Abstract
Burkholderia glumae and B. gladioli are seed-borne rice pathogens that cause bacterial panicle blight (BPB) disease, resulting in huge rice yield losses worldwide. However, the excessive use of chemical pesticides in agriculture has led to an increase in environmental toxicity. Microbe-mediated nanoparticles (NPs) have recently gained significant attention owing to their promising application in plant disease control. In the current study, we biologically synthesize zinc oxide nanoparticles (ZnONPs) from a native Bacillus cereus RNT6 strain, which was taxonomically identified using 16S rRNA gene analysis. The biosynthesis of ZnONPs in the reaction mixture was confirmed by using UV–Vis spectroscopy. Moreover, XRD, FTIR, SEM-EDS, and TEM analysis revealed the functional groups, crystalline nature, and spherical shape of ZnONPs with sizes ranging from 21 to 35 nm, respectively. Biogenic ZnONPs showed significant antibacterial activity at 50 µg mL−1 against B. glumae and B. gladioli with a 2.83 cm and 2.18 cm zone of inhibition, respectively, while cell numbers (measured by OD600) of the two pathogens in broth culture were reduced by 71.2% and 68.1%, respectively. The ultrastructure studies revealed the morphological damage in ZnONPs-treated B. glumae and B. gladioli cells as compared to the corresponding control. The results of this study revealed that ZnONPs could be considered as promising nanopesticides to control BPB disease in rice.
Keywords: antibacterial activity, biosynthesis, nanopesticides, rice pathogen, ZnONPs
1. Introduction
Rice (Oryza sativa L.) is an important cereal crop consumed by more than half of the world’s population and is a good source of proteins, carbohydrates, vitamins, and minerals. Almost 85% of rice is cultivated in Asia, whereby China is the world’s leading producer of rice [1]. The global food security challenge in the modern world is continuously increasing due to the constant increase in the world’s population, which is expected to be 9 billion by 2050 [2]. Over the last few decades, rice crops have been threatened by different bacterial phytopathogens, and plant diseases are limiting crop yields across the world. The most important rice bacterial pathogens, Burkholderia glumae and B. gladioli, are deemed to be the leading threat to rice cultivation that cause bacterial panicle blight (BPB) disease, which poses a severe threat to rice production [3,4].
The rice BPB disease was first reported in Japan; however, it is currently spread in most rice-producing countries, including China, the United States, Malaysia, Sri Lanka, Thailand, South Korea, and Panama [5,6]. The BPB has severely reduced yields of infected rice fields as it causes severe damage, e.g., floret sterility, grain abortion, and milling quality reduction [7,8,9]. It is widely accepted that the rice yield losses associated with BPB disease cannot be mitigated with applications of chemical pesticides [10]. The lack of effective alternative control for BPB may be due to the increased bacterial resistance, high genetic diversity, and climatic conditions in the current global warming scenario [11]. There is an urgent need to develop unique techniques for managing the rice BPB disease caused by B. glumae and B. gladioli.
In recent years, nanotechnology has emerged as a promising research area with a wide range of applications in the agriculture sector, especially in plant disease management [12,13,14]. Moreover, NPs are low-cost, efficient alternatives to parent materials with high reaction rates, superior efficiency, and a large surface/volume ratio [15,16]. In the past few years, NPs have been synthesized through physical and chemical methods, which had less biocompatibility, higher production rates, required the use of hazardous chemicals, and had high energy requirements [17]. In the previous literature, the use of zinc oxide nanoparticles (ZnONPs) as an antibacterial agent is a well-known effective approach and has received considerable attention in plant disease management due to their unique physico-chemical properties [18,19,20,21]. Moreover, numerous studies have reported that ZnONPs significantly increase plant growth and inhibit the growth of broad-spectrum pathogens [22,23]. An earlier study reported that green ZnONPs significantly inhibited the growth of rice pathogens Xanthomonas oryzae pv. oryzae [24]. Similarly, Rajabairavi et al. [25] revealed the antibacterial effect of microbe-mediated ZnONPs against two bacterial pathogens, Pseudomonas aeruginosa and Enterobacter aerogens. Conversely, some studies reported the adverse effects of chemically produced ZnONPs on plant soil microbial communities [26,27]. Another study performed by Zhou et al. [28] showed the adverse impact of chemically produced ZnONPs on soil microorganism biomass and rac-metalaxyl transformation. However, microbe-based synthesis of ZnONPs has enormous potential benefits in terms of long-term stability, nontoxicity, eco-friendliness, and ease of scaling-up as compared to those NPs fabricated via chemical and physical methods [22]. Nevertheless, the ameliorative effect of green ZnONPs against B. glumae and B. gladioli is not being investigated. Furthermore, a literature review is available for the potential application of biogenic ZnONPs as a nanopesticide. To the best of our knowledge, this is the first report of biogenic ZnONPs as nanopesticides against rice pathogens B. glumae and B. gladioli.
The objectives of the current study are to synthesize the biogenic ZnONPs by using a culture supernatant of native B. cereus strain RNT6, to evaluate their antibacterial activity against rice bacterial panicle blight disease pathogens B. glumae and B. gladioli, and to observe the antagonistic cellular interactions of ZnONPs with rice pathogens by electron microscopy analysis.
2. Materials and Methods
2.1. Collection of Bacterial Strains
The bacterial strain B. cereus RNT6 was isolated (source of biogenic ZnONPs) from soil collected from rice fields of Zhejiang University, Hangzhou, China, by using the dilution plate method [29]. In brief, 1 g of soil samples were added to test tubes containing 9 mL of sterilized saline solution (0.85% NaCl, w/v) and mixing of suspension was performed through vortex mixture (Vortex QL-861). Afterward, the 10−4 and 10−6 dilutions were spread on the nutrient agar (NA) media plates and incubated at 30 ± 2 °C for 24 h. The bacterial isolates were purified by repeated streaking for maximum purity. In this experiment, bacterial phytopathogens B. glumae and B. gladioli that cause bacterial panicle blight disease were obtained from the laboratory collection of the Institute of Biotechnology, Zhejiang University, Zhejiang, China.
2.2. Taxonomic Identification RNT6 Strain
The genomic DNA of B. cereus RNT6 was isolated using the CTAB method and quantified through Nano Drop™ 2000/2000c (Thermo-Fisher Scientific, Waltham, MA, USA). The amplification of the 16S rRNA gene was carried out with the universal primer pair (fD1 and rD1) according to the method of Weisburg et al. [30]. The amplicon was sequenced commercially from Tsingke, Beijing, China, by Sanger-sequencing technique. The 16S rRNA gene sequence of bacterial isolate RNT6 was compared with different databases for similarity search using BLASTn server at National Center for Biotechnology Information (NCBI), RDP database and EzBioCloud server. The phylogenetic tree of B. cereus RNT6 was formed with the MEGA 7.0 software package through the maximum likelihood method.
