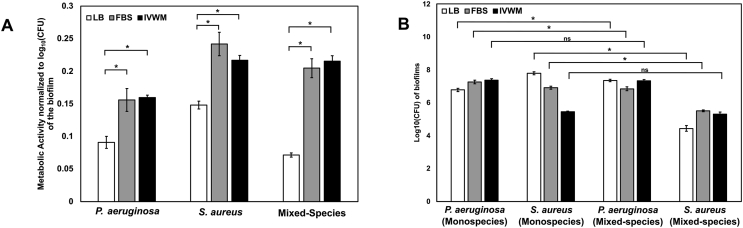
Fig. 3.
IVWM supports the formation of metabolically-active biofilms of P. aeruginosa and S. aureus, and indicates the coexistence of both species under mixed-species conditions. Pre-formed 24-h biofilms of Pseudomonas aeruginosa and Staphylococcus aureus were quantified for metabolic activity by the XTT assay and for viability using the CFU technique. Assays were performed under monospecies and mixed-species conditions in LB, FBS and IVWM. (A) Metabolic activity normalized to log10 (CFU) of the biofilm for P. aeruginosa and S. aureus biofilms under monospecies and mixed-species states (B) Log10 (CFU) of viable biofilm cells of P. aeruginosa and S. aureus biofilms under monospecies and mixed-species states in LB, FBS and IVWM. Error bars represent SEM, n = 3 (biological replicates). A p-value of <0.05 was considered significant (∗).
To examine the formation and metabolic activity of mixed-species biofilms, P. aeruginosa and S. aureus were grown together under different media conditions, and metabolic activity (measured by XTT) was normalized to the log10 (CFU) of the total living cells of both species in the biofilm. Mixed-species biofilms in FBS and IVWM demonstrated robust metabolic activity normalized to total viable cells, significantly increased as compared to LB media. Host components, including matrix and chemical factors, are well-known to influence the coexistence of P. aeruginosa and S. aureus biofilms [13,17,18,31,[99], [100], [101]]. To explore possible interspecies interactions, we compared colony-forming units (CFUs) of P. aeruginosa and S. aureus, across monospecies and mixed-species biofilms, under different media conditions (Fig. 3B). In LB medium, S. aureus demonstrated a notable decrease in viable cell counts under mixed-species conditions in LB; recovery of viable S. aureus cells was 40% less in mixed-species conditions as compared to monospecies biofilms. This points to the presence of interspecies interactions, an effect possibly mediated by the widely-reported killing of S. aureus by P. aeruginosa [13,14,18,76]. While biofilms grown in FBS also demonstrated a decrease in viable S. aureus counts in mixed-species conditions, the recovery was better than that in LB medium, with the S. aureus viable counts in mixed-species being only 20% less than that in monospecies. On the other hand, the IVWM was seen to support both, the viability of P. aeruginosa biofilms, and nearly complete recovery of S. aureus viable cells under mixed-species conditions (Fig. 3B). This indicates that the IVWM enables the co-existence of S. aureus and P. aeruginosa biofilms. This is in accordance with previous work that reports that the presence of host components support the concomitant growth of P. aeruginosa and S. aureus, and ‘rescue’ of S. aureus from P. aeruginosa killing [14,76]. Notably, in the IVWM, despite this ‘rescue’ of S. aureus, based on viable cell counts, P. aeruginosa is seen to outnumber S. aureus in the mixed-species biofilm (Fig. 3B).
It is important to note that the XTT assay was performed with the widely-described LB:XTT:Menadione solution in the ratio of 79:20:1 (see Materials and Methods) for all three biofilm conditions (LB, FBS, IVWM). This means that biofilms grown in LB were provided fresh nutrients for an additional 4-h growth period (duration of exposure to the LB:XTT:Menadione mix), whereas for biofilms in FBS and IVWM this involved a change of growth media. Given that growth is necessary for the metabolic activity readout, this could result in a shorter lag phase for biofilms grown in LB, as compared to those in FBS and IVWM. As seen in Fig. 1, for planktonic conditions, given the lag phase for P. aeruginosa and S. aureus in LB media is around 3 h, this could lead to 3–4 doublings in the 4-h time-point for the XTT assay. For biofilm conditions as well, this 4-h period of growth could potentially affect biofilm metabolic activity and viable cell counts [102]. Based on results seen in Fig. 3A and B, biofilms in LB display lower metabolic activity and similar CFU counts as compared with biofilms in FBS and IVWM. Nevertheless, for future work it would be relevant to consider performing the XTT:Menadione assay in the original growth conditions (FBS, IVWM). However, given that serum albumin is known to affect XTT readings [103], and so could other components of the IVWM, these factors would need to be optimized.
Overall, our results indicate that the IVWM supports the biofilm formation and metabolic activity of P. aeruginosa and S. aureus biofilms. Further, under mixed-species conditions, the IVWM supports the co-existence of both pathogens, with P. aeruginosa observed in larger numbers.
Antibiotic susceptibility of P. aeruginosa and S. aureus biofilms in the in vitro wound milieu (IVWM)
To study the effects of the IVWM on the antibiotic tolerance of P. aeruginosa and S. aureus biofilms, pre-formed biofilms were grown for 24 h (also in LB and FBS) and exposed to varying concentrations of tobramycin and vancomycin respectively, following which they were assayed for antibiotic susceptibility (Table 2). The Minimum Biofilm Eradication Concentration (MBEC) was considered to be the concentration resulting in 80% of biofilm eradication (MBEC80).
Table 2.
Minimum Biofilm Eradication Concentration (MBEC)∗ for P. aeruginosa and S. aureus under different conditions (∗The concentration resulting in 80% reduction in biofilm metabolic activity (by the XTT assay) was considered as the MBEC80 value for that particular antibiotic).
| LB | FBS | IVWM | |
|---|---|---|---|
|
P. aeruginosa (PAO1) Tobramycin |
1 μg/mL | 8 μg/mL | 8 μg/mL |
|
S. aureus (AH 133) Vancomycin |
>512 μg/mL | 16 μg/mL | >512 μg/mL |
For biofilms grown in LB media, the MBEC80 of P. aeruginosa for tobramycin was similar to the previously reported value of 1 μg/mL [35]. On the other hand, the MBEC80 of pre-formed P. aeruginosa biofilms grown in FBS and IVWM was determined to be 8 μg/mL, an 8-fold increase compared to LB. The presence of serum is known to increase antimicrobial tolerance [[104], [105], [106]], owing to the serum-binding properties of certain antibiotics, including binding of tobramycin and serum albumin [107]. However, this increased tolerance could also be due to the inherent biofilm properties under these conditions, including nutrient distribution and variations in microbial metabolism [108]. The similarity of MBEC80 in FBS and IVWM (which contains 70% FBS) could possibly indicate the dominant role of serum in influencing the antibiotic tolerance of P. aeruginosa biofilms.
When grown in LB media, pre-formed S. aureus biofilms displayed increased tolerance, even at higher concentrations of vancomycin (MBEC80>512 μg/mL). This is in accordance with previously reported values, across different laboratory media conditions [73,104,109,110]. Interestingly, we find that biofilms grown in FBS demonstrate increased susceptibility to vancomycin, with a 32-fold reduction in the MBEC80 (16 μg/mL) in FBS, as compared with LB. As stated previously, S. aureus biofilms in FBS were observed as thin layers on the round-bottom surface of the microtiter wells (data not shown), albeit demonstrating the presence of robust metabolic activity. We speculate that inhibitory effects of serum on S. aureus [80], along with the formation of thin biofilms, that could increase antibiotic exposure, could lead to this increased susceptibility.
