Summary
Fixation facilitates imaging of subcellular localization and cell morphology, yet it remains unknown how fixation affects cellular dimensions and intracellular fluorescence patterns, particularly during long-term storage. Here, we characterized the effects of multiple fixatives on several bacterial species. Fixation generally reduced cell length by 5–15%; single-cell tracking in microfluidics revealed that the length decrease was an aggregate effect of many steps in the fixation protocol and that fluorescence of cytoplasmic GFP but not membrane-bound MreB-msfGFP was rapidly lost with formaldehyde-based fixatives. Cellular dimensions were preserved in formaldehyde-based fixatives for ≥4 days, but methanol caused length to decrease. Although methanol preserved cytoplasmic fluorescence better than formaldehyde-based fixatives, some Escherichia coli cells were able to grow directly after fixation. Moreover, methanol fixation caused lysis in a subpopulation of cells, with virtually all Bacillus subtilis cells lysing after one day. These findings highlight tradeoffs between maintenance of fluorescence and membrane integrity for future applications of fixation.
Subject areas: Biological Sciences, Microbiology, Cell Biology
Graphical abstract
Highlights
-
•
Effects of fixation on cellular dimensions and fluorescence patterns were quantified
-
•
Length and cytoplasmic GFP exhibited rapid dynamics during formaldehyde fixation
-
•
Methanol preserved fluorescence but did not fully inhibit growth and caused lysis
-
•
Extended storage faces tradeoffs of fluorescence maintenance and membrane integrity
Biological Sciences; Microbiology; Cell Biology
Introduction
Bacterial cell shape is a critical physiological parameter that is connected with a variety of behaviors including adhesion, motility, immune system evasion, and antibiotic tolerance (Young, 2006). Long-term evolution of the Gram-negative bacterium Escherichia coli selected for cells with increased cellular volume coupled to increased fitness relative to the parental strain (Lenski and Travisano, 1994), suggesting that cell size is at least correlated with fitness and may be directly selected on. Cell shape and size are defined by the cell wall, a rigid macromolecule formed from glycan strands cross-linked by short peptides (Holtje, 1998). The cell wall is important for maintaining the mechanical integrity of bacterial cells: it resists swelling due to large internal turgor pressures (∼1 atm (Deng et al., 2011)), and thus cells that lack a cell wall become osmotically sensitive. Although the cell wall is generally associated with determination of cell shape, recent studies have shown that proteins involved in a variety of other processes such as metabolism (Elbaz and Ben-Yehuda, 2010), DNA replication (Bazill and Retief, 1969; Harry et al., 1999; Hirota et al., 1968; Mann et al., 2017; Yoshikawa, 1970), and translation (Peters et al., 2016) can also play major roles in morphogenesis. Cell shape can therefore be a critical parameter that is informative of the internal cellular state. For example, filamentation can be an indicator of defects in cell division or DNA damage (Schoemaker et al., 1984), and increases in cell width are associated with nutritional transitions (Grover et al., 1980; Woldringh et al., 1980; Zaritsky et al., 1993).
Achieving a deeper understanding of bacterial cell biology will likely benefit from extensive, unbiased screens of genetic libraries and chemical perturbations. Measurements of morphology and the intracellular patterns of DNA and membrane staining have been used to define cytological profiles that revealed the mechanisms of action of novel antibiotics (Martin et al., 2020; Nonejuie et al., 2016). Although high-throughput systems biology has driven advances that enable rapid imaging of large strain libraries (Campos et al., 2018; Kuwada et al., 2015; Shi et al., 2017; Ursell et al., 2017; Werner et al., 2009), in the time required for imaging cells can continue to grow or change physiology, motivating fixation methods to “freeze” the physiological state of cells for sequential imaging.
Fixation is ubiquitous in cell biology. In the late 19th century, formaldehyde was discovered to be an antibacterial agent that hardened tissues and yielded excellent histological staining (Fox et al., 1985). Fixatives denature proteins via coagulation or cross-linking, thereby terminating biochemical reactions (Kiernan, 2000). In addition to preserving cells in a particular state, fixation also allows cells to be permeabilized to introduce fluorescent antibodies targeting a protein of interest for immunofluorescence microscopy, which has revealed intracellular organization patterns in bacteria such as localization of the divisome (Den Blaauwen et al., 1999; Pende et al., 2018) and of sporulation proteins (Pogliano et al., 1995). By tagging native proteins, immunofluorescence microscopy avoids the need to construct a version of the protein of interest fused to a fluorescent protein and can be used to validate localization conclusions based on fluorescent protein fusions that may not be completely functional. Importantly, while cells are thought to remain stable after fixation for at least one week (Levin, 2002), the impacts of fixation on cell morphology and on fluorescence levels remain mostly unknown on both short and long timescales. Multiple studies reported decreased cell numbers and altered fluorescence histograms after storage of fixed marine bacteria (Kamiya et al., 2007; Troussellier et al., 1995), and another study found fixative-dependent effects on Yersinia pestis cell morphology (Wang et al., 2016). Thus, it is critical to evaluate whether and how fixed cells change over time, especially given that certain labeling protocols (e.g. 16S rRNA fluorescence in situ hybridization) require overnight incubation (Schramm et al., 2002). Moreover, even careful fixation can alter samples and potentially introduce artifacts; for example, the mesosome was purported to be an organelle within Gram-positive bacteria (Van Iterson, 1961) but was later shown to be an artifact of fixation (Ryter, 1988).
Fixatives also increase the mechanical strength or stability of tissues (Lee et al., 1989; Talman and Boughner, 1995), yet the mechanical and morphological effects of fixatives on bacterial cells have not been studied. Gram-negative bacteria, which have a relatively thin cell wall compared with Gram-positive bacteria (Holtje, 1998), possess an outer membrane external to the cell wall. The outer leaflet of the Gram-negative outer membrane contains mainly lipopolysaccharide (LPS) molecules instead of phospholipids, and ionic bonds between LPS molecules as well as protein components endow the outer membrane with mechanical stiffness comparable to that of the cell wall (Rojas et al., 2018). The outer membrane also acts as a barrier to many chemicals (Ruiz et al., 2006). In Gram-positive bacteria, the cell wall is intercalated with teichoic acids that could similarly affect envelope stiffness and permeability. Thus, fixation could increase or decrease envelope stiffness depending on its relative effects on the molecules that stabilize each layer, impacting cell size by changing the mechanical balance with turgor pressure and decreasing intracellular fluorescence if some cytoplasmic molecules are lost during fixation. Overall, despite the importance of fixation for drawing meaningful biological conclusions about bacterial structure and physiology, its quantitative effects on bacterial cells have yet to be sufficiently quantified.
To close this knowledge gap, here we systematically tested the effects of several common fixatives on three model rod-shaped bacteria: Gram-negative Escherichia coli MG1655 and Salmonella enterica serovar Typhimurium 12023 and Gram-positive Bacillus subtilis 168. We found that cell size generally decreased as a result of fixation. By tracking single cells in a microfluidic flow cell throughout the fixation protocol, we determined that most steps exert measurable effects on cell length and that cytoplasmic fluorescence rapidly decreased during exposure to the fixative while MreB-msfGFP levels were maintained. We measured the fluorescence, dimensions, and integrity of each species over several days after fixation and discovered that losses in fluorescence and disrupted cell envelope integrity were dependent on the particular fixative used. These findings constitute an important resource for predicting and evaluating the degree to which fixation affects quantitative cell biology measurements.