2.3. Extracellular Biosynthesis of ZnONPs
The RNT6 strain was grown in 250 mL Erlenmeyer flasks containing 100 mL of nutrient broth (NB) at 30 ± 2 °C for 24 h. After the incubation period, the bacterial culture of strain RNT6 was centrifuged at 5000× g for 15 min, and the collected supernatant was used for ZnONPs synthesis according to the method [31] with slight modifications. For the green synthesis of ZnONPs, 100 mL supernatant of B. cereus RNT6 was mixed with an equal amount of 0.1 M ZnSO4⋅7H2O in 250 mL Erlenmeyer flasks and heated on a water bath up to 80 °C for 15 min. A white precipitate starts to appear at the bottom of the flask, indicating the formation of ZnONPs. Consequently, the ZnONPs were collected by centrifuged at 15,000× g for 10 min and washed twice with distilled water and followed by ethanol to remove the remaining Zn2+ on the surface to obtain purified ZnONPs. The collected ZnONPs were then freeze-dried using Alpha 1-2 LDplus 101521 (Fisher Scientific, Waltham, MA, USA).
2.4. Characterization of ZnONPs
The ZnONPs were characterized by a UV-Vis spectrophotometer (Shimadzu-Kyoto, Japan) at a 250–800 nm wavelength. The size distribution of biogenic ZnONPs in water suspension with three different concentrations (10, 25, and 50 mg mL−1) were analyzed using a Malvern Zetasizer (Nano ZS90, Malvern, UK) according to Ahmed et al. [32]. The crystalline nature of green ZnONPs was confirmed by using XRD analysis as described by Mahdi et al. [33]. The XRD diffraction of the ZnONPs was characterized by X-ray diffractometer (Siemens-D5000, Germany). Fourier transforms infrared (FTIR) analysis was carried out according to Hossain et al. [34] to identify functional groups present in the ZnONPs. The dried sample of ZnONPs was used for the FTIR spectrum (Vector22, Bruker, Germany) in the range of 4000–500 cm−1. The shape, size and surface morphology of ZnONPs were observed by transmission electron microscopy (TEM) (JEM 1230-JEOL, Akishima, Japan) and scanning electron microscopy (SEM) (SU8010, Hitachi, Japan). In addition, energy dispersive spectroscopy (EDS) (Oxford Instruments, UK) was used for the conformation of elemental composition of the green ZnONPs.
2.5. In Vitro Antibacterial Activity of ZnONPs
The antibacterial activity of ZnONPs against rice the pathogenic bacteria B. glumae and B. gladioli was estimated through agar well diffusion according to Ibrahim et al. [35]. Briefly, 40 µL of ZnONPs with different suspensions (10, 25, and 50 μg mL−1) was added to a 5-mm-diameter well on Luria-Bertani (LB) agar plates; uniform swabbing was performed by bacterial culture (108 CFU mL−1) and incubated at 30 ± 2 °C for 24 h. The diameter of the inhibition zone was calculated edge to edge across the center of disk.
Likewise, the antibacterial activity of ZnONPs against the two rice pathogenic bacteria was determined in liquid broth by measuring the optical density at 600 nm in 96-well microtiter plates (Corning, NY, USA) as per the method of Hossain et al. [36]. In general, 20 µL of pathogenic bacterial culture (108 CFU mL−1) was added to 200 µL of LB broth containing different concentrations of ZnONPs (10, 25, and 50 μg mL−1) and incubated at 30 ± 2 °C for 24 h. The wells treated with 20 µL of B. glumae and B. gladioli culture without ZnONPs was used as control. Afterward, the bactericidal effect of ZnONPs on bacterial growth in the LB broth was analyzed by using a scanning microplate spectrophotometer (Thermo-Fisher Scientific, Waltham, MA, USA) at the wavelength of 600 nm.
2.6. Biofilm Inhibition Assay
The inhibition of B. glumae and B. gladioli biofilm by biogenic ZnONPs was measured in 96-well microtiter plates as previously described by Ogunyemi et al. [24]. The microtiter plate wells were filled with 200 mL of LB broth containing 100 mL overnight bacterial culture (108 CFU mL−1) and ZnONPs at different suspensions (10, 25, and 50 μg mL−1) and incubated at 30 ± 2 °C for 24 h. Bacterial culture without ZnONPs was used as the control. Afterward, cultures were smoothly discarded and washed with sterile distilled water to remove the loosely attached cells and air-dried. Afterward, 100 μL of 1% (w/v) aqueous crystal violet solution was added for staining and kept for 30 min at room temperature. The samples intensity was found out by using a microplate spectrophotometer (Thermo-Fisher Scientific, Waltham, MA, USA) at awavelength of 570 nm after the dye was dissolved by the addition of 100 mL of 33% acetic acid.
2.7. Live/Dead Cell Staining
The live/dead cell staining was used to observe the membrane damage in bacterial cells by using Invitrogen, BacLight bacterial viability kit (Thermo-Fisher Scientific, Waltham, MA, USA), according to Radzig et al. [37]. Briefly, the bacterial culture (108 CFU mL−1) was centrifuged at 8000× g for 5 min and the supernatant was discarded. Afterward, the bacterial cells were suspended in a 50 μg mL−1 concentration of ZnONPs at 30 ± 2 °C for 8 h. The live bacterial cells without ZnONPs were used as the control. Afterward, the staining reagent mixture, a red fluorescent propidium iodide (PI) stain and a green fluorescent (SYTO 9) stain, was added to the reaction mixture and incubated in the dark at room temperature for 15 min. The fluorescence emission of bacteria was assessed by means of scanning confocal laser microscopy (SCLM) (Leica-SP8, Heidelberg, Germany).
2.8. Flow Cytometry Observation
Flow cytometry was used to detect, identify, and count dead bacterial cells after incubation with the ZnONPs according to the method of Cai et al. [38]. The bacterial culture (108 CFU mL−1) was centrifuged at 8000× g for 5 min, and pellets were treated 50 μg mL−1 concentration of ZnONPs at 30 ± 2 °C for 8 h. Afterward, bacterial cells were stained with red fluorescent PI stain in the dark for 20 min, andyhe cellular damage of B. glumae and B. gladioli cells was detected by using flow cytometry (Gallios-Beckman Coulter, Krefeld, Germany).
2.9. Determination of the Reactive Oxygen Species
ROS production in rice pathogenic bacteria B. glumae and B. gladioli was assessed by using dichlorofluorescein-diacetate (DCFH-DA) (Sigma-Aldrich, Saint Louis, MO, USA) as previously described by the method of Ogunyemi et al. [22]. In brief, 1 mL overnight culture of bacteria (108 CFU mL−1) was centrifuged at 8000× g for 5 min. Afterward, the bacterial pellets were treated with 50 μg mL−1 concentration of ZnONPs at 30 ± 2 °C for 8 h. The nanoparticle-treated samples were gently rinsed thrice with sterilized distilled water; 10 mM DCFH-DA was added to the reaction mixture and incubated at room temperature for 10 min in a dark condition, and fluorescence was detected by using the scanning confocal laser microscopy (SCLM, Leica SP8, Germany).
