Abstract
The circadian clock matches various biological processes to diurnal environmental cycles, such as light and temperature. Accumulating evidence shows that chromatin modification is crucial for robust circadian oscillation in plants, although chromatin modifiers involved in regulating core clock gene expression have been limitedly investigated. Here, we report that the Jumonji C domain-containing histone demethylase JMJ29, which belongs to the JHDM2/KDM3 group, shapes rhythmic changes in H3K4me3 histone marks at core clock loci in Arabidopsis. The evening-expressed JMJ29 protein interacts with the Evening Complex (EC) component EARLY FLOWERING 3 (ELF3). The EC recruits JMJ29 to the CCA1 and PRR9 promoters to catalyze the H3K4me3 demethylation at the cognate loci, maintaining a low-level expression during the evening time. Together, our findings demonstrate that interaction of circadian components with chromatin-related proteins underlies diurnal fluctuation of chromatin structures to maintain circadian waveforms in plants.
Keywords: circadian clock, evening complex, histone methylation, JMJ29, CCA1, PRR9
1. Introduction
The circadian clock is an internal timekeeping mechanism that generates biological rhythms with a period of ~24 h and synchronizes plant growth and development with environmental cycles. Multiple transcriptional feedback loops establish the basic architecture of the plant circadian clock. In Arabidopsis, the two morning-expressed single-MYB transcription factors, CIRCADIAN CLOCK–ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), repress transcription of the evening-expressed TIMING OF CAB EXPRESSION 1 (TOC1)/PSEUDO RESPONSE REGULATOR 1 (PRR1), that in turn represses CCA1 and LHY expression, forming the central loop [1,2,3,4]. The central loop is further interconnected with additional transcriptional loops. In the morning, a subset of the PRR family, including PRR7 and PRR9, associate with the CCA1 and LHY promoters to repress expression [5,6], and PRR5 also binds to the CCA1 promoter late in the day [7]. Evening-expressed clock components, such as TOC1 and the Evening Complex (EC), contribute to repressing the morning genes [8,9,10].
Additional layers of regulation add complexity to precise circadian oscillation. In particular, the intimate association between chromatin modification and the circadian clock is beginning to emerge [11]. Rhythmic expression of core clock genes is linked to the diurnal changes in histone 3 acetylation (H3ac), e.g., H3K56ac and H3K9/14ac, and histone 3 lysine 4 trimethylation (H3K4me3) levels at the gene promoters in Arabidopsis [11,12,13,14]. The two active histone marks have different modes of action in clock gene control: temporal H3ac deposition facilitates open chromatin formation to allow peak expression of clock genes [15,16]; whereas the H3K4me3 mark blocks access of clock repressor at the core clock gene promoters [16], shaping elaborate circadian waveforms. Despite the molecular connection between chromatin modification and the circadian control, a limited number of chromatin modifiers responsible for circadian histone modification have been demonstrated.
As for the diurnal accumulation of H3ac, HISTONE DEACETYLASE 9 (HDA9) regulates circadian oscillation by repressing TOC1 expression. HDA9 associates with an EC component, ELF3, and the EC enables HDA9 to bind to the TOC1 promoter, stimulating H3 deacetylation to inhibit TOC1 expression [17]. The temporal binding of HDA9 is defined by the EC that acts at nighttime, and thus, HDA9 facilitates declining phase of TOC1 expression during the night period [17]. A couple of HDACs also participate in circadian oscillation. PRR5, PRR7, and PRR9 interact with HDA6 and HDA19 to robustly repress CCA1 and LHY [18,19]. In support, treatment with the potent HDAC inhibitor TSA promotes CCA1 expression and possibly lengthens circadian period [19].
The H3K4me3 level at core clock genes is delicately regulated by antagonistic actions of histone methyltransferases and histone demethylases. The SET DOMAIN GROUP 2 (SDG2)/ARABIDOPSIS TRITHORAX–RELATED 3 (ATXR3) protein globally deposits the H3K4me3 mark at multiple clock-associated loci to increase gene expression [16,20]. The SDG2/ATXR3-deficient mutants exhibit a reduced H3K4me3 accumulation, increased clock repressor binding, and thereby a low-amplitude expression of core clock genes [20]. Several Jumonji C (JmjC) domain-containing histone demethylases are known to counterbalance clock gene expression. The dusk-expressed JMJ30/JMJD5 gene regulates expression of CCA1 and LHY, which reciprocally repress JMJ30/JMJD5 [21]. Moreover, CCA1 and LHY also form a feedback relationship with JMJ14 and SDG2, which facilitates rhythmic H3K4me3 dynamics for the clock genes as well as their output genes [20].
The coordinated control of H3ac and H3K4me3 accumulation has also been demonstrated in the regulation of circadian oscillation [11]. The morning-expressed CCA1 and LHY transcription repressors interact with the protein complex containing the LYSINE-SPECIFIC HISTONE DEMETHYLASE 1 HOMOLOG 1 (LDL1) and LDL2 H3K4 demethylases and the HDA6 histone deacetylase [18]. The LDL1/2-HDA6 complex is recruited to CCA1/LHY-target genes, such as TOC1 [18]. In the morning, the TOC1 gene is suppressed synergistically by H3 deacetylation and H3K4 demethylation. By contrast, in the evening, because CCA1 and LHY are expressed at a low level, the binding of LDL1/2-HDA6 complex to the TOC1 promoter is impaired, increasing TOC1 expression [18]. Overall, interplays between chromatin modifiers and core circadian clock components fine-tune circadian oscillation and thereby plant growth and development.
Despite the advances in the understanding of epigenetic regulation of circadian oscillation, chromatin modifiers contributing to the rhythms of chromatin structures and their action mechanisms remain to be fully elucidated. Here, we report that JMJ29 participates in the circadian regulation of morning-expressed CCA1 and PRR9 genes. JMJ29 physically associates with an EC component ELF3, and both EC and JMJ29 bind to the CCA1 and PRR9 promoters, maintaining their expression at a low level during evening time. Our findings suggest that temporal regulation of histone methylation underlies robust circadian rhythmicity.
2. Materials and Methods
2.1. Plant Growth Conditions
The Arabidopsis thaliana ecotype Col-0 was used. Plants were grown under neutral day conditions (NDs; 12 h light/12 h dark cycles) with cool white fluorescence (120 μmol photons m−2 s−1) at 22–23 °C. The jmj29-1, elf3-1, pJMJ29:JMJ29-GFP/jmj29-1, and pELF3:ELF3-MYC/elf3-1 mutants were previously described [22,23]. To investigate the biological function of JMJ29, T-DNA insertional jmj29-1 (Salk_021597) mutant seeds were obtained from the Arabidopsis Biological Research Center.
2.2. RNA Extraction and Quantitative RT-PCR (RT-qPCR)
Total RNAs were isolated using TRI agent (TAKARA Bio, Singa, Japan). Two μg of total RNAs were pretreated with an RNAse-free DNAse and then used to synthesize first-strand cDNA using Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (Dr. protein, Seoul, South Korea) and oligo (dT18). Synthesized cDNAs were used for PCR amplification.
Quantitative RT-PCR reactions were conducted in 96-well blocks using the Step-One Plus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The PCR primer sequences used in this study are listed in Supplementary Table S1. EUKARYOTIC TRANSLATION INITIATION FACTOR 4A1 (eIF4A) gene (At3g13920) was used as a reference gene for relative normalization. RT-qPCR was performed with SYBR Master Mix (Enzynomics, Seoul, South Korea) using the △△Ct method. The specificity of the RT-qPCR reactions was determined by melt curve analysis of the amplified products using the standard method installed in the system.