Results
Fixation impacts cellular dimensions in a variable manner across fixatives and bacterial species
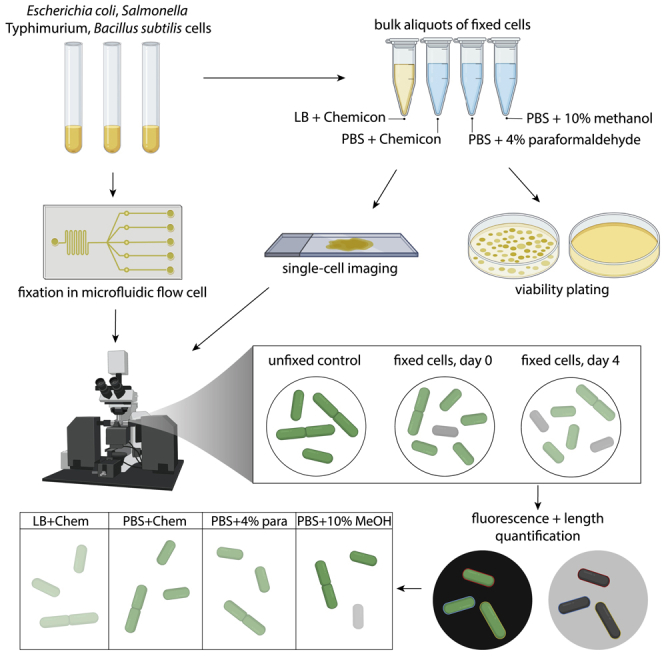
There are a variety of ways in which fixation could impact bacterial morphology, for instance by disrupting the membrane or by changing turgor pressure. We sought to determine the extent to which various common fixatives preserve cellular dimensions in three model bacterial species that differ in the composition of their cell envelopes. E. coli and S. Typhimurium have an outer membrane and a thin (∼2-4 nm (Gan et al., 2008)) cell wall, while B. subtilis lacks an outer membrane but has a thick (∼30 nm (Misra et al., 2013)) cell wall. Some fixatives have been reported to affect membrane structure (Pogliano et al., 1999), leading us to hypothesize that Gram-negative and Gram-positive species might be differentially affected by fixatives due to their different envelope architectures. Each of the strains we investigated also expressed cytoplasmic green fluorescent protein (GFP, transparent methods), enabling us to measure any loss of fluorescent proteins during fixation.
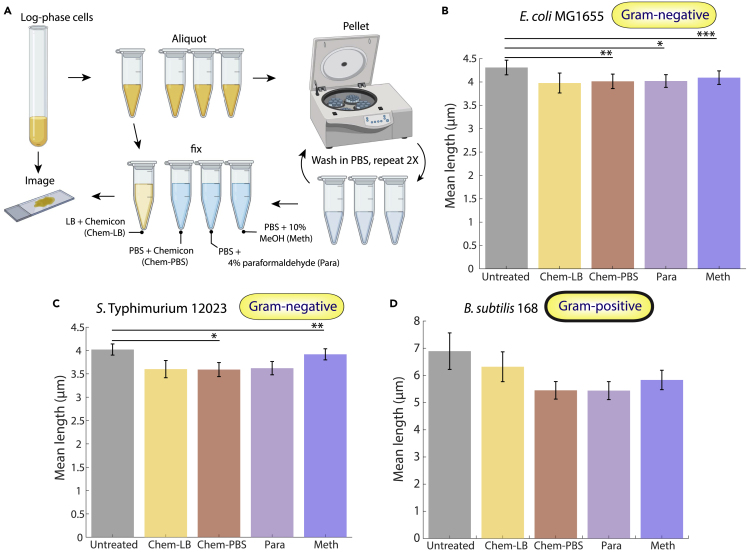
For each species, we grew cultures into log-phase growth and split them into several aliquots (Figure 1A). One aliquot was spotted immediately onto 1% agarose pads with lysogeny broth (LB) and imaged to determine the size and dimensions of live cells (transparent methods). We fixed cells in another aliquot by adding 5X Chemicon, a commercial fixation solution containing proprietary amounts of formaldehyde, methanol, and various buffering agents, for a final concentration of 1X Chemicon in LB; this aliquot was to test the necessity of removing the culture medium prior to fixation. The remaining aliquots were spun down and washed twice with phosphate-buffered saline (PBS) before resuspension in PBS with one of three common fixatives: 1X Chemicon, 4% paraformaldehyde, or 10% methanol (transparent methods). Immediately after this protocol, we acquired phase-contrast images of hundreds of cells for each condition on LB agarose pads, segmented cells, and quantified cellular dimensions (transparent methods).
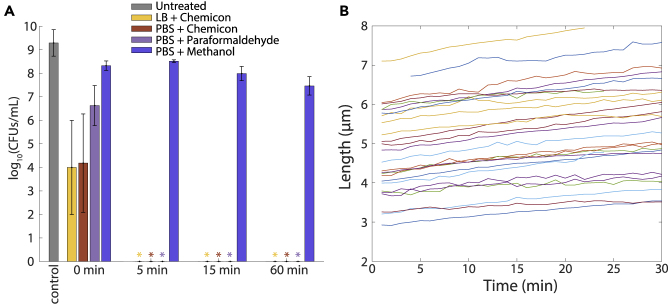
Figure 1.
Fixation generally results in decreases in bacterial cell size
(A) Schematic of fixation protocol. A log-phase culture grown in LB was split into several aliquots: one was immediately imaged, one was fixed in Chemicon without washing and then imaged, and the other three were washed twice in PBS and then fixed in Chemicon, paraformaldehyde, or methanol before imaging (transparent methods). Fixed cells were imaged and segmented to quantify cellular dimensions and average fluorescence and compared to unfixed cells.
(B–D) Mean lengths of E. coli (B), S. Typhimurium (C), and B. subtilis (D) cells before (untreated) and after fixation. The fixation protocol resulted in similar length decreases for E. coli and S. Typhimurium (Gram-negative, thin cell wall) and larger decreases in B. subtilis (Gram-positive, thick cell wall), likely due to disruption of chaining. Measurements are means of ≥3 experiments, and error bars represent 1 standard error of the mean. Each replicate experiment involved n ≥ 152 cells. ∗: p < 0.05; ∗∗: p < 0.01; ∗∗∗: p < 0.001 by paired t-test.
The mean cell length and volume of unfixed E. coli MG1655 cells were 4.33 ± 0.16 μm (Figure 1B) and 7.30 ± 1.80 μm3 (Figure S1A), respectively. Length and volume measurements were highly correlated as expected (Figure S1A); hence, we focused on cell length for simplicity. In all four fixation conditions, cell length decreased (4–7%), with the greatest decrease in Chemicon (Figure 1B). We observed similar levels of shrinkage in S. Typhimurium (3–11%), with Chemicon again resulting in the largest decrease (Figure 1C). For B. subtilis, the decrease was much more pronounced for all fixation conditions following washing (15–20%) compared with direct fixation with Chemicon in LB (8%). The enhanced decrease was due at least in part to the disruption of B. subtilis chaining during centrifugation; washing of live cells in PBS resulted in a similar ∼15–20% decrease in cell length (Figure S1B). Thus, fixation variably impacts measurements of cell morphology, although in a mostly systematic manner.