2.10. Morphological Observation with Electron Microscopy
The morphological damage of B. glumae and B. gladioli cells in relation to the ZnONPs were observed by using SEM and TEM analysis referring to Cheng et al. [39]. Briefly, 1 mL overnight culture of bacteria (108 CFU mL−1) was centrifuged at 8000× g for 5 min. Afterward, the bacterial pellets were suspended at (50 μg mL−1) concentration of ZnONPs at 30 ± 2 °C for 8 h. The bacterial cells without ZnONPs were used as a control. After incubation, the bacteria were washed twice with PBS and fixed in 2.5% (v/v) glutaraldehyde in 0.1 M PBS overnight. The samples were fixed with (1% w/v, osmium tetroxide) for one hour at room temperature, and then specimens were dehydrated for 15 min in a series of gradients ethanol concentration (30 to 100%). Finally, the samples of B. glumae and B. gladioli cells were observed by using TEM (JEM1230, JEOL, Akishima, Japan) and SEM (TM1000, Hitachi, Japan) analysis.
2.11. Statistical Analysis
All data reported in this study are the means of four replicates (n = 4 ± standard deviations). The experimental data were analyzed with Statistic (version 8.1) software, and the means were compared by least significant difference (Fisher’s LSD) at the probability level (p ≤ 0.05) according to Steel and Torrie [40].
3. Results and Discussion
3.1. Molecular Identification Phylogenetic Analysis of RNT6 Strain
The sequence similarity searching and phylogenetic analyses revealed the taxonomic identity of the RNT6 strain up to species level, and the name “Bacillus cereus RNT6 (MT173798)” was assigned to the strain. The 16S rRNA gene sequence of RNT6 strain displayed similarity with B. cereus XS 24-5 (MT000038; 99.86%), B. cereus ATCC 14579T (AE016877; 99.44%), and B. cereus B005 (DQ289984) when searching for sequence homologues using BLASTn of NCBI, EzBioCloud, and RDP servers, respectively. The identity of the RNT6 strain was further evidenced by a phylogenetic tree, where B. cereus RNT6 (MT173798) shared the branch nodes with B. cereus XS 24-5 (MT000038), and B. cereus ATCC 14579T (AE016877) and Escherichia coli U 5/41 (NR 024570) appeared as an outgroup (Figure 1).
Figure 1.
Phylogenetic analysis of B. cereus RNT6 with the type strains and closest Genbank matches of genus Bacillus.
3.2. Biosynthesis and Characterization of ZnONPs
In the current study, the extracellular biosynthesis of ZnONPs was synthesized by using the B. cereus RNT6. The maximum precipitate clustered in the bottom of the 100 mL flask was observed at a 0.1 M ZnSO4⋅7H2O concentration. These results are in agreement with the study of Malaikozhundan et al. [41], who reported the biosynthesis of ZnONPs by using B. thuringiensis as a capping and stabilizing agent. Moreover, the reduction of Zn2+ to ZnONPs was observed through UV–Vis spectroscopy of the reaction mixture in the range of 300–700 nm. The biosynthesis of ZnONPs in reaction mixtures was confirmed through the UV-Vis characteristic peak measured at 382 nm (Figure 2). The results of UV–Vis analysis were consistent with Yusof et al. [42], who reported an absorption peak at 351 nm for ZnONPs synthesized using Lactobacillus plantarum TA4. The zeta potential is an important parameter that used to measure the average size and stability of NP dispersions in liquid phase. In the present study, the zeta potential and hydrodynamic diameter of three different concentrations of biogenic ZnONPs are given in Table 1. The zeta potential value of NPs > 25 mV usually has a high degree of stability [43]. The higher stability of biogenic ZnONPs has been suggested as being due to their smaller size, specific surface area and shape, and functional groups of bacteria, which did not exist in the chemically synthesized NPs. However, the biogenic ZnONPs size increased in the liquid medium as compared to the solid state, which is consistent with previously reported studies [44,45].
Figure 2.
The UV–Vis spectra of reaction mixture containing stabilized biogenic zinc oxide nanoparticles (ZnONPs).
Table 1.
Zeta potential (mV) and size of three different concentration of biogenic ZnONP suspensions in water.
| ZnONPs Concentrations | Zeta Potential (mV) | Size (nm) |
|---|---|---|
| 10 mg mL−1 | 29.5 ± 0.7 | 120.1 ± 6.7 |
| 25 mg mL−1 | 30.5 ± 1.1 | 111.9 ± 8.8 |
| 50 mg mL−1 | 31.5 ± 0.9 | 135.3 ± 5.3 |
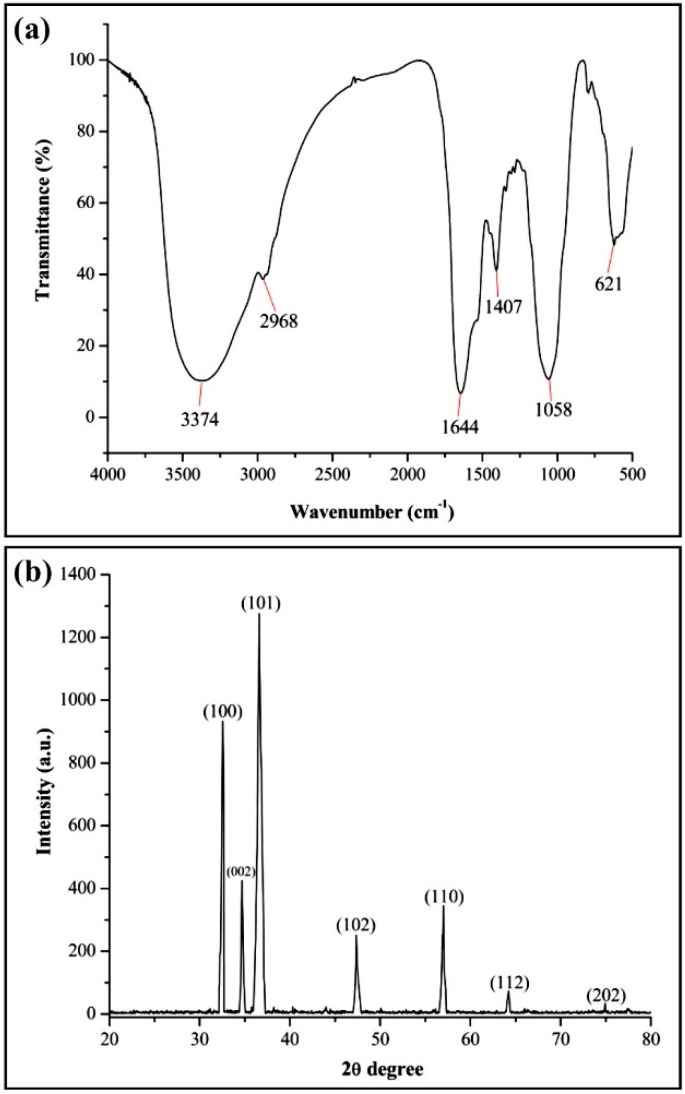
The FTIR analysis was used to reveal the occurrence of different functional groups that are accountable for the long-term stabilization of ZnONPs [46]. In addition, these functional groups also demonstrated the presence of macromolecules including proteins, sugars, lipids, nucleic acids, and carbohydrates that ensure the stability of biogenic ZnONPs [47]. In this study, the FTIR spectra of biogenic ZnONPs presented different absorption peaks at 3374, 2968, 1644, 1407, and 1058 cm−1 including a weak peak at 621 cm−1 (Figure 3). The peak at 3374 cm−1 represents the hydroxyl (O-H) group of alcohol and the peak at 2968 cm−1 was related to the C-H stretching of alkane group. The consistent peaks at 1644 and 1407 cm−1 were due to the C=N stretching of imine/oxime group and S=O stretching of sulfate group, respectively. The absorption peak at 1058 cm−1 revealed the presence of the S=O stretching of sulfoxide group. Our results are supported by the findings of Selvarajan and Mohanasrinivasan [31], who synthesized ZnONPs by Lactobacillus plantarum VITES07.
