Abstract
The ubiquitin–proteasome system plays an important role in the cell under normal physiological conditions but also during viral infections. Indeed, many auxiliary proteins from the (HIV-1) divert this system to its own advantage, notably to induce the degradation of cellular restriction factors. For instance, the HIV-1 viral infectivity factor (Vif) has been shown to specifically counteract several cellular deaminases belonging to the apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like (APOBEC3 or A3) family (A3A to A3H) by recruiting an E3-ubiquitin ligase complex and inducing their polyubiquitination and degradation through the proteasome. Although this pathway has been extensively characterized so far, Vif has also been shown to impede A3s through degradation-independent processes, but research on this matter remains limited. In this review, we describe our current knowledge regarding the degradation-independent inhibition of A3s, and A3G in particular, by the HIV-1 Vif protein, the molecular mechanisms involved, and highlight important properties of this small viral protein.
Keywords: HIV, Vif, APOBEC3G, ubiquitin, proteasome, translation, encapsidation, deamination, RNP granules
1. Introduction
Once the human immunodeficiency virus type 1 (HIV-1) has entered the cell, it must overcome, or disarm, a network of restriction factors (including A3 enzymes) that block specific steps of the replication cycle [1], and hijack the intracellular machinery to its own benefit. Restriction factors play an important role in innate immune responses and protect the host from viral pathogens [1,2,3]. Members of the apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like (APOBEC3 or A3) family are innate immune effectors restricting many exogenous viruses, such as HIV-1, endogenous retroelements [4,5,6,7], and several non-retroviral viruses [8]. The human genome encodes seven A3 genes (A3A, B, C, D, F, G, and H) resulting from gene duplications and rearrangements during mammalian evolution [9,10,11]. A3G was the first to be discovered and is the most functionally characterized enzyme. A3G was demonstrated to inhibit HIV-1 replication in the absence of the viral infectivity factor (Vif) [12] thanks to its cytidine deaminase activity that converts cytidine (C) to uridine (U) present in single-stranded DNA generated during reverse transcription of the viral genome. Completion of proviral DNA synthesis results in guanine (G) to adenine (A) mutations in the plus strand DNA, eventually leading to disruption of protein synthesis (through nonsense and/or missense mutations) and interruption of HIV-1 replication [13,14,15,16]. Amongst the seven A3 proteins expressed in human cells, A3D, F, and H have also been identified as anti-HIV-1 factors [17]. While the deaminase activity is essential for the A3 antiviral function, it has been reported that A3s could repress HIV-1 replication in a deaminase-independent manner at different stages of the replication cycle (reverse transcription, integration, and maturation) [8,18,19,20,21].
To counteract restriction factors’ antiviral activities, HIV-1 expresses several accessory proteins, and Vif was shown to efficiently degrade A3G in virus-producing cells and inhibit its incorporation into nascent viral particles [12,14,15,22,23]. This process requires the recruitment of an E3-ubiquitin ligase complex by Vif, leading to the proteasomal degradation of A3G [12,24,25,26]. Interestingly, the Vif–A3 interplay is the consequence of an evolutionary arms race between lentiviruses and their hosts [27,28], i.e., interactions involving Vif and A3 proteins are species-specific and not all A3s are submitted to proteasomal degradation and are capable of inhibiting viral replication. For instance, A3B does not restrict HIV-1 and is not degraded by the HIV-1 Vif protein [29,30], at least for some HIV-1 Vif isolates (IIIB, JR-CSF, 89.6), while Vif from HIV-1 HXB2, ELI-1, and YU-2 shows some degradative effects [31]. SIV Vif proteins behave similarly [29], SIVmac239 Vif being the most potent at inhibiting A3B through the canonical polyubiquitination mechanism [29]. Although the degradation-dependent mechanism has been extensively studied and documented in the literature for the last twenty years [12,14,24,25,26,32], degradation-independent mechanisms mediated by Vif could also account for the reduced levels of A3 proteins in cells and in viral particles. Our review mainly focuses on the latter mechanism, highlighting their importance to overcome the potent antiviral activity of A3s in general, and of A3G in particular. We will also describe processes developed by other viruses which do not encode a functional Vif protein to overcome A3 enzymatic activity.
2. Degradation-Dependent Inhibition of APOBEC3G by Vif: A Brief Overview
2.1. The Players: APOBEC3G and Vif
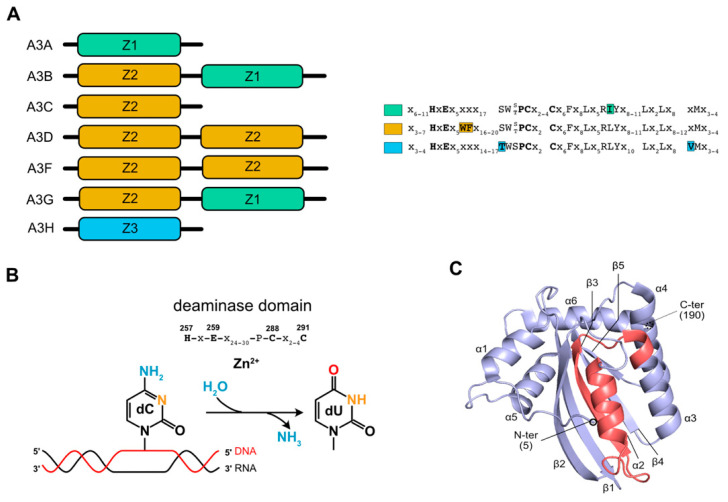
The seven members of the A3 family of proteins contain one (A3A, A3C, and A3H) or two (A3B, A3D, A3F, and A3G) copies of a catalytic deaminase (CD) domain that is distinguished by the presence of a signature motif, His-X-Glu-X23-28-Pro-Cys-X2-4-Cys (Figure 1A). When the enzyme contains two CDs, CD2 is catalytically active [33] while CD1 is involved in nucleic acids binding [34,35,36], its oligomerization and movements along the substrate to increase the deamination efficiency [37,38,39]. CDs and adjacent domains are organized into three phylogenetically distinct groups, named zinc-coordinating domains, Z1, Z2, and Z3 [11,40]. The mechanism of catalysis involves the histidine and the two cysteine residues that coordinate a zinc ion that enables the glutamic acid to deprotonate a water molecule, and generates the zinc hydroxide ion that will react with the carbon C4 of the cytosine (Figure 1B). The CDs’ tridimensional structure is conserved between A3s and is composed of five β strands and six α helices organized around the deaminase domain (Figure 1C) (for more details regarding the structure of A3 proteins, please refer to these recent reviews [33,41,42]).
Figure 1.
APOBEC3 structural and catalytic features. (A) Schematic structural domains of A3 proteins are represented. Characteristic phylogenetic ZDD domains are represented in color, with their amino acid sequences on the side. (B) A3G catalytic mechanism allowing cytidine deamination on a single-stranded DNA target (red strand) is represented. Amino acids constituting the catalytic site are mentioned above. These amino acids accommodate a zinc ion and the use of a water molecule leading to the deamination of the cytidine. (C) A3C crystal structure (hA3C) (PDB: 3VOW) represented in ribbons. Catalytic (CD) and ZDD domains are represented in purple and pink, respectively. Secondary structure motifs are annotated.
Amongst A3 proteins, human A3G was the first to be identified and is the most potent inhibitor of HIV-1 replication in the absence of Vif protein [12], whereas A3D, A3F, and A3H (haplotype II, V, VII) have been shown to exhibit various degrees of antiviral activities [17,43]. A3D, A3F, A3G, and A3H are localized in the cytoplasm, A3A and A3C are distributed throughout the cell, while A3B is mainly nuclear [17,44]. When non-permissive cells are infected with HIV-1 ∆Vif viruses, A3G is efficiently incorporated into budding virions, a fundamental aspect of its antiviral function. A3G packaging is determined by specific interactions between its N-terminal CD domain and the nucleocapsid region of the HIV-1 Pr55Gag precursor [45,46,47,48,49,50], but also depends on its association to RNAs, mainly the HIV-1 genomic and 7SL RNAs [35,36,49,50,51,52,53]. Following its packaging, A3G induces dC-to-dU deamination in the (−) strand DNA of HIV-1 during reverse transcription, resulting in G-to-A substitutions in the (+) strand DNA and eventually blocking viral replication due to the incorporation of missense or stop-codons [14,15,54].
The local nucleotide sequence preference for deamination is 5’-CC-3’ (the substrate C is underlined) for A3G and 5’-TC-3’ for other A3s [13,17,55,56]. In addition to G-to-A hypermutation, deaminase-independent activities could also contribute to the overall antiviral activity of A3G [18,21]. Indeed, A3G may bind to viral RNA and sterically interfere with the progression of reverse transcription [19,57,58,59] or bind the reverse transcriptase itself [60].