2.3. Yeast Two Hybrid (Y2H) Assays
The BD Matchmaker system (Clontech, Mountain View, CA, USA) was employed to conduct Y2H assays. The pGADT7 vector was used for the GAL4 activation domain (AD) fusion, and the pGBKT7 vector was used for GAL4 binding domain (BD) fusion. PCR products were subcloned into the pGBKT7 and pGADT7 vectors. The recombinant expression constructs were co-transformed into yeast pJG69-4A cells harboring the LacZ and His reporter genes, and transformed cells were selected by incubating on SD/-Leu/-Trp medium and SD/-Leu/-Trp/-His/-Ade.
2.4. Bimolecular fluorescence Complementation (BiFC) Assays
The full-size ELF3, ELF4, and LUX coding sequences were fused in-frame to the 5′ end of a gene sequence encoding the N-terminal half of EYFP in the pSATN-nEYFP-C1 vector (E3081). The JMJ29 gene was fused to the 3′ end of a gene sequence encoding the C-terminal half of EYFP in the pSATN-cEYFP-N1 vector (E3084).
For protoplast isolation, 14-day-old seedlings grown under the ND conditions were harvested in 20 mL 0.5 M mannitol solution (90Φ plate) and incubated for 1 h at room temperature (RT). Then, the 0.5 M mannitol solution was replaced with 20 mL enzyme solution (2% Viscozyme L, 1% Celluclast 1.5 L, 1% Pectinex Ultra SP-L in MMC, adjusted to pH 5.8 by NaOH and sterilized through a 0.2 μm syringe filtering) and incubated in the dark for 12–14 h at RT. The protoplasts were collected by centrifugation at 100 g for 7 min and washed twice with the W5 solution (0.1% glucose, 0.08% KCl, 0.9% NaCl, 1.84% CaCl2, and 2 mM MES, adjusted to pH 5.7). For BiFC assays, 10 μg nYFP- and 10 μg cYFP-fusion plasmid DNAs were used to transfect 1.2 × 106 protoplasts in 300 μL MMg solution (4 mM MES containing 0.4 M D-mannitol and 15 mM MgCl2, adjusted to pH 5.7 with 2 M NaOH). The recombinant plasmids were co-transfected with a nuclear marker (mCherry-NLS plasmid). After 14 h incubation, the protoplasts were harvested and observed using Confocal Quantitative Image Cytometer CQ1 (YOKOGAWA).
2.5. Co-Immunoprecipitation Assays
35S:ELF3-HA and 35S:JMJ29-GFP constructs were co-transfected into protoplasts isolated from 14-day-old Arabidopsis seedlings, and the transfected protoplasts were incubated in the dark for 16 h at RT. The protoplasts were collected by centrifugation at 300 g for 5 min. About 2 × 106 protoplasts were homogenized in IP buffer (25 mM Tris-HCl, 150 mM NaCl, 0.5 % Triton X-100, 1 mM EDTA, and 1 % protease inhibitor, adjusted to pH 7.5), incubated for 30 min on ice, and centrifuged at 15,000 g for 10 min at 4 °C to collect supernatant. Then, 50 μL of Protein A/G Agarose Beads (SC-2003, Santa Cruz Biotechnology, Santa Cruz, CA, USA) were added, and the mixture was incubated for 3 h at 4 °C with gentle shaking (50 rpm). The beads were removed by centrifugation at 14,000 g for 5 min at 4 °C, and antibodies were then added to the supernatant. After overnight incubation at 4 °C, 50 μL of protein A-Sepharose (P9424, Sigma-Aldrich, USA) was added and incubated for an additional 6 h at 4 °C. The beads were collected by centrifugation at 100 g for 3 min at 4 °C and washed five times with IP buffer. The proteins were eluted by boiling in SDS-PAGE sample buffer for 5 min and subjected to western blot analysis. The anti-HA (ab9110, Abcam, Cambridge, UK) and anti-GFP (ab290, Abcam, Cambridge, UK) were used for immunoprecipitation and western blot analyses.
2.6. Chromatin Immunoprecipitation (ChIP) Assays
The pJMJ29:JMJ29-GFP/jmj29-1, pELF3:ELF3-MYC/elf3-1, jmj29-1, and elf3-1 plants [22,23] were used for ChIP assays. Plant materials used for the experiments were grown under NDs for 2 weeks. Harvested plant materials were fixed in 1% formaldehyde for 20 min with vacuum infiltration and ground in liquid nitrogen. Chromatin solubilized by the nuclei lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA pH 8.0, 1% SDS, 1 mM PMSF, and 1× PIs) was sonicated at 4 °C to generate ~500 bp fragments using a Bioruptor Pico (Diagenode). The chromatin solutions were ultrasonicated for 10 cycles (30 s ON and 30 s OFF for each cycle on full power). The antibodies, including anti-GFP (ab290, Abcam, Cambridge, UK), anti-MYC (05-724, Millipore, Billerica, USA), anti-H3 (04-928, Millipore, Billerica, MA, USA), anti-H3K4me3 (07-473, Millipore, Billerica, USA), and anti-H3K9me2 (ab1220, Abcam, Cambridge. UK), and agarose A/G beads (SC-2003, Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used for ChIP. The fragmented DNAs were purified using a DNA elution kit. Precipitated DNA level was quantified by quantitative real-time PCR using specific primer sets listed in Supplementary Table S2. ChIP-qPCR values were normalized as a percentage of the input DNA.
2.7. Transient Gene Expression Assays
For transient expression assays using Arabidopsis protoplasts, reporter and effector plasmids were employed [17,24,25]. The reporter plasmid contained a minimal 35S promoter sequence as well as the GUS gene [26]. The core elements on the CCA1 and PRR9 promoters were subcloned into the reporter plasmid. To construct the p35S:JMJ29, p35S:ELF3, p35S:ELF4, and p35S:LUX effector plasmids, coding regions were subcloned into the effector vector containing the CaMV 35S promoter. Recombinant reporter and effector plasmids were co-expressed into Arabidopsis protoplasts by a polyethylene glycol (PEG) method [27]. GUS activities were measured based on the fluorometric method. A CaMV 35S promoter-luciferase construct was also co-transformed as an internal control to normalize GUS activity. The luciferase assay was performed using the D-luciferin (BT11-1000Na, BioThema, Handen, Sweden).
2.8. Bioluminescence Assays
Seeds were sown in 1/2 MS-solid medium and stratified in darkness at 4 °C for 3 days, and germinated seedlings were entrained for 3 weeks under ND conditions at 22–23 °C. To examine circadian oscillations in protoplasts, mesophyll protoplasts were isolated from the lower epidermal layer of 3-week-old Arabidopsis seedlings. Whole seedlings were soaked in 10 mL of an enzyme solution (400 mM mannitol, 20 mM KCl, 20 mM MES-KOH [pH 5.7], 10 mM CaCl2, 1% Cellulase R10, 0.5% Macerozyme R10, and 0.1% bovine serum albumin) for 16 h in the dark. The isolated protoplasts were filtered through sterile 100 mm stainless mesh and resuspended in W5 solution. The pCCA1:LUC and p35S:ELF3 plasmids were prepared by PEG purification. Then, the recombinant constructs were transiently introduced into Arabidopsis protoplasts via PEG-mediated transformation [23].