When grown in IVWM, 24-h old S. aureus biofilms showed increased tolerance to vancomycin, with an MBEC80>512 μg/mL. This value is 32-fold higher than that observed for biofilms grown in FBS alone, which is important to note given that the IVWM comprises 70% FBS, along with additional matrix and host factors. Host factors and matrix components have been shown to influence the susceptibility of S. aureus to vancomycin. Notably, upregulation of fibrinogen-and fibronectin-binding proteins [92], or pretreatment with fibronectin [111], has been shown to reduce the susceptibility of S. aureus biofilms. It is important to note that these matrix factors are constituents of the IVWM, and are either absent or present in low levels in FBS. On the other hand, while host components such as lactoferrin are known to have antimicrobial properties against S. aureus [[112], [113], [114], [115], [116]], the cumulative effect of such components in a complex wound milieu such as in the IVWM is not known.
Under in vivo conditions, wound biofilms are known to display increased tolerance to antimicrobial treatments [86,117,118], and our results show that the IVWM recapitulates the increased antibiotic tolerance of both P. aeruginosa and S. aureus biofilms.
3D biofilm structure of P. aeruginosa and S. aureus biofilms under different conditions
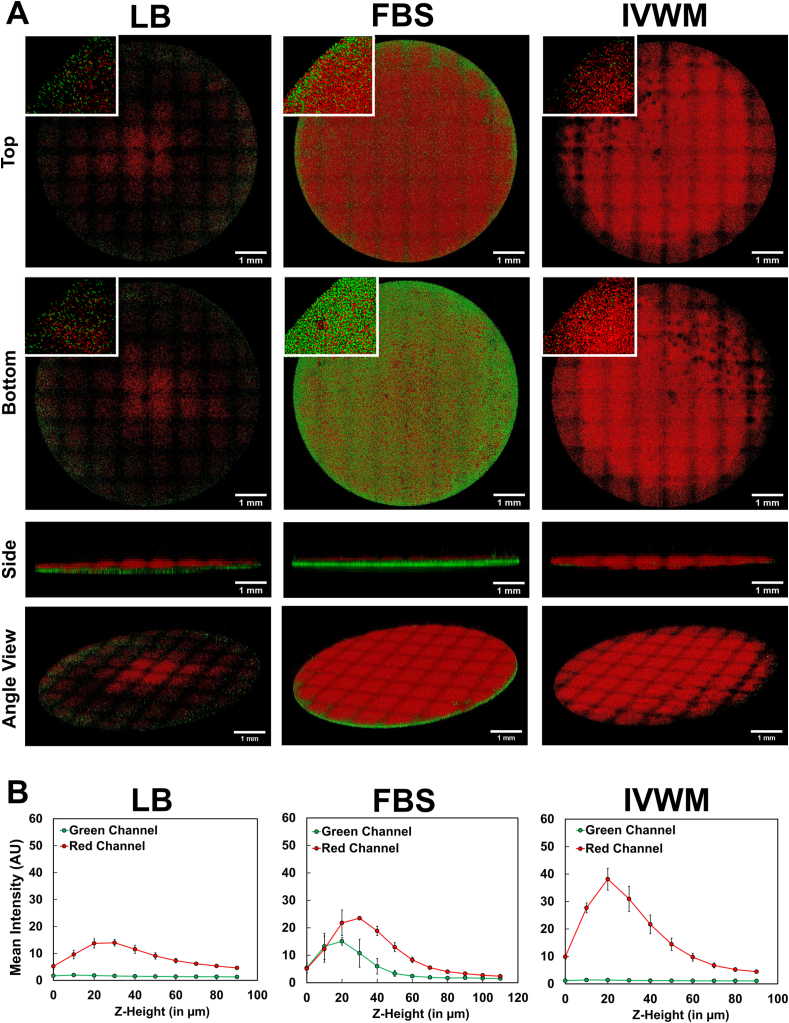
In order to visualize the 3D structure of the biofilms under different conditions, undisturbed 24-h old biofilms of P. aeruginosa (PAO1-mCherry) and S. aureus (AH133-GFP) were imaged (as monospecies and mixed-species) using confocal microscopy via the tile scan approach. The tile scan approach uses an 8x8 grid set to scan the entire well, and thereby enables visualization of the 3D structure across the entire biomass. To reduce the role of surface attachment [119], given that polystyrene is not a biotic surface, and to better understand the role of the media conditions, biofilms were grown in tissue-culture treated 96 well plates.
When grown alone in LB medium, S. aureus showed the presence of dense biomass after 24 h, with an average thickness of 92 ± 3 μm (Fig. 4A). This biomass likely represents biofilm, as well as attached and unattached planktonic cells (as seen on the surface). In FBS, S. aureus biomass was observed to be thinner as compared with LB, with an average thickness of 63 ± 4 μm (Fig. 4A). Notably, in the IVWM, S. aureus formed biofilms that were significantly thinner than those seen in both FBS and LB, with an average thickness of 40 ± 3 μm (Fig. 4A). This indicates that refined protein-based media, such as LB broth, likely overestimate biofilm thickness in comparison to that observed in host-relevant media conditions (such as FBS and IVWM).
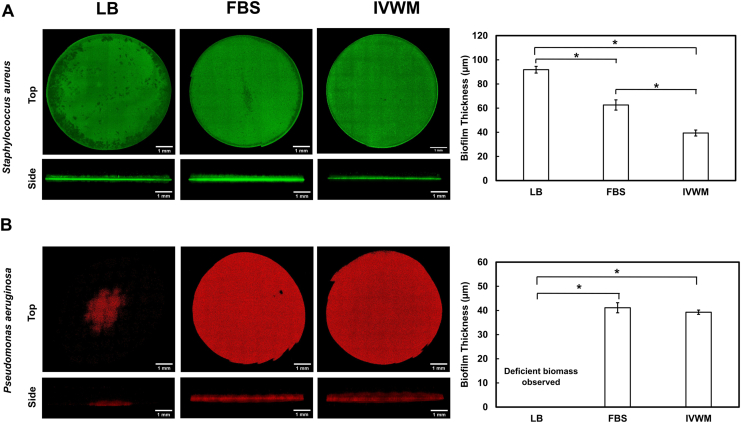
Fig. 4.
3D biofilm structure of P. aeruginosa and S. aureus single-species biofilms in IVWM are distinct from that in LB and FBS. Tile-scan confocal microscopy showing 3D structure and thickness of (A) P. aeruginosa (PAO1-mCherry) and (B) S. aureus (AH133-GFP) biofilms in LB, FBS and IVWM. To reduce the role of surface attachment, and better explore the role of the different media conditions, biofilms were grown in tissue-culture treated microtiter plates. The poor biomass formed by P. aeruginosa in LB, is likely due to the reduced surface attachment to the tissue-culture treated surfaces, and absence of host and matrix proteins in the media. For all images, given that the wells were not rinsed prior to imaging, the observed biomass represents not only biofilm, but also attached or unattached single bacterial cells within, and on top of, the dense bacterial mats (visible in the side view images). The grids are a result from stitching of the tiles in the tile-scan processing. Error bars represent SEM, n = 3 (biological replicates). A p-value of <0.05 was considered significant (∗).
Notably, we find that while biofilms in LB appear to be visually thick and dense, when examined for metabolic activity normalized for biomass (Fig. 3A), they displayed lower activity. On the other hand, S. aureus biofilms in FBS displayed significantly high metabolic activity normalized to the living biofilm mass (Fig. 3A), but when observed visually, these biofilms were seen as thin layers on the bottom and sides of the microtiter well (data not shown). This could possibly be the reason that certain previous studies using biomass staining protocols (and not metabolic activity) [21,80,97], report reduced or absent S. aureus biofilm formation in the presence of FBS or plasma. Further, studies that have specifically explored the relationship between biomass and metabolic activity, have found an inverse relationship [120] (could be an effect of mature biofilms having cellular niches with low metabolic activity), or a poor correlation between the two biofilm features [[121], [122], [123]].