HIV-1 overcomes A3G restriction by expressing Vif, which targets A3G for polyubiquitination and proteasomal degradation (see below) [12,24,25,26]. Initially considered as an accessory protein, Vif is in fact an important virulence factor whose absence considerably reduces viral infectivity [28,61]. Vif is a small, unstructured, and multimeric protein of 23 kDa expressed from singly-spliced viral mRNAs during the last stages of HIV-1 infection [62,63,64,65]. Notably, an intronic G run (GI3-2) has been shown to be critical for the splicing regulation of Vif mRNA in non-permissive cells in order to maintain an appropriate ratio of Vif to A3G protein levels [66,67,68], but this ratio is highly dependent on the physiological environment. Vif is mainly localized in the cytoplasm of infected cells but is also found in viral particles [69,70,71,72,73]. It possesses various properties linked to its functional domains and RNA chaperone activities (Figure 2): for instance, Vif was shown to influence HIV-1 particle morphology, maturation of Pr55Gag, reverse transcription, and HIV-1 genomic RNA dimerization [68,74,75,76,77,78]. However, one of the principal functions of Vif is to induce the polyubiquitination and subsequent proteasomal degradation of A3G, thereby reducing the pool of cytosolic A3G available for incorporation into viral particles [25,79,80].
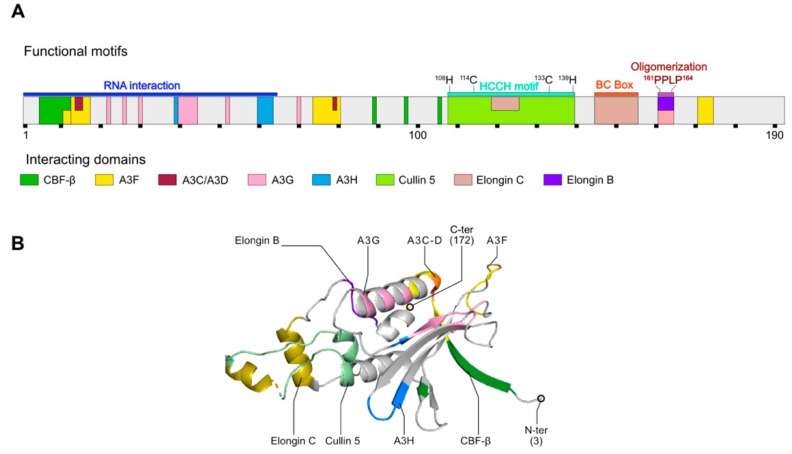
Figure 2.
HIV-1 Vif protein structure and interactions with E3-ubiquitin ligase complex members. (A) Vif mono-dimensional structure (Genbank M19921; UniProtKB—P12504 (Vif_HV1N5)) is represented using 10 amino acids per scaling unit (black squares under the structure). Functional motifs are represented with colored rectangles above the structure. Interaction regions with A3 proteins or members of the E3-ubiquitin ligase are represented in color inside the structure. (B) Vif 3D structure (PDB: 4N9F; positions 3-172) is represented in ribbons. N- and C- terminal extremities (N-ter and C-ter, respectively) as well as interaction regions with several partners are colored and annotated on the structure. Adapted from [81].
2.2. Recruitment of an Ubiquitin Ligase Complex by Vif
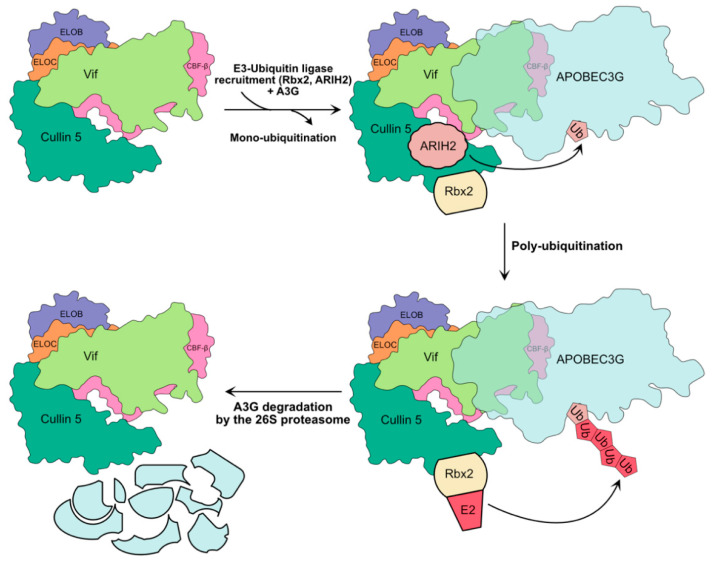
The most studied mechanism allowing the restriction of A3G is the recruitment by Vif of an E3-ubiquitin ligase complex [25,26,82], which is composed of the scaffold protein Cullin 5, Elongin B/C adaptor proteins, RBX2 catalytic units (RING-box protein 2), transcription factor CBF-β (Core-Binding Factor β), which plays a critical role in stabilizing Vif and its assembly with the ligase, and ARIH2 (Ariadne homolog 2) [24,25,26,32,83,84] (Figure 3). This last partner has been recently identified to act in an E1-E2-E3/E3 cascade to target A3G for degradation and has been shown to be essential for efficient HIV-1 infection in primary CD4+ T cells [83]. This allows the 48K poly-ubiquitinylation of A3G and its redirection towards the proteasome, which ensures its degradation. Many studies have mapped the domains and peptide sequences involved in the interaction between Vif, A3G, and the ligase complex (Figure 2, and for recent reviews on these different domains see [33,41,81]). The crystal structure of the human Vif-EloB/C-Cul5-CBF-β (PDB: 4N9F) [85] and simian Vif-EloB/C-CBF-β (PDB: 6P59) [86] have been solved and allowed to validate most of the protein–protein interactions previously determined by mutagenesis and biochemical assays. Very recently, a complex structure of the CBF-β-fused A3FCTD with HIV-1 Vif has been obtained by cryo-EM and this structure may provide a structural insight into A3-Vif interaction [87], although further investigations are required to understand their interaction.
Figure 3.
Hijacking of ubiquitin–proteasome pathway by Vif. HIV-1 first recruits the core components of the E3-ubiquitin ligase complex, namely Elongin B (ELOB) and ELOC, CBF-β, and Cullin 5. This complex is represented in accordance with its known 3D structure (PDB: 4N9F). The recruitment of A3G and catalytic subunits Rbx2 and ARIH2 leads to ARIH-2 mediated mono-ubiquitination of A3G. Subsequent recruitment of an E2 ubiquitin-conjugating enzyme by RBX2 leads to the poly-ubiquitination of A3G and its degradation by the 26S proteasome.
3. Degradation-Independent Inhibition of APOBEC3G by Vif
Although the primary mechanism of A3G neutralization in HIV-1 infected cells involves its ubiquitination and subsequent degradation by the 26S proteasome, other mechanisms have been shown to reduce A3G packaging into viral particles (through a direct or an indirect effect of Vif) or to interfere with the deaminase activity of encapsidated A3G. The following section will focus on these different mechanisms.
3.1. Transcriptional Inhibition of APOBEC3G, Hijacking of CBF-β by Vif
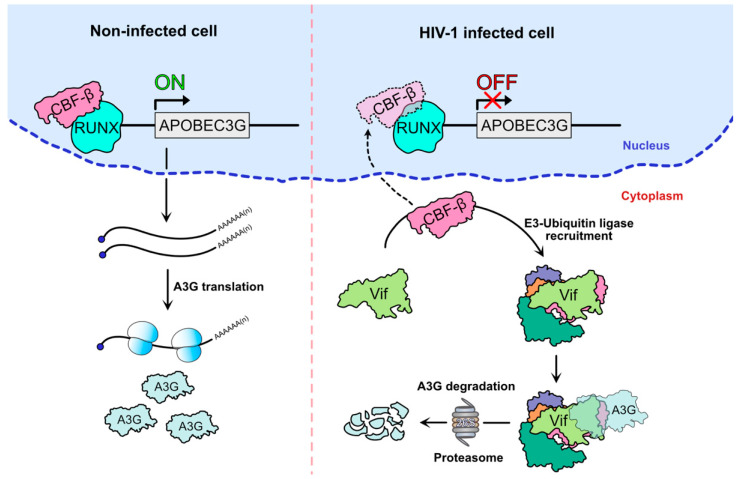
The recruitment by Vif of the E3-ubiquitin ligase complex mediating the degradation of A3G may have an additional indirect effect on A3G expression. Indeed, the recruitment of CBF-β is mandatory for Vif stability and for the assembly of the E3-ubiquitin ligase complex responsible for A3G and other A3 proteins’ degradation [32,84]. The interaction was detected by mass spectrometry after the co-immunoprecipitation of tagged Vif construct in HEK 293 and Jurkat cells [32]. CBF-β is a non-DNA binding subunit that heterodimerizes with Runt-related transcription factor (RUNX) proteins to form the CBF family of transcription factors, which are important for cell differentiation and proliferation, hematopoiesis, and bone development [88]. Interestingly, the binding of Vif to CBF-β is mutually exclusive with RUNX heterodimerization and has been shown to impact the expression of genes whose regulatory domains are associated with RUNX1, which includes A3 genes, suggesting that Vif inhibits transcription by competing with RUNX for CBF-β binding [89,90] (Figure 4). Mutational analysis [91,92,93,94] and structural study of the pentameric E3-ubiquitin ligase complex (Vif-EloB/C-Cul5-CBF-β) [85] identified several Vif residues important for its binding to CBF-β. For instance, the N-terminal 5WQVMIVW11, present in an anti-parallel β-strand of Vif, is a major interaction domain (Figure 2), along with residues W89, T96, and L106. Thus, the sequestration of CBF-β by Vif in the cytoplasm provides a dual hijacking mechanism by reducing A3G expression at both transcriptional and post-translational (degradation of A3G through the proteasome) levels.
Figure 4.