Figure 3.
Characterization of biogenic ZnONPs synthesized from B. cereus RNT6. (a) FTIR spectra, (b) XRD analysis.
The XRD data of biogenic ZnONPs revealed the typical diffraction peaks at 32.50º, 34.74°, 36.69°, 47.33°, 56.90°, 64.32°, and 74.90°, which corresponded to (100), (002), (101), (102) (110) (112), and (202) diffraction planes of ZnONPs, respectively (Figure 3b). These XRD spectra peaks revealed the crystalline structure of the biogenic ZnONPs [48]. A similar XRD spectrum was described by Mahdi et al. [33], who produced green ZnONPs by utilizing two bacterial strains, namely, Bacillus sp. PTCC 1538 and Lactococcus lactis. Hence, the size, shape, and surface morphology of ZnONPs were observed through electron microscopy analysis [24]. The SEM and TEM analysis showed that ZnONPs have spherical shapes with the size ranging from 21 to 35 nm (Figure 4). Similarly, Mashrai et al. [49] biosynthesized spherical shape ZnONPs with a particle size of 20 nm using Candida albicans as eco-friendly reducing and capping agent. Furthermore, the EDS analysis revealed the presence of Zinc (57.05%), oxygen (30.30%), phosphorus (10.90%) silicon (0.39%), sulfur (0.83%), and aluminum (0.52%) in biogenic ZnONPs (Figure 4c). Our EDS results corroborate with the study of Ogunyemi et al. [22], who observed the elemental percentage, e.g., zinc (80.35%) and oxygen (19.65%) of ZnONPs.
Figure 4.
Characterization of biogenic ZnONPs through SEM-EDS and TEM analysis to observe the shape, size, and elemental composition of nanoparticles (NPs) (scale bar = 200 nm). (a) SEM analysis of ZnONPs, (b) TEM analysis of ZnONPs, and (c) EDS analysis of ZnONPs.
3.3. In Vitro Antibacterial Activity
The growth of rice bacterial pathogens, B. glumae and B. gladioli, was inhibited significantly after the application of biogenic ZnONPs as compared to the control treatment. In the literature, various studies reported that ZnONPs effectively controlled the bacterial phytopathogens [34,50,51]. In this study, we described for the first time the antibacterial potential of biogenic ZnONPs against B. glumae and B. gladioli, which causes rice bacterial panicle blight disease. Furthermore, the zone of inhibition diameters were found to be (2.38 ± 0.15, 2.65 ± 0.05, and 2.83 ± 0.08 cm) at three different concentrations of ZnONPs (10, 25, and 50 µg mL−1), respectively, against B. glumae, whereas, for B. gladioli, ZnONPs revealed the diameters of zone of inhibition as 1.65 ± 0.05 cm at a 10 µg mL−1 concentration, 1.85 ± 0.13 cm at a 25 µg mL−1 concentration, and 2.18 ± 0.10 mm at a 50 µg mL−1 concentration. The maximum concentration, 50 µg mL−1, of ZnONPs demonstrated the maximum zone of inhibition diameter (Figure 5 and Table 2). Likewise, the findings of antibacterial activity in liquid broth indicated that biogenic ZnONPs significantly suppressed the growth of B. glumae and B. gladioli. The three different concentrations of ZnONPs suspension (10, 25, and 50 µg mL−1) caused (60.0, 66.2, and 71.2%, respectively,) reduction in the OD600 value of B. glumae and (52.1, 63.7, and 68.1%, respectively) reduction in the OD600 value of B. gladioli (Figure 5 and Table 2). Our results are constant with previous study of Jayaseelan et al. [46], who observed the growth inhibition of the bacterial pathogen Pseudomonas aeruginosa in solid and liquid medium by ZnONPs. Similarly, Ogunyemi et al. [22] reported the in vitro antibacterial activity (zone of inhibition and MIC at OD600) of ZnONPs against rice bacterial pathogen, while a comparatively greater inhibition effect of biogenic ZnONPs against B. glumae and B. gladioli was observed in our study as compared with the study of Ogunyemi et al. [22]. In another recent study, Yusof et al. [42] synthesized ZnONPs by using the supernatant of Lactobacillus plantaruma and observed their antibacterial activity against bacterial pathogens at 1000 μg mL−1, a high dose of ZnONPs, as compared with our study. Similarly, Venkataraju et al. [52] described the antibacterial activity of chemically produced ZnONPs (10 mg mL−1) against bacterial pathogens Pseudomonas aeruginosa and Escherichia coli; however, a comparatively high dose of ZnONPs was used as compared to the current study.
Figure 5.
In vitro antibacterial activity of biogenic ZnONPs at three different concentrations (10, 25, and 50 µg mL−1) against B. glumae and B. gladioli. (a) The zone of inhibition was observed by well diffusion assay, (b) the inhibition of B. glumae and B. gladioli growth in liquid medium, (c) the inhibition of biofilm formation.
Table 2.
In vitro antibacterial activity of biogenic ZnONPs against rice pathogens against B. glumae and B. gladioli.
| ZnONPs Concentration |
Zone of Inhibition (cm) |
Bacterial Growth (OD600) |
Biofilm Formation (OD570) |
|---|---|---|---|
| B. glumae | |||
| Control | 0.00 ± 0.00 | 0.80 ± 0.04 | 1.18 ± 0.01 |
| 10 µg mL−1 | 2.38 ± 0.15 ** | 0.32 ± 0.02 ** | 0.51 ± 0.01 ** |
| 25 µg mL−1 | 2.65 ± 0.05 ** | 0.27 ± 0.01 ** | 0.50 ± 0.01 ** |
| 50 µg mL−1 | 2.83 ± 0.08 ** | 0.23 ± 0.01 ** | 0.41 ± 0.01 ** |
| B. gladioli | |||
| Control | 0.00 ± 0.00 | 0.69 ± 0.02 | 0.98 ± 0.01 |
| 10 µg mL−1 | 1.65 ± 0.05 ** | 0.33 ± 0.04 ** | 0.50 ± 0.02 ** |
| 25 µg mL−1 | 1.85 ± 0.13 ** | 0.25 ± 0.02 ** | 0.48 ± 0.04 ** |
| 50 µg mL−1 | 2.18 ± 0.10 ** | 0.22 ± 0.01 ** | 0.40 ± 0.02 ** |
Data are presented as the mean of four replicates (n = 4 ± SE). ** Significant at p ≤ 0.05.