When grown in LB media, P. aeruginosa displayed deficient biomass growth or minimal biofilm formation after 24 h (Fig. 4B) seen as clumps of bacteria in the center of the well. We can speculate that this is possibly due to the tissue-culture treatment of the wells, which is known to reduce the surface attachment of bacteria. This is notably in contrast to that seen in Fig. 3B, where in the presence of non-treated wells, P. aeruginosa forms robust biofilms that demonstrate metabolic activity and recovery of viable cells. On the other hand, in the presence of host factors such as in FBS and IVWM, P. aeruginosa showed the presence of dense biomass, and biofilms were observed as mat-like structures, with an average thickness of 41 ± 2 μm in FBS and 39 ± 1 μm in the IVWM (Fig. 4B). Given the absence of surface attachment, this highlights the role of host components (as in FBS and in the IVWM) in P. aeruginosa biofilm formation.
While S. aureus biofilms are thinner in the IVWM as compared to that in FBS (Fig. 4A), they display comparable metabolic activity (Fig. 3A) and high MBEC80 in the IVWM (>512 μg/mL) (Table 2). On the other hand, S. aureus biofilms in FBS display thicker biomass and high metabolic activity (Figs. 3A and 4A), and are yet observed to have an MBEC80 of 16 μg/mL. Previous studies have correlated the thickness of biofilms to antibiotic tolerance, and have reported that thicker biofilms are typically observed to display increased antibiotic tolerance, possibly due to reduced antibiotic penetration [124,125]. However, the presence of nutrient conditions, metabolic gradients, and host components have also been observed to influence antibiotic tolerance in P. aeruginosa and S. aureus biofilms [21,[125], [126], [127], [128], [129]]. It is also important to note that the assay formats are distinct (tissue-culture treatments, biofilm washes), which impact biofilm formation and features under these conditions, and thereby limit a comparative analysis.
3D biofilm structure of P. aeruginosa and S. aureus mixed-species biofilms
To examine mixed-species biofilms, P. aeruginosa (PAO1-mCherry) and S. aureus (AH133-GFP) were inoculated in a 1:1 ratio, followed by in situ visualization of 24-h biomass (Fig. 5). In LB media, under mixed-species conditions, P. aeruginosa displayed minimal biomass after 24 h, similar to that observed in the monospecies state (Fig. 5A and B). Notably, under mixed-species conditions, S. aureus was also seen to form sparse biofilms. Based on known interspecies interactions between the two pathogens, it is likely that under mixed-species conditions in LB media, inoculated P. aeruginosa results in killing of S. aureus, and thereby impairs the formation of S. aureus biofilms [87,130]. It is important to note that the reduced surface attachment (resulting from the tissue-culture treated wells) could contribute to the existence of P. aeruginosa in the planktonic state, which is known to mediate this killing effect.
Fig. 5.
3D visualization of biofilm structure of mixed-species P. aeruginosa and S. aureus biofilms in IVWM shows distinct predominance of P. aeruginosa. (A) Tile scan confocal microscopy of P. aeruginosa (PAO1-mCherry) and S. aureus (AH133-GFP) mixed-species biofilms in LB, FBS and IVWM grown in tissue-culture treated microtiter plates. (B) Mean intensity of fluorescence (representing PAO1-mCherry and SA-GFP) across the Z-height of the biofilm (with the bottom as Z = 0). Mean intensities were calculated using the LAS software as the average fluorescence intensity of each channel in each Z-plane and plotted as the mean intensity versus Z-position. This was averaged across 3 biological replicates, and therefore represents variation seen across biological replicates, as well as variation across the Z-plane. Given that the wells were not rinsed prior to imaging, the observed biomass represents not only biofilm, but also attached or unattached single bacterial cells within, and on top of, the dense bacterial mats (visible in the side view images). The inset images are zoomed in ~8 times. The grids are a result from stitching of the tiles in the tile-scan processing. Error bars represent SEM, n = 3 (biological replicates).
In FBS, the two pathogens were observed to form a robust mixed-species biofilm (Fig. 5A and B), consisting of a thick mat of S. aureus (thickness 20 ± 2 μm) and P. aeruginosa (thickness 27 ± 1 μm). This supports observations in Fig. 3 that demonstrate the presence of high metabolic activity and coexistence of both pathogens in FBS in the mixed-species biofilm state.
On the other hand, in the IVWM, the mixed-species biofilm showed a distinct predominance of P. aeruginosa (average thickness 29 ± 3 μm), with the presence of S. aureus observed to be sparsely scattered in the P. aeruginosa biomass (Fig. 5A and B). The IVWM contains additional host factors (as compared with FBS) that could possibly counter the protective effect of P. aeruginosa killing of S. aureus, or the effect could be mediated by the presence of factors such as lactoferrin known to exhibit antimicrobial activity against S. aureus biofilms [49,116,[131], [132], [133]]. These observations correspond to that seen in Fig. 3B, where P. aeruginosa is seen to outnumber S. aureus in the mixed-species biofilm formed in the IVWM.
It is important to note that the wells were not rinsed prior to imaging, that the observed biomass (Fig. 4, Fig. 5) represents not only biofilm, but also attached or unattached single bacterial cells within, and on top of, the dense bacterial mats. This is evident in the lateral (side) views in Fig. 4, Fig. 5, where fluorescence signal scattered across the surface of the biofilm structure likely depicts unattached cells. Further, in the inset images in Fig. 5, the red and green pixels represent likely both single cells and clusters of cells, depending on the biofilm density.
Overall, our results indicate that the IVWM supports the formation of dense, mat-like biofilms of P. aeruginosa and S. aureus when grown alone, and under mixed-species conditions, the biofilm shows a distinct predominance of P. aeruginosa. This is similar to in vivo conditions where in spite of the well-established co-existence of the two pathogens, P. aeruginosa appears to outcompete S. aureus [17,19].
Conclusions
Based on previous reports of clinical wound fluid composition, we have developed an in vitro wound milieu (IVWM), consisting of fetal bovine serum, and additional host matrix and biochemical factors. Our results indicate that the IVWM recapitulates key in vivo biofilm features such as biomass formation, metabolic activity, interspecies coexistence and interactions, antibiotic tolerance and three-dimensional structure. Notably, under both planktonic and biofilm states, the IVWM supports a distinct predominance of P. aeruginosa under mixed-species conditions. This is important to explore further, particularly under clinical and in vivo conditions, given that P. aeruginosa-S. aureus interactions have been largely studied in in vitro systems [13,18,130,131]. Notably, we find that this is distinct from that observed in serum alone, underscoring the importance of developing and studying biofilms in composite wound-like media conditions.
While S. aureus and P. aeruginosa are two of the most important bacterial pathogens implicated in wound biofilms, an important next step to this work would be to leverage the IVWM to evaluate the structure, function and interspecies interactions of biofilms formed by a diverse range of wound pathogens. In the wound bed, bacterial species such as Enterococcus, Proteus, Streptococcus, Citrobacter, Morganella, Propionibacterium and Corynebacterium, as well as fungal species such as Candida albicans, Candida parapsilosis, Malasezzia restricta and Curvularia lunata are also known to contribute to the infected wound state [134]. In addition to different pathogenic microbial species, the impact of IVWM on biofilm features and interactions across clinical isolates and commensal microbes could also be explored.