APOBEC3G mRNA transcriptional inhibition after CBF-β recruitment by HIV-1 Vif. The binding of CBF-β to the RUNX complex drives the transcription of the A3G gene and maintains a robust antiviral state in the absence of HIV-1 infection (left). In the presence of Vif (right), Vif hijacks CBF-β and prevents its binding to RUNX, thus simultaneously repressing A3G transcription and promoting its ubiquitination and proteasomal degradation.
3.2. Translational Inhibition of APOBEC3G mRNA by Vif
A second mechanism suggesting that Vif could reduce the intracellular level of A3G by affecting its translation was proposed shortly after the discovery of A3G [79,95]. First, the use of proteasome inhibitors on H9 infected cells or HEK 293 cells transiently co-transfected with Vif and A3G-HA expression vectors rescued only partial A3G expression, without altering the A3G mRNA levels. Second, ex-vivo pulse-chase experiments showed that Vif impaired the radiolabeling of the A3G-HA protein in a dose-dependent manner, reducing A3G translation by 30–40% [79,95]. Third, in vitro-coupled transcription/translation assays in rabbit reticulocyte lysates confirmed that Vif impaired A3G translation by approximatively 70–75% [79]. However, these first studies were performed using expression vectors lacking the authentic 5′ and 3′ untranslated regions (UTRs) of A3G mRNA, which could play a key role in A3G translation [96,97]. Thus, they may not faithfully recapitulate events occurring with endogenous A3G mRNA. Interestingly, we showed that Vif could bind A3G mRNA, and more specifically its UTRs [98]. Within the 5’UTR, two stem-loop structures (SL2–SL3) (Figure 5) were required for Vif to inhibit A3G translation [98,99] and viruses produced after transfection with expression vectors of A3G mRNAs lacking this motif showed reduced infectivity, which was mainly due to an increase in A3G encapsidation into viral particles [99]. Moreover, our results show that translational inhibition of A3G by Vif is independent of the proteasomal degradation pathway as some mutations in the N-terminus of Vif impede translation inhibition without affecting A3G degradation (K26R, this mutant still binds A3G [100]), while others present a complete inhibition of A3G degradation and translational reduction (H42/43N, this mutant is defective in A3G binding [101]), raising the possibility that Vif/A3G interaction might be required for translational regulation [99].
Figure 5.
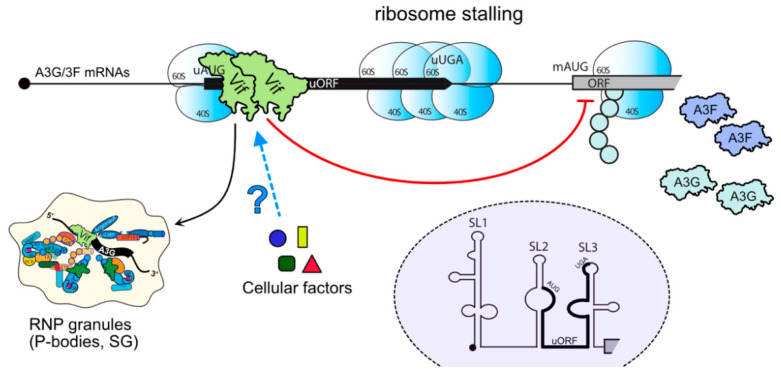
APOBEC3G mRNA translation inhibition mediated by HIV-1 Vif. Vif associates with the 5’UTR of A3G mRNAs, in the vicinity of A3G mRNA uORF. This association leads to A3G mRNA translation inhibition, possibly by ribosome stalling, and its relocation to RNP granules (P-bodies, stress granules, etc.). A3F mRNA could be regulated similarly as their 5’UTRs are phylogenetically conserved [102]. The involvement of other cellular factors in this process is still unknown but it could favor, or inhibit, A3G translation. The secondary structure of the 5’UTR of A3G mRNA as well as the position of the uORF (within SL2–SL3) are highlighted (purple dashed circle).
While mechanisms leading to A3G translation inhibition still remain unclear, we recently uncovered the importance of a short and conserved upstream ORF (uORF) located within SL2–SL3 of the 5’UTR of A3G and A3F mRNAs (Figure 5) [102]. Interestingly, the uORF represses A3G translation itself (40%), as do many uORF located in 5’UTR of mRNAs [103,104,105], but it is also mandatory for the Vif-mediated repression of A3G translation and for the redirection of A3G mRNA into stress granules in the presence of Vif [102]. The fact that Vif was previously shown in vitro to interact with the lower and upper stems of SL3 [98] suggests that Vif may block ribosome scanning (stalling). Given the proportion of genes that have one or more uORFs in their 5’UTR [103,104,105], one cannot rule out the possibility that Vif might be able to regulate the translation or the trafficking of such mRNAs in the cell. Notably, A3G was shown by immunofluorescence studies to localize to P-bodies and stress granules [31,106,107,108,109,110]; on the contrary, Vif was observed in P-bodies only in the presence of A3G, suggesting that Vif could selectively remove A3G from and/or restrict its localization to P-bodies to induce its degradation [107].
Taken together, translational inhibition of A3G by Vif seems to be a multi-layer process, involving direct (ribosome stalling at the 5’UTR) and indirect (shuttling of A3G mRNA to ribonucleoprotein granules) blockages to delay or prevent mRNA translation. Whereas additional experiments will be needed to clearly decipher this mechanism, one can imagine that Vif interacts with components of the eukaryotic translation initiation machinery to reduce the translation rate of A3G and/or recruit cellular factors involved in the negative regulation of the translation (Figure 5).
3.3. Packaging Inhibition of APOBEC3G by Vif
Packaging of A3G/3F proteins into HIV-1 particles depends on the nucleocapsid (NC) domain of Pr55Gag, and the current view is that its packaging is also reliant upon its capacity to bind RNA [35,45,46,47,48,50,53,111]. A3C has evolved to use distinct mechanisms for retrovirus targeting by interacting with the matrix (MA) domain of Pr55Gag for its encapsidation [112]. A3D–H have been shown to be efficiently packaged into viral particles [17] and a correlation exists between the multimerization state of these A3s and their packaging (and restrictive) capacities [113,114]. Several studies have suggested that Vif prevents A3G packaging into HIV-1 particles independently of the reduction in its intracellular concentration [22,25,95,115]. Indeed, comparison of wild-type and ∆vif HIV-1 virions produced from transfected cells expressing a similar level of A3G showed that wild-type virions incorporated less A3G than ∆vif virions, suggesting that Vif can directly exclude A3G from progeny virions [25,95]. Moreover, A3G exclusion requires biologically active Vif. Indeed, the Vif C114 and C133 are critical residues for Cullin 5 interaction, as the HCCH motif and for Vif-mediated A3G degradation. Interestingly, the mutation of these residues (C114/S and C133/S) impaired Vif’s ability to inhibit A3G packaging [95]. Additional experiments further highlighted that A3G packaging inhibition was not linked to its Vif-induced intracellular degradation. In fact, an A3G C97/A mutant, which was defective for multimerization but resistant to Vif-mediated degradation, showed a decreased packaging yield in the presence of Vif [113,116]. Likewise, a recent study using antibody antigen-binding fragments (Fabs) directed against the ubiquitin transfer reaction and Vif-E3 assembly showed that the ubiquitination of A3C, A3F, and A3G was inhibited, although not sufficiently to restore their packaging into viral particles and antiviral activity [117]. These studies suggest that the inhibition of A3G packaging and A3G degradation are two distinct properties of Vif. Although a clear mechanism has not been described yet, it has been proposed that the RNA-binding capacities of A3G and Vif could account for this packaging restriction. Indeed, A3G and Vif share similar RNA-binding domains on HIV-1 genomic RNA, such as stem-loops in the 5’UTR packaging signal [51,71,118], and one can imagine that at early stages of assembly, Vif and A3G compete for their own packaging through HIV-1 RNA-binding, or that Vif prevents A3G encapsidation by masking a domain on A3G that mediates the interaction with the assembling virion [63]. As previously mentioned, it is well-established that Vif and A3G could be encapsidated through their capacity to bind the nucleocapsid (NC) domain of Pr55Gag [45,46,47,111,119,120,121]. It is therefore conceivable that Vif may interfere with these A3G–Gag interactions. Notably, this inhibition may also favor reverse transcription as A3G was reported to reduce tRNA annealing to the primer binding site (PBS), which is required for the initiation of reverse transcription [122,123]. Interestingly, A3 (F/G/H) evolved to partially mimic the RNA-binding specificity of the HIV-1 NC (by targeting G-rich and A-rich sequences) in order to ensure their concomitant packaging with RNA into nascent virions [53]. Similar to a potential competition between Vif and A3G, it is conceivable that Gag may compete with A3 proteins for their encapsidation; thus, Gag–RNA occlusion could be seen as a countermeasure against A3s by retroviruses lacking a Vif protein. Alternatively, Vif could direct A3G away from sites of virus assembly by inducing the formation of the high molecular weight form of A3G [124,125], thus sequestering A3G in packaging-incompetent complexes.