Cell size is affected by many steps in the fixation protocol
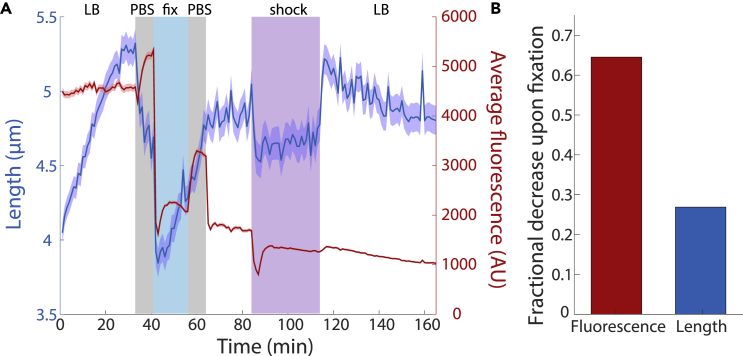
Since fixation protocols generally involve several resuspensions in solutions such as PBS that can have differing osmolality, it was unclear whether the length decreases we observed were directly due to fixation or to physiological changes such as a reduction in turgor pressure. To analyze the shape dynamics of individual E. coli cells during fixation, we carried out the entire Chemicon fixation protocol in a microfluidic flow cell (Video S1, transparent methods). We first equilibrated log-phase cells in the flow cell in LB for 30 min (after 1 hr of outgrowth from stationary phase in a test tube) so that they achieved exponential growth, and then rapidly (in <10 s) switched the medium to 1X PBS for 8 min to mimic the time scale of washing in a centrifuge. Immediately after the cells were transferred to PBS, they displayed a rapid decrease in length (Figure 2A), presumably due to hyperosmotic shock since LB has lower osmolality (240–260 mOsm/kg (Rojas et al., 2014)) than PBS (∼290 mOsm/kg). Consistent with a reduction in turgor pressure, cell width also decreased upon the transfer to PBS (Figure S2A). Following the shock and initial rapid decrease in length, the cytoplasm gradually shrank by ∼6–10% (Figure 2A), consistent with other reports that transfer into a medium lacking carbon causes shrinkage of the cytoplasm away from the cell wall (Shi et al., 2020). After the PBS wash, cells were transferred into PBS with 1X Chemicon for 15 min of fixation. Cells initially shrank even further, reaching a ∼27% decrease relative to pre-wash (Figure 2B), but then began to increase in length after ∼6 min and partially recovered to near their starting length at the end of the preceding PBS wash (Figure 2A). Upon transfer from the fixative back to PBS for 8 min to mimic a second wash, cell length increased rapidly back to approximately the mean length at the end of the first wash (Figure 2A), presumably due to the hypoosmotic shock induced by transfer from 1X Chemicon to PBS. The final single-cell lengths were consistently lower by ∼5–10% than the lengths before fixation, consistent with our bulk culture measurements on LB agarose pads (Figure 1B) and thus indicating that imaging on agarose pads does not itself affect cell size in a fixation-dependent manner.
Figure 2.
Cell length varies throughout the fixation protocol
(A) Mean cell length (blue) and fluorescence (red) for E. coli MG1655 cells expressing cytoplasmic GFP tracked in a microfluidic flow cell (transparent methods) and subjected to the fixation protocol followed by osmotic shocks. Cells were equilibrated for 30 min in LB to achieve exponential growth and then switched to 1X PBS for 8 min to mimic washing. Cells were then fixed in PBS with 1X Chemicon for 15 min and transferred back to PBS for a second wash. To ensure successful fixation, cells were switched to LB for 20 min and no growth occurred. Finally, cells were exposed to a hyperosmotic shock with LB+1 M sorbitol for 30 min, which demonstrated that the membranes still acted as a permeability barrier (Figure S2A). Length varied over many steps of the protocol, while fluorescence decreased dramatically primarily upon fixation. The slow decrease after fixation is likely due to photobleaching. Lines are mean values, and shaded regions represent 1 standard error of the mean (n = 150 cells).
(B) The fractional decrease in cell length and fluorescence between t = 30 min (just before the first PBS wash) and 43 min (just after introduction of the fixative).
E. coli MG1655 cells were equilibrated in a CellAsic B04A chip and then sequentially exposed to a PBS wash for 8 min, Chemicon fixative for 15 min, and a second PBS wash for 8 min, and then returned to LB for 20 min during which time little to no growth was observed. Cells were then exposed to a hyperosmotic shock with 1 M sorbitol for 30 min, demonstrating that the envelope still acted as a semi-impermeable barrier capable of maintaining turgor.
To isolate the effects of Chemicon on cell length, we repeated the above protocol with LB substituted for PBS, avoiding the wash prior to Chemicon addition. We observed a ∼10% decrease in length during fixation that rapidly reversed after cells were returned to LB, indicating that Chemicon addition causes a hyperosmotic shock relative to LB (Figure S2B). These data are consistent with the length decrease in bulk culture measurements, given that we did not wash cells fixed with Chemicon+LB in bulk culture prior to imaging (and hence they would resemble the cells during fixation in the microfluidic flow cell). Taken together, the overall length decrease is a function of both washing in PBS and the added fixative solution.
To ensure that E. coli cells were actually fixed by Chemicon (and therefore dead), after the second PBS wash in our initial experiment, we switched the medium in the flow cell to LB for 20 min. We observed an initial, rapid increase in mean cell length (Figure 2A), due to the hypoosmotic shock from the lower osmolality of LB compared with PBS. Subsequently, no cells grew, suggesting that all cells had been effectively fixed.
To determine the extent to which the inner and outer membranes still acted as a permeability barrier after fixation, we applied a hyperosmotic shock with LB+1 M sorbitol to post-fixation cells in the microfluidic device (transparent methods). Cells rapidly shrank by ∼10% (Figure 2A), similar to the response of growing E. coli cells (Rojas et al., 2014). We then switched the cells back to LB, which caused a hypoosmotic shock that fully recovered mean cell length back to the mean post-fixation length (Figure 2A). Thus, fixation with Chemicon does not completely disrupt the ability of the E. coli membranes to act as a permeability barrier.
Chemicon fixative solution causes a rapid decrease in cytoplasmic fluorescence
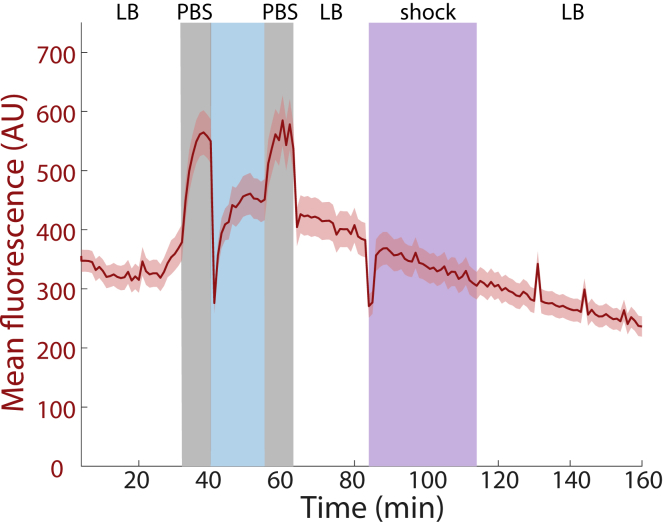
In our microfluidic flow-cell experiments with E. coli cells expressing cytoplasmic GFP, during both PBS washes the average fluorescence intensity (total fluorescence per unit area) increased, by up to ∼2-fold (Figure 2A). The increase was coincident with cell shrinking during the first wash but not the second wash (Figure 2A), and no such large increases were observed when the cells were washed in LB rather than PBS (Figure S2B). To investigate whether imaging in PBS resulted in a fluorescence increase relative to imaging in LB, we quantified the fluorescence of live cells and fixed cells without and with washing in PBS. After background normalization, PBS washing of live cells resulted in a 10–15% increase in fluorescence (Figure S3A). PBS washing of fixed cells resulted in a larger, ∼2-fold increase (Figure S3B). These increases were approximately consistent with the magnitudes of fluorescence increase in the microfluidic flow cell during the washes before and after fixation (Figure 2A), suggesting that imaging in PBS versus LB itself affects fluorescence levels in a manner independent of the difference in backgrounds.