Further, while the IVWM developed in this study recapitulates key factors in the wound state, it is certainly not representative of all aspects of the wound milieu. However, its ease of formulation, use of widely-available components, and compatibility with standard biofilm assays lends itself well for further adaptations and modifications, such as the inclusion of additional factors such as glucose and matrix-metalloproteinases [[135], [136], [137], [138], [139]]. Finally, given the chemical composition of the milieu, examining biofilm formation, features, structure and susceptibility under wound-relevant microaerophilic or anaerobic conditions, would provide interesting insights.
Taken together, the IVWM holds potential as a tractable approach to study wound biofilms under host-relevant conditions, particularly for high-throughput applications such as screening of novel and combination antimicrobial treatments. In doing so, it could bridge the gap between reductionist in vitro systems and complex in vivo models, and provide host-relevant insights in laboratory biofilm studies.
Funding
This study was funded by the Ramalingaswami Re-entry Fellowship (BT/HRD/35/02/2006 to KSK) and Har Gobind Khorana-Innovative Young Biotechnologist Award (BT/12/IYBA/2019/05 to KSK).
Preprint available
This manuscript is submitted as a preprint to bioRxiv https://doi.org/10.1101/2021.01.07.425734.
CRediT authorship contribution statement
Snehal Kadam: Conceptualization, Methodology, Investigation, Validation, Formal analysis, Data curation, Visualization, Writing – original draft, Writing – review & editing. Vandana Madhusoodhanan: Conceptualization, Methodology, Investigation, Validation. Radhika Dhekane: Investigation. Devyani Bhide: Investigation, Validation. Rutuja Ugale: Investigation. Utkarsha Tikhole: Investigation. Karishma S. Kaushik: Conceptualization, Methodology, Formal analysis, Project administration, Supervision, Writing – original draft, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Drs. Derek Fleming and Kendra Rumbaugh (Texas Tech University Health Science Center, Lubbock) for the bacterial strains, Dr. Avinash Sharma (National Center for Microbial Research, NCCS, Pune) for erythromycin, and Dr. Amit Agarwal for 96-well black plates. We acknowledge Sujaya Ingle, NCL-Innovation Park, for technical assistance with confocal microscopy and Dr. Kaspar Kragh for inputs on the tile-scan approach. We also thank Drs. Ameeta Ravikumar, Shadab Ahmed, Tuli Dey and Geetanjali Tomar (Institute of Bioinformatics and Biotechnology, SPPU) for use of certain laboratory equipment.
References
- 1.Gonzalez A.C.D.O., Andrade Z.D.A., Costa T.F., Medrado A.R.A.P. Wound healing - a literature review. An Bras Dermatol. 2016;91:614–620. doi: 10.1590/abd1806-4841.20164741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo S., DiPietro L.A. Factors affecting wound healing. J Dent Res. 2010;89:219–229. doi: 10.1177/0022034509359125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Junker J.P.E., Caterson E.J., Eriksson E. The microenvironment of wound healing. J Craniofac Surg. 2013;24:12–16. doi: 10.1097/SCS.0b013e31827104fb. [DOI] [PubMed] [Google Scholar]
- 4.Uluer E.T., Vatansever H.S., Kurt F.Ő. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2017. Wound healing and microenvironment; pp. 67–77. Wound heal. [DOI] [Google Scholar]
- 5.Manuela B., Milad K., Anna-Lena S., Julian-Dario R., Ewa Klara S. Acute and chronic wound fluid inversely influence wound healing in an in-vitro 3D wound model. J Tissue Repair Regen. 2017;1:1. doi: 10.14302/issn.2640-6403.jtrr-17-1818. [DOI] [Google Scholar]
- 6.Drinkwater S.L., Smith A., Burnand K.G. What can wound fluids tell us about the venous ulcer microenvironment? Int J Low Extrem Wounds. 2002;1:184–190. doi: 10.1177/153473460200100307. [DOI] [PubMed] [Google Scholar]
- 7.Staiano-Coico L., Higgins P., Schwartz S.B., Zimm A., Goncalves J. Wound fluids: a reflection of the state of healing. Ostomy/Wound Manag. 2000 Jan;46(1A Suppl):85S–93S. quiz 94S-95S. [PubMed] [Google Scholar]
- 8.Schneider L.A., Korber A., Grabbe S., Dissemond J. Influence of pH on wound-healing: a new perspective for wound-therapy? Arch Dermatol Res. 2007;298:413–420. doi: 10.1007/s00403-006-0713-x. [DOI] [PubMed] [Google Scholar]
- 9.Edwards R., Harding K.G. Bacteria and wound healing. Curr Opin Infect Dis. 2004 Apr;17(2):91–96. doi: 10.1097/00001432-200404000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Percival S.L., McCarty S.M., Lipsky B. Biofilms and wounds: an overview of the evidence. Adv Wound Care. 2015;4:373–381. doi: 10.1089/wound.2014.0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper R.A., Bjarnsholt T., Alhede M. Biofilms in wounds: a review of present knowledge. J Wound Care. 2014;23:570–582. doi: 10.12968/jowc.2014.23.11.570. [DOI] [PubMed] [Google Scholar]
- 12.Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013;121:1–51. doi: 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- 13.Cendra M. del M., Blanco-Cabra N., Pedraz L., Torrents E. Optimal environmental and culture conditions allow the in vitro coexistence of Pseudomonas aeruginosa and Staphylococcus aureus in stable biofilms. Sci Rep. 2019;9 doi: 10.1038/s41598-019-52726-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLeon S., Clinton A., Fowler H., Everett J., Horswill A.R., Rumbaugh K.P. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun. 2014;82:4718–4728. doi: 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fazli M., Bjarnsholt T., Kirketerp-Møller K., Jørgensen B., Andersen A.S., Krogfelt K.A. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J Clin Microbiol. 2009 Dec;47(12):4084–4089. doi: 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phalak P., Chen J., Carlson R.P., Henson M.A., Phalak P., Chen J. Metabolic modeling of a chronic wound biofilm consortium predicts spatial partitioning of bacterial species. BMC Syst Biol. 2016 Sep 7;10(1):90. doi: 10.1186/s12918-016-0334-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hotterbeekx A., Kumar-Singh S., Goossens H., Malhotra-Kumar S. In vivo and in vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front Cell Infect Microbiol. 2017;7:106. doi: 10.3389/fcimb.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wijesinghe G., Dilhari A., Gayani B., Kottegoda N., Samaranayake L., Weerasekera M. Influence of laboratory culture media on in vitro growth, adhesion, and biofilm formation of Pseudomonas aeruginosa and Staphylococcus aureus. Med Princ Pract. 2019;28:28–35. doi: 10.1159/000494757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pastar I., Nusbaum A.G., Gil J., Patel S.B., Chen J., Valdes J. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PloS One. 2013;8 doi: 10.1371/journal.pone.0056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slade E.A., Thorn R.M.S., Young A., Reynolds D.M. An in vitro collagen perfusion wound biofilm model; with applications for antimicrobial studies and microbial metabolomics. BMC Microbiol. 2019;19:310. doi: 10.1186/s12866-019-1682-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werthén M., Henriksson L., Jensen P.Ø., Sternberg C., Givskov M., Bjarnsholt T. An in vitro model of bacterial infections in wounds and other soft tissues. APMIS. 2010;118:156–164. doi: 10.1111/j.1600-0463.2009.02580.x. [DOI] [PubMed] [Google Scholar]