3.4. Inhibition of Intravirion Deaminase Activity by Vif
While downregulation of A3G by Vif within the cell is very efficient (see above), A3G can still be found in viral particles during wild-type HIV-1 infection, even though to a lesser extent than in ∆vif viruses (4-11 A3G/∆vif(−) virion versus < 1 A3G/wild-type virion) [115,126,127,128,129]; thus, only a few A3G molecules are sufficient to potently inhibit HIV-1 replication. However, the efficiency of packaged A3G enzymes into wild-type particles indicated lower deaminase activity compared to those from ∆vif virions (65% reduction) [130], suggesting that virion-associated Vif molecules inhibit the intrinsic deaminase activity of A3G [131]. Indeed, earlier work performed in Escherichia coli showed that A3G-mediated hypermutation was inhibited by Vif [132], and this was correlated to a direct binding of Vif to A3G, as the use of a degradation-resistant A3G mutant (D128K, a species-specific determinant of Vif sensitivity) rendered A3G resistant to Vif inhibition [132,133]. In addition, enzymatic studies showed that Vif can attenuate the processivity of A3G by disrupting its scanning on viral ssDNA [133]. Taken together, these studies strongly argue in favor of a direct effect of Vif on A3G enzymatic activity, either through Vif–A3G binding (thus disrupting the binding of A3G to its substrate or A3G movements such as sliding or jumping), or by competing for its binding to the ssDNA substrate. Indeed, Vif is a well-known nucleic acids-binding protein [70,75,76,77,98,118,134,135,136]. Vif-binding sites that were identified in the 5’-end region of HIV-1 genomic RNA correspond to hypermutated sites in ∆vif virions [118], and Vif also binds with a significant affinity (Kd < 40 nM) to the consensus A3G ssDNA target sites [134]. Thus, Vif could interfere with A3G by masking its access to the neosynthesized viral DNA. Alternatively, in the target cell, it might locally limit the RNase H activity of HIV-1 reverse transcriptase that is necessary to allow the deaminase activity of A3G, which is otherwise inhibited upon binding to HIV-1 genomic RNA [125].
4. Degradation-Independent Inhibition of APOBEC3 Proteins by Other Viruses
The tumultuous relationship between A3s and viral proteins is also witnessed in other animal species and for various viruses. Indeed, viral proteins other than Vif have been shown to counteract A3 proteins in lentiviruses: this is the case for feline immunodeficiency virus (FIV) protease [137], HIV-1 protease (against A3H variant SV200) [138], and reverse transcriptase for A3G [139]. In other retroviruses, mechanisms developed to counteract A3 proteins have been adapted to compensate for the absence of a functional Vif-like gene. Foamy or spumaretroviruses are also restricted by A3 proteins, and these viruses use the accessory Bet protein, an equivalent of Vif, to inhibit A3 antiviral activity [140,141,142]. The interaction of feline foamy virus (FFV) and prototype/human foamy virus (PFV) Bet protein with feline A3 and A3F/3G proteins, respectively, reduces their packaging into viral particles without impacting their expression level [140,141]. Instead, Bet prevents A3G dimerization [143,144] and traps A3 (A3G/3C) in insoluble complexes, rendering them unavailable for virion packaging. Murine leukemia virus (MLV) possesses two mechanisms to overcome the restriction by mouse A3. On the one hand, its P50 protein, produced from an alternatively spliced gag RNA [145,146], prevents mouse A3 packaging by interacting with its C-terminus CD2 domain, but impacts neither its degradation nor its deaminase activity [147]. Considering that the C-terminus of mouse A3 is necessary for its incorporation into viruses, it is not surprising that P50 binding to this domain affects A3 packaging. On the other hand, glyco-Gag is known to be important for MLV-induced pathogenesis, and its abrogation leads to decreased virus replication and pathogenesis [148,149,150]. Glyco-Gag uses a unique mechanism to counteract the antiviral action of A3 by affecting the capsid stability and by protecting the reverse transcription complex in the viral cores from A3 proteins [151,152]. Human T-cell lymphotropic virus type 1 (HTLV-1) is also sensitive to human A3G, even though its cytidine deaminase activity is inoperant [153,154]. This virus has evolved a cis-acting mechanism to prevent A3G restriction. A peptide motif in the C-terminus of the NC domain inhibits A3G packaging into nascent virions [155]. In this case, the physical interaction between HTLV-1 NC and RNA would occlude A3G packaging. In addition, A3 proteins have been reported to inhibit several human pathogenic viruses other than retroviruses, such as the hepatitis B virus (HBV) [156], papillomavirus [157], and herpesvirus [158]. These viruses have evolved new strategies to antagonize the antiviral activities of A3s, either by enhancing the externalization of A3G by HBx (HBV), or by sequestering A3A/3B away from their site of activity thanks to the viral proteins BORF2 (Epstein–Barr virus, EBV), ICP6 (herpes simplex virus 1, HSV), and ORF6 (Kaposi’s sarcoma-associated herpesvirus) [159]. Notably, these viral proteins do not induce the degradation of A3 proteins.
5. Conclusions
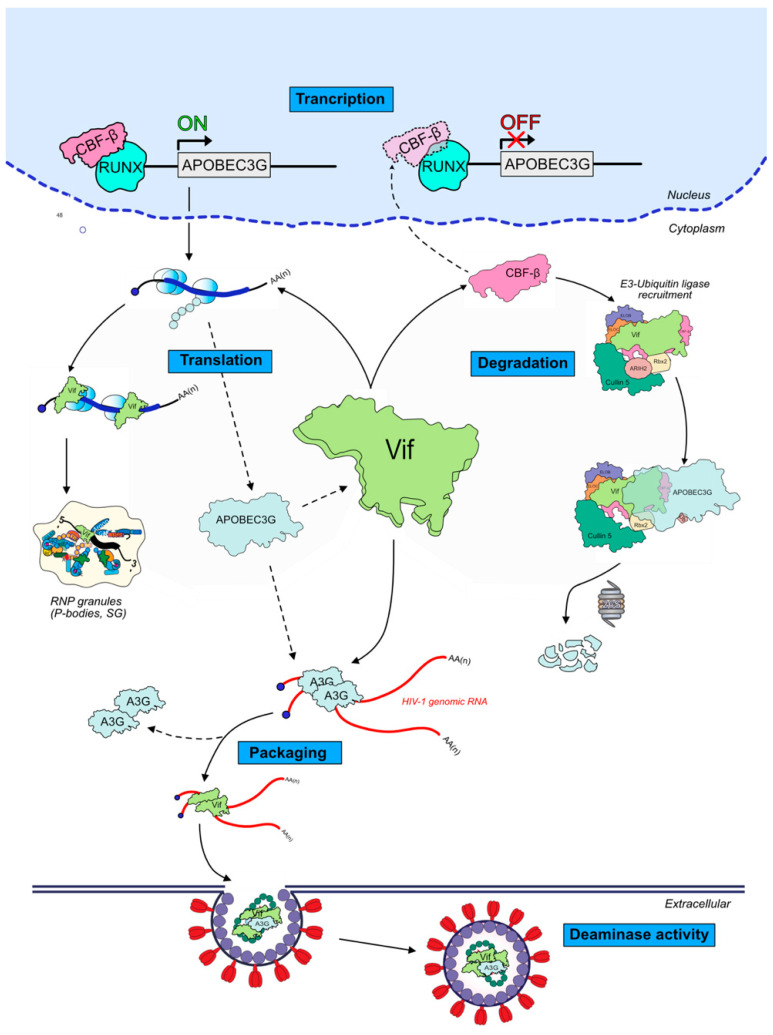
The Vif protein is of major importance for an efficient viral infection in non-permissive cells by antagonizing A3 proteins antiviral activity. While mechanisms leading to A3G degradation through the recruitment of an E3-ubiquitin ligase complex by Vif are well understood, little is known concerning other degradation-independent routes used by Vif to inhibit A3G and other A3 proteins. These mechanisms are diverse, and aim to inhibit or reduce A3G transcription, translation, virion encapsidation, and enzymatic activity (Figure 6). Interestingly, Vif-proteins and Vif-RNA interactions are common characteristics between all these important steps, thus targeting them through their interacting interfaces may constitute promising antiviral strategies. A better knowledge of how the virus hijacks different cellular processes and which components are involved in viral replication is crucial in this attempt.
Figure 6.
Summary of the different modes of inhibition of APOBEC3G by HIV-1 Vif protein. Other A3 proteins have also been shown to be regulated at different levels (see text for details): transcription (potentially all A3s); translation (A3G and A3F); degradation (all A3s, depending on considered HIV isolates and hosts); and packaging (A3D, A3F, A3G, and A3H).
Acknowledgments
This work was supported by the CNRS and grants and fellowships from SIDACTION and the French National Agency for Research on AIDS and Viral Hepatitis (ANRS) to JCP and BS, respectively, and by a doctoral fellowship from the French Ministry of Higher Education and Research to CV.