Luminescence rhythms were monitored using the Tristar2 LB 942 Multimode Microplate Reader (Berthold Technologies, Wildbad, Germany). The circadian period was estimated using the Fast Fourier Transform-Nonlinear Least Squares (FFT-NLLS) suite of programs available in the Biodare2 software.
3. Results
3.1. Altered Circadian Oscillation in jmj29 Mutant
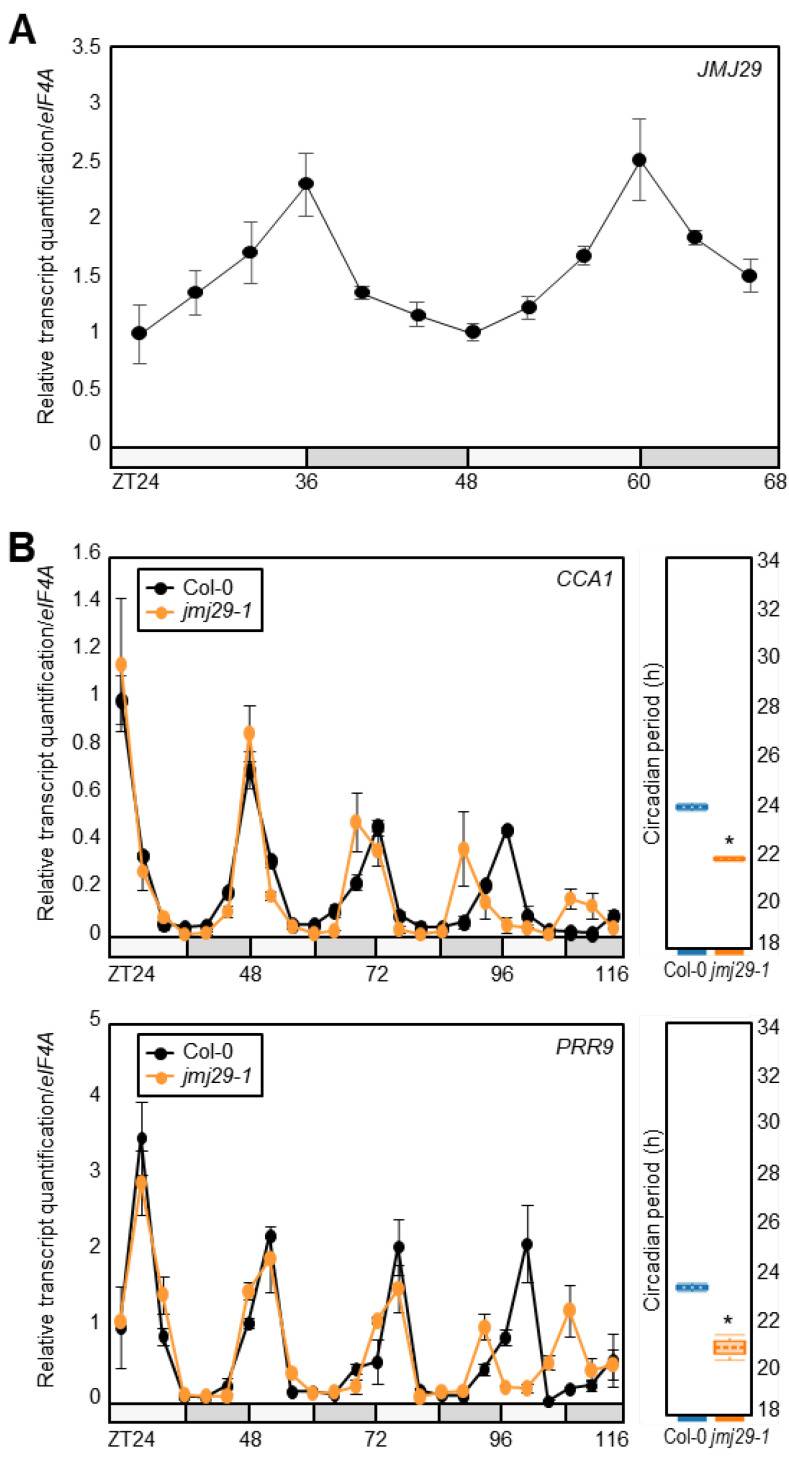
Accumulating evidence has supported that histone demethylase activity is important for the rhythmic oscillation of histone methylation at the core clock promoters [16]. Although several JMJ proteins have been characterized as crucial regulators of circadian oscillation [20,21,28], we wanted to identify additional catalytic enzymes responsible for the H3K4me3 dynamics. Among others, we noticed the JMJ29 gene, which encodes the JHDM2/KDM3 group Jumonji-C domain-containing histone demethylase [22,29], because JMJ29 expression displayed circadian rhythm with a peak at dusk (Figure 1A) [30].
Figure 1.
Circadian regulation of JMJ29. (A) Analysis of JMJ29 circadian expression. Seedlings grown under neutral day conditions (ND) for 14 days were transferred to continuous light conditions (LL) at Zeitgeber Time 0 (ZT0). (B) Altered circadian oscillation in jmj29-1. Two-week-old seedlings grown under ND were transferred to LL at ZT0. Whole seedlings (n > 15) were harvested from ZT24 to ZT116 to analyze transcript levels of CCA1 and PRR9. Period estimates were calculated using FFT-NLLS (Biodare2). Statistical significance was determined by Student’s t-test (* p < 0.05). In (A) and (B), two independent experiments were averaged. Gene expression values were normalized relative to eIF4A expression and represented as n-fold relative to the value of the ZT24. Bars indicate the standard error of the mean. The white and gray boxes indicate the subjective day and night, respectively.
We then obtained a JMJ29-deficient jmj29-1 mutant [22] and analyzed rhythmic expression of the core clock oscillator genes, CCA1 and PRR9 (Figure 1B). Quantitative real-time RT-PCR (RT-qPCR) analysis revealed that jmj29-1 mutation led to alterations in circadian oscillation. Circadian period of clock gene expression was shortened in jmj29-1 mutants, compared with wild type (Figure 1B). Consistently, expression of a circadian output gene, COLD, CIRCADIAN RHYTHM, AND RNA BINDING (CCR2), also displayed advanced rhythmic phase, possibly due to a period shortening (Supplementary Figure S1). These results suggest that JMJ29 is involved in sustaining circadian oscillation under free-running conditions.