- 22.Bowler P.G., Jones S.A., Walker M., Parsons D. Microbicidal properties of a silver-containing Hydrofiber® dressing against a variety of burn wound pathogens. J Burn Care Rehabil. 2004;25:192–196. doi: 10.1097/01.BCR.0000112331.72232.1B. [DOI] [PubMed] [Google Scholar]
- 23.Said J., Dodoo C.C., Walker M., Parsons D., Stapleton P., Beezer A.E. An in vitro test of the efficacy of silver-containing wound dressings against Staphylococcus aureus and Pseudomonas aeruginosa in simulated wound fluid. Int J Pharm. 2014;462:123–128. doi: 10.1016/j.ijpharm.2013.12.037. [DOI] [PubMed] [Google Scholar]
- 24.Leonhard M., Zatorska B., Moser D., Tan Y., Schneider-Stickler B. Evaluation of combined growth media for in vitro cultivation of oropharyngeal biofilms on prosthetic silicone. J Mater Sci Mater Med. 2018;29:45. doi: 10.1007/s10856-018-6051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price B.L., Lovering A.M., Bowling F.L., Dobson C.B. Development of a novel collagen wound model to simulate the activity and distribution of antimicrobials in soft tissue during diabetic foot infection. Antimicrob Agents Chemother. 2016;60:6880–6889. doi: 10.1128/AAC.01064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scalise A., Bianchi A., Tartaglione C., Bolletta E., Pierangeli M., Torresetti M. Microenvironment and microbiology of skin wounds: the role of bacterial biofilms and related factors. Semin Vasc Surg. 2015;28:151–159. doi: 10.1053/j.semvascsurg.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y., Dowd S.E., Smith E., Rhoads D.D., Wolcott R.D. In vitro multispecies Lubbock chronic wound biofilm model. Wound Repair Regen. 2008;16:805–813. doi: 10.1111/j.1524-475X.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- 28.Thaarup I.C., Bjarnsholt T. Current in vitro biofilm-infected chronic wound models for developing new treatment possibilities. Adv Wound Care. 2021;10:91–102. doi: 10.1089/wound.2020.1176. [DOI] [PubMed] [Google Scholar]
- 29.Trengove N.J., Langton S.R., Stacey M.C. Biochemical analysis of wound fluid from nonhealing and healing chronic leg ulcers. Wound Repair Regen. 1996;4:234–239. doi: 10.1046/j.1524-475X.1996.40211.x. [DOI] [PubMed] [Google Scholar]
- 30.Cutting K.F. Wound exudate: composition and functions. Br J Community Nurs. 2003;8:S4–S9. doi: 10.12968/bjcn.2003.8.sup3.11577. [DOI] [PubMed] [Google Scholar]
- 31.Alves P.M., Al-Badi E., Withycombe C., Jones P.M., Purdy K.J., Maddocks S.E. Interaction between Staphylococcus aureus and Pseudomonas aeruginosa is beneficial for colonisation and pathogenicity in a mixed biofilm. Pathog Dis. 2018;76:fty003. doi: 10.1093/femspd/fty003. [DOI] [PubMed] [Google Scholar]
- 32.Irie Y., Borlee B.R., O’Connor J.R., Hill P.J., Harwood C.S., Wozniak D.J. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2012;109 doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malone C.L., Boles B.R., Lauderdale K.J., Thoendel M., Kavanaugh J.S., Horswill A.R. Fluorescent reporters for Staphylococcus aureus. J Microbiol Methods. 2009;77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stauffer E., Dolan J.A., Newman R. Elsevier; 2008. Chemistry and physics of fire and liquid fuels. Fire debris anal; pp. 85–129. [DOI] [Google Scholar]
- 35.Kaushik K.S., Stolhandske J., Shindell O., Smyth H.D., Gordon V.D. Tobramycin and bicarbonate synergise to kill planktonic Pseudomonas aeruginosa, but antagonise to promote biofilm Survival. Npj Biofilms Microbiomes. 2016;2:16006. doi: 10.1038/npjbiofilms.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kragh K.N., Alhede M., Kvich L., Bjarnsholt T. Into the well—a close look at the complex structures of a microtiter biofilm and the crystal violet assay. Biofilms. 2019;1:100006. doi: 10.1016/j.bioflm.2019.100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White R., Cutting K.F. World Wide Wounds; 2006. Modern exudate management : a review of wound treatments. [Google Scholar]
- 38.Karayiannakis A.J., Zbar A., Polychronidis A., Simopoulos C. Serum and drainage fluid vascular endothelial growth factor levels in early surgical wounds. Eur Surg Res. 2003;35:492–496. doi: 10.1159/000073388. [DOI] [PubMed] [Google Scholar]
- 39.Brackman G., Coenye T. In vitro and in vivo biofilm wound models and their application. Adv Exp Med Biol. 2016;15–32 doi: 10.1007/5584_2015_5002. [DOI] [PubMed] [Google Scholar]
- 40.James T.J., Hughes M.A., Cherry G.W., Taylor R.P. Simple biochemical markers to assess chronic wounds. Wound Repair Regen. 2000;8:264–269. doi: 10.1046/j.1524-475X.2000.00264.x. [DOI] [PubMed] [Google Scholar]
- 41.Hunt T.K., Conolly W.B., Aronson S.B., Goldstein P. Anaerobic metabolism and wound healing: an hypothesis for the initiation and cessation of collagen synthesis in wounds. Am J Surg. 1978;135:328–332. doi: 10.1016/0002-9610(78)90061-2. [DOI] [PubMed] [Google Scholar]
- 42.Löffler M., Zieker D., Weinreich J., Löb S., Königsrainer I., Symons S. Wound fluid lactate concentration: a helpful marker for diagnosing soft-tissue infection in diabetic foot ulcers? Preliminary findings. Diabet Med. 2011;28:175–178. doi: 10.1111/j.1464-5491.2010.03123.x. [DOI] [PubMed] [Google Scholar]
- 43.Britland S., Ross-Smith O., Jamil H., Smith A.G., Vowden K., Vowden P. The lactate conundrum in wound healing: clinical and experimental findings indicate the requirement for a rapid point-of-care diagnostic. Biotechnol Prog. 2012;28:917–924. doi: 10.1002/btpr.1561. [DOI] [PubMed] [Google Scholar]
- 44.Birgens H.S. Lactoferrin in plasma measured by an ELISA technique: evidence that plasma lactoferrin is an indicator of neutrophil turnover and bone marrow activity in acute leukaemia. Scand J Haematol. 1985;34:326–331. doi: 10.1111/j.1600-0609.1985.tb00757.x. [DOI] [PubMed] [Google Scholar]
- 45.Bennett R.M., Kokocinski T. Lactoferrin content of peripheral blood cells. Br J Haematol. 1978;39:509–521. doi: 10.1111/j.1365-2141.1978.tb03620.x. [DOI] [PubMed] [Google Scholar]
- 46.Venge P., Foucard T., Henriksen J., Håkansson L., Kreuger A. Serum-levels of lactoferrin, lysozyme and myeloperoxidase in normal, infection-prone and leukemic children. Clin Chim Acta. 1984;136:121–130. doi: 10.1016/0009-8981(84)90283-3. [DOI] [PubMed] [Google Scholar]
- 47.Malmquist J., Thorell J.I., Wollheim F.A. Lactoferrin and lysozyme in arthritic exudates. Acta Med Scand. 1977;202:313–318. doi: 10.1111/j.0954-6820.1977.tb16834.x. [DOI] [PubMed] [Google Scholar]
- 48.Takayama Y. Springer Netherlands; 2013. Lactoferrin and its role in wound healing. [DOI] [Google Scholar]