After the switch to Chemicon solution in the microfluidic flow cell, there was a rapid (within 2 min), dramatic decrease in GFP fluorescence of ∼60% (Figures 2A and 2B). This decrease was followed by partial fluorescence recovery during the first 5–10 min, possibly due to maturation of fluorophores, followed by a slight decrease for the remainder of the time in Chemicon (Figure 2A). After the second wash, we surmised that cells were no longer producing GFP as fixation should render them metabolically inactive and that the slow decrease in fluorescence, at a rate similar to that at the end of Chemicon exposure (Figure 2A), was likely due to photobleaching. Relative to live cells, the decrease in fluorescence in E. coli due to fixation was >60% (Figure 2B), similar to measurements from bulk cultures in Chemicon (Figures 3A and S3B).
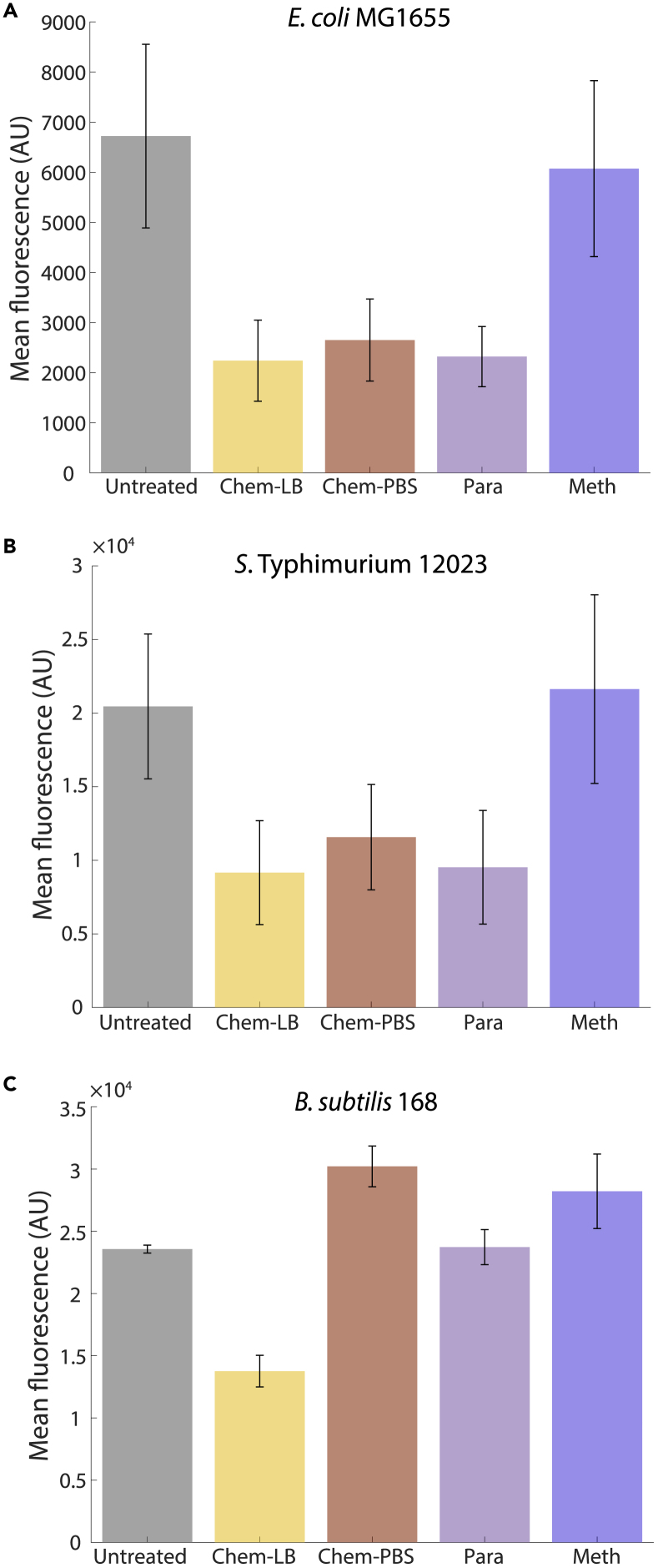
Figure 3.
Methanol generally maintains cytoplasmic fluorescence levels, while the effects of formaldehyde-based fixatives are dependent on the species and on washing prior to fixation
(A and B) For E. coli (A) and S. Typhimurium (B), there was a ~60% loss in intracellular fluorescence directly after the fixation protocol with any formaldehyde-based fixative (but not methanol), presumably due to the sharp decrease we observed 2 min after switching to Chemicon solution in a microfluidic flow cell (Figure 2A).
(C) B. subtilis cells maintained cytoplasmic fluorescence in all fixatives, as long as cells were washed prior to fixation.
Measurements are averages over 3 experiments, and error bars represent 1 standard error of the mean. Each replicate experiment involved n ≥ 152 cells.
Bulk cultures yielded similar fluorescence decreases in Chemicon+LB and paraformaldehyde+PBS as in Chemicon+PBS, while fluorescence levels were largely maintained with methanol (Figure 3A). S. Typhimurium cells experienced similar decreases as E. coli (Figure 3B). B. subtilis cells maintained cytoplasmic GFP fluorescence in all fixation conditions except direct fixation in LB (Figure 3C), indicating that washing is important for fluorescence quantification in B. subtilis. Thus, the large decrease in cytoplasmic GFP fluorescence with formaldehyde-based fixatives likely occurs very rapidly upon introduction of the fixative, while methanol fixation largely avoids fluorescence loss in all three species.
MreB levels do not decrease during fixation
While cytoplasmic GFP levels decreased by >2-fold upon fixation for E. coli and S. Typhimurium (Figures 2A, 2B, and 3A), many proteins with nonuniform localization patterns bind to the membrane or are membrane associated, which may prevent protein loss during fixation. MreB is an essential protein that forms short, membrane-bound filaments and is required for rod-shaped growth in E. coli and many other rod-shaped bacteria (Shi et al., 2018). To study the effects of fixation on MreB levels, we employed an E. coli MG1655 strain that expresses a sandwich fusion of MreB to msfGFP (MreBsw-msfGFP) from the chromosome as the sole copy of mreB (Ursell et al., 2014); this strain has approximately wild-type-like growth rate and cell shape (Ouzounov et al., 2016). We performed the fixation protocol in a microfluidic flow cell as above and found that MreBsw-msfGFP fluorescence dynamics exhibited a major difference compared with wild-type cells expressing cytoplasmic GFP (Figure 2A): the decrease in MreBsw-msfGFP fluorescence immediately after Chemicon introduction was not as large, and more importantly, fluorescence levels recovered back to pre-fixation levels with 5 min (Figure 4). These data indicate that the decrease in cytoplasmic GFP is likely due to loss of GFP molecules rather than denaturation and that loss of membrane-bound proteins such as MreBsw-msfGFP may be much less than cytoplasmic proteins.
Figure 4.
Levels of fluorescent protein fused to MreB were not strongly affected by fixation
Mean fluorescence of E. coli MG1655 cells expressing MreBsw-msfGFP subjected to the fixation protocol as in Figure 2A. msfGFP levels were much less affected by fixation than cytoplasmic GFP (Figure 2A). The slow decrease after fixation is likely due to photobleaching. Lines are mean values, and shaded regions represent 1 standard error of the mean (n = 51 cells).