Author Contributions
J.-C.P. and B.S. conceived the review topic; J.-C.P. drafted the manuscript and B.S. generated the figures. All authors corrected, edited, and approved the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the French National Agency for Research on AIDS and Viral Hepatitis (#ECTZ134179) and SIDACTION (#20-1-AEQ-12613-1) to JCP and SGM (grants) and BS (post-doctoral fellowships), and by a doctoral fellowship from the French Ministry of Research and Higher Education (CV). The APC was funded by the CNRS.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Seissler T., Marquet R., Paillart J.C. Hijacking of the ubiquitin/proteasome pathway by the hiv auxiliary proteins. Viruses. 2017;9:322. doi: 10.3390/v9110322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colomer-Lluch M., Ruiz A., Moris A., Prado J.G. Restriction Factors: From Intrinsic Viral Restriction to Shaping Cellular Immunity Against HIV-1. Front. Immunol. 2018;9:2876. doi: 10.3389/fimmu.2018.02876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Urbano V., De Crignis E., Re M.C. Host Restriction Factors and Human Immunodeficiency Virus (HIV-1): A Dynamic Interplay Involving All Phases of the Viral Life Cycle. Curr. HIV Res. 2018 doi: 10.2174/1570162X16666180817115830. [DOI] [PubMed] [Google Scholar]
- 4.Harris R.S., Dudley J.P. APOBECs and virus restriction. Virology. 2015;479–480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salter J.D., Bennett R.P., Smith H.C. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem. Sci. 2016;41:578–594. doi: 10.1016/j.tibs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hulme A.E., Bogerd H.P., Cullen B.R., Moran J.V. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu Y.L., Witkowska H.E., Hall S.C., Santiago M., Soros V.B., Esnault C., Heidmann T., Greene W.C. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. USA. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu W.K., Byun H., Dudley J.P. The role of APOBECs in viral replication. Microorganisms. 2020 doi: 10.3390/microorganisms8121899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Münk C., Willemsen A., Bravo I.G. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol. Biol. 2012 doi: 10.1186/1471-2148-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris R.S., Liddament M.T. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 11.Langlois M.A., Beale R.C.L., Conticello S.G., Neuberger M.S. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005;33:1913–1923. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 13.Yu Q., König R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J.M., Landau N.R. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 14.Harris R.S., Bishop K.N., Sheehy A.M., Craig H.M., Petersen-Mahrt S.K., Watt I.N., Neuberger M.S., Malim M.H. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/S0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 15.Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H., Yang B., Pomerantz R.J., Zhang C., Arunachalam S.C., Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hultquist J.F., Lengyel J.A., Refsland E.W., LaRue R.S., Lackey L., Brown W.L., Harris R.S. Human and Rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H Demonstrate a Conserved Capacity To Restrict Vif-Deficient HIV-1. J. Virol. 2011;85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman E.N.C., Holmes R.K., Craig H.M., Klein K.C., Lingappa J.R., Malim M.H., Sheehy A.M. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 19.Bishop K.N., Verma M., Kim E.Y., Wolinsky S.M., Malim M.H. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X.Y., Guo F., Zhang L., Kleiman L., Cen S. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J. Biol. Chem. 2007;282:32065–32074. doi: 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- 21.Hakata Y., Miyazawa M. Deaminase-independent mode of antiretroviral action in human and mouse APOBEC3 proteins. Microorganisms. 2020;8:1–23. doi: 10.3390/microorganisms8121976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mariani R., Chen D., Schröfelbauer B., Navarro F., König R., Bollman B., Münk C., Nymark-McMahon H., Landau N.R. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/S0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 23.Conticello S.G., Harris R.S., Neuberger M.S. The Vif Protein of HIV Triggers Degradation of the Human Antiretroviral DNA Deaminase APOBEC3G. Curr. Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 24.Marin M., Rose K.M., Kozak S.L., Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 25.Sheehy A.M., Gaddis N.C., Malim M.H. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 26.Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X.F. Induction of APOBEC3G Ubiquitination and Degradation by an HIV-1 Vif-Cul5-SCF Complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 27.Uriu K., Kosugi Y., Ito J., Sato K. The Battle between Retroviruses and APOBEC3 Genes: Its Past and Present. Viruses. 2021;13 doi: 10.3390/v13010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano Y., Aso H., Soper A., Yamada E., Moriwaki M., Juarez-Fernandez G., Koyanagi Y., Sato K. A conflict of interest: The evolutionary arms race between mammalian APOBEC3 and lentiviral Vif. Retrovirology. 2017 doi: 10.1186/s12977-017-0355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Land A.M., Wang J., Law E.K., Aberle R., Kirmaier A., Krupp A., Johnson W.E., Harris R.S. Degradation of the cancer genomic DNA deaminase APOBEC3B by SIV Vif. Oncotarget. 2015;6:39969–39979. doi: 10.18632/oncotarget.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J., Shaban N.M., Land A.M., Brown W.L., Harris R.S. Simian Immunodeficiency Virus Vif and Human APOBEC3B Interactions Resemble Those between HIV-1 Vif and Human APOBEC3G. J. Virol. 2018;92 doi: 10.1128/JVI.00447-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marin M., Golem S., Rose K.M., Kozak S.L., Kabat D. Human Immunodeficiency Virus Type 1 Vif Functionally Interacts with Diverse APOBEC3 Cytidine Deaminases and Moves with Them between Cytoplasmic Sites of mRNA Metabolism. J. Virol. 2008;82:987–998. doi: 10.1128/JVI.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jäger S., Kim D.Y., Hultquist J.F., Shindo K., Larue R.S., Kwon E., Li M., Anderson B.D., Yen L., Stanley D., et al. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012;481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delviks-Frankenberry K.A., Desimmie B.A., Pathak V.K. Structural insights into APOBEC3-mediated lentiviral restriction. Viruses. 2020;12:587. doi: 10.3390/v12060587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro F., Bollman B., Chen H., König R., Yu Q., Chiles K., Landau N.R. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 35.Svarovskaia E.S., Xu H., Mbisa J.L., Barr R., Gorelick R.J., Ono A., Freed E.O., Hu W.S., Pathak V.K. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 2004;279:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- 36.Wang T., Tian C., Zhang W., Luo K., Sarkis P.T.N., Yu L., Liu B., Yu Y., Yu X.-F. 7SL RNA Mediates Virion Packaging of the Antiviral Cytidine Deaminase APOBEC3G. J. Virol. 2007 doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chelico L., Prochnow C., Erie D.A., Chen X.S., Goodman M.F. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J. Biol. Chem. 2010;285:16195–16205. doi: 10.1074/jbc.M110.107987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Y., Chelico L. Intensity of deoxycytidine deamination of HIV-1 proviral DNA by the retroviral restriction factor APOBEC3G is mediated by the noncatalytic domain. J. Biol. Chem. 2011;286:11415–11426. doi: 10.1074/jbc.M110.199604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holden L.G., Prochnow C., Chang Y.P., Bransteitter R., Chelico L., Sen U., Stevens R.C., Goodman M.F., Chen X.S. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LaRue R.S., Andrésdóttir V., Blanchard Y., Conticello S.G., Derse D., Emerman M., Greene W.C., Jónsson S.R., Landau N.R., Löchelt M., et al. Guidelines for Naming Nonprimate APOBEC3 Genes and Proteins. J. Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Azimi F.C., Lee J.E. Structural perspectives on HIV-1 Vif and APOBEC3 restriction factor interactions. Protein Sci. 2020 doi: 10.1002/pro.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y., Knecht K.M., Shen Q., Xiong Y. Multifaceted HIV-1 Vif interactions with human E3 ubiquitin ligase and APOBEC3s. FEBS J. 2020 doi: 10.1111/febs.15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X., Abudu A., Son S., Dang Y., Venta P.J., Zheng Y.-H. Analysis of Human APOBEC3H Haplotypes and Anti-Human Immunodeficiency Virus Type 1 Activity. J. Virol. 2011;85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muckenfuss H., Hamdorf M., Held U., Perkovic M., Löwer J., Cichutek K., Flory E., Schumann G.G., Münk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 45.Alce T.M., Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 gag nucleocapsid protein. J. Biol. Chem. 2004;279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- 46.Schäfer A., Bogerd H.P., Cullen B.R. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Luo K., Liu B., Xiao Z., Yu Y., Yu X., Gorelick R., Yu X.-F. Amino-Terminal Region of the Human Immunodeficiency Virus Type 1 Nucleocapsid Is Required for Human APOBEC3G Packaging. J. Virol. 2004;78:11841–11852. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zennou V., Perez-Caballero D., Göttlinger H., Bieniasz P.D. APOBEC3G Incorporation into Human Immunodeficiency Virus Type 1 Particles. J. Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burnett A., Spearman P. APOBEC3G Multimers Are Recruited to the Plasma Membrane for Packaging into Human Immunodeficiency Virus Type 1 Virus-Like Particles in an RNA-Dependent Process Requiring the NC Basic Linker. J. Virol. 2007;81:5000–5013. doi: 10.1128/JVI.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Apolonia L., Schulz R., Curk T., Rocha P., Swanson C.M., Schaller T., Ule J., Malim M.H. Promiscuous RNA Binding Ensures Effective Encapsidation of APOBEC3 Proteins by HIV-1. PLoS Pathog. 2015;11:1–22. doi: 10.1371/journal.ppat.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khan M.A., Kao S., Miyagi E., Takeuchi H., Goila-Gaur R., Opi S., Gipson C.L., Parslow T.G., Ly H., Strebel K. Viral RNA Is Required for the Association of APOBEC3G with Human Immunodeficiency Virus Type 1 Nucleoprotein Complexes. J. Virol. 2005;79:5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strebel K., Khan M.A. APOBEC3G encapsidation into HIV-1 virions: Which RNA is it? Retrovirology. 2008;5 doi: 10.1186/1742-4690-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.York A., Kutluay S.B., Errando M., Bieniasz P.D. The RNA Binding Specificity of Human APOBEC3 Proteins Resembles That of HIV-1 Nucleocapsid. PLoS Pathog. 2016;12:1–24. doi: 10.1371/journal.ppat.1005833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lecossier D., Bouchonnet F., Clavel F., Hance A.J. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 55.Suspène R., Sommer P., Henry M., Ferris S., Guétard D., Pochet S., Chester A., Navaratnam N., Wain-Hobson S., Vartanian J.P. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 2004;32:2421–2429. doi: 10.1093/nar/gkh554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bishop K.N., Holmes R.K., Sheehy A.M., Davidson N.O., Cho S.J., Malim M.H. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 57.Chaurasiya K.R., McCauley M.J., Wang W., Qualley D.F., Wu T., Kitamura S., Geertsema H., Chan D.S.B., Hertz A., Iwatani Y., et al. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nat. Chem. 2014;6:28–33. doi: 10.1038/nchem.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gillick K., Pollpeter D., Phalora P., Kim E.-Y., Wolinsky S.M., Malim M.H. Suppression of HIV-1 Infection by APOBEC3 Proteins in Primary Human CD4 + T Cells Is Associated with Inhibition of Processive Reverse Transcription as Well as Excessive Cytidine Deamination. J. Virol. 2013;87:1508–1517. doi: 10.1128/JVI.02587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iwatani Y., Chan D.S.B., Wang F., Maynard K.S., Sugiura W., Gronenborn A.M., Rouzina I., Williams M.C., Musier-Forsyth K., Levin J.G. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pollpeter D., Parsons M., Sobala A.E., Coxhead S., Lang R.D., Bruns A.M., Papaioannou S., McDonnell J.M., Apolonia L., Chowdhury J.A., et al. Deep sequencing of HIV-1 reverse transcripts reveals the multifaceted antiviral functions of APOBEC3G. Nat. Microbiol. 2018;3:220–233. doi: 10.1038/s41564-017-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strebel K., Daugherty D., Clouse K., Cohen D., Folks T., Martin M.A. The HIV A (sor) gene product is essential for virus infectivity. Nature. 1988;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 62.Yang S., Sun Y., Zhang H. The multimerization of human immunodeficiency virus type I Vif protein: A requirement for Vif function in the viral life cycle. J. Biol. Chem. 2001;276:4889–4893. doi: 10.1074/jbc.M004895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Batisse J., Guerrero S.X., Bernacchi S., Richert L., Godet J., Goldschmidt V., Mely Y., Marquet R., de Rocquigny H., Paillart J.-C. APOBEC3G Impairs the Multimerization of the HIV-1 Vif Protein in Living Cells. J. Virol. 2013;87:6492–6506. doi: 10.1128/JVI.03494-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang B., Gao L., Li L., Lu Z., Fan X., Patel C.A., Pomerantz R.J., DuBois G.C., Zhang H. Potent suppression of viral infectivity by the peptides that inhibit multimerization of human immunodeficiency virus type 1 (HIV-1) Vif proteins. J. Biol. Chem. 2003;278:6596–6602. doi: 10.1074/jbc.M210164200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barraud P., Paillart J.-C., Marquet R., Tisne C. Advances in the Structural Understanding of Vif Proteins. Curr. HIV Res. 2008;6:91–99. doi: 10.2174/157016208783885056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Widera M., Hillebrand F., Erkelenz S., Vasudevan A.A.J., Münk C., Schaal H. A functional conserved intronic G run in HIV-1 intron 3 is critical to counteract APOBEC3G-mediated host restriction. Retrovirology. 2014;11 doi: 10.1186/s12977-014-0072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Widera M., Erkelenz S., Hillebrand F., Krikoni A., Widera D., Kaisers W., Deenen R., Gombert M., Dellen R., Pfeiffer T., et al. An Intronic G Run within HIV-1 Intron 2 Is Critical for Splicing Regulation of vif mRNA. J. Virol. 2013;87:2707–2720. doi: 10.1128/JVI.02755-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akari H., Fujita M., Kao S., Khan M.A., Shehu-Xhilaga M., Adachi A., Strebel K. High Level Expression of Human Immunodeficiency Virus Type-1 Vif Inhibits Viral Infectivity by Modulating Proteolytic Processing of the Gag Precursor at the p2/Nucleocapsid Processing Site. J. Biol. Chem. 2004 doi: 10.1074/jbc.M312426200. [DOI] [PubMed] [Google Scholar]
- 69.Simon J.H., Carpenter E.A., Fouchier R.A., Malim M.H. Vif and the p55(Gag) polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J. Virol. 1999 doi: 10.1128/JVI.73.4.2667-2674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang H., Pomerantz R.J., Dornadula G., Sun Y. Human Immunodeficiency Virus Type 1 Vif Protein Is an Integral Component of an mRNP Complex of Viral RNA and Could Be Involved in the Viral RNA Folding and Packaging Process. J. Virol. 2000;74:8252–8261. doi: 10.1128/JVI.74.18.8252-8261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khan M.A., Aberham C., Kao S., Akari H., Gorelick R., Bour S., Strebel K. Human Immunodeficiency Virus Type 1 Vif Protein Is Packaged into the Nucleoprotein Complex through an Interaction with Viral Genomic RNA. J. Virol. 2001;75:7252–7265. doi: 10.1128/JVI.75.16.7252-7265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu H., Wu X., Newman M., Shaw G.M., Hahn B.H., Kappes J.C. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J. Virol. 1995;69:7630–7638. doi: 10.1128/JVI.69.12.7630-7638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Camaur D., Trono D. Characterization of human immunodeficiency virus type 1 Vif particle incorporation. J. Virol. 1996;70:6106–6111. doi: 10.1128/JVI.70.9.6106-6111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Höglund S., öhagen Å., Lawrence K., Gabuzda D. Role of vif during packing of the core of HIV-1. Virology. 1994;201:349–355. doi: 10.1006/viro.1994.1300. [DOI] [PubMed] [Google Scholar]
- 75.Henriet S., Sinck L., Bec G., Gorelick R.J., Marquet R., Paillart J.C. Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription. Nucleic Acids Res. 2007;35:5141–5153. doi: 10.1093/nar/gkm542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sleiman D., Bernacchi S., Guerrero S.X., Brachet F., Larue V., Paillart J.C., Tisné C. Characterization of RNA binding and chaperoning activities of HIV-1 Vif protein Importance of the C-terminal unstructured tail. RNA Biol. 2014;11:906–920. doi: 10.4161/rna.29546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Batisse J., Guerrero S., Bernacchi S., Sleiman D., Gabus C., Darlix J.L., Marquet R., Tisné C., Paillart J.C. The role of Vif oligomerization and RNA chaperone activity in HIV-1 replication. Virus Res. 2012;169:361–376. doi: 10.1016/j.virusres.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 78.von Schwedler U., Song J., Aiken C., Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 1993 doi: 10.1128/JVI.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stopak K., De Noronha C., Yonemoto W., Greene W.C. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell. 2003;12:591–601. doi: 10.1016/S1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 80.Mehle A., Strack B., Ancuta P., Zhang C., McPike M., Gabuzda D. Vif Overcomes the Innate Antiviral Activity of APOBEC3G by Promoting Its Degradation in the Ubiquitin-Proteasome Pathway. J. Biol. Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 81.Verriez C., Marquet R., Paillart J.C., Stupfler B. Les APOBEC3: Histoire d’une famille de protéines antivirales et mutagènes. Virol. Montrouge Fr. 2020;24:381–418. doi: 10.1684/vir.2020.0870. [DOI] [PubMed] [Google Scholar]