3.2. JMJ29 Binds to the CCA1 and PRR9 Promoters
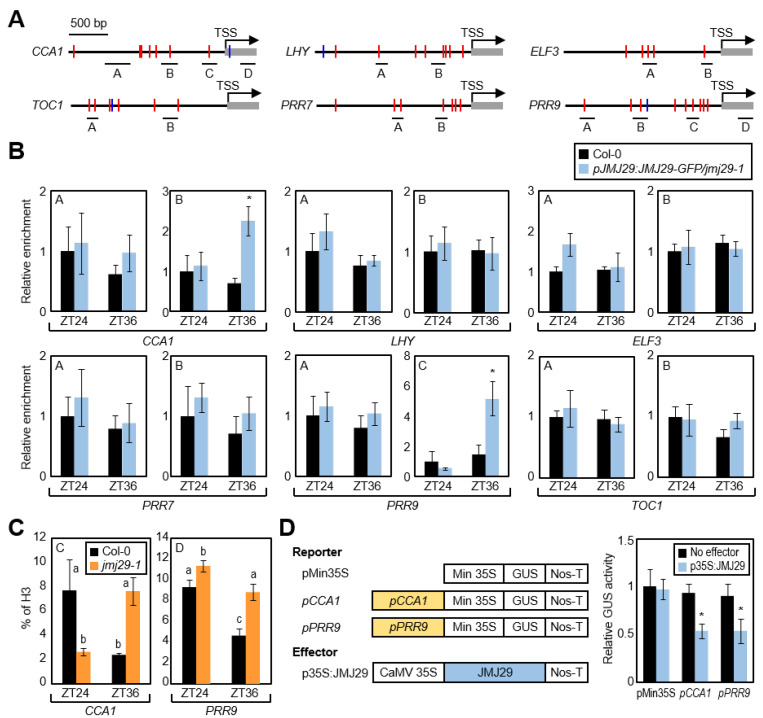
We then asked whether JMJ29 regulates circadian clock activity through direct binding to the core clock genes. To this end, we performed chromatin immunoprecipitation (ChIP) assays using the pJMJ29:JMJ29-GFP/jmj29-1 transgenic plants [16,20,21]. ChIP enrichment analysis in the promoter regions of core clock genes, which include key clock-related cis-elements, such as E-boxes and LUX-binding sites (LBSs) [8,9] (Figure 2A), showed that JMJ29 was associated specifically at the CCA1 and PRR9 promoters (Figure 2B and Supplementary Figure S2), and none of the other examined regions were targeted by JMJ29 (Figure 2B). We further explored whether JMJ29 rhythmically bound to the CCA1 and PRR9 promoters. The JMJ29 protein accumulation was likely to be correlated with the oscillation pattern of JMJ29 transcripts (Figure 1A), and consistently, binding of JMJ29 to the CCA1 and PRR9 loci was observed preferentially at dusk (Figure 2B), when expression of the two morning genes was low [2,3,14].
Figure 2.
Binding of JMJ29 to the CCA1 and PRR9 promoters. (A) Promoter analysis of core clock genes. Underbars represent the regions of PCR amplification after chromatin immunoprecipitation (ChIP). E-boxes and LBSs are represented by red and blue lines, respectively. (B) Binding of JMJ29 to clock gene promoters. Two-week-old seedlings grown under ND condition were subjected to LL at ZT0. Plants were harvested at ZT24 and ZT36 for ChIP with anti-GFP antibody, and statistical significance of the measurements was analyzed with Student’s t-test (* p < 0.05; difference between Col-0 and pJMJ29:JMJ29-GFP/jmj29-1 transgenic plants). (C) H3K4me3 levels at the CCA1 and PRR9 loci in jmj29-1 mutant. Two-week-old seedlings grown in ND condition were subjected to LL at ZT0. Plants were harvested at ZT24 and ZT36 for ChIP with anti-H3 and anti-H3K4me3 antibodies, and H3K4me3 levels were normalized to total histone H3 protein levels. Statistical significance of the measurements was analyzed by one-way ANOVA with Fisher’s post hoc test, * p < 0.05. (D) Transient expression assays. The recombinant reporter and effector constructs were co-expressed transiently in Arabidopsis protoplasts. The GUS activity was determined fluorometrically. Statistical significance of the measurements was determined by Student’s t-test (* p < 0.05). In (B), (C), and (D), biological triplicates were averaged. Bars indicate the standard error of the mean. Min 35S, minimal 35S promoter; CaMV, Cauliflower mosaic virus; Nos-T, nopaline synthase terminator.
JMJ29 that belongs to the JHDM2/KDM3 group is known to primarily catalyze H3K9 demethylation [22]. However, circadian expression of CCA1 and PRR9 is known to be independent of H3K9me1/2 levels [12]. Further, JMJ29 did not significantly influence H3K9me2 accumulation at the CCA1 and PRR9 loci (Supplementary Figure S3). Thus, we suspected that JMJ29 might contribute to changes in H3K4me3 accumulation locally at the CCA1 and PRR9 promoters. ChIP assays with an anti-H3K4me3 antibody showed that, in wild-type seedlings, reduction of H3K4me3 accumulation at the CCA1 and PRR9 loci occurred around dusk (Figure 2C) when JMJ29 was actively expressed (Figure 1A). However, the reduction of H3K4me3 levels at dusk was diminished in the jmj29-1 mutant (Figure 2C). Although histone modification at the CCA1 and PRR9 loci was altered in jmj29-1, their expression was not immediately changed (Figure 1B), possibly due to extensive transcriptional feedback loops. Thus, the JMJ29 protein promotes H3K4me3 demethylation at the CCA1 and PRR9 loci and subsequently influences their expression at later stages of free running.
Since H3K4me3 is responsible for gene activation [16,31], JMJ29 most likely inhibits the expression of CCA1 and PRR9. To test this hypothesis, we carried out transient expression assays using Arabidopsis protoplasts. The CCA1 and PRR9 promoters were fused to the 35S minimal promoter in the reporter plasmid. The recombinant reporter plasmid and the effector construct expressing the JMJ29 gene were co-transfected into Arabidopsis protoplasts (Figure 2D). Co-expression of these constructs repressed the GUS activity by ~50% (Figure 2D). Although regulation of CCA1 and PRR9 expression by JMJ29 was not immediately observed in wild-type seedlings (Figure 1B), overexpression of JMJ29 clearly led to the repression of CCA1 and PRR9 (Figure 2D). These results indicate that dusk-expressed JMJ29 is recruited to the CCA1 and PRR9 promoters and represses expression by promoting H3K4me3 demethylation.
3.3. Protein–Protein Interaction between JMJ29 and ELF3
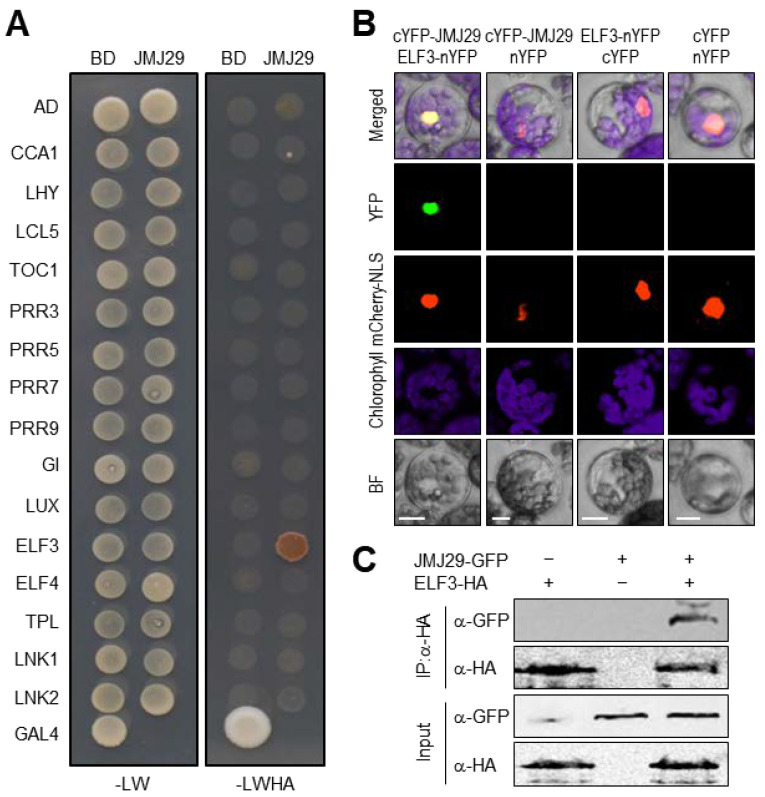
Given that JMJ29 has no selective DNA-binding domain responsible for the recognition of specific cis-elements like other JMJ proteins [22], we hypothesized that additional molecular component(s) were required for JMJ29 regulation of CCA1 and PRR9 expression. To examine this hypothesis, we conducted yeast-two-hybrid assays and tested whether JMJ29 interacts with core clock proteins. The JMJ29-GAL4 DNA BD fusion construct was co-expressed with a construct expressing a core clock protein fused to GAL4 AD in yeast cells. As a result, JMJ29 specifically interacted with ELF3 (Figure 3A), but none of the other clock proteins were associated (Figure 3A).