The treatment time necessary for fixation varies across fixatives
Since E. coli cells still exhibited morphological dynamics throughout and after exposure to Chemicon (Figure 2A), we next assessed cell viability over various treatment times in fixative. We treated E. coli cells with Chemicon in LB or PBS-washed cells with 1X Chemicon, 4% paraformaldehyde, or 10% methanol and then plated on LB without fixative to test for viable cells (defined as the ability to form a colony; note that any remaining fixative was diluted substantially) at fixation durations of 0 min (exposure to the fixative and then plating within 1 min), 5 min, 15 min, and 60 min. The viability of cells treated with Chemicon or paraformaldehyde was severely attenuated (by >100-fold) within 1 min, and no cells were viable after 5 min (Figure 5A). By contrast, methanol reduced viability by only 1–2 orders of magnitude even for 1 hr of treatment (Figure 5A). Similar behavior was observed for S. Typhimurium and B. subtilis (Figure S4).
Figure 5.
Some E. coli cells are still able to grow after one hour of methanol fixation, while other fixatives halt growth almost immediately
(A) Colony forming units (CFUs) of E. coli cells treated for varying amounts of time with different fixatives. Cells plated immediately after resuspension in Chemicon or paraformaldehyde (0 min) exhibited reductions of multiple orders of magnitude in colony forming ability, while 60 min of methanol fixation still permitted growth of >107 CFUs/mL. Asterisks indicate that no colonies were observed. Values are means and error bars represent 1 standard deviation of n = 3 independent replicates.
(B) Single-cell growth at room temperature was impacted by 15 min of methanol fixation. Thirty three of 64 cells exhibited growth (26 representative cells shown here) but with a substantial decrease in growth rate relative to untreated log-phase cells (Figure S5B).
To interrogate the physiological state induced by methanol treatment, we exposed E. coli cells to 10% methanol for 15 min and then placed them on LB-agarose pads for 2 hr of single-cell imaging. 33 of 64 cells exhibited growth (Figure 5B, Video S2), roughly consistent with our plating results (Figure 5A). Of the cells that grew, their growth (Figure 5B) was considerably slower than untreated cells, which have doubling times of ∼60 min at room temperature (Barber, 1908), indicating that the methanol also affected the ability of cells to grow. Even after 2 hr of methanol treatment, some cells were able to grow and divide (Figure S5A), at a rate only slightly lower than those that grew after 15 min of treatment (Figure S5B). Thus, although typical methanol treatments reported in the literature are for 15–60 min, E. coli cells are not necessarily fully killed by this treatment interval.
Time-lapse imaging on a 1% agarose pad. Frames were acquired every 5 min for 130 min.
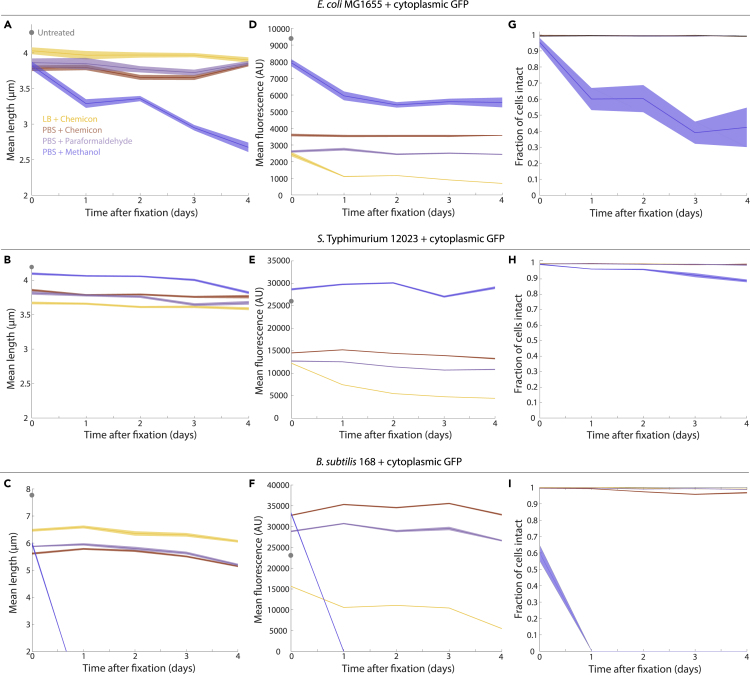
Fluorescent protein concentration in fixed cells decreases over days in a fixative-dependent manner
Since our microfluidic experiments revealed that fluorescence initially decreased rapidly upon exposure to the fixative but then stabilized for at least tens of minutes (aside from photobleaching, Figure 2A), we next asked whether fluorescence levels in fixed cells remain stable over long periods (days) relevant to laboratory experiments. We stored samples of E. coli, S. Typhimurium, and B. subtilis after 15 min of fixation in Chemicon+LB, or Chemicon, 4% paraformaldehyde, or 10% methanol in PBS at 4°C; all samples except Chemicon+LB were washed and resuspended in PBS prior to storage. We imaged an aliquot every 24 hr to quantify cell size and fluorescence. For E. coli (Figure 6A) and S. Typhimurium (Figure 6B), the mean length of intact cells changed only slightly over 4 days in any of the fixatives, while B. subtilis cell length decreased by ∼0.5 μm (Figure 6C).
Figure 6.
After multiple days of storage, cell length, intracellular fluorescence, and envelope integrity are affected in a fixative-dependent manner
(A–C) The mean cell length of E. coli (A), S. Typhimurium (B), and B. subtilis (C) cells over several days of storage at 4°C. E. coli and S. Typhimurium cells treated with formaldehyde-based fixatives approximately maintained cell length, while B. subtilis cells generally decreased somewhat in length and E. coli cells decreased substantially in methanol. No B. subtilis cells were intact 1 day after fixation in methanol (I), explaining the sharp decrease in length.
(D–F) For E.coli (D) and S. Typhimurium (E), intracellular GFP fluorescence intensity was maintained after the initial ~2-fold decrease in Chemicon or paraformaldehyde (Figures 3A and 3B). For S. Typhimurium, fluorescence was maintained at pre-fixation levels, while E. coli cells lost fluorescence over time after methanol fixation. For B. subtilis (F), fluorescence levels continued to decrease after Chemicon fixation in LB.
(G–I) Fixation in methanol led to an increasing number of lysed cells over time, particularly for E. coli (G) and B. subtilis (I), indicating cell membrane damage and loss of cellular contents.
Measurements are averages over cells and error bars represent 1 standard error of the mean, with n ≥ 152 cells each. Gray circles are untreated cells.
For some fixatives, cytoplasmic GFP fluorescence intensity decreased over time, suggesting loss of intracellular material. For E. coli (Figure 6D) and S. Typhimurium (Figure 6E), methanol preserved fluorescence intensity better than formaldehyde or Chemicon-based fixation, consistent with our observation that many cells remain alive after methanol exposure (Figure 5B); for E. coli, methanol fixation resulted in a ∼25% decrease after 1 day, but the overall decrease relative to untreated cells was much less than in formaldehyde or Chemicon-based fixation (Figure 6D). For formaldehyde-based fixation, fluorescence intensity dropped the most (∼2-fold) on day 1 (Figures 6D and 6E); as we observed in our microfluidic experiments (Figure 2A), this drop likely occurred almost immediately upon fixation. For all three species (Figures 6D–6F), fluorescence in Chemicon+LB continued to drop over time. To test whether washing Chemicon+LB-fixed cells in PBS would rescue the fluorescence decrease, we fixed E. coli cells as described above and split the solution into two aliquots. One aliquot was washed in PBS. Over 4.5 days, the msfGFP intensity of PBS-washed cells still decreased, remaining approximately twice that of unwashed cells (Figure S6).