3.4. Inhibition of Biofilm Formation
Biofilm formation is a one of the major mechanisms of bacterial pathogens to adapt to various stresses and become a part of infections [53,54]. Moreover, biofilm formation most probably protects the bacterial pathogens from the immune attacks and also contribute to the survival of bacterial pathogens during the saprophytic life and latent infections [55,56]. In this study, we demonstrate that the biogenic ZnONPs significantly reduced the biofilm formation as compared to control treatment. To check the inhibition of biofilm formation, we considered three different suspensions (10, 25, and 50 µg mL−1) of ZnONPs. Our results revealed a 56.7, 57.6, and 65.2% reduction in the biofilm formation of B. glumae and a 48.9, 51.0, and 59.1% reduction of B. gladioli at 10, 25, and 50 µg mL−1 of ZnONPs, respectively (Table 2). All readings were taken at optimum OD570. Moreover, the purple biofilm circle can be clearly seen on the test tube wall, as shown in Figure 5. Our results are similar to previous studies regarding biofilm inhibition toward bacterial phytopathogens by using different nanoparticles [35,57]. Similarly, Lee et al. [58] reported the inhibition of biofilm production of Pseudomonas aeruginosa by using ZnONPs.
3.5. Live/Dead Cell Staining
The results of this study demonstrated the antibacterial mechanism of ZnONPs against B. glumae and B. gladioli after application of biogenic ZnONPs. In agreement with the previous studies, ZnONPs directly changed the cellular mechanism, including cell membrane permeability, the transport of electrons, respiration, and osmoregulation, due to attachment of Zn2+ ions with a negatively charged bacterial cell surface that are responsible for the release of intracellular content [18,19]. Here, the staining results of the non-treated control in live bacteria evidently exhibited intact membranes that could be induced from the green fluoresce (Figure 6). Hence, the red fluorescence significantly increased after being treated with a 50 μg mL−1 concentration of ZnONPs at 30 ± 2 °C for 8 h compared with the control treatment. Overall, the results of live/dead cell staining revealed that the ZnONPs had a strong bactericidal effect against rice pathogens. Similarly, Cheng et al. [39] observed the red fluorescence in phytopathogen Ralstonia solanacearum cells after treatment by green silver NPs by using live/dead cell staining.
Figure 6.
Antibacterial potential of biogenic ZnONPs against B. glumae and B. gladioli cells after 8 h treatment with (50 µg mL−1) and without (control) biogenic ZnONPs. (a) Live/dead cell staining, with green fluorescence representing the live bacteria, while red fluorescence presents dead bacteria (scale bar = 20 μm]. (b) Formation of reactive oxygen species (ROS) in B. glumae and B. gladioli cells. (c) Flow cytometry images of B. glumae and B. gladioli cells.
3.6. ROS Production and Flow Cytometry Observation
ROS play an important role as cell signaling molecules for many biological processes. However, the generation of ROS can also damage cellular organelles, which can disrupt normal cellular functions [59,60]. In an earlier report, Premanathan et al. [61] reported the induction of damage in bacterial pathogens, Pseudomonas aeruginosa and Escherichia coli, due to the generation of lipid peroxidation. Inspired by previous studies, we hypothesized that ZnONPs could produce ROS that interrupted the cell membrane and infiltrated the bacterial cells, which eventually leads to the leakage of genetic material resulting in B. glumae and B. gladioli cell death. In the present study, the fluorescence intensity of B. glumae and B. gladioli significantly increased after treatment with 50 μg mL−1 ZnONPs as compared with the non-treated control (Figure 6). Similarly, Ogunyemi et al. [22] have found that three metal oxide nanoparticles (ZnO, MgO, and MnO2) can produce ROS in the rice bacterial pathogen, Xanthomonas oryzae pv. oryzae, which causes cell damage, degeneration of cell walls and cell membranes, and ultimately leads to bacterial death. Overall, our findings indicated that the antibacterial effects of ZnONPs on pathogenic bacteria B. glumae and B. gladioli are partially attributed to the production of ROS. A flow cytometry test was conducted to analyze the number of dead bacteria cells to make a quick and accurate evaluation of bacterial damage. The results of this study demonstrated that biogenic ZnONPs at a 50 μg mL−1 suspension significantly increased the cellular damage of B. glumae and B. gladioli (viz., 29.9 and 29.6%), respectively, while the bacterial death ratio of the negative control (without ZnONPs) was found to be 5.52 and 5.61%, respectively (Figure 6). Likewise, Abdallah, et al. [23] observed the death of bacterial cell after treatment with green chitosan and zinc oxide NPs by using flow cytometry analysis.
3.7. Effect of ZnONPs on Bacterial Morphology
The antibacterial potential of biogenic ZnONPs has gained tremendous attention due to their unique properties such as their ability to damage cell walls, the interrupted production of ATP, ribosomal damage, the blockage of cell transport, replication, and the leakage of genetic material [18]. Here, SEM and TEM analysis showed the ultrastructural changes in bacterial cells. After treatment with ZnONPs, the SEM micrograph showed severely disrupted and shrinkage structure of B. glumae and B. gladioli as compared with the control treatment (Figure 7). Correspondingly, TEM analysis showed that biogenic ZnONPs highly damaged cell membrane, ribosome, proteins, and cytoplasmic material that cause the leakage of nucleic acid contents, which eventually leads to the death of both pytopathogens (Figure 7). In a previous study, Ogunyemi et al. [24] observed the ruptured and collapsed cell membrane with the leakage of the cytoplasm material after treatment with plan-tmediated ZnONPs. Overall, the results of our study revealed that direct interaction between biogenic ZnONPs and B. glumae and B. gladioli caused the leakage of nucleic acid contents that results in bacterial death.
Figure 7.
Electron microscopic SEM and TEM images of rice bacterial pathogens B. glumae and B. gladioli cells after 8 h treatment with (50 µg mL−1) and without (control) biogenic ZnONPs. (a) SEM images reveal a highly ruptured structure and small cellular holes; (b) TEM images show integrated bacterial membranes, disorganized cytoplasm, and leakage of bacterial genetic material.
4. Conclusions
In the present study, we reported the potential antibacterial activity of biogenic ZnONPs against B. glumae and B. gladioli for the first time. Biogenic ZnONPs were synthesized by using B. cereus RNT6 strain and characterized with standard material characterization techniques. We confirmed the spherical shapes with the size ranging from 21 to 35 nm and stabilized through coating proteins in the bacterial supernatant. The biogenic ZnONPs demonstrated potential antibacterial activity against B. glumae and B. gladioli, which was revealed in terms of inhibitory zone, decrease in bacterial growth, and biofilm formation. Furthermore, ultrastructure studies revealed a wide variation in cell wall morphology and the large amount of damage to the structure of bacterial phytopathogens treated with ZnONPs (50 μg mL−1). Overall, the results of the current study revealed that green ZnONPs have a tendency to protect rice plants as bactericidal agents and can be utilize to produce nanopesticide formulations. However, more research insights into field conditions are needed to better understand the interactions of ZnONPs with rice plants.