Figure 3.
Interaction of ELF3 with JMJ29. (A) Yeast-two-hybrid (Y2H) assays. Y2H assays were performed with the JMJ29 protein fused to the GAL4 BD, and clock proteins fused to the GAL4 AD. -LWHA represents Leu, Trp, His, and Ade drop-out plates. -LW represents Leu and Trp drop-out plates. GAL4 was used as a positive control. The experiments were repeated three times with similar results, and one representative experiment is shown. (B) Bimolecular fluorescence complementation (BiFC) assays. Partial YFP fragment fusion constructs containing either JMJ29 or ELF3 were transiently co-expressed in Arabidopsis protoplasts. The mCherry-NLS construct was used as a positive control of nuclear localization. The number of protoplast cells yielding detectable fluorescent signals was counted, and the representative image is shown only when we observed positive interactions over negative controls. Scale bars = 20 μm. (C) Coimmunoprecipitation (Co-IP) assays. 35S:ELF3-HA and 35S:JMJ29-GFP constructs are transiently co-expressed in Arabidopsis protoplasts. Epitope-tagged proteins were detected immunologically using corresponding antibodies. The experiments were repeated three times with similar results, and one representative experiment is shown. IP, immunoprecipitation.
To support the in vivo interaction of JMJ29 with ELF3, we conducted bimolecular fluorescence complementation (BiFC) analysis in Arabidopsis protoplasts. The JMJ29 gene was fused to the sequence encoding the C-terminal half of YFP (cYFP), and the ELF3 gene was fused to the sequence encoding the N-terminal half of YFP (nYFP). The fusion constructs were then transiently co-expressed in Arabidopsis protoplasts. Co-expression of the JMJ29-ELF3 combination generated yellow fluorescence exclusively in the nucleus (Figure 3B), while expression with empty vectors did not show visible fluorescence (Figure 3B). Consistent with the fact that ELF3 is a component of the EC [32,33], JMJ29 also physically associated with other EC components, ELF4 and LUX (Supplementary Figure S4). The discrepancy of interactions between JMJ29 and EC components in yeast and plant cells might be due to the requirement of plant-specific molecular factors in the protein-protein interactions. We also confirmed in planta interactions of JMJ29 and ELF3 (Figure 3C). These results indicated that JMJ29 interacts with the EC to directly repress CCA1 and PRR9.
3.4. The EC Facilitates Transcriptional Repression of CCA1 and PRR9
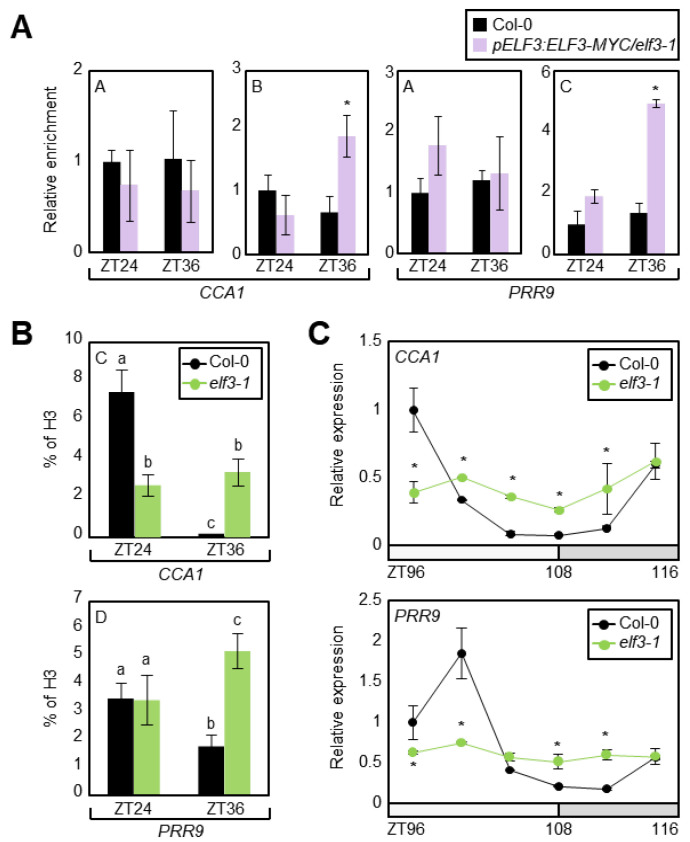
Considering that the JMJ29 protein is involved in the temporal regulation of CCA1 and PRR9, we needed to confirm whether the EC function is also relevant in rhythmic H3K4me3 accumulation at the CCA1 and PRR9 promoters. We first employed pELF3:ELF3-MYC/elf3-1 transgenic plants [23] and performed ChIP assays using an anti-MYC antibody. In agreement with the temporal association of JMJ29 to the CCA1 and PRR9 loci (Figure 2B), ELF3 also bound to the CCA1 and PRR9 promoters in the regions where JMJ29 was recruited, preferentially at dusk (Figure 2A, 2B, and Figure 4A).
Figure 4.
Repression of CCA1 and PRR9 expression by EC via H3K4me3 demethylation. (A) Binding of ELF3 to the CCA1 and PRR9 promoters. Two-week-old plants grown in ND condition were subjected to LL at ZT0. Plants were harvested at ZT24 and ZT36 for ChIP assays. Enrichment of target chromatin regions was analyzed by ChIP-qPCR with the primer sets shown in Figure 2A. Biological triplicates were averaged and statistically analyzed with Student’s t-test (* p < 0.05). (B) H3K4me3 levels at the CCA1 and PRR9 loci in elf3-1 mutant. Plants were harvested at ZT24 and ZT36 for ChIP with anti-H3 and anti-H3K4me3 antibodies, and H3K4me3 levels were normalized to total histone H3 protein levels. Biological triplicates were averaged, and statistical significance of the measurements was analyzed by one-way ANOVA with Fisher’s post hoc test (p < 0.05). (C) Transcript levels of CCA1 and PRR9 in elf3-1 mutant. Two-week-old seedlings grown under ND were transferred to LL at ZT0. Whole seedlings (n > 15) were harvested from ZT96 to ZT116. Gene expression values were normalized to the eIF4A expression. Two technical replicates were averaged and statistically analyzed with Student’s t-test (*p < 0.05). Bars indicate the standard error of the mean. The white and gray boxes indicate the subjective day and night.
Since the EC recruits JMJ29 to repress gene expression via H3K4me3 demethylation, we analyzed H3K4me3 levels at the CCA1 and PRR9 loci in the elf3-1 mutant. ChIP-qPCR analysis for H3K4me3 accumulation revealed that the reduction of H3K4me3 levels at the CCA1 and PRR9 loci around dusk was impaired in elf3-1 (Figure 4B), similar to that in jmj29-1 (Figure 2C). We also examined expression of CCA1 and PRR9 in elf3-1 and found that their expression was arrhythmic and increased around dusk (Figure 4C), when JMJ29 and EC were active (Figure 1A) [33]. The expression amplitude of CCA1 and PRR9 in elf3-1 was a bit different from previous reports [8,34], probably because of our growth conditions.