While methanol exposure better preserved cytoplasmic fluorescence, we also observed an increasing fraction of “ghosts” (lysed cells, as visually assessed by the obvious loss of phase intensity) over time (Figures 6G–6I and S7), suggesting that methanol causes significant membrane damage. For B. subtilis, ∼40% of cells lysed during fixation, and there were no cells with intact membranes one day later. Envelope integrity was more sensitive to methanol in E. coli than in S. Typhimurium (Figures 6G and 6H), consistent with the greater decrease in cell length (Figures 6A and 6B) and fluorescence (Figures 6D and 6E) in E. coli compared with S. Typhimurium cells over time. All lysed cells lacked cytoplasmic fluorescence (Figure S7). While there was a small number of ghosts in all fixatives (Figure S8), paraformaldehyde and Chemicon largely preserved the integrity of E. coli and S. Typhimurium cells over the course of 5 days (Figures 6H and 6I).
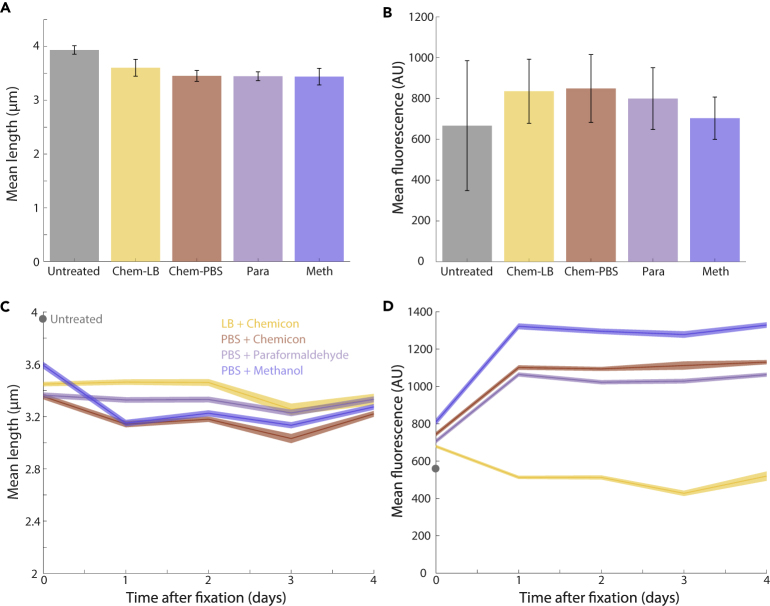
To test whether the fluorescence of membrane-associated proteins decreased in intensity over time after fixation, we processed MreBsw-msfGFP cells with the same growth and fixation protocols as above. Similar length decreases were observed upon fixation (Figures 7A and 7B) as for wild-type cells (Figures 1B and 5B). However, in all but Chemicon+LB, msfGFP fluorescence actually increased over time (Figures 7C and 7D), consistent with the modest increase while exposed to Chemicon in our microfluidic experiments (Figure 4) and similar to previous observations of FtsZ-GFP in fixed versus live B. subtilis cells (Arjes et al., 2014). Moreover, cell length was essentially maintained in all fixatives, including methanol (Figure 7C); interestingly, the contrast with the large methanol-induced length decrease of wild-type MG1655 cells (Figure 6A) suggests that the fluorescent fusion to MreB may be altering cellular mechanical properties.
Figure 7.
Cell length and fluorescence in E. coli MG1655 MreBsw-msfGFP cells are approximately maintained over several days
(A) Directly after fixation, cell length was similar to wild-type E. coli cells expressing cytoplasmic GFP (Figure 1B).
(B) MreBsw-msfGFP fluorescence increased somewhat after fixation, by contrast to cytoplasmic GFP (Figures 2A, 2B, and 3A) but consistent with our microfluidic measurements (Figure 4).
(C) Cell length was approximately constant over several days in every fixative; there was a slight decrease in methanol during the first day but not nearly to the extent as wild-type cells (Figure 6A), suggesting that expression of MreBsw-msfGFP affects cell mechanics.
(D) The small increase in MreBsw-msfGFP fluorescence was accentuated after days of storage in all fixatives as long as cells were washed in PBS prior to fixation.
Measurements in (A,B) are averages of 2 experiments, and error bars represent 1 standard error of the mean, with n ≥ 144 cells each. Measurements in (C,D) are averages over cells, and error bars represent 1 standard error of the mean, with n ≥ 303 cells each.
Thus, the choice of fixative is a balance between preservation of fluorescence, for which methanol is optimal (although it may not actually fix the physical state of all cells), and maintenance of cellular integrity, for which methanol is by far the worst.
Discussion
Our study provides several key points of guidance for using fixed samples in bacterial cell biology. Fixation causes small but potentially important changes in cellular dimensions (Figures 1B–1D), and the precise details of each step in the protocol such as the length of each wash step (Figure 2A) impact the magnitude of size changes. Thus, it is preferable to complete wash steps as quickly as possible and at the very least to maintain the same durations across samples for each step in the protocol in order to compare measurements. In bacteria such as B. subtilis with a propensity for chaining, the centrifugation required for washing breaks up chains and hence results in larger decreases in cellular dimensions (Figures 1D and S1B). Cytoplasmic fluorescence can decrease drastically during fixation, with a rapid, virtually unavoidable ∼2-fold fluorescence decrease in formaldehyde-based fixatives in E. coli and S. Typhimurium (Figures 2A, 2B, 3A, and 3B). We suspect that the membrane is leaky for a short period of time after the start of fixation during which cytoplasmic GFP is lost, after which the permeability barrier is restored. Notably, B. subtilis cells did not exhibit fluorescence decreases in any fixative as long as cells were washed in PBS first (Figure 3C), suggesting that differences in envelope structure can substantially impact fixation and that washing is a critical step of fixation.
Our observation that cytoplasmic GFP fluorescence can change on both short (Figure 2A) and long (Figures 6D–6F) time scales suggests that some cellular material such as cytoplasmic fluorescent proteins can be lost during and after fixation, which needs to be accounted for in any fluorescence quantification based on fixed cells. By contrast, the fluorescence of MreB-msfGFP, which is largely membrane bound, was not reduced by fixation (Figures 7C and 7D). A previous study found that some molecules in the membrane remain mobile even after fixation (Tanaka et al., 2010). The observation that cells lose integrity in methanol over several days after during fixation (Figures 6G–6I) suggests that the cell envelope becomes mechanically compromised, particularly for B. subtilis (Figure 6I). Aldehyde fixatives were previously found to perform better than alcohols in preserving surface ultrastructure in atomic force microscopy (Chao and Zhang, 2011), suggesting that methanol may be generally problematic for envelope integrity. Certain cells (for instance, at a particular stage in the cell cycle) may be more prone to lysis—which would explain the decrease in E. coli cell length over time in methanol (Figure 6A) if longer cells are more prone to lysis—making it preferable to avoid fixatives that enhance integrity loss.
For morphological screens, recent high-throughput imaging protocols (Kuwada et al., 2015; Shi et al., 2017) have enabled rapid image acquisition for hundreds of strains without fixation. Nonetheless, in some cases, it may be challenging to screen large libraries without fixation, as long imaging sequences allow for variable growth during the imaging process. Fixation would ideally enable comparisons at the same time point, and many plates can be fixed for rapid, sequential imaging. For some studies, it may be impossible to perform imaging measurements at remote locations of sample collection (e.g., in the field (Kamiya et al., 2007)), in which case the choice of fixative and protocol should be selected to balance cell shape/integrity and fluorescence maintenance. In the future, identification of storage methods that enhance retention of cellular contents and maintenance of envelope integrity would go a long way toward overcoming the complex effects of fixation over time, although samples should still be imaged as soon as possible. Regardless, when working with a new organism, measurements similar to those we have undertaken here should be an important control for determining the morphological and cytological impact of fixation.