Author Contributions
Conceptualization, T.A. and B.L.; methodology, T.A., M.N. and Z.W.; software, H.J. and Z.W.; validation, J.L., M.S. and H.J.; formal analysis, T.A.; investigation, K.S.A., F.A. and I.M.; resources, J.L.; data curation, K.S.A., F.A. and M.N.; writing—original draft preparation, T.A.; writing—review and editing, K.S.A., F.A. and B.L.; visualization, K.S.A., F.A. and M.S.; supervision, B.L.; project administration, B.L.; funding acquisition, B.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work is financially supported by Shanghai-Agriculture Applied Technology Development Program (2019-02-08-00-08-F01150), National Natural Science Foundation of China (32072472, 31872017) Zhejiang Provincial Natural Science Foundation of China (LZ19C140002), Zhejiang Provincial Project (2019C02006, 2017C02002, 2020C02006).
Data Availability Statement
The data presented in this study are available within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ahmed T., Noman M., Shahid M., Shahid M.S., Li B. Antibacterial potential of green magnesium oxide nanoparticles against rice pathogen Acidovorax oryzae. Mater. Lett. 2021;282:128839. doi: 10.1016/j.matlet.2020.128839. [DOI] [Google Scholar]
- 2.Edwards J., Johnson C., Santos-Medellín C., Lurie E., Podishetty N.K., Bhatnagar S., Eisen J.A., Sundaresan V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA. 2015;112:E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nandakumar R., Shahjahan A., Yuan X., Dickstein E., Groth D., Clark C., Cartwright R., Rush M. Burkholderia glumae and B. gladioli cause bacterial panicle blight in rice in the southern United States. Plant Dis. 2009;93:896–905. doi: 10.1094/PDIS-93-9-0896. [DOI] [PubMed] [Google Scholar]
- 4.Ortega L., Rojas C. Bacterial Panicle Blight and Burkholderia glumae: From pathogen biology to disease control. Phytopathology. 2020 doi: 10.1094/PHYTO-09-20-0401-RVW. [DOI] [PubMed] [Google Scholar]
- 5.Pedraza L.A., Bautista J., Uribe-Vélez D. Seed-born Burkholderia glumae infects rice seedling and maintains bacterial population during vegetative and reproductive growth stage. Plant Pathol. J. 2018;34:393. doi: 10.5423/PPJ.OA.02.2018.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shew A.M., Durand-Morat A., Nalley L.L., Zhou X.-G., Rojas C., Thoma G. Warming increases Bacterial Panicle Blight (Burkholderia glumae) occurrences and impacts on USA rice production. PLoS ONE. 2019;14:e0219199. doi: 10.1371/journal.pone.0219199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nandakumar R., Bollich P., Shahjahan A., Groth D., Rush M. Evidence for the soilborne nature of the rice sheath rot and panicle blight pathogen, Burkholderia gladioli 1. Can. J. Plant Pathol. 2008;30:148–154. doi: 10.1080/07060660809507505. [DOI] [Google Scholar]
- 8.Jeong Y., Kim J., Kim S., Kang Y., Nagamatsu T., Hwang I. Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis. 2003;87:890–895. doi: 10.1094/PDIS.2003.87.8.890. [DOI] [PubMed] [Google Scholar]
- 9.Ham J.H., Melanson R.A., Rush M.C. Burkholderia glumae: Next major pathogen of rice? Mol. Plant Pathol. 2011;12:329–339. doi: 10.1111/j.1364-3703.2010.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedraza-Herrera L.A., Bautista J.P., Cruz-Ramírez C.A., Uribe-Vélez D. IBUN2755 Bacillus strain controls seedling root and bacterial panicle blight caused by Burkholderia glumae. Biol. Control. 2021;153:104494. doi: 10.1016/j.biocontrol.2020.104494. [DOI] [Google Scholar]
- 11.Pérez C., Saavedra E. Avances en el manejo integrado de la bacteria Burkholderia glumae en el cultivo de arroz en el Caribe colombiano. Rev. Colomb. Cienc. Anim.-RECIA. 2011:111–124. doi: 10.24188/recia.v3.n1.2011.344. [DOI] [Google Scholar]
- 12.Noman M., Ahmed T., Hussain S., Niazi M.B.K., Shahid M., Song F. Biogenic copper nanoparticles synthesized by using a copper-resistant strain Shigella flexneri SNT22 reduced the translocation of cadmium from soil to wheat plants. J. Hazard. Mater. 2020:123175. doi: 10.1016/j.jhazmat.2020.123175. [DOI] [PubMed] [Google Scholar]
- 13.Noman M., Shahid M., Ahmed T., Tahir M., Naqqash T., Muhammad S., Song F., Abid H.M.A., Aslam Z. Green copper nanoparticles from a native Klebsiella pneumoniae strain alleviated oxidative stress impairment of wheat plants by reducing the chromium bioavailability and increasing the growth. Ecotoxicol. Environ. Saf. 2020;192:110303. doi: 10.1016/j.ecoenv.2020.110303. [DOI] [PubMed] [Google Scholar]
- 14.Abd-Elsalam K.A., Prasad R. Nanobiotechnology Applications in Plant Protection. Springer; Cham, Switzerland: 2018. [Google Scholar]
- 15.Hofmann T., Lowry G.V., Ghoshal S., Tufenkji N., Brambilla D., Dutcher J.R., Gilbertson L.M., Giraldo J.P., Kinsella J.M., Landry M.P. Technology readiness and overcoming barriers to sustainably implement nanotechnology-enabled plant agriculture. Nat. Food. 2020;1:416–425. doi: 10.1038/s43016-020-0110-1. [DOI] [Google Scholar]
- 16.Ahmed T., Shahid M., Noman M., Niazi M.B.K., Zubair M., Almatroudi A., Khurshid M., Tariq F., Mumtaz R., Li B. Bioprospecting a native silver-resistant Bacillus safensis strain for green synthesis and subsequent antibacterial and anticancer activities of silver nanoparticles. J. Adv. Res. 2020;24:475–483. doi: 10.1016/j.jare.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmed T., Noman M., Luo J., Muhammad S., Shahid M., Ali M.A., Zhang M., Li B. Bioengineered chitosan-magnesium nanocomposite: A novel agricultural antimicrobial agent against Acidovorax oryzae and Rhizoctonia solani for sustainable rice production. Int. J. Biol. Macromol. 2020;168:834–845. doi: 10.1016/j.ijbiomac.2020.11.148. [DOI] [PubMed] [Google Scholar]