- 82.Mehle A., Goncalves J., Santa-Marta M., McPike M., Gabuzda D. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004;18:2861–2866. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hüttenhain R., Xu J., Burton L.A., Gordon D.E., Hultquist J.F., Johnson J.R., Satkamp L., Hiatt J., Rhee D.Y., Baek K., et al. ARIH2 Is a Vif-Dependent Regulator of CUL5-Mediated APOBEC3G Degradation in HIV Infection. Cell Host Microbe. 2019;26:86–99.e7. doi: 10.1016/j.chom.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang W., Du J., Evans S.L., Yu Y., Yu X.F. T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2012;481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 85.Guo Y., Dong L., Qiu X., Wang Y., Zhang B., Liu H., Yu Y., Zang Y., Yang M., Huang Z. Structural basis for hijacking CBF-β and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–233. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 86.Binning J.M., Chesarino N.M., Emerman M., Gross J.D. Structural Basis for a Species-Specific Determinant of an SIV Vif Protein toward Hominid APOBEC3G Antagonism. Cell Host Microbe. 2019 doi: 10.1016/j.chom.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hu Y., Desimmie B.A., Nguyen H.C., Ziegler S.J., Cheng T.C., Chen J., Wang J., Wang H., Zhang K., Pathak V.K., et al. Structural basis of antagonism of human APOBEC3F by HIV-1 Vif. Nat. Struct. Mol. Biol. 2019 doi: 10.1038/s41594-019-0343-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Bruijn M.F.T.R., Speck N.A. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004 doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- 89.Kim D.Y., Kwon E., Hartley P.D., Crosby D.C., Mann S., Krogan N.J., Gross J.D. CBFβ Stabilizes HIV Vif to Counteract APOBEC3 at the Expense of RUNX1 Target Gene Expression. Mol. Cell. 2013;49:632–644. doi: 10.1016/j.molcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anderson B.D., Harris R.S. Transcriptional regulation of APOBEC3 antiviral immunity through the CBF-b/RUNX axis. Sci. Adv. 2015;1 doi: 10.1126/sciadv.1500296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fribourgh J.L., Nguyen H.C., Wolfe L.S., DeWitt D.C., Zhang W., Yu X.-F., Rhoades E., Xiong Y. Core Binding Factor Beta Plays a Critical Role by Facilitating the Assembly of the Vif-Cullin 5 E3 Ubiquitin Ligase. J. Virol. 2014;88:3309–3319. doi: 10.1128/JVI.03824-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matsui Y., Shindo K., Nagata K., Io K., Tada K., Iwai F., Kobayashi M., Kadowaki N., Harris R.S., Takaori-Kondo A. Defining HIV-1 Vif residues that interact with CBFβ by site-directed mutagenesis. Virology. 2014;449:82–87. doi: 10.1016/j.virol.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou X., Han X., Zhao K., Du J., Evans S.L., Wang H., Li P., Zheng W., Rui Y., Kang J., et al. Dispersed and Conserved Hydrophobic Residues of HIV-1 Vif Are Essential for CBF Recruitment and A3G Suppression. J. Virol. 2014;88:2555–2563. doi: 10.1128/JVI.03604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Desimmie B.A., Smith J.L., Matsuo H., Hu W.S., Pathak V.K. Identification of a tripartite interaction between the N-terminus of HIV-1 Vif and CBFβ that is critical for Vif function. Retrovirology. 2017;14 doi: 10.1186/s12977-017-0346-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kao S., Khan M.A., Miyagi E., Plishka R., Buckler-White A., Strebel K. The Human Immunodeficiency Virus Type 1 Vif Protein Reduces Intracellular Expression and Inhibits Packaging of APOBEC3G (CEM15), a Cellular Inhibitor of Virus Infectivity. J. Virol. 2003;77:11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jackson R.J., Hellen C.U.T., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilkie G.S., Dickson K.S., Gray N.K. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 98.Mercenne G., Bernacchi S., Richer D., Bec G., Henriet S., Paillart J.C., Marquet R. HIV-1 Vif binds to APOBEC3G mRNA and inhibits its translation. Nucleic Acids Res. 2009;38:633–646. doi: 10.1093/nar/gkp1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guerrero S., Libre C., Batisse J., Mercenne G., Richer D., Laumond G., Decoville T., Moog C., Marquet R., Paillart J.C. Translational regulation of APOBEC3G mRNA by Vif requires its 5′UTR and contributes to restoring HIV-1 infectivity. Sci. Rep. 2016;6 doi: 10.1038/srep39507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Russell R.A., Pathak V.K. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 2007 doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mehle A., Wilson H., Zhang C., Brazier A.J., McPike M., Pery E., Gabuzda D. Identification of an APOBEC3G Binding Site in Human Immunodeficiency Virus Type 1 Vif and Inhibitors of Vif-APOBEC3G Binding. J. Virol. 2007;81:13235–13241. doi: 10.1128/JVI.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Libre C., Seissler T., Guerrero S., Batisse J., Verriez C., Stupfler B., Gilmer O., Weber M., Cimarelli A., Etienne L., et al. A conserved uORF impacts APOBEC3G translation and is essential for translational inhibition by the HIV-1 Vif protein. BioRivx. 2021 doi: 10.1101/2021.01.13.426487. [DOI] [Google Scholar]
- 103.Calvo S.E., Pagliarini D.J., Mootha V.K. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Renz P.F., Valdivia Francia F., Sendoel A. Some like it translated: Small ORFs in the 5′UTR. Exp. Cell Res. 2020;396 doi: 10.1016/j.yexcr.2020.112229. [DOI] [PubMed] [Google Scholar]
- 105.Johnstone T.G., Bazzini A.A., Giraldez A.J. Upstream ORF s are prevalent translational repressors in vertebrates. EMBO J. 2016;35:706–723. doi: 10.15252/embj.201592759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gallois-Montbrun S., Kramer B., Swanson C.M., Byers H., Lynham S., Ward M., Malim M.H. Antiviral Protein APOBEC3G Localizes to Ribonucleoprotein Complexes Found in P Bodies and Stress Granules. J. Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wichroski M.J., Robb G.B., Rana T.M. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2:374–383. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Martin K.L., Johnson M., D’Aquila R.T. APOBEC3G Complexes Decrease Human Immunodeficiency Virus Type 1 Production. J. Virol. 2012;86:8916. doi: 10.1128/JVI.01199-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gallois-Montbrun S., Holmes R.K., Swanson C.M., Fernández-Ocaña M., Byers H.L., Ward M.A., Malim M.H. Comparison of Cellular Ribonucleoprotein Complexes Associated with the APOBEC3F and APOBEC3G Antiviral Proteins. J. Virol. 2008;82:5636–5642. doi: 10.1128/JVI.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kozak S.L., Marin M., Rose K.M., Bystrom C., Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 2006;281:29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- 111.Cen S., Guo F., Niu M., Saadatmand J., Deflassieux J., Kleiman L. The interaction between HIV-1 gag and APOBEC3G. J. Biol. Chem. 2004;279:33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- 112.Wang T., Zhang W., Tian C., Liu B., Yu Y., Ding L., Spearman P., Yu X.F. Distinct viral determinants for the packaging of human cytidine deaminases APOBEC3G and APOBEC3C. Virology. 2008;377:71–79. doi: 10.1016/j.virol.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li J., Chen Y., Li M., Carpenter M.A., McDougle R.M., Luengas E.M., Macdonald P.J., Harris R.S., Mueller J.D. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J. Mol. Biol. 2014;426:1296–1307. doi: 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Adolph M.B., Ara A., Feng Y., Wittkopp C.J., Emerman M., Fraser J.S., Chelico L. Cytidine deaminase efficiency of the lentiviral viral restriction factor APOBEC3C correlates with dimerization. Nucleic Acids Res. 2017;45:3378–3394. doi: 10.1093/nar/gkx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kao S., Miyagi E., Khan M.A., Takeuchi H., Opi S., Goila-Gaur R., Strebel K. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology. 2004;1 doi: 10.1186/1742-4690-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Opi S., Kao S., Goila-Gaur R., Khan M.A., Miyagi E., Takeuchi H., Strebel K. Human Immunodeficiency Virus Type 1 Vif Inhibits Packaging and Antiviral Activity of a Degradation-Resistant APOBEC3G Variant. J. Virol. 2007;81:8236–8246. doi: 10.1128/JVI.02694-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Binning J.M., Smith A.M., Hultquist J.F., Craik C.S., Caretta Cartozo N., Campbell M.G., Burton L., La Greca F., McGregor M.J., Ta H.M., et al. Fab-based inhibitors reveal ubiquitin independent functions for HIV Vif neutralization of APOBEC3 restriction factors. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Henriet S., Richer D., Bernacchi S., Decroly E., Vigne R., Ehresmann B., Ehresmann C., Paillart J.C., Marquet R. Cooperative and specific binding of Vif to the 5′ region of HIV-1 genomic RNA. J. Mol. Biol. 2005;354:55–72. doi: 10.1016/j.jmb.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 119.Douaisi M., Dussart S., Courcoul M., Bessou G., Vigne R., Decroly E. HIV-1 and MLV Gag proteins are sufficient to recruit APOBEC3G into virus-like particles. Biochem. Biophys. Res. Commun. 2004;321:566–573. doi: 10.1016/j.bbrc.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 120.Bardy M., Gay B., Pébernard S., Chazal N., Courcoul M., Vigne R., Decroly E., Boulanger P. Interaction of human immunodeficiency virus type 1 Vif with Gag and Gag-Pol Precursors: Co-encapsidation and interference with viral protease-mediated Gag processing. J. Gen. Virol. 2001;82:2719–2733. doi: 10.1099/0022-1317-82-11-2719. [DOI] [PubMed] [Google Scholar]
- 121.Syed F., McCrae M.A. Interactions in vivo between the Vif Protein of HIV-1 and the precursor (Pr55GAG) of the virion nucleocapsid proteins. Arch. Virol. 2009;154:1797–1805. doi: 10.1007/s00705-009-0520-8. [DOI] [PubMed] [Google Scholar]
- 122.Guo F., Cen S., Niu M., Yang Y., Gorelick R.J., Kleiman L. The Interaction of APOBEC3G with Human Immunodeficiency Virus Type 1 Nucleocapsid Inhibits tRNA3Lys Annealing to Viral RNA. J. Virol. 2007;81:11322–11331. doi: 10.1128/JVI.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guo F., Cen S., Niu M., Saadatmand J., Kleiman L. Inhibition of tRNA(Lys)-Primed Reverse Transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 2006;80:11710–11722. doi: 10.1128/JVI.01038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Goila-Gaur R., Khan M.A., Miyagi E., Kao S., Opi S., Takeuchi H., Strebel K. HIV-1 Vif promotes the formation of high molecular mass APOBEC3G complexes. Virology. 2008;372:136–146. doi: 10.1016/j.virol.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Soros V.B., Yonemoto W., Greene W.C. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 2007;3:0152–0167. doi: 10.1371/journal.ppat.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nowarski R., Britan-Rosich E., Shiloach T., Kotler M. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat. Struct. Mol. Biol. 2008;15:1059–1066. doi: 10.1038/nsmb.1495. [DOI] [PubMed] [Google Scholar]