- 49.Ammons M.C., Copié V. Lactoferrin: a bioinspired, anti-biofilm therapeutic. Biofouling. 2013;29:443–455. doi: 10.1080/08927014.2013.773317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quintieri L., Caputo L., Monaci L., Cavalluzzi M.M., Denora N. Lactoferrin-derived peptides as a control strategy against skinborne staphylococcal biofilms. Biomedicines. 2020;8:323. doi: 10.3390/biomedicines8090323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh P.K., Parsek M.R., Greenberg E.P., Welsh M.J. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 52.Tang L., Wu J.J., Ma Q., Cui T., Andreopoulos F.M., Gil J. Human lactoferrin stimulates skin keratinocyte function and wound re-epithelialization. Br J Dermatol. 2010;163:38–47. doi: 10.1111/j.1365-2133.2010.09748.x. [DOI] [PubMed] [Google Scholar]
- 53.Chen Z.J., Yang J.P., Wu B.M., Tawil B. A novel three-dimensional wound healing model. J Dev Biol. 2014;2:198–209. doi: 10.3390/jdb2040198. [DOI] [Google Scholar]
- 54.Ko Y.P., Flick M.J. Fibrinogen is at the interface of host defense and pathogen virulence in Staphylococcus aureus infection. Semin Thromb Hemost. 2016;42:408–421. doi: 10.1055/s-0036-1579635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clark R.A.F., An J.Q., Greiling D., Khan A., Schwarzbauer J.E. Fibroblast migration on fibronectin requires three distinct functional domains. J Invest Dermatol. 2003;121:695–705. doi: 10.1046/j.1523-1747.2003.12484.x. [DOI] [PubMed] [Google Scholar]
- 56.Kaur J., Jain A. StatPearls [Internet] StatPearls Publishing; Treasure Island (FL): 2021 Jan. Fibrinogen. 2020 May 23. PMID: 30725869. [Google Scholar]
- 57.To W.S., Midwood K.S. Plasma and cellular fibronectin: distinct and independent functions during tissue repair. Fibrogenesis Tissue Repair. 2011;4:1–17. doi: 10.1186/1755-1536-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bruhn H.D., Heimburger N. Factor-VIII-Related antigen and cold-insoluble globulin in leukemias and carcinomas. Pathophysiol Haemostasis Thrombosis. 1976;5:189–192. doi: 10.1159/000214134. [DOI] [PubMed] [Google Scholar]
- 59.Ai S., Kuzuya M., Iguchi A. Neutrophil elastase in pressure ulcer fluid degrades fibronectin in the exudates. Geriatr Gerontol Int. 2004;4:141–145. doi: 10.1111/j.1447-0594.2004.00243.x. [DOI] [Google Scholar]
- 60.Hayman E.G., Ruoslahti E. Distribution of fetal bovine serum fibronectin and endogenous rat cell fibronectin in extracellular matrix. J Cell Biol. 1979;83(1):255–259. doi: 10.1083/jcb.83.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wysocki A. Wound fluids and the pathogenesis of chronic wounds. J WOCN. 1996;23:283–290. doi: 10.1016/s1071-5754(96)90047-9. [DOI] [PubMed] [Google Scholar]
- 62.Wysocki A.B., Grinnell F. Fibronectin profiles in normal and chronic wound fluid. Lab Invest. 1990 Dec;63(6):825–831. [PubMed] [Google Scholar]
- 63.Kadam S., Nadkarni S., Lele J., Sakhalkar S., Mokashi P., Kaushik K.S. Bioengineered platforms for chronic wound infection studies: how can we make them more human-relevant? Front Bioeng Biotechnol. 2019;7 doi: 10.3389/fbioe.2019.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Birkenhauer E., Neethirajan S., Weese J.S. Collagen and hyaluronan at wound sites influence early polymicrobial biofilm adhesive events. BMC Microbiol. 2014;14:191. doi: 10.1186/1471-2180-14-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Velnar T., Bailey T., Smrkolj V. vol. 37. 2009. (The wound healing process: an overview of the cellular and molecular mechanisms). [DOI] [PubMed] [Google Scholar]
- 66.Tarlton J.F., Bailey A.J., Crawford E., Jones D., Moore K., Harding K.D. Prognostic value of markers of collagen remodeling in venous ulcers. Wound Repair Regen. 1999;7:347–355. doi: 10.1046/j.1524-475X.1999.00347.x. [DOI] [PubMed] [Google Scholar]
- 67.Metcalf D.G., Haalboom M., Bowler P.G., Gamerith C., Sigl E., Heinzle A. Elevated wound fluid pH correlates with increased risk of wound infection. Wound Med. 2019;26:100166. doi: 10.1016/j.wndm.2019.100166. [DOI] [Google Scholar]
- 68.Kaushik K.S., Ratnayeke N., Katira P., Gordon V.D. The spatial profiles and metabolic capabilities of microbial populations impact the growth of antibiotic-resistant mutants. J R Soc Interface. 2015;12:20150018. doi: 10.1098/rsif.2015.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rumbaugh K., Watters C., Yuan T. Beneficial and deleterious bacterial-host interactions in chronic wound pathophysiology. Chron Wound Care Manag Res. 2015;2:53. doi: 10.2147/CWCMR.S60317. [DOI] [Google Scholar]
- 70.Shukla V.K., Shukla D., Tiwary S.K., Agrawal S., Rastogi A. Evaluation of pH measurement as a method of wound assessment. J Wound Care. 2007;16:291–294. doi: 10.12968/jowc.2007.16.7.27062. [DOI] [PubMed] [Google Scholar]
- 71.She P., Wang Y., Liu Y., Tan F., Chen L., Luo Z. Effects of exogenous glucose on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. Microbiologyopen. 2019;8 doi: 10.1002/mbo3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seidl K., Goerke C., Wolz C., Mack D., Berger-Bächi B., Bischoff M. Staphylococcus aureus CcpA affects biofilm formation. Infect Immun. 2008;76:2044–2050. doi: 10.1128/IAI.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X., Thomsen T.R., Winkler H., Xu Y. Influence of biofilm growth age, media, antibiotic concentration and exposure time on Staphylococcus aureus and Pseudomonas aeruginosa biofilm removal in vitro. BMC Microbiol. 2020;20:264. doi: 10.1186/s12866-020-01947-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gries C.M., Biddle T., Bose J.L., Kielian T., Lo D.D. Staphylococcus aureus fibronectin binding protein a mediates biofilm development and infection. Infect Immun. 2020;88 doi: 10.1128/IAI.00859-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crabbé A., Liu Y., Matthijs N., Rigole P., De La Fuente-Nùñez C., Davis R. Antimicrobial efficacy against Pseudomonas aeruginosa biofilm formation in a three-dimensional lung epithelial model and the influence of fetal bovine serum. Sci Rep. 2017;7 doi: 10.1038/srep43321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith A.C., Rice A., Sutton B., Gabrilska R., Wessel A.K., Whiteley M. Albumin inhibits Pseudomonas aeruginosa quorum sensing and alters polymicrobial interactions. Infect Immun. 2017;85 doi: 10.1128/IAI.00116-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hammond A., Dertien J., Colmer-Hamood J.A., Griswold J.A., Hamood A.N. Serum inhibits P. aeruginosa biofilm formation on plastic surfaces and intravenous catheters. J Surg Res. 2010;159:735–746. doi: 10.1016/j.jss.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 78.Thompson K.M., Abraham N., Jefferson K.K. Staphylococcus aureus extracellular adherence protein contributes to biofilm formation in the presence of serum. FEMS Microbiol Lett. 2010;305:143–147. doi: 10.1111/j.1574-6968.2010.01918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yin S., Jiang B., Huang G., Gong Y., You B., Yang Z. Burn serum increases Staphylococcus aureus biofilm formation via oxidative stress. Front Microbiol. 2017;8:1191. doi: 10.3389/fmicb.2017.01191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abraham N.M., Jefferson K.K. A low molecular weight component of serum inhibits biofilm formation in staphylococcus aureus. Microb Pathog. 2010;49:388–391. doi: 10.1016/j.micpath.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Langevin P.B., Gravenstein N., Doyle T.J., Roberts S.A., Skinner S., Langevin S.O. Growth of Staphylococcus aureus in diprivan and intralipid: implications on the pathogenesis of infections. Anesthesiology. 1999;91:1394–1400. doi: 10.1097/00000542-199911000-00032. [DOI] [PubMed] [Google Scholar]