- 18.Yusof H.M., Mohamad R., Zaidan U.H. Microbial synthesis of zinc oxide nanoparticles and their potential application as an antimicrobial agent and a feed supplement in animal industry: A review. J. Anim. Sci. Biotechnol. 2019;10:57. doi: 10.1186/s40104-019-0368-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sirelkhatim A., Mahmud S., Seeni A., Kaus N.H.M., Ann L.C., Bakhori S.K.M., Hasan H., Mohamad D. Review on zinc oxide nanoparticles: Antibacterial activity and toxicity mechanism. Nano-Micro Lett. 2015;7:219–242. doi: 10.1007/s40820-015-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abebe B., Zereffa E.A., Tadesse A., Murthy H.A. A review on enhancing the antibacterial activity of ZnO: Mechanisms and microscopic investigation. Nanoscale Res. Lett. 2020;15:1–19. doi: 10.1186/s11671-020-03418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudkov S., Burmistrov D., Serov D., Rebezov M., Semenova A., Lisitsyn A. A Mini Review of Antibacterial properties of ZnO nanoparticles. Front. Phys. 2021;9:641481. doi: 10.3389/fphy.2021.641481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogunyemi S.O., Zhang M., Abdallah Y., Ahmed T., Qiu W., Ali M., Yan C., Yang Y., Chen J., Li B. The Bio-Synthesis of Three Metal Oxide Nanoparticles (ZnO, MnO2, and MgO) and Their Antibacterial Activity Against the Bacterial Leaf Blight Pathogen. Front. Microbiol. 2020;11:3099. doi: 10.3389/fmicb.2020.588326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdallah Y., Liu M., Ogunyemi S.O., Ahmed T., Fouad H., Abdelazez A., Yan C., Yang Y., Chen J., Li B. Bioinspired green synthesis of chitosan and zinc oxide nanoparticles with strong antibacterial activity against rice pathogen Xanthomonas oryzae pv. oryzae. Molecules. 2020;25:4795. doi: 10.3390/molecules25204795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogunyemi S.O., Abdallah Y., Zhang M., Fouad H., Hong X., Ibrahim E., Masum M.M.I., Hossain A., Mo J., Li B. Green synthesis of zinc oxide nanoparticles using different plant extracts and their antibacterial activity against Xanthomonas oryzae pv. oryzae. Artif. Cells Nanomed. Biotechnol. 2019;47:341–352. doi: 10.1080/21691401.2018.1557671. [DOI] [PubMed] [Google Scholar]
- 25.Rajabairavi N., Raju C.S., Karthikeyan C., Varutharaju K., Nethaji S., Hameed A.S.H., Shajahan A. Recent Trends in Materials Science and Applications. Springer; Heidelberg, Germany: 2017. Biosynthesis of novel zinc oxide nanoparticles (ZnO NPs) using endophytic bacteria Sphingobacterium thalpophilum; pp. 245–254. [Google Scholar]
- 26.Ge Y., Schimel J.P., Holden P.A. Evidence for negative effects of TiO2 and ZnO nanoparticles on soil bacterial communities. Environ. Sci. Technol. 2011;45:1659–1664. doi: 10.1021/es103040t. [DOI] [PubMed] [Google Scholar]
- 27.Khan S.T. Sustainable Agriculture Reviews 41. Springer; Heidelberg, Germany: 2020. Interaction of Engineered Nanomaterials with Soil Microbiome and Plants: Their Impact on Plant and Soil Health; pp. 181–199. [Google Scholar]
- 28.Zhou Q., Zhang X., Wu Z. Impact of TiO2 and ZnO Nanoparticles on Soil Bacteria and the Enantioselective Transformation of Racemic-Metalaxyl in Agricultural Soil with Lolium perenne: A Wild Greenhouse Cultivation. J. Agric. Food Chem. 2020;68:11242–11252. doi: 10.1021/acs.jafc.0c03959. [DOI] [PubMed] [Google Scholar]
- 29.Somasegaran P., Hoben H.J. Handbook for Rhizobia. Springer; Heidelberg, Germany: 1994. Quantifying the growth of rhizobia; pp. 47–57. [Google Scholar]
- 30.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/JB.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selvarajan E., Mohanasrinivasan V. Biosynthesis and characterization of ZnO nanoparticles using Lactobacillus plantarum VITES07. Mat. Lett. 2013;112:180–182. doi: 10.1016/j.matlet.2013.09.020. [DOI] [Google Scholar]
- 32.Ahmed T., Ren H., Noman M., Shahid M., Liu M., Ali M.A., Zhang J., Tian Y., Qi X., Li B. Green synthesis and characterization of zirconium oxide nanoparticles by using a native Enterobacter sp. and its antifungal activity against bayberry twig blight disease pathogen Pestalotiopsis versicolor. NanoImpact. 2021;21:100281. doi: 10.1016/j.impact.2020.100281. [DOI] [PubMed] [Google Scholar]
- 33.Mahdi Z.S., Talebnia Roshan F., Nikzad M., Ezoji H. Biosynthesis of zinc oxide nanoparticles using bacteria: A study on the characterization and application for electrochemical determination of bisphenol A. Inorg. Nano-Met. Chem. 2020:1–9. doi: 10.1080/24701556.2020.1835962. [DOI] [Google Scholar]
- 34.Hossain A., Abdallah Y., Ali M., Masum M., Islam M., Li B., Sun G., Meng Y., Wang Y., An Q. Lemon-fruit-based green synthesis of zinc oxide nanoparticles and titanium dioxide nanoparticles against soft rot bacterial pathogen Dickeya dadantii. Biomolecules. 2019;9:863. doi: 10.3390/biom9120863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ibrahim E., Fouad H., Zhang M., Zhang Y., Qiu W., Yan C., Li B., Mo J., Chen J. Biosynthesis of silver nanoparticles using endophytic bacteria and their role in inhibition of rice pathogenic bacteria and plant growth promotion. RSC Adv. 2019;9:29293–29299. doi: 10.1039/C9RA04246F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hossain A., Hong X., Ibrahim E., Li B., Sun G., Meng Y., Wang Y., An Q. Green synthesis of silver nanoparticles with culture supernatant of a bacterium Pseudomonas rhodesiae and their antibacterial activity against soft rot pathogen Dickeya dadantii. Molecules. 2019;24:2303. doi: 10.3390/molecules24122303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radzig M., Nadtochenko V., Koksharova O., Kiwi J., Lipasova V., Khmel I. Antibacterial effects of silver nanoparticles on gram-negative bacteria: Influence on the growth and biofilms formation, mechanisms of action. Colloids Surf. B Biointerfaces. 2013;102:300–306. doi: 10.1016/j.colsurfb.2012.07.039. [DOI] [PubMed] [Google Scholar]
- 38.Cai L., Chen J., Liu Z., Wang H., Yang H., Ding W. Magnesium oxide nanoparticles: Effective agricultural antibacterial agent against Ralstonia solanacearum. Front. Microbiol. 2018;9:790. doi: 10.3389/fmicb.2018.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng H.-J., Wang H., Zhang J.-Z. Phytofabrication of silver nanoparticles using three flower extracts and their antibacterial activities against pathogen Ralstonia solanacearum strain YY06 of bacterial wilt. Front. Microbiol. 2020;11:2110. doi: 10.3389/fmicb.2020.02110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steel R.G.D., Torrie J.H. Principles and Procedures of Statistics. McGraw Hill; New York, NJ, USA: 1960. [Google Scholar]