Limitations of the study
It is possible that other fixatives may be less disruptive than those in this study; we have provided a framework for addressing their efficacy. The difference in dynamics of cytoplasmic GFP versus MreB-msfGFP during fixation suggests that the degree of fluorescence loss may be dependent on intracellular compartmentalization. However, fluorescence dynamics should ideally be tested for each protein of interest as there may be protein-specific determinants of loss. Notably, any loss will particularly impact the detection of proteins expressed at low abundance. For high-throughput imaging, as long as the effects of fixation on cellular dimensions are consistent across all strains, these effects can be normalized across a library. However, mutants with altered envelope mechanics may respond differently to fixation. While such variability would be problematic for interpreting shape measurements, it could be useful as a screen for mechanical phenotypes. For instance, the lack of length decreases during long-term methanol exposure in MreBsw-msfGFP cells in contrast to wild-type cells suggests a previously unappreciated mechanical phenotype of this strain.
Resource availability
Lead contact
Kerwyn Casey Huang (kchuang@stanford.edu).
Materials availability
All materials are available from the corresponding author upon reasonable request.
Data and code availability
All data and code are available from the corresponding author upon reasonable request.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The authors thank the Huang Lab for helpful discussions and biorender.com for schematics in Figure 1A. The authors acknowledge support from the Stanford Bioengineering Research Experience for Undergraduates program (to L.Z.), NIH Ruth Kirschstein Award F32-AI133917 (to M.R.), NIH RM1 Award GM135102 (to K.C.H.), and the Allen Discovery Center at Stanford on Systems Modeling of Infection (to K.C.H.). K.C.H. is a Chan Zuckerberg Biohub Investigator.
Author contributions
L.Z., M.R.., and K.C.H. conceptualized the study. L.Z., M.R.., and K.C.H. designed the experiments. L.Z. performed osmotic shock, fixation, and single-cell imaging experiments. L.Z. and M.R. analyzed data. L.Z., M.R.., and K.C.H. wrote the manuscript. All authors reviewed the manuscript before submission.
Declaration of interests
The authors declare no competing interests.
Published: April 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102348.
Supplemental information
References
- Arjes H.A., Kriel A., Sorto N.A., Shaw J.T., Wang J.D., Levin P.A. Failsafe mechanisms couple division and DNA replication in bacteria. Curr. Biol. 2014;24:2149–2155. doi: 10.1016/j.cub.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber M.A. The rate of multiplication of Bacillus coli at different temperatures. J. Infect. Dis. 1908;5:379–400. [Google Scholar]
- Bazill G.W., Retief Y. Temperature-sensitive DNA synthesis in a mutant of Bacillus subtilis. J. Gen. Microbiol. 1969;56:87–97. doi: 10.1099/00221287-56-1-87. [DOI] [PubMed] [Google Scholar]
- Campos M., Govers S.K., Irnov I., Dobihal G.S., Cornet F., Jacobs-Wagner C. Genomewide phenotypic analysis of growth, cell morphogenesis, and cell cycle events in Escherichia coli. Mol. Syst. Biol. 2018;14:e7573. doi: 10.15252/msb.20177573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y., Zhang T. Optimization of fixation methods for observation of bacterial cell morphology and surface ultrastructures by atomic force microscopy. Appl. Microbiol. Biotechnol. 2011;92:381–392. doi: 10.1007/s00253-011-3551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Blaauwen T., Buddelmeijer N., Aarsman M.E., Hameete C.M., Nanninga N. Timing of FtsZ assembly in Escherichia coli. J. Bacteriol. 1999;181:5167–5175. doi: 10.1128/jb.181.17.5167-5175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Sun M., Shaevitz J.W. Direct measurement of cell wall stress stiffening and turgor pressure in live bacterial cells. Phys. Rev. Lett. 2011;107:158101. doi: 10.1103/PhysRevLett.107.158101. [DOI] [PubMed] [Google Scholar]
- Elbaz M., Ben-Yehuda S. The metabolic enzyme ManA reveals a link between cell wall integrity and chromosome morphology. PLoS Genet. 2010;6:e1001119. doi: 10.1371/journal.pgen.1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C.H., Johnson F.B., Whiting J., Roller P.P. Formaldehyde fixation. J. Histochem. Cytochem. 1985;33:845–853. doi: 10.1177/33.8.3894502. [DOI] [PubMed] [Google Scholar]
- Gan L., Chen S., Jensen G.J. Molecular organization of Gram-negative peptidoglycan. Proc. Natl. Acad. Sci. U S A. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover N.B., Zaritsky A., Woldringh C.L., Rosenberger R.F. Dimensional rearrangement of rod-shaped bacteria following nutritional shift-up. I. Theory. J. Theor. Biol. 1980;86:421–439. doi: 10.1016/0022-5193(80)90343-4. [DOI] [PubMed] [Google Scholar]
- Harry E.J., Rodwell J., Wake R.G. Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Mol. Microbiol. 1999;33:33–40. doi: 10.1046/j.1365-2958.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- Hirota Y., Ryter A., Jacob F. Thermosensitive mutants of E. coli affected in the processes of DNA synthesis and cellular division. Cold Spring Harb. Symp. Quant. Biol. 1968;33:677–693. doi: 10.1101/sqb.1968.033.01.077. [DOI] [PubMed] [Google Scholar]
- Holtje J.V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya E., Izumiyama S., Nishimura M., Mitchell J.G., Kogure K. Effects of fixation and storage on flow cytometric analysis of marine bacteria. J. Oceanogr. 2007;63:101–112. [Google Scholar]
- Kiernan J.A. Formaldehyde, formalin, paraformaldehyde and glutaraldehyde: what they are and what they do. Microsc. Today. 2000;1:8–12. [Google Scholar]
- Kuwada N.J., Traxler B., Wiggins P.A. Genome-scale quantitative characterization of bacterial protein localization dynamics throughout the cell cycle. Mol. Microbiol. 2015;95:64–79. doi: 10.1111/mmi.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M., Haberer S.A., Boughner D.R. The bovine pericardial xenograft: I. Effect of fixation in aldehydes without constraint on the tensile viscoelastic properties of bovine pericardium. J. Biomed. Mater. Res. 1989;23:457–475. doi: 10.1002/jbm.820230502. [DOI] [PubMed] [Google Scholar]
- Lenski R.E., Travisano M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl. Acad. Sci. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin P.A. Light microscopy techniques for bacterial cell biology. Methods Microbiol. 2002;31:115–132. [Google Scholar]