The transcriptional repression of CCA1 and PRR9 by EC was also confirmed by transient expression assays (Supplementary Figure S5). The recombinant reporter plasmid containing the CCA1 or PRR9 promoter and the effector construct expressing ELF3, ELF4, or LUX were co-transfected into Arabidopsis protoplasts. GUS activity measurement showed that the EC components could bind to the CCA1 and PRR9 promoters and repressed expression (Supplementary Figure S5). These results indicate that the EC is responsible for the H3K4me3-dependent regulation of CCA1 and PRR9.
3.5. The EC and JMJ29 Are Interdependent in Regulating CCA1 and PRR9 Expression
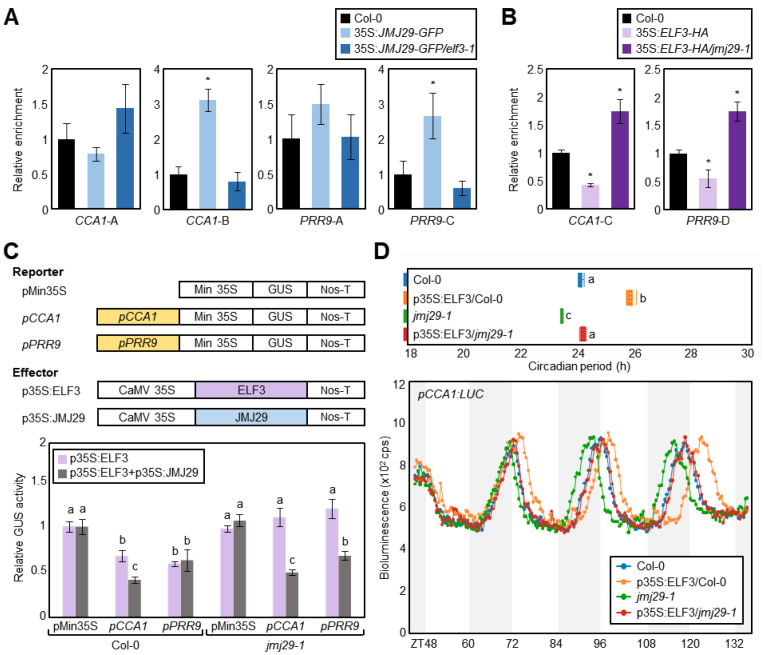
Our results demonstrated that the EC and JMJ29 act together in the repression of CCA1 and PRR9. We thus asked whether ELF3 repression of CCA1 and PRR9 required JMJ29. The importance of EC function for JMJ29 binding to the CCA1 and PRR9 promoters was tested using wild-type and elf3-1 mutant protoplasts transiently expressing 35S:JMJ29-GFP. ChIP analysis with the anti-GFP antibody showed that JMJ29 binding to the CCA1 and PRR9 loci was reduced in the elf3-1 mutant background compared with the wild-type background (Figure 5A). Since JMJ29 protein catalyzes H3K4me3 demethylation at the cognate regions (Figure 2C), H3K4me3 levels at the CCA1 and PRR9 loci were also measured in wild-type and jmj29-1 protoplasts transiently expressing 35S:ELF3-HA. H3K4me3 levels were reduced by overexpression of ELF3 in wild-type protoplasts, but ELF3-induced H3K4me3 demethylation was compromised in the jmj29-1 mutant protoplasts (Figure 5B).
Figure 5.
Interdependency of EC and JMJ29 in repression of CCA1 and PRR9. (A) Binding of JMJ29 to the CCA1 and PRR9 loci in elf3-1. 35S:JMJ29-GFP construct was transfected in protoplasts isolated from two-week-old seedlings grown under NDs. The protoplasts were harvested at ZT36 for ChIP using anti-GFP antibody. (B) H3K4me3 levels in 35S:ELF3-HA/jmj29-1. 35S:ELF3-HA construct was transfected in protoplasts isolated from two-week-old seedlings grown under NDs. The protoplasts were harvested at ZT36 for ChIP using anti-H3K4me3 antibody. In (A) and (B), the indicated genomic regions (see Figure 2A) were analyzed by ChIP-qPCR. Biological triplicates were averaged and statistically analyzed with Student’s t-test (* p < 0.05). Bars indicate the standard error of the mean. (C) Transient expression assays using Arabidopsis protoplasts. The recombinant reporter and effector constructs were co-expressed transiently in Arabidopsis protoplasts, and then GUS activity was determined fluorometrically. Biological triplicates were averaged, and statistical significance of the measurements was analyzed by one-way ANOVA with Fisher’s post hoc test (p < 0.05). Bars indicate the standard error of the mean. Min 35S, minimal 35S promoter; CaMV, Cauliflower mosaic virus; Nos-T, nopaline synthase terminator. (D) Impact of ELF3 overexpression in circadian period. The 35S:ELF3 effector plasmid was transfected into wild-type and jmj29-1 mutant protoplasts. Biological triplicates were averaged, and statistical significance of the measurements was analyzed by one-way ANOVA (p < 0.05). Bars indicate the standard error of the mean.
We also performed transient gene expression assays using Arabidopsis protoplasts. Reporter GUS activity measurement indicated that ELF3 significantly repressed CCA1 and PRR9 promoter activities in wild-type protoplasts, but the ELF3 function disappeared in the jmj29-1 mutant (Figure 5C). The impaired ELF3 function was restored by complementation of JMJ29 (Figure 5C). In further support, overexpression of ELF3 led to prolonged circadian period in wild-type protoplasts, but the ELF3 function was impaired in jmj29-1 mutant protoplasts (Figure 5D). These results demonstrate that both EC and JMJ29 are simultaneously involved in repressing CCA1 and PRR9 expression in circadian cycles.
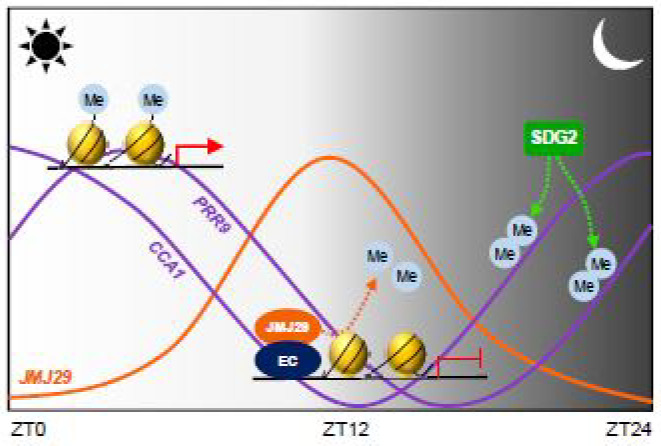
In summary, the CCA1 and PRR9 genes are regulated by rhythmic changes in H3K4me3 levels. SDG2 is possibly responsible for the raising phase of their expression [20]. After peak expression, JMJ29 is recruited via the interaction with ELF3, a component of the EC. The EC-JMJ29 induces H3K4me3 demethylation at the CCA1 and PRR9 loci to reach their basal expression during evening time (Figure 6).
Figure 6.