- 41.Malaikozhundan B., Vaseeharan B., Vijayakumar S., Thangaraj M.P. Bacillus thuringiensis coated zinc oxide nanoparticle and its biopesticidal effects on the pulse beetle, Callosobruchus maculatus. J. Photoch. Photobio. B. 2017;174:306–314. doi: 10.1016/j.jphotobiol.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 42.Yusof H.M., Mohamad R., Zaidan U.H., Samsudin A.A. Biosynthesis of zinc oxide nanoparticles by cell-biomass and supernatant of Lactobacillus plantarum TA4 and its antibacterial and biocompatibility properties. Sci. Rep. 2020;10:1–13. doi: 10.1038/s41598-020-76402-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shnoudeh A.J., Hamad I., Abdo R.W., Qadumii L., Jaber A.Y., Surchi H.S., Alkelany S.Z. Synthesis, characterization, and applications of metal nanoparticles. JBNB Elsevier. 2019:527–612. [Google Scholar]
- 44.Khan A.R., Wakeel A., Muhammad N., Liu B., Wu M., Liu Y., Ali I., Zaidi S.H.R., Azhar W., Song G. Involvement of ethylene signaling in zinc oxide nanoparticle-mediated biochemical changes in Arabidopsis thaliana leaves. Environ. Sci. Nano. 2019;6:341–355. [Google Scholar]
- 45.Bian S.-W., Mudunkotuwa I.A., Rupasinghe T., Grassian V.H. Aggregation and dissolution of 4 nm ZnO nanoparticles in aqueous environments: Influence of pH, ionic strength, size, and adsorption of humic acid. Langmuir. 2011;27:6059–6068. doi: 10.1021/la200570n. [DOI] [PubMed] [Google Scholar]
- 46.Jayaseelan C., Rahuman A.A., Kirthi A.V., Marimuthu S., Santhoshkumar T., Bagavan A., Gaurav K., Karthik L., Rao K.B. Novel microbial route to synthesize ZnO nanoparticles using Aeromonas hydrophila and their activity against pathogenic bacteria and fungi. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2012;90:78–84. doi: 10.1016/j.saa.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Ali M., Ahmed T., Wu W., Hossain A., Hafeez R., Islam Masum M., Wang Y., An Q., Sun G., Li B. Advancements in plant and microbe-based synthesis of metallic nanoparticles and their antimicrobial activity against plant pathogens. Nanomaterials. 2020;10:1146. doi: 10.3390/nano10061146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohamed A.A., Fouda A., Abdel-Rahman M.A., Hassan S.E.-D., El-Gamal M.S., Salem S.S., Shaheen T.I. Fungal strain impacts the shape, bioactivity and multifunctional properties of green synthesized zinc oxide nanoparticles. Biocatal. Agric. Biotechnol. 2019;19:101103. doi: 10.1016/j.bcab.2019.101103. [DOI] [Google Scholar]
- 49.Mashrai A., Khanam H., Aljawfi R.N. Biological synthesis of ZnO nanoparticles using C. albicans and studying their catalytic performance in the synthesis of steroidal pyrazolines. Arab. J. Chem. 2017;10:S1530–S1536. [Google Scholar]
- 50.Elsharkawy M., Derbalah A., Hamza A., El-Shaer A. Zinc oxide nanostructures as a control strategy of bacterial speck of tomato caused by Pseudomonas syringae in Egypt. Environ. Sci. Pollut. Res. 2020;27:19049–19057. doi: 10.1007/s11356-018-3806-0. [DOI] [PubMed] [Google Scholar]
- 51.Happy A., Soumya M., Kumar S.V., Rajeshkumar S., Sheba R.D., Lakshmi T., Nallaswamy V.D. Phyto-assisted synthesis of zinc oxide nanoparticles using Cassia alata and its antibacterial activity against Escherichia coli. Biochem. Biophys. Rep. 2019;17:208–211. doi: 10.1016/j.bbrep.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venkataraju J.L., Sharath R., Chandraprabha M., Neelufar E., Hazra A., Patra M. Synthesis, characterization and evaluation of antimicrobial activity of zinc oxide nanoparticles. J. Biochem Technol. 2014;3:151–154. [Google Scholar]
- 53.Rajkumari J., Busi S., Vasu A.C., Reddy P. Facile green synthesis of baicalein fabricated gold nanoparticles and their antibiofilm activity against Pseudomonas aeruginosa PAO1. Microb. Pathog. 2017;107:261–269. doi: 10.1016/j.micpath.2017.03.044. [DOI] [PubMed] [Google Scholar]
- 54.Malafaia C.B., Jardelino A.C.S., Silva A.G., de Souza E.B., Macedo A.J., dos Santos Correia M.T., Silva M.V. Effects of Caatinga plant extracts in planktonic growth and biofilm formation in Ralstonia solanacearum. Microb. Ecol. 2018;75:555–561. doi: 10.1007/s00248-017-1073-0. [DOI] [PubMed] [Google Scholar]
- 55.Okshevsky M., Gautier E., Farner J.M., Schreiber L., Tufenkji N. Biofilm formation by marine bacteria is impacted by concentration and surface functionalization of polystyrene nanoparticles in a species-specific manner. Environ. Microbiol. Rep. 2020;12:203–213. doi: 10.1111/1758-2229.12824. [DOI] [PubMed] [Google Scholar]
- 56.Mahamuni-Badiger P.P., Patil P.M., Badiger M.V., Patel P.R., Thorat-Gadgil B.S., Pandit A., Bohara R.A. Biofilm formation to inhibition: Role of zinc oxide-based nanoparticles. Mater. Sci. Eng. C. 2020;108:110319. doi: 10.1016/j.msec.2019.110319. [DOI] [PubMed] [Google Scholar]
- 57.Alavi M., Karimi N. Antiplanktonic, antibiofilm, antiswarming motility and antiquorum sensing activities of green synthesized Ag–TiO2, TiO2–Ag, Ag–Cu and Cu–Ag nanocomposites against multi-drug-resistant bacteria. Artif. Cells Nanomed. Biotechnol. 2018;46:S399–S413. doi: 10.1080/21691401.2018.1496923. [DOI] [PubMed] [Google Scholar]
- 58.Lee J.-H., Kim Y.-G., Cho M.H., Lee J. ZnO nanoparticles inhibit Pseudomonas aeruginosa biofilm formation and virulence factor production. Microbiol. Res. 2014;169:888–896. doi: 10.1016/j.micres.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 59.Padmavathy N., Vijayaraghavan R. Interaction of ZnO nanoparticles with microbes—A physio and biochemical assay. J. Biomed. Nanotechnol. 2011;7:813–822. doi: 10.1166/jbn.2011.1343. [DOI] [PubMed] [Google Scholar]
- 60.Leung Y.H., Ng A.M., Xu X., Shen Z., Gethings L.A., Wong M.T., Chan C.M., Guo M.Y., Ng Y.H., Djurišić A.B. Mechanisms of antibacterial activity of MgO: Non-ROS mediated toxicity of MgO nanoparticles towards Escherichia coli. Small. 2014;10:1171–1183. doi: 10.1002/smll.201302434. [DOI] [PubMed] [Google Scholar]
- 61.Premanathan M., Karthikeyan K., Jeyasubramanian K., Manivannan G. Selective toxicity of ZnO nanoparticles toward Gram-positive bacteria and cancer cells by apoptosis through lipid peroxidation. Nanomed. Nanotechnol. Biol. Med. 2011;7:184–192. doi: 10.1016/j.nano.2010.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available within the article.