- Mann K.M., Huang D.L., Hooppaw A.J., Logsdon M.M., Richardson K., Lee H.J., Kimmey J.M., Aldridge B.B., Stallings C.L. Rv0004 is a new essential member of the mycobacterial DNA replication machinery. PLoS Genet. 2017;13:e1007115. doi: 10.1371/journal.pgen.1007115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.K., 2nd, Sheehan J.P., Bratton B.P., Moore G.M., Mateus A., Li S.H., Kim H., Rabinowitz J.D., Typas A., Savitski M.M. A dual-mechanism antibiotic kills Gram-negative bacteria and avoids drug resistance. Cell. 2020;181:1518–1532.e4. doi: 10.1016/j.cell.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra G., Rojas E.R., Gopinathan A., Huang K.C. Mechanical consequences of cell-wall turnover in the elongation of a Gram-positive bacterium. Biophysical J. 2013;104:2342–2352. doi: 10.1016/j.bpj.2013.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonejuie P., Trial R.M., Newton G.L., Lamsa A., Ranmali Perera V., Aguilar J., Liu W.T., Dorrestein P.C., Pogliano J., Pogliano K. Application of bacterial cytological profiling to crude natural product extracts reveals the antibacterial arsenal of Bacillus subtilis. J. Antibiot. (Tokyo) 2016;69:353–361. doi: 10.1038/ja.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzounov N., Nguyen J.P., Bratton B.P., Jacobowitz D., Gitai Z., Shaevitz J.W. MreB orientation correlates with cell diameter in Escherichia coli. Biophysical J. 2016;111:1035–1043. doi: 10.1016/j.bpj.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende N., Wang J., Weber P.M., Verheul J., Kuru E., Rittmann S.K.R., Leisch N., VanNieuwenhze M.S., Brun Y.V., den Blaauwen T. Host-polarized cell growth in animal symbionts. Curr. Biol. 2018;28:1039–1051 e1035. doi: 10.1016/j.cub.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.M., Colavin A., Shi H., Czarny T.L., Larson M.H., Wong S., Hawkins J.S., Lu C.H., Koo B.-M., Marta E. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell. 2016;165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano K., Harry E., Losick R. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol. Microbiol. 1995;18:459–470. doi: 10.1111/j.1365-2958.1995.mmi_18030459.x. [DOI] [PubMed] [Google Scholar]
- Pogliano J., Osborne N., Sharp M.D., Abanes-De Mello A., Perez A., Sun Y.L., Pogliano K. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas E.R., Billings G., Odermatt P.D., Auer G.K., Zhu L., Miguel A., Chang F., Weibel D.B., Theriot J.A., Huang K.C. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature. 2018;559:617–621. doi: 10.1038/s41586-018-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas E., Theriot J.A., Huang K.C. Response of Escherichia coli growth rate to osmotic shock. Proc. Natl. Acad. Sci. U S A. 2014;111:7807–7812. doi: 10.1073/pnas.1402591111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N., Wu T., Kahne D., Silhavy T.J. Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem. Biol. 2006;1:385–395. doi: 10.1021/cb600128v. [DOI] [PubMed] [Google Scholar]
- Ryter A. Contribution of new cryomethods to a better knowledge of bacterial anatomy. Ann. Inst. Pasteur Microbiol. 1988;139:33–44. doi: 10.1016/0769-2609(88)90095-6. [DOI] [PubMed] [Google Scholar]
- Schoemaker J.M., Gayda R.C., Markovitz A. Regulation of cell division in Escherichia coli: SOS induction and cellular location of the SulA protein, a key to Lon-associated filamentation and death. J. Bacteriol. 1984;158:551–561. doi: 10.1128/jb.158.2.551-561.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm A., Fuchs B.M., Nielsen J.L., Tonolla M., Stahl D.A. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol. 2002;4:713–720. doi: 10.1046/j.1462-2920.2002.00364.x. [DOI] [PubMed] [Google Scholar]
- Shi H., Bratton B.P., Gitai Z., Huang K.C. How to build a bacterial cell: MreB as the foreman of E. coli construction. Cell. 2018;172:1294–1305. doi: 10.1016/j.cell.2018.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Colavin A., Lee T.K., Huang K.C. Strain Library Imaging Protocol for high-throughput, automated single-cell microscopy of large bacterial collections arrayed on multiwell plates. Nat. Protoc. 2017;12:429–438. doi: 10.1038/nprot.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Westfall C.S., Kao J., Odermatt P.D., Cesar S., Sievert M., Moore J., Gonzalez C.G., Zhang L., Elias J.E. bioRxiv; 2020. Starvation Induces Shrinkage of the Bacterial Cytoplasm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talman E.A., Boughner D.R. Glutaraldehyde fixation alters the internal shear properties of porcine aortic heart valve tissue. Ann. Thorac. Surg. 1995;60:S369–S373. doi: 10.1016/0003-4975(95)00250-o. [DOI] [PubMed] [Google Scholar]
- Tanaka K.A., Suzuki K.G., Shirai Y.M., Shibutani S.T., Miyahara M.S., Tsuboi H., Yahara M., Yoshimura A., Mayor S., Fujiwara T.K. Membrane molecules mobile even after chemical fixation. Nat. Methods. 2010;7:865–866. doi: 10.1038/nmeth.f.314. [DOI] [PubMed] [Google Scholar]
- Troussellier M., Courties C., Zettelmaier S. Flow cytometric analysis of coastal lagoon bacterioplankton and picophytoplankton: fixation and storage effects. Estuarine Coastal Shelf Sci. 1995;40:621–633. [Google Scholar]
- Ursell T., Lee T.K., Shiomi D., Shi H., Tropini C., Monds R.D., Colavin A., Billings G., Bhaya-Grossman I., Broxton M. Rapid, precise quantification of bacterial cellular dimensions across a genomic-scale knockout library. BMC Biol. 2017;15:17. doi: 10.1186/s12915-017-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell T.S., Nguyen J., Monds R.D., Colavin A., Billings G., Ouzounov N., Gitai Z., Shaevitz J.W., Huang K.C. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc. Natl. Acad. Sci. U S A. 2014;111:E1025–E1034. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Iterson W. Some features of a remarkable organelle in Bacillus subtilis. J. Biophys. Biochem. Cytol. 1961;9:183–192. doi: 10.1083/jcb.9.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Stanciu C.E., Ehrhardt C.J., Yadavalli V.K. Evaluation of whole cell fixation methods for the analysis of nanoscale surface features of Yersinia pestis KIM. J. Microsc. 2016;263:260–267. doi: 10.1111/jmi.12387. [DOI] [PubMed] [Google Scholar]
- Werner J.N., Chen E.Y., Guberman J.M., Zippilli A.R., Irgon J.J., Gitai Z. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc. Natl. Acad. Sci. U S A. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh C.L., Grover N.B., Rosenberger R.F., Zaritsky A. Dimensional rearrangement of rod-shaped bacteria following nutritional shift-up. II. Experiments with Escherichia coli B/r. J. Theor. Biol. 1980;86:441–454. doi: 10.1016/0022-5193(80)90344-6. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H. Temperature-sensitive mutants of Bacillus subtilis. I. Multiforked replication and sequential transfer of DNA by a temperature-sensitive mutant. Proc. Natl. Acad. Sci. U S A. 1970;65:206–213. doi: 10.1073/pnas.65.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K.D. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritsky A., Woldringh C.L., Helmstetter C.E., Grover N.B. Dimensional rearrangement of Escherichia coli B/r cells during a nutritional shift-down. J. Gen. Microbiol. 1993;139:2711–2714. doi: 10.1099/00221287-139-11-2711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
E. coli MG1655 cells were equilibrated in a CellAsic B04A chip and then sequentially exposed to a PBS wash for 8 min, Chemicon fixative for 15 min, and a second PBS wash for 8 min, and then returned to LB for 20 min during which time little to no growth was observed. Cells were then exposed to a hyperosmotic shock with 1 M sorbitol for 30 min, demonstrating that the envelope still acted as a semi-impermeable barrier capable of maintaining turgor.
Time-lapse imaging on a 1% agarose pad. Frames were acquired every 5 min for 130 min.
Data Availability Statement
All data and code are available from the corresponding author upon reasonable request.