- 127.Xu H., Chertova E., Chen J., Ott D.E., Roser J.D., Hu W.S., Pathak V.K. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology. 2007;360:247–256. doi: 10.1016/j.virol.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 128.Kao S., Goila-Gaur R., Miyagi E., Khan M.A., Opi S., Takeuchi H., Strebel K. Production of infectious virus and degradation of APOBEC3G are separable functional properties of human immunodeficiency virus type 1 Vif. Virology. 2007;369:329–339. doi: 10.1016/j.virol.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sadler H.A., Stenglein M.D., Harris R.S., Mansky L.M. APOBEC3G Contributes to HIV-1 Variation through Sublethal Mutagenesis. J. Virol. 2010;84:7396–7404. doi: 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Britan-Rosich E., Nowarski R., Kotler M. Multifaceted counter-APOBEC3G mechanisms employed by HIV-1 Vif. J. Mol. Biol. 2011;410:1065–1076. doi: 10.1016/j.jmb.2011.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang Y., Kinlock B.L., Shao Q., Turner T.M., Liu B. HIV-1 Vif inhibits G to A hypermutations catalyzed by virus-encapsidated APOBEC3G to maintain HIV-1 infectivity. Retrovirology. 2014;11 doi: 10.1186/s12977-014-0089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Santa-Marta M., Da Silva F.A., Fonseca A.M., Goncalves J. HIV-1 Vif can directly inhibit apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G-mediated cytidine deamination by using a single amino acid interaction and without protein degradation. J. Biol. Chem. 2005;280:8765–8775. doi: 10.1074/jbc.M409309200. [DOI] [PubMed] [Google Scholar]
- 133.Feng Y., Love R.P., Chelico L. HIV-1 viral infectivity factor (Vif) alters processive single-stranded DNA scanning of the retroviral restriction factor APOBEC3G. J. Biol. Chem. 2013;288:6083–6094. doi: 10.1074/jbc.M112.421875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bernacchi S., Henriet S., Dumas P., Paillart J.C., Marquet R. RNA and DNA binding properties of HIV-1 Vif protein: A fluorescence study. J. Biol. Chem. 2007;282:26361–26368. doi: 10.1074/jbc.M703122200. [DOI] [PubMed] [Google Scholar]
- 135.Bernacchi S., Mercenne G., Tournaire C., Marquet R., Paillart J.C. Importance of the proline-rich multimerization domain on the oligomerization and nucleic acid binding properties of HIV-1 Vif. Nucleic Acids Res. 2011;39:2404–2415. doi: 10.1093/nar/gkq979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dettenhofer M., Cen S., Carlson B.A., Kleiman L., Yu X.-F. Association of Human Immunodeficiency Virus Type 1 Vif with RNA and Its Role in Reverse Transcription. J. Virol. 2000;74:8938–8945. doi: 10.1128/JVI.74.19.8938-8945.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yoshikawa R., Takeuchi J.S., Yamada E., Nakano Y., Misawa N., Kimura Y., Ren F., Miyazawa T., Koyanagi Y., Sato K. Feline Immunodeficiency Virus Evolutionarily Acquires Two Proteins, Vif and Protease, Capable of Antagonizing Feline APOBEC3. J. Virol. 2017;91 doi: 10.1128/JVI.00250-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ebrahimi D., Richards C.M., Carpenter M.A., Wang J., Ikeda T., Becker J.T., Cheng A.Z., McCann J.L., Shaban N.M., Salamango D.J., et al. Genetic and mechanistic basis for APOBEC3H alternative splicing, retrovirus restriction, and counteraction by HIV-1 protease. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-06594-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ikeda T., Symeonides M., Albin J.S., Li M., Thali M., Harris R.S. HIV-1 adaptation studies reveal a novel Env-mediated homeostasis mechanism for evading lethal hypermutation by APOBEC3G. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Löchelt M., Romen F., Bastone P., Muckenfuss H., Kirchner N., Kim Y.B., Truyen U., Rösler U., Battenberg M., Saib A., et al. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. USA. 2005;102:7982–7987. doi: 10.1073/pnas.0501445102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Russell R.A., Wiegand H.L., Moore M.D., Schäfer A., McClure M.O., Cullen B.R. Foamy Virus Bet Proteins Function as Novel Inhibitors of the APOBEC3 Family of Innate Antiretroviral Defense Factors. J. Virol. 2005;79:8724–8731. doi: 10.1128/JVI.79.14.8724-8731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chareza S., Slavkovic Lukic D., Liu Y., Räthe A.M., Münk C., Zabogli E., Pistello M., Löchelt M. Molecular and functional interactions of cat APOBEC3 and feline foamy and immunodeficiency virus proteins: Different ways to counteract host-encoded restriction. Virology. 2012;424:138–146. doi: 10.1016/j.virol.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 143.Jaguva Vasudevan A.A., Perkovic M., Bulliard Y., Cichutek K., Trono D., Haussinger D., Munk C. Prototype Foamy Virus Bet Impairs the Dimerization and Cytosolic Solubility of Human APOBEC3G. J. Virol. 2013;87:9030–9040. doi: 10.1128/JVI.03385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Perković M., Schmidt S., Marino D., Russell R.A., Stauch B., Hofmann H., Kopletz F., Kloke B.P., Zielonka J., Ströver H., et al. Species-specific inhibition of APOBEC3C by the prototype foamy virus protein bet. J. Biol. Chem. 2009;284:5819–5826. doi: 10.1074/jbc.M808853200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Déjardin J., Bompard-Maréchal G., Audit M., Hope T.J., Sitbon M., Mougel M. A Novel Subgenomic Murine Leukemia Virus RNA Transcript Results from Alternative Splicing. J. Virol. 2000;74:3709–3714. doi: 10.1128/JVI.74.8.3709-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Houzet L., Battini J.L., Bernard E., Thibert V., Mougel M. A new retroelement constituted by a natural alternatively spliced RNA of murine replication-competent retroviruses. EMBO J. 2003;22:4866–4875. doi: 10.1093/emboj/cdg450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhao W., Akkawi C., Mougel M., Ross S.R. Murine Leukemia Virus P50 Protein Counteracts APOBEC3 by Blocking Its Packaging. J. Virol. 2020;94 doi: 10.1128/JVI.00032-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fujisawa R., McAtee F.J., Zirbel J.H., Portis J.L. Characterization of glycosylated Gag expressed by a neurovirulent murine leukemia virus: Identification of differences in processing in vitro and in vivo. J. Virol. 1997;71:5355–5360. doi: 10.1128/JVI.71.7.5355-5360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Corbin A., Prats A.C., Darlix J.L., Sitbon M. A nonstructural gag-encoded glycoprotein precursor is necessary for efficient spreading and pathogenesis of murine leukemia viruses. J. Virol. 1994;68:3857–3867. doi: 10.1128/JVI.68.6.3857-3867.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schwartzberg P., Colicelli J., Goff S.P. Deletion mutants of Moloney murine leukemia virus which lack glycosylated gag protein are replication competent. J. Virol. 1983;46:538–546. doi: 10.1128/JVI.46.2.538-546.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stavrou S., Nitta T., Kotla S., Ha D., Nagashima K., Rein A.R., Fan H., Ross S.R. Murine leukemia virus glycosylated Gag blocks apolipoprotein B editing complex 3 and cytosolic sensor access to the reverse transcription complex. Proc. Natl. Acad. Sci. USA. 2013;110:9078–9083. doi: 10.1073/pnas.1217399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jaguva Vasudevan A.A., Balakrishnan K., Franken A., Krikoni A., Häussinger D., Luedde T., Münk C. Murine leukemia virus resists producer cell APOBEC3A by its Glycosylated Gag but not target cell APOBEC3A. Virology. 2021;557:1–14. doi: 10.1016/j.virol.2021.01.017. [DOI] [PubMed] [Google Scholar]
- 153.Strebel K. APOBEC3G & HTLV-I: Inhibition without deamination. Retrovirology. 2005;2 doi: 10.1186/1742-4690-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sasada A., Takaori-Kondo A., Shirakawa K., Kobayashi M., Abudu A., Hishizawa M., Imada K., Tanaka Y., Uchiyama T. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology. 2005;2 doi: 10.1186/1742-4690-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Derse D., Hill S.A., Princler G., Lloyd P., Heidecker G. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl. Acad. Sci. USA. 2007;104:2915–2920. doi: 10.1073/pnas.0609444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Turelli P., Mangeat B., Jost S., Vianin S., Trono D. Inhibition of Hepatitis B Virus Replication by APOBEC3G. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 157.Warren C.J., Westrich J.A., Van Doorslaer K., Pyeon D. Roles of APOBEC3A and APOBEC3B in human papillomavirus infection and disease progression. Viruses. 2017;9:233. doi: 10.3390/v9080233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cheng A.Z., Yockteng-Melgar J., Jarvis M.C., Malik-Soni N., Borozan I., Carpenter M.A., McCann J.L., Ebrahimi D., Shaban N.M., Marcon E., et al. Epstein–Barr virus BORF2 inhibits cellular APOBEC3B to preserve viral genome integrity. Nat. Microbiol. 2019;4:78–88. doi: 10.1038/s41564-018-0284-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cheng A.Z., Moraes S.N., Shaban N.M., Fanunza E., Bierle C.J., Southern P.J., Bresnahan W.A., Rice S.A., Harris R.S. APOBECs and Herpesviruses. Viruses. 2021;13:390. doi: 10.3390/v13030390. [DOI] [PMC free article] [PubMed] [Google Scholar]