- 82.LaBauve A.E., Wargo M.J. Growth and laboratory maintenance of Pseudomonas aeruginosa. Curr Protoc Microbiol. 2012 doi: 10.1002/9780471729259.mc06e01s25. 0 6:Unit; PMID: 22549165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harriott M.M., Noverr M.C. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother. 2009;53:3914–3922. doi: 10.1128/AAC.00657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ehrenkranz N.J., Elllott D.F., Zarco R. 1971. Serum bacteriostasis of Staphylococcus aureus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Korgaonkar A., Trivedi U., Rumbaugh K.P., Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A. 2013;110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dalton T., Dowd S.E., Wolcott R.D., Sun Y., Watters C., Griswold J.A. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PloS One. 2011;6 doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Filkins L.M., Graber J.A., Olson D.G., Dolben E.L., Lynd L.R., Bhuju S. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol. 2015;197:2252–2264. doi: 10.1128/JB.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Di Martino P. Effects of antibiotics on adherence of Pseudomonas aeruginosa and Pseudomonas fluorescens to human fibronectin. Chemotherapy. 2001;47:344–349. doi: 10.1159/000048541. [DOI] [PubMed] [Google Scholar]
- 89.Gagnière H., Di Martino P. Effects of antibiotics on Pseudomonas aeruginosa NK125502 and Pseudomonas fluorescens MF0 biofilm formation on immobilized fibronectin. J Chemother. 2004;16:244–247. doi: 10.1179/joc.2004.16.3.244. [DOI] [PubMed] [Google Scholar]
- 90.Rebiere-Huët J., Di Martino P., Hulen C. Inhibition of Pseudomonas aeruginosa adhesion to fibronectin by PA-IL and monosaccharides: involvement of a lectin-like process. Can J Microbiol. 2004;50:303–312. doi: 10.1139/w04-015. [DOI] [PubMed] [Google Scholar]
- 91.Kwiecinski J., Kahlmeter G., Jin T. Biofilm formation by staphylococcus aureus isolates from skin and soft tissue infections. Curr Microbiol. 2015;70:698–703. doi: 10.1007/s00284-014-0770-x. [DOI] [PubMed] [Google Scholar]
- 92.Cardile A.P., Sanchez C.J., Samberg M.E., Romano D.R., Hardy S.K., Wenke J.C. Human plasma enhances the expression of Staphylococcal microbial surface components recognizing adhesive matrix molecules promoting biofilm formation and increases antimicrobial tolerance in Vitro. BMC Res Notes. 2014;7:457. doi: 10.1186/1756-0500-7-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mccourt J., O’Halloran D.P., Mccarthy H., O’Gara J.P., Geoghegan J.A. Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol Lett. 2014;353:157–164. doi: 10.1111/1574-6968.12424. [DOI] [PubMed] [Google Scholar]
- 94.Herrmann M., Vaudaux P.E., Pittet D., Auckenthaler R., Lew P.D., Perdreau F.S. Fibronectin, fibrinogen, and laminin act as mediators of adherence of clinical staphylococcal isolates to foreign. Material. J Infect Dis. 1988;158:693–701. doi: 10.1093/infdis/158.4.693. [DOI] [PubMed] [Google Scholar]
- 95.Akiyama H., Ueda M., Kanzaki H., Tada J., Arata Jirô. Biofilm formation of Staphylococcus aureus strains isolated from impetigo and furuncle: role of fibrinogen and fibrin. J Dermatol Sci. 1997;16:2–10. doi: 10.1016/S0923-1811(97)00611-7. [DOI] [PubMed] [Google Scholar]
- 96.Herman-Bausier P., El-Kirat-Chatel S., Foster T.J., Geoghegan J.A., Dufrêne Y.F. Staphylococcus aureus fibronectin-binding protein a mediates cell- cell adhesion through low-affinity homophilic bonds. mBio. 2015;6:1–10. doi: 10.1128/mBio.00413-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Millenbaugh N., Watters C., Burton T., Kirui D. Enzymatic degradation of in vitro Staphylococcus aureus biofilms supplemented with human plasma. Infect Drug Resist. 2016;9:71. doi: 10.2147/IDR.S103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Proctor R.A., Mosherj D.F., Olbrantzj P.J. vol. 257. 1982. (Fibronectin binding to Staphylococcus aureus). [PubMed] [Google Scholar]
- 99.Magalhães A.P., Jorge P., Pereira M.O. Pseudomonas aeruginosa and Staphylococcus aureus communication in biofilm infections: insights through network and database construction. Crit Rev Microbiol. 2019;45:712–728. doi: 10.1080/1040841X.2019.1700209. [DOI] [PubMed] [Google Scholar]
- 100.Beaudoin T., Yau Y.C.W., Stapleton P.J., Gong Y., Wang P.W., Guttman D.S. Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. Npj Biofilms Microbiomes. 2017;3 doi: 10.1038/s41522-017-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Armbruster C.R., Wolter D.J., Mishra M., Hayden H.S., Radey M.C., Merrihew G. Staphylococcus aureus protein a mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa. mBio. 2016;7 doi: 10.1128/mBio.00538-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Benov L. Effect of growth media on the MTT colorimetric assay in bacteria. PloS One. 2019;14 doi: 10.1371/journal.pone.0219713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Funk D., Schrenk H.H., Frei E. Serum albumin leads to false-positive results in the XTT and the MTT assay. Biotechniques. 2007 Aug;43(2) doi: 10.2144/000112528. 178, 180, 182. [DOI] [PubMed] [Google Scholar]
- 104.Li J., Xie S., Ahmed S., Wang F., Gu Y., Zhang C. Antimicrobial activity and resistance: influencing factors. Front Pharmacol. 2017;8:364. doi: 10.3389/fphar.2017.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morrison J.M., Chojnacki M., Fadrowski J.J., Bauza C., Dunman P.M., Dudas R.A. Serum-Associated antibiotic tolerance in pediatric clinical isolates of Pseudomonas aeruginosa. J Pediatric Infect Dis Soc. 2019;XX:1–9. doi: 10.1093/jpids/piz094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Blanchard C., Barnett P., Perlmutter J., Dunman P.M. Identification of acinetobacter baumannii serum-associated antibiotic efflux pump inhibitors. Antimicrob Agents Chemother. 2014;58:6360–6370. doi: 10.1128/AAC.03535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Raza M., Wei Y., Jiang Y., Ahmad A., Raza S., Ullah S. Molecular mechanism of tobramycin with human serum albumin for probing binding interactions: multi-spectroscopic and computational approaches. New J Chem. 2017;41:8203–8213. doi: 10.1039/c7nj02054f. [DOI] [Google Scholar]
- 108.Stokes J.M., Lopatkin A.J., Lobritz M.A., Collins J.J. Bacterial metabolism and antibiotic efficacy. Cell Metabol. 2019;30:251–259. doi: 10.1016/j.cmet.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mottola C., Matias C.S., Mendes J.J., Melo-Cristino J., Tavares L., Cavaco-Silva P. Susceptibility patterns of Staphylococcus aureus biofilms in diabetic foot infections. BMC Microbiol. 2016;16 doi: 10.1186/s12866-016-0737-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Antunes A.L.S., Bonfanti J.W., Perez L.R.R., Pinto C.C.F., de Freitas A.L.P., Macedo A.J. High vancomycin resistance among biofilms produced by Staphylococcus species isolated from central venous catheters. Mem Inst Oswaldo Cruz. 2011;106:51–55. doi: 10.1590/S0074-02762011000100008. [DOI] [PubMed] [Google Scholar]