Proposed working model of the EC-JMJ29 complex in circadian control. Circadian expression of CCA1 and PRR9 are associated with H3K4me3 levels. SDG2 possibly catalyzes the H3K4me3 deposition at the CCA1 and PRR9 promoters at dawn, facilitating raising phase of gene expression. After peak expression, the declining phase of CCA1 and PRR9 is shaped by the Evening Complex (EC) and JMJ29. The EC recruits JMJ29 to the CCA1 and PRR9 promoters and removes H3K4me3 at dusk, maintaining low levels of expression during evening time.
4. Discussion
Rhythmic transcriptional regulation of core clock genes is intimately connected with chromatin status, which is under the extensive control of DNA methylation, histone modifications, and histone variant exchange [35,36,37,38]. To date, changes in histone modifications, such as H3K56ac, H3K9/14ac, and H3K4me3, at clock gene promoters are known to be particularly important for circadian dynamics in Arabidopsis [13,16]. Notably, the circadian accumulation of H3K4me3 at core clock gene loci is mediated by antagonistic actions of histone methyltransferases and histone demethylases. The SDG2/ATXR3 histone methyltransferase deposits H3K4me3 globally at core clock gene loci [20]. After peak expression, multiple histone demethylases, which belong to the KDM1/LSD1 or JmjC family [39], regulate a specific subset of clock genes, returning to the basal expression level of their target genes [39]. For example, LDL1, LDL2, JMJ14, and JMJ30 allow the diurnal repression of CCA1 and LHY, possibly through controlling rhythmic H3K4me3 accumulation [20,21,39]. Here, we show that JMJ29 regulates circadian oscillations by repressing CCA1 and PRR9 expression. JMJ29 interacts with the EC component ELF3, and the EC rhythmically recruits JMJ29 to the CCA1 and PRR9 promoters at dusk, when EC and JMJ29 are active. Then, the JMJ29 protein catalyzes H3K4me3 demethylation to maintain low levels of CCA1 and PRR9 expression during evening time.
However, several questions remain to be elucidated. Although JMJ29 is a member of the JHDM2/KDM3 group JmjC domain-containing histone demethylases [29], which were identified to predominantly catalyze the H3K9me1/2 demethylation in eukaryotes [40,41,42,43], our results showed that JMJ29 was also able to remove H3K4me3, especially at core clock gene loci. JMJ29 most likely has a pervasive role in H3K9me1/2 demethylation to activate gene expression, but the JMJ29 protein is also relevant in transcriptional repression locally at certain chromatin regions via H3K4me3 demethylation. It is currently unclear what structural and biochemical features of JMJ29 enable it to have the dual functions. It is also a reasonable question, to determine which catalytic activity of JMJ29 is predominant at a given sequence context. Additionally, even though EC has a broader spectrum of clock gene binding, binding of JMJ29 is limited to the CCA1 and PRR9 promoters. EC also interacts with various chromatin modifiers and remodelers, in addition to JMJ29. While H3K4me3 demethylation at the core clock genes by ELF3 was dependent on JMJ29, regulation of circadian gene expression by ELF3 was further elaborated by additional epigenetic and molecular factors [17,44,45]. Thus, detailed molecular mechanisms should be elucidated to give a comprehensive view of circadian-chromatin networks.
In addition to circadian oscillation, the JMJ29 protein was also implicated in plant growth and development. For instance, JMJ29 participates in trichrome development. JMJ29 directly binds to the GLABRA 3 (GL3) promoter and activates gene expression via H3K9me2 demethylation [22]. JMJ29 may also indirectly affect expression of GL1 and GL2 [22]. Consistently, the trichome density on leaves and inflorescent stems were reduced in jmj29 mutants with a low expression of GL1, GL2, and GL3 [22]. Based on our study, it is likely that trichome development might be under the circadian control, and JMJ29 relays diurnal cues to induce proper developmental processes. Altogether, the JMJ29 protein is likely to integrate temporal information into circadian oscillation programs and also a variety of output processes optimizing plant growth and development.
5. Conclusions
Diurnal changes in H3K4me3 levels shape the rhythmic oscillation of core clock gene expression. The global deposition of H3K4me3 at the clock gene loci is known to be catalyzed by SDG2/ATXR3. The removal of H3K4me3 at core clock genes is mediated by multiple histone demethylases, each of which has a limited target gene spectrum. The JMJ29 protein interacts with the EC, and the EC-JMJ29 complex is able to associate the CCA1 and PRR9 promoters at dusk. The JMJ29 protein promotes H3K4me3 demethylation and maintains their expression at low levels during evening time.
Acknowledgments
We thank Keqiang Wu (National Taiwan University, Taiwan) for kindly providing pJMJ29:JMJ29-GFP/jmj29-1 seeds.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12040529/s1, Figure S1: Circadian expression of CCR2 in jmj29-1. Figure S2: Binding of JMJ29 to coding regions of CCA1 and PRR9. Figure S3: H3K9me2 accumulation at the CCA1 and PRR9 loci. Figure S4: Interaction of JMJ29 with ELF4 and LUX. Figure S5: Transient expression assays using Arabidopsis protoplasts. Table S1: List of primers used in this study. Table S2: List of primers used in chromatin immunoprecipitation (ChIP) assays.
Author Contributions
Conceptualization, P.J.S.; formal analysis, H.G.L.; investigation, H.G.L.; writing—original draft preparation, H.G.L. and P.J.S.; writing—review and editing, P.J.S.; supervision, P.J.S.; funding acquisition, P.J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Basic Science Research (NRF-2019R1A2C2006915) and Basic Research Laboratory (NRF-2020R1A4A2002901) programs provided by the National Research Foundation of Korea and by the Creative-Pioneering Researchers Program (0409-20200281) of Seoul National University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lu S.X., Knowles S.M., Andronis C., Ong M.S., Tobin E.M. CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009;150:834–843. doi: 10.1104/pp.108.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.R., Carre I.A., Coupland G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell. 2002;2:629–641. doi: 10.1016/S1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- 3.Alabadi D., Oyama T., Yanovsky M.J., Harmon F.G., Mas P., Kay S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 4.Mas P., Alabadi D., Yanovsky M.J., Oyama T., Kay S.A. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell. 2003;15:223–236. doi: 10.1105/tpc.006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams S., Manfield I., Stockley P., Carre I.A. Revised morning loops of the Arabidopsis circadian clock based on analyses of direct regulatory interactions. PLoS ONE. 2015;10:e0143943. doi: 10.1371/journal.pone.0143943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamichi N., Kita M., Ito S., Yamashino T., Mizuno T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005;46:686–698. doi: 10.1093/pcp/pci086. [DOI] [PubMed] [Google Scholar]
- 7.Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.H., Sakakibara H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon L.E., Knox K., Kozma-Bognar L., Southern M.M., Pokhilko A., Millar A.J. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 2011;21:120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizuno T., Nomoto Y., Oka H., Kitayama M., Takeuchi A., Tsubouchi M., Yamashino T. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol. 2014;55:958–976. doi: 10.1093/pcp/pcu030. [DOI] [PubMed] [Google Scholar]