- 111.Chuard C. Decreased susceptibility to antibiotic killing of a stable small colony variant of Staphylococcus aureus in fluid phase and on fibronectin- coated surfaces. J Antimicrob Chemother. 1997;39:603–608. doi: 10.1093/jac/39.5.603. [DOI] [PubMed] [Google Scholar]
- 112.Aguila A., Herrera A.G., Morrison D., Cosgrove B., Perojo A., Montesinos I. Bacteriostatic activity of human lactoferrin against Staphylococcus aureus is a function of its iron-binding properties and is not influenced by antibiotic resistance. FEMS Immunol Med Microbiol. 2001;31:145–152. doi: 10.1111/j.1574-695x.2001.tb00511.x. [DOI] [PubMed] [Google Scholar]
- 113.Jenssen H., Hancock R.E.W. Antimicrobial properties of lactoferrin. Biochimie. 2009;91:19–29. doi: 10.1016/j.biochi.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 114.Sinha M., Kaushik S., Kaur P., Sharma S., Singh T.P. Antimicrobial lactoferrin peptides: the hidden players in the protective function of a multifunctional protein. Int J Pept. 2013;2013 doi: 10.1155/2013/390230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stallmann H.P., Faber C., Bronckers A.L.J.J., De Blieck-Hogervorst J.M.A., Brouwer C.P.J.M., Amerongen A.V.N. Histatin and lactoferrin derived peptides: antimicrobial properties and effects on mammalian cells. Peptides. 2005;26:2355–2359. doi: 10.1016/j.peptides.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 116.Flores-Villaseñor H., Canizalez-Román A., Reyes-Lopez M., Nazmi K., De La Garza M., Zazueta-Beltrán J. Bactericidal effect of bovine lactoferrin, LFcin, LFampin and LFchimera on antibiotic-resistant Staphylococcus aureus and Escherichia coli. Biometals. 2010;23:569–578. doi: 10.1007/s10534-010-9306-4. Biometals. [DOI] [PubMed] [Google Scholar]
- 117.Davis S.C., Ricotti C., Cazzaniga A., Welsh E., Eaglstein W.H., Mertz P.M. Microscopic and physiologic evidence for biofilm-associated wound colonization in vivo. Wound Repair Regen. 2008;16:23–29. doi: 10.1111/j.1524-475X.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- 118.Watters C., Deleon K., Trivedi U., Griswold J.A., Lyte M., Hampel K.J. Pseudomonas aeruginosa biofilms perturb wound resolution and antibiotic tolerance in diabetic mice. Med Microbiol Immunol. 2013;202:131–141. doi: 10.1007/s00430-012-0277-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gross M., Cramton S.E., Götz F., Götz G., Peschel A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun. 2001;69:3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kuhn D.M., Chandra J., Mukherjee P.K., Ghannoum M.A. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect Immun. 2002 Feb;70(2):878–888. doi: 10.1128/IAI.70.2.878-888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Alonso B., Lucio J., Pérez-Granda M.J., Cruces R., Sánchez-Carrillo C., Bouza E. Does biomass production correlate with metabolic activity in Staphylococcus aureus? J Microbiol Methods. 2016;131:110–112. doi: 10.1016/j.mimet.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 122.Xu Z., Liang Y., Lin S., Chen D., Li B., Li L. Crystal violet and XTT assays on Staphylococcus aureus biofilm quantification. Curr Microbiol. 2016 Oct;73(4):474–482. doi: 10.1007/s00284-016-1081-1. [DOI] [PubMed] [Google Scholar]
- 123.Marcos-Zambrano L.J., Escribano P., Bouza E., Guinea J. Production of biofilm by Candida and non-Candida spp. isolates causing fungemia: comparison of biomass production and metabolic activity and development of cut-off points. Int J Med Microbiol. 2014 Nov;304(8):1192–1198. doi: 10.1016/j.ijmm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 124.Hall C.W., Mah T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 125.Høiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 126.Lebeaux D., Ghigo J.-M., Beloin C. Biofilm-Related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev. 2014;78:510–543. doi: 10.1128/mmbr.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tewari A., Jain B., Dhamannapatil P.S., Saxena M.K. vol. 14. 2018. (Biofilm resistance to antimicrobial agents and novel approaches to combat biofilm mediated resistance in bacteria). [Google Scholar]
- 128.Batoni G., Maisetta G., Esin S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim Biophys Acta Biomembr. 2016;1858:1044–1060. doi: 10.1016/j.bbamem.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 129.Sharma D., Misba L., Khan A.U. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Contr. 2019;8:1–10. doi: 10.1186/s13756-019-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tognon M., Köhler T., Luscher A., Van Delden C. Transcriptional profiling of Pseudomonas aeruginosa and Staphylococcus aureus during in vitro co-culture. BMC Genom. 2019;20:30. doi: 10.1186/s12864-018-5398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gounani Z., Şen Karaman D., Venu A.P., Cheng F., Rosenholm J.M. Coculture of P. aeruginosa and S. aureus on cell derived matrix - an in vitro model of biofilms in infected wounds. J Microbiol Methods. 2020;175:105994. doi: 10.1016/j.mimet.2020.105994. [DOI] [PubMed] [Google Scholar]
- 132.Bruni N., Capucchio M.T., Biasibetti E., Pessione E., Cirrincione S., Giraudo L. Antimicrobial activity of lactoferrin-related peptides and applications in human and veterinary medicine. Molecules. 2016;21:752. doi: 10.3390/molecules21060752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nibbering P.H., Ravensbergen E., Welling M.M., Van Berkel L.A., Van Berkel P.H.C., Pauwels E.K.J. Human lactoferrin and peptides derived from its N terminus are highly effective against infections with antibiotic-resistant bacteria. Infect Immun. 2001;69:1469–1476. doi: 10.1128/IAI.69.3.1469-1476.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bowler P.G., Duerden B.I., Armstrong D.G. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev. 2001;14:244–269. doi: 10.1128/CMR.14.2.244-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Armstrong D.G., Jude E.B. The role of matrix metalloproteinases in wound healing. J Am Podiatr Med Assoc. 2002;92:12–18. doi: 10.7547/87507315-92-1-12. [DOI] [PubMed] [Google Scholar]
- 136.Ravanti L., Kähäri V.M. Matrix metalloproteinases in wound repair (review) Int J Mol Med. 2000;6:391–407. [PubMed] [Google Scholar]
- 137.Ayuk S.M., Abrahamse H., Houreld N.N. The role of matrix metalloproteinases in diabetic wound healing in relation to photobiomodulation. J Diabetes Res. 2016;2016:2897656. doi: 10.1155/2016/2897656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brem H., Tomic-Canic M. Cellular and molecular basis of wound healing in diabetes. J Clin Invest. 2007 May;117(5):1219–1222. doi: 10.1172/JCI32169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tomic-Canic M., Burgess J.L., O’Neill K.E., Strbo N., Pastar I. Skin microbiota and its interplay with wound healing. Am J Clin Dermatol. 2020;21:36–43. doi: 10.1007/s40257-020-00536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]