- 11.Perales M., Mas P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell. 2007;19:2111–2123. doi: 10.1105/tpc.107.050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song H.R., Noh Y.S. Rhythmic oscillation of histone acetylation and methylation at the Arabidopsis central clock loci. Mol. Cells. 2012;34:279–287. doi: 10.1007/s10059-012-0103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baerenfaller K., Shu H., Hirsch-Hoffmann M., Futterer J., Opitz L., Rehrauer H., Hennig L., Gruissem W. Diurnal changes in the histone H3 signature H3K9ac|H3K27ac|H3S28p are associated with diurnal gene expression in Arabidopsis. Plant Cell Environ. 2016;39:2557–2569. doi: 10.1111/pce.12811. [DOI] [PubMed] [Google Scholar]
- 14.Hemmes H., Henriques R., Jang I.C., Kim S., Chua N.H. Circadian clock regulates dynamic chromatin modifications associated with Arabidopsis CCA1/LHY and TOC1 transcriptional rhythms. Plant Cell Physiol. 2012;53:2016–2029. doi: 10.1093/pcp/pcs148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farinas B., Mas P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011;66:318–329. doi: 10.1111/j.1365-313X.2011.04484.x. [DOI] [PubMed] [Google Scholar]
- 16.Malapeira J., Khaitova L.C., Mas P. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA. 2012;109:21540–21545. doi: 10.1073/pnas.1217022110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee K., Mas P., Seo P.J. The EC-HDA9 complex rhythmically regulates histone acetylation at the TOC1 promoter in Arabidopsis. Commun. Biol. 2019;2:1–10. doi: 10.1038/s42003-019-0377-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung F.Y., Chen F.F., Li C., Chen C., Chen J.H., Cui Y., Wu K. The LDL1/2-HDA6 Histone Modification Complex Interacts With TOC1 and Regulates the Core Circadian Clock Components in Arabidopsis. Front. Plant Sci. 2019;10:233. doi: 10.3389/fpls.2019.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L., Kim J., Somers D.E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA. 2013;110:761–766. doi: 10.1073/pnas.1215010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song Q., Huang T.Y., Yu H.H., Ando A., Mas P., Ha M., Chen Z.J. Diurnal regulation of SDG2 and JMJ14 by circadian clock oscillators orchestrates histone modification rhythms in Arabidopsis. Genome Biol. 2019;20:1–12. doi: 10.1186/s13059-019-1777-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu S.X., Knowles S.M., Webb C.J., Celaya R.B., Cha C., Siu J.P., Tobin E.M. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 2011;155:906–915. doi: 10.1104/pp.110.167015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung F.Y., Chen J.H., Feng Y.R., Lai Y.C., Yang S., Wu K. Arabidopsis JMJ29 is involved in trichome development by regulating the core trichome initiation gene GLABRA3. Plant J. 2020;103:1735–1743. doi: 10.1111/tpj.14858. [DOI] [PubMed] [Google Scholar]
- 23.Ezer D., Jung J.H., Lan H., Biswas S., Gregoire L., Box M.S., Charoensawan V., Cortijo S., Lai X., Stockle D., et al. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants. 2017;3:1–12. doi: 10.1038/nplants.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamioka M., Takao S., Suzuki T., Taki K., Higashiyama T., Kinoshita T., Nakamichi N. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell. 2016;28:696–711. doi: 10.1105/tpc.15.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H.G., Won J.H., Choi Y.R., Lee K., Seo P.J. Brassinosteroids regulate circadian oscillation via the BES1/TPL-CCA1/LHY module in Arabidopsis thaliana. iScience. 2020;23:101528. doi: 10.1016/j.isci.2020.101528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore I., Galweiler L., Grosskopf D., Schell J., Palme K. A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl. Acad. Sci. USA. 1998;95:376–381. doi: 10.1073/pnas.95.1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abel S., Theologis A. Transient transformation of Arabidopsis leaf protoplasts: A versatile experimental system to study gene expression. Plant J. 1994;5:421–427. doi: 10.1111/j.1365-313X.1994.00421.x. [DOI] [PubMed] [Google Scholar]
- 28.Zheng S., Hu H., Ren H., Yang Z., Qiu Q., Qi W., Liu X., Chen X., Cui X., Li S., et al. The Arabidopsis H3K27me3 demethylase JUMONJI 13 is a temperature and photoperiod dependent flowering repressor. Nat. Commun. 2019;10:1303. doi: 10.1038/s41467-019-09310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu F., Li G., Cui X., Liu C., Wang X.J., Cao X. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J. Integr. Plant Biol. 2008;50:886–896. doi: 10.1111/j.1744-7909.2008.00692.x. [DOI] [PubMed] [Google Scholar]
- 30.Lee H.G., Lee K., Jang K., Seo P.J. Circadian expression profiles of chromatin remodeling factor genes in Arabidopsis. J Plant Res. 2015;128:187–199. doi: 10.1007/s10265-014-0665-8. [DOI] [PubMed] [Google Scholar]
- 31.Liu C., Lu F., Cui X., Cao X. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010;61:395–420. doi: 10.1146/annurev.arplant.043008.091939. [DOI] [PubMed] [Google Scholar]
- 32.Huang H., Nusinow D.A. Into the Evening: Complex Interactions in the Arabidopsis Circadian Clock. Trends Genet. 2016;32:674–686. doi: 10.1016/j.tig.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farre E.M., Kay S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolmos E., Herrero E., Bujdoso N., Millar A.J., Toth R., Gyula P., Nagy F., Davis S.J. A reduced-function allele reveals that EARLY FLOWERING3 repressive action on the circadian clock is modulated by phytochrome signals in Arabidopsis. Plant Cell. 2011;23:3230–3246. doi: 10.1105/tpc.111.088195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfluger J., Wagner D. Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 2007;10:645–652. doi: 10.1016/j.pbi.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pikaard C.S., Mittelsten Scheid O. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 2014;6:a019315. doi: 10.1101/cshperspect.a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Probst A.V., Desvoyes B., Gutierrez C. Similar yet critically different: The distribution, dynamics and function of histone variants. J. Exp. Bot. 2020;71:5191–5204. doi: 10.1093/jxb/eraa230. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H., Lang Z., Zhu J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mo.l Cell Biol. 2018;19:489–506. doi: 10.1038/s41580-018-0016-z. [DOI] [PubMed] [Google Scholar]
- 39.Prakash S., Singh R., Lodhi N. Histone demethylases and control of gene expression in plants. Cell Mol. Biol. 2014;60:97–105. [PubMed] [Google Scholar]
- 40.Brauchle M., Yao Z., Arora R., Thigale S., Clay I., Inverardi B., Fletcher J., Taslimi P., Acker M.G., Gerrits B., et al. Protein complex interactor analysis and differential activity of KDM3 subfamily members towards H3K9 methylation. PLoS ONE. 2013;8:e60549. doi: 10.1371/journal.pone.0060549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dutta A., Choudhary P., Caruana J., Raina R. JMJ27, an Arabidopsis H3K9 histone demethylase, modulates defense against Pseudomonas syringae and flowering time. Plant J. 2017;91:1015–1028. doi: 10.1111/tpj.13623. [DOI] [PubMed] [Google Scholar]
- 42.Saze H., Shiraishi A., Miura A., Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319:462–465. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- 43.Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 44.Park H.J., Baek D., Cha J.Y., Liao X., Kang S.H., McClung C.R., Lee S.Y., Yun D.J., Kim W.Y. HOS15 interacts with the histone deacetylase HDA9 and the evening complex to epigenetically regulate the floral activator GIGANTEA. Plant Cell. 2019;31:37–51. doi: 10.1105/tpc.18.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tong M., Lee K., Ezer D., Cortijo S., Jung J., Charoensawan V., Box M.S., Jaeger K.E., Takahashi N., Mas P., et al. The evening complex establishes repressive chromatin domains via H2A.Z deposition. Plant Physiol. 2020;182:612–625. doi: 10.1104/pp.19